Abstract
Mitochondrial activity and metabolic reprogramming influence the phenotype of cancer cells and resistance to targeted therapy. We previously established that an insulin-like growth factor 1 (IGF-1)-inducible mitochondrial UTP carrier (PNC1/SLC25A33) promotes cell growth. This prompted us to investigate whether IGF signaling is essential for mitochondrial maintenance in cancer cells and whether this contributes to therapy resistance. Here we show that IGF-1 stimulates mitochondrial biogenesis in a range of cell lines. In MCF-7 and ZR75.1 breast cancer cells, IGF-1 induces peroxisome proliferator–activated receptor γ coactivator 1β (PGC-1β) and PGC-1α–related coactivator (PRC). Suppression of PGC-1β and PRC with siRNA reverses the effects of IGF-1 and disrupts mitochondrial morphology and membrane potential. IGF-1 also induced expression of the redox regulator nuclear factor-erythroid-derived 2-like 2 (NFE2L2 alias NRF-2). Of note, MCF-7 cells with acquired resistance to an IGF-1 receptor (IGF-1R) tyrosine kinase inhibitor exhibited reduced expression of PGC-1β, PRC, and mitochondrial biogenesis. Interestingly, these cells exhibited mitochondrial dysfunction, indicated by reactive oxygen species expression, reduced expression of the mitophagy mediators BNIP3 and BNIP3L, and impaired mitophagy. In agreement with this, IGF-1 robustly induced BNIP3 accumulation in mitochondria. Other active receptor tyrosine kinases could not compensate for reduced IGF-1R activity in mitochondrial protection, and MCF-7 cells with suppressed IGF-1R activity became highly dependent on glycolysis for survival. We conclude that IGF-1 signaling is essential for sustaining cancer cell viability by stimulating both mitochondrial biogenesis and turnover through BNIP3 induction. This core mitochondrial protective signal is likely to strongly influence responses to therapy and the phenotypic evolution of cancer.
Keywords: cancer biology, cancer therapy, cell metabolism, cell signaling, cell surface receptor, drug resistance, insulin-like growth factor (IGF), mitochondria, mitophagy
Introduction
Mitochondrial function is implicated in aging, certain degenerative diseases, and, increasingly, in cancer progression and immune responses (reviewed in Refs. 1–5). Mitochondrial biogenesis requires expression of genes encoded by both the mitochondrial and nuclear genomes, and it is controlled by the peroxisome proliferator–activated receptor γ coactivator-1 (PGC-1)3 transcriptional response pathways, which can be activated by mitogenic signals and energetic stress (1, 6–8). Induction of PGC-1α, increased mitochondrial biogenesis, and oxidative phosphorylation (OXPHOS) have recently been associated with certain cancer phenotypes, enabling cell migration in metastasis (9) and the outgrowth and survival of oncogene-depleted pancreatic cancer cells (9, 10). Moreover, increased mitochondrial biogenesis has been associated with adaptive resistance to tyrosine kinase inhibitors, and active AKT at the mitochondria has been implicated in stimulating glycolysis (11, 12). Mitochondrial dysfunction triggers cellular antioxidant and redox protective systems, many of which are transcriptionally regulated by NRF-2/NFE2L2 (1, 2, 33). Damaged or aged mitochondria are marked for degradation in autophagosomes by a process called mitophagy, which is initiated by cargo receptor proteins such as PINK1/Parkin (in response to mitochondrial depolarization) or by mitophagy receptor proteins such as Bcl-2/adenovirus E1B 19-kDa–interacting protein 3 (BNIP3) or BNIP-L/Nix, induced by hypoxia, nutrient deprivation, or oncogenes (34–39). A balance between mitochondrial biogenesis and mitophagy maintains a healthy pool of mitochondria (1, 2). Thus, mitochondrial metabolism is emerging as a potentially important area to target in cancer (13).
IGF-1 signaling through the PI3K/AKT/mTORC1 signaling pathway has a well-documented role in cell growth and also in oncogenic transformation and cancer progression. IGF-1 signaling is strongly linked with glycolysis (14) and the expression of glucose transporters, but the IGF signaling pathway is also linked with enhanced mitochondrial respiration in normal tissues, including the liver (reviewed in Ref. 15), and with PGC-1β expression in breast cancer cells (16). Moreover, the anti-tumorigenic activity of metformin has been linked to reduced IGF-1 signaling (17, 18). We previously characterized an insulin/IGF-1-responsive mitochondrial carrier protein (PNC1/SLC25A33) (19), a conserved orthologue of the yeast Rim2 carrier (20), that is essential for mitochondrial maintenance. Suppression of PNC1 leads to impaired OXPHOS, leakage of ROS from the respiratory chain, and a profound ROS-dependent epithelial-to-mesenchymal transition in non-invasive MCF-7 cells (21). All of these studies suggest that IGF-1–promoted mitochondrial protection has an underappreciated function in maintaining cancer cell survival and growth and predict that suppressed IGF signaling could cause mitochondrial dysfunction. Moreover, IGF-1 promoted mitochondrial protection may contribute to therapy resistance.
To test these concepts, we investigated the mechanism of IGF actions in mitochondrial maintenance in a panel of cancer cell lines that included an MCF-7 cell line that has acquired resistance to an IGF-1R tyrosine kinase inhibitor. We found that IGF-1/PI3K signaling promotes both mitochondrial biogenesis and mitophagy and that inhibition of IGF-1R activity or expression leads to mitochondrial dysfunction.
Results
IGF-1 stimulates mitochondrial biogenesis
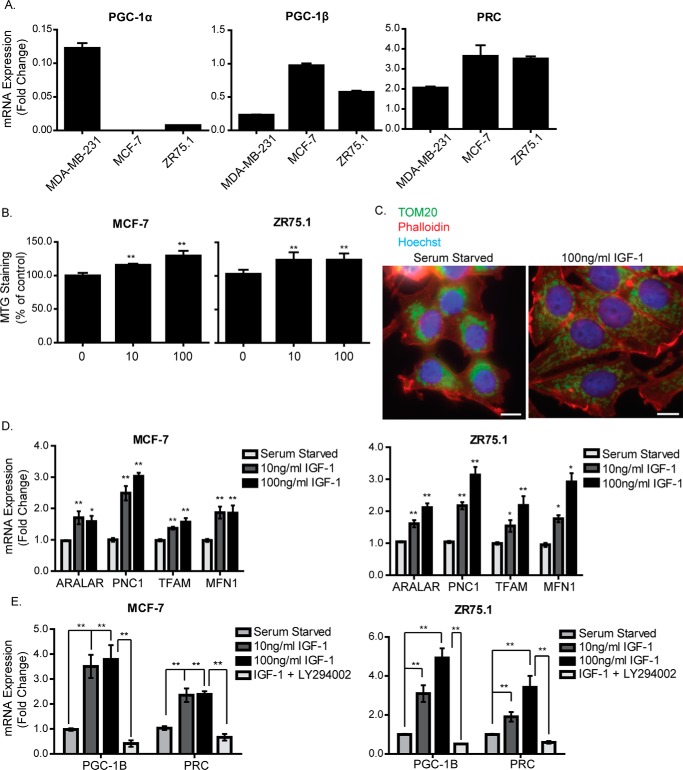
We first surveyed the expression of the PGC-1 family transcriptional co-activators PGC-1α, PGC-1β, and PRC in cell lines and publicly available datasets. This indicated that PGC-1α levels are generally low or undetectable compared with PGC-1β and PRC, which are both widely expressed in breast and ovarian cell lines (supplemental Fig. 1, A–C). We confirmed this pattern in MCF-7 and ZR75.1 breast cancer cells, both of which have a luminal phenotype and express high levels of PGC-1β and PRC and low levels of PGC-1α. In contrast, the mesenchymal MDA-MB-231 breast cell line expresses high levels of PGC-1α and low levels of PGC-1β and PRC (Fig. 1A).
Figure 1.
IGF-1 induces mitochondrial biogenesis and expression of mitochondrial genes. A, RNA expression levels of PGC-1α, PGC-1β, and PRC in MCF-7, MDA-MB-231, and ZR75.1 cells cultured in complete medium were determined by qPCR, normalized to the housekeeping gene UBC, and presented as -fold change compared with MDA-MB-231 cells set at a value of 1. B, mitochondrial mass in MCF-7 and ZR75.1 cells. Cells were serum-starved for 4 h and stimulated with 10 ng/ml or 100 ng/ml IGF-1 for a further 20 h. Mitochondrial mass was measured by FACS using the MTG probe. C, immunofluorescence staining of TOM20 in MCF-7 cells following stimulation with IGF-1. MCF-7 cells were serum-starved for 4 h, followed by stimulation with 100 ng/ml IGF-1 for a further 20 h, after which they were prepared for immunofluorescence using an antibody specific for the mitochondrial outer membrane protein TOM20 (green). Nuclei were stained with Hoechst dye (blue), and actin was stained with phalloidin (red). The original magnification was ×40. Scale bars = 25 μm. D, expression levels of Aralar, PNC1, TFAM, and MFN1 mRNA in MCF-7 and ZR75.1 cells following stimulation with IGF-1. Cells were serum-starved for 4 h and stimulated with 10 ng/ml or 100 ng/ml IGF-1 for a further 20 h. The expression level of each gene was normalized to the housekeeping gene UBC and is shown as -fold change of the control (serum-starved cells) set at 1. E, qPCR showing the mRNA expression levels of PGC-1β and PRC in MCF-7 cells following stimulation with IGF-1. Cells were serum-starved for 4 h and stimulated with 10 ng/ml or 100 ng/ml IGF-1 for a further 4 h. Cells stimulated with 100 ng IGF-1 were treated with 20 μm LY294002 30 min prior to stimulation. The levels of PGC-1β and PRC mRNA were normalized to the housekeeping gene UBC and are presented as -fold change of the control (serum-starved) set at 1. In B, D, and E, data are presented from three independent experiments as mean ± S.E. Statistical analysis was performed using Student's t test (*, p < 0.05; **, p < 0.01).
We then investigated the effects of IGF-1 on mitochondrial biogenesis by first measuring mitochondrial mass using MitoTracker Green. As can be seen in Fig. 1B, IGF-1 stimulated an increase in mitochondrial mass in MCF-7 and ZR75.1 cells. This was evident at 100 ng/ml IGF-1, a concentration that may activate both IGF-1R and insulin receptor (IR), and at a lower concentration of 10 ng/ml, which would selectively activate the IGF-1R. This IGF-1-stimulated increase in mitochondrial mass was also observed in a range of different cell lines, including fibroblasts and HeLa and PC12 cells (supplemental Fig. 2A), and in response to ectopic expression of the IGF-1R in the TOV112D ovarian cancer cell line (supplemental Fig. 2B). It was accompanied by increased expression of VDAC and CoxIV in MCF-7 cells (supplemental Fig. 2C). The effect of IGF-1 stimulation on cellular mitochondrial distribution in MCF-7 cells was observed using TOM20 as a mitochondrial marker (Fig. 1C). In serum-starved cells, mitochondria were clustered and aggregated around the nucleus, whereas in IGF-1-stimulated cells, mitochondria were more evenly spread throughout the cell and clearly evident at the cell periphery. In both MCF-7 cells and ZR75.1 cells, IGF-1 induced transcription of two nuclearly encoded mitochondrial carrier proteins, Aralar and PNC1/SLC25A33, as well as transcription factor A, mitochondrial (TFAM), and MFN1 (which encodes a protein important in mitochondrial fusion) (Fig. 1D).
We next investigated whether IGF-1 induced expression of PGC-1α, PGC-1β, or PRC mRNA in MCF-7 and ZR75.1 cells, noting that PGC-1β has been shown previously to be induced by IGF-1 (16). We observed that, although PGC-1α mRNA was not detectable in cells cultured in complete medium, both PGC-1β and PRC were significantly induced following 4-h stimulation with either 10 ng/ml or 100 ng/ml IGF-1 (Fig. 1E). The PI3K inhibitor LY294002 suppressed induction of PGC-1β and PRC by IGF-1 (Fig. 1E).
Interestingly, we also observed that PGC-1α expression was induced whereas PGC-1β was suppressed by serum starvation in MDA-MB-231 cells (supplemental Fig. 2D). Because IGF-1 induces mitochondrial mass in MDA-MB-231 cells, this suggest that the effects of IGF-1 do not require PGC-1α and may be mediated through PGC-1β or PRC. In support of this, PGC-1β but not PGC-1α expression correlated with IGF-1R expression in mouse embryonic fibroblasts derived from IGF-1R knockout mice (R− cells) that were reconstituted with IGF-1R (R+ cells) (supplemental Fig. 2E). Moreover, either PI3K inhibition or ROS scavenging was sufficient to reverse the effects of IGF-1 on mitochondrial mass in MDA-MB-231 cells (supplemental Fig. 2F). All of these observations indicate that PGC-1β and PRC are induced by IGF-1 to promote mitochondrial biogenesis, so we focused subsequent studies on the mechanisms of IGF-1 signaling in mitochondrial biogenesis.
PGC-1β and PRC are required for IGF-1–stimulated mitochondrial biogenesis
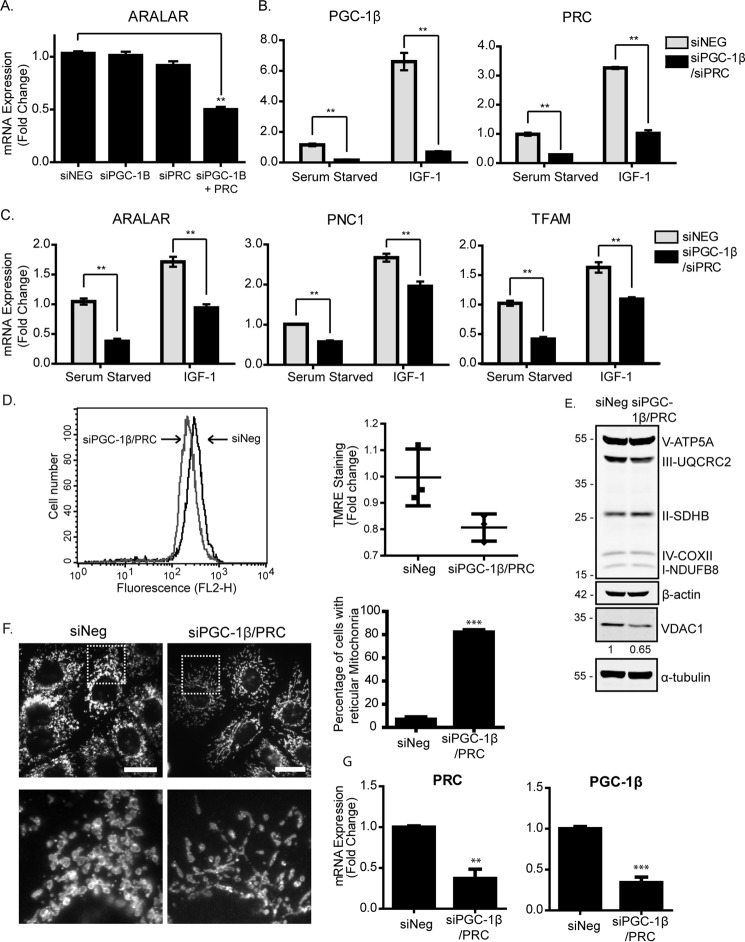
To test whether IGF-1–promoted mitochondrial biogenesis required either PGC-1β, PRC, or both, we used siRNA to suppress their expression. As can be seen in Fig. 2A for MCF-7 cells, suppression of PGC-1β or PRC alone had little to no effect on transcription of Aralar, but simultaneous suppression of PGC-1β and PRC caused a significant reduction in expression. This indicates that PGC-1β and PRC act redundantly to support mitochondrial biogenesis. Next, we tested suppression of PGC-1β and PRC in cells stimulated with IGF-1 (we suppressed each gene with siRNA for 24 h, followed by serum starvation for 4 h and subsequent stimulation with IGF-1 for 5 h). This demonstrated that simultaneous suppression of PGC-1β and PRC reduced the levels of both PGC-1β and PRC in serum-starved cells and, in addition, blocked the induction by IGF-1 observed in siNeg controls (Fig. 2B). Aralar mRNA expression in both serum-starved and IGF-1–stimulated cells was also significantly reduced by suppression of PGC-1β and PRC, as were TFAM and PNC1 (Fig. 2C).
Figure 2.
IGF-1 promotes mitochondrial biogenesis through induction of PGC-1β and PRC. A, expression of Aralar mRNA in MCF-7 cells following single or simultaneous suppression of PGC-1β and PRC. MCF-7 cells were transfected with PGC-1β or PRC siRNA for 48 h before being harvested for qPCR. Aralar expression levels were normalized to the housekeeping gene UBC and are shown as -fold change of the siRNA negative control (siNeg) set at 1. The data for this and all qPCR experiments are presented as mean ± S.E. from three independent experiments. B, expression of PGC-1β and PRC mRNA in MCF-7 cells following simultaneous suppression of PGC-1β and PRC. MCF-7 cells were transfected with PGC-1β or PRC siRNA for 48 h prior to serum starvation for 4 h and stimulation with 10 ng/ml or 100 ng/ml IGF-1 for a further 4 h. Gene expression was analyzed by qPCR, and mRNA levels were normalized to the housekeeping gene UBC and are presented as -fold change of the serum-starved controls set at 1. C, expression of Aralar, TFAM, and PNC1 mRNA in MCF-7 cells following simultaneous suppression of PGC-1β and PRC. MCF-7 cells were treated, and the results were analyzed as described in B. D, analysis of mitochondrial membrane potential by flow cytometry using the TMRE probe. MCF-7 cells were transfected with PGC-1β and PRC siRNA (gray) for 48 h compared with siNeg (black). A representative histogram of TMRE fluorescence is shown, with the scatter plot showing the geometric mean from three independent experiments. Statistical analysis using Student's t test indicated no significance. E, Western blot showing OXPHOS and VDAC1 levels in MCF-7 cells transfected with PGC-1β and PRC siRNA for 48 h. Cells were lysed and prepared for Western blotting with anti-OXPHOS, anti-VDAC1, and anti-α-tubulin antibodies. Quantification was performed by densitometry relative to α-tubulin and normalized to the siNeg control. The -fold change for VDAC1 is indicated below the Western blot. F, immunofluorescence of MCF-7 cells following transfection with PGC-1β and PRC siRNA for 72 h. The cells were stained with an antibody specific for the mitochondrial outer membrane protein TOM20 (white). Scale bars = 20 μm. The enlarged images below are six times larger. The number of rounded and reticular mitochondria was counted in a total of 100 fields per condition (10–20 cells/field) from three individual experiments and is presented in the bar chart as a percentage of total cells counted. G, levels of PRC and PG1-β mRNA suppression in MCF-7 cells used in D–F, determined by qPCR. In all panels, data are presented from three independent experiments as mean ± S.E. Statistical analysis was performed using Student's t test (*, p < 0.05; **, p < 0.01; ***, p < 0.005).
We also investigated the effects of PGC-1β and PRC suppression on mitochondrial mass, morphology, and membrane potential. In MCF-7 cells transfected with both PGC-1β and PRC siRNA, the mitochondrial membrane potential was reduced compared with the control cells, as indicated by reduced TMRE staining, although this was not statistically significant (Fig. 2D). VDAC1 was slightly reduced by suppression of PGC-1β and PRC, supporting a reduction in mitochondrial mass, but the protein levels of the OXPHOS subunits were not significantly altered (Fig. 2E). Mitochondrial morphology was examined by immunofluorescence using an anti-TOM20 antibody. In MCF-7 cells with PGC-1β and PRC suppressed, mitochondria appeared to be less abundant and to exhibit a more elongated, reticular shape than control cells, which exhibited rounded mitochondria (Fig. 2F). These reticular mitochondria were ∼10-fold more evident in cells with PGC-1β and PRC suppressed than in controls (Fig. 2F). These experiments were carried out in medium containing either high glucose (250 mm) or low glucose (5.5 mm), but we did not observe a significant difference in gene or protein expression and mitochondrion morphology between these culture conditions. Taken together, the data indicate that PGC-1β and PRC expression is required for IGF-1–promoted mitochondrial biogenesis, for expression of mitochondrial and nuclear genes, and to maintain mitochondrial morphology and membrane potential.
Mitochondrial dysfunction and reduced biogenesis upon suppression of IGF-1R kinase activity
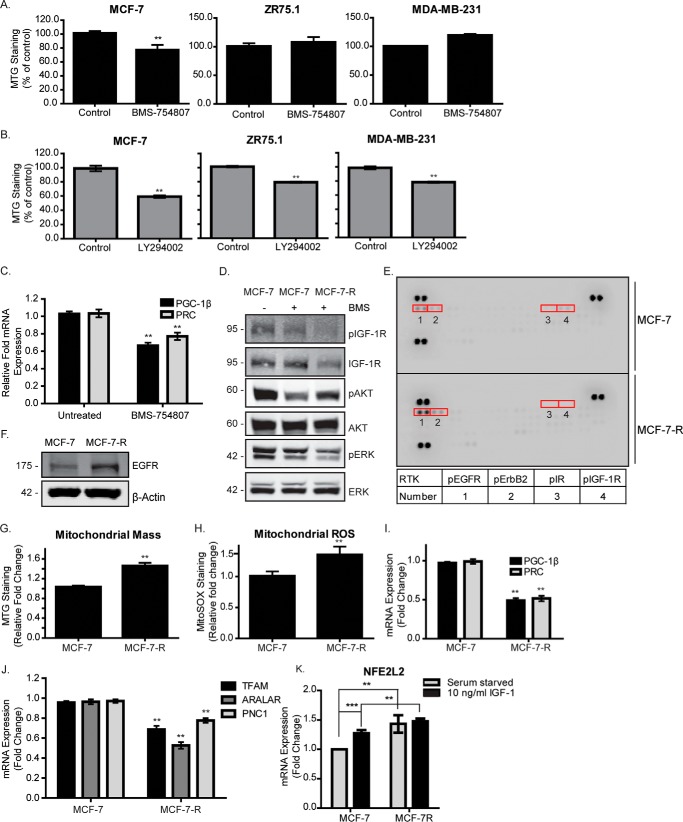
If IGF-1 signaling is required for mitochondrial biogenesis, then inhibition of this pathway could lead to mitochondrial dysfunction. Therefore, we next investigated whether IGF-1R kinase inhibition would impair mitochondrial biogenesis and cause mitochondrial dysfunction. To test this, we made use of the observation that the three breast cancer cell lines used in this study are known to exhibit different degrees of sensitivity to the IGF-1R tyrosine kinase inhibitor (TKI) BMS-754807. MCF-7 cells are the most sensitive, with an approximate IC50 of 16 nm, whereas MDA-MB-231 and ZR75.1 cells are more resistant, with IC50 values of 1.6 μm and 2 μm, respectively. BMS-754807 suppressed phosphorylation of the IGF-1R in all three of these cell lines (supplemental Fig. 3A). AKT phosphorylation on Ser-473 was also suppressed in MCF-7 cells but not in MDA-MB-231 and ZR75.1 cells (supplemental Fig. 3A). The effects of BMS-754807 on mitochondrial mass were tested in MDA-MB-231, MCF-7, and ZR75.1 cells exposed to the drug for 24 h at 50 nm for MCF-7 cells, 1 μm for MDA-MB-231 cells, or 2 μm for ZR75.1 cells. As can be seen in Fig. 3A, MCF-7 cells exposed to BMS-754807 exhibited a significant decrease in mitochondrial mass, but no significant effect was observed in ZR75.1 or MDA-MB-231 cells. PI3K inhibition caused decreased mitochondrial mass in all three cell lines (Fig. 3B), whereas inhibition of the MAPK signaling pathway had no effect on mitochondrial mass (supplemental Fig. 3B).
Figure 3.
IGF-1R inhibition is associated with mitochondrial dysfunction. A, mitochondrial mass in MCF-7, ZR75.1, and MDA-MB-231 cells following exposure to BMS-754807 for 24 h. B, mitochondrial mass in MCF-7, ZR75.1 and MDA-MB-231 cells following exposure to LY294002 for 24 h. Mitochondrial mass was measured by FACS using the MTG probe. C, expression levels of PGC-1β and PRC in MCF-7 cells following exposure to BMS-754807 for 24 h. The levels of PGC-1β and PRC were determined by qPCR normalized to the housekeeping gene UBC and are shown as -fold change of the untreated control set at 1. D, representative Western blot showing the activity of the IGF-1R, PI3K, and MAPK signaling pathways, as measured by phosphorylation levels of IGF-1R, AKT, and ERK, respectively. MCF-7 and MCF-7-R cells were cultured in complete medium supplemented with or without 500 nm BMS-754807 (BMS), as indicated, followed by lysis and immunoblotting with anti-phospho and non-phospho antibodies against IGF-1R, ERK, and Akt. E, phospho-RTK array showing the phosphorylation levels of 42 different receptor tyrosine kinases in MCF-7 and MCF-7-R cells. The arrays were exposed to 100 μg of MCF-7 and MCF-7-R whole-cell lysates and probed with an HRP-conjugated pan phosphotyrosine antibody. F, EGFR expression in MCF-7 and MCF-7-R cells. MCF-7 and MCF-7-R cell lysates were prepared and analyzed by Western blotting using anti-EGFR and anti-β-actin antibodies. G, mitochondrial mass in MCF-7 and MCF-7-R cells, measured by flow cytometry using the MitoTracker Green probe. H, mitochondrial ROS in MCF-7 and MCF-7-R cells, measured by flow cytometry using the MitoSOX Red probe. I, expression levels of PGC-1β and PRC in MCF-7 and MCF-7-R cells. The levels of PGC-1β and PRC were determined by qPCR, normalized to the housekeeping gene UBC, and are presented as -fold change of the levels in MCF-7 cells set at 1. J, qPCR showing the levels of TFAM, Aralar, and PNC1 in MCF-7 compared with MCF-7-R cells. The expression level of each gene was normalized to the housekeeping gene UBC, and the levels in MCF-7-R cells are presented as -fold change of the levels in the MCF-7 cells set at 1. K, qPCR showing the expression levels of NFE2L2 in MCF-7 and MCF-7-R cells that were serum-starved and stimulated with IGF-1 or not. The levels of NFE2L2 were normalized to the housekeeping gene UBC and are shown as -fold change compared with serum-starved MCF-7 cells. Data are presented from three independent experiments as mean ± S.D. Statistical analysis was performed using Student's t test (*, p < 0.05; **, p < 0.01).
PGC-1β and PRC expression were significantly reduced in cells exposed to either BMS-754807 or LY294002 (Fig. 3C and supplemental Fig. 2C). A corresponding decrease in levels of nuclear respiratory factor 2 (NRF-2α/GABPA), PNC1, Aralar, Cox1, and CytC was also observed when IGF-1R or PI3K activity was suppressed (supplemental Fig. 3, D and E). Altogether, these data indicate that acute inhibition of IGF-1R kinase in TKI-sensitive cells or of PI3K, irrespective of sensitivity to IGF-1R TKI, results in decreased mitochondrial biogenesis associated with decreased expression of PGC-1β and PRC.
Mitochondrial reprogramming and a reliance on oxidative phosphorylation have been associated with resistance to MAPK and PI3K inhibitors and with tumor recurrence following oncogene ablation (10, 22). It was thus interesting to investigate the impact of chronic IGF-1R inhibition on mitochondrial function and cell phenotype in an MCF-7 cell line variant (MCF-7R) we had selected for acquired resistance to the kinase inhibitor BMS-754807 (23). As can be seen in Fig. 3D, MCF-7-R cells displayed amplified AKT phosphorylation on Ser-473 in the presence of 500 nm BMS-754807 compared with parental cells exposed to the drug for a short time. This reactivation of AKT has been described in cells resistant to PI3K inhibitors (11), but MCF-7-R cell lysates also exhibited amplified phosphorylation of both EGFR and ERBB2 on human phospho-RTK arrays (Fig. 3E) and higher total levels of EGFR than MCF-7 parental cells (Fig. 3F). This, together with the slightly reduced proliferation rates observed in MCF-7-R cells compared with parental cells (supplemental Fig. 3F), initially suggested that these RTKs could contribute to compensatory signaling in the presence of IGF-1R inhibition.
We next investigated mitochondrial mass. As can be seen in Fig. 3G, and in contrast to observations in MCF-7 cells with short-term exposure to BMS-754807, MCF-7-R cells exhibited significantly greater mitochondrial mass. MCF-7-R cells also displayed higher levels of mitochondrial superoxide radicals, as determined by FACS analysis using MitoSOX Red (Fig. 3H), indicating mitochondrial dysfunction. The levels of PGC-1β, PRC, TFAM, Aralar, and PNC1 were all lower in MCF-7-R cells compared with MCF-7 cells (Fig. 3, I and J). We also assessed NFE2L2/NRF-2, which promotes antioxidant cell protection responses and possibly promotes mitochondrial biogenesis through induction of nuclear respiratory factor 1 (NRF-1/αPAL) or PGC-1α (24). However, although NFE2L2 was clearly induced by IGF-1 in MCF-7 cells and constitutively elevated in MCF-7-R cells (Fig. 3K), neither PGC-1α nor NRF-1 mRNA could be detected at significant levels (data not shown). Taken together, these data demonstrate that the increased mitochondrial mass observed in MCF-7-R cells cannot be entirely due to increased mitochondrial biogenesis and also indicate that compensatory RTK activity is not sufficient to overcome the loss of IGF-1R activity in promoting mitochondrial biogenesis.
The mitophagy mediators BNIP3 and BNIP3L and mitophagy are reduced in MCF-7 cells with acquired resistance to the IGF-1R kinase inhibitor (MCF-7-R)
MCF-7-R cells exhibited increased mitochondrial mass in the presence of decreased mitochondrial biogenesis. This change in overall mitochondrial mass could reflect alterations in the balance between mitochondrial biogenesis and mitophagy (25), so we next asked whether mitophagy was disrupted in MCF-7-R cells. Reduced mitochondrial clearance could promote the accumulation of dysfunctional mitochondria.
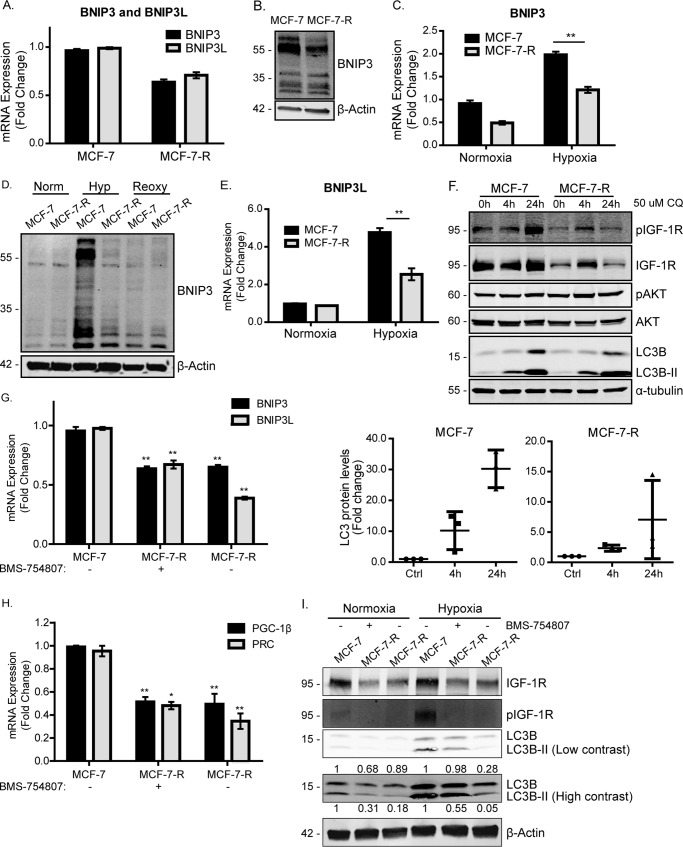
We first investigated the expression of BNIP3 and BNIP3L (Nip-like protein X) (26), which are transcriptional targets of HIF-1α (27) and can interact with LC3-related molecules at phagophore membranes to target mitochondria for degradation in autophagosomes (1, 25–28). BNIP3 suppression has also been associated with accumulation of dysfunctional mitochondria and with increased ROS production and mitochondrial mass under normoxic conditions (29). As can be seen in Fig. 4A, MCF-7-R cells exhibited lower levels of both BNIP3 and BNIP3L mRNA (Fig. 4A) and BNIP3 protein (Fig. 4B) than MCF-7 cells. BNIP3 migrates in SDS-PAGE as an ∼25- to 30-kDa monomer but may also form homodimers or heterodimers with other mitochondrial proteins (30). The reduced expression of both BNIP3 and BNIP3L in MCF-7-R cells suggested impaired BNIP3/BNIP3L-mediated mitophagy, which could contribute to increased mitochondrial mass in MCF-7-R cells.
Figure 4.
The mitophagy mediators BNIP3 and BNIP3L are reduced in MCF-7-R cells. A, BNIP3 and BNIP3L expression in MCF-7 and MCF-7-R cells. The levels of BNIP3 and BNIP3L were determined by qPCR, normalized to the housekeeping gene UBC. The levels in MCF-7-R cells are presented as -fold change of the levels in the MCF-7 cells set at 1. B, BNIP3 protein expression in MCF-7 and MCF-7-R cells. MCF-7 cells were maintained in complete medium, whereas MCF-7-R cells were maintained in complete medium supplemented with 500 nm BMS-754807. Cells were lysed and prepared for immunoblotting using anti-BNIP3 and anti-β-actin antibodies. C, BNIP3 mRNA in MCF-7 and MCF-7-R cells under normoxic or hypoxic (1% O2) conditions. Cells were exposed to hypoxia for 20 h. BNIP3 levels were determined by qPCR and normalized to the housekeeping gene UBC. The levels are presented as -fold change of expression in MCF-7 cells under normoxic conditions set at 1. D, BNIP3 protein expression in MCF-7 and MCF-7-R cells in normoxia (Norm), hypoxia (Hyp, 1% O2), or reoxygenation (Reoxy). Cells were maintained under normoxia, exposed to hypoxia for 20 h, or exposed to hypoxia for 20 h followed by normoxia for a further 5 h. Cells were then lysed and prepared for Western blotting using anti-BNIP3 and anti-β-Actin antibodies. E, qPCR showing BNIP3L mRNA in MCF-7 and MCF-7-R cells under normoxic or hypoxic (1% O2) conditions. Cells were exposed to hypoxia for 20 h. BNIP3L levels, normalized to UBC expression, are presented as -fold change relative to normoxia control samples for each cell line. F, LC3B levels in MCF-7 and MCF-7-R cells in the presence of 50 μm chloroquine (CQ) for 4 or 24 h. Cells were lysed and prepared for immunoblotting using anti-phospho-IGF-1R, anti-phospho-AKT, anti-AKT, anti-LC3B antibodies, and anti-α-tubulin antibodies. G, BNIP3 and BNIP3L expression in MCF-7 and MCF-7-R cells maintained in medium in the presence of 500 nm BMS-754807 or not, as indicated. The levels of BNIP3 and BNIP3L were normalized to the housekeeping gene UBC, and the levels are shown as -fold change of the levels in control (Ctrl) MCF-7 cells. H, PGC-1β and PRC expression in MCF-7 and MCF-7-R cells in the presence of 500 nm BMS-754807 or not, as indicated. PGC-1β and PRC levels, determined by qPCR, were normalized to the housekeeping gene UBC, and the levels are presented as -fold change of the levels in MCF-7 cells. The data for all PCR experiments are presented mean ± S.E. from three independent experiments. Statistical analysis was performed using Student's t test (*, p < 0.05; **, p < 0.01). I, LC3B protein levels in MCF-7 and MCF-7-R cells. MCF-7 and MCF-7-R cells were maintained in complete medium with or without 500 nm BMS-754807 as indicated. Cells were lysed and prepared for immunoblotting using anti-IGF-1R, anti-phospho-IGF-1R, anti-LC3B, and anti-β-actin antibodies. LCB3 expression was quantified by densitometry relative to actin and normalized to the MCF-7 cell controls. The -fold change in LC3B and LCB3II expression is indicated above and below the Western blot, respectively.
We next compared the levels of BNIP3 and BNIP3L expression in response to hypoxia in MCF-7 and MCF-7-R cells (1% O2 for 20 h). BNIP3 was induced in both MCF-7 and MCF-7-R cells upon exposure to hypoxia (Fig. 4C). However, the induction of BNIP3 was ∼2-fold lower in MCF-7-R cells than in MCF-7 cells. Similarly, the induction of BNIP3 protein by hypoxia was less in MCF-7-R cells than in MCF-7 cells (Fig. 4D). Similar results were observed for BNIP3L mRNA expression (Fig. 4E).
To assess levels of cellular autophagy, we examined the LC3B protein and its processed lower-migrating form, LC3B-II. It has been shown previously that hypoxia increases LC3B-II protein levels in an HIF-1/BNIP3-dependent manner and that suppression of BNIP3/BNIP3L in hypoxia reduces LC3B levels and activation of mitophagy (27, 31). We observed that MCF-7-R cells expressed overall lower levels of LC3B than parental controls (Fig. 4F), again indicating reduced autophagy. In the presence of the lysosomal inhibitor chloroquine, which suppresses autophagic flux, LC3B accumulated at a significantly greater rate in MCF-7 cells than in MCF-7-R cells (Fig. 4F). Interestingly, when BMS-754807 was removed from MCF-7-R cells for 14 days (MCF-7-R-CM), these cells maintained low BNIP3, BNIP3L (Figs. 4F), PGC-1β/PRC (Fig. 4H), and LC3B levels (Fig. 4I), indicating that mitochondrial metabolism is irreversibly reprogrammed.
Overall, the data indicate that MCF-7-R cells exhibit impaired mitophagy and impaired induction of BNIP3 necessary for initiating mitophagy. This suggests that IGF-1 signaling is required for efficient induction of BNIP3-dependent mitophagy.
BNIP3 expression and accumulation in mitochondria is induced by IGF-1
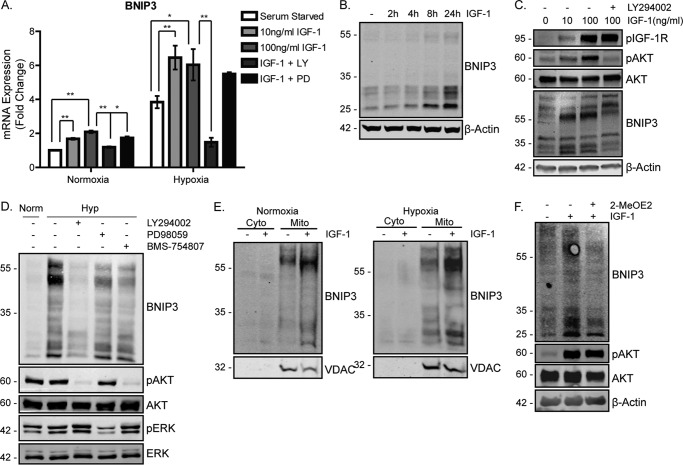
Because mitochondrial BNIP3/BNIP3L expression was impaired in MCF-7-R cells, we asked whether IGF-1 signaling generally stimulates expression of BNIP3. To test this, MCF-7 and ZR75.1 cells were serum-starved and then stimulated with IGF-1 for 20 h. As can be seen in Fig. 5A for MCF-7 cells, BNIP3 mRNA expression was significantly induced by IGF-1 under both normoxic and hypoxic conditions. BNIP3 mRNA expression was dependent on PI3K signaling because LY294002 suppressed IGF-1-induction, whereas the MAPK inhibitor PD90859 had little effect. IGF-1-mediated induction of BNIP3 protein was evident from 8 h following stimulation, and this was reduced by PI3K inhibition (Fig. 3C and supplemental Fig. 3C). MCF-7 cells exposed to hypoxia showed an increase in BNIP3 expression, as expected, but this was substantially attenuated by PI3K inhibition and less by MAPK and IGF-1R inhibition (Fig. 5D). Similar IGF-1-mediated induction of BNIP-3 was observed in ZR75.1 cells (supplemental Fig. 4, A and B). Subcellular fractionation of MCF-7 cell lysates indicated that the BNIP3 protein was located in mitochondrially enriched protein fractions under normoxic conditions and that IGF-1 significantly increased the levels of BNIP3 protein in mitochondria under both normoxic and hypoxic conditions (Fig. 5E).
Figure 5.
BNIP3 is induced by IGF-1 in a PI3K-dependent manner. A, BNIP3 mRNA expression following stimulation with IGF-1 in MCF-7 cells. Cells were serum-starved for 4 h, stimulated with 10 ng/ml or 100 ng/ml IGF-1, and placed under normoxic or hypoxic conditions for a further 20 h. Cells stimulated with 100 ng/ml IGF-1 were pretreated with LY294002 or PD98059 prior to being stimulated and placed under hypoxic conditions. The levels of BNIP3 were determined by qPCR, normalized to the housekeeping gene UBC, and are presented as -fold change of the levels in the cells under serum-starved normoxic conditions. Data are presented as mean ± S.E. from three independent experiments. The graph represents the mean ± S.D. derived from three individual experiments. Statistical significance was determined using Student's t test (*, p < 0.05; **p < 0.01). B, BNIP3 protein expression following stimulation with IGF-1 in MCF-7 cells over time. Cells were serum-starved for 4 h and stimulated with 10 ng/ml IGF-1 from 4–24 h. β-Actin was used as a loading control. C, BNIP3 protein expression dependence on PI3K activity. Cells were serum-starved for 4 h and stimulated with 10 ng/ml IGF-1 for a further 20 h. LY294002 was added 30 min prior to IGF-1 stimulation. Cells lysates were probed with anti-phospho-IGF-1R, anti-phospho-AKT, anti-AKT, anti-BNIP3, and anti-β-actin antibodies. D, BNIP3 expression in MCF-7 cells following exposure to hypoxia (Hyp). Cells were maintained in complete medium and exposed to DMSO (−), LY294002, PD98059, or BMS-754807 and placed under hypoxia for 20 h. Cells lysates were prepared for immunoblotting using anti-BNIP3, anti-phospho-AKT, anti-AKT, anti-phospho-ERK and anti-ERK antibodies. Norm, normoxia. E, subcellular localization of BNIP3 in MCF-7 cells under normoxic or hypoxic conditions. Cells were serum-starved and stimulated with 100 ng/ml IGF-1 for 20 h Cells were then lysed and subjected to subcellular fractionation before being harvested for lysis and Western blotting with anti-BNIP3 and anti-VDAC antibodies. Cyto indicates cytoplasmic fraction, and mito indicates mitochondria-enriched fraction. F, BNIP3 expression following IGF-1 stimulation in the presence of Myc inhibitor. MCF-7 cells were serum-starved for 4 h and stimulated with 100 ng/ml IGF-1 in the presence or absence of 2-MeOE2 as indicated. Cell lysates were prepared for immunoblotting using anti-BNIP3, anti-phospho-AKT, anti-AKT, and anti-β-actin antibodies. For all Western blots, a representative is presented from a minimum of three independent experiments that showed similar results.
IGF-1/PI3K signaling is known to regulate HIF-1α levels and expression of several of its target genes, but, unlike hypoxia, which inhibits degradation of HIF-1α, IGF-1/PI3K/mammalian target of rapamycin (mTOR) signaling increases HIF-1α synthesis (32). Because BNIP3 is an HIF-1α target gene, we next asked whether IGF-1–mediated induction of BNIP3 protein requires HIF-1α. To test this, cells were serum-starved for 4 h, preincubated with an HIF-1α inhibitor (2-MeOE2) for 30 min, and then stimulated with IGF-1. As can be seen in Fig. 5F and supplemental Fig. 3D, IGF-1–induced BNIP3 expression was significantly reduced by preincubation with 2-MeOE2, indicating that HIF-1α transcriptional activity contributes to induction of BNIP3 expression by IGF-1.
We also investigated whether BNIP3L mRNA was transcriptionally induced by IGF-1. However, BNIP3L was induced by serum starvation, and IGF-1 stimulation caused a slight decrease in BNIP3L mRNA expression (supplemental Fig. 4E). It has been reported that BNIP3 is induced to higher levels by hypoxia than BNIP3L because of the differential dependence of the two genes on the two transactivation domains in HIF-1α (33), which may explain this result. Other mitophagy regulators were also assessed. FUNDC1 mRNA levels were unaltered, whereas PINK1 mRNA was induced by serum starvation and suppressed by IGF-1 (supplemental Fig. 4E). Taken together, the data indicate that IGF-1 induces the mitophagy mediator BNIP3 in a HIF-1α-dependent manner and that reduced BNIP3 expression in MCF-7-R cells may impair mitophagy under both normoxic and hypoxic conditions.
Suppressed IGF-1R activity is associated with reduced mitophagy, mitochondrial dysfunction, and increased sensitivity to glycolysis inhibition
Having observed significantly reduced expression of BNIP3 in MCF-7-R cells compared with parental cells under both normoxic and hypoxic conditions, as well as increased mitochondrial mass, it was next important to assess how effectively these cells activated mitophagy and the clearance of mitochondria in response to hypoxia.
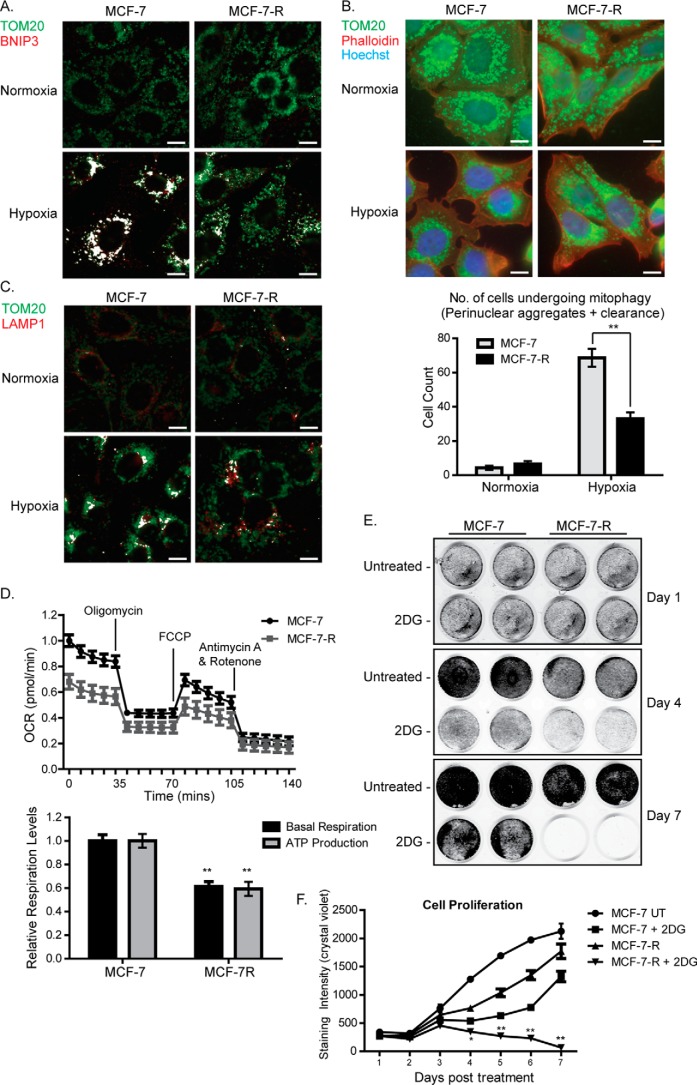
As shown in Fig. 6A, using an anti-TOM20 antibody (green) as a mitochondrial marker and an anti-BNIP3 antibody (red), we compared the co-localization of BNIP3 with mitochondria in MCF-7 and MCF-7-R cells; co-localization of TOM20 with BNIP3 is represented in white. Under normoxic conditions, very little co-localization of TOM20 with BNIP3 was observed, which could be expected with low expression of BNIP3. In MCF-7 cells exposed to hypoxia, a significant increase in TOM20 and BNIP3 co-localization was evident, which is likely due to increased expression of BNIP3 (Fig. 6A). Although co-localization of TOM20 with BNIP3 was also observed to be increased after exposure to hypoxia in MCF-7-R cells, this was much lower than in MCF-7 cells. This suggests that BNIP3-mediated clearance of mitochondria is less effective in MCF-7-R cells than MCF-7 cells.
Figure 6.
MCF-7-R cells display reduced levels of mitophagy, reduced mitochondrial activity, and increased sensitivity to glycolysis inhibition. A, localization of BNIP3 at mitochondria of MCF-7 and MCF-7-R cells. Cells were exposed to hypoxia for 24 h and prepared for immunofluorescence with an antibody specific for BNIP3 (red) and a TOM20 antibody (green) as a mitochondrial marker. Co-localization, shown in white, was assessed using the Colocalization plugin for ImageJ described under “Materials and Methods.” B, immunofluorescence depicting levels of mitophagy in MCF-7 and MCF-7-R cells. Cells were exposed to hypoxia for 24 h and then fixed and stained for TOM20 (green) or phalloidin (actin, red). Nuclei were stained with Hoechst (blue). The graph shows quantification of the number of cells that were undergoing mitophagy following exposure to hypoxia as described in the text. The graph represents the average ± S.E. of three individual experiments. Statistical significance was determined using Student's t test (*, p < 0.05; **, p < 0.01). C, co-localization of TOM20 with LAMP1 in MCF-7 and MCF-7-R cells. Cells were exposed to hypoxia for 24 h, and then they were fixed and stained with anti-OM20 (green) or -LAMP1 (red) antibodies. D, mitochondrial stress test showing mitochondrial activity in MCF-7 and MCF-7-R cells. The top plot shows the OCR, measured using a Seahorse XFp analyzer, over a course of 2 h under basal conditions and following addition of the indicated uncouplers. The bar chart shows basal respiration and ATP production, which were calculated as described under “Materials and Methods.” The data represent the mean ± S.E. derived from three independent experiments. FCCP, carbonyl cyanide p-trifluoromethoxyphenylhydrazone. E, growth rates of MCF-7 parental and MCF-7-R cells following exposure to 25 mm 2-DG. Cells were initially plated at a density of 2.5 × 104/well of a 24-well plate, and cell density was assessed every 24 h using crystal violet staining and quantified using Odyssey scanning. Representative cell culture plates are shown in the top panel, and cell density from three independent experiments are shown in the graph as mean ± S.D. Statistical analysis for each experiment was performed using Student's t test (*, p < 0.05; **, p < 0.01). Scale bars = 25 μm.
We next measured the clearance of mitochondria in response to hypoxia in both cell lines. Generally, mitophagy occurs in three visible stages: first, the mitochondria isolate and begin to migrate toward the nucleus, then they start to form large aggregates around the nucleus, and finally they are cleared by the autophagosomes, resulting in a reduction in overall mitochondrial mass (34). To estimate the extent of mitophagy in parental and resistant cells, we analyzed the morphology of 100 randomly chosen cells from each population and sorted them into three categories: 1) cells that have not entered mitophagy (mitochondria spread evenly throughout the cell), 2) cells in which the mitochondria have started to form perinuclear aggregates, and 3) cells that have started to clear mitochondria (significantly reduced mitochondrial staining). We then combined the numbers of cells assigned to categories 2 and 3 to give an estimation of cells undergoing mitophagy. As can be seen from the representative image and graph in Fig. 6B, ∼65% of MCF-7 parental cells had entered mitophagy following 24 h of hypoxia exposure, whereas only 30% of MCF-7-R cells had entered mitophagy and were apparently successfully clearing mitochondria. To further evaluate the levels of mitophagy in MCF-7-R cells, we measured the co-localization of TOM20 with LAMP1, a glycoprotein located on the outer membrane of lysosomes, under normoxia and hypoxia. Co-localization of TOM20 with LAMP1 would indicate delivery of the mitochondria to the lysosomes for degradation. As can be seen in Fig. 6C, co-localization (represented in white) of TOM20 with LAMP1 was greatly increased in MCF-7 cells following exposure to hypoxia. Although also increased in MCF-7-R cells, the levels were clearly lower than in MCF-7 cells. Altogether, the data indicate that MCF-7-R cells exhibit a reduced ability to enter mitophagy under hypoxic conditions.
Reduced clearance of mitochondria by impaired BNIP3-mediated mitophagy would be expected to cause an accumulation of dysfunctional mitochondria, which is consistent with the increased mitochondrial mass observed in MCF-7-R cells. To assess mitochondrial function, we measured respiration in MCF-7-R cells using the Seahorse Mito Stress Test assay. Both basal respiration and mitochondrial ATP production were significantly reduced in MCF-7-R cells compared with parental cells (Fig. 6D). Together, these results indicate that reduced BNIP3 expression and reduced clearance of mitochondria are associated with increased mitochondrial mass but reduced mitochondrial function.
Although MCF-7-R cells clearly exhibit mitochondrial dysfunction and increased ROS production, these cells survive and proliferate well with dysfunctional mitochondria (Fig. 3F and supplemental Fig. 3F). We therefore hypothesized that MCF-7-R cells may be more dependent on glycolysis than OXPHOS for ATP production and survival and would thus be more sensitive to glycolytic inhibitors than parental MCF-7 cells. To test this, we investigated the effects of 2-deoxyglucose (2-DG) on the proliferation of MCF-7 parental and MCF-7-R cells over 7 days. As can be seen in Fig. 6, E and F, MCF-7 parental cells continue to proliferate in the presence of 2-DG, albeit at a slower rate. However, MCF-7-R cells did not survive in the presence of 2-DG, and no viable cells were detected after 7 days in culture. MCF-7-R cells also displayed decreased colony-forming ability when cultured over time in the presence of lower concentrations (2–10 mm) of 2-DG compared with parental cells (supplemental Fig. 4A). Taken together, these data indicate that MCF-7-R cells are exquisitely sensitive to inhibition of glycolysis. The increased sensitivity of MCF-7-R cells to 2-DG was not associated with impaired PI3K signaling. As can be seen in supplemental Fig. 4B, 2-DG increased AKT phosphorylation in MCF-7 parental cells. It has been reported (35) that co-exposure of MCF-7 cells to 2-DG and BMS-754807 suppresses the effect of 2-DG on AKT phosphorylation. Interestingly, 2-DG induced phosphorylation of AKT in MCF-7-R cells even in the presence of BMS-754807 and suppressed IGF-1R activity, confirming that IGF-1R activity is not required for AKT activation in these cells. We conclude that the increased sensitivity of MCF-7-R cells to 2-DG is due to the inability of MCF-7-R cells to compensate for the inhibition of glycolytic ATP flux in the presence of an already impaired OXPHOS pathway.
Discussion
Our findings provide strong evidence for an essential IGF-1/PI3K signal that stimulates both mitochondrial biogenesis and mitophagy, principally through induction of the PGC-1β and PRC transcriptional activators and through induction of BNIP3, an important mediator of mitophagy. In this way, IGF-1 signaling enables cells to maintain a pool of healthy mitochondria to facilitate cellular bioenergetic efficiency and mitochondrial homeostasis. This IGF-promoted mitochondrial protective activity has an obvious function in normal cells, but its activity in cancer cells may be particularly important for tumor growth and acquired resistance to cancer therapies, where accumulating evidence supports a key role for metabolic reprogramming and mitochondrial metabolism (10, 11, 13, 22, 36).
Mitochondrial biogenesis is generally controlled by the activity of the PGC-1 coactivators PGC-1α, PGC-1β, and PRC, which co-activate transcription factors, including NRF-1/NRF-2 and ERRα (37). Consistent with a previous study (16), we observed that IGF-1 stimulates PGC-1β expression. IGF-1 also induces PRC expression, and PGC-1β and PRC act in a redundant manner to support the transcription of mitochondrial genes and maintain mitochondrial morphology and mass. Acute inhibition of IGF-1R or PI3K reduced expression of both PGC-1β and PRC and caused decreased mitochondrial mass and biogenesis. Mitochondrial biogenesis was also reduced in an MCF-7 cell line variant (MCF-7-R) that was derived for adaptive resistance to the IGF-1R kinase inhibitor BMS-754807 (23). These cells exhibited increased mitochondrial mass, high ROS levels, and decreased expression of markers of mitochondrial biogenesis, although NFE2L2/NRF-2 levels were increased, which may contribute to survival and antioxidant protection.
BNIP3 mRNA expression and BNIP3 protein levels were reduced in MCF-7-R cells, indicating impaired mitophagy and the accumulation of dysfunctional mitochondria. Although EGF and IGF signaling have been linked previously to mitochondrial biogenesis via mTOR, cMyc, and PGC-1α (16, 38), overall mitochondrial protection was apparently not rescued by the clearly increased EGFR and Her2 activity observed in MCF-7-R cells. Although it is possible that some component of the increased mitochondrial mass observed in MCF-7-R cells could be due to an alternative compensatory mitochondrial biogenesis program mediated by NFE2L2, we could not detect any increase in NRF-1/α PAL that might mediate this. Thus, our data support the overall conclusion of an essential basal function for the IGF signaling pathway in supporting both mitochondrial biogenesis and mitochondrial turnover.
Our observations with MDA-MB-231 cells, which, unlike most breast cancer cell lines, express high levels of PGC-1α and low levels of PGC-1β and PRC, suggest that IGF-1–promoted mitochondrial biogenesis does not require PGC-1α expression or activity. A number of recent studies have proposed that enhanced OXPHOS potential in cancer cells is mediated by PGC-1α induction (9, 10), but we propose that it is very likely that PGC-1β and PRC also contribute to mitochondrial protection in these scenarios. Indeed, the mitobiogenesis signature described in melanoma cells (12) includes all three PGC co-activators and other genes that are induced by IGF-1, including MFN1 and TFAM. Thus, PGC-1α expression in cancer cells is likely associated with metabolic stress and is not essential for basal mitochondrial maintenance. Our observation that either inhibition of PI3K or ROS scavenging was sufficient to block IGF-1–promoted mitochondrial biogenesis in MDA-MB-231 cells supports this conclusion.
Our observation of reduced BNIP3 and mitophagy in MCF-7-R cells led us to investigate whether IGF-1 induces BNIP3 expression and thereby induces mitophagy potential in concert with induction of mitochondrial biogenesis. Mitophagy requires the interaction of adaptor proteins in the outer mitochondrial membrane with LC3B-II at the phagophore membrane for subsequent autophagosomal degradation of mitochondria (39). The observation that IGF-1 induces BNIP3 mRNA and protein expression and mitophagy could at first be considered somewhat surprising because BNIP3 can be induced by FOXO1, which is suppressed by IGF-1, and because nutrient deprivation is a well-described activator of mitophagy via suppressed TOR activity. There is, however, strong evidence for IGF signaling in supporting both mitochondrial biogenesis (PGC-1β and PRC) and turnover (BNIP3) from studies on longevity (40). SIRT1 has been reported to induce mitochondrial biogenesis by deacetylating and thus activating PGC-1α in times of nutrient deprivation (41) while also stimulating mitophagy (42). Similarly, AMPK regulates mitochondrial biogenesis through activation of PGC-1α (43), whereas loss of AMPK, and thus its downstream target ULK1, impairs mitophagy during starvation (44). SKN-1, the Caenorhabditis elegans NFE2L2/NRF-2 homologue, drives the expression of various mitochondrial genes but also modulates the expression of DCT-1, the C. elegans homologue of BNIP3 (45). Finally, increased OXPHOS has been shown to promote turnover of mitochondria via Rheb and BNIP3L-mediated mitophagy to maintain optimal efficiency of mitochondrial ATP production (46). Although loss of mTOR signaling has been strongly implicated in the initiation of mitophagy, mTOR was active in this setting, and BNIP3L-mediated mitophagy was shown to occur independently of mTOR activity (46). Were IGF-1 signaling to increase mitochondrial biogenesis without also stimulating mitophagy, this could result in a continuous increase in OXPHOS and, thus, increased ROS production and potential cellular damage and accumulation of old or damaged mitochondria, resulting in elevated ROS production that could damage cells.
We propose that IGF/PI3K signaling enhances mitochondrial homeostasis by increasing the pool of new mitochondria and increasing OXPHOS to produce ATP required for proliferation while simultaneously degrading old, damaged mitochondria through increasing BNIP3 expression. Our data are also consistent with previous studies showing that BNIP3 suppression reduces mitophagy and causes an accumulation of dysfunctional mitochondria, even under normoxic conditions, illustrating a role for BNIP3 in basal cellular mitophagy (47). In a broader context, impaired mitochondrial function is associated with numerous pathological conditions and with aging (47). Enhancing mitochondrial function may be an important mechanism by which IGF-1 promotes tissue protection and regeneration (15).
Our findings directly suggest that IGF-1-mediated mitochondrial protection plays a major role in metabolic reprogramming and resistance to targeted cancer therapies. Sustained or elevated IGF-1R signaling would maintain healthy mitochondria to sustain increased growth while also removing dysfunctional mitochondria to prevent accumulation of ROS, thereby enhancing stress resistance and cell survival. This idea is supported by recent observations on the role of mitochondrial biogenesis in melanoma cells exposed to MAPK inhibitors (12), where many IGF-1–responsive genes are induced and where suppression of PRC, TFAM, or estrogen related receptor alpha (ESSRA) were all shown to augment cell killing in the presence of resistance to MAPK inhibitors.
We also hypothesize that co-targeting of the IGF-1R and glycolysis may represent a novel strategy to overcome resistance to IGF-1R therapies. Although MCF-7-R cells exhibited a 1.4-fold increase in mitochondrial mass, overall mitochondrial function was significantly reduced, and MCF-7-R cells were exceptionally sensitive to glycolysis inhibition with 2-DG. We conclude that, although MCF-7 cells may up-regulate OXPHOS in response to 2-DG treatment to compensate for the loss of ATP production through glycolysis, MCF-7-R cells cannot sufficiently increase OXPHOS because of the presence of dysfunctional mitochondria. Co-inhibiting mitochondria and glycolysis has been shown previously enhance cancer cell death (48, 49). Thus, suppression of OXPHOS potential through inhibition of the IGF–PI3K pathway with concomitant suppression of glycolysis could be a highly effective anticancer therapy.
Materials and methods
Antibodies
Rabbit polyclonal anti-IGF-1R (c20, sc-713), rabbit TOM2O (sc-11415), and goat VDAC1 (N-18, sc-8828) antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Mouse anti-AKT (2920), rabbit anti-phospho-AKT (Ser-473, 4060s), mouse anti-ERK (4696), rabbit anti-phospho-ERK (Thr-202/Tyr-204, 4377), rabbit anti-IGF-1R PY1135/136 (3024s), rabbit anti-p70S6K (2708), rabbit anti-phospho-p70S6K (Thr-389, 9205), rabbit anti-poly-ADP ribose polymerase (PARP) (9542), rabbit anti-phospho-H2AX (Ser-139, 2577), and rabbit anti-LC3B (3868) antibodies were from Cell Signaling Technology (Beverly, MA). Mouse MitoProfile Total OXPHOS human (ab110411) and mouse anti-BNIP3 (ab10433) antibodies were purchased from Abcam (Cambridge, UK).
Cell lines and cell culture
MCF-7 and MDA-MB-231 cells were obtained from the ATCC (Manassas, VA). ZR75.1 and TOV112D cells were a kind gift from Almac Diagnostics. MCF-7, MDA-MB-231, R−, and R+ cells were all maintained in Dulbecco's modified Eagle's medium (Biowhittaker, Verviers, Belgium) supplemented with 10% (v/v) heat-inactivated FBS, 10 mm l-glutamine, and 5 mg/ml penicillin/streptomycin (complete medium) at 37 °C in a humidified 95% air, 5% CO2 atmosphere. ZR75.1 and TOV112D cells were maintained in Roswell Park Memorial Institute medium. For analysis of cell signaling in response to IGF-1, MCF7, MDA-MB-231, and ZR75.1 cells were seeded at a density of 3 × 105 cells/well of a 6-well plate. R− cells were seeded at a density of 1.5 × 105 cells/well of a 6-well tissue culture dish. Approximately 16 h after seeding, the cells were washed three times with serum-free medium (DMEM with 10 mm l-glutamine) and maintained in serum-free medium for a total of 4 h before stimulation with IGF-1 (10 ng/ml or 100 ng/ml). All cells were lysed in either Nonidet P-40 or RIPA lysis buffer.
BMS-754807–resistant MCF-7 cells (MCF-7-R cells) were derived from MCF-7 cells that were cultured in increasing stepwise increments of BMS-754807, starting at 100 nm BMS-754807, until cells were proliferating successfully in the presence of each increased drug concentration and a resistant pool had been generated; they have been described previously (23). Cells were then permanently maintained in DMEM containing 500 nm BMS-754807.
Transfection with siRNA and chemical inhibitors
To suppress PGC-1β and PRC expression, siRNA pools targeting human PGC-1β (L-008556-00-0005) and human PRC (L-016647-00-0005) were obtained from Dharmacon. Silencer negative control 1 was purchased from Thermo Fisher. Reverse siRNA transfections were carried out using Lipofectamine RNAiMAX transfection reagent (Invitrogen) according to the instructions of the manufacturer.
BMS-754807 was purchased from Active Biochem (Hong Kong, China). LY294002 and PD98059 were purchased from Merck Millipore. 2-Deoxyglucose was purchased from Sigma-Aldrich. 10058-F4 and 2-MeOE2 were purchased from Selleckchem (Munich, Germany).
Proliferation and wound healing assays
For proliferation assays, MCF-7 and MCF-7-R cells were seeded at a density of 2 × 105 cells/well of a 24-well plate in triplicate cells. After 24 h, cells were drugged as required. Each day, cells were fixed with using 96% ethanol for 10 min and stained with 0.05% crystal violet in 0.1% ethanol for 30 mins. Cell intensity was measured by infrared scanning using Odyssey scanning equipment.
To measure wound healing, cells were seeded in 6-well plates to near confluency. After 24 h, the monolayer was scratched using a sterile p10 tip (time 0 h). The cells were left to migrate into the wound for 16–24 h. Pictures were taken of the wound at 0 h and 24 h using a ×10 objective.
Immunofluorescence and flow cytometry
MCF-7 cells were seeded in a 6-well plate with untreated sterile coverslips at 2.5 × 105 cells/well. After 18 h, the cells were held in either 1% O2 or normoxia for an additional 18 h. Cells were fixed with 4% paraformaldehyde (PFA)/PHEM and permeabilized with 0.1% Triton X/PHEM. Nonspecific binding was blocked using 5% goat serum/PHEM, and cells were stained with anti TOM20 1:600 (Santa Cruz Biotechnology, sc-11415), anti-BNIP3 (1:200, Abcam, ab10433), and anti-LAMP1 (1:100, Santa Cruz Biotechnology, sc-20011). Cells were photographed at ×40 or ×100 magnification by epifluorescence. A background removal correction was performed on each image, using a rolling ball radius of 50 pixels. The resulting 8-bit .tiff files were analyzed using the Colocalization plugin in ImageJ to generate a mask of co-localization using a threshold of 50 for each image, with a ratio threshold of 50%.
For MCF-7 cells transfected with PGC-1β and PRC siRNA, the cells were reseeded onto untreated sterile coverslips in a 6-well plate 24 h after transfection and, 72 h after transfection, were fixed and permeabilized as described above. Blocking was performed with 5% donkey serum/PHEM. The anti-TOM20 antibody was used at 1:500. Images were taken at ×100 magnification.
For flow cytometry analysis, the following probes were used: 100 nm MitoTracker Green (MTG) (Molecular Probes) for mitochondrial mass, 5 μm MitoSOX Red (Molecular Probes) for mitochondrial superoxide detection, and 500 nm of TMRE (Sigma) for mitochondrial membrane potential analysis. Cells were seeded at a density of 3 × 105 cells/well of a 6-well plate, and 24 h later, the cells were incubated in the presence of the relevant probe for 30 min with MitoSOX and MTG and 20 min with TMRE at 37 °C in the dark; thereafter, cells were collected and washed in PBS. Cells were then resuspended in PHEM buffer and analyzed by FACS using FACSCalibur (BD Biosciences) and Cellquest Pro software (BD Biosciences). The fluorescent intensity of cells of 10,000 events was recorded in the FL-1 channel for MTG or in the FL-2 channel for TMRE and MitoSOX. The geometric mean of three technical replicates of the fluorescence of cells was calculated.
Cell lysis, SDS-PAGE, and Western blotting
Cells were lysed with RIPA lysis buffer (50 mm Tris, 150 mm NaCl, 0.1% SDS, 0.5% sodium deoxycholate, and 1% Nonidet P-40 (pH 7.4)) for 20 min on ice. All protein samples for Western blot analysis were separated using 4–16% SDS-PAGE gels. Following SDS-PAGE, proteins were transferred electrophoretically to a nitrocellulose membrane using a Bio-Rad Mini Trans-Blot electrophoretic transfer cell. Membranes were then blocked for 1 h at room temperature in TBS containing 0.05% Tween 20 (TBS-T) and 5% nonfat dried milk (w/v) or 5% BSA as indicated. Following overnight incubation with primary antibody, membranes were probed with IRdye700- or IRdye800-conjugated secondary antibodies, and proteins were detected using the Odyssey image scanner system (LI-COR Biosciences, Cambridge, UK). Molecular masses in kilodaltons are indicated on the left of each Western blot.
Quantitative PCR
RNA was extracted using the PureLink RNA Mini Kit (Thermo Scientific, Bremen, Germany) according to the instructions of the manufacturer. cDNA synthesis was performed with the QuantiTect reverse transcription kit (Qiagen) using 1 μg of RNA. The primers used for QPCR are shown in supplemental Table 1. Quantitative PCR was performed using the Roche FastStart essential DNA Green Master Kit using a Lightcycler® 96 Instrument (Roche Diagnostics). Ubiquitin C (UBC) was used as housekeeping control, and the 2−ΔΔCT method was used to analyze data and determine relative mRNA expression levels. Gene expression levels were normalized to control samples that were set at a value of 1 and expressed as -fold change. Data are presented from three independent experiments as mean ± S.E.
Mitochondrial membrane protein enrichment
Mitochondria were isolated using the Mitochondria Isolation Kit For Cultured Cells (Thermo Scientific). Cells were seeded onto 2 × 15 cm dishes and allowed to grow to ∼70% confluence to ensure high mitochondrial yield. Cells were pelleted at 850 × g for 2 min and resuspended in mitochondrial isolation reagent A. Cells were vortexed at medium speed for 5 s and incubated on ice for 2 min. Mitochondrial isolation reagent B was then added, and the solution was vortexed at maximum speed for 5 s before being incubated on ice for 5 min. Mitochondrial isolation C reagent was added, and the tube was inverted several times. The tube was then centrifuged at 700 × g for 10 min at 4 °C. The supernatant was removed to a fresh tube to which mitochondrial isolation reagent C was then added. Mitochondria were pelleted from the supernatant (cytosol) through centrifugation at 12,000 × g for 15 min at 4 °C. Pellets were then washed once in mitochondrial isolation reagent C, and the mitochondria were pelleted through centrifugation at 12,000 × g for 5 min. The mitochondrial fraction was lysed for 30 min in RIPA lysis buffer. For immunoblotting, an antibody specific to the mitochondrial outer membrane protein voltage-dependent anion channel (VDAC) was used to confirm pure subcellular fractions.
Hypoxia exposure
Hypoxia experiments were performed using a hypoxia chamber (Stemcell Technologies). The chamber was flushed with nitrogen for 10–15 min until the oxygen concentration within the chamber reached 1%. Oxygen concentration was measured using an Optical Fluorescence O2 analyzer (Luxcel Biosciences, Cork, Ireland).
Oxygen consumption measurements
MCF-7 and MCF-7-R cells were cultured in DMEM supplemented with 10% FBS, 1% l-glutamine, and 1% penicillin and streptomycin, and MCF-7-R cells were cultured in the presence of 500 nm BMS-754807. The oxygen consumption rate was measured in these cells using a Seahorse XFp instrument and Seahorse XFp Cell Mito Stress Test Kit (Seahorse Biosciences). 2 × 104 cells/well were seeded in Seahorse XFp FluxPak plates. After 18 h, the growth medium was removed and replaced with XF assay medium (modified DMEM supplemented with 2 mm l-glutamine, 1 mm sodium pyruvate, and 10 mm glucose), freshly made up and adjusted to pH 7.4. The plate was allowed to equilibrate at 37 °C and ambient CO2 pressure for 1 h. The OCR was measured using a Seahorse XFp analyzer. 20 measures were taken at regular intervals over the course of 2 h. The OCR was measured under basal conditions and after serial addition of each 50 μm oligomycin, 50 μm carbonyl cyanide p-trifluoromethoxyphenylhydrazone, and 25 μm rotenone/antimycin A. The OCR was standardized for total protein concentration after the assay was completed. Sequential addition of oligomycin, carbonyl cyanide p-trifluoromethoxyphenylhydrazone, and antimycin plus rotenone allowed for the measurement of basal respiration, ATP production, proton leak, maximal respiration, and spare capacity.
Receptor tyrosine kinase array
The human phospho-RTK array (R&D Systems, Minneapolis, MN) was used according to the instructions of the manufacturer. Cells (1 × 107)) were lysed in the supplied lysis buffer 17, and 800 μg of the lysate was incubated for 1 h at room temperature with the array membrane. The membrane was washed with the supplied wash buffer and incubated with HRP-tagged mouse anti-phosphotyrosine antibody, and the signal was generated using ECL Plus reagent (RPN2133, GE Healthcare). Chemiluminescence was detected using FujiFilm Chemiluminescence LAS-3000, which was also used to quantitate the signals.
Author contributions
A. L., M. C., S. R., C. F., and C. H. O. contributed to conception and design, acquisition of data, analysis and interpretation of data, and drafting of the article. A. V. Z., D. B. P., and S. D. H. contributed to conception and design and critical review of the article. R. O. C. contributed to conception design, interpretation of data, and drafting of the article.
Supplementary Material
Acknowledgments
We thank Orla Cox for assistance with immunofluorescence and colleagues in the Cell Biology Laboratory for helpful discussions.
This work was funded by Science Foundation Ireland PI Awards 11/PI/1139 and 06/IN.1/B107; the Health Research Board; the Higher Education Authority of Ireland Ph.D. Program in Molecular Cell Biology (to A. L.); and a Ph.D. Studentship from the Irish Cancer Society (to M. C.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supplemental Figs. 1–5 and Table 1.
- PGC
- peroxisome proliferator–activated receptor γ coactivator
- OXPHOS
- oxidative phosphorylation
- IGF
- insulin-like growth factor
- ROS
- reactive oxygen species
- PRC
- PGC-1α–related coactivator
- VDAC
- voltage-dependent anion channel
- siNeg
- siRNA negative control
- TKI
- tyrosine kinase inhibitor
- IGF-1R
- insulin-like growth factor 1 receptor
- NRF
- nuclear respiratory factor
- EGFR
- EGF receptor
- mTOR
- mechanistic target of rapamycin
- 2-DG
- 2-deoxyglucose
- 2-MeOE2
- 2-methoxyestradiol
- MTG
- MitoTracker Green
- PHEM
- PIPES HEPES EGTA Magnesium
- RTK
- receptor tyrosine kinase
- TMRE
- tetramethylrhodamine ethyl ester
- UBC
- ubiquitin C
- OCR
- oxygen consumption rate
- qPCR
- quantitative PCR.
References
- 1. Vyas S., Zaganjor E., and Haigis M. C. (2016) Mitochondria and Cancer. Cell 166, 555–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Palikaras K., Lionaki E., and Tavernarakis N. (2015) Coordination of mitophagy and mitochondrial biogenesis during ageing in C. elegans. Nature 521, 525–528 [DOI] [PubMed] [Google Scholar]
- 3. Raimundo N. (2014) Mitochondrial pathology: stress signals from the energy factory. Trends Mol. Med. 20, 282–292 [DOI] [PubMed] [Google Scholar]
- 4. Marín de Mas I., Aguilar E., Jayaraman A., Polat I. H., Martín-Bernabé A., Bharat R., Foguet C., Milà E., Papp B., Centelles J. J., and Cascante M. (2014) Cancer cell metabolism as new targets for novel designed therapies. Future Med. Chem. 6, 1791–1810 [DOI] [PubMed] [Google Scholar]
- 5. Moreno-Sánchez R., Marín-Hernández A., Saavedra E., Pardo J. P., Ralph S. J., and Rodríguez-Enríquez S. (2014) Who controls the ATP supply in cancer cells? Biochemistry lessons to understand cancer energy metabolism. Int. J. Biochem. Cell Biol. 50, 10–23 [DOI] [PubMed] [Google Scholar]
- 6. Gleyzer N., Vercauteren K., and Scarpulla R. C. (2005) Control of mitochondrial transcription specificity factors (TFB1M and TFB2M) by nuclear respiratory factors (NRF-1 and NRF-2) and PGC-1 family coactivators. Mol. Cell. Biol. 25, 1354–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Deblois G., St-Pierre J., and Giguère V. (2013) The PGC-1/ERR signaling axis in cancer. Oncogene 32, 3483–3490 [DOI] [PubMed] [Google Scholar]
- 8. Baldelli S., Aquilano K., and Ciriolo M. R. (2013) Punctum on two different transcription factors regulated by PGC-1α: nuclear factor erythroid-derived 2-like 2 and nuclear respiratory factor 2. Biochim. Biophys. Acta 1830, 4137–4146 [DOI] [PubMed] [Google Scholar]
- 9. LeBleu V. S., O'Connell J. T., Gonzalez Herrera K. N., Wikman H., Pantel K., Haigis M. C., de Carvalho F. M., Damascena A., Domingos Chinen L. T., Rocha R. M., Asara J. M., and Kalluri R. (2014) PGC-1α mediates mitochondrial biogenesis and oxidative phosphorylation in cancer cells to promote metastasis. Nat. Cell Biol. 16, 992–1003, 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Viale A., Pettazzoni P., Lyssiotis C. A., Ying H., Sánchez N., Marchesini M., Carugo A., Green T., Seth S., Giuliani V., Kost-Alimova M., Muller F., Colla S., Nezi L., Genovese G., et al. (2014) Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature 514, 628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chae Y. C., Vaira V., Caino M. C., Tang H. Y., Seo J. H., Kossenkov A. V., Ottobrini L., Martelli C., Lucignani G., Bertolini I., Locatelli M., Bryant K. G., Ghosh J. C., Lisanti S., Ku B., et al. (2016) Mitochondrial Akt regulation of hypoxic tumor reprogramming. Cancer Cell 30, 257–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang G., Frederick D. T., Wu L., Wei Z., Krepler C., Srinivasan S., Chae Y. C., Xu X., Choi H., Dimwamwa E., Ope O., Shannan B., Basu D., Zhang D., Guha M., et al. (2016) Targeting mitochondrial biogenesis to overcome drug resistance to MAPK inhibitors. J. Clin. Invest. 126, 1834–1856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Weinberg S. E., and Chandel N. S. (2015) Targeting mitochondria metabolism for cancer therapy. Nat. Chem. Biol. 11, 9–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Elstrom R. L., Bauer D. E., Buzzai M., Karnauskas R., Harris M. H., Plas D. R., Zhuang H., Cinalli R. M., Alavi A., Rudin C. M., and Thompson C. B. (2004) Akt stimulates aerobic glycolysis in cancer cells. Cancer Res. 64, 3892–3899 [DOI] [PubMed] [Google Scholar]
- 15. Sádaba M. C., Martín-Estal I., Puche J. E., and Castilla-Cortázar I. (2016) Insulin-like growth factor 1 (IGF-1) therapy: mitochondrial dysfunction and diseases. Biochim. Biophys. Acta 1862, 1267–1278 [DOI] [PubMed] [Google Scholar]
- 16. Chang C. Y., Kazmin D., Jasper J. S., Kunder R., Zuercher W. J., and McDonnell D. P. (2011) The metabolic regulator ERRα, a downstream target of HER2/IGF-1R, as a therapeutic target in breast cancer. Cancer Cell 20, 500–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cao H., Dong W., Qu X., Shen H., Xu J., Zhu L., Liu Q., and Du J. (2016) Metformin enhances the therapy effects of anti-IGF-1R mAb figitumumab to NSCLC. Sci Rep. 6, 31072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li L., Wang Y., Peng T., Zhang K., Lin C., Han R., Lu C., and He Y. (2016) Metformin restores crizotinib sensitivity in crizotinib-resistant human lung cancer cells through inhibition of IGF1-R signaling pathway. Oncotarget 7, 34442–34452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Floyd S., Favre C., Lasorsa F. M., Leahy M., Trigiante G., Stroebel P., Marx A., Loughran G., O'Callaghan K., Marobbio C. M., Slotboom D. J., Kunji E. R., Palmieri F., and O'Connor R. (2007) The insulin-like growth factor-I-mTOR signaling pathway induces the mitochondrial pyrimidine nucleotide carrier to promote cell growth. Mol. Biol. Cell 18, 3545–3555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Van Dyck E., Jank B., Ragnini A., Schweyen R. J., Duyckaerts C., Sluse F., and Foury F. (1995) Overexpression of a novel member of the mitochondrial carrier family rescues defects in both DNA and RNA metabolism in yeast mitochondria. Mol. Gen. Genet. 246, 426–436 [DOI] [PubMed] [Google Scholar]
- 21. Favre C., Zhdanov A., Leahy M., Papkovsky D., and O'Connor R. (2010) Mitochondrial pyrimidine nucleotide carrier (PNC1) regulates mitochondrial biogenesis and the invasive phenotype of cancer cells. Oncogene 29, 3964–3976 [DOI] [PubMed] [Google Scholar]
- 22. Gopal Y. N., Rizos H., Chen G., Deng W., Frederick D. T., Cooper Z. A., Scolyer R. A., Pupo G., Komurov K., Sehgal V., Zhang J., Patel L., Pereira C. G., Broom B. M., Mills G. B., et al. (2014) Inhibition of mTORC1/2 overcomes resistance to MAPK pathway inhibitors mediated by PGC1α and oxidative phosphorylation in melanoma. Cancer Res. 74, 7037–7047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. O'Flanagan C.H., O'Shea S., Lyons A., Fogarty F. M., McCabe N., Kennedy R. D., and O'Connor R. (2016) IGF-1R inhibition sensitizes breast cancer cells to ATM-Related Kinase (ATR) inhibitor and cisplatin. Oncotarget 7, 56826–56841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dominy J.E., and Puigserver P. (2013) Mitochondrial biogenesis through activation of nuclear signaling proteins. Cold Spring Harb. Perspect. Biol. 5, a015008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hamacher-Brady A., and Brady N. R. (2016) Mitophagy programs: mechanisms and physiological implications of mitochondrial targeting by autophagy. Cell. Mol. Life Sci. 73, 775–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhang J., and Ney P. A. (2011) Mechanisms and biology of B-cell leukemia/lymphoma 2/adenovirus E1B interacting protein 3 and Nip-like protein X. Antioxid. Redox Signal. 14, 1959–1969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang H., Bosch-Marce M., Shimoda L. A., Tan Y. S., Baek J. H., Wesley J. B., Gonzalez F. J., and Semenza G. L. (2008) Mitochondrial autophagy is an HIF-1-dependent adaptive metabolic response to hypoxia. J. Biol. Chem. 283, 10892–10903 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28. Zhang P., Zhang X., Hao X., Wang Y., Hui Y., Wang H., Hu D., and Zhou J. (2009) Rac1 activates HIF-1 in retinal pigment epithelium cells under hypoxia. Graefes Arch. Clin. Exp. Ophthalmol. 247, 633–639 [DOI] [PubMed] [Google Scholar]
- 29. Chourasia A. H., Tracy K., Frankenberger C., Boland M. L., Sharifi M. N., Drake L. E., Sachleben J. R., Asara J. M., Locasale J. W., Karczmar G. S., and Macleod K. F. (2015) Mitophagy defects arising from BNip3 loss promote mammary tumor progression to metastasis. EMBO Rep. 16, 1145–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhu Y., Massen S., Terenzio M., Lang V., Chen-Lindner S., Eils R., Novak I., Dikic I., Hamacher-Brady A., and Brady N. R. (2013) Modulation of serines 17 and 24 in the LC3-interacting region of Bnip3 determines pro-survival mitophagy versus apoptosis. J. Biol. Chem. 288, 1099–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bellot G., Garcia-Medina R., Gounon P., Chiche J., Roux D., Pouysségur J., and Mazure N. M. (2009) Hypoxia-induced autophagy is mediated through hypoxia-inducible factor induction of BNIP3 and BNIP3L via their BH3 domains. Mol. Cell. Biol. 29, 2570–2581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fukuda R., Hirota K., Fan F., Jung Y. D., Ellis L. M., and Semenza G. L. (2002) Insulin-like growth factor 1 induces hypoxia-inducible factor 1-mediated vascular endothelial growth factor expression, which is dependent on MAP kinase and phosphatidylinositol 3-kinase signaling in colon cancer cells. J. Biol. Chem. 277, 38205–38211 [DOI] [PubMed] [Google Scholar]
- 33. Dayan F., Roux D., Brahimi-Horn M. C., Pouyssegur J., and Mazure N. M. (2006) The oxygen sensor factor-inhibiting hypoxia-inducible factor-1 controls expression of distinct genes through the bifunctional transcriptional character of hypoxia-inducible factor-1α. Cancer Res. 66, 3688–3698 [DOI] [PubMed] [Google Scholar]
- 34. Lee J. Y., Nagano Y., Taylor J. P., Lim K. L., and Yao T. P. (2010) Disease-causing mutations in parkin impair mitochondrial ubiquitination, aggregation, and HDAC6-dependent mitophagy. J. Cell Biol. 189, 671–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Xia L. M., Huang W. J., Wang B., Liu M., Zhang Q., Yan W., Zhu Q., Luo M., Zhou Z. Z., and Tian D. A. (2009) Transcriptional up-regulation of FoxM1 in response to hypoxia is mediated by HIF-1. J. Cell. Biochem. 106, 247–256 [DOI] [PubMed] [Google Scholar]
- 36. Caino M. C., and Altieri D. C. (2016) Molecular pathways: mitochondrial reprogramming in tumor progression and therapy. Clin Cancer Res. 22, 540–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Scarpulla R. C. (2011) Metabolic control of mitochondrial biogenesis through the PGC-1 family regulatory network. Biochim. Biophys. Acta 1813, 1269–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tazearslan C., Huang J., Barzilai N., and Suh Y. (2011) Impaired IGF1R signaling in cells expressing longevity-associated human IGF1R alleles. Aging Cell 10, 551–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kim I., Rodriguez-Enriquez S., and Lemasters J. J. (2007) Selective degradation of mitochondria by mitophagy. Arch. Biochem. Biophys. 462, 245–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Palikaras K., Lionaki E., and Tavernarakis N. (2015) Coupling mitogenesis and mitophagy for longevity. Autophagy 11, 1428–1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gerhart-Hines Z., Rodgers J. T., Bare O., Lerin C., Kim S. H., Mostoslavsky R., Alt F. W., Wu Z., and Puigserver P. (2007) Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1α. EMBO J. 26, 1913–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lee I. H., Cao L., Mostoslavsky R., Lombard D. B., Liu J., Bruns N. E., Tsokos M., Alt F. W., and Finkel T. (2008) A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc. Natl. Acad. Sci. U.S.A. 105, 3374–3379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jäger S., Handschin C., St-Pierre J., and Spiegelman B. M. (2007) AMP-activated protein kinase (AMPK) action in skeletal muscle via direct phosphorylation of PGC-1α. Proc. Natl. Acad. Sci. U.S.A. 104, 12017–12022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Egan D. F., Shackelford D. B., Mihaylova M. M., Gelino S., Kohnz R. A., Mair W., Vasquez D. S., Joshi A., Gwinn D. M., Taylor R., Asara J. M., Fitzpatrick J., Dillin A., Viollet B., Kundu M., et al. (2011) Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science 331, 456–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Palikaras K., Lionaki E., and Tavernarakis N. (2015) Balancing mitochondrial biogenesis and mitophagy to maintain energy metabolism homeostasis. Cell Death Differ. 22, 1399–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Melser S., Chatelain E. H., Lavie J., Mahfouf W., Jose C., Obre E., Goorden S., Priault M., Elgersma Y., Rezvani H. R., Rossignol R., and Bénard G. (2013) Rheb regulates mitophagy induced by mitochondrial energetic status. Cell Metab. 17, 719–730 [DOI] [PubMed] [Google Scholar]
- 47. Palikaras K., Lionaki E., and Tavernarakis N. (2016) Mitophagy: In sickness and in health. Mol. Cell. Oncol. 3, e1056332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cheng G., Zielonka J., Dranka B. P., McAllister D., Mackinnon A. C. Jr., Joseph J., and Kalyanaraman B. (2012) Mitochondria-targeted drugs synergize with 2-deoxyglucose to trigger breast cancer cell death. Cancer Res. 72, 2634–2644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wu Y., Sarkissyan M., Mcghee E., Lee S., and Vadgama J. V. (2015) Combined inhibition of glycolysis and AMPK induces synergistic breast cancer cell killing. Breast Cancer Res. Treat. 151, 529–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.