Abstract
The transcription factor Gbx2 (gastrulation brain homeobox 2) is a direct target of the LIF/STAT3 signaling pathway, maintains mouse embryonic stem cell (mESC) self-renewal, and facilitates mouse epiblast stem cell (mEpiSC) reprogramming to naïve pluripotency. However, the mechanism by which Gbx2 mediates its effects on pluripotency remains unknown. Here, using an RNA-Seq approach, we identified Klf4 (Kruppel-like factor 4) as a direct target of Gbx2. Functional studies indicated that Klf4 mediates the self-renewal–promoting effects of Gbx2, because knockdown of Klf4 expression abrogated the ability of Gbx2 to maintain the undifferentiated state of mESCs. We also found that Gbx2 largely depends on Klf4 to reprogram mEpiSCs to a mESC-like state. In summary, our study has uncovered a mechanism by which Gbx2 maintains and induces naïve pluripotency. These findings expand our understanding of the pluripotency control network and may inform the development of culture conditions for improved ESC maintenance and differentiation.
Keywords: cell biology, differentiation, embryonic stem cell, Kruppel-like factor 4 (KLF4), reprogramming, Gbx2, self-renewal
Introduction
Mouse embryonic stem cells (mESCs)3 are isolated from the inner cell mass of the preimplantation embryos (1, 2). They can be maintained indefinitely as self-renewing populations while retaining the pluripotency to differentiate into the cells of all three primary germ layers when cultured under appropriate conditions in vitro (3). In the past, the maintenance of mESCs rely on feeder cells in vitro, including mitotically inactivated fibroblasts or buffalo rat liver cell (1, 2, 4, 5). Now, however, the simultaneous addition of serum and leukemia inhibitory factor (LIF) can support the long-term self-renewal of mESCs in feeder-free conditions (6–8). LIF is able to activate three intracellular signaling pathways: the JAK/STAT3 pathway, the PI3K/AKT pathway, and the SH2 domain-containing tyrosine phosphatase/MAPK pathway (4, 9–11). LIF mainly through the JAK/STAT3 signaling pathway maintains mESC naïve pluripotent state (12–14). LIF initiates signaling cascade by binding to a heterodimer receptor complex, LIF receptor, and subunit glycoprotein 130 (6, 15), which will activate JAKs family members. JAKs stimulate cytoplasmic STAT3 (11, 16, 17). STAT3 then enters into the nucleus, where they act as a transcription factors to induce the expressions of the target genes (18).
Many downstream targets of the LIF/STAT3 signaling have been identified, such as Gbx2 (19), Klf4 (20, 21), Sp5 (22), and Tfcp2l1 (23, 24), whereas current exploring of these genes-mediated mechanisms in promoting mESC self-renewal is very limited. For example, elevated expression of Gbx2 (Gastrulation brain homeobox 2), a LIF/STAT3 signaling downstream target, not only can allow long-term expansion of the undifferentiated mESCs in the absence of LIF but also is sufficient to improve the efficiency of reprogramming from mouse epiblast stem cells (mEpiSCs) to ground state mESC (19). The exact molecular mechanism of Gbx2 exerting these effects, however, remains elusive. Here, we identified that Klf4, as a key direct target of Gbx2, is critical for Gbx2 to promote mESC self-renewal and convert mEpiSCs to mESC state.
Results
Identification of downstream targets of Gbx2 in 46C mESCs
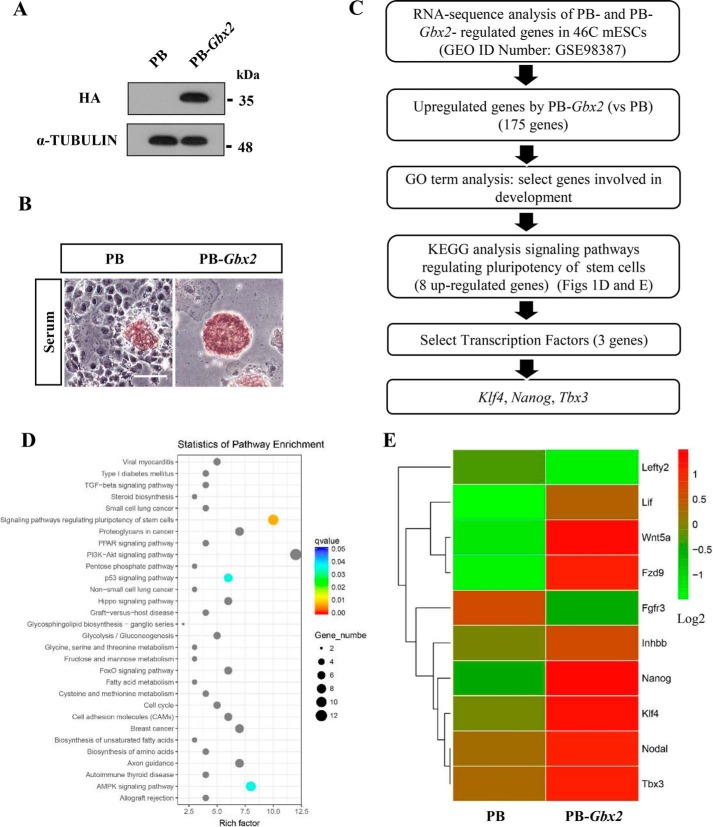
Overexpression of HA-tagged Gbx2 through PiggyBac system (PB-Gbx2) is able to promote mESC self-renewal in the absence of LIF; these cells could be continually passaged while retaining typical mESC morphology and positive alkaline phosphatase (AP) activity, whereas empty vector (PB) control cells differentiated (Fig. 1, A and B, and supplemental Fig. S1, A and B), indicating that overexpressing Gbx2 can recapitulate the effect of LIF to support mESC self-renewal. These results are consistent with our previous report (19). To screen out the functional targets of Gbx2, we performed a RNA-sequence analysis in PB and PB-Gbx2 46C mESCs and got 175 genes up-regulated by 2-fold or greater in PB-Gbx2 cells compared with PB cells (GEO accession no. GSE98387) (Fig. 1C). To survey possible biological roles of these differentially expressed genes, we next performed gene ontology analysis and filtered out those involved in development. The Kyoto Encyclopedia of Genes and Genomes method was then carried out to select those enriched in signaling pathways regulating pluripotency of stem cells. These gave a short list of eight candidates (Wnt5a, Klf4, Nanog, Nodal, Inhbb, Tbx3, Lif, and Fzd9) (Fig. 1, D and E). Among these genes, three transcription factors were finally identified: Klf4, Nanog, and Tbx3. Previous reports have shown that these targets are closely relevant to mESC pluripotency (20, 21, 25–29).
Figure 1.
RNA-sequence analysis of the genes up-regulated by Gbx2 in 46C mESCs. A, HA-tagged Gbx2 was introduced into 46C mESCs, and the protein level of HA-tagged Gbx2 was determined by Western blot. α-Tubulin is used as a loading control. B, AP staining of 46C mESCs overexpressing Gbx2 cultured in the absence of LIF for 8 days. Bar, 100 μm. C, flow chart illustrating the approach used to identify the candidate genes regulated by Gbx2 in 46C mESCs. Cells were maintained in serum medium supplemented with LIF. D, the Kyoto Encyclopedia of Genes and Genomes analysis of signal pathways classified and enriched by Gbx2-induced genes. The data are presented in a scatter diagram. E, heat map showed the expression pattern of the indicated genes in PB and PB-Gbx2 mESCs cultured in serum/LIF condition. Genes were ranked according to the level of log2 fold change. Red, expression above average; green, below average.
Klf4 is a direct downstream target of Gbx2
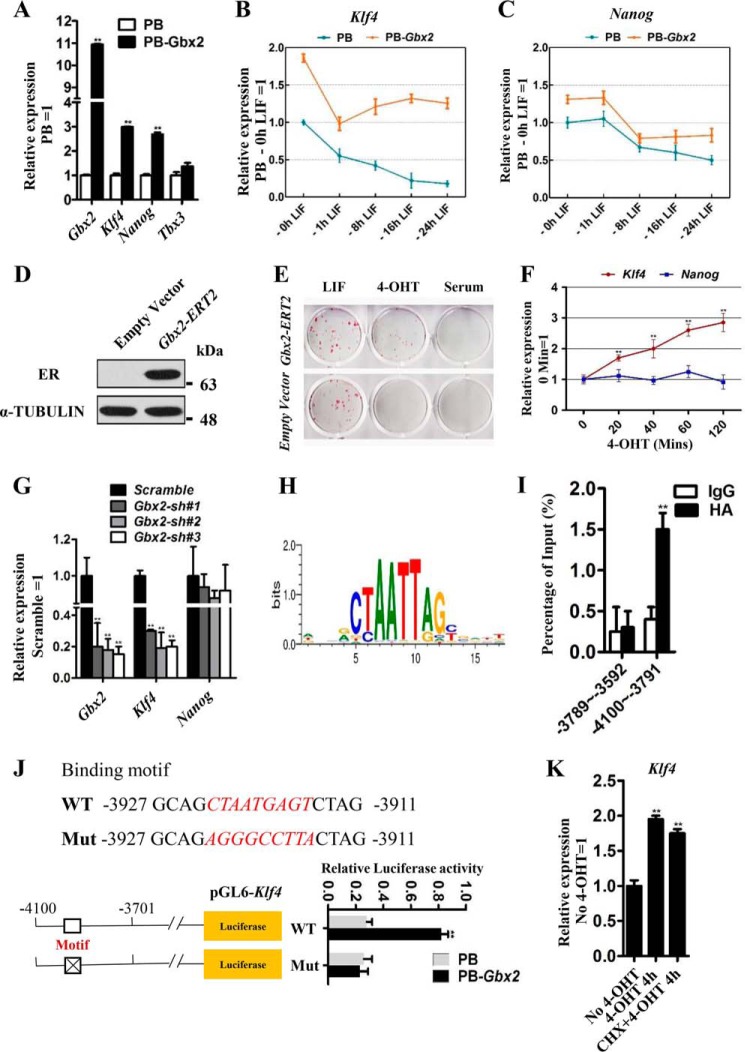
To confirm that the expression levels of these three candidates are really regulated by PB-Gbx2, we used quantitative real-time PCR (qRT-PCR) to detect their mRNA levels and found that only the Nanog and Klf4 transcripts obviously increased upon Gbx2 overexpression (Fig. 2A). To further investigate the effect of Gbx2 on Nanog and Klf4 expression, we used three different approaches. First, PB or PB-Gbx2 mESCs were routinely cultured in LIF/serum condition, and then LIF was withdrawn from 1 to 24 h. qRT-PCR analysis revealed that enforced Gbx2 could inhibit the further down-regulation of Klf4 caused by LIF withdrawn, but not Nanog (Fig. 2, B and C). Second, we introduced a Gbx2-ERT2 transgene into 46C mESCs (Fig. 2D). Administration of 4-hydroxytamoxifen (4-OHT) to Gbx2-ERT2-expressing cells results in the translocation of GBX2-ERT2 into the nucleus to activate Gbx2 targets to support the mESC pluripotency (Fig. 2E). As a result, 4-OHT treatment gradually up-regulated Klf4 expression, but not Nanog, in mESCs overexpressing Gbx2-ERT2 within 120 min (Fig. 2F). Finally, to test whether Gbx2 knockdown has an effect on Klf4 expression, we infected 46C mESCs with lentiviruses encoding three shRNAs specific for Gbx2 mRNA (Gbx2 sh#1, Gbx2 sh#2, and Gbx2 sh#3). Stable knockdown (80–90%) of Gbx2 transcript levels was observed following drug selection (Fig. 2G). As expected, the transcript of Klf4 decreased in Gbx2 shRNA cells compared with scramble control mESCs (Fig. 2G and supplemental Fig. S2). Together, these data suggest that Gbx2 engages in Klf4 expression to support the mESC pluripotency, and the latter is a downstream target of Gbx2.
Figure 2.
Gbx2 directly regulates Klf4 expression in mESCs. A, qRT-PCR analysis of Nanog, Klf4, and Tbx3 expression levels in PB and PB-Gbx2 46C mESCs. The data are presented as the means ± S.D. of three independent experiments. **, p < 0.01 versus PB. B and C, qRT-PCR analysis of Klf4 and Nanog gene expression in PB or PB-Gbx2 mESCs after withdrawal of LIF for 0, 1, 8, 16, and 24 h. D, Western blot analysis of ER protein in empty vector and Gbx2-ERT2 transfected mESCs cultured in LIF/serum. E, AP staining of 46C mESCs overexpressing Gbx2-ERT2 or empty vector. The cells were cultured in serum medium supplemented with LIF or 1 μm 4-OHT for 8 days. F, qRT-PCR analysis of the expression levels of Klf4 and Nanog in Gbx2-ERT2 cells treated with 4-OHT for the indicated time. The data are presented as the means ± S.D. of three independent experiments. **, p < 0.01 versus 0 min. G, qRT-PCR analysis of Gbx2, Klf4, and Nanog gene expression in 46C mESCs infected with scramble or Gbx2 shRNA lentivirus. The data are presented as the means ± S.D. of three independent experiments. **, p < 0.01 versus scramble. H, the Gbx2-binding consensus motifs defined from the JASPAR CORE database. I, HA-Gbx2 transfected 46C mESCs were processed for ChIP with antibody for the HA epitope or IgG as control. Fold enrichments of precipitated DNA relative to the total input chromatin were assessed by qRT-PCR. −3789 to −3593 and −4100 to −3791 indicate two fragments of Klf4 gene promoter. The data are presented as the means ± S.D. of three independent experiments. **, p < 0.01 versus IgG. J, sequence of the Gbx2 consensus motif within the Klf4 promoter. The WT and mut sequences were used in the luciferase activity assay. The data are presented as the means ± S.D. of three independent experiments. **, p < 0.01 versus PB. K, qRT-PCR analysis of Klf4 in Gbx2-ERT2 mESCs deprived of LIF overnight and 4-OHT stimulation for 4 h in the presence or absence of the protein-synthesis inhibitor CHX. CHX (50 mg/ml) was added to the medium 1 h before 4-OHT treatment. The data are presented as the means ± S.D. of three independent experiments. **, p < 0.01 versus No 4-OHT.
To determine whether Klf4 is a direct target of Gbx2, we analyzed the Gbx2-binding consensus motifs (Fig. 2H) and predicted two potential binding sites within Klf4 promoter regions (from −5000 to +1) from the JASPAR CORE database (motif 1, −3677TGATCTAATTAGATCCT−3661; and motif 2, −3927GCAGCTAATGAGTCTAG−3911). We then performed ChIP-qRT-PCR in PB-Gbx2 46C mESCs and found the direct interaction of Gbx2 with the Klf4 promoter containing the sequence of motif 2 (Fig. 2I). To further determine whether Gbx2 is a functional activator of the Klf4 promoter, Gbx2 was co-transfected with reporter vectors that drive the expression of luciferase under the control of Klf4 promoter fragments (pGL6-Klf4), comprising our validated Gbx2 consensus motif 2 or mutated motif 2 sequences (Mut) (Fig. 2J). Under these conditions, we observed a 3.6-fold increase in wild-type promoter activity relative to the mutant sequence in PB-Gbx2 cells (Fig. 2J). These results further verify direct transcriptional activation of Klf4 by Gbx2. Moreover, the induction of Klf4 expression by Gbx2-ERT2 cells was similar with or without cycloheximide (CHX) (Fig. 2K), an inhibitor of protein biosynthesis, which excludes the possible regulation of Klf4 by other Gbx2-induced targets. In conclusion, these results collectively indicate that Klf4 is a direct target of Gbx2.
Klf4 mediates the effect of Gbx2 in promoting mESC self-renewal
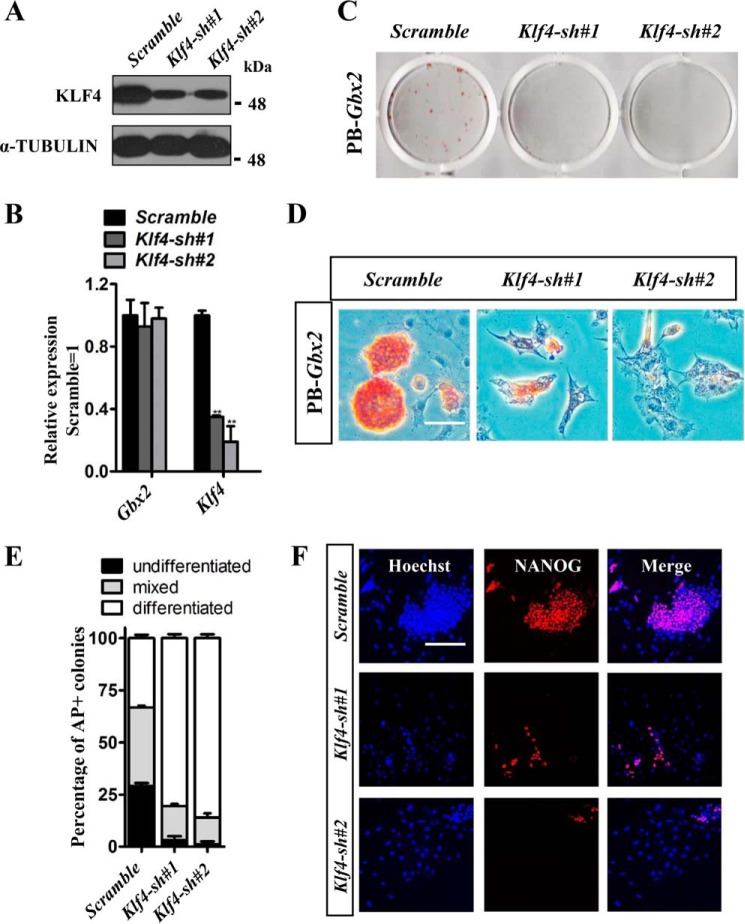
Both Gbx2 and Klf4 are able to promote 46C mESC self-renewal in the absence of LIF when overexpressed (Fig. 1B and supplemental Fig. S3, A and B). To examine whether Gbx2 depends on Klf4 to maintain mESC pluripotency, Klf4 transcript was knocked down (Klf4 sh#1 and Klf4 sh#2) in PB-Gbx2 46C mESCs and validated by Western blot and qRT-PCR methods (Fig. 3, A and B). Knockdown of Klf4 led to PB-Gbx2 mESC differentiation in the absence of LIF, as indicated by the flat cell morphology, decreased AP activity (Fig. 3, C–E), and low expression level of pluripotency marker NANOG (Fig. 3F). Conversely, they exhibited high-level expression of the endoderm markers, such as FoxA2, Gata4, and Gata6 (supplemental Fig. S4), whereas PB-Gbx2 cells infected with scramble control lentivirus kept undifferentiated state (Fig. 3, C–F), suggesting that deletion of Klf4 abolished the defining ability of Gbx2 to confer LIF-independent self-renewal. These results together indicate that the function of Gbx2 for the acquisition of naïve pluripotency in mESCs is dependent on Klf4.
Figure 3.
Knockdown of Klf4 abrogates the self-renewal-promoting effect of Gbx2 in mESCs. A and B, Western blot or qRT-PCR analysis of Klf4 and Gbx2 expression in PB-Gbx2 46C mESCs infected with Klf4 shRNA lentivirus. The data are presented as the means ± S.D. of three independent experiments. **, p < 0.01 versus scramble. C and D, AP staining of colonies arising from Klf4 knockdown mESCs carrying PB-Gbx2 transgenes cultured in serum medium without LIF for 7 days. Bar, 100 μm. E, quantification of AP positive colonies in Fig. 3D. The data represent means ± S.D. from triplicate experiments. F, immunofluorescence staining of NANOG in Klf4 knockdown 46C mESCs overexpressing PB-Gbx2. Bar, 100 μm.
Klf4 is critical for Gbx2 to reprogram mEpiSCs to naïve pluripotent state
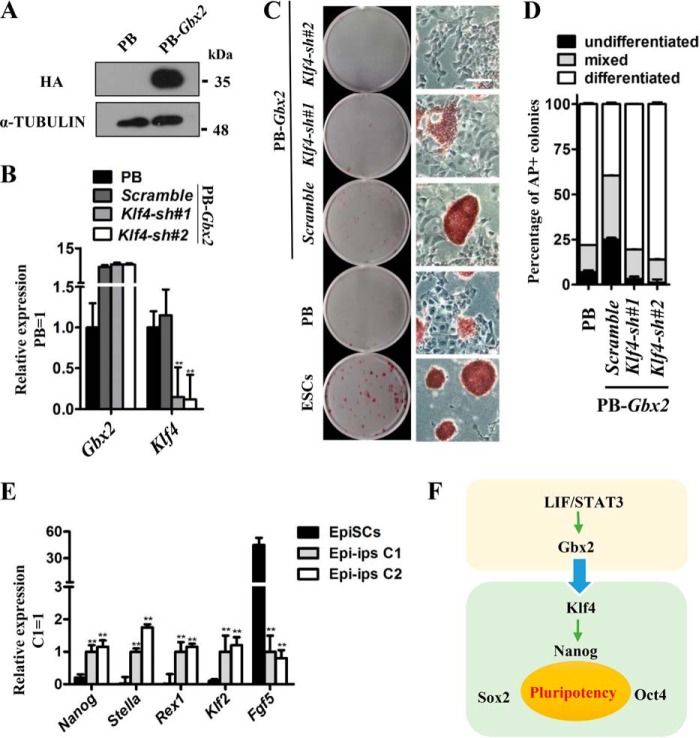
Recent reports have established that the reprogramming efficiency from primed-state mEpiSCs to naïve-state mESCs can be facilitated by activation of STAT3 signaling or its downstream targets, such as Gbx2 (19), Tfcp2l1 (22), and Klf4 (25). These promote us to examine whether Klf4 has ability to mediate the function of Gbx2 in reprogramming mEpiSCs toward naïve pluripotency. First, we overexpressed HA-tagged Gbx2 (PB-Gbx2) in CD1 mEpiSCs (Fig. 4A) and then infected these cells with Klf4 shRNA lentivirus. After selection, the mRNA level of Klf4 decreased significantly in PB-Gbx2 CD1 EpiSCs when compared with PB cells or PB-Gbx2 cells infected with scramble lentivirus (Fig. 4B). These cells were cultured in the conventional mESC medium supplemented with LIF plus 2i. After 12 days, we observed that the PB-Gbx2 cells expressing scramble shRNA formed many dome-shaped colonies and showed strong AP activity (Fig. 4, C and D). Two ESC-like colonies were picked up and could be continuously expanded in LIF/serum. They expressed high levels of the naïve pluripotency markers Nanog, Stella, Klf2, and Rex1 but a low level of the mEpiSC-specific marker Fgf5 (Fig. 4E). However, Klf4 shRNA cells died or differentiated; similar results were observed in PB mEpiSCs (Fig. 4, C and D). Therefore, knockdown of Klf4 is capable of eliminating the reprogramming-promoting effect of Gbx2. Previous report shows that Klf4 interacts directly with other reprogramming factors (Oct4 and Sox2) to induce induced pluripotent stem cells (iPS cells) (30, 31). To investigate whether KLF4 protein and GBX2 protein interact during the mEpiSC reprogramming, we overexpressed Flag-tagged Klf4 and HA-tagged Gbx2 in one CD1 mEpiSC line and then performed co-immunoprecipitation experiments after the cells were transferred into reprograming conditions (LIF/2i) for 2 days. The results show that there is no direct interaction between GBX2 and KLF4 at protein levels (supplemental Fig. S5). Overall, these data thus imply that Gbx2 is sufficient to convert mEpiSCs to naïve pluripotent state via up-regulating the Klf4 expression at transcriptional level but not through direct interaction with KLF4 protein.
Figure 4.
Klf4 is indispensable for Gbx2 to reprogram mEpiSCs to naïve pluripotent state. A, HA-tagged Gbx2 (PB-Gbx2) was introduced into CD1 mEpiSCs and the protein level of HA-tagged Gbx2 was determined by Western blot. The cells were maintained in medium supplemented with activin A, basic FGF, and IWR-1. B, qRT-PCR analysis of Gbx2 and Klf4 expression levels in Klf4 knockdown mEpiSCs overexpressing PB-Gbx2. The data are presented as the means ± S.D. of three independent experiments. **, p < 0.01 versus PB. C, AP staining of colonies generated from Klf4 knockdown mEpiSCs carrying PB-Gbx2 transgene. The cells were cultured in serum medium supplemented with LIF/2i for 12 days. Bar, 100 μm. D, quantification of the AP-positive colonies in C. E, comparison of gene marker expression in PB-Gbx2/scramble Epi-iPS cells and CD1 mEpiSCs. Nanog, Stella, Rex1, and Klf2 are mESC markers, whereas Fgf5 is a mEpiSC marker. The data are presented as the means ± S.D. of three independent experiments. **, p < 0.01 versus mEpiSCs. Epi-iPS, induced pluripotent stem cells generated from mEpiSCs; C1, clone 1; C2, clone 2. F, schematic diagram of Gbx2 input to the pluripotency network. Gbx2 and Klf4 integrate the LIF/STAT3 signal pathway into the core transcription factor network of mESCs.
Discussion
LIF/STAT3 signaling pathway maintains mESC pluripotency through activation of many downstream transcription factors including Gbx2 (19), Tfcp2l1 (24), Klf4 (21), etc. As one of the direct targets of STAT3, Gbx2 plays pivotal roles during embryogenesis and is required for the development of the anterior hindbrain, spinal cord, inner ear, heart, and neural crest cells (32, 33). Previous research shows that the overexpression of Gbx2 can facilitate the maintenance of mESC self-renewal and the reprogramming of mEpiSCs to naïve pluripotent state (19). However, how Gbx2 input is integrated with the pluripotency gene regulatory network has remained unclear.
To resolve this issue, we sought the key target of Gbx2 and identified the transcription factor Klf4. Gbx2 markedly increases the Klf4 expression level, and this is likely the key contribution of Gbx2 to the maintenance of mESC self-renewal, because knockdown of Klf4 is able to abrogate the self-renewal-promoting effect of Gbx2 in mESCs (Fig. 3, A–F). This is unsurprising, given that Klf4, a member of the Kruppel-like factor family, has been shown to be a direct target of STAT3 (20, 21). Klf4 transfectants were able to produce undifferentiated colonies in the absence of LIF (supplemental Figs. S3, A and B), consistent with previous reports (20, 21). Klf4 functions may largely rely on Nanog, because Klf4 directly binds to the promoter region of Nanog and regulates its expression (34). On the other hand, knockdown of Nanog expression induces differentiation of mESCs that overexpress Klf4 (34). Accordingly, we also observed that enforced Gbx2 could slightly up-regulate Nanog expression (Fig. 2A). Nanog is a key naïve pluripotency factor and has ability to support mESC self-renewal when overexpressed (11, 35). Therefore, Klf4 may function as a bridge in linking the LIF/STAT3/Gbx2 signaling into the core pluripotency network (Nanog, Oct4, and Sox2) to guard the ground state of mESCs (Fig. 4F).
Apart from its role in promoting mESC self-renewal, Gbx2 also acts as a transcription factor capable of reprogramming mEpiSCs to naïve pluripotency state (19). mEpiSCs are isolated from the epiblasts of early postimplantation mouse embryos and have a different and even opposite expression pattern with mESCs (36, 37). Gbx2 is highly expressed in the mESCs but is barely detectable in mEpiSCs (19), similar to the dynamic change of Klf4 expression (25). Klf4, one of the four canonical Yamanaka factors that direct somatic cell reprogramming (30), has been shown to be capable of reprogramming mEpiSCs to an ESC state (25). Gbx2 binds to the promoter regions of Klf4 and positively regulates its transcription (Fig. 2, H–J). Thus, knockdown of Klf4 eliminates the reprograming function of Gbx2 in mEpiSCs (Fig. 4, B–D), indicating that Klf4 is essential for Gbx2 to induce the generation of naïve pluripotency. Notably, human ESCs share many similar features with the primed mEpiSCs (4), which are truly differentiated from the ground state mESCs (38). Although overexpression of STAT3 or its target Gbx2 fails to promote human ESC self-renewal (39, 40), the reinforcement of STAT3 or its other target Klf4 in combination with Klf2 or Oct4 was capable of reprogramming human ESCs into naïve-like pluripotency (41, 42). It will be of great interesting to investigate whether Gbx2 and the Gbx2/Klf4 axis contribute to establish and maintain the naïve pluripotent state of human ESCs.
In summary, our study reveals an unrecognized mechanism of Gbx2 in mESC self-renewal. Gbx2 may exert this function by acting as a fast responding mediator to the LIF/STAT3 signal changes, eventually modulating the expression of Klf4, a well established key factor in maintenance of mESC stemness. In the future, understanding how Gbx2 and Klf4 cooperate with other downstream targets of STAT3 to maintain the pluripotent state of mESCs might greatly facilitate the development of better culture conditions for the maintenance and derivation of authentic ESCs from species other than mice and rats.
Experimental procedures
Cell culture
46C mESCs were routinely maintained on 0.1% gelatin-coated plates in DMEM (HyClone) containing 10% FBS (HyClone), 1× sodium pyruvate (Gibco), 1× non-essential amino acids (Gibco), 2 mm GlutaMAX (Gibco), 0.1 mm β-mercaptoethanol, and 100 units/ml LIF (Millipore). Mouse CD1 mEpiSCs (43) were maintained in the mESC medium without LIF but supplemented with 10 ng/ml activin A (Peprotech), 10 ng/ml basic FGF (Peprotech), and 2 μm IWR-1 (Sigma).
Overexpression and knockdown plasmid construction
The coding regions of Gbx2 and Klf4 were cloned from mESC cDNA and inserted into the PiggyBac vector (PB). The shRNA-expressing plasmids were generated according to pLKO.1-TRC protocol (Addgene). The target-specific shRNA sequences for Gbx2 and Klf4 used in this study are listed in supplemental Table S1.
Alkaline phosphatase staining and qRT-PCR
The AP activity assay and qRT-PCR analysis were performed according to our previous report (24). The primers used for the qRT-PCR analysis are listed in supplemental Table S2.
Western blotting
Western blotting was performed according to a standard protocol. The primary antibodies used for probing were α-tubulin (SC-8035, Santa Cruz, 1:1000), HA (C29F4, Cell Signaling Technology, 1:1000), KLF4 (D1F2, Cell Signaling Technology, 1:1000), and ER (MC-20, Santa Cruz, 1:500).
Immunofluorescence staining
Immunostaining was performed via standard protocols. Primary antibody used was against NANOG (14295-1-AP, Proteintech, 1:500). Hoechst 33342 (Hoechst) was used for nuclear staining.
Chromatin immunoprecipitation assay
ChIP was performed using ChIP assay kit (P2078; Beyotime, Haimen, China) according to the manufacturer's protocol. Anti-HA antibody (C29F4; Cell Signaling Technology) was used for immunoprecipitation and IgG (2729S; Cell Signaling Technology) as a control antibody. ChIP enrichment was performed by qRT-PCR. The primer sequences and locations in regions of Klf4 gene are listed in supplemental Table S3.
Luciferase assay
The Klf4 gene promoter regions (from −4100 to −3701) carrying the wild-type binging sites or mutant sequences were analyzed by amplified PCR and then cloned into pGL6-basic plasmid, named pGL6-Klf4. The primers sequences are listed in supplemental Table S4. Plasmids were co-transfected with a Renilla luciferase plasmid into 46C mESCs overexpressing Gbx2 or empty vector. After 48 h, luciferase activities were measured with the dual luciferase assay kit (Transgen, Beijing, China).
Reprogramming
For reprogramming, 1 × 105 transfectants were seeded on a 0.1% gelatin-coated 6-well plate and cultured in serum medium supplemented with LIF/2i (3 μm CHIR99021 and 1.5 μm PD0325901). Medium were refreshed every other day.
Co-immunoprecipitation
Cell extracts were prepared using Nonidet P-40 lysis buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 0.5% Nonidet P-40, and protease inhibitors). The supernatant was collected and incubated with 10 μl of anti-FLAG affinity gel (SG4110-16; GNI, Tokyo, Japan) for 2 h at 4 °C. The beads were then washed six times with lysis buffer and resuspended in 100 μl of 1× SDS sample buffer for Western blotting analysis.
Accession numbers
Data and details of the method for the RNA-sequence analysis are available in the Gene Expression Omnibus under accession number GSE98387.
Statistical analysis
All data are reported as the mean ± S.D. A Student's t test was used to determine the significance of differences in comparisons. Values of p < 0.05 were considered statistically significant.
Author contributions
M. W., L. T., and S. Y. conceived and designed the experiments. M. W. and L. T. performed the experiments. D. L. analyzed the data. Q.-L. Y. contributed reagents or materials. M. W. and S. Y. wrote the paper.
Supplementary Material
This work was supported by Natural Science Foundation of China Grants 31501191 and 31671535, Anhui Province Grant 1508085SQC204, and Scientific Research Startup Fund of Anhui University Grants J01006068 and J01006045. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supplemental Tables S1–S4 and Figs. S1–S5.
Data and details of the method used for the RNA-sequence analysis are available in the Gene Expression Omnibus (GEO) database under accession number GSE98387.
- mESC
- mouse embryonic stem cell
- mEpiSC
- mouse epiblast stem cell
- LIF
- leukemia inhibitory factor
- PB
- PiggyBac system
- AP
- alkaline phosphatase
- qRT-PCR
- quantitative real-time PCR
- 4-OHT
- 4-hydroxytamoxifen
- CHX
- cycloheximide.
References
- 1. Evans M. J., and Kaufman M. H. (1981) Establishment in culture of pluripotential cells from mouse embryos. Nature 292, 154–156 [DOI] [PubMed] [Google Scholar]
- 2. Martin G. R. (1981) Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl. Acad. Sci. U.S.A. 78, 7634–7638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Smith A. G. (2001) Embryo-derived stem cells: of mice and men. Annu. Rev. Cell Dev. Biol. 17, 435–462 [DOI] [PubMed] [Google Scholar]
- 4. Ye S., Liu D., and Ying Q. L. (2014) Signaling pathways in induced naive pluripotency. Curr. Opin. Genet. Dev. 28, 10–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Smith A. G., and Hooper M. L. (1987) Buffalo rat liver cells produce a diffusible activity which inhibits the differentiation of murine embryonal carcinoma and embryonic stem cells. Dev. Biol. 121, 1–9 [DOI] [PubMed] [Google Scholar]
- 6. Ohtsuka S., Nakai-Futatsugi Y., and Niwa H. (2015) LIF signal in mouse embryonic stem cells. Jak-Stat 4, e1086520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Williams R. L., Hilton D. J., Pease S., Willson T. A., Stewart C. L., Gearing D. P., Wagner E. F., Metcalf D., Nicola N. A., and Gough N. M. (1988) Myeloid leukaemia inhibitory factor maintains the developmental potential of embryonic stem cells. Nature 336, 684–687 [DOI] [PubMed] [Google Scholar]
- 8. Smith A. G., Heath J. K., Donaldson D. D., Wong G. G., Moreau J., Stahl M., and Rogers D. (1988) Inhibition of pluripotential embryonic stem cell differentiation by purified polypeptides. Nature 336, 688–690 [DOI] [PubMed] [Google Scholar]
- 9. Hirai H., Karian P., and Kikyo N. (2011) Regulation of embryonic stem cell self-renewal and pluripotency by leukaemia inhibitory factor. Biochem. J. 438, 11–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Burdon T., Chambers I., Stracey C., Niwa H., and Smith A. (1999) Signaling mechanisms regulating self-renewal and differentiation of pluripotent embryonic stem cells. Cells Tissues Organs 165, 131–143 [DOI] [PubMed] [Google Scholar]
- 11. Huang G., Ye S., Zhou X., Liu D., and Ying Q. L. (2015) Molecular basis of embryonic stem cell self-renewal: from signaling pathways to pluripotency network. Cell Mol. Life Sci. 72, 1741–1757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Burdon T., Stracey C., Chambers I., Nichols J., and Smith A. (1999) Suppression of SHP-2 and ERK signalling promotes self-renewal of mouse embryonic stem cells. Dev. Biol. 210, 30–43 [DOI] [PubMed] [Google Scholar]
- 13. Matsuda T., Nakamura T., Nakao K., Arai T., Katsuki M., Heike T., and Yokota T. (1999) STAT3 activation is sufficient to maintain an undifferentiated state of mouse embryonic stem cells. EMBO J. 18, 4261–4269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Niwa H., Burdon T., Chambers I., and Smith A. (1998) Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes Dev. 12, 2048–2060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fukada T., Hibi M., Yamanaka Y., Takahashi-Tezuka M., Fujitani Y., Yamaguchi T., Nakajima K., and Hirano T. (1996) Two signals are necessary for cell proliferation induced by a cytokine receptor gp130: involvement of STAT3 in anti-apoptosis. Immunity 5, 449–460 [DOI] [PubMed] [Google Scholar]
- 16. Levy D. E., and Darnell J. E. Jr. (2002) Stats: transcriptional control and biological impact. Nat. Rev. Mol. Cell Biol. 3, 651–662 [DOI] [PubMed] [Google Scholar]
- 17. Wang X., Crowe P. J., Goldstein D., and Yang J. L. (2012) STAT3 inhibition, a novel approach to enhancing targeted therapy in human cancers (review). Int. J. Oncol. 41, 1181–1191 [DOI] [PubMed] [Google Scholar]
- 18. Sasse J., Hemmann U., Schwartz C., Schniertshauer U., Heesel B., Landgraf C., Schneider-Mergener J., Heinrich P. C., and Horn F. (1997) Mutational analysis of acute-phase response factor/Stat3 activation and dimerization. Mol. Cell. Biol. 17, 4677–4686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tai C. I., and Ying Q. L. (2013) Gbx2, a LIF/Stat3 target, promotes reprogramming to and retention of the pluripotent ground state. J. Cell Sci. 126, 1093–1098 [DOI] [PubMed] [Google Scholar]
- 20. Hall J., Guo G., Wray J., Eyres I., Nichols J., Grotewold L., Morfopoulou S., Humphreys P., Mansfield W., Walker R., Tomlinson S., and Smith A. (2009) Oct4 and LIF/Stat3 additively induce Kruppel factors to sustain embryonic stem cell self-renewal. Cell Stem Cell 5, 597–609 [DOI] [PubMed] [Google Scholar]
- 21. Niwa H., Ogawa K., Shimosato D., and Adachi K. (2009) A parallel circuit of LIF signalling pathways maintains pluripotency of mouse ES cells. Nature 460, 118–122 [DOI] [PubMed] [Google Scholar]
- 22. Ye S., Zhang D., Cheng F., Wilson D., Mackay J., He K., Ban Q., Lv F., Huang S., Liu D., and Ying Q. L. (2016) Wnt/β-catenin and LIF-Stat3 signaling pathways converge on Sp5 to promote mouse embryonic stem cell self-renewal. J. Cell Sci. 129, 269–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Martello G., Bertone P., and Smith A. (2013) Identification of the missing pluripotency mediator downstream of leukaemia inhibitory factor. EMBO J. 32, 2561–2574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ye S., Li P., Tong C., and Ying Q. L. (2013) Embryonic stem cell self-renewal pathways converge on the transcription factor Tfcp2l1. EMBO J. 32, 2548–2560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Guo G., Yang J., Nichols J., Hall J. S., Eyres I., Mansfield W., and Smith A. (2009) Klf4 reverts developmentally programmed restriction of ground state pluripotency. Development 136, 1063–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Festuccia N., Osorno R., Halbritter F., Karwacki-Neisius V., Navarro P., Colby D., Wong F., Yates A., Tomlinson S. R., and Chambers I. (2012) Esrrb is a direct Nanog target gene that can substitute for Nanog function in pluripotent cells. Cell Stem Cell 11, 477–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chambers I., Colby D., Robertson M., Nichols J., Lee S., Tweedie S., and Smith A. (2003) Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell 113, 643–655 [DOI] [PubMed] [Google Scholar]
- 28. Jiang J., Chan Y. S., Loh Y. H., Cai J., Tong G. Q., Lim C. A., Robson P., Zhong S., and Ng H. H. (2008) A core Klf circuitry regulates self-renewal of embryonic stem cells. Nat. Cell Biol. 10, 353–360 [DOI] [PubMed] [Google Scholar]
- 29. Russell R., Ilg M., Lin Q., Wu G., Lechel A., Bergmann W., Eiseler T., Linta L., Kumar P. P., Klingenstein M., Adachi K., Hohwieler M., Sakk O., Raab S., Moon A., et al. (2015) A dynamic role of TBX3 in the pluripotency circuitry. Stem Cell Reports 5, 1155–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Takahashi K., and Yamanaka S. (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 [DOI] [PubMed] [Google Scholar]
- 31. Wei Z., Yang Y., Zhang P., Andrianakos R., Hasegawa K., Lyu J., Chen X., Bai G., Liu C., Pera M., and Lu W. (2009) Klf4 interacts directly with Oct4 and Sox2 to promote reprogramming. Stem Cells 27, 2969–2978 [DOI] [PubMed] [Google Scholar]
- 32. Roeseler D. A., Sachdev S., Buckley D. M., Joshi T., Wu D. K., Xu D., Hannink M., and Waters S. T. (2012) Elongation factor 1 α1 and genes associated with Usher syndromes are downstream targets of GBX2. PLoS One 7, e47366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chapman G., Remiszewski J. L., Webb G. C., Schulz T. C., Bottema C. D., and Rathjen P. D. (1997) The mouse homeobox gene, Gbx2: genomic organization and expression in pluripotent cells in vitro and in vivo. Genomics 46, 223–233 [DOI] [PubMed] [Google Scholar]
- 34. Zhang P., Andrianakos R., Yang Y., Liu C., and Lu W. (2010) Kruppel-like factor 4 (Klf4) prevents embryonic stem (ES) cell differentiation by regulating Nanog gene expression. J. Biol. Chem. 285, 9180–9189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pan G., and Thomson J. A. (2007) Nanog and transcriptional networks in embryonic stem cell pluripotency. Cell Res. 17, 42–49 [DOI] [PubMed] [Google Scholar]
- 36. Brons I. G., Smithers L. E., Trotter M. W., Rugg-Gunn P., Sun B., Chuva de Sousa Lopes S. M., Howlett S. K., Clarkson A., Ahrlund-Richter L., Pedersen R. A., and Vallier L. (2007) Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature 448, 191–195 [DOI] [PubMed] [Google Scholar]
- 37. Tesar P. J., Chenoweth J. G., Brook F. A., Davies T. J., Evans E. P., Mack D. L., Gardner R. L., and McKay R. D. (2007) New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature 448, 196–199 [DOI] [PubMed] [Google Scholar]
- 38. Nichols J., and Smith A. (2009) Naive and primed pluripotent states. Cell Stem Cell 4, 487–492 [DOI] [PubMed] [Google Scholar]
- 39. Wang Z., Oron E., Nelson B., Razis S., and Ivanova N. (2012) Distinct lineage specification roles for NANOG, OCT4, and SOX2 in human embryonic stem cells. Cell Stem Cell 10, 440–454 [DOI] [PubMed] [Google Scholar]
- 40. Dahéron L., Opitz S. L., Zaehres H., Lensch M. W., Andrews P. W., Itskovitz-Eldor J., and Daley G. Q. (2004) LIF/STAT3 signaling fails to maintain self-renewal of human embryonic stem cells. Stem Cells 22, 770–778 [DOI] [PubMed] [Google Scholar]
- 41. Chen H., Aksoy I., Gonnot F., Osteil P., Aubry M., Hamela C., Rognard C., Hochard A., Voisin S., Fontaine E., Mure M., Afanassieff M., Cleroux E., Guibert S., Chen J., et al. (2015) Reinforcement of STAT3 activity reprogrammes human embryonic stem cells to naive-like pluripotency. Nat. Commun. 6, 7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hanna J., Cheng A. W., Saha K., Kim J., Lengner C. J., Soldner F., Cassady J. P., Muffat J., Carey B. W., and Jaenisch R. (2010) Human embryonic stem cells with biological and epigenetic characteristics similar to those of mouse ESCs. Proc. Natl. Acad. Sci. U.S.A. 107, 9222–9227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kim H., Wu J., Ye S., Tai C. I., Zhou X., Yan H., Li P., Pera M., and Ying Q. L. (2013) Modulation of β-catenin function maintains mouse epiblast stem cell and human embryonic stem cell self-renewal. Nat. Commun. 4, 2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.