Abstract
MukB is a structural maintenance of chromosome-like protein required for DNA condensation. The complete condensin is a large tripartite complex of MukB, the kleisin, MukF, and an accessory protein, MukE. As found previously, MukB DNA condensation is a stepwise process. We have defined these steps topologically. They proceed first via the formation of negative supercoils that are sequestered by the protein followed by hinge–hinge interactions between MukB dimers that stabilize topologically isolated loops in the DNA. MukB itself is sufficient to mediate both of these topological alterations; neither ATP nor MukEF is required. We show that the MukB hinge region binds DNA and that this region of the protein is involved in sequestration of supercoils. Cells carrying mutations in the MukB hinge that reduce DNA condensation in vitro exhibit nucleoid decondensation in vivo.
Keywords: chromosomes, DNA, DNA structure, DNA topology, nucleic acid enzymology, DNA enzymes, nucleic acids, nucleic acid enzymes
Introduction
Chromosomal DNA must be condensed in an orderly manner that allows access by the enzymes that transcribe, repair, and replicate the chromosomes. DNA condensation is achieved by a combination of a number of factors that include the action of members of the structural maintenance of chromosome (SMC)2 family of proteins. SMC proteins (1) are characterized by head domains that are separated by a long coiled-coil region that folds back on itself to bring the head domains, which generally bind DNA and possess ATPase activity, together. The apex of the coiled-coil region is referred to as the hinge region, which mediates dimerization. The coiled-coil region can be 40–50 nm in length. In eukaryotes, there are three versions of SMC proteins, cohesin, condensin, and the SMC5/6 complex. Bacteria either contain a true SMC protein (e.g. in Bacillus subtilis) or SMC-like proteins called MukB (e.g. in Escherichia coli) or MksB (e.g. in Pseudomonas aeruginosa). SMC proteins interact with other accessory proteins: the kleisin, which bridges the head domains of the SMC proteins to form a topologically closed ring of proteins; and a third protein that interacts with the kleisin.
MukB is encoded in an operon along with its kleisin, MukF, and an additional accessory protein, MukE (2, 3). Mutations in mukB, mukF, and mukE cause nucleoid decondensation, chromosome segregation defects, and the generation of anucleate cells (2, 3). MukB binds double-stranded DNA and can alter DNA topology when bound to DNA in the presence of a topoisomerase, forming right-handed knots and negative supercoils in DNA rings (4). Interestingly, whereas MukF and MukE form a tripartite complex with MukB (5), and are required for localization of MukB in the cell (6–8), they inhibit the DNA binding and topology modification activities of MukB in vitro (9) while stimulating the ATPase activity (10). ATP hydrolysis causes a rearrangement of the MukBEF complex, loosening the contacts between MukF and one MukB head domain in the MukB dimer (11). These observations suggest that MukF and MukE may be involved in the loading of MukB to DNA rather than in DNA condensation per se.
How these SMC complexes act to cohese and condense DNA has been under active investigation for some time. The eukaryotic cohesin and condensin both trap DNA helices topologically within the tripartite protein complex (1). Recent studies also reported the same mechanism held true for the B. subtilis SMC protein (12) and MukB (13). Several models have been elaborated as to how this topological trapping of DNA can lead to chromosome condensation (1).
We have investigated the mechanism of DNA condensation by MukB. We found that unlike either the eukaryotic or B. subtilis condensins, MukB did not entrap DNA topologically. As shown previously (14), DNA condensation was a stepwise process. We identified two topological alterations. First, the protein forms and sequesters negative supercoils and then forms larger loops that are mediated by hinge–hinge interactions between MukB dimers. These large loops of DNA could be supercoiled by DNA gyrase. MukB alone was sufficient to organize these topological alterations in the DNA that led to condensation; neither MukEF nor ATP was required.
Results
MukB protects negative supercoils and forms topologically stable loops in the DNA
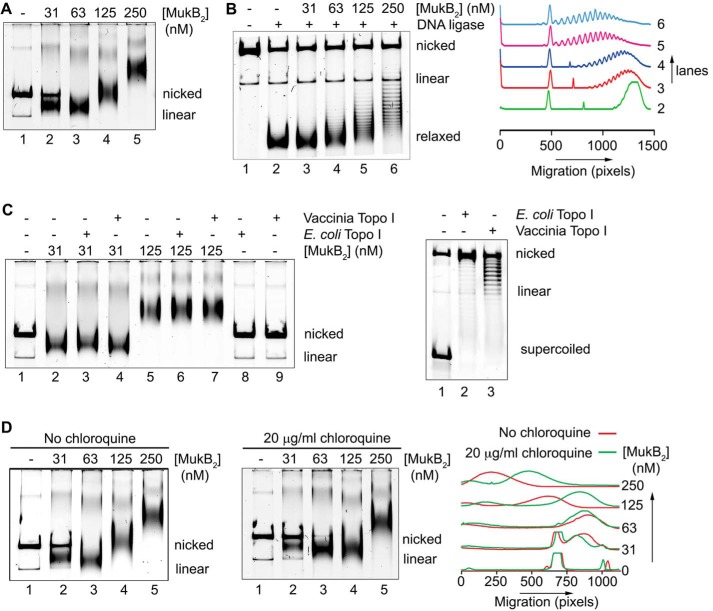
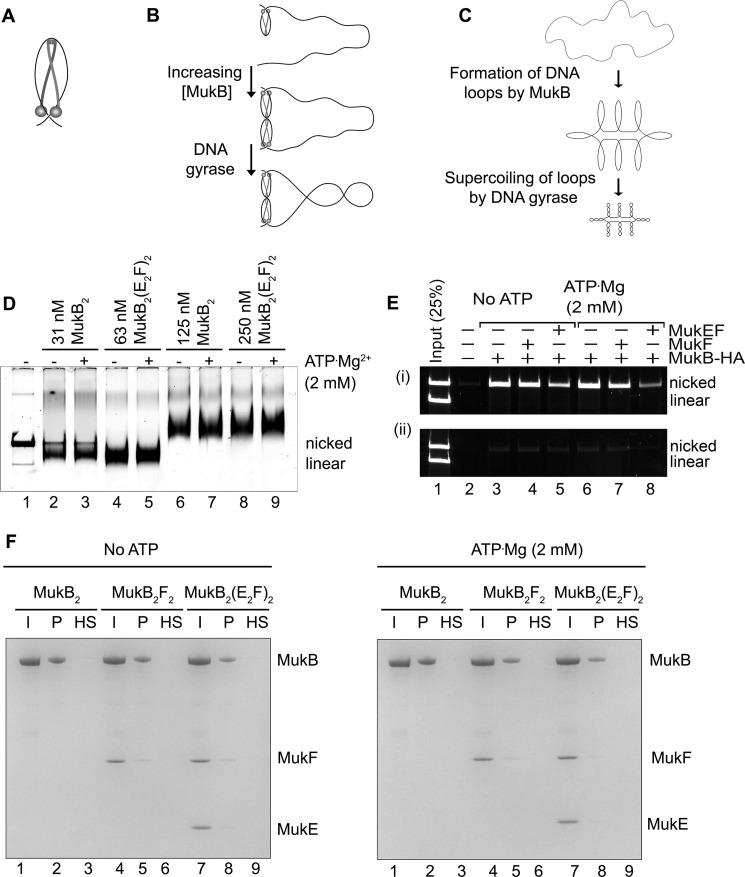
We asked whether we could detect direct alteration of DNA topology by MukB in the absence of a topoisomerase. Because agarose gel electrophoresis of DNA rings is very sensitive to DNA topology (15), we used this technique as the basis of our assay. The substrate was an 11-kbp dsDNA plasmid that had been relaxed by the introduction of a single nick in one of the DNA strands. Remarkably, under conditions previously established for examining MukB binding to DNA (4, 9), protein-DNA complexes formed at low concentrations of MukB moved faster during electrophoresis than the nicked DNA (Fig. 1A). As the concentration of MukB was increased, the mobility of the protein-DNA complex became retarded compared with that of the DNA alone (Fig. 1A). We refer to the protein-DNA complexes formed at 31 and 63 nm MukB as the fast-moving complexes (FMcx) and those formed at 125 and 250 nm MukB as the slow-moving complexes (SMcx). It is important to note that these reaction mixtures contained only MukB and the relaxed DNA. There was neither topoisomerase, ATP, nor MukEF present. The fact that the FMcx had a greater mobility in the gel than that of the DNA alone suggested that the DNA must be condensed. We investigated the nature of this condensation using a number of biochemical assays, as well as scanning force microscopy (SFM).
Figure 1.
MukB protects negative supercoils and stabilizes loop formation in DNA that is not constrained topologically. A, gel mobility shift analysis of MukB binding to DNA. MukB was incubated with the nicked DNA substrate for 5 min at 37 °C and then analyzed by agarose gel electrophoresis as described under “Experimental procedures.” Nicked, nicked 11-kbp DNA substrate; Linear, linear form of the DNA substrate. B, MukB forms negative supercoils in the nicked DNA substrate. MukB, E. coli DNA ligase, and NAD were incubated with the nicked DNA substrate for 30 min at 37 °C. Left panel, products were deproteinized and then analyzed by electrophoresis through agarose gels containing 10 μg/ml chloroquine in both the gel and the running buffer as described under “Experimental procedures.” Right panel, densitometric lane traces of the gel shown in the left panel. C, MukB protects negative supercoils in the nicked DNA substrate. Left panel, MukB was incubated with the nicked DNA for 5 min at 37 °C. Either Vacinnia DNA topoiosmerase or E. coli DNA topoisomerase I were added to 3.1 and 6.2 nm, respectively, as indicated, and the incubation continued for 30 min. The products were then analyzed by gel electrophoresis as described in A above. Right panel, concentrations of topoisomerase (Topo) I used was more than sufficient to completely relax the supercoiled form of the plasmid in the absence of bound MukB. D, MukB stabilizes topologically isolated loops in the DNA. Left and middle panels, MukB was incubated with the nicked DNA substrate for 5 min at 37 °C and then analyzed by electrophoresis through agarose gels either in the absence or presence of 20 μg/ml chloroquine, respectively, in the gel and running buffer. Right panel, densitometric lane traces of the gels shown in the left and middle panels.
Because MukB stabilizes negative supercoils (4) and supercoiling of the DNA increases its electrophoretic mobility, we asked whether MukB was inducing supercoils in the nicked DNA. The MukB-DNA complex was incubated with E. coli DNA ligase, which uses NAD as a cofactor rather than ATP (16), to seal the nick. After deproteinization, DNA products were analyzed by electrophoresis through an agarose gel containing chloroquine (Fig. 1B). Chloroquine unwinds the DNA helix, and thus in a relaxed, closed circular DNA (the product of sealing the nick) it will induce positive supercoils, and the mobility of the DNA will increase (Fig. 1B, compare lanes 1 and 2). DNA sealed in the presence of MukB contained negative supercoils, as indicated by the ladder of topoisomers with retarded mobility compared with that of the relaxed DNA alone (Fig. 1B, compare lane 2 to lanes 4–6).
To determine whether these negative supercoils were merely stabilized by MukB or actually protected by it, we exposed the protein-DNA complexes to two different type I topoisomerases that relax negative supercoils: E. coli topoisomerase I, which acts on single-stranded DNA (17), and vaccinia virus topoisomerase, which acts on dsDNA (18). Both the SMcx and FMcx were examined (Fig. 1C). In neither case did the topoisomerase reduce the mobility of the protein-DNA complex, arguing that the supercoils were unavailable to be relaxed. We conclude that the supercoils could not be relaxed because the helical crossover that forms them was completely protected by binding to MukB. Controls showing that the amount of topoisomerase used was sufficient to relax all the supercoiled DNA are shown in Fig. 1C (right panel).
Thus, it seemed likely that the FMcx was probably dominated by the formation of protected supercoils. The addition of more MukB to the reaction causes the mobility of the complex to slow. This effect could simply be because of the accretion of additional mass to the complex via the binding of more MukB. However, we wondered whether the effect could also be attributed to another distinct layer of topology imposed by the increasing amounts of bound MukB. To probe this possibility, we compared the mobility of the protein-DNA complexes in gels that either did or did not contain chloroquine (Fig. 1D). As expected, the mobility of the FMcx was reduced compared with the nicked DNA in the chloroquine-containing gel (Fig. 1D, compare lanes 1–3 in each panel). This effect is because chloroquine is a small molecule and can unwind even the MukB-protected supercoils, confirming that this FMcx is dominated by negative supercoiling.
Remarkably, the presence of chloroquine in the gel caused the SMcx to now move faster than the nicked DNA (Fig. 1D, compare lanes 4 and 5 in each panel). This inversion of DNA mobility can only be explained by the presence of relaxed loops in the SMcx that become positively supercoiled in the presence of chloroquine. In order for these loops to exist, they must be stabilized by MukB.
To confirm our interpretation, we reasoned that these loops should be stable enough to be supercoiled by the action of DNA gyrase (19). We therefore designed an experimental scheme so that gyrase would only be acting when the DNA was still nicked and thus not topologically constrained by virtue of the continuity of both DNA strands. MukB protein-DNA complexes were formed and then incubated with DNA gyrase and ATP (19). Novobiocin was then added to inactivate gyrase (20) followed by DNA ligase to seal the DNA and lock in any changes in DNA topology. After deproteinization, the DNA products were analyzed by gel electrophoresis in either the absence or presence of chloroquine (Fig. 2, A, top and bottom panels, respectively, and B). As before (Fig. 1B), the presence of MukB alone caused the accumulation of negative supercoils (Fig. 2, A and B, lanes 2–6). However, the addition of DNA gyrase to SMcxs clearly caused the accumulation of additional supercoils (Fig. 2, A and B, compare lane 6 to lane 9). Controls demonstrating the inhibition of DNA gyrase by novobiocin are shown in Fig. 8C (lanes 11 and 12).
Figure 2.
Loops formed by MukB in the DNA are topologically isolated. A, DNA loops stabilized by MukB can be supercoiled by DNA gyrase. MukB was incubated with the nicked DNA substrate, 2 mm ATP, and 20 nm DNA gyrase for 30 min at 37 °C. Novobiocin was added to 10 μm and the incubation continued for 5 min. Bacteriophage T4 DNA ligase was then added and the incubation continued for 30 min. The products were deproteinized before analysis by electrophoresis through agarose gels either in the absence (top panel) or presence (bottom panel) of 10 μg/ml chloroquine in both the gel and the running buffer. Relaxed, the nicked DNA sealed by DNA ligase to give a closed DNA ring that does not contain supercoils. B, densitometric lane traces of the gels shown in A. C, MukB stabilizes topologically isolated loops in linear DNA. MukB was incubated with either nicked DNA (lanes 1–5) or linear DNA (blunt-end cut, lanes 6–10) for 5 min at 37 °C. Protein-DNA complexes were then analyzed as in Fig. 1D above.
Figure 8.
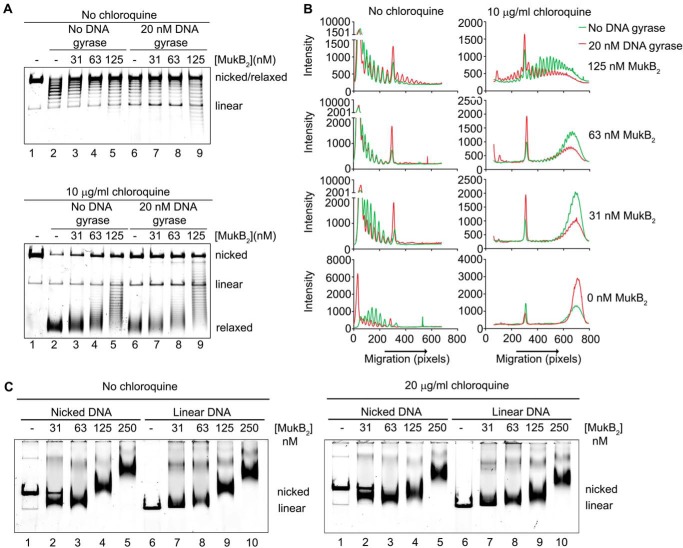
MukB KE,RE variant is defective in the induction of negative supercoils and stabilizing loops in the DNA. A, either wild type or MukB KE,RE was incubated with the nicked DNA substrate, NAD, and E. coli DNA ligase for 30 min at 37 °C. The products were deproteinized and then analyzed by electrophoresis through an agarose gel containing 10 μg/ml chloroquine in both the gel and the running buffer. B, densitometric lane traces of the gels shown in A. C, MukB KE,RE is defective in stabilizing loops in the DNA. The indicated concentrations of either wild type or MukB KE,RE were incubated with the nicked DNA substrate, DNA gyrase, novobiocin, and DNA ligase as described in the legend to Fig. 2A. The order of addition of components is outlined at the top of the gel. B, MukB; N, novobiocin; G, DNA gyrase; L, DNA ligase. Electrophoresis was with 10 μg/ml chloroquine in both the gel and the running buffer. D, densitometric lane traces of the gel shown in C.
We considered that some curvature of the DNA might be required for the action of MukB, as has been suggested for the yeast condensin (21). Accordingly, we asked whether we could detect topological loop stabilization when MukB was bound to linear DNA (Fig. 2C). This proved to be the case. MukB-linear DNA complexes formed at 125 and 250 nm MukB clearly displayed increased mobility in gels containing chloroquine compared with those that did not (Fig. 2C, compare lanes 4 and 5 and 9 and 10 in the presence and absence of chloroquine). We conclude that at higher concentrations MukB was stabilizing topologically isolated loops in dsDNA. These observations are consistent with the results of single molecule analyses of the effect of MukB binding on the length of DNA that were interpreted as indicating MukB-mediated loop formation (14).
Scanning force microscopy of MukB-DNA complexes: The MukB hinge region binds DNA
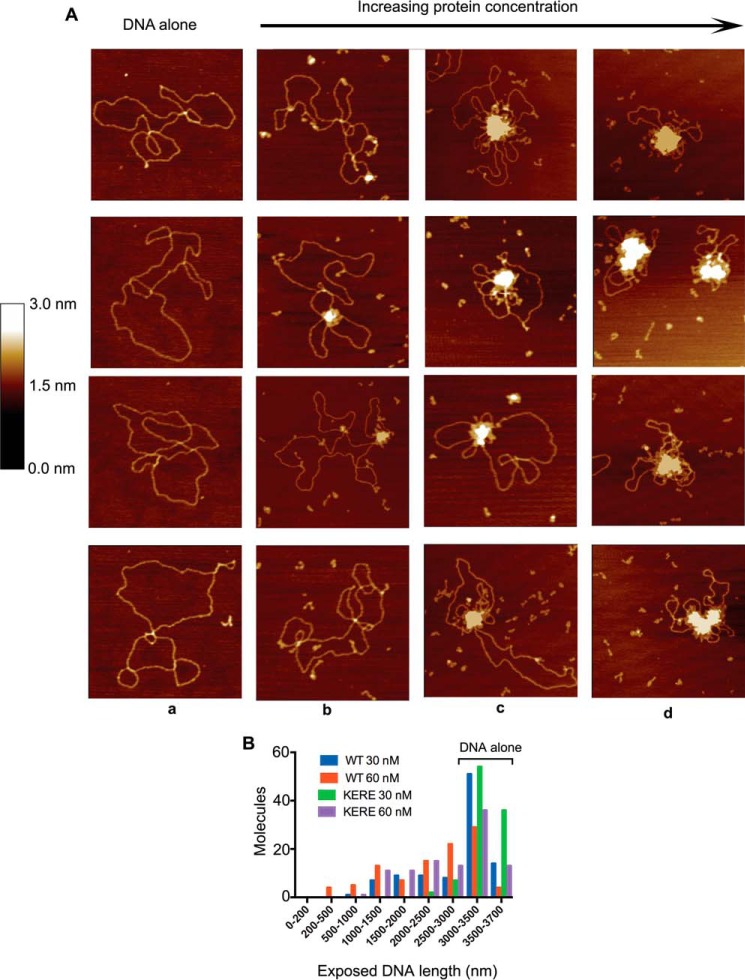
MukB-DNA complexes were examined using SFM. As the MukB concentration increased, the protein-DNA complexes presented increasingly aggregated masses with unbound DNA extruding from the central mass (Fig. 3A). The extent of exposed DNA in the protein-DNA complex decreased as the concentration of MukB increased, suggesting that the SFM images corresponded to that of progressively condensed DNA. To assess this possibility, we used as a measure of DNA condensation the length of dsDNA not present in the aggregated mass. Unconstrained plasmid DNA was 3430 ± 174 nm long. As the concentration of MukB increased from 15 to 60 nm, the length of exposed DNA decreased (Fig. 3B), suggesting that the SFM images report on the structure of the same MukB-DNA complex that has an electrophoretic mobility greater than that of the nicked DNA alone (the FMcx). Therefore, under these conditions the DNA becomes condensed. Such aggregation of MukB-DNA complexes is consistent with cooperative binding of MukB to the DNA (14). It was not possible to examine MukB-DNA complexes formed at higher concentrations of MukB with the SFM because these appeared as highly aggregated forms.
Figure 3.
Progressive DNA condensation of DNA by MukB as visualized by scanning force microscopy. A, protein-DNA complexes formed with the nicked DNA substrate either in the absence of MukB (column a) or in the presence of 15 (column b), 30 (column c), or 60 nm MukB (column d) were imaged in the SFM as described under “Experimental procedures.” Images shown are for wild-type MukB. Images for MukB KE,RE were identical in appearance. B, extent of exposed DNA in the MukB-DNA complexes, measured as described under “Experimental procedures,” is presented as a function of MukB concentration. WT, wild type; KERE, MukB K761E/R765E protein variant. A total of 100 molecules was measured from three independent experiments.
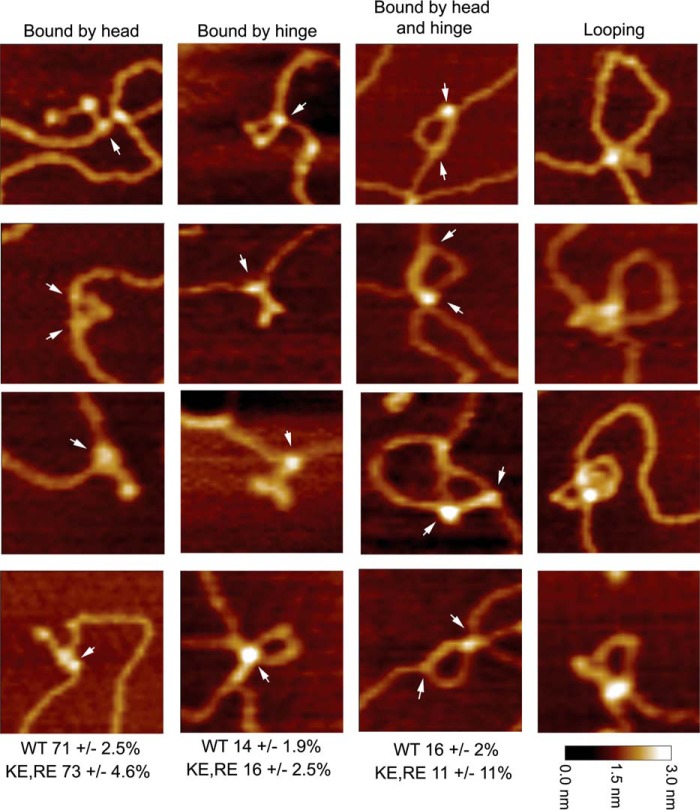
At low concentrations of MukB, individual MukB molecules bound to the DNA could be observed (Fig. 4). Head and hinge domains could not be distinguished per se; however, we know that MukB is a V-shaped molecule with the hinge domain at the apex and the head domains on the opposite ends (22). Bound MukB molecules could be divided into three classes (Fig. 4): bound by the head domain; bound by the hinge domain; and bound by both the head and hinge domains. Anchoring of DNA loops by bound MukB was also observed. MukB binding via the hinge domain was unexpected because it had been reported previously that the isolated hinge domain did not bind DNA (23, 24). Binding of isolated MukB hinge to DNA was confirmed as described below.
Figure 4.
Modes of MukB binding to DNA as visualized by scanning force microscopy. One hundred molecules were counted from three independent experiments where MukB at either 7.5 or 15 nm was bound to the nicked DNA substrate, and the modes of binding were characterized as either MukB bound to the DNA by a head domain, MukB bound to the DNA by the hinge domain, or MukB bound to the DNA by both the hinge and head domains. The fraction of molecules bound in each mode for the wild type and the MukB KE,RE variant is given below each column. Images where the DNA is looped with MukB bound at the apex are also shown. Images shown are for wild-type MukB. Images for MukB KE,RE were identical in appearance. Arrows indicate points of contact between MukB and the DNA.
Model for DNA condensation by MukB
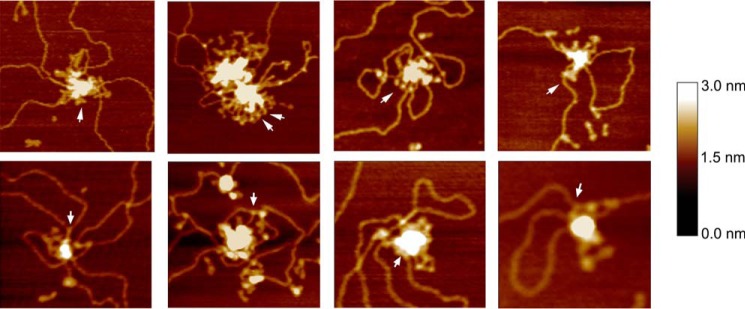
Individual MukB molecules could often be observed protruding from the aggregated mass in the SFM images (Fig. 5). In many instances, it seemed like the hinge domain was in the root of the aggregate with the head domains, occasionally bound to DNA, protruding (see arrows in Fig. 5). This possible feature of the aggregation, MukB protection of negative supercoils and formation of topologically stable loops, and hinge domain binding of DNA suggested a model for how MukB condenses DNA (Fig. 6, A–C). We propose that MukB initially binds DNA such that a negative supercoil is protected by interactions with the DNA-binding domains on the head domains and the hinge domain (Fig. 6A). In this fashion, extraneous topoisomerases might not be able to relax the supercoil, whereas chloroquine would be able to bind the DNA and untwist it. As more MukB binds the DNA, interactions between the hinge domains of MukB dimers become more likely leading to the formation of loops (Fig. 6B). These loops are topologically isolated and can be supercoiled by DNA gyrase (Fig. 6C). Aspects of this model are similar to models proposed recently describing the action of the yeast condensin (21), MukB (25), and the B. subtilis SMC protein (26). Using single molecule analyses of the effect of MukB on the length of DNA with one end tethered to a glass slide and the other held in magnetic tweezers, Cui et al. (14) proposed a stepwise condensation of DNA and inferred from their results that one aspect of the condensation would be the formation of loops in the DNA.
Figure 5.
Close-up views of MukB-DNA aggregates. Close-up SFM views of MukB aggregates on the DNA showing individual MukB molecules protruding from the aggregated mass (arrows).
Figure 6.
Model for MukB condensation of DNA. A, MukB protects a negative supercoil when bound to DNA. We propose that DNA binding by both the head domain and hinge domain is necessary for efficient negative supercoiling. The disposition of the DNA on the MukB hinge should not be taken literally. Whereas the amino acid residues mutated in the KE,RE variant are on the top of the hinge, we do not know precisely how DNA is bound to the hinge. One presumes that the DNA binds in such a manner that it does not interfere with MukB oligomerization via hinge–hinge interactions. B, MukB can form stable, topologically isolated loops in the DNA via hinge–hinge interactions between dimers. C, progressive DNA condensation by MukB is driven by the formation of topological loops in the DNA that can be further negatively supercoiled by DNA gyrase. D, effect of MukEF and ATP on MukB-DNA complexes. The indicated concentrations of MukB or the MukBEF complex in the presence or absence of ATP were incubated for 5 min at 37 °C. Protein-DNA complexes were then analyzed as described in the legend to Fig. 1A. Note that twice the concentration of MukBEF was required to observe similar DNA-binding activity compared with MukB alone. E, MukB does not trap DNA topologically. The indicated concentrations of MukB-HA, MukF, and MukEF, either in the presence or absence of ATP, were incubated with an equal mixture of nicked and linear DNA for 5 min at 37 °C. Panel i, anti-HA antibody attached to magnetic beads was then added and immediately thereafter NaCl was added to 0.5 m, the beads were washed in 0.75 m NaCl after pulldown, and the bound DNA was released by proteinase K digestion and analyzed by agarose gel electrophoresis. Panel ii shows the same experiment except that 0.5 m NaCl was added immediately before addition of the antibody beads. F, MukB does not trap DNA topologically. Tailed, nicked plasmid DNA attached to magnetic beads was incubated with the indicated concentrations of MukB, MukF, and MukEF either in the presence or absence of ATP in standard reaction buffer (low salt) for 5 min at 37 °C. The beads were then pulled down and resuspended in buffer containing 0.5 m NaCl. The beads were pulled down again, and the samples were resuspended in 1× SDS-PAGE loading dye. After heating to 95 °C, the proteins released were analyzed by SDS-PAGE. I, input; P, DNA bound to the beads after the first pulldown in low salt; HS, DNA bound to the beads after washing the beads in high salt.
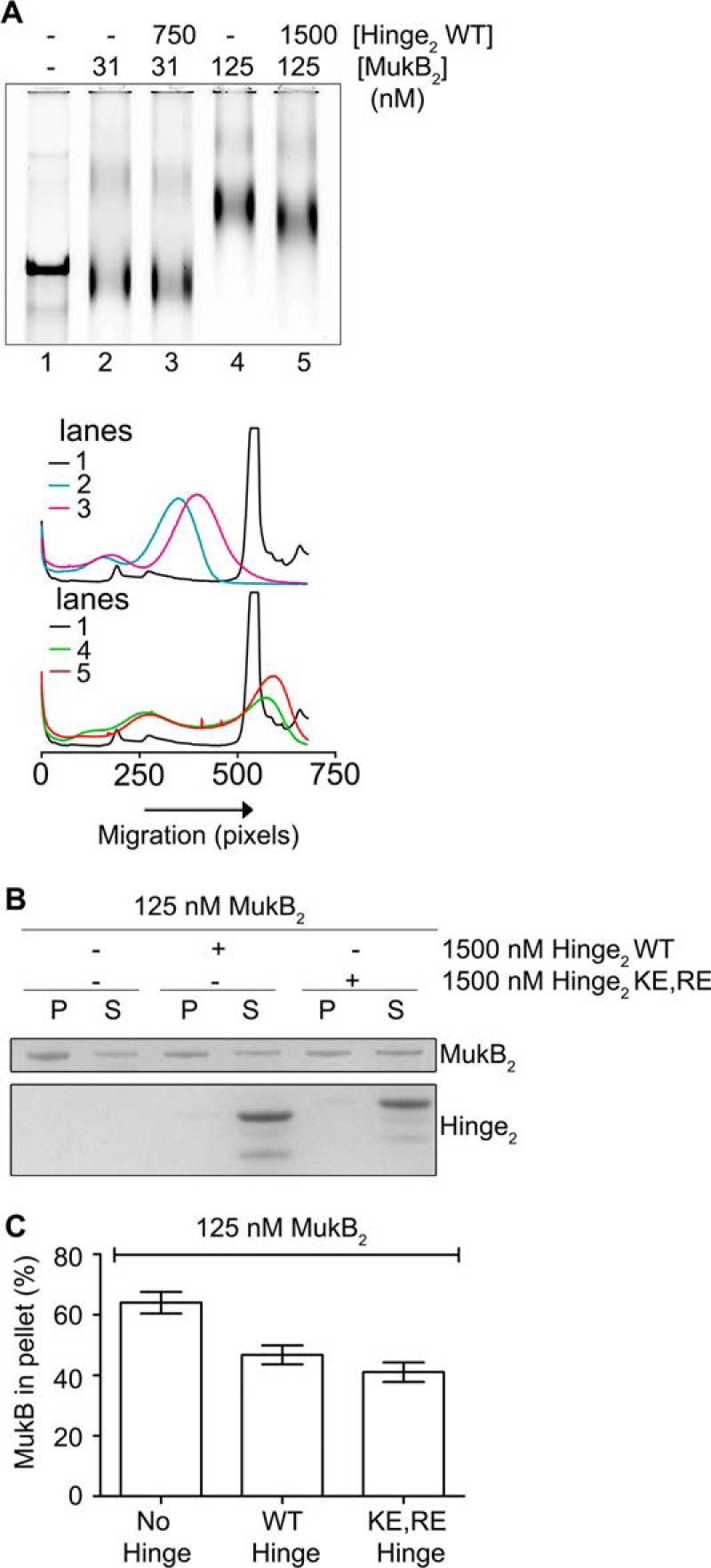
Because our model seems counter to the topological entrapment model for SMC proteins binding DNA (1), we addressed this issue directly. First, we asked whether MukF or MuKEF and ATP had an effect on MukB DNA condensation. MukEF appeared to stabilize the FMcx somewhat, but it also inhibited MukB binding overall. The mobility of the SMcx was unaffected, and ATP had little effect on either complex with or without MukEF being present (Fig. 6D). Thus, MukB alone is sufficient for DNA condensation, as has been demonstrated previously (14). Second, we used the assay developed by Uhlmann's laboratory for topological trapping (27). Here, MukB-HA bound to a mixture of circular and linear DNA is captured by anti-HA antibodies bound to beads in the presence of 500 mm NaCl and washed with 750 mm NaCl. Persistence of the circular, but not linear, DNA after the high salt wash argues that the DNA had been bound topologically by the protein. Our initial result appeared to support a similar conclusion (Fig. 6E, panel i). Consistent with previous studies, the presence of MukEF and ATP decreased MukB DNA binding (9, 14). However, we were concerned that this result could be artifactual, dependent on the antibodies bound to the beads providing the topological seal that prevented dissociation of the circular DNA. This proved to be the case. If the high salt was added immediately before binding of MukB by the antibodies, no DNA was retained, neither circular nor linear (Fig. 6E, panel ii). Finally, we used an additional assay. Here, we bound tailed, nicked circular DNA to a magnetic bead, then bound MukB to the DNA in the presence or absence of MukEF and ATP, pulled down the beads to isolate the bound DNA, washed the beads with 500 mm NaCl, and then pulled them down again. After high salt washing, no MukB remained bound to the DNA (Fig. 6F). We conclude that MukB does not trap DNA topologically as proposed for the eukaryotic SMC proteins, lending support to our model for MukB binding. This is consistent with the observation that MukB can stably bind linear DNA (9).
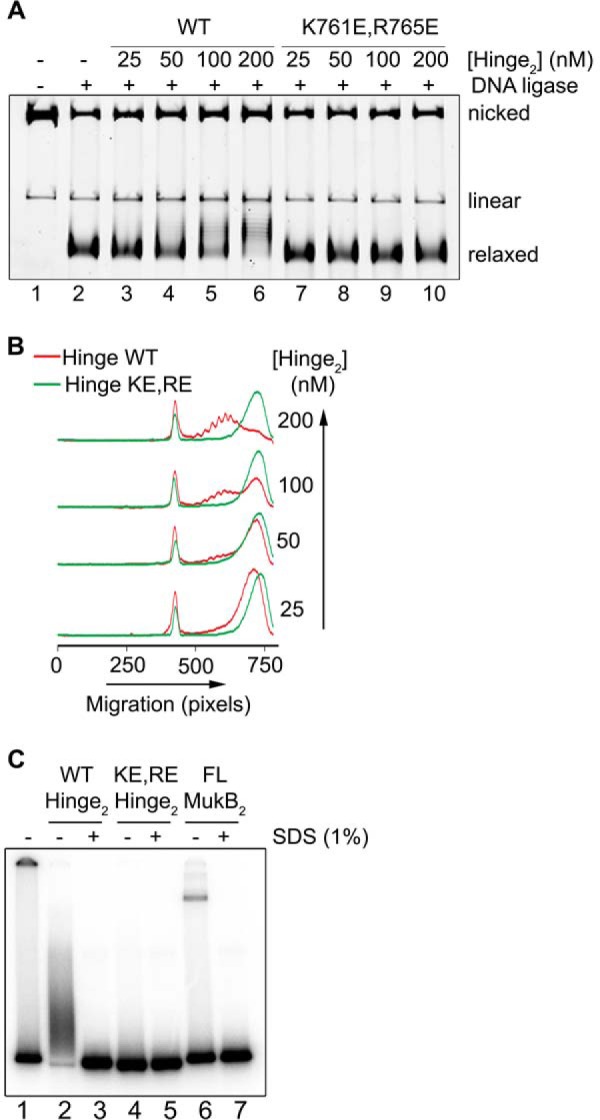
MukB K761E/R765E, mutated in the hinge domain, is defective in DNA condensation
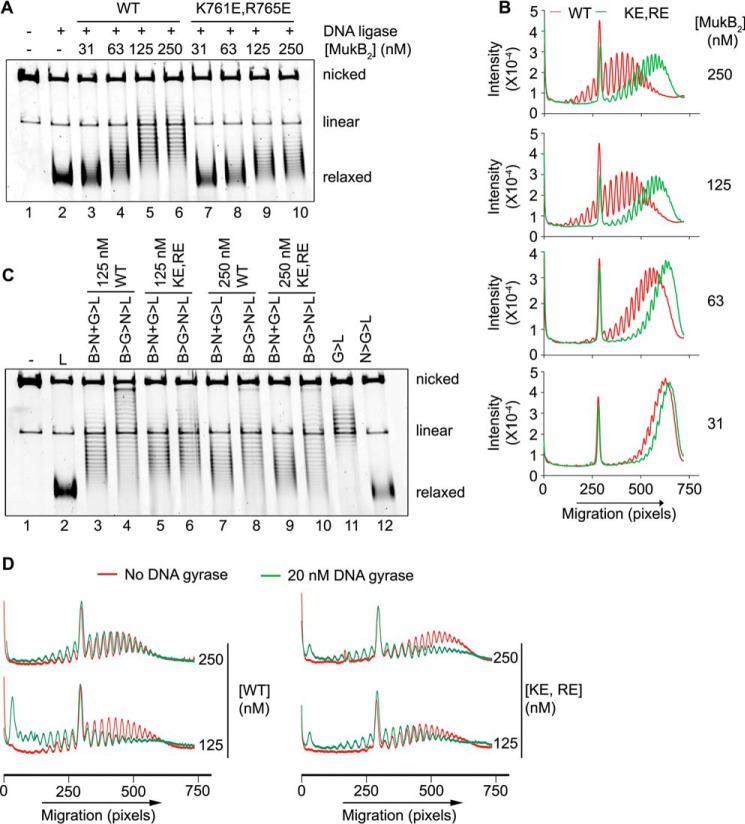
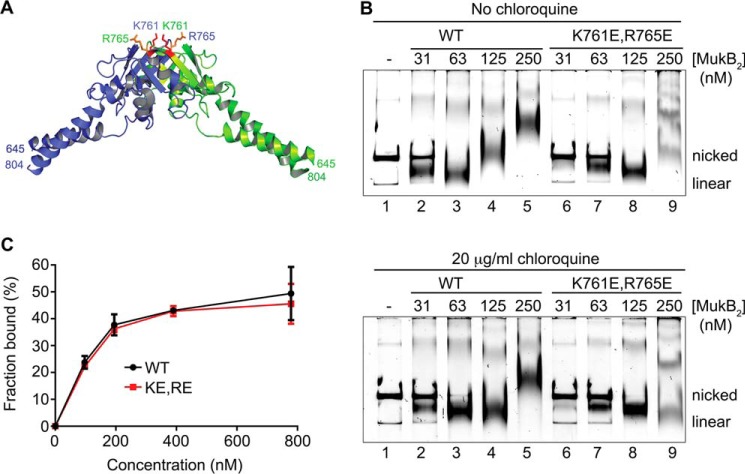
Our model and the SFM data suggest that the MukB hinge domain should bind DNA. The structure of the hinge domains of the B. subtilis SMC protein and MukB is somewhat similar, although the B. subtilis structure showed a clear electropositive patch that was demonstrated to bind DNA (28). We examined the surface charge distribution of the MukB hinge and found several small electropositive patches. Charge reversal amino acid substitutions were made, and one, MukB K761E/R765E (named MukB KE,RE), which is located at the top of the hinge (Fig. 7A), distinct from the amino acid residues that interact with ParC (29), was found to have a DNA condensation defect as assayed by both SFM (Fig. 3B) and gel electrophoresis (Fig. 7B). MukB KE,RE was about 50% as effective as the wild type in both assays. Although the mutant protein had a tendency to aggregate at high concentrations, both the FMcx and SMcx were observed.
Figure 7.
MukB K761E/R765E variant protein can form both the FMcx and SMcx. A, schematic of the structure of the hinge domain of MukB (23) with the Lys-761 and Arg-765 amino acids shown in stick representation. B, MukB KE,RE can form both the FMcx and SMcx protein-DNA complexes. Either wild type or MukB KE,RE was incubated with the nicked DNA for 5 min at 37 °C, and the products were analyzed by electrophoresis through agarose gels that either did or did not contain 20 μg/ml chloroquine in the gel and running buffer. C, wild type and MukB KE,RE binding to a 5′-end-labeled 50-bp duplex DNA is identical. Nitrocellulose filter-binding assays were performed as described under “Experimental procedures.”
The defect in DNA condensation by MukB KE,RE could be a result of defective DNA binding; however, DNA-binding activity, as measured in a nitrocellulose filter-binding assay with a duplex 50-mer oligonucleotide, was identical for both the wild-type and variant proteins (Fig. 7C). This assay presumably measures the DNA-binding activity of the head domains but not the hinge domain. Binding of the hinge domain to DNA as measured by SFM was also identical to that of the wild-type protein (Fig. 3). Nevertheless, a DNA-binding defect was apparent for the variant protein as measured by the extent of supercoiling imparted to the nicked DNA after it was sealed by DNA ligase (Fig. 8, A and B, compare lanes 5 and 6 to lanes 9 and 10). Similarly, although MukB KE,RE could form loops that could be supercoiled by DNA gyrase, it was less efficient compared with the wild type (Fig. 8, C and D).
Thus, the MukB KE,RE was deficient in protecting supercoils and forming loops in the DNA. One possible explanation for these observations was that the mutations might cause an alteration of the disposition of the hinge and/or coiled-coil regions that affected the ability of the protein to condense DNA. Long-range transmission of the effect of mutations in the hinge region of the B. subtilis SMC protein on the ATPase activity of the protein have been reported (30). To investigate this issue, we purified the isolated hinge domain and examined its activities.
Isolated hinge domain interferes with MukB-mediated DNA condensation
Neither gel-shift assays (23, 24), nitrocellulose filter DNA-binding assays (data not shown), nor DNA pulldown assays (see below) could score binding by the isolated hinge domain; we therefore turned to a more sensitive assay of DNA binding, assessing the changes in topology of a DNA ring by virtue of exposure to the protein. Isolated hinge domain induced negative supercoils in the nicked DNA that were evident after sealing the nick (Fig. 9, A and B). Thus, the hinge domain likely interacts with DNA, although this interaction could be unstable. This supercoiling activity was absent when the KE,RE hinge domain was used (Fig. 9, A and B).
Figure 9.
MukB hinge domain fragment binds DNA. A, wild-type, but not KE,RE, MukB hinge domain induces supercoils in the nicked DNA substrate. Either wild-type or KE,RE hinge domain fragment was incubated with the nicked DNA substrate for 5 min at 37 °C, NAD and E. coli DNA ligase were then added, and the products were analyzed as in the legend to Fig. 1B. B, densitometric lane traces of the gel shown in A. C, wild-type, but not KE,RE, hinge domain fragment cross-links to DNA. The indicated concentrations of wild-type, KE,RE hinge domain fragment, or full-length MukB were incubated with a 5′-32P-labeled 100-mer DNA duplex for 5 min at 37 °C. Formaldehyde was then added to 0.7%, and the incubation continued for 30 min on ice. The HCHO was then neutralized by the addition of Tris base, and the DNA-protein complexes were analyzed by electrophoresis through 4–12% gradient SDS-PAGE. One percent SDS was added to the reactions shown in lanes 3, 5, and 7 prior to the addition of HCHO.
To probe this issue further, we performed DNA cross-linking experiments with the wild-type and mutant hinge domain fragments and a 5′-32P-end-labeled 100-mer DNA duplex. Wild-type hinge fragment cross-linked to the DNA, and this cross-linking was sensitive to prior treatment of the hinge fragment with SDS (Fig. 9C, compare lanes 2 and 3), similar to the cross-linking observed with the full-length wild-type protein (Fig. 9C, compare lanes 6 and 7). However, the KE,RE hinge fragment exhibited little cross-linking to the DNA (Fig. 9C, lanes 4 and 5). Thus, the hinge domain fragment clearly binds DNA.
On the one hand, the assay presented in Fig. 9 apparently confirmed the SFM data suggesting that the hinge region bound DNA; however, on the other hand, the absence of supercoiling by the KE,RE hinge is inconsistent with its apparent binding as assayed by SFM. It is possible that DNA binding of the KE,RE hinge is stabilized in the full-length variant protein; however, given that the defects of MukB KE,RE–less negative supercoiling imparted to the DNA–and the KE,RE hinge fragment correspond, we conclude that the defect in supercoiling contributes to the observed defect in DNA condensation by the variant protein.
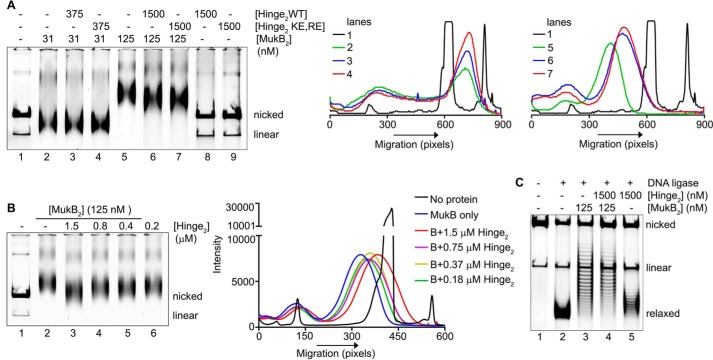
To assess whether interfering with hinge–hinge interactions also affected DNA condensation, we added wild-type and KE,RE hinge fragment to the DNA condensation gel assay. The presence of the hinge domains had little effect on the FMcx (Fig. 10A, lanes 2–4); however, they increased the mobility of the SMcx (Fig. 10A, lanes 5–7). This effect was confirmed by the results of a titration of the hinge domain, showing that at lower concentrations of the hinge domain than those used in Fig. 10A, intermediate effects on the mobility of the SMcx were observed (Fig. 10B). The hinge domains had no effect on the mobility of the DNA (Fig. 10A, compare lanes 8 and 9 to lane 1), so the observed increase in the mobility of the SMcx was not a result of additional supercoiling of the DNA. This was confirmed by examining the extent of supercoiling induced when the nicked DNA was sealed in the presence of MukB either in the presence or absence of the wild-type hinge fragment (Fig. 10C). Instead, this observation suggested that the presence of the hinge fragment was destabilizing MukB binding to the DNA and that destabilization did not require hinge fragment DNA binding.
Figure 10.
Hinge domain fragments interfere with MukB-mediated loop formation in the DNA. A, left panel, MukB was incubated with the nicked DNA substrate for 5 min at 37 °C. Either wild-type or KE,RE hinge domain fragment was then added, and the incubation continued for an additional 30 min. Reaction products were then analyzed by agarose gel electrophoresis. Middle panel, densitometric lane traces of lanes 1–4 of the gel shown in A. Right panel, densitometric lane traces of lanes 1 and 5–7 of the gel shown in A. B, left panel, as in A, except that the concentration of wild-type hinge domain fragment in the reaction was varied. Right panel, densitometric lane traces of the gel shown in the left panel. C, hinge domain fragment does not interfere with MukB-induced supercoiling in the slow-moving protein-DNA complex. MukB and wild-type hinge domain fragment were incubated with the nicked DNA substrate for 5 min at 37 °C. NAD and E. coli DNA ligase were then added, and the products were analyzed as in the legend to Fig. 1B.
To examine this question, we used a version of the nicked DNA substrate with a 5′-biotinylated single-stranded DNA tail so that protein-DNA complexes could be isolated using streptavidin-coated magnetic beads. We confirmed that this DNA behaved similarly to the nicked DNA substrate, forming both the FMcx and SMcx, and that hinge fragment affected the mobility of the SMcx (Fig. 11A).
Figure 11.
Hinge domain fragment destabilizes bound MukB. A, tailed, nicked DNA substrate is equivalent to the nicked DNA substrate. Top panel, MukB in the presence or absence of the wild-type hinge domain fragment was incubated with the biotinylated, tailed, nicked DNA substrate for 5 min at 37 °C and then analyzed by agarose gel electrophoresis. Bottom panel, densitometric tracings of the lanes of the gel shown in the top panel. B and C, hinge domain fragment destabilizes bound MukB. MukB was incubated on a rotator with tailed, nicked DNA substrate that had been bound to magnetic beads for 5 min at 37 °C. Either wild-type or KE,RE hinge domain fragment was then added, and the incubation continued for 30 min. The beads were then pulled down on a magnet, and the supernatant was removed. The protein present in the pellet (P) and supernatant (S) fractions were then assayed by SDS-PAGE. B, example of the SDS gel. C, distribution of MukB in the pellet and supernatant. The mean and standard deviation is shown for three independent experiments.
Pulldown assays showed that either the wild-type or KE,RE hinge fragment reduced the amount of MukB in the SMcx by one-quarter and one-third, respectively (Fig. 11, B and C). Similar amounts of MukB were destabilized from the FMcx (data not shown). We conclude that the hinge fragment was disrupting interactions between MukB dimers in the protein-DNA complexes that led to their destabilization. This effect is likely independent of the ability of the hinge domain to bind DNA. Because the mobility of the slow-moving complex is dominated by loop formation, this destabilization results in an observable change in mobility.
Wild-type and KE,RE hinge fragments consistently had different mobilities in SDS-PAGE (Fig. 11C). Proteolysis followed by mass spectrometry showed identical coverage of peptides on both the C and N termini of the protein fragments, indicating that the difference in mobility was not because of proteolytic clipping. Thus the K761E/R765E mutations may also affect the flexibility of the hinge. This change in hinge flexibility could also be responsible for some of the defects of the variant MukB KE,RE protein.
Hinge mutations disrupt DNA condensation in vivo
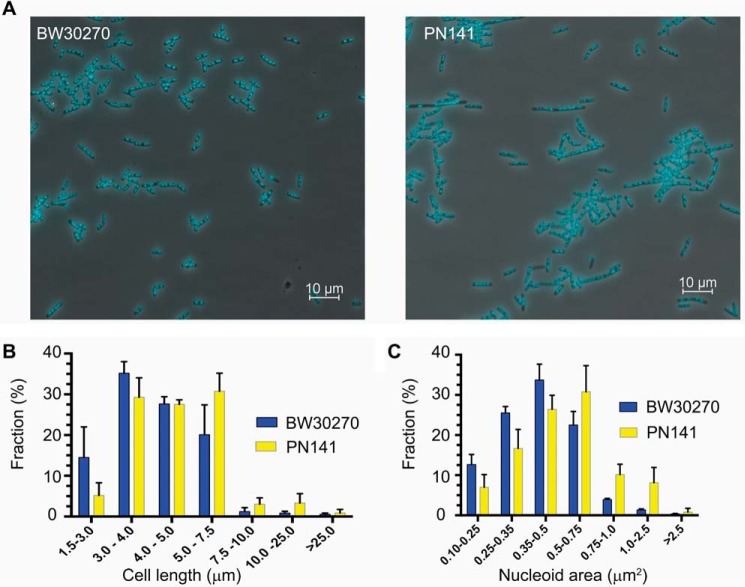
To determine the effect of the hinge mutations on MukB-mediated condensation of DNA in vivo, we engineered an E. coli strain where the wild-type mukB allele had been replaced with the mukBK761E/R765E allele. PN141 cells carrying the mukBK761E/R765E allele and wild-type BW2070 were grown in LB medium, stained with DAPI (without fixation), and examined (Fig. 12). Mutant cells displayed slightly increased cell length (Fig. 12B) and nucleoid area (Fig. 12C), with median cell length increasing by 13% (from 4.0 to 4.53 μm) and median nucleoid area increasing by 25% (from 0.4 to 0.5 μm2), suggesting a delay in cell division and nucleoid decondensation. However, because these differences were small and not statistically significant, we examined nucleoid compaction in the mutant strain by velocity sedimentation of isolated nucleoids directly (Fig. 13). Spermidine nucleoids were isolated from wild-type and PN141 cells grown to mid-log phase in MOPS medium and sedimented through 10–40% sucrose gradients. Nucleoids isolated from PN141 sedimented significantly more slowly than those isolated from the wild-type strain (Fig. 13). Thus, we conclude that the KE,RE MukB hinge mutation causes nucleoid decondensation.
Figure 12.
mukBK761E/R765E mutant cells are longer, and their nucleoids are larger than wild type. A, fields of DAPI-stained wild-type (BW30270) and mutant PN141 (BW30270mukBK761E/R765E) E. coli cells. B, the mutant strain is somewhat longer than the wild type. Cell length distribution of BW30270 and PN141 grown to early log phase in LB medium. A total of 1641 and 1717 cells of the wild type and PN141, respectively, were measured from three independent experiments. C, mutant strain has a somewhat larger average nucleoid area than the wild type. A total of 1630 and 1610 nucleoids in wild-type and PN141 cells, respectively, was measured from three independent experiments. The very diffuse nucleoids present in the filamented cells of PN141 were excluded from the latter measurement.
Figure 13.
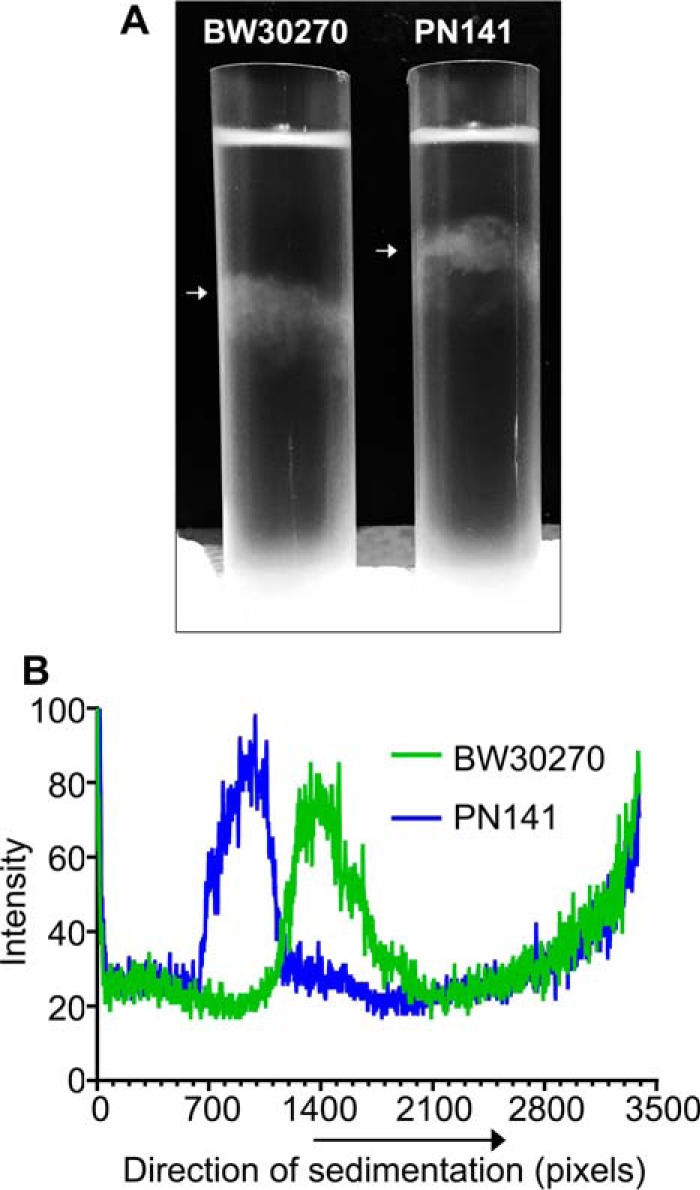
Nucleoids in the mukBK761E/R765E mutant strain are decondensed. Wild-type (BW30270) and hinge mutant (PN141, BW30270mukBK761E/R765E) E. coli cells were grown to mid-log phase in MOPS medium. Cells were harvested, and spermidine nucleoids were prepared and sedimented through sucrose gradients as described under “Experimental procedures.” A, photograph of typical sucrose gradients showing the positions of the sedimented nucleoids (white arrows). B, densitometric trace of OD260 absorbing material in each gradient.
Discussion
Chromosomes must be condensed efficiently and in an orderly fashion so as to both fit in their specialized compartments and to allow for proper chromosome replication, gene expression, and segregation. We applied several novel biochemical assays supported by molecular imaging and studies in vivo to examine how the bacterial SMC-like protein MukB condenses DNA.
MukB forms and stabilizes two distinct types of topological domains to condense DNA
Of the two types of MukB-DNA complexes formed, the FMcx was dominated by the formation of negative supercoils, as shown by decreased mobility in chloroquine-containing gels. Surprisingly, although the decrease in mobility of the SMcx compared with that of the FMcx could be accounted for by the addition of more MukB, we found that under these conditions relaxed DNA loops were being formed in the DNA that could be supercoiled by DNA gyrase. Thus, MukB was sufficient to establish two distinct types of topological domains in a nominally unconstrained DNA in the absence of either ATP or its accessory proteins.
SFM imaging of these protein-DNA complexes demonstrated progressive condensation of the DNA as MukB concentrations increased, as had been observed previously (14). Moreover, these analyses pointed to the likelihood that the MukB hinge region both bound DNA and seeded aggregation of MukB dimers. Accordingly, we developed a model for MukB DNA condensation that incorporated these features whereby condensation of the DNA occurs by the protection of negative supercoils by individual MukB dimers locally and the formation of loops in the DNA anchored by MukB dimers that interact via their hinge domains. These loops are themselves topologically isolated and can thus be supercoiled further.
MukB DNA condensation
A current model for how SMC proteins bind DNA invokes topological entrapment of the DNA by the tripartite complex formed by the dimer of the SMC proteins, the kleisin, and additional accessory proteins (1). Yeast cohesin has been shown to become trapped around circular but not linear small and large minichromosomes if all three interfaces in the tripartite complex are cross-linked (32, 33). In an assay where the cohesin was loaded onto DNA, the protein immunoprecipitated, the precipitate washed in high salt, and the DNA contents analyzed, only circular DNA survived the process (27). Similar types of assays have led to similar conclusions about the manner of DNA binding for the yeast condensin (34), the Smc5/6 complex (35), the B. subtilis SMC protein (12), and MukB (13).
There are, however, counterarguments to the topological trapping model. In single molecule experiments, Sun et al. (21) measuring compaction of DNA by the yeast cohesin against applied force concluded that DNA condensation proceeded via supercoiling-dependent loop formation that did not require ATP. Eng et al. (36) demonstrated interallelic complementation between two inactive (non-DNA binding) yeast cohesin complexes and argued that cohesion of sister chromatids was elaborated via protein–protein interactions between two distinct cohesin complexes, one bound to each of the sisters. Recently, in single molecule analysis of the manner by which the Schizosaccharomyces pombe cohesin diffused along a DNA molecule, Stigler et al. (37) found that although the protein could be pushed along by motor proteins, it could not bypass nucleosomes, and its diffusion was hindered by bound EcoRI and dCas9. These authors concluded that although the binding of the cohesin to DNA was topological, the central pore of the complex was not as large as predicted, with the protein assuming a more rod-like structure.
When tested directly in two different assays, we found no evidence that MukB, with or without MukF, MukE, or ATP, trapped DNA topologically. Furthermore, significant curvature of the DNA was not required for MukB DNA condensation. Our model for DNA condensation is more in line with the “handcuff” model proposed by Koshland and co-workers (36), where protein–protein interactions between MukB molecules are responsible for tethering distal parts of the chromosome to form stable loops in the DNA. Binding of MukB to the chromosome and the linking of distal loops are likely to elaborate a structure with a central core with loops extending from the core (Fig. 6C). Previous models describing a similar chromosomal arrangement have suggested MukB stabilization of loops in supercoiled DNA (14, 25). We point out, however, that MukB loading to the DNA is likely continuous with DNA replication, during which the two sister chromosomes are not topologically closed and thus cannot be supercoiled. Our model therefore suggests a manner by which overall chromosomal supercoiling can be maintained. Topological isolation of the loops formed by MukB allows supercoiling by DNA gyrase of the nascent sisters, thus ensuring continuous readout of the information imprinted in a supercoiled chromosome. Our model also is consistent with the order of genetic markers along the long axis of the cell and the occupancy of individual replichores in opposite cell halves (38, 39). Indeed, the latter organizing principle is dependent on active MukB (40). Our model is also consistent with ChIP data that show MukB distributed more or less evenly across the genome (41). In addition, a structure of this type is consistent with proposed nucleoid structures whether it be a constrained ring polymer (42, 43) or not (44, 45).
Role of the MukB hinge domain in DNA condensation
We have shown that the MukB hinge domain does bind DNA, an activity previously unappreciated (23, 24). This activity of the hinge clearly contributes to DNA condensation directly in that condensed MukB-DNA complexes formed with the wild-type protein were supercoiled to a greater extent than those formed with the KE,RE variant. The discovery of the hinge DNA-binding activity brings the action of MukB more in line with that of other SMC proteins where the hinge is known to bind DNA (1).
However, we believe that the most important activity of the hinge domain is in its requirement for DNA loop formation by MukB. Both the wild-type and KE,RE hinge fragment interfered with DNA loop formation by causing the dissociation of MukB. This interference was clearly not because the hinge fragments either competed or interfered with binding of the MukB head domains to DNA. Neither version of the hinge domain was recovered in the pellet of the DNA-binding pulldown assays, and the KE,RE hinge fragment may not bind DNA at all. Thus, the destabilization likely comes from disruption of hinge–hinge interactions between MukB dimers that themselves are stabilizing to the MukB on the DNA. DNA binding of the hinge region in the full-length MukB KE,RE protein may result in additional stabilizing interactions between the protein and the DNA.
The picture developed thus differs from other models of aggregate formation by SMC proteins that most often show aggregation via head–head domain interactions fueled by binding of ATP. ATP was not required nor did it affect the DNA condensation that we observed, implying that even when head domains are in the open configuration (i.e. not joined together by the binding of ATP), the protein can condense DNA effectively.
DNA condensation, MukEF, and ATP
Our observations that neither ATP nor MukEF were required to observe MukB-mediated condensation of DNA are consistent with previous observations reporting: (i) MukB did not require ATP to bind plasmid DNA and alter its topology (4); (ii) preformed MukBEF complex is defective in binding plasmid DNA (9); and (iii) adding MukEF to a MukB-DNA complex displaced MukB from the DNA (9). As mentioned above, MukE and MukF are required for and are present in MukB foci in vivo (6–8). MukB focus formation in vivo also requires its ATP-binding and hydrolysis activities (8). Strains carrying the MukB hinge mutations exhibited nucleoid decondensation. Thus, our model for MukB DNA condensation is reflective of what occurs in vivo. One possibility is that the effects of MukEF and ATP relate more to turnover of MukB on the DNA than to DNA condensation per se. We report the effect of the MukB-topoisomerase IV interaction on MukB-mediated DNA condensation in the accompanying article (31).
Experimental procedures
DNAs and proteins
The nicked DNA substrate was prepared as described in Bahng et al. (48). Biotinylated, tailed, and nicked DNA substrate was prepared as described in Yeeles and Marians (46) using the oligonucleotide 5′-TTTTTTTTTTTTCCCCCTCGAGCCCCTTTTTTTTTTTTGGGGCTCGAGGGGGTTTTTGTTTTCCCAGTCACGAC-3′ to prime synthesis of the complementary strand of M13mp18 viral DNA by bacteriophage T4 DNA polymerase. MukB was prepared as described previously (47). MukB K761E/R765E was prepared as for the wild type from E. coli strain BL21(DE3)pET11d-mukBK761E/R765E.
Hinge domain fragments were prepared as follows. A DNA fragment (synthesized by Gene Script) encoding amino acids 645–804 of MukB and provided cloned into pUC57 was cloned into the NdeI and XhoI restriction sites of the pET28a vector (Novagen) to create the overexpression vectors for both the WT and K761E, R765E variant of the hinge domain. E. coli BL21(DE3) Rosetta cells (Novagen) carrying these plasmids were grown in LB medium at 37 °C to an A600 = 0.4. isopropyl 1-thio-β-d-galactopyranoside (0.4 mm) was added and growth continued for 3 h. After harvesting, cells were lysed by sonication in buffer A (50 mm Tris-HCl (pH 8.0), 5% glycerol, 500 mm NaCl, and 2 mm DTT) with 10 mm imidazole-HCl followed by centrifugation at 20,000 × g for 30 min. The supernatant was mixed with Ni-NTA resin (Invitrogen) pre-equilibrated with buffer A containing 10 mm imidazole-HCl. The resin was washed with buffer A containing 75 mm imidazole HCl, and the hinge fragments were eluted with the same buffer containing 250 mm imidazole HCl. Fractions containing the hinge fragments were pooled and dialyzed against buffer B (50 mm Tris-HCl (pH 8.0), 150 mm NaCl, 2.5 mm CaCl2, 2 mm DTT, and 20 mm imidazole HCl). One unit of thrombin per mg of protein was added followed by incubation for 8–10 h at 4 °C. PMSF was then added to a final concentration of 1 mm, and the NaCl concentration was increased to 500 mm. The sample was mixed with Ni-NTA resin pre-equilibrated with buffer A containing 20 mm imidazole HCl for 1 h, and the flow-through was collected, dialyzed against buffer C (50 mm Tris-HCl (pH 8.0), 5% glycerol, 10 mm DTT, 100 mm NaCl, and 1 mm EDTA), and loaded on to a Q-Sepharose column (GE Healthcare) equilibrated with the same buffer. The column was washed with buffer C, and protein was eluted with a linear gradient of 100–800 mm NaCl in buffer C. Fractions were pooled based on SDS-PAGE analysis, dialyzed first against buffer C, and then storage buffer (50 mm HEPES-KOH (pH 7.5), 150 mm NaCl, 0.1 mm EDTA, 2 mm DTT. and 40% glycerol). Purified hinge fragments were stored at −80 °C.
Agarose gel assays
All biochemical gel assays were repeated at least three times.
DNA condensation
Reaction mixtures (20 μl) containing 50 mm HEPES-KOH (pH 7.5), 20 mm KCl, 10 mm DTT, 0.5 mm Mg(OAc)2, 7.5% glycerol, 0.34 nm nicked or ScaI-cut (linearized) pCG09, and either wild-type or the KE,RE variant MukB at the concentrations indicated in the figures were incubated for 5 min at 37 °C. Reaction products were analyzed by electrophoresis through 0.8% agarose gels at 23 V for 18 h at 4 °C using 50 mm Tris-HCl (pH 7.8 at 23 °C), 40 mm NaOAc, 1 mm EDTA, and 1.5 mm Mg(OAc)2 as the electrophoresis buffer. Chloroquine di-phosphate (20 μg/ml) was added to the gel and the electrophoresis buffer as indicated in the figures. Gels were stained with SYBR gold (Invitrogen) and scanned using a Typhoon FLA 9500 scanner. Profiles of gel lanes were taken using Multi Gauge Software (Fujifilm).
Negative supercoil induction
To assay the introduction of negative supercoils, 26 μm NAD and E. coli DNA ligase (New England Biolabs, 0.4 units) were included in the DNA condensation reaction mixtures that were incubated for 30 min at 37 °C. Reactions were stopped by the addition of EDTA and KCl to final concentrations of 50 and 300 mm, respectively, followed by incubation for 5 min at room temperature. Proteinase K and SDS were then added to 0.2 mg/ml and 0.2%, respectively, and the incubation was continued for 30 min at 37 °C. DNA products were resolved by electrophoresis through 0.8% agarose gels at 30 V for 15 h at room temperature using 50 mm Tris-HCl (pH 7.8 at 23 °C), 40 mm NaOAc, 1 mm EDTA, and 10 μg/ml chloroquine di-phosphate as the running buffer. Gels were stained and visualized as above.
DNA loop supercoiling by DNA gyrase
In DNA loop supercoiling assays, 2 mm ATP, an additional 2 mm Mg(OAc)2, and 20 nm DNA gyrase were included in DNA condensation reaction mixtures that were incubated for 30 min at 37 °C. Novobiocin was then added to 10 μm to inhibit DNA gyrase, and the incubation was continued at room temperature for 5 min. Bacteriophage T4 DNA ligase (New England Biolabs, 0.2 units) was then added, and the incubation continued for 30 min at 37 °C. The reactions were terminated and analyzed as above for negative supercoil induction.
MukB DNA-binding assays
Nitrocellulose filter binding
As described in Bahng et al. (48), data shown are from three independent experiments.
Magnetic bead pulldown
The biotinylated, tailed, nicked DNA was attached to M280 beads using the Kilobase Binder Kit (Invitrogen). Reaction mixtures were as for the DNA condensation assays except that beads carrying 50 ng of DNA were used and 0.05% Nonidet P-40 was also included. Reactions were incubated for 30 min at 37 °C on a rotator. DNA beads were then pulled down with a magnet. The supernatant was removed, and the beads were resuspended in the same volume of reaction buffer without protein and DNA. SDS sample loading buffer was added, and the reaction mixtures were heated at 95 °C for 5 min. The reaction products were resolved by 4–12% gradient SDS-PAGE (Invitrogen) and visualized by Coomassie staining. Intensity of the bands was quantified using MultiGauge Software (Fujifilm). Data shown are from three independent experiments.
DNA entrapment assayed by MukB-HA immunoprecipitation
DNA entrapment was assayed by MukB-HA immunoprecipitation as per Murayama and Uhlmann (27). Reaction mixtures (20 μl) containing 50 mm HEPES-KOH (pH 8.0), 20 mm KCl, 10 mm DTT, 7.5% glycerol, 0.05% Nonidet P-40, 100 ng each of NtBspQ1-nicked pET11b (nicked) and HpaI-cut pET11b (linear) DNA, the indicated concentrations of MukB-HA, MukF, and MukE, and either 0.5 mm Mg(OAc)2 or 2.5 mm Mg(OAc)2 and 2 mm ATP were incubated for 5 min at 37 °C. In the experiment shown in Fig. 6E, panel i, anti-HA antibody conjugated to magnetic beads (Pierce) that was sufficient to bind all of the MukB-HA was added; the reaction was mixed quickly by vortex genie for 2 s, and 500 μl of overnight binding buffer (50 mm HEPES-KOH (pH 8.0), 500 mm NaCl, 7.5% glycerol, 0.05% Nonidet P-40, and either 0.5 mm Mg(OAc)2 or 2.5 mm Mg(OAc)2 and 2 mm ATP, as appropriate) was then added, and the incubation continued overnight on a rotator at 4 °C. In the experiment shown in Fig. 6E, panel ii, the order of addition of the overnight binding buffer and the anti-HA antibody beads was reversed. The magnetic beads were then pulled down and washed by resuspension followed by re-pelleting twice in 400 μl and once in 200 μl of 50 mm HEPES-KOH (pH 8.0), 750 mm NaCl, 0.05% Nonidet P-40, and either 0.5 mm Mg(OAc)2 or 2.5 mm Mg(OAc)2 and 2 mm ATP as appropriate. The beads were then resuspended once in 200 μl of 50 mm HEPES-KOH (pH 8.0), 100 mm NaCl, and 0.05% Nonidet P-40, and pelleted. The beads were then resuspended in 15 μl of 50 mm HEPES-KOH (pH 8.0), 1 mm EDTA, 50 mm NaCl, 1% SDS, and 1 mg/ml proteinase K, and incubated at 50 °C for 30 min. The beads were pelleted, and DNA present in the supernatants was analyzed by electrophoresis through 0.9% agarose gels at 60 V for 150 min at room temperature using 50 mm Tris-HCl (pH 7.8 at 23 °C), 40 mm NaOAc, 1 mm EDTA, as the running buffer. Gels were stained and visualized as above. Shown is a typical gel from three independent experiments.
DNA entrapment assayed by DNA pulldown
Reaction mixtures (20 μl) as for the DNA condensation assay, but also containing 0.05% Nonidet P-40 and tailed, nicked DNA (50 ng) immobilized on M280 streptavidin beads as above, as well as the indicated concentrations of MukB, MukF, and MukE either in the presence or absence of ATP, were incubated for 5 min at 37 °C. The beads were pulled down and washed twice by resuspension and pulldown in reaction buffer (200 μl) containing 500 mm NaCl. The final washed pellet was resuspended in 25 μl of SDS-PAGE sample buffer and heated at 95 °C for 5 min, and the protein present was analyzed by SDS-PAGE. Shown are typical gels from three independent experiments.
DNA binding by cross-linking
Standard MukB DNA-binding reactions containing 1500 nm wild-type MukB hinge domain fragment, KE,RE hinge domain fragment, or full-length MukB (125 nm) and 1 nm 5′-32P duplex 100-mer (5′-GAGTGCGGATCCTGACGACATGCCGTGCTACGACTGACGACATGCGAAGTACTGCAGCCCGGGTACGACGATGGATCGCATGCAGTGGATCCACGCATCA-3′) were incubated for 5 min at 37 °C. HCHO/MeOH mix (33–10%) was added to a final concentration of 0.7% HCHO and the incubation continued on ice for 30 min. Tris-HCl (pH 7.5) and SDS-polyacrylamide gel loading buffer were then added to 0.5 m and 1×, respectively, and the reaction products were resolved by electrophoresis through a 4–12% gradient BisTris polyacrylamide gel using MES-SDS (Invitrogen) as the electrophoresis buffer. Gels were dried, exposed to a phosphorimager screen, and scanned using a Fujifilm FLA 7000 scanner. Images were processed using ImageQuant TL (GE Healthcare).
Scanning force microscopy
DNA condensation by MukB and MukB K761E/R765E
Either wild type or MukB K761E/R765E at 15, 30, and 60 nm were incubated in standard MukB DNA-binding reactions as above. The samples were then diluted 5-fold with deposition buffer (10 mm HEPES-NaOH (pH 8), 20 mm MgCl2), and 20 μl were deposited on freshly cleaved mica. After 1 min of incubation at room temperature, the mica was washed with Milli-Q water and dried with filtered air. Samples were imaged at room temperature and humidity with a Nanoscope IV (Digital Instruments) operating in tapping mode. Type NHC-W silicon tips of the resonance frequency at 310–372 kHz were obtained from Nanosensors (Veeco Instruments, Europe). Images were collected at 2 × 2 μm, with a standard resolution of 512 lines and 512 rows, and processed only by flattening to remove background slope. Detailed methods for SFM sample preparation can be found in Ref. 49. The degree of DNA condensation was quantified using the SFMetricsV4c1 analysis tool (50). The length of DNA protruding from MukB multimers was manually traced. As a point of reference, the total length of 50 unambiguous individual circular pCG09 molecules was measured in the same manner, giving an average length of 3430 ± 174 nm. The length of DNA loops protruding from a MukB multimer complex was summed. Data shown are from three independent experiments.
DNA binding of MukB and MukB K761E/R765E
Either wild type or MukB K761E/R765E at 7.5 and 15 nm were incubated in standard MukB DNA-binding reactions as above. Samples were then diluted and imaged as above. Images were quantified by visual inspection where individual MukB molecules were identified as bound to a DNA strand with the head domain (appearing as one large or two small globular domains), with the hinge domain (appearing as a singular small globular domain), with both head and hinge domains, or as a “looping” mode where the head domain was bound to two DNA regions bringing them together, forming a DNA loop. All DNA-bound MukB molecules in an image were counted and categorized. The frequency of the different forms was expressed as percentage of total molecules counted. Data shown are from three independent experiments.
E. coli strains
BW30270 is an rph+ derivative of MG1655 (E. coli Genetic Stock Center). PN141 (BW30270mukBK761E/R765E) was engineered using CRISPR-Cas9 technology as described (51). A guide RNA plasmid was constructed by inserting an EcoRI- and SpeI-digested DNA fragment (GeneScript) 5′-GCTCGTCCACTAGTGGAAGGCTTGACCGATTGCCGTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGCTAGTCCGTTATCAACTTGAAAAAGTGGCACCGAGTCGGTGCTTTTTTTGAATTCTCAGTGCC-3′, where the underlined sequence represents the mukB N20 sequence, into similarly digested pTargetF to create pTargetF-mukB2172. An editing template PCR fragment, 5′-TGAGCGAGAGGCGATTGTTGAACGCGATGAAGTGGGCGCGCGCAAAAACGCCGTCGATGAAGAGATCGAACGTTTAAGCCAGCCTGGCGGCTCTGAAGATCAGCGTCTGAACGCGCTGGCGGAGCGTTTTGGTGGTGTGCTGCTGTCAGAAATTTATGACGACGTTAGCCTGGAAGATGCGCCGTACTTCTCAGCGCTGTATGGCCCGTCACGCCACGCCATCGTGGTGCCAGATCTGTCACAGGTAACTGAACACCTGGAAGGCTTGACCGATTGCCCAGAAGATCTCTATCTGATCGAAGGAGATCCGCAGTCATTCGATGACAGCGTGTTCAGCGTTGATGAGCTGGAAAAAGCGGTAGTGGTGGAAATCGCCGATGAACAGTGGCGTTATTCACGTTTCCCGGAAGTGCCGCTGTTTGGTCGTGCTGCGCGTGAAAGCCGTATTGAAAGCCTCCATGCCGAGCGTGAAGTGCTTTCCGAACGCTTCGCCACGCTCTCCTTTGATGTACAGAAAACTCAGCGTCTGCATCAGGCGTTCAGCCGCTTTATCGGCAGTCATCTGGCGGTTGCGTTTGAGTCTGACCCGGAAGCAGAAATCCGTCAACTGAACAGCCGTCGCGTCGAACTGGAGCGGGCGTTAAGTAATCATGAAAATGATAACCAGCAGCAGCGTATTCAGTTTGAGCAGGCGAAAGAGGG-3′, where the bold nucleotide represents the change in the photospacer adjacent motif, and the underlined nucleotide triplets are the amino acid substitutions generated by overlap PCR using the following primers for mukB: 5′-TGAGCGAGAGGCGATTGTTG-3′; 5′-GATCAGATAGAGATCTTCTGGGCAATCGGTCAAGCCTTCCAGGTGTTCAGTTACCTGTGA; TCACAGGTAACTGAACACCTGGAAGGCTTGACCGATTGCCCAGAAGATCTCTATCTGATC; CTGTACATCAAAGGAGAGCGTGGC-3′; 5′-GCCACGCTCTCCTTTGATGTACAG-3′; and 5′-CCCTCTTTCGCCTGCTCAAA-3′. BW30270 was transformed with plasmid pCas, supplying Cas9. Competent cells were prepared from BW30270(pCas9) grown in the presence of 1 mm l-arabinose to induce Cas9 expression. These cells were transformed with pTargetF-mukB2172 (100 ng) and editing template DNA (400 ng). After loss of both plasmids, surviving cells were screened by DNA sequencing of mukB. Whole-genome sequencing was then performed to ensure that there were no off-target mutations introduced during the strain construction.
Isolation of spermidine nucleoids
BW30270 and PN141 were grown at 37 °C in MOPS EZ Rich Defined Medium with 2% glucose to an OD600 = 0.5. Cells (25 ml) were pelleted by centrifugation, lysed, and sedimented through 10–40% sucrose gradients as described by Murphy and Zimmerman (52) with the exception that the gradients were centrifuged for 20 min.
Cell growth and fluorescent microscopy
For DAPI staining, strains were grown at 37 °C in LB medium with 0.5% glucose, 20 μg/ml thymine, and 20 μg/ml thiamine to OD600 = 0.2. Five milliliters of cell culture were incubated with 1 μg/ml DAPI for 10 min at 37 °C. Cells were pelleted, washed with 5 ml of M9 medium, pelleted, and resuspended in 0.5 ml of M9 medium, and 100 μl of this suspension were spread on freshly-coated poly-l-lysine slides for 2 min at room temperature. The slides were drained and washed with 1× PBS, air-dried, covered with 2 drops fluorescent mounting medium and a coverslip, and observed in a Nikon Eclipse Ti microscope using a 100 × CFI Plan Apo DM 1.4 numerical aperture objective. Images were captured with an Andor Neo sCMOS camera. Cell length and nucleoid area measurements were made using the measurement tools in version 4.4 of Nikon Elements Software.
Author contributions
S. B. developed the gel-based assay for DNA condensation. R. K., M. G., S. B., P. N., C. L. W., and K. J. M. conceived and designed the experiments. R. K., M. G., P. N., and S. B. performed the experiments. All authors analyzed the data. K. J. M. wrote the manuscript.
Acknowledgments
We thank James Berger and Chris Lima for discussion. The Memorial Sloan Kettering Cancer Center was recipient of National Institutes of Health Cancer Center Core Support Grant P30CA008748 from NCI.
This work was supported, in whole or in part, by National Institutes of Health Grant GM34558 (to K. J. M.) and grants from the Dutch Technology Foundation (STW) Project NWOnano11425 and Cancer Genomics Netherlands (NWO gravitation) (to C. L. W.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- SMC
- structural maintenance of chromosome protein
- FMcx
- fast-moving complex
- SMcx
- slow-moving complex
- SFM
- scanning-force microscope
- MukB KE,RE
- the MukB K761E/R765E protein
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- Ni-NTA
- nickel-nitrilotriacetic acid.
References
- 1. Uhlmann F. (2016) SMC complexes: from DNA to chromosomes. Nat. Rev. Mol. Cell Biol. 17, 399–412 [DOI] [PubMed] [Google Scholar]
- 2. Niki H., Jaffé A., Imamura R., Ogura T., and Hiraga S. (1991) The new gene mukB codes for a 177-kd protein with coiled-coil domains involved in chromosome partitioning of E. coli. EMBO J. 10, 183–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yamanaka K., Ogura T., Niki H., and Hiraga S. (1996) Identification of two new genes, mukE and mukF, involved in chromosome partitioning in Escherichia coli. Mol. Gen. Genet. 250, 241–251 [DOI] [PubMed] [Google Scholar]
- 4. Petrushenko Z. M., Lai C. H., Rai R., and Rybenkov V. V. (2006) DNA reshaping by MukB. Right-handed knotting, left-handed supercoiling. J. Biol. Chem. 281, 4606–4615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yamazoe M., Onogi T., Sunako Y., Niki H., Yamanaka K., Ichimura T., and Hiraga S. (1999) Complex formation of MukB, MukE and MukF proteins involved in chromosome partitioning in Escherichia coli. EMBO J. 18, 5873–5884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ohsumi K., Yamazoe M., and Hiraga S. (2001) Different localization of SeqA-bound nascent DNA clusters and MukF-MukE-MukB complex in Escherichia coli cells. Mol. Microbiol. 40, 835–845 [DOI] [PubMed] [Google Scholar]
- 7. She W., Wang Q., Mordukhova E. A., and Rybenkov V. V. (2007) MukEF is required for stable association of MukB with the chromosome. J. Bacteriol. 189, 7062–7068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Badrinarayanan A., Reyes-Lamothe R., Uphoff S., Leake M. C., and Sherratt D. J. (2012) In vivo architecture and action of bacterial structural maintenance of chromosome proteins. Science 338, 528–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Petrushenko Z. M., Lai C. H., and Rybenkov V. V. (2006) Antagonistic interactions of kleisins and DNA with bacterial condensin MukB. J. Biol. Chem. 281, 34208–34217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen N., Zinchenko A. A., Yoshikawa Y., Araki S., Adachi S., Yamazoe M., Hiraga S., and Yoshikawa K. (2008) ATP-induced shrinkage of DNA with MukB protein and the MukBEF complex of Escherichia coli. J. Bacteriol. 190, 3731–3737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Woo J. S., Lim J. H., Shin H. C., Suh M. K., Ku B., Lee K. H., Joo K., Robinson H., Lee J., Park S. Y., Ha N. C., and Oh B. H. (2009) Structural studies of a bacterial condensin complex reveal ATP-dependent disruption of intersubunit interactions. Cell 136, 85–96 [DOI] [PubMed] [Google Scholar]
- 12. Wilhelm L., Burmann F., Minnen A., Shin H. C., Toseland C. P., Oh B. H., and Gruber S. (2015) SMC condensin entraps chromosomal DNA by an ATP hydrolysis dependent loading mechanism in Bacillus subtilis. Elife 4, e06659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Niki H., and Yano K. (2016) In vitro topological loading of bacterial condensin MukB on DNA, preferentially single-stranded DNA rather than double-stranded DNA. Sci. Rep. 6, 29469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cui Y., Petrushenko Z. M., and Rybenkov V. V. (2008) MukB acts as a macromolecular clamp in DNA condensation. Nat. Struct. Mol. Biol. 15, 411–41818376412 [Google Scholar]
- 15. Michieletto D., Marenduzzo D., and Orlandini E. (2015) Topological patterns in two-dimensional gel electrophoresis of DNA knots. Proc. Natl. Acad. Sci. U.S.A. 112, E5471–E5477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lehman I. R. (1974) DNA ligase: structure, mechanism, and function. Science 186, 790–797 [DOI] [PubMed] [Google Scholar]
- 17. Liu L. F., Depew R. E., and Wang J. C. (1976) Knotted single-stranded DNA rings: a novel topological isomer of circular single-stranded DNA formed by treatment with Escherichia coli omega protein. J. Mol. Biol. 106, 439–452 [DOI] [PubMed] [Google Scholar]
- 18. Shuman S., and Prescott J. (1990) Specific DNA cleavage and binding by vaccinia virus DNA topoisomerase I. J. Biol. Chem. 265, 17826–17836 [PubMed] [Google Scholar]
- 19. Gellert M., Mizuuchi K., O'Dea M. H., and Nash H. A. (1976) DNA gyrase: an enzyme that introduces superhelical turns into DNA. Proc. Natl. Acad. Sci. U.S.A. 73, 3872–3876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gellert M., O'Dea M. H., Itoh T., and Tomizawa J. (1976) Novobiocin and coumermycin inhibit DNA supercoiling catalyzed by DNA gyrase. Proc. Natl. Acad. Sci. U.S.A. 73, 4474–4478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sun M., Nishino T., and Marko J. F. (2013) The SMC1-SMC3 cohesin heterodimer structures DNA through supercoiling-dependent loop formation. Nucleic Acids Res. 41, 6149–6160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Matoba K., Yamazoe M., Mayanagi K., Morikawa K., and Hiraga S. (2005) Comparison of MukB homodimer versus MukBEF complex molecular architectures by electron microscopy reveals a higher-order multimerization. Biochem. Biophys. Res. Commun. 333, 694–702 [DOI] [PubMed] [Google Scholar]
- 23. Ku B., Lim J. H., Shin H. C., Shin S. Y., and Oh B. H. (2010) Crystal structure of the MukB hinge domain with coiled-coil stretches and its functional implications. Proteins 78, 1483–1490 [DOI] [PubMed] [Google Scholar]
- 24. Li Y., Schoeffler A. J., Berger J. M., and Oakley M. G. (2010) The crystal structure of the hinge domain of the Escherichia coli structural maintenance of chromosomes protein MukB. J. Mol. Biol. 395, 11–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rybenkov V. V., Herrera V., Petrushenko Z. M., and Zhao H. (2014) MukBEF, a chromosomal organizer. J. Mol. Microbiol. Biotechnol. 24, 371–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kim H., and Loparo J. J. (2016) Multistep assembly of DNA condensation clusters by SMC. Nat. Commun. 7, 10200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Murayama Y., and Uhlmann F. (2014) Biochemical reconstitution of topological DNA binding by the cohesin ring. Nature 505, 367–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hirano M., and Hirano T. (2002) Hinge-mediated dimerization of SMC protein is essential for its dynamic interaction with DNA. EMBO J. 21, 5733–5744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hayama R., Bahng S., Karasu M. E., and Marians K. J. (2013) The MukB-ParC interaction affects the intramolecular, not intermolecular, activities of topoisomerase IV. J. Biol. Chem. 288, 7653–7661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Soh Y. M., Bürmann F., Shin H. C., Oda T., Jin K. S., Toseland C. P., Kim C., Lee H., Kim S. J., Kong M. S., Durand-Diebold M. L., Kim Y. G., Kim H. M., Lee N. K., Sato M., et al. (2015) Molecular basis for SMC rod formation and its dissolution upon DNA binding. Mol. Cell 57, 290–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kumar R., Nurse P., Bahng S., Lee C. M., and Marians K. J. (2017) The MukB–topoisomerase IV interaction is required for proper chromosome compaction. J. Biol. Chem. 292, 16921–16932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Haering C. H., Farcas A. M., Arumugam P., Metson J., and Nasmyth K. (2008) The cohesin ring concatenates sister DNA molecules. Nature 454, 297–301 [DOI] [PubMed] [Google Scholar]
- 33. Farcas A. M., Uluocak P., Helmhart W., and Nasmyth K. (2011) Cohesin's concatenation of sister DNAs maintains their intertwining. Mol. Cell 44, 97–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cuylen S., Metz J., and Haering C. H. (2011) Condensin structures chromosomal DNA through topological links. Nat. Struct. Mol. Biol. 18, 894–901 [DOI] [PubMed] [Google Scholar]
- 35. Kanno T., Berta D. G., and Sjögren C. (2015) The Smc5/6 complex is an ATP-dependent intermolecular DNA linker. Cell Rep. 12, 1471–1482 [DOI] [PubMed] [Google Scholar]
- 36. Eng T., Guacci V., and Koshland D. (2015) Interallelic complementation provides functional evidence for cohesin-cohesin interactions on DNA. Mol. Biol. Cell 26, 4224–4235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Stigler J., Çamdere G. Ö., Koshland D. E., and Greene E. C. (2016) Single-molecule imaging reveals a collapsed conformational state for DNA-bound cohesin. Cell Rep. 15, 988–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang X., Liu X., Possoz C., and Sherratt D. J. (2006) The two Escherichia coli chromosome arms locate to separate cell halves. Genes Dev. 20, 1727–1731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nielsen H. J., Ottesen J. R., Youngren B., Austin S. J., and Hansen F. G. (2006) The Escherichia coli chromosome is organized with the left and right chromosome arms in separate cell halves. Mol. Microbiol. 62, 331–338 [DOI] [PubMed] [Google Scholar]
- 40. Danilova O., Reyes-Lamothe R., Pinskaya M., Sherratt D., and Possoz C. (2007) MukB colocalizes with the oriC region and is required for organization of the two Escherichia coli chromosome arms into separate cell halves. Mol. Microbiol. 65, 1485–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nolivos S., Upton A. L., Badrinarayanan A., Müller J., Zawadzka K., Wiktor J., Gill A., Arciszewska L., Nicolas E., and Sherratt D. (2016) MatP regulates the coordinated action of topoisomerase IV and MukBEF in chromosome segregation. Nat. Commun. 7, 10466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jun S., and Mulder B. (2006) Entropy-driven spatial organization of highly confined polymers: lessons for the bacterial chromosome. Proc. Natl. Acad. Sci. U.S.A. 103, 12388–12393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Youngren B., Nielsen H. J., Jun S., and Austin S. (2014) The multifork Escherichia coli chromosome is a self-duplicating and self-segregating thermodynamic ring polymer. Genes Dev. 28, 71–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fisher J. K., Bourniquel A., Witz G., Weiner B., Prentiss M., and Kleckner N. (2013) Four-dimensional imaging of E. coli nucleoid organization and dynamics in living cells. Cell 153, 882–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hadizadeh Yazdi N., Guet C. C., Johnson R. C., and Marko J. F. (2012) Variation of the folding and dynamics of the Escherichia coli chromosome with growth conditions. Mol. Microbiol. 86, 1318–1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yeeles J. T., and Marians K. J. (2011) The Escherichia coli replisome is inherently DNA damage tolerant. Science 334, 235–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hayama R., and Marians K. J. (2010) Physical and functional interaction between the condensin MukB and the decatenase topoisomerase IV in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 107, 18826–18831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bahng S., Hayama R., and Marians K. J. (2016) MukB-mediated catenation of DNA is ATP and MukEF independent. J. Biol. Chem. 291, 23999–24008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ristic D., Sanchez H., and Wyman C. (2011) Sample preparation for SFM imaging of DNA, proteins, and DNA-protein complexes. Methods Mol. Biol. 783, 213–231 [DOI] [PubMed] [Google Scholar]
- 50. Sánchez H., and Wyman C. (2015) SFMetrics: an analysis tool for scanning force microscopy images of biomolecules. BMC Bioinformatics 16, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Jiang Y., Chen B., Duan C., Sun B., Yang J., and Yang S. (2015) Multigene editing in the Escherichia coli genome via the CRISPR-Cas9 system. Appl. Environ. Microbiol. 81, 2506–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Murphy L. D., and Zimmerman S. B. (1997) Isolation and characterization of spermidine nucleoids from Escherichia coli. J. Struct. Biol. 119, 321–335 [DOI] [PubMed] [Google Scholar]