Abstract
Yeast Prx1 is a mitochondrial 1-Cys peroxiredoxin that catalyzes the reduction of endogenously generated H2O2. Prx1 is synthesized on cytosolic ribosomes as a preprotein with a cleavable N-terminal presequence that is the mitochondrial targeting signal, but the mechanisms underlying Prx1 distribution to distinct mitochondrial subcompartments are unknown. Here, we provide direct evidence of the following dual mitochondrial localization of Prx1: a soluble form in the intermembrane space and a form in the matrix weakly associated with the inner mitochondrial membrane. We show that Prx1 sorting into the intermembrane space likely involves the release of the protein precursor within the lipid bilayer of the inner membrane, followed by cleavage by the inner membrane peptidase. We also found that during its import into the matrix compartment, Prx1 is sequentially cleaved by mitochondrial processing peptidase and then by octapeptidyl aminopeptidase 1 (Oct1). Oct1 cleaved eight amino acid residues from the N-terminal region of Prx1 inside the matrix, without interfering with its peroxidase activity in vitro. Remarkably, the processing of peroxiredoxin (Prx) proteins by Oct1 appears to be an evolutionarily conserved process because yeast Oct1 could cleave the human mitochondrial peroxiredoxin Prx3 when expressed in Saccharomyces cerevisiae. Altogether, the processing of peroxiredoxins by Imp2 or Oct1 likely represents systems that control the localization of Prxs into distinct compartments and thereby contribute to various mitochondrial redox processes.
Keywords: hydrogen peroxide; mitochondria; oxidative stress; peroxiredoxin; protein import; protein sorting; yeast; inner membrane peptidase complex; IMP complex; mitochondrial processing peptidase (MPP); octapeptidylpeptidase 1, Oct1
Introduction
The mitochondrion is the home of several redox processes that are confined into distinct subcompartments. The two lipid membranes that spatially define this organelle are required to create enclosed compartments that provide specialized spaces with the appropriate conditions for some of its functions. For instance, processes, such as the Krebs cycle, mitochondrial DNA (mtDNA) replication, and protein synthesis, occur in the matrix, whereas the more oxidizing environment of the intermembrane space is appropriate for disulfide bond formation (1, 2). Therefore, mitochondrial protein import into these distinct compartments should be precisely regulated (3).
Distinct redox processes located in distinct mitochondrial subcompartments can generate superoxide radicals by the monoelectronic reduction of O2, which can then be dismutated into H2O2 and O2. The leakage of electrons in the respiratory chain is the best-studied process of mitochondrial ROS (mROS)4 generation, but it is currently clear that the superoxide radicals and H2O2 are also generated by other processes in the mitochondria (4, 5).
The increased production of mROS can cause wide-ranging damage to macromolecules, which can result in mitochondrial dysfunction and eventually cell death. In addition to these deleterious effects, mROS are currently known to play roles as signaling molecules in processes, such as immunity, differentiation, and autophagy (6). Therefore, the levels of mROS are tightly controlled, and peroxidases play regulatory roles by decreasing the steady-state levels of H2O2 within the mitochondria (7).
Peroxiredoxin (Prx) enzymes are frequently found in the mitochondria and are extremely effective at scavenging peroxides (7–9). These thiol-dependent peroxidases reduce peroxides using a fully conserved Cys residue, designated the “peroxidatic” Cys (CP). All Prxs share the first step in the catalytic cycle in which peroxides oxidize the CP-S− to cysteine sulfenic acid (CP-SOH). The next step in the catalytic cycle varies, depending on the number of the catalytic Cys residues. In 2-Cys Prxs, the sulfenic acid undergoes a condensation reaction with a second conserved Cys residue, named the “resolving” Cys (CR), and forms an intermolecular or intramolecular disulfide bond. In most cases, this disulfide is subsequently reduced by thioredoxin (Trx) (10). In contrast, 1-Cys Prxs do not have a CR, and the sulfenic acid is directly reduced by a thiol-disulfide oxidoreductase (Trx or glutaredoxin (Grx)) or low-molecular-weight reductants (glutathione or ascorbate) (11). Remarkably, the type of Prx (1-Cys or 2-Cys) present at the mitochondria of distinct organisms is not conserved. For instance, although the yeast mitochondria contain a 1-Cys Prx (Prx1), mammalian mitochondria contain two types of 2-Cys Prxs (Prx3 and Prx5). Although their relevance in controlling the levels of H2O2 is well established (7), not much is known about the mechanisms by which they are sorted into the distinct mitochondrial subcompartments.
Thiol-dependent peroxidases such as Prx and glutathione peroxidase can mediate H2O2 signaling by the so-called protein-oxidation relay mechanism (12). For instance, Yap1 is a transcription factor that is redox-regulated in yeast in a process mediated by Gpx3/Orp1 (13). Similarly, the transcription factor STAT3 is activated by oxidation in mammalian cells in a process mediated by Prx2 (14). So far, no protein oxidation relay mechanism was described in mitochondria. Notably, through an alternative mechanism, H2O2 can also transduce signal by regulating the import of proteins into mitochondria through a process that involves hyperoxidation of mammalian Prx3 (15).
Among the five Prxs from Saccharomyces cerevisiae, Prx1 is the only one that is located in the mitochondria (16). The mechanism by which Prx1 is reduced remains controversial. Glutaredoxin 2 (Grx2), thioredoxin 3 (Trx3), thioredoxin reductase 2 (Trr2), and ascorbate have been proposed as possible reductants (16–20). As several genes encoding mitochondrial proteins, PRX1 gene is under catabolic repression by glucose (21). Even under fermentative conditions, ΔPRX1 strain displays increased sensitivity to oxidative insults (16, 19). Furthermore, ΔPRX1 is sensitive to Ca2+-induced inner mitochondrial membrane permeabilization (22). Therefore, although yeast contains eight thiol peroxidases and two catalases, phenotypes can still be associated with the single ΔPRX1 mutant.
Prx1 mRNA is translated on cytosolic ribosomes as a nucleus-encoded mitochondrial precursor protein and contains a cleavable N-terminal extension that is termed presequence (16). The presequence is predicted to direct the import of nearly all mitochondrial matrix proteins and a small number of inner membrane and intermembrane space proteins (23). Presequences in precursor proteins are recognized sequentially by the translocase of the outer membrane (TOM complex) and translocase of the inner membrane (TIM23 complex), which are located in the outer and inner membranes, respectively (23–25). In the matrix, the presequences are proteolytically removed by the matrix-located mitochondrial processing peptidase (MPP) (26).
A global mass spectrometric analysis of mature mitochondrial N termini in the yeast mitochondrial proteome enabled the determination of the R-2 consensus motif (Arg-Xaa-Xaa) as the predominant MPP cleavage site that separates the presequence from the mature portion of the mitochondrial proteins (27, 28). Notably, a systematic profiling of the cleavage motifs revealed that several mitochondrial proteins can undergo a second proteolytic event after the MPP digestion. For instance, the Icp55 and Oct1 proteases remove one or eight (octapeptide) amino acid residues, respectively, from the precursor protein intermediates that are processed by MPP (27–30). The function of the MPP/Oct1 (or Icp55) double digestion remains elusive. Nevertheless, the double digestion provides increased stability to some of the Oct1 (or Icp55) targets in the matrix compartment in comparison with the single digestion by MPP (27–30).
As an alternative pathway, some precursor proteins are processed by the inner membrane peptidase (IMP complex), resulting in their sorting to the intermembrane space. The IMP complex is localized in the mitochondrial inner membrane and contains two proteolytically active subunits, Imp1 and Imp2, and the auxiliary protein Som1 (31, 32). During the import by the TIM23 complex, the hydrophobic signal of the precursor protein is arrested at the inner membrane and laterally integrated into the lipid bilayer. Then, the catalytic subunits Imp1 and Imp2 cleave the hydrophobic sorting signals present after the presequence of the precursor proteins (28, 30), which releases the processed protein into the intermembrane space (33, 34).
In this study, we characterized the mitochondrial import pathway of yeast Prx1, Trr2, and Trx3, which were implicated in the reduction of this peroxidase. These three proteins co-localized in the mitochondrial matrix compartment and were loosely associated with the inner membrane facing the matrix. In addition, Prx1 displayed a dual localization and was also present in the mitochondrial intermembrane space, likely in its soluble form. The Prx1 import mechanism to the intermembrane space probably involves the release of the Prx1 preprotein within the lipid phase of the inner membrane during the translocation by the TIM23 complex. Then, Imp2 cleaves the hydrophobic signal of Prx1, thereby releasing the mature protein into the intermembrane space. Alternatively, the Prx1 import into the matrix involves hydrolysis by MPP and Oct1. Remarkably, the processing of Prxs by Oct1 appears to be an evolutionarily conserved process as the yeast Oct1 cleaved the human mitochondrial peroxiredoxin Prx3 that was expressed in S. cerevisiae.
Results
Intramitochondrial localization of Trr2, Trx3, and Prx1
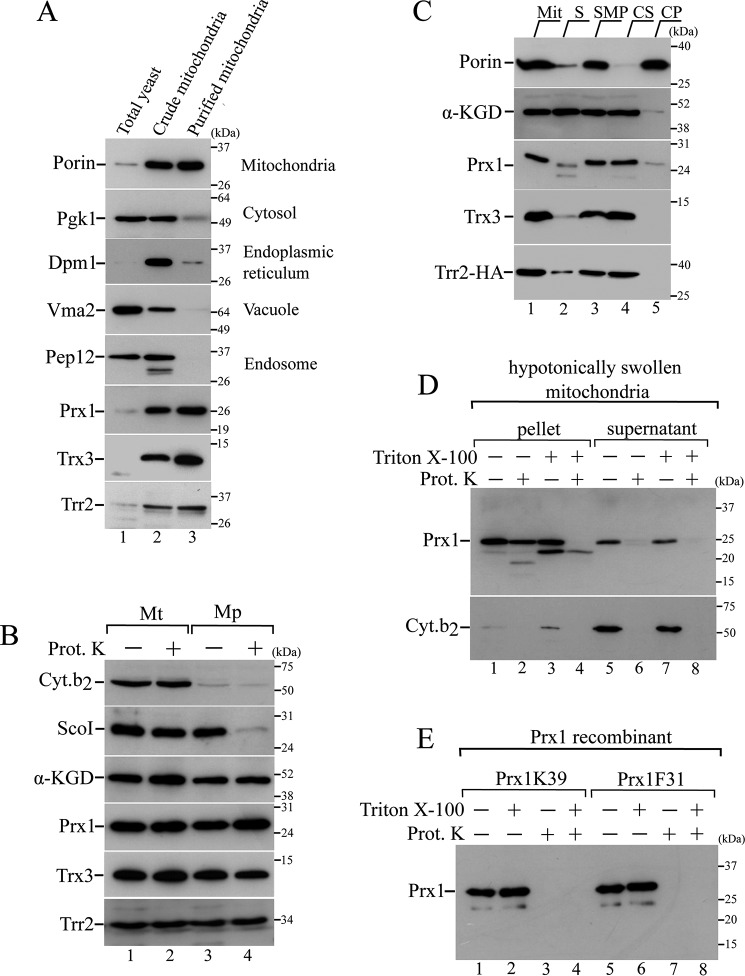
Trr2, Trx3, and Prx1 are synthesized as precursor proteins on cytosolic ribosomes and subsequently imported into the mitochondria (16, 35). We investigated the mechanisms underlying the import and intramitochondrial sorting to obtain insights regarding the roles of these proteins in redox homeostasis. Initially, the presence of Trr2, Trx3, and Prx1 in crude mitochondrial fractions was ascertained by an immunoblotting analysis (Fig. 1A, lane 2). Because this crude mitochondrial preparation presented contamination by vacuoles, endoplasmic reticulum, cytosol, and endosome proteins, further processing through a three-step sucrose density gradient was performed. This additional purification step generated a highly pure mitochondrial fraction (Fig. 1A, lane 3). The Trr2, Trx3, and Prx1 proteins displayed a porin-like fractioning profile (Fig. 1A, lane 3), as described previously (16, 35).
Figure 1.
Intramitochondrial localization of Trr2, Trx3, and Prx1. A, total yeast extract fractions (lane 1), crude mitochondrial fractions (lane 2), and highly pure mitochondrial fractions (lane 3) from BY4741 strain were analyzed by Western blotting using antibodies against markers for distinct cellular compartments as described on the right side of the gel. B, mitochondria were converted into mitoplasts by hypotonic shock (swelling). Equivalent amounts of mitochondria (Mt) and mitoplasts (Mp) were incubated in the presence or absence of 0.1 mg/ml proteinase K (Prot. K). Specific antibody markers are as follows: α-ketoglutarate dehydrogenase (α-KGD, a soluble matrix protein); Sco1 (an inner membrane protein that faces the intermembrane space); and cytochrome b2 (a soluble intermembrane protein). C, highly pure mitochondria (Mit, lane 1) were sonicated and separated into the soluble protein fraction (S, lane 2) and submitochondrial membranes fraction (SMP, lane 3). SMP fractions were then treated with 100 mm Na2CO3 and centrifuged to obtain the soluble (carbonate supernatant, CS, lane 4) and the insoluble (carbonate precipitate, CP, lane 5) fractions. Proteins from these different fractions were analyzed by Western blotting. D, mitochondria were initially converted into mitoplasts (hypotonic shock), followed by a centrifugation to separate the mitoplasts (pellet) from the supernatant (corresponding to the intermembrane space proteins). Both fractions were treated for 1 h with proteinase K (0.2 mg/ml) in the absence or presence of nonionic detergent Triton X-100 (0.5% v/v). E, recombinant Prx1 proteins (Prx1K39 and Prx1F31, representing the enzymes that were processed and unprocessed by Oct1, respectively) were incubated with proteinase K (0.2 mg/ml) in the absence or presence of Triton X-100 (0.5% v/v). These results are representative of at least two independent biological replicates, each one in two technical replicates.
To determine the intramitochondrial localization of the Trr2, Trx3, and Prx1 proteins, we performed a subfractionation protocol with the highly pure mitochondria preparation. The outer membrane of the purified mitochondria was disrupted by incubation in a hypotonic buffer, which generated the so-called mitoplasts, i.e. mitochondria devoid of outer membrane. In parallel, both the mitochondria and mitoplast were treated with proteinase K. In whole mitochondria, proteinase K can only digest proteins that are associated with the surface of the outer membrane, although in mitoplasts, the proteinase K can digest proteins in the intermembrane space and those associated with the surface of the inner membrane that faces the intermembrane space (36). Finally, both the mitochondria and mitoplasts that were incubated in the presence or absence of proteinase K were centrifuged, and equivalent samples of mitochondria and mitoplasts were analyzed by immunoblotting.
The efficient conversion of the mitochondria to mitoplasts by hypotonic shock (swelling) was evident by the substantial loss of the soluble intermembrane space marker cytochrome b2 (Fig. 1B, lanes 3 and 4) (37). This finding is also supported by the decreased levels of Sco1, which is an inner membrane protein that faces the intermembrane space (38), as a result of the proteinase K treatment of the mitoplasts but not the mitochondria (Fig. 1B, compare lane 2 with lane 4). Finally, the matrix protein α-ketoglutarate dehydrogenase (α-KGD) (39) was protected against proteinase K in the mitochondria and mitoplasts, which confirmed the integrity of the inner membrane in the mitoplasts (Fig. 1B, lanes 1–4). Similar to the α-KGD protein profile, Trr2, Trx3, and Prx1 were protected from the digestion in the mitochondria and mitoplasts, indicating their matrix compartment localization (Fig. 1B, lanes 1–4).
We then investigated the solubility of Prx1, Trx3, and Trr2 in the matrix compartment. Therefore, we sonicated the mitochondria and centrifuged the obtained extract, which resulted in a soluble protein fraction, and the compartmentalized membranous product, called submitochondrial particle (SMP) (Fig. 1C, lanes 2 and 3). The SMP fraction was then alkali-treated with sodium carbonate (Na2CO3) and centrifuged, resulting in carbonate supernatant and carbonate-precipitated fractions (Fig. 1C, lanes 4 and 5) (36). Trr2, Trx3, and Prx1 were mainly detected in the SMP, indicating that these proteins are associated with the mitochondrial membranes (Fig. 1C, lane 3). All of these proteins, however, were solubilized when mitochondria were treated with sodium carbonate, indicating that they are peripheral membrane proteins (Fig. 1C, lane 4). A different profile was observed for porin, which is an integral mitochondrial membrane protein that is not released from mitochondrial membranes by alkali treatment with sodium carbonate (Fig. 1C, lane 5) (36).
Surprisingly, sonic disruption of mitochondria resulted in only partial recovery of α-KGD, a marker for soluble matrix protein (39), in the soluble protein fraction with a significant portion in the SMP fraction (Fig. 1C, lanes 2 and 3). This unexpected result might be due to slight variations in the sonication parameters that can affect the formation of the pelleted SMP vesicle's (40) soluble matrix material. In fact, previous results from our group using similar sonication conditions also displayed a partial recovery of α-KGD in the SMP fraction (41). Taken together, our results indicated that Trr2, Trx3, and Prx1 proteins are peripherally attached to the mitochondrial inner membrane, facing the matrix side.
We also analyzed the supernatants obtained after each step in the mitochondrial subfractionation. As expected, the intermembrane space marker Cyt.b2 was detected only in the supernatants of the mitoplasts that were not treated with proteinase K (Fig. 1D, lanes 5 and 7, and supplemental Fig. S1A, lane 7); and the matrix marker α-KGD was not detected in the supernatant (supplemental Fig. S1A), which again demonstrated the mitochondrial inner membrane integrity. Unexpectedly, Prx1 was also detected in the mitoplast-derived supernatant (Fig. 1D, lanes 5 and 7, and supplemental Fig. S1A, lane 7), which provided direct evidence for the dual mitochondrial distribution of this protein (matrix and the intermembrane space). This finding will be further analyzed below.
However, in contrast to Cyt.b2, Prx1 was resistant against proteolytic degradation (supplemental Fig. S1A, lane 8) at the standard proteinase K concentration (0.1 mg/ml) employed in this study. Doubling proteinase K concentration (0.2 mg/ml) was sufficient to enable Prx1 digestion (Fig. 1D, lane 6), independently of non-ionic detergent Triton X-100 treatment (Fig. 1D, lane 8). Surprisingly, the incubation of mitoplasts with Triton X-100 led to the appearance of a faster migration Prx1 band, whose meaning is currently unknown (Fig. 1D, lanes 3 and 4).
We also investigated the digestion of recombinant Prx1 to proteinase K, analyzing two isoforms (recombinant Prx1F31 and Prx1K39, representing the processed and unprocessed forms of Prx1 by Oct1 protease, as is discussed below). Both of them were efficiently degraded by proteinase K in the presence or absence of Triton X-100 (Fig. 1F). Taken together, these results indicated that Prx1 is less efficiently degraded by proteinase K in comparison with Cyt.b2. Nevertheless, at 0.2 mg/ml concentration, proteinase K treatment could fully digest Prx1 in the supernatant (corresponding to the intermembrane space) fraction (Fig. 1D, lanes 6 and 8) but not in the pellet compartment (Fig. 1D, lane 4). Further evidence for the Prx1 localization on the intermembrane space will be presented below.
Mitochondrial import of Prx1 involves cleavage by three peptidases, MPP, Oct1, and Imp2
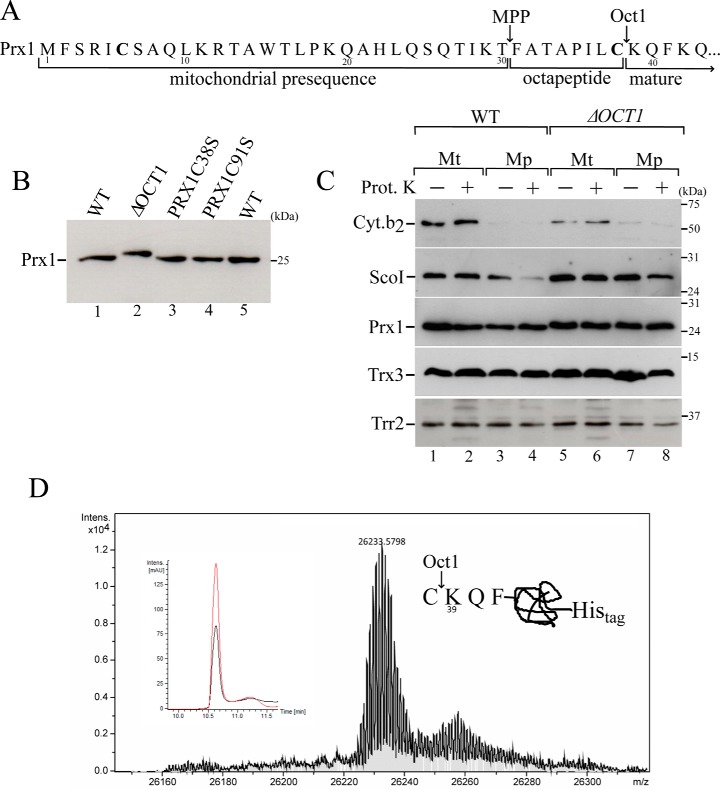
Previously, it was hypothesized that the mature protein Prx1 could be processed by the protease MPP with a predicted cleavage site between amino acids 21 and 22 (16). Subsequently, a global analysis of mature mitochondrial N termini in yeast by mass spectrometry enabled the identification of the following two peptidases that act on Prx1: MPP and Oct1 (27). Accordingly, Prx1 is initially cleaved by MPP between amino acids 30 and 31, and then Oct1 removes the next eight amino acids from the Prx1 N terminus. Thus, in its mature form, Prx1 starts at lysine 39 (Lys-39) of the precursor protein (Fig. 2A).
Figure 2.
Mitochondrial processing of Prx1 by Oct1 protease. A, scheme of the Prx1 N-terminal amino acid sequence. Arrows indicate the MPP and Oct1p cleavage sites based on a global mass spectrometric analysis (27). Cysteines present in the Prx1 presequence are highlighted in bold. B, total cell-free extracts from the wild type (WT, BY4741), OCT1 null mutant (ΔOCT1), and PRX1 null mutant expressing cysteine mutant versions of PRX1 (PRX1C38S and PRX1C91S) were analyzed by Western blotting. C, intramitochondrial localization of Prx1 in the wild-type and ΔOCT1 mitochondria (Mt). Mp, mitoplasts. Mitochondria isolated from cells grown on YPGal were processed according to legend of Fig. 1B. D, mass spectrometry analysis of C-terminal His-tagged Prx1 present in the matrix fraction. C-terminal His-tagged Prx1 expressed in the ΔPRX1 strain was purified by nickel affinity chromatography. The obtained mass was 26,233.57, which was consistent with the incorporation of two oxygen atoms to the Prx1 polypeptide. The inset depicts the corresponding reverse phase chromatogram. The red and black lines correspond to UV chromatogram at 280 and 254 nm, respectively. The scheme at the right side of the mass spectrum represents the cleavage site. These results are representative of at least two independent biological replicates, each one in two technical replicates. See Table 2 for strains and genotypes.
To confirm that Prx1 is an Oct1 substrate, a Western blot analysis was performed with yeast cell extracts in which the OCT1 gene was deleted (ΔOCT1). Prx1 from the ΔOCT1 strain exhibited a slightly slower migrating band than the corresponding band from the WT strain (Fig. 2B, compare lane 1 with 2), indicating a higher molecular weight and the proteolysis of Prx1 by Oct1. The full sequence of Prx1 contains three Cys residues at positions 6, 38, and 91. Therefore, the Cys-6 residue is removed as a part of the proteolysis by MPP. Then, the Cys-38 residue is removed through proteolysis by Oct1 (Fig. 2A). However, Cys-38 was reported to be involved in an intermolecular disulfide bond in vivo (19), which could represent an immature form of this peroxidase. In any case, Cys-91 that is the CP residue remains in the mature protein after the MPP and Oct1 double digestion. Mutant versions of Prx1, in which Cys-38 and Cys-91 were individually replaced by Ser (C38S and C91S mutants), were digested to the same extent by Oct1 as the wild-type protein (Fig. 2B, compare lanes 3 and 4 with lane 1), suggesting that these Cys residues do not affect the proteolysis by Oct1. However, other steps in the Prx1 import into the mitochondria might be affected by the Cys-38 replacement, for example the formation of a mixed disulfide intermediate with another protein.
To confirm whether Cys-38 is removed during Oct1 proteolysis, we purified Prx1 from the matrix compartment and analyzed its molecular mass by mass spectrometry. The calculated mass for an isoform of a C-terminally His-tagged Prx1 starting at Lys-39 is 26,201.57, which is consistent with the mass obtained (26,233.57) in the mass spectrometry analysis (Fig. 2D). The additional 32 atomic mass units likely reflected the incorporation of two oxygen atoms due to an unexpected oxidation. Thus, the Prx1 that is sorted into matrix indeed starts at Lys-39.
The OCT1 deletion did not affect the mitochondrial import of Prx1 into the matrix (Fig. 2C). Based on a global analysis of the mature mitochondrial N termini in yeast, the Trr2 and Trx3 proteins are not cleaved by Oct1 (27), and as expected, these proteins did not show differences in the molecular sizes between the WT and ΔOCT1 strains (Fig. 2C).
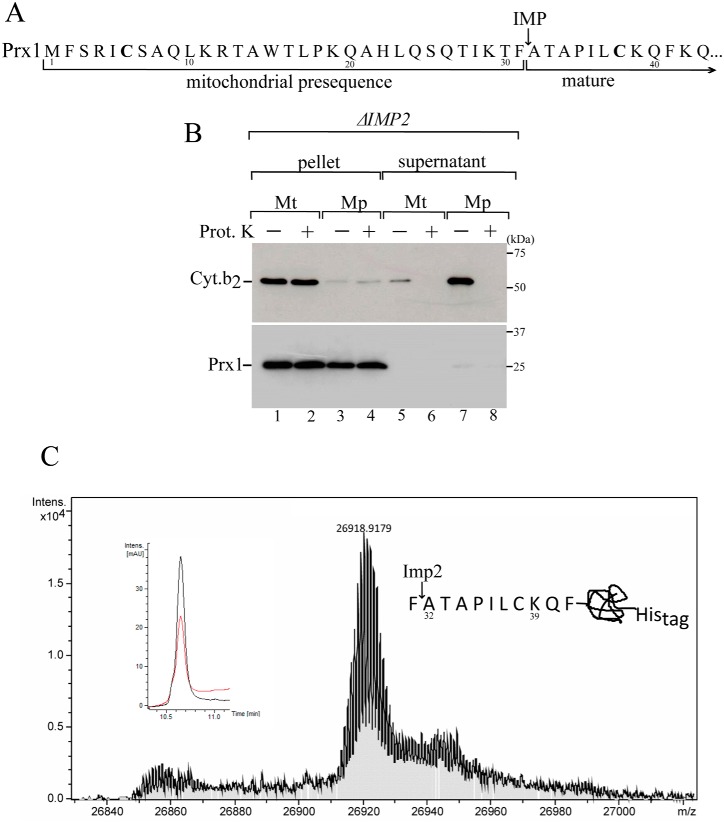
As mentioned above, our data indicated that Prx1 is present in both the matrix and the intermembrane space. Therefore, to investigate the mechanism by which Prx1 is directed into the intermembrane space, we visually inspected the N-terminal sequence of Prx1, and we found a motif that resembles a cleavage site that could be recognized by Imp2 (Fig. 3A).
Figure 3.
Mitochondrial processing of Prx1 by the IMP complex. A, Prx1 N-terminal amino acid sequence highlighting the likely Imp2 cleavage site. B, intramitochondrial localization of Prx1 in the ΔIMP2 mitochondria. Fractionation was performed as described in the legend of supplemental Fig. S1A. See Table 2 for strains and genotypes. C, mass spectrometry analysis of Prx1 sorted into the intermembrane space. C-terminal His-tagged Prx1 expressed in the ΔPRX1 strain was purified by nickel affinity chromatography. The obtained mass was 26918.92, which was consistent with the incorporation of three oxygen atoms to the Prx1 polypeptide. The insets depict the corresponding reverse phase chromatogram. The red and black lines correspond to UV chromatogram at 280 and 254 nm, respectively. The scheme at the right side of the mass spectrum represents the cleavage site. These results are representative of at least two independent biological replicates, each one in two technical replicates. See Table 2 for strains and genotypes.
However, it is not possible at this moment to define a consensus sequence for Imp2, as the only substrate identified so far for this protease is Cyc1 (31). The presence of an alanine at position −1 and of a small uncharged or small polar residue at position −3 relative to the cleavage site is required for Imp2 to hydrolyze Cyc1 (42). Nevertheless, the absence of Prx1 in the supernatant of the mitochondrial subfractionation from the ΔIMP2 strain indicated the involvement of the Imp2 protease in sorting Prx1 into the intermembrane space (Fig. 3B).
To determine the cleavage site of Imp2 protease, we purified the Prx1 that was sorted into the intermembrane space and analyzed its molecular weight by mass spectrometry. The mass obtained was 26,918.92, which is consistent with the mass obtained for an isoform of a C-terminally His-tagged Prx1 starting at Ala-32 (Fig. 3C).
Prx1 processed by Oct1 does not display an increased half-life in the mitochondrial matrix
The physiological meaning of the second proteolytic event following the MPP hydrolysis remains elusive. Nonetheless, Oct1 cleaves several unstable precursors that are generated by MPP into more stable proteins (29).
It is well known that deletion of the OCT1 gene results in a respiratory deficiency due to the loss of mitochondrial DNA (Rho0) (43). Therefore, to properly control the effects of the absence of Oct1 in yeast cells, the OCT1 gene was transformed in the ΔOCT1 background under the control of its endogenous promoter and terminator, thereby generating the ΔOCT1 + OCT1 strain.
To monitor the Prx1 stability, isolated mitochondria from ΔOCT1 and ΔOCT1 + OCT1 yeast cells were analyzed at different time points. Porin, an integral mitochondrial outer membrane protein that is not an Oct1 substrate, was analyzed as a control. Prx1 was equally stable in isolated mitochondria from ΔOCT1 and ΔOCT1 + OCT1 yeast cells (Fig. 4). However, Prx1 degradation rate was considerably faster than the corresponding rate for porin (Fig. 4). These results indicated that hydrolysis by Oct1 does not stabilize Prx1 into the mitochondria at least in our experimental conditions.
Figure 4.
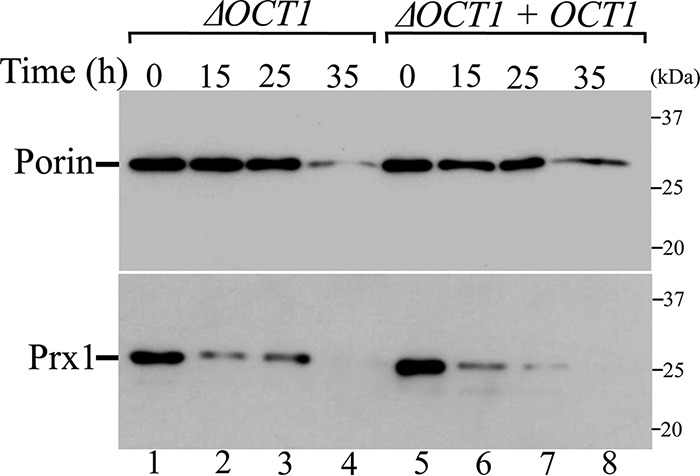
Analysis of half-life of Prx1 in the presence or absence of Oct1 protease in organello. Mitochondria from ΔOCT1 and ΔOCT1 + OCT1 (re-expressing the OCT1 gene) yeast cells were incubated at 37 °C for the indicated time. Mitochondria were reisolated and analyzed by SDS-PAGE and immunoblotting. These results are representative of at least two independent biological replicates, each one in two technical replicates. See Table 2 for strains and genotypes.
We also compared the peroxidase activities of the intermediate and mature forms of Prx1 in vitro by producing the following two recombinant forms of Prx1: Prx1F31 (representing the protein cleaved by MPP but unprocessed by Oct1) and Prx1K39 (representing the protein processed by MPP and Oct1). The thioredoxin-dependent peroxidase activity of Prx1 was determined spectrophotometrically by a coupled assay that monitored the disappearance of NADPH at 340 nm using different H2O2 concentrations and fixed concentrations of Trx3 (direct reductant) and Trr2. Both isoforms displayed similar specificity constants (kcat/Km, H2O2) for the H2O2 reduction (Table 1 and supplemental Fig. S2).
Table 1.
Thioredoxin-dependent peroxidase activity of processed (Prx1K39) and unprocessed (Prx1F31) Prx1 by Oct1 protease
The rates were monitored by decreases in absorbance due to the oxidation of NADPH in a reaction mixture containing 200 μm NADPH, 0.1 μm Trr2, 5 μm Trx3, 5 μm peroxiredoxin, and 100 μm DTPA. Assays were carried out at 37 °C. H2O2 concentrations ranged from 10 to 500 μm. Therefore, the specificity constant is apparent for H2O2. All recombinant proteins are His-tagged proteins. These enzymatic parameters were calculated from three independent biological replicates, each in three technical replicates. Average and standard deviations were calculated using the GraphPad Prism 6 Software (GraphPad Software, Inc.) by non- linear regression (Michaelis-Menten equation).
| Substrate | Km, app | kcat, app | kcat/Km, app |
|---|---|---|---|
| μm | min−1 | m−1 min−1 | |
| Prx1K39 | |||
| H2O2 | 5.2 ± 0.4 | 1.98 ± 0.02 | (3.8 ± 0.3) 105 |
| Prx1F31 | |||
| H2O2 | 4.6 ± 0.4 | 1.58 ± 0.02 | (3.5 ± 0.3) 105 |
Mammalian mitochondrial Prx3 is processed by Oct1 from yeast
Mammalian mitochondria have two Prxs, i.e. Prx3 and Prx5. Although Prx3 is restricted to the mitochondria, Prx5 is also found in other compartments, including peroxisomes and the cytosol. Because Prx3 is present in the matrix at high concentrations and rapidly reacts with H2O2, it has been proposed to be a major sink for H2O2 within the mitochondria, whereas Prx5 can contribute to the same process in a more discrete manner (7, 44).
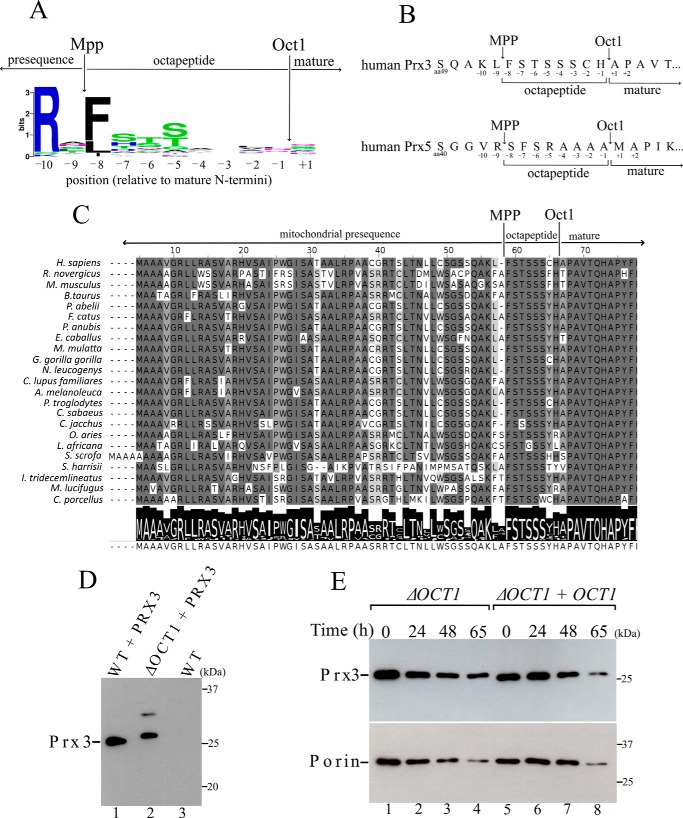
Based on previous studies that experimentally determined the N-terminal sequences of mammalian proteins (7, 45), we analyzed the N-terminal sequences of the two mammalian mitochondrial Prxs, and according to the Oct1 consensus motif (Fig. 5A), the most conserved amino acid residue at position −8 is a Phe in Prx3 (Fig. 5B). Furthermore, Ser and Thr residues are frequently found at positions −5, −6, and −7 (Fig. 5A) and are also present in the amino acid sequence of Prx3 (Fig. 5B). Furthermore, these features are conserved among mammalian Prx3 (Fig. 5C), indicating that these enzymes are processed by Oct1.
Figure 5.
Human mitochondrial Prx3 is cleaved by yeast Oct1. A, consensus analysis of the amino acid presequences. Arrows depict the cleavage sites of MPP and Oct1 (23). Amino acid frequency blots were generated using the WebLogo program. B, schematic representation of the amino acids in the N terminus of the human mitochondrial peroxiredoxins Prx5 and Prx3. The possible octapeptides that could be cleaved by Oct1 are highlighted. C, alignment of the N-terminal amino acid sequence of Prx3 from mammals. Arrows indicate the likely cleavage sites by MPP and Oct1 proteases. D, total cell-free extracts from the wild-type (WT) and OCT1 null mutant (ΔOCT1) expressing the non-tagged human Prx3. An upper band with lower intensity was observed only in the ΔOCT1 strain, whose meaning is unknown. Possibly an artifactual intermolecular disulfide Prx3 was generated in the denaturing conditions of the SDS-PAGE. E, stability of Prx3 in mitochondria isolated from ΔOCT1 and ΔOCT1 + OCT1 (re-expressing the OCT1 gene) yeast cells. These results are representative of at least two independent biological replicates, each one in two technical replicates. See Table 2 for strains and genotypes.
To test this hypothesis, we transformed the WT and ΔOCT1 yeast strains with a plasmid expressing human mitochondrial Prx3. Prx3 from the ΔOCT1 strain exhibited a slower migrating band, indicating a higher molecular weight than the corresponding protein band from the WT strain (Fig. 5D). Thus, Oct1 from yeast is able to cleave human mitochondrial Prx3, indicating that this process is conserved throughout evolution. As yeast Prx1, the levels of human Prx3 in the ΔOCT1 strain were the same as those in the ΔOCT1 + OCT1 strain (Fig. 5E).
Discussion
Mitochondrial dysfunction is associated with multiple degenerative or acute diseases and aging (46, 47). Understanding how compartmentalization affects the malfunctioning of mitochondrial processes is a challenge (48). Impairments in the mitochondria can be related to oxidative insults as this organelle is a major source of H2O2 in cells, but not much is known about the functional role of Prx1 in maintaining the mitochondrial redox homeostasis at the distinct organellar subcompartments. Here, we describe that Prx1 is located in the following two submitochondrial compartments: the matrix and intermembrane space.
In the first case, Prx1 is peripherally attached to the mitochondrial inner membrane from the matrix side along with Trr2 and Trx3 proteins. The Trr2, Trx3, and Prx1 co-localization might suggest a possible in vivo interaction among these three proteins during H2O2 reduction. However, it is currently premature to assume that the mitochondrial Trx system is responsible for the reduction of Prx1-SOH because the possible mechanisms underlying these processes are controversial, and several reductants have been proposed to be involved (16–20).
The matrix localization of Prx1 is dependent on Oct1, but not much is known about this protease and its physiological significance. A global analysis of the N-proteome of yeast mitochondria indicated that Oct1 processes Prx1 after the MPP proteolysis, and consequently, the mature Prx1 sequence starts at Lys-39 (27) rather than at Gln-21 as was previously considered (16). Through a mass spectrometry analysis, we unequivocally demonstrated here that Oct1 cleaved Prx1 in vivo and that the Prx1 mature form initiates from Lys-39. Therefore, this observation challenged previous investigations, which considered that the mature form of Prx1 would start at Gln-21 and, therefore, contain Cys-38. Some observations indicated that an intermolecular disulfide bond at Cys-38 would be formed in vitro (16) and in vivo (19). A possible mechanism to reconcile these observations is that Cys-38 might have a regulatory role during the import of Prx1 into the yeast mitochondria and is then removed in the mature form. The proteolysis of Prx1 by Oct1 does not appear to be affected by Cys-38; however, this residue may participate in other steps of the import process. For instance, the cytosolic thioredoxin system facilitates the import of some proteins into the intermembrane space by keeping them in their reduced state (49).
In the absence of Oct1, deleterious effects associated with mitochondrial DNA loss (rho0) arise in ΔOCT1 cells, generating a respiratory-deficient phenotype (43, 50). Besides Prx1, other Oct1 substrates play a role to ameliorate stressful conditions, helping to stabilize the mitochondrial functioning. For instance, some of the Oct1 substrates are components of the mitochondrial genetic system (Mrps28, Mrp21, Rim1, Tuf1, and Rms24), and their decreased levels in ΔOCT1 cells likely affect protein translation in this organelle. Furthermore, other Oct1 substrates participate in the Krebs cycle and the mitochondrial respiratory chain; therefore, OCT1 inactivation likely increases the leakage of electrons and, consequently, augments mROS production.
There is a functional interaction between Oct1 and Yfh1, whose human ortholog is frataxin (51). Patients with a recessive autosomal mutation in frataxin gene display a progressive loss of movements, cardiomyopathy, and diabetes, which combined is a clinical condition called Friedreich's ataxia (52). The molecular mechanisms involved in Friedreich's ataxia include a mitochondrial iron accumulation, followed by an increase in hydroxyl radical generation. Accordingly, lack of Yfh1 results in iron accumulation in yeast mitochondria (53–56). Remarkably, the ΔOCT1 mutant presented an opposed effect, reduction in mitochondrial iron levels (57). Furthermore, high expression of the YFH1 gene partially complements the respiratory-deficient phenotype of a yeast strain expressing a thermal-sensitive allele of OCT1 (57). At this moment, the mechanisms underlying the functional interaction between Oct1 and Yfh1 are not known but probably involve mitochondrial iron metabolism. Nevertheless, Oct1 is required for the maturation of several iron-containing proteins in the mitochondria (57).
Remarkably, yeast Oct1 can cleave human Prx3, indicating that the processing of mitochondrial peroxiredoxins is highly conserved. Indeed, the consensus sequence is also conserved across mammalian Prxs. Therefore, it is tempting to speculate that HMIP (human Oct1 ortholog) and human mitochondrial peroxiredoxins contribute to the functional effects of frataxin deficiency and the clinical manifestations of Friedreich's ataxia.
An additional contribution of this work is the demonstration that Prx1 presents a double localization as it is also present in the intermembrane space in a process that is mediated by Imp2. Imp2 is one of the two catalytic subunits of the mitochondrial IMP complex that recognizes hydrophobic sorting motifs in the mitochondrial precursor proteins, thereby releasing the mature proteins into the intermembrane space after proteolysis (31, 32). Six substrates of the IMP complex have been identified in yeast (Cox2, Mcr1, Gut2, Cyb2, Ptc5, and Mgr2) and five are cleaved by Imp1, whereas only Cyc1 was shown to be processed by Imp2 thus far (58). Currently, a prediction of the IMP cleavage site in the client proteins is premature. Our mass spectrometry analysis contributes to the understanding of IMP complex, showing that the Prx1 that is processed by Imp2 starts at Ala-32. Thus, the Prx1 cleavage site contains lysine, phenylalanine, and alanine at positions −3, −1, and +1, respectively, indicating a distinct selectivity in relation to Cyc1 (42). The identification of more substrates of the IMP complex is required to establish a consensus sequence.
It is likely that following the lateral release of Prx1 preprotein into the inner membrane, the presequence is removed by Imp2 protease and does not require MPP hydrolysis. A similar mechanism occurs in Cox2 and Mcr1, which are cleaved in a single step at the outer face of the inner membrane and require Som1 for maturation (32, 59, 60).
Corroborating our findings, an isoform of Prx1 starting at residue Ala-32 was found in the N-proteome of yeast mitochondria (27). However, Prx1 that starts at Lys-39 (located in the mitochondrial matrix) was more frequently found than the Prx1 that starts at Ala-32. It is likely that during translocation by the Tim23 complex, the Prx1 precursor can be preferentially imported toward the matrix with a few molecules of Prx1 being laterally released into the inner membrane. This hypothesis is consistent with the small size of the hydrophobic sorting signal that is present in the Prx1 presequence and is responsible for arresting the translocation by the Tim23 complex. Alternatively, the concentration of Prx1 in the matrix and intermembrane space could be the same, but because the volume of the intermembrane space is considerably lower than that of the matrix, more molecules of Prx1 could be found in the latter compartment (1).
The intermembrane space is known to be an oxidative compartment, which is appropriate for disulfide bond formation (1). Although the repertoire of antioxidant enzymes in the mitochondrial matrix is well characterized, the set of antioxidant enzymes in the intermembrane remains poorly defined. Some of the antioxidants described in this compartment include glutathione, cytochrome c peroxidase (Ccp1), and superoxide dismutase (Sod1) (61). Recently, glutathione peroxidase 3 (Gpx3) was also found in this mitochondrial compartment (62). Currently, the Prx1 reductant in the intermembrane space remains to be identified, and a possible candidate could be the Trx system as Trx1 and Trr1 were identified in a proteomic study (63).
The mammalian IMP2 gene could complement the ΔIMP2 mutant yeast (64, 65). Mutations in the mammalian homolog of Imp2, IMMP2L, have been implicated in the pathogenesis of Tourette syndrome (66, 67). Therefore, Prx1 is a novel example of a protein that is encoded by a single gene but is transported to two different locations within the same organelle in vivo. The transport of this peroxiredoxin through distinct mitochondrial subcompartments is likely relevant as they are conserved in mammalian cells and are implicated in pathological processes.
Experimental procedures
Yeast strains
The genotypes and sources of yeast strains used in this study are listed in Table 2. Yeast cells were grown on YPD (1% (w/v) yeast extract, 2% (w/v) peptone, 2% (w/v) glucose), or YPGal (1% (w/v) yeast extract, 2% (w/v) peptone, 2% (w/v) galactose). For the yeast strains transformed with the expression plasmids, the cells were grown in a selective medium (0.67% (w/v) yeast nitrogen base without amino acids, 0.13% (w/v) amino acid mix without amino acids produced by selective gene present in expression plasmid, and 2% (w/v) galactose as the carbon source).
Table 2.
Genotypes and sources of the yeast strains
| Strain | Genotype | Source |
|---|---|---|
| BY4741WT | BY4741 MATa; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0 | Invitrogen |
| BY4741ΔPRX1 | BY4741 MATa; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0; PRX1::kanMX4 | Invitrogen |
| BY4741ΔOCT1 | BY4741 MATa; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0; OCT1::kanMX4 | Invitrogen |
| BY4741ΔIMP2 | BY4741 MATa; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0; IMP2::kanMX4 | Invitrogen |
| BY4741ΔOCT1/OCT1 | BY4741 MATa; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0; OCT1::kanMX4; his3::OCT1 | This study |
| BY4741ΔPRX1/PRX1C38S | BY4741 MATa; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0; PRX1::kanMX4; leu2::PRX1C38S | This study |
| BY4741ΔPRX1/PRX1C91S | BY4741 MATa; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0; PRX1::kanMX4; leu2::PRX1C91S | This study |
| BY4741ΔPRX1/PRX1–6xHis | BY4741 MATa; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0; PRX1::kanMX4; leu2::PRX1–6xHis | This study |
| BY4741ΔTRR2/TRR2-HA | BY4741 MATa; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0; TRR2::kanMX4; leu2::TRR2-HA | This study |
| BY4741WT/PRX3 | BY4741 MATa; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0; his3::PRX3 | This study |
| BY4741ΔOCT1/PRX3 | BY4741 MATa; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0; OCT1::kanMX4; his3::PRX3 | This study |
| BY4741ΔOCT1/PRX3/OCT1 | BY4741 MATa; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0; OCT1::kanMX4; his3:: PRX3; leu2::OCT1 | This study |
Plasmid and strain constructions
For the generation of a yeast strain expressing a C-terminally His-tagged Prx1, the PRX1-coding sequence was amplified from the genomic DNA with the primers Prx1His-F 5′-TCAGGATCCATGTTTAGTAGAATTTGTAGC-3′ and Prx1His-R 5′-TCAGTCGACTTAATGGTGATGGTGATGGTGACCACCACCTTTCGACTTGGTGAATCTTAA-3′. The purified PCR product with 831 bp was digested with BamHI and SalI and cloned into YEp531 (68) carrying the constitutive TEF1 promoter that was previously cloned as a SacI-BamHI insert. The recombinant plasmid called Yep351TEF/PRX1His was used to transform the ΔPRX1 strain.
For the generation of a yeast strain expressing a C-terminally HA-tagged Trr2, ∼500 bp of the 5′-UTR plus the TRR2-coding sequence minus the termination codon was amplified from the genomic DNA with the primers Trr2 5′-TGGATCCTATTTCTTGAACCCAACTTGAAGGC-3′ and Trr2HA 5′-TTAAAGCTTTCAAGCGTAGTCTGGGACGTCGTATGGGTACTCTTGGGCACTTAGGTACCGTTC-3′. The purified PCR product with 1609 bp was digested with BamHI and PstI and cloned into YCplac111 (69) to yield Ycplac111/TRR2HA, which was used to transform the ΔTRR2 strain.
For the re-expression of OCT1 into the ΔOCT1 background, the yeast cells were transformed with the plasmid pRS413/OCT1, which was kindly provided by Chris Meisinger (University of Freiburg, Germany). Alternatively, the open reading frame, which included the endogenous promoter and terminator regions, was cloned into YCplac111 (69) to yield Ycplac111/OCT1.
For the generation of a yeast strain expressing Prx1C38S or Prx1C91S, PRX1 was amplified from the genomic DNA with the primers Prx1F 5′-TGAGCTCTCAAAGAAGAAGAATTATGGGCAT-3′ and Prx1R 5′-GGCCTGCAGAGTTTATTACATGCATTTTCATAT-3′. The purified PCR product with 1477 bp was digest with SacI and PstI and cloned into YCplac111 (69) to yield Ycplac111/PRX1. Ycplac111/PRX1 was used as a template for the site-directed mutagenesis (QuikChange XL site-directed mutagenesis kit; Agilent Technologies).
To construct strains expressing the human mitochondrial peroxiredoxin Prx3, PRX3 was commercially synthesized by GenScript USA, Inc., and was cloned into pRS413. The pRS413/PRX3 recombinant plasmid, containing the coding sequence PRX3 under the constitutive TEF1 promoter control, was used to transform the yeast cells.
Isolation of highly pure mitochondria
Mitochondria were isolated from yeast cells grown at 30 °C on fermentable YPGal medium as described above. The cells were harvested at an optical density of 1.5–2, and the mitochondria were isolated by differential centrifugation as described previously (70). Spheroplasts were prepared by the enzymatic digestion of the cell wall using Zymolyase-20T (MPBiomedicals, Irvine, CA) at 3 mg of Zymolyase/g wet weight. The formation of spheroplasts was monitored by the absorbance of the yeast suspension prior to and following the Zymolyase treatment. The crude mitochondrial fraction was resuspended in an SEM buffer (250 mm sucrose, 1 mm EDTA, and 10 mm MOPS-KOH, pH 7.2) to a final concentration of 10 mg of protein/ml. To obtain highly pure mitochondria, the crude mitochondrial fraction was loaded onto a three-step sucrose gradient (1.5 ml 60%, 4 ml 32%, 1.5 ml 23%, and 1.5 ml 15% sucrose in an EM buffer containing 1 mm EDTA and 10 mm MOPS/KOH, pH 7.2) (70). The purity of the isolated mitochondria was assessed by immunoblotting using various cellular marker proteins.
Submitochondrial fractionation
Highly pure mitochondria (0.4 mg of protein) were initially suspended in 210 μl of the SEM buffer or 210 μl of the hypotonic buffer (10 mm MOPS/KOH, pH 7.2) in the absence or presence of 0.1 mg/ml proteinase K. The samples were then incubated on ice for 1 h. In this step, the hypotonic buffer induces the rupture (swelling) of the outer membrane, converting the mitochondria into mitoplasts. Thus, proteinase K degrades the proteins that are released from intermembrane space and those associated with the inner membrane facing the intermembrane space. The reactions were stopped by the addition of 10 μl of 50 mm PMSF, followed by centrifugation for 30 min at 4 °C and 20,000 × g. The supernatant was collected, and the pellet was suspended in 100 μl of the SEM buffer. Both the pellet and supernatant were then precipitated by the addition of 10 μl of 50% trichloroacetic acid (TCA), followed by centrifugation for 10 min at 4 °C and 20,000 × g. The proteins were solubilized in Laemmli buffer, separated by SDS-PAGE, and analyzed by Western blotting using antibodies against marker proteins for the distinct mitochondrial subcompartments and against Prx1, Trx3, and Trr2.
To remove the peripherally associated proteins from the mitochondrial membranes, 200 μl of mitochondria at a 10 mg/ml concentration were sonicated for 10 s in a Sonics Vibra-Cell VCX 130, followed by centrifugation for 1 h at 4 °C and 30,000 × g. The supernatant was collected, and the pellet was resuspended in 200 μl of the SEM buffer. The pellet fraction was then extracted with alkaline Na2CO3 at a final concentration of 100 mm and centrifuged for 1 h at 4 °C and 30,000 × g. The supernatant was collected, and the pellet was resuspended in 200 μl of the SEM buffer. All samples were then analyzed by SDS-PAGE and Western blotting.
Susceptibility of Prx1 degradation by proteinase K
The susceptibility of native mitochondrial Prx1 to externally added proteinase K was performed as follows: 40 μl of mitochondria at a 10 mg/ml concentration were suspended in 210 μl of the hypotonic buffer (10 mm MOPS/KOH, pH 7.2) and incubated on ice for 1 h. The suspension was centrifuged at 20,000 × g, at 4 °C for 30 min. The supernatant was collected, and the pellet consisting of mitoplasts was suspended in the starting volume of the SEM buffer. The samples were treated on ice for 1 h with 0.2 mg/ml proteinase K (twice the concentration in the standard conditions) in the presence or absence of 0.5% (v/v) Triton X-100. After incubation, the proteinase K-treated and -untreated samples were dissolved in Laemmli buffer, separated by SDS-PAGE, and analyzed by Western blotting.
Alternatively, the accessibility of recombinant Prx1 to proteinase K degradation was performed as follows. Recombinant Prx1 isoforms (Prx1K39 and Prx1F31) (10 μg) were incubated for 1 h on ice with proteinase K (0.2 mg/ml) in the absence or presence of 0.5% (v/v) Triton X-100. The protease activity was heat-inactivated (5 min at 95 °C), and protein content was analyzed using SDS-PAGE and Western blotting.
Stabilities of mitochondrial proteins
The stabilities of mitochondrial proteins were analyzed as described previously (27). Briefly, isolated mitochondria (0.2 mg of protein) were solubilized in 200 μl of the SEM buffer and incubated at 37 °C. The samples were collected at various time points, and the mitochondria were pelleted by centrifugation, resuspended in Laemmli buffer, and analyzed by SDS-PAGE and immunoblotting.
Assay of enzymatic activities
The peroxidase activities of both Prx1 isoforms (Prx1K39 and Prx1F31) were spectrophotometrically determined by the thioredoxin system coupled assay, following the disappearance of the NADPH absorbance at 340 nm (17). The standard reaction mixture contained 50 mm HEPES, pH 7.4, 200 μm NADPH, 0.1 μm Trr2, 5 μm Trx3, 5 μm peroxiredoxin, and 100 μm diethylenetriaminepentaacetic acid (DTPA). The reactions were initiated by the addition of distinct H2O2 concentrations that ranged from 10 to 500 μm. Recombinant Trx3 and Trr2 were produced as described before (35), and they are N-terminally His-tagged proteins. Enzymatic parameters were determined by non-linear regression (Michaelis-Menten equation), employing the GraphPad Prism 6 software (GraphPad Software, Inc.).
Prx1 purification and mass spectrometry analysis
Mitochondria were isolated from yeast cells expressing a C-terminally His-tagged Prx1. The mitochondria (40 mg of protein) were divided into aliquots of 0.8 mg of protein and converted to mitoplasts by incubation with 420 μl of the hypotonic buffer (10 mm MOPS/KOH, pH 7.2) for 1 h at 4 °C. The samples were centrifuged for 30 min at 4 °C and 20,000 × g. The supernatant, containing the intermembrane space proteins, was collected and mixed with the purification buffer (500 mm NaCl, 20 mm sodium phosphate buffer, pH 7.4, and 10 mm imidazole). The pellets (mitoplasts) were resuspended in the purification buffer, and the Prx1 that was weakly associated with the mitochondrial inner membrane was extracted by the addition of 1% of Triton X-100, followed by incubation for 30 min at 4 °C with vigorous shaking. C-terminally His-tagged Prx1 of both compartments (intermembrane space and matrix) was purified by nickel affinity chromatography using nickel-nitrilotriacetic acid-agarose column (Qiagen). The purity of the purified proteins was ascertained by SDS-PAGE. The proteins were analyzed by mass spectrometry in a UHR-ESI-Q-TOF-MS from Bruker Daltonics MaXis 3G (Bruker, Billerica, MA) in the positive mode coupled with an HPLC system from Shimadzy CBM-20A (Tokyo, Japan).
Miscellaneous procedures
Proteins were separated by SDS-PAGE on different concentrations of polyacrylamide and transferred to nitrocellulose membranes (Amersham BiosciencesTM ProtranTM Premium, GE Healthcare). The membranes were probed with primary antibodies (Table 3) against the appropriate proteins, followed by a second reaction with secondary antibodies conjugated to horseradish peroxidase. Proteins were detected with the ECLTM prime Western blotting detection reagent (GE Healthcare). The protein concentrations were estimated by the method proposed by Bradford (71). The antibodies utilized were either commercially acquired or produced in 2-month-old male rabbits in the Animal Care Facility of the Butantan Institute. Anti-Cyt.b2, anti-αKGD, and anti-Sco1 were kindly provided by Dr. A. Tzagoloff (Columbia University). For more details, please see Table 3.
Table 3.
Antibodies used in this study
| Antibody | Incubation buffer | Dilution | Source |
|---|---|---|---|
| Anti-αKGD | Milk 5% | 1:200 | Dr. A. Tzagoloff (Columbia University)a |
| Anti-cytochrome b2 | Milk 5% | 1:200 | Dr. A. Tzagoloff (Columbia University)a |
| Anti-ScoI | Milk 5% | 1:200 | Dr. A. Tzagoloff (Columbia University)a |
| Anti-Prx1 | Milk 5% | 1:2000 | This studyb |
| Anti-Trx3 | Milk 5% | 1:1000 | This studyb |
| Anti-Trr2 | Milk 5% | 1:1000 | This studyb |
| Anti-HA | Milk 5% | 1:2000 | Sigma (H6908) |
| Anti-Prx3 | TBST | 1:4000 | AbFrontierLF (MA0044) |
| Anti-Porin | Milk 5% | 1:10,000 | Thermo Fisher Scientific (16G9E6BC4) |
| Anti-Pgk1 | Milk 5% | 1:10,000 | Nordic Immunology (NE130/7S) |
| Anti-Dpm1 | Milk 5% | 1:1000 | Thermo Fisher Scientific (5C5A7) |
| Anti-Vma2 | Milk 5% | 1:1000 | Thermo Fisher Scientific (11D11B2) |
| Anti-Pep12 | Milk 5% | 1:1000 | Thermo Fisher Scientific (2C3G4) |
| Anti-rabbit IgG HRP-linked | Milk 5% | 1:10,000 | Cell Signaling Technology |
| Anti-mouse IgG HRP-linked | Milk 5% | 1:10,000 | Cell Signaling Technology |
a This was produced in the laboratory of Dr. A. Tzagoloff after immunization of rabbits with recombinant proteins.
b Antibodies were produced in 2-month-old male rabbits in the Animal Care Facility of the Butantan Institute.
Author contributions
F. G., M. H. B., and L. E. S. N. designed the research; F. G., F. R. P., E. T. T., T. G. P. A., and H. T. performed the research; M. H. B. and M. D. contributed new reagents/analytic tools; F. G., T. G. P. A., M. H. B., M. D., and L. E. S. N. analyzed the data; and F. G., M. H. B., M. D., and L. E. S. N. wrote the paper.
Supplementary Material
Acknowledgments
We thank Dr. Paolo Di Mascio and Emerson Finco Marques (Instituto Quimica, Universidade de São Paulo) for their support in the mass spectrometry analysis. We also thank Chris Meisinger (University of Freiburg, Germany), Patrycja Mulica (University of Freiburg, Germany), and Nora Vögtle (University of Freiburg, Germany) for helpful discussion and for providing material for this research.
This work was supported in part by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) Grant 13/07937-8 (Redox Processes in Biomedicine) (to L. E. S. N.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supplemental Figs. S1 and S2.
- mROS
- mitochondrial reactive oxygen species
- IMP
- inner membrane peptidase
- MPP
- mitochondrial processing peptidase
- Prx
- peroxiredoxin
- DTPA
- diethylenetriaminepentaacetic acid
- α-KGD
- α-ketoglutarate dehydrogenase
- SMP
- submitochondrial particle
- Cyt.b2
- cytochrome b2
- Trx
- thioredoxin.
References
- 1. Herrmann J. M., and Riemer J. (2010) The intermembrane space of mitochondria. Antioxid. Redox Signal. 13, 1341–1358 [DOI] [PubMed] [Google Scholar]
- 2. Ryan M. T., and Hoogenraad N. J. (2007) Mitochondrial-nuclear communications. Annu. Rev. Biochem. 76, 701–722 [DOI] [PubMed] [Google Scholar]
- 3. Harbauer A. B., Zahedi R. P., Sickmann A., Pfanner N., and Meisinger C. (2014) The protein import machinery of mitochondria–A regulatory hub in metabolism, stress, and disease. Cell Metab. 19, 357–372 [DOI] [PubMed] [Google Scholar]
- 4. Murphy M. P. (2009) How mitochondria produce reactive oxygen species. Biochem. J. 417, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kowaltowski A. J., de Souza-Pinto N. C., Castilho R. F., and Vercesi A. E. (2009) Mitochondria and reactive oxygen species. Free Radic. Biol. Med. 47, 333–343 [DOI] [PubMed] [Google Scholar]
- 6. Sena L. A., and Chandel N. S. (2012) Physiological roles of mitochondrial reactive oxygen species. Mol. Cell 48, 158–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cox A. G., Winterbourn C. C., and Hampton M. B. (2010) Mitochondrial peroxiredoxin involvement in antioxidant defence and redox signalling. Biochem. J. 325, 313–325 [DOI] [PubMed] [Google Scholar]
- 8. Knoops B., Goemaere J., Van der Eecken V., and Declercq J.-P. (2011) Peroxiredoxin 5: structure, mechanism, and function of the mammalian atypical 2-Cys peroxiredoxin. Antioxid. Redox Signal. 15, 817–829 [DOI] [PubMed] [Google Scholar]
- 9. Rhee S. G., and Kil I. S. (2016) Mitochondrial H2O2 signaling is controlled by the concerted action of peroxiredoxin III and sulfiredoxin: linking mitochondrial function to circadian rhythm. Free Radic. Biol. Med. 100, 73–80 [DOI] [PubMed] [Google Scholar]
- 10. Rhee S. G., Woo H. A., Kil I. S., and Bae S. H. (2012) Peroxiredoxin functions as a peroxidase and a regulator and sensor of local peroxides. J. Biol. Chem. 287, 4403–4410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fisher A. B. (2017) Peroxiredoxin 6 in the repair of peroxidized cell membranes and cell signaling. Arch. Biochem. Biophys. 617, 68–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gutscher M., Sobotta M. C., Wabnitz G. H., Ballikaya S., Meyer A. J., Samstag Y., and Dick T. P. (2009) Proximity-based protein thiol oxidation by H2O2-scavenging peroxidases. J. Biol. Chem. 284, 31532–31540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Delaunay A., Pflieger D., Barrault M. B., Vinh J., and Toledano M. B. (2002) A thiol peroxidase is an H2O2 receptor and redox-transducer in gene activation. Cell 111, 471–481 [DOI] [PubMed] [Google Scholar]
- 14. Sobotta M. C., Liou W., Stöcker S., Talwar D., Oehler M., Ruppert T., Scharf A. N., and Dick T. P. (2015) Peroxiredoxin-2 and STAT3 form a redox relay for H2O2 signaling. Nat. Chem. Biol. 11, 64–70 [DOI] [PubMed] [Google Scholar]
- 15. Kil I. S., Ryu K. W., Lee S. K., Kim J. Y., Chu S. Y., Kim J. H., Park S., and Rhee S. G. (2015) Circadian oscillation of sulfiredoxin in the mitochondria. Mol. Cell 59, 651–663 [DOI] [PubMed] [Google Scholar]
- 16. Pedrajas J. R., Miranda-Vizuete A., Javanmardy N., Gustafsson J.-A., and Spyrou G. (2000) Mitochondria of Saccharomyces cerevisiae contain one-conserved cysteine type peroxiredoxin with thioredoxin peroxidase activity. J. Biol. Chem. 275, 16296–16301 [DOI] [PubMed] [Google Scholar]
- 17. Pedrajas J. R., Padilla C. A., McDonagh B., and Bárcena J. A. (2010) Glutaredoxin participates in the reduction of peroxides by the mitochondrial 1-Cys peroxiredoxin in Saccharomyces cerevisiae. Antioxid. Redox Signal. 13, 249–258 [DOI] [PubMed] [Google Scholar]
- 18. Pedrajas J. R., McDonagh B., Hernández-Torres F., Miranda-Vizuete A., González-Ojeda R., Martínez-Galisteo E., Padilla C. A., and Bárcena J. A. (2016) Glutathione is the resolving thiol for thioredoxin peroxidase activity of 1-Cys peroxiredoxin without being consumed during the catalytic cycle. Antioxid. Redox Signal. 24, 115–128 [DOI] [PubMed] [Google Scholar]
- 19. Greetham D., and Grant C. M. (2009) Antioxidant activity of the yeast mitochondrial one-Cys peroxiredoxin is dependent on thioredoxin reductase and glutathione in vivo. Mol. Cell. Biol. 29, 3229–3240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Monteiro G., Horta B. B., Pimenta D. C., Augusto O., and Netto L. E. (2007) Reduction of 1-Cys peroxiredoxins by ascorbate changes the thiol-specific antioxidant paradigm, revealing another function of vitamin C. Proc. Natl. Acad. Sci. U.S.A. 104, 4886–4891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Monteiro G., Pereira G. A., and Netto L. E. (2002) Regulation of mitochondrial thioredoxin peroxidase I expression by two different pathways: one dependent on cAMP and the other on heme. Free Radic. Biol. Med. 32, 278–288 [DOI] [PubMed] [Google Scholar]
- 22. Monteiro G., Kowaltowski A. J., Barros M. H., and Netto L. E. (2004) Glutathione and thioredoxin peroxidases mediate susceptibility of yeast mitochondria to Ca2+-induced damage. Arch. Biochem. Biophys. 425, 14–24 [DOI] [PubMed] [Google Scholar]
- 23. Schmidt O., Pfanner N., and Meisinger C. (2010) Mitochondrial protein import: from proteomics to functional mechanisms. Nat. Rev. Mol. Cell Biol. 11, 655–667 [DOI] [PubMed] [Google Scholar]
- 24. Neupert W., and Herrmann J. M. (2007) Translocation of proteins into mitochondria. Annu. Rev. Biochem. 76, 723–749 [DOI] [PubMed] [Google Scholar]
- 25. Chacinska A., Koehler C. M., Milenkovic D., Lithgow T., and Pfanner N. (2009) Importing mitochondrial proteins: machineries and mechanisms. Cell 138, 628–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Taylor A. B., Smith B. S., Kitada S., Kojima K., Miyaura H., Otwinowski Z., Ito A., and Deisenhofer J. (2001) Crystal structures of mitochondrial processing peptidase reveal the mode for specific cleavage of import signal sequences. Structure 9, 615–625 [DOI] [PubMed] [Google Scholar]
- 27. Vögtle F. N., Wortelkamp S., Zahedi R. P., Becker D., Leidhold C., Gevaert K., Kellermann J., Voos W., Sickmann A., Pfanner N., and Meisinger C. (2009) Global analysis of the mitochondrial N-proteome identifies a processing peptidase critical for protein stability. Cell 139, 428–439 [DOI] [PubMed] [Google Scholar]
- 28. Mossmann D., Meisinger C., and Vögtle F. N. (2012) Processing of mitochondrial presequences. Biochim. Biophys. Acta 1819, 1098–1106 [DOI] [PubMed] [Google Scholar]
- 29. Vögtle F.-N., Prinz C., Kellermann J., Lottspeich F., Pfanner N., and Meisinger C. (2011) Mitochondrial protein turnover: role of the precursor intermediate peptidase Oct1 in protein stabilization. Mol. Biol. Cell 22, 2135–2143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Teixeira P. F., and Glaser E. (2013) Processing peptidases in mitochondria and chloroplasts. Biochim. Biophys. Acta 1833, 360–370 [DOI] [PubMed] [Google Scholar]
- 31. Nunnari J., Fox T. D., and Walter P. (1993) A mitochondrial protease with two catalytic subunits of nonoverlapping specificities. Science 262, 1997–2004 [DOI] [PubMed] [Google Scholar]
- 32. Jan P. S., Esser K., Pratje E., and Michaelis G. (2000) Som1, a third component of the yeast mitochondrial inner membrane peptidase complex that contains Imp1 and Imp2. Mol. Gen. Genet. 263, 483–491 [DOI] [PubMed] [Google Scholar]
- 33. Glick B. S., Brandt A., Cunningham K., Müller S., Hallberg R. L., and Schatz G. (1992) Cytochromes c1 and b2 are sorted to the intermembrane space of yeast mitochondria by a stop-transfer mechanism. Cell 69, 809–822 [DOI] [PubMed] [Google Scholar]
- 34. Herrmann J. M., and Hell K. (2005) Chopped, trapped or tacked–protein translocation into the IMS of mitochondria. Trends Biochem. Sci. 30, 205–211 [DOI] [PubMed] [Google Scholar]
- 35. Pedrajas J. R., Kosmidou E., Miranda-Vizuete A., Gustafsson J. A., Wright A. P., and Spyrou G. (1999) Identification and functional characterization of a novel mitochondrial thioredoxin system in Saccharomyces cerevisiae. J. Biol. Chem. 274, 6366–6373 [DOI] [PubMed] [Google Scholar]
- 36. Boldogh I. R., and Pon L. A. (2007) Purification and subfractionation of mitochondria from the yeast Saccharomyces cerevisiae. Methods Cell Biol. 80, 45–64 [DOI] [PubMed] [Google Scholar]
- 37. Bömer U., Meijer M., Guiard B., Dietmeier K., Pfanner N., and Rassow J. (1997) The sorting route of cytochrome b2 branches from the general mitochondrial import pathway at the preprotein translocase of the inner membrane. J. Biol. Chem. 272, 30439–30446 [DOI] [PubMed] [Google Scholar]
- 38. Glerum D. M., Shtanko A., and Tzagoloff A. (1996) SCO1 and SCO2 act as high copy suppressors of a mitochondrial copper recruitment defect in Saccharomyces cerevisiae. J. Biol. Chem. 271, 20531–20535 [DOI] [PubMed] [Google Scholar]
- 39. Repetto B., and Tzagoloff A. (1990) Structure and regulation of KGD2, the structural gene for yeast dihydrolipoyl transsuccinylase. Mol. Cell. Biol. 10, 4221–4232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Penefsky H. S., and Tzagoloff A. (1971) Extraction of water-soluble enzymes and proteins from membranes. Methods Enzymol. 22, 204–219 [Google Scholar]
- 41. Moda B. S., Ferreira-Júnior J. R., and Barros M. H. (2016) Partial suppression of the respiratory defect of qrs1/her2 glutamyl-tRNA amidotransferase mutants by overexpression of the mitochondrial pentatricopeptide Msc6p. Curr. Genet. 62, 607–617 [DOI] [PubMed] [Google Scholar]
- 42. Luo W., Fang H., and Green N. (2006) Substrate specificity of inner membrane peptidase in yeast mitochondria. Mol. Genet. Genomics 275, 431–436 [DOI] [PubMed] [Google Scholar]
- 43. Isaya G., Miklos D., and Rollins R. A. (1994) MIP1, a new yeast gene homologous to the rat mitochondrial intermediate peptidase gene, is required for oxidative metabolism in Saccharomyces cerevisiae. Mol. Cell. Biol. 14, 5603–5616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Murphy M. P. (2012) Mitochondrial thiols in antioxidant protection and redox signaling: distinct roles for glutathionylation and other thiol modifications. Antioxid. Redox Signal. 16, 476–495 [DOI] [PubMed] [Google Scholar]
- 45. Vaca Jacome A. S., Rabilloud T., Schaeffer-Reiss C., Rompais M., Ayoub D., Lane L., Bairoch A., Van Dorsselaer A., and Carapito C. (2015) N-terminome analysis of the human mitochondrial proteome. Proteomics 15, 2519–2524 [DOI] [PubMed] [Google Scholar]
- 46. Green D. R., Galluzzi L., and Kroemer G. (2011) Mitochondria and the autophagy-inflammation-cell death axis in organismal aging. Science 333, 1109–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Andreux P. A., Houtkooper R. H., and Auwerx J. (2013) Pharmacological approaches to restore mitochondrial function. Nat. Rev. Drug Discov. 12, 465–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang X., and Chen X. J. (2015) A cytosolic network suppressing mitochondria-mediated proteostatic stress and cell death. Nature 524, 481–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Durigon R., Wang Q., Ceh Pavia E., Grant C. M., and Lu H. (2012) Cytosolic thioredoxin system facilitates the import of mitochondrial small Tim proteins. EMBO Rep. 13, 916–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Branda S. S., and Isaya G. (1995) Prediction and identification of new natural substrates of the yeast mitochondrial intermediate peptidase. J. Biol. Chem. 270, 27366–27373 [DOI] [PubMed] [Google Scholar]
- 51. Campuzano V., Montermini L., Moltò M. D., Pianese L., Cossée M., Cavalcanti F., Monros E., Rodius F., Duclos F., Monticelli A., Zara F., Cañizares J., Koutnikova H., Bidichandani S. I., Gellera C., et al. (1996) Friedreich's ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science 271, 1423–1427 [DOI] [PubMed] [Google Scholar]
- 52. Harding A. E. (1981) Friedreich's ataxia: a clinical and genetic study of 90 families with an analysis of early diagnostic criteria and intrafamilial clustering of clinical features. Brain 104, 589–620 [DOI] [PubMed] [Google Scholar]
- 53. Babcock M., de Silva D., Oaks R., Davis-Kaplan S., Jiralerspong S., Montermini L., Pandolfo M., and Kaplan J. (1997) Regulation of mitochondrial iron accumulation by Yfh1p, a putative homolog of frataxin. Science 276, 1709–1712 [DOI] [PubMed] [Google Scholar]
- 54. Koutnikova H., Campuzano V., Foury F., Dollé P., Cazzalini O., and Koenig M. (1997) Studies of human, mouse and yeast homologues indicate a mitochondrial function for frataxin. Nat. Genet. 16, 345–351 [DOI] [PubMed] [Google Scholar]
- 55. Foury F., and Cazzalini O. (1997) Deletion of the yeast homologue of the human gene associated with Friedreich's ataxia elicits iron accumulation in mitochondria. FEBS Lett. 411, 373–377 [DOI] [PubMed] [Google Scholar]
- 56. Wilson R. B., and Roof D. M. (1997) Respiratory deficiency due to loss of mitochondrial DNA in yeast lacking the frataxin homologue. Nat. Genet. 16, 352–357 [DOI] [PubMed] [Google Scholar]
- 57. Branda S. S., Yang Z. Y., Chew A., and Isaya G. (1999) Mitochondrial intermediate peptidase and the yeast frataxin homolog together maintain mitochondrial iron homeostasis in Saccharomyces cerevisiae. Hum. Mol. Genet. 8, 1099–1110 [DOI] [PubMed] [Google Scholar]
- 58. Poveda-Huertes D., Mulica P., and Vögtle F. N. (2017) The versatility of the mitochondrial presequence processing machinery: cleavage, quality control and turnover. Cell Tissue Res. 367, 73–81 [DOI] [PubMed] [Google Scholar]
- 59. Hahne K., Haucke V., Ramage L., and Schatz G. (1994) Incomplete arrest in the outer membrane sorts NADH-cytochrome b5 reductase to two different submitochondrial compartments. Cell 79, 829–839 [DOI] [PubMed] [Google Scholar]
- 60. Haucke V., Ocana C. S., Hönlinger A., Tokatlidis K., Pfanner N., and Schatz G. (1997) Analysis of the sorting signals directing NADH-cytochrome b5 reductase to two locations within yeast mitochondria. Mol. Cell. Biol. 17, 4024–4032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Toledano M. B., Delaunay-Moisan A., Outten C. E., and Igbaria A. (2013) Functions and cellular compartmentation of the thioredoxin and glutathione pathways in yeast. Antioxid. Redox Signal. 18, 1699–1711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kritsiligkou P., Chatzi A., Charalampous G., Mironov A. Jr., Grant C. M., and Tokatlidis K. (2017) Unconventional targeting of a thiol peroxidase to the mitochondrial intermembrane space facilitates oxidative protein folding. Cell Rep. 18, 2729–2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Vögtle F.-N., Burkhart J. M., Rao S., Gerbeth C., Hinrichs J., Martinou J.-C., Chacinska A., Sickmann A., Zahedi R. P., and Meisinger C. (2012) Intermembrane space proteome of yeast mitochondria. Mol. Cell. Proteomics 11, 1840–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Petek E., Windpassinger C., Vincent J. B., Cheung J., Boright A. P., Scherer S. W., Kroisel P. M., and Wagner K. (2001) Disruption of a novel gene (IMMP2L) by a breakpoint in 7q31 associated with Tourette syndrome. Am. J. Hum. Genet. 68, 848–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Burri L., Strahm Y., Hawkins C. J., Gentle I. E., Puryer M. A., Verhagen A., Callus B., Vaux D., and Lithgow T. (2005) Mature DIABLO/Smac is produced by the IMP protease complex on the mitochondrial inner membrane. Mol. Biol. Cell 16, 2926–2933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Bertelsen B., Melchior L., Jensen L. R., Groth C., Glenthøj B., Rizzo R., Debes N. M., Skov L., Brøndum-Nielsen K., Paschou P., Silahtaroglu A., and Tümer Z. (2014) Intragenic deletions affecting two alternative transcripts of the IMMP2L gene in patients with Tourette syndrome. Eur. J. Hum. Genet. 22, 1283–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Patel C., Cooper-Charles L., McMullan D. J., Walker J. M., Davison V., and Morton J. (2011) Translocation breakpoint at 7q31 associated with tics: further evidence for IMMP2L as a candidate gene for Tourette syndrome. Eur. J. Hum. Genet. 19, 634–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hill J. E., Myers A. M., Koerner T. J., and Tzagoloff A. (1986) Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast 2, 163–167 [DOI] [PubMed] [Google Scholar]
- 69. Gietz R. D., and Sugino A. (1988) New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74, 527–534 [DOI] [PubMed] [Google Scholar]
- 70. Meisinger C., Pfanner N., and Truscott K. N. (2006) Isolation of yeast mitochondria. Methods Mol. Biol. 313, 33–39 [DOI] [PubMed] [Google Scholar]
- 71. Kruger N. J. (1994) The Bradford method for protein quantitation. Methods Mol. Biol. 32, 9–15 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.