Abstract
Both chemical and mechanical stimuli can dramatically influence cell behavior. By optimizing the signals cells experience, it may be possible to control the behavior of therapeutic cell populations. In this work, biomimetic geometries of adhesive ligands, which recapitulate the morphology of mature cells, are used to direct human mesenchymal stem cell (HMSC) differentiation toward a desired lineage. Specifically, adipocytes cultured in 2D are imaged and used to develop biomimetic virtual masks used in laser scanning lithography to form patterned fibronectin surfaces. The impact of adipocyte-derived pattern geometry on HMSC differentiation is compared to the behavior of HMSCs cultured on square and circle geometries, as well as adipocyte-derived patterns modified to include high stress regions. HMSCs on adipocyte mimetic geometries demonstrate greater adipogenesis than HMSCs on the other patterns. Greater than 45% of all HMSCs cultured on adipocyte mimetic patterns underwent adipogenesis as compared to approximately 19% of cells on modified adipocyte patterns with higher stress regions. These results are attributed to variations in cytoskeletal tension experienced by cells on the different protein micropatterns. The effects of geometry on adipogenesis are mitigated by the incorporation of a cytoskeletal protein inhibitor; exposure to this inhibitor leads to increased adipogenesis on all patterns examined.
Keywords: stem cell engineering, micropatterns, laser scanning lithography, adipogenesis, biomimetic
Graphical abstract
1. INTRODUCTION
Adult mesenchymal stem cells (MSCs) are of great interest for regenerative medicine. These cells have been isolated from numerous tissues1 and have the unique ability to self-renew and differentiate into cells of the mesoderm lineage including adipocytes and osteoblasts.2 The therapeutic potential of MSCs is currently being investigated in numerous clinical trials for a wide variety of diseases.3 They have shown promise in the treatment of conditions ranging from postinfarction left ventricular dysfunction4–6 to osteogenesis imperfecta.7,8 Developing a better understanding of the factors that influence MSC differentiation will enable the design of superior stem cell therapies, including tissue engineering scaffolds that may be used to deliver exogenous stem cells or recruit endogenous cells.9–11
Typical stem cell differentiation approaches can be divided into chemical, genetic, and biomechanical methods.9 Chemical approaches to MSC differentiation have been the most commonly implemented and studied, in which hormone, growth factor, and extracellular matrix (ECM) composition is varied to specifically influence certain lineage-specific differentiation pathways.2,12,13 The differentiated cells that result following these in vitro chemical treatments do not display the same biochemical and biomechanical properties as mature cells found in vivo.14 Therefore, it is unclear whether MSCs differentiated via chemical approaches in vitro possess the same regenerative capabilities as those MSCs residing in the body. As an alternative to chemical differentiation approaches, biomechanical approaches have recently gained significant interest.5,15–19 By attempting to mimic the in vivo stem cell microenvironment, these approaches to MSC differentiation can be combined with the use of appropriate chemical cues to yield optimized MSC differentiation.
Intracellular and extracellular forces generated by the actomyosin contractile apparatus of the cell and transmitted via focal adhesions act on the cell, the ECM, and the surrounding cells, influencing intra- and extracellular signaling processes, including proliferation and differentiation.20,21 Several studies have related the biomechanical and morphological properties of stem cells to their differentiation capabilities.22,23 Biomechanical approaches have been implemented to influence MSC behavior in many recent studies, including varying the mechanical properties of cell culture substrates15,17,24,25 and controlling cell size and shape.5,16,18,19 In the case of cell shape, aspect ratio and curvature of MSCs appear to have a significant impact on the differentiation of individual cells.16,26 Inhibition of key contractile proteins has mitigated or eliminated the influence of geometry on cell differentiation, suggesting that cytoskeletal tension is a key regulator of the effects of shape on MSC differentiation.5,16
Thus far, arbitrary geometries have been implemented to probe the role of shape in MSC differentiation. To realize the true potential of geometric control on MSC differentiation, it is important to consider whether a better-informed geometry can lead to superior MSC differentiation. We hypothesized that human MSCs (HMSCs) restricted to mimicking the overall cell shape of fully differentiated cells would preferentially undergo differentiation to these specific lineages. In this work, we have tested this hypothesis by examining the effects of biomimetic protein micropatterns on HMSCs. We recently developed a cell-derived patterning technique that allows for direct recapitulation of the shape, size, and cytoskeletal architecture of user-chosen cells of interest. In this technique, cells of interest cultured in 2D are fixed and stained via immunocytochemistry for important cytoskeletal components (e.g., focal adhesions, actin, and nuclei). Images of these cells are used to generate biomimetic virtual masks used in laser scanning lithography, yielding a large array of biomimetic protein patterns.27 Building off of this biomimetic patterning approach, here we have developed adipocyte mimetic protein patterns and modified adipocyte patterns that were designed to induce regions of high intracellular tension. We have compared the behavior of HMSCs cultured on these patterns to the behavior of HMSCs on simple square and circle patterns of matched area. We found that HMSCs cultured on adipocyte mimetic patterns tend to more preferentially undergo adipogenesis as compared to those cultured on other patterns examined in the presence of mixed adipogenic and osteogenic soluble factors. To the best of our knowledge, this is the first study that examines the role of biomimetic, single-cell geometries on HMSC differentiation.
2. EXPERIMENTAL SECTION
2.1. Materials
Bone marrow-derived human mesenchymal stem cells (HMSCs), human subcutaneous preadipocytes (HPAd), human preadipocyte growth media, HMSC growth media, adipogenic media, and osteogenic media were obtained from Lonza (Walkersville, MD). Trypsin (0.05%)/ethylenediaminetetraacetic acid (0.53 mM EDTA) and phosphate buffered saline (10×) were purchased from Mediatech (Manassas, VA). Paraformaldehyde (16% aqueous solution) was obtained from Electron Microscopy Sciences (Hatfield, PA). Triton X-100, 5-bromo-4-chloro-3-indolyl phosphate (BCIP)/nitro blue tetrazolium (NBT) solution, human plasma fibronectin, mitomycin C, cytochalasin D, 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI), 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), sucrose (C12H22O11), and mouse anti-vinculin antibody were purchased from Sigma-Aldrich (St. Louis, MO). Magnesium chloride (MgCl2), sodium chloride (NaCl), and round borosilicate cover glass (35 mm, no. 1) were obtained from Fisher Scientific (Fair Lawn, NJ). Rabbit anti-peroxisome proliferator-activated receptor γ (PPARγ) antibody was obtained from Cell Signaling Technology (Danvers, MA), while mouse anti-Runt-related transcription factor 2 (RUNX2) antibody was obtained from Abcam (Cambridge, MA). In addition to goat serum, all Alexa Fluor (AF) conjugated antibodies and phalloidin were purchased from Invitrogen (Eugene, OR). Oil red O was purchased from Alfa Aesar (Ward Hill, MA). Gold (Au) and titanium (Ti) pellets (99.999% pure) for electron beam evaporation were obtained from KAMIS (Mahopac Falls, NY). Ethanol (200 proof), sulfuric acid (H2SO4), and hydrogen peroxide (H2O2) were obtained from VWR International (King of Prussia, Radnor, and West Chester, PA, respectively). Isopropyl alcohol was purchased from EMD Millipore (Billerica, MA). HS-(CH2)11(O(CH2)2)6OH (OEG6) was purchased from ProChimia (Sopot, Poland). Ultrapure deionized water (18.2 MΩ, Millipore Super-Q Water System) and ultrapure nitrogen gas (Matheson Tri-Gas, Basking Ridge, NJ) were used in all experiments. Borosilicate cover glass (no. 1) bottom chamber slides were obtained from Nalge Nunc International (Rochester, NY).
2.2. Cell Culture
2.2.1. Human Subcutaneous Preadipocyte Culture
HPAd were treated with 10 µg mL−1 mitomycin C for 2 h at 37 °C/5% CO2 to arrest proliferation. This treatment has previously been shown not to affect HMSC viability or differentiation.5 Coverglass bottom chamber slides were coated with fibronectin at a concentration of 12.5 µg mL−1 in 1× PBS (pH 7.4) for 1 h at room temperature followed by three 1× PBS rinses. Mitomycin C treated cells were subsequently trypsinized and seeded at passage 2 at a density of 2.5 × 103 cells cm−2 on these fibronectin-coated chamber slides in adipocyte differentiation media as recommended by Lonza at 37 °C/5% CO2. Controls in preadipocyte growth media were seeded similarly. Media was changed every 3–4 days.
2.2.2. Human Mesenchymal Stem Cell Culture
HMSCs expanded in MSC growth media as recommended by Lonza were treated with mitomycin C as described for HPAd. Patterned samples were seeded with these cells (passage 3) in differentiation media (approximately 2.7 × 104 cells sample−1). Differentiation media contained 1:1 (v/v) adipogenic induction media and osteogenic differentiation media. Differentiation media was replaced every 2–3 days. Nonpatterned metalized coverglass surfaces were coated with fibronectin and seeded similarly with HMSCs in growth media or differentiation media. For experiments conducted in the presence of the F-actin inhibitor, 0.2 µM cytochalasin D in differentiation media was added with daily media changes, as previously described.16
2.3. Surface Preparation
2.3.1. Developing Virtual Masks
HPAd were cultured in differentiation media for 31 days; F-actin, vinculin, and nuclei were fluorescently labeled and confocal microscopy and DIC images were collected (see section 2.4.1. Microscopy and section 2.4.2. Preadipocyte Characterization). After identifying the adipocyte of interest, an adipocyte mimetic virtual mask was developed for this cell geometry using NIH Fiji and similar MATLAB scripts to those that have been previously reported.27,28 The confocal image of the cell was processed in Fiji to create an image of the cell shape. To do this, we outlined the cell in Fiji and all surrounding cell debris and cells in the image were removed; a corresponding binary image was developed using the Auto Local Threshold function in Fiji. This binary image was provided as input into the MATLAB scripts, which subsequently divided the cell shape into adjacent quadrilaterals filling the entire cell area. The quadrilaterals were interpreted as regions of interest (ROIs) for laser irradiation by Zeiss AIM software. Other virtual masks studied in this work were developed similarly.
2.3.2. Protein Pattern Fabrication
Arrays of fibronectin micropatterns were fabricated using laser scanning lithography (LSL).27,29 Initially, 35 mm round cover glass were cleaned for 1 h in a 3:1 (v/v) mixture of H2SO4 and H2O2 (Piranha Solution), followed by repetitive rinsing in water, soaking in water for 18 h, and subsequent drying with nitrogen. An electron beam evaporator (Sharon Vacuum, Brockton, MA) was used to deposit 2 nm of Ti followed by 8 nm of Au with a deposition rate of 0.2–0.4 Å s−1 on the clean cover glass. These metalized surfaces were subsequently used to deposit OEG6 self-assembled monolayers (SAMs). OEG6 SAMs were deposited for 24 h by covering the thin metal substrates with a 4 mM solution of the OEG6-alkanethiol in ethanol. Following SAM deposition, the substrates were rinsed in ethanol and dried with nitrogen. LSL patterning was carried out in an inert nitrogen environment using a Zeiss 5LIVE laser scanning confocal microscope equipped with Zeiss AIM software. The “Edit Bleach” function in the Zeiss AIM software was utilized along with the virtual mask ROIs (see section 2.3.1. Developing Virtual Masks). DIC imaging of the substrate was used to identify areas of the substrate to pattern. A 532 nm laser was applied with a laser fluence of 7.2 nJ µm−2 (corresponding to a 1.28 µs pixel dwell time and 40× objective with numerical aperture of 1.3; calculated as previously reported30) with 3 patterning iterations in order to desorb the OEG6-alkanethiol from the ROI defined regions to yield bare Au patterns. Arrays of 200 patterns were fabricated per sample. Following LSL, the sample was rinsed in ethanol, dried with nitrogen, and exposed to fibronectin, which was able to adsorb only in the patterned regions. Human plasma fibronectin (10 µg mL−1 in pH 7.4, 1× PBS) was applied to the patterned samples for 20 min, followed by a pH 7.4, 1× PBS rinse. These protein patterns were either visualized using DIC imaging or seeded with HMSCs. Nonpatterned Au surface controls were functionalized with fibronectin directly on the Ti–Au surface using the same procedure and treated similarly to patterned samples.
2.4. Pattern and Cell Characterization
2.4.1. Microscopy
Confocal and DIC microscopy was performed with a Zeiss 5LIVE laser scanning confocal microscope with Zeiss AIM software and a 40× oil immersion objective (numerical aperture of 1.3). Nuclei (DAPI) were imaged with a 405 nm excitation and a band-pass (BP) 415–480 nm emission filter. Vinculin and RUNX2 (labeled with goat anti-mouse AF 488) were imaged with a 488 nm excitation and a BP 500–525 nm emission filter. PPARγ (labeled with goat anti-rabbit AF 532) was imaged with a 532 nm excitation and a BP 560–675 nm emission filter. F-actin (labeled with AF 633 phalloidin) was imaged with a 633 nm excitation and a long pass 650 nm emission filter. Phase contrast imaging was done on a Zeiss Axio Observer.A1m microscope.
2.4.2. Preadipocyte Characterization
Following culture in differentiation media, HPAd were fixed with 4% paraformaldehyde and permeabilized using a cytoskeletal buffer (298.5 mM C12H22O11, 49.75 mM NaCl, 9.95 mM HEPES, 6.37 mM MgCl2 with 0.5% triton X-100 in water, pH 6.8).31 After blocking with 10% goat serum, the cells were incubated with a mouse anti-vinculin primary antibody at 4 °C for 18–24 h. Secondary antibody staining with goat anti-mouse AF 488 in 1% goat serum was performed at room temperature for 30 min, along with F-actin staining using AF 532 phalloidin (0.14 µM in 1% goat serum), followed by a 5 min DAPI counter stain (2 µM in 1× PBS). These cells were imaged via confocal microscopy and their lipid content was imaged using DIC microscopy.
2.4.3. HMSC Characterization
Characterization of HMSCs on patterned samples and nonpatterned controls was carried out after 7 days in culture. Cells were fixed with 4% paraformaldehyde. For immunocytochemistry, cells were permeabilized with 0.25% triton-X 100 and blocked with 10% goat serum. These cells were incubated with rabbit anti-PPARγ and mouse anti-RUNX2 primary antibodies in 1% goat serum at 4 °C for 18–24 h. Secondary antibody staining with goat anti-rabbit AF 532 and goat anti-mouse AF 488 was carried out in 1% goat serum at room temperature for 30 min. AF 633 phalloidin (0.26 µM in 1% goat serum) was used to stain F-actin during the secondary antibody incubation for 1.5 h at room temperature. Nuclear counterstaining with DAPI (2 µM in 1× PBS) was carried out for 5 min at room temperature. Cells were imaged via confocal microscopy. DIC imaging of the cells was also performed.
Nuclear to cytoplasmic ratios of PPARγ and RUNX2 were evaluated to assess activity of these transcription factors. Fluorescence intensity was assumed to correlate with expression levels of each protein. NIH Fiji software was utilized to sum the transcription factor staining z-slices taken for a given cell, yielding a separate 3D projection for either PPARγ or RUNX2. Corrected total cell fluorescence (CTCF) and a corrected nuclear fluorescence (CNF) were calculated using formulas 1 and 2 for each transcription factor by subtracting background staining. The nuclear to cytoplasmic ratio of the transcription factor was calculated using formulas 3–5. In these formulas, mean gray value (MGV) and area are obtained from NIH Fiji. MGV is the sum of the gray values (pixel intensity) of all the pixels in a selected area divided by the number of pixels.
| (1) |
| (2) |
| (3) |
| (4) |
| (5) |
NIH Fiji was utilized to quantify the diameter of lipid droplets present in DIC images of HMSCs. The projections of the lipid droplets in all cells were found to be elliptical. When the droplet projections were not perfect circles, the area of an ellipse was used to calculate the diameter of a circle with equivalent area. This equivalent diameter was taken as the lipid droplet diameter. The maximum cell height for each cell was also evaluated in NIH Fiji by using the orthogonal views feature for each F-actin fluorescence z-slice and measuring the maximum cell height.
Histochemical staining of lipid droplets and alkaline phosphatase of fixed cells was also performed. Fixed cells were treated with 60% isopropyl alcohol for 5 min, followed by room temperature incubation with oil red O lipid stain (1.8 mg mL−1 in 60% isopropyl alcohol) for 20 min. After removing oil red O solution and rinsing with 1× PBS, the cells were incubated with BCIP/NBT solution at 37 °C for 30 min to stain alkaline phosphatase. A DAPI counterstain was performed similar to immunocytochemistry. These cells were imaged with phase contrast microscopy.
2.5. Statistical Analysis
One-way analysis of variance (ANOVA) and posthoc analysis based on Tukey’s honestly significant difference (HSD) criterion were performed in MATLAB R2013b (MathWorks, Natick, MA) via the built-in anova1 and multcompare functions. Data are reported as mean ± standard deviation. For patterned samples, analysis was only conducted on cells that were restricted to the protein patterns. At least three repeats were performed for each experiment described.
3. RESULTS AND DISCUSSION
3.1. Developing Biomimetic Virtual Masks
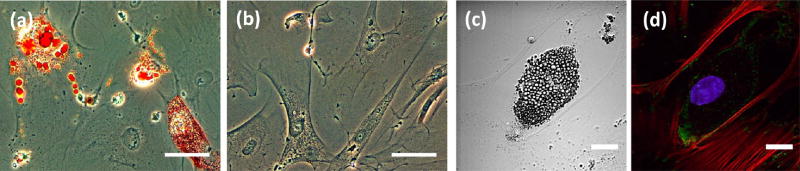
We used laser scanning lithography (LSL) to develop protein micropattern arrays, which were used to culture individual HMSCs. Previously, we reported on the development of the LSL technique as a facile method for patterning photoresists for subsequent use in soft lithography32 and have recently demonstrated uses of LSL in direct surface patterning of adhesive peptides and proteins.27,30,33 In contrast to traditional micropatterning techniques often employed in cell studies, such as microcontact printing, LSL does not require the use of a physical mask to generate micropatterns. Instead, LSL utilizes a virtual mask, a computerized representation of regions of interest (ROIs), that guides the movement of a laser and the laser shutter in order to expose only the ROIs to the laser used in patterning.29 The geometry of the micropatterns examined in this work ranged from common geometric shapes to those that mimicked the shape of mature adipocytes cultured in vitro. To develop the virtual masks for these patterns, we first examined human subcutaneous preadipocytes seeded on fibronectin in both adipogenic differentiation media and preadipocyte growth media. These cells were stained with a common lipid stain, oil red O, as seen in Figure 1. Oil red O staining was clearly visible in cells treated with differentiation media indicating the presence of mature adipocytes (Figure 1a). The low seeding density employed enabled imaging of individual cells, allowing us to focus on the shape adopted by individual differentiated cells devoid of significant cell-to-cell contact. As expected, preadipocytes in growth media did not stain positive for lipid inclusions as seen in Figure 1b.
Figure 1.
Human subcutaneous preadipocyte culture. (a, b) Phase contrast imaging of cells after 31 days in culture in (a) adipogenic differentiation media or in (b) preadipocyte growth media (red = oil red O lipid stain; scale bar = 100 µm). (c, d) Adipocyte of interest; (c) DIC image showing dense lipid droplets, (d) confocal image (red = F-actin, green = vinculin, blue = nucleus); scale bar = 25 µm.
Confocal and differential interference contrast (DIC) microscopy enabled the identification of a differentiated adipocyte of interest, and this image was then used to develop an adipocyte mimetic virtual mask. As with previous studies,34 DIC imaging distinguished mature adipocytes from undifferentiated cells via clear visualization of intracellular lipid content. Confocal imaging of F-actin and vinculin, along with the cell nuclei, allowed further visualization of the cellular structure. The average area of the differentiated preadipocytes imaged was 3678 ± 1278 µm2 with an average aspect ratio of 2.249 ± 0.789 and average circularity of 0.653 ± 0.165, where a circularity of 1 represents a perfect circle. An adipocyte representative of these average measurements was selected as the basis for the virtual mask in this study and is shown in Figure 1c, d. The aspect ratio of this cell was 1.846 along with a circularity of 0.727. In addition, this adipocyte had a total cell area of 4116 µm2. This cell area is significantly greater than the area of protein patterns that have been used in previous studies exploring the effects of geometric constraint on HMSC adipogenesis, which have typically been less than or equal to 2500 µm2.5,16 In general, smaller patterns have tended to favor adipogenesis; in fact, HMSCs confined to geometric patterns of 5000 µm2 or greater have previously been shown to lead to significant or complete osteogenesis of the differentiated cells.5,16,35 However, we hypothesized that properties of shape other than area may profoundly influence intracellular tension and thus, differentiation. Therefore, we chose to utilize a cell shape with a large area for our patterns.
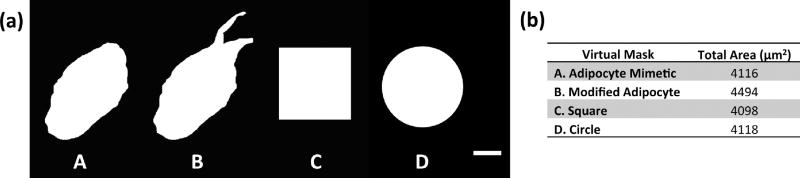
The adipocyte mimetic virtual mask was developed using the cell shape of the adipocyte in Figure 1c, d. NIH Fiji software was used to prepare a binary image of the entire cell area as seen in Figure 2. This binary image was then used as input for a MATLAB based segmentation algorithm;27,28 this algorithm segments the cell shape into adjacent horizontal quadrilateral regions to control laser scanning. In addition to the adipocyte mimetic pattern, we examined several other patterns in our study, as seen in Figure 2a. We developed a modified adipocyte virtual mask, based on the adipocyte mimetic mask, in which we added two projections (left projection with a high aspect ratio of 4.694; right projection with a moderate aspect ratio of 1.752) to the adipocyte mimetic mask, raising the overall aspect ratio of the mask to 2.193 and drastically reducing the overall circularity to 0.399. These modifications resemble lamellipodia and create regions of high tension, as has been demonstrated in studies examining traction forces generated in relationship to cell shape and aspect ratio.36,37 Square and circle virtual masks with similar areas to the adipocyte mimetic virtual mask were also developed. Figure 2b summarizes the areas of each of these four virtual masks.
Figure 2.
Virtual masks. (a) Virtual masks for patterning (A = adipocyte mimetic, B = modified adipocyte, C = square, D = circle; scale bar = 25 µm). (b) Virtual mask areas.
3.2. Biomimetic Protein Patterning
Metalized surfaces were prepared for patterning by depositing a binding layer of titanium (2 nm Ti), followed by a thin layer of gold (8 nm Au) on clean cover glass via electron beam evaporation. An oligo(ethylene glycol) (OEG6)-terminated alkanethiol was deposited on these metalized surfaces, forming a self-assembled monolayer (SAM). The terminal oligo(ethylene glycol) renders the surface protein resistant, and therefore, resistant to cell adhesion. These OEG6-alkanethiol SAM surfaces were subsequently used in LSL patterning to produce protein micropattern arrays. In the LSL technique, local heating of the Au-coated surface leads to disruption of the semicovalent Ausulfur bond which destabilizes the SAM and leads to localized desorption of the OEG6-alkanethiol in the irradiated regions, leaving behind bare Au.29,30 When exposed to a protein, the protein will adsorb only to the laser-exposed, bare Au regions, with undetectable adsorption to the OEG-terminated regions.30
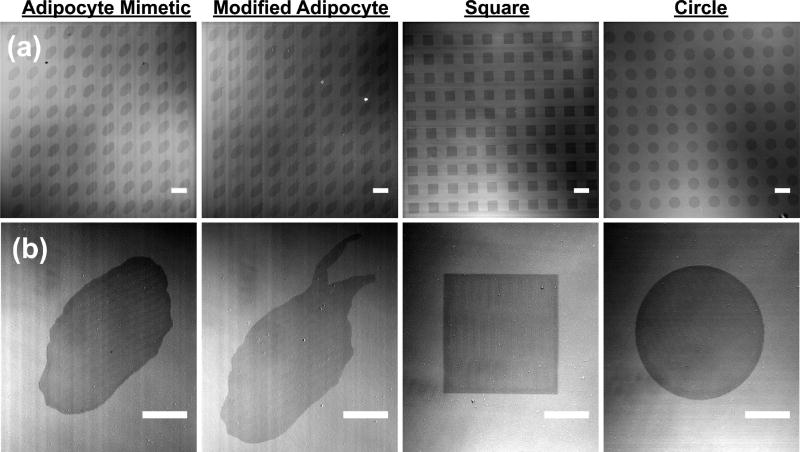
In our work, human plasma fibronectin was adsorbed directly to the bare Au micropatterns after LSL patterning. Thin Au films have strong absorption in the 520 to 560 nm wavelength range.38,39 Therefore, we used a 532 nm laser focused through a 40× oil immersion objective on a laser scanning confocal microscope for LSL patterning. The scanning of the laser across the surface was dictated by the virtual masks we developed, shown in Figure 2. We developed arrays of fibronectin patterns on the Au-coated glass surfaces, as shown in Figure 3 for each of these four geometries. Figure 3b shows a high-magnification image of the fibronectin functionalized patterns whose arrays are shown in Figure 3a. DIC microscopy was able to capture the contrast between the patterned and nonpatterned areas as seen in Figure 3. As LSL is based on thermal desorption, the radiation of heat from the laser focal point inevitably leads to some pattern enlargement. This enlargement can be controlled by laser scanning conditions (e.g., number of scanning iterations) or properties of the metal surface (e.g., thickness).30 Using a laser fluence of approximately 7.2 nJ µm−2 and 3 patterning iterations we obtained an acceptable average pattern enlargement of 2.8 ± 0.5%, 2.4 ± 0.2%, 3.1 ± 0.3%, and 2.5 ± 0.2% compared to the virtual mask for the adipocyte mimetic, modified adipocyte, square, and circle patterns, respectively.
Figure 3.
Fibronectin micropatterns. (a) DIC images of fibronectin functionalized micropattern arrays (scale bar = 100 µm). (b) DIC images of single fibronectin functionalized micropatterns (scale bar = 25 µm). Left to right: adipocyte mimetic, modified adipocyte, square, and circle patterns.
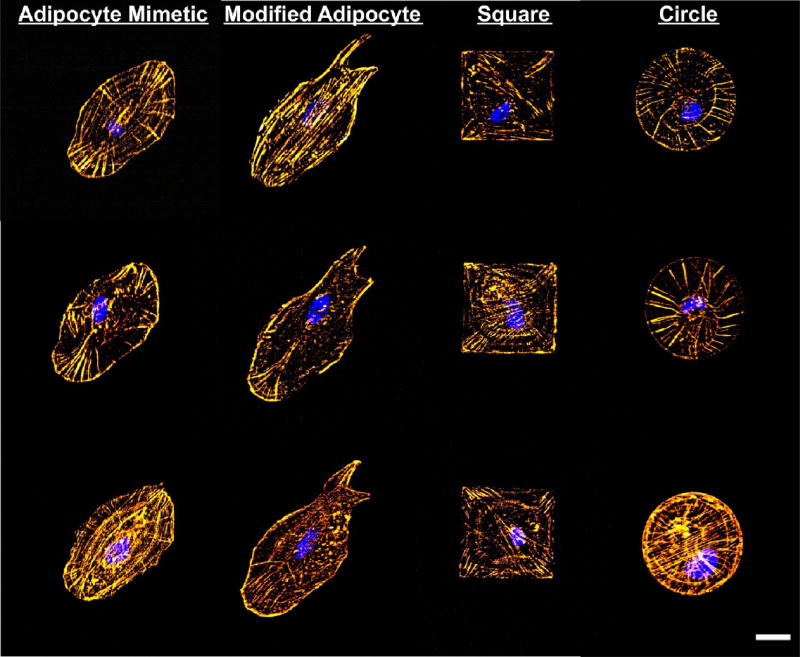
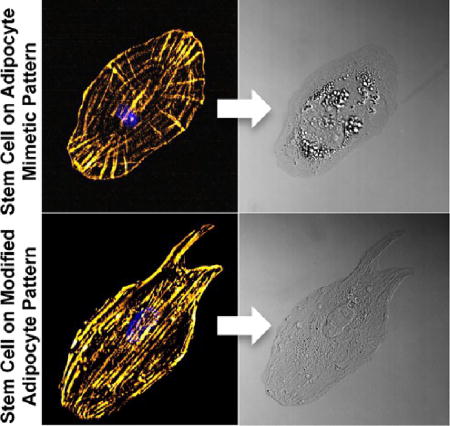
After fabricating arrays of these fibronectin micropatterns, we seeded bone marrow-derived HMSCs on the patterned surfaces, which selectively adhered to the fibronectin patterns. The cells were cultured in a mixed differentiation media composed of a 1:1 (v/v) adipogenic:osteogenic media mixture for 7 days before fixing and staining. Representative 3D projections of the confocal microscopy z-slices of HMSCs that were fixed and stained for F-actin and nuclei on these patterns are shown in Figure 4 (1 cell per pattern) following 7 days of culture. HMSCs were entirely confined to the patterned region in the case of the adipocyte mimetic, square, and circle patterns. On the modified adipocyte patterns, cells ranged from being perfectly confined to the pattern to bridging the two lamellipodia-reminiscent projections. The proximity of the projections (especially the region closest to the base on the projections) appeared to enable this bridging. It is also likely that the nonpatterned region lying between the two projections had decreased OEG6-alkanethiol SAM fidelity due to heat radiation during the LSL process. Only cells that lacked bridging of the lamellipodia-reminiscent projections were used in subsequent cell differentiation analysis. HMSCs on the different pattern types appeared to take on different actin cytoskeletal morphologies. For example, there is prominent elongated actin staining in the HMSCs that have filled the exact shape of the modified adipocyte pattern as well as in the corners of the HMSCs on the square patterns. In contrast, HMSCs on the adipocyte mimetic and circle patterns exhibit actin staining that appears as concentric contours rather than elongated prominent actin fibers that are seen predominantly in the HMSCs on the modified adipocyte and square patterns.
Figure 4.
Actin cytoskeleton of stem cells on fibronectin patterns. Left to right: adipocyte mimetic, modified adipocyte, square, and circle patterns. Gold = F-actin, blue = nucleus, scale bar = 25 µm.
3.3. Stem Cell Behavior on Patterned Surfaces
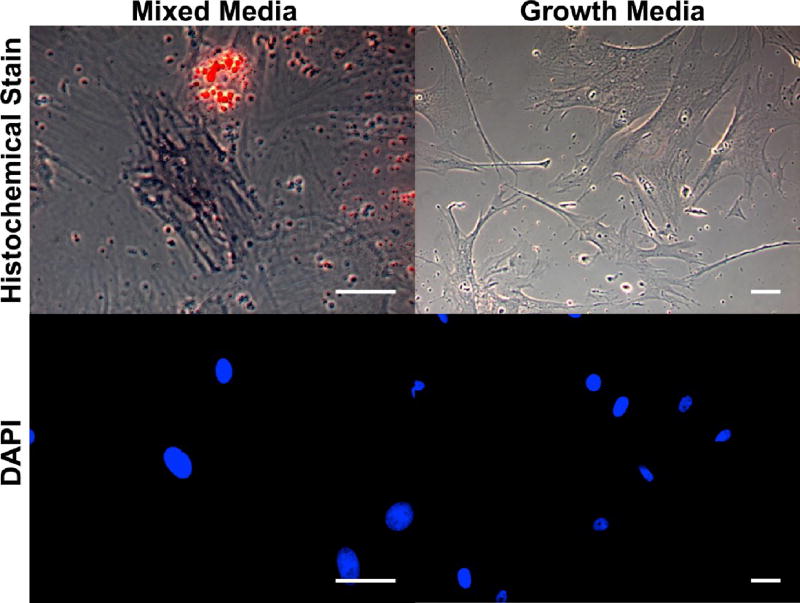
Prior to characterization of HMSC differentiation on the patterned surfaces, HMSCs were seeded on nonpatterned controls in order to assess their differentiation potential in mixed 1:1 (v/v) adipogenic:osteogenic differentiation media after 7 days in culture. These controls were fabricated with 2 nm of Ti followed by 8 nm Au on which fibronectin was directly adsorbed to provide a surface highly similar to that presented within the micropatterns. Cells on these control surfaces in adipogenic:osteogenic differentiation media demonstrated the ability to undergo adipogenesis (staining positively for lipids via oil red O staining) and osteogenesis (staining positively for alkaline phosphatase, an early marker of osteogenesis2) as shown in Figure 5. Approximately 10% of cells on these control surfaces stained positively for oil red O, 20% stained positively for alkaline phosphatase, and the remaining cells were not positive for either stain. Cells cultured in growth media on nonpatterned fibronectin controls did not stain positively for these markers of differentiation.
Figure 5.
HMSCs on fibronectin functionalized substrates (2 nm titanium/8 nm gold) following 7 days in culture. Top row: histochemical stain (red = oil red O, purple = alkaline phosphatase) of cells in 1:1 (v/v) adipogenic:osteogenic differentiation media (left) and growth media (right). Bottom row: corresponding fluorescence images of nuclei. Scale bars = 100 µm.
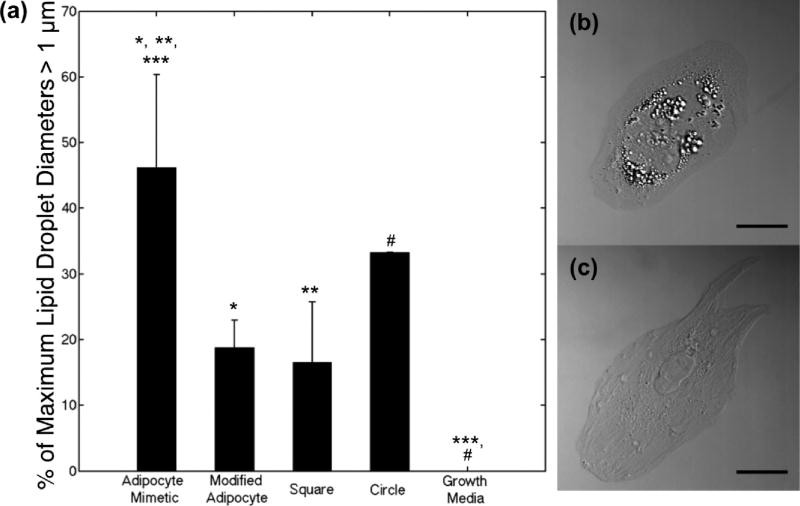
Adipogenesis and osteogenesis of HMSCs on the fibronectin micropatterns was quantified by assessing lipid content and alkaline phosphatase staining, respectively, following 7 days in mixed 1:1 (v/v) adipogenic:osteogenic differentiation media. As has previously been observed, alkanethiol-based micropatterns tend to lose stability following 1 week, restricting measurements to this time period.16 Lipid droplets are ubiquitous in a large variety of mammalian cells, but lipid droplets greater than 1 µm in diameter are characteristic of adipocytes.40 Therefore, we quantified the number of cells on patterns with lipid droplet diameters greater than 1 µm, equating this to HMSCs undergoing adipogenic differentiation. DIC z-slices were imaged through the entire cell height of each HMSC. It was found that individual cells on patterns had similarly sized lipid droplet throughout the entire volume of the cell by examining each z-slice. Figure 6 shows the results of our analysis for z-slices taken at the base of the cell closest to the patterned surface (representative of the behavior of the entire cell), along with representative DIC images of HMSCs on the adipocyte mimetic (Figure 6b) and modified adipocyte (Figure 6c) patterns. Alkaline phosphatase staining was not observed in any of the HMSCs cultured on the protein micropatterns. This was an interesting result as cells cultured in the presence of differentiation media and no patterns were shown to stain positively for both alkaline phosphatase and lipids (as seen in Figure 5).
Figure 6.
Lipid droplet analysis of HMSCs following 7 days in culture. (a) Percentage of HMSCs displaying lipid droplets with maximum diameter greater than 1 µm on adipocyte mimetic (31 cells), modified adipocyte (33 cells), square (23 cells), and circle (33 cells) patterns, as well as cells cultured in growth media (no patterns, 32 cells). Results for at least 3 samples per pattern type are shown. Data are shown as mean ± standard deviation; significance was calculated using one-way ANOVA with a Tukey posthoc analysis (p < 0.05: adipocyte mimetic compared to modified adipocyte (*), square (**), and growth media (***); circle compared to growth media (#)). Representative DIC image of HMSC on an (b) adipocyte mimetic and a (c) modified adipocyte fibronectin pattern (scale bar = 25 µm).
Figure 6 shows that approximately 46% of HMSCs displaying lipid droplets on adipocyte mimetic patterns displayed a maximum lipid droplet diameter of greater than 1 µm as compared to 19% of cells on modified adipocyte patterns with higher stress regions. Cells on square patterns had a similar response to that of cells on the modified adipocyte patterns, with approximately 17% of these cells displaying maximum lipid droplet diameters greater than 1 µm, while cells on circle patterns had a greater adipogenic response than cells on the modified adipocyte and square patterns, but less than the adipocyte mimetic patterns (approximately 33% displayed lipid droplets with diameters greater than 1 µm). Control cells cultured in growth media on fibronectin-coated substrates that displayed lipid droplets had a maximum lipid droplet diameter of 0.73 ± 0.16 µm (n = 32). The average area of these nonpatterned cells was 6270 ± 1894 µm2. Nonpatterned control cells with areas of less than 4,500 µm2 (similar to the patterns explored in this work) had comparable maximum lipid droplet diameters of 0.77 ± 0.13 µm, suggesting that cell area alone had little impact on adipogenesis. HMSCs on adipocyte mimetic patterns displayed the maximum adipogenic response of the patterns examined. Previous studies have shown adipogenesis to occur preferentially on circle geometries while increasing aspect ratios have reduced adipogenesis and increased osteogenesis.16 Our results, however, suggest that the adipocyte mimetic pattern we have examined, despite having an aspect ratio greater than our circle pattern (1.846 versus 1), may lead to improved adipogenesis.
We can attribute the differences in adipogenic differentiation to variations in the cytoskeletal tension experienced by HMSCs on the different patterns examined in this work, which can be related to the actin cytoskeletal structure.20,21 As seen in Figure 4, the actin cytoskeleton of HMSCs on the various patterns varies considerably. The elongated fibers observed in HMSCs completely confined to the modified adipocyte pattern, suggest overall greater cytoskeletal tension than HMSCs on the adipocyte mimetic pattern. It has previously been shown that the adipogenic response of HMSCs cultured on protein micropatterns can be affected by the presence of actin inhibitors, including cytochalasin D.5,16 We found that exposing the cells to cytochalasin D increased adipogenesis (i.e., percentage of cells on patterns with maximum lipid droplet diameters greater than 1 µm) for cells cultured on both the adipocyte mimetic and the modified adipocyte pattern as compared to cells cultured without the inhibitor. The impact of the inhibitor appeared to be more significant for cells on the modified adipocyte pattern, where cytoskeletal tension should be enhanced, with the inhibitor treatment increasing adipogenesis by 72.5 ± 15.8% as compared to a 24.9 ± 7.6% increase in adipogenesis for HMSCs treated with cytochalasin D on the adipocyte mimetic pattern. In the presence of this actin inhibitor, we expect that the elongated actin fibers originating in the projections of the modified adipocyte patterns would not be able to form, lowering the cytoskeletal tension experienced by cells on these patterns and increasing adipogenesis. 3D projections of the confocal microscopy z-slices of HMSCs that were treated with cytochalasin D on the adipocyte mimetic and modified adipocyte patterns confirm the lack of elongated F-actin fiber formation, as shown in Figure S1.
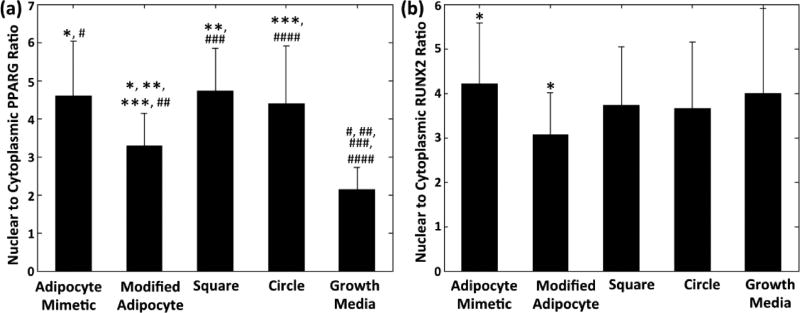
Expression of the transcription factors PPARγ and RUNX2 was examined on HMSCs cultured on our fibronectin patterns in the presence of mixed adipogenic:osteogenic differentiation media and compared to cells cultured in growth media on nonpatterned fibronectin-coated controls. PPARγ is a key regulator of several adipogenesis associated cellular activities and is activated early in adipogenic differentiation,41,42 while RUNX2 activity is associated with osteogenesis and seen to increase early in osteogenic differentiation.43 As a measure of activity, we quantified the nuclear to cytoplasmic intensity of these transcription factors following 7 days in culture as shown in Figure 7.
Figure 7.
PPARγ and RUNX2 nuclear localization. (a) PPARγ nuclear:cytoplasmic ratio (p < 0.05: adipocyte mimetic (*), square (**), and circle (***) compared to modified adipocyte; adipocyte mimetic (#), modified adipocyte (##), square (###), and circle (####) compared to growth media). (b) RUNX2 nuclear:cytoplasmic ratio (p < 0.05: adipocyte mimetic (*) compared with modified adipocyte). Data for HMSCs on adipocyte mimetic (n = 35), modified adipocyte (n = 38), square (n = 24), and circle (n = 34) patterns, as well as cells cultured in growth media (no patterns, n = 33) is shown. Data is shown as mean ± standard deviation; significance was calculated using one-way ANOVA with a Tukey posthoc analysis.
PPARγ nuclear localization was found to be significantly greater in HMSCs cultured on any of the patterns explored in the mixed differentiation media compared to HMSCs cultured in growth media. There was no statistical difference in PPARγ nuclear localization for HMSCs restricted to the adipocyte mimetic, square, and circle patterns. However, cells cultured on the modified adipocyte patterns were found to have a significantly lower level of PPARγ nuclear localization compared to the other patterns examined. Transcription factors other than PPARγ, which are important in lipid droplet formation, may potentially be regulated by cytoskeletal tension and cause the differences in adipogenesis observed here on the various protein patterns explored. It is also possible that PPARγ nuclear localization levels may vary between pattern conditions at earlier times than the 7 days examined here. RUNX2 nuclear localization was found not to differ for cells cultured in growth media at 7 days compared to those cultured in mixed differentiation medium on any of the four patterns examined. These results support the lack of alkaline phosphatase staining observed in the HMSCs cultured on the fibronectin patterns.
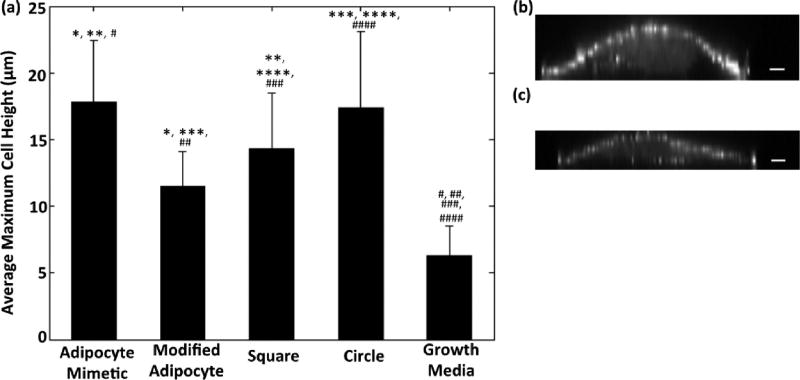
Finally, we compared the maximum cell height of HMSCs on patterns in adipogenic:osteogenic differentiation media following 7 days in culture to that of cells in growth media on nonpatterned fibronectin controls (Figure 8). Our results agree with previous work, which has shown that adipogenesis of adipose-derived stem cells is positively correlated with increasing cell height.23 HMSCs in differentiation media on all patterns we examined were found to have greater cell heights compared to cells in growth media. HMSCs on the adipocyte mimetic and circle patterns had the greatest cell heights (with no statistical difference between the two groups), whereas cells on the modified adipocyte and square patterns had the lowest cell heights. In general, HMSCs on fibronectin patterns that promoted greater adipogenesis as quantified by lipid droplet diameter had larger maximum cell heights.
Figure 8.
Average maximum HMSC height at 7 days in culture. (a) HMSCs on adipocyte mimetic (n = 33), modified adipocyte (n = 35), square (n = 24), and circle (n = 33) patterns, as well as cells cultured in growth media (no patterns, n = 35) (p < 0.05: modified adipocyte (*) and square (**) compared with adipocyte mimetic; adipocyte mimetic (#), modified adipocyte (##), square (###), and circle (####) compared to growth media; modified adipocyte (***) and square (****) compared to circle). (b) YZ profile of HMSC on adipocyte mimetic fibronectin pattern (scale bar = 5 µm). (c) YZ profile of HMSC on modified adipocyte fibronectin pattern (scale bar = 5 µm). Data are shown as mean ± standard deviation; significance was calculated using one-way ANOVA with a Tukey posthoc analysis.
4. CONCLUSION
Our work suggests that HMSCs restricted to adopting the shape of a differentiated adipocyte may preferentially undergo adipogenesis. We used LSL to generate arrays of adipocyte mimetic, modified adipocyte, square, and circle fibronectin patterns. HMSCs were cultured on these patterns, and differentiation was evaluated by quantifying lipid droplet diameters, staining for alkaline phosphatase and lipids, and examining PPARγ and RUNX2 nuclear localization. Overall, cells on adipocyte mimetic fibronectin patterns showed a greater occurrence of large lipid droplets indicative of adipogenesis, compared with cells on the other patterns examined. We attributed our findings to variations in cytoskeletal tension experienced by cells on these different shapes. This study demonstrates that the use of biomimetic geometries can greatly enhance HMSC differentiation toward a desired lineage. Despite their tremendous promise, currently, only one Food and Drug Administration (FDA) approved stem cell product exists.44 The results of this study have implications that could potentially help increase the number of FDA approved products, by leading to the design of improved HMSC culture techniques and therapies involving injection of HMSCs following in vitro culture, as well as the design of appropriate HMSC scaffolds.
Supplementary Material
Acknowledgments
The authors acknowledge the use of facilities in the Department of Bioengineering and the Nanofabrication Cleanroom at Rice University. A. Shukla acknowledges funding from the National Institute of General Medical Sciences of the NIH under Award F32GM103194. J.L. West and M.E. Dickinson acknowledge support from the NIH under Award R01HL097520 and R01EB016629. J.C.C. acknowledges support from NIH Award T32HL007676. J.H.S. acknowledges support through postdoctoral fellowships from the NIH Nanobiology Training Program of the Keck Center of the Gulf Coast Consortia (T32DK070121). The content of this publication is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
ASSOCIATED CONTENT
Supporting Information
Actin cytoskeleton micrographs of stem cells on fibronectin patterns treated with actin polymerization inhibitor, cytochalasin D (PDF)
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
References
- 1.da Silva Meirelles L, Chagastelles PC, Nardi NB. Mesenchymal Stem Cells Reside in Virtually All Post-Natal Organs and Tissues. J. Cell Sci. 2006;119:2204–2213. doi: 10.1242/jcs.02932. [DOI] [PubMed] [Google Scholar]
- 2.Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S, Marshak DR. Multilineage Potential of Adult Human Mesenchymal Stem Cells. Science. 1999;284:143–147. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 3.Sharma RR, Pollock K, Hubel A, McKenna D. Mesenchymal Stem or Stromal Cells: A Review of Clinical Applications and Manufacturing Practices. Transfusion. 2014;54:1418–1437. doi: 10.1111/trf.12421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bolli R, Chugh AR, D’Amario D, Loughran JH, Stoddard MF, Ikram S, Beache GM, Wagner SG, Leri A, Hosoda T, Sanada F, Elmore JB, Goichberg P, Cappetta D, Solankhi NK, Fahsah I, Rokosh DG, Slaughter MS, Kajstura J, Anversa P. Cardiac Stem Cells in Patients With Ischaemic Cardiomyopathy (SCIPIO): Initial Results of a Randomised Phase 1 Trial. Lancet. 2011;378:1847–1857. doi: 10.1016/S0140-6736(11)61590-0. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 5.McBeath R, Pirone DM, Nelson CM, Bhadriraju K, Chen CS. Cell Shape, Cytoskeletal Tension, and RhoA Regulate Stem Cell Lineage Commitment. Dev. Cell. 2004;6:483–495. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- 6.Heldman AW, DiFede DL, Fishman JE, Zambrano JP, Trachtenberg BH, Karantalis V, Mushtaq M, Williams AR, Suncion VY, NcNiece IK, Ghersin E, Soto V, Lopera G, Miki R, Wilens H, Hendel R, Mitrani R, Pattany P, Feigenbaum G, Oskouei B, Byrnes J, Lowery MH, Sierra J, Pujol MV, Delgado C, Gonzalez PJ, Rodriguez JE, Bagno LL, Rouy D, Altman P, Foo CW, da Silva J, Anderson E, Schwarz R, Mendizabal A, Hare JM. Transendocardial Mesenchymal Stem Cells and Mononuclear Bone Marrow Cells for Ischemic Cardiomyopathy: The TAC-HFT Randomized Trial. JAMA. 2014;311:62–73. doi: 10.1001/jama.2013.282909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horwitz EM, Gordon PL, Koo WKK, Marx JC, Neel MD, McNall RY, Muul L, Hofmann T. Isolated Allogeneic Bone Marrow-Derived Mesenchymal Cells Engraft and Stimulate Growth in Children With Osteogenesis Imperfecta: Implications for Cell Therapy of Bone. Proc. Natl. Acad. Sci. U. S. A. 2002;99:8932–8937. doi: 10.1073/pnas.132252399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Götherström C, Westgren M, Shaw SWS, Åström E, Biswas A, Byers PH, Mattar CNZ, Graham GE, Taslimi J, Ewald U, Fisk NM, Yeoh AEJ, Lin J-L, Cheng P-J, Choolani M, Le Blanc K, Chan JKY. Pre- and Postnatal Transplantation of Fetal Mesenchymal Stem Cells in Osteogenesis Imperfecta: A Two-Center Experience. Stem Cells Transl. Med. 2014;3:255–264. doi: 10.5966/sctm.2013-0090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abdallah BM, Kassem M. Human Mesenchymal Stem Cells: From Basic Biology to Clinical Applications. Gene Ther. 2008;15:109–16. doi: 10.1038/sj.gt.3303067. [DOI] [PubMed] [Google Scholar]
- 10.Zhao J, Zhang N, Prestwich GD, Wen X. Recruitment of Endogenous Stem Cells for Tissue Repair. Macromol. Biosci. 2008;8:836–842. doi: 10.1002/mabi.200700334. [DOI] [PubMed] [Google Scholar]
- 11.Ko IK, Lee SJ, Atala A, Yoo JJ. In situ Tissue Regeneration Through Host Stem Cell Recruitment. Exp. Mol. Med. 2013;45:e57. doi: 10.1038/emm.2013.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chamberlain G, Fox J, Ashton B, Middleton J. Concise Review: Mesenchymal Stem Cells: Their Phenotype, Differentiation Capacity, Immunological Features, and Potential for Homing. Stem Cells. 2007;25:2739–2749. doi: 10.1634/stemcells.2007-0197. [DOI] [PubMed] [Google Scholar]
- 13.Jaiswal N, Haynesworth SE, Caplan AI, Bruder SP. Osteogenic Differentiation of Purified, Culture-Expanded Human Mesenchymal Stem Cells In Vitro. J. Cell. Biochem. 1997;64:295–312. [PubMed] [Google Scholar]
- 14.Hwang NS, Zhang C, Hwang Y-S, Varghese S. Mesenchymal Stem Cell Differentiation and Roles in Regenerative Medicine. Wiley Interdiscip. Rev. Syst. Biol. Med. 2009;1:97–106. doi: 10.1002/wsbm.26. [DOI] [PubMed] [Google Scholar]
- 15.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix Elasticity Directs Stem Cell Lineage Specification. Cell. 2006;126:77–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 16.Kilian KA, Bugarija B, Lahn BT, Mrksich M. Geometric Cues for Directing the Differentiation of Mesenchymal Stem Cells. Proc. Natl. Acad. Sci. U. S. A. 2010;107:4872–4877. doi: 10.1073/pnas.0903269107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang C, Tibbitt MW, Basta L, Anseth KS. Mechanical Memory and Dosing Influence Stem Cell Fate. Nat. Mater. 2014;13:645–652. doi: 10.1038/nmat3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ruiz SA, Chen CS. Emergence of Patterned Stem Cell Differentiation Within Multicellular Structures. Stem Cells. 2008;26:2921–2927. doi: 10.1634/stemcells.2008-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wan LQ, Kang SM, Eng G, Grayson WL, Lu XL, Huo B, Gimble J, Guo XE, Mow VC, Vunjak-Novakovic G. Geometric Control of Human Stem Cell Morphology and Differentiation. Integr. Biol. 2010;2:346–353. doi: 10.1039/c0ib00016g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eyckmans J, Boudou T, Yu X, Chen; Christopher S. A Hitchhiker’s Guide to Mechanobiology. Dev. Cell. 2011;21:35–47. doi: 10.1016/j.devcel.2011.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choi CK, Breckenridge MT, Chen CS. Engineered Materials and the Cellular Microenvironment: A Strengthening Interface Between Cell Biology and Bioengineering. Trends Cell Biol. 2010;20:705–14. doi: 10.1016/j.tcb.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Treiser MD, Yang EH, Gordonov S, Cohen DM, Androulakis IP, Kohn J, Chen CS, Moghe PV. Cytoskeleton-Based Forecasting of Stem Cell Lineage Fates. Proc. Natl. Acad. Sci. U. S. A. 2010;107:610–615. doi: 10.1073/pnas.0909597107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.González-Cruz RD, Fonseca VC, Darling EM. Cellular Mechanical Properties Reflect the Differentiation Potential of Adipose-Derived Mesenchymal Stem Cells. Proc. Natl. Acad. Sci. U. S. A. 2012;109:E1523–E1529. doi: 10.1073/pnas.1120349109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee J, Abdeen AA, Kilian KA. Rewiring Mesenchymal Stem Cell Lineage Specification by Switching the Biophysical Microenviron-ment. Sci. Rep. 2014;4 doi: 10.1038/srep05188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guvendiren M, Burdick JA. Stiffening Hydrogels to Probe Short- and Long-Term Cellular Responses to Dynamic Mechanics. Nat. Commun. 2012;3:792. doi: 10.1038/ncomms1792. [DOI] [PubMed] [Google Scholar]
- 26.Yao X, Peng R, Ding J. Effects of Aspect Ratios of Stem Cells on Lineage Commitments With and Without Induction Media. Biomaterials. 2013;34:930–939. doi: 10.1016/j.biomaterials.2012.10.052. [DOI] [PubMed] [Google Scholar]
- 27.Slater JH, Culver JC, Long BL, Hu CW, Hu J, Birk TF, Qutub AA, Dickinson ME, West JL. Recapitulation and Modulation of the Cellular Architecture of a User-Chosen Cell of Interest Using Cell-Derived, Biomimetic Patterning. ACS Nano. 2015;9:6128. doi: 10.1021/acsnano.5b01366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Culver JC, Hoffmann JC, Poché RA, Slater JH, West JL, Dickinson ME. Three-Dimensional Biomimetic Patterning in Hydrogels to Guide Cellular Organization. Adv. Mater. 2012;24:2344–2348. doi: 10.1002/adma.201200395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Slater JH, West JL. In: Methods in Cell Biology. Matthieu P, Manuel T, editors. Academic Press; Waltham, MA: 2014. pp. 93–217. Chapter 11. [Google Scholar]
- 30.Slater JH, Miller JS, Yu SS, West JL. Fabrication of Multifaceted Micropatterned Surfaces with Laser Scanning Lithography. Adv. Funct. Mater. 2011;21:2876–2888. doi: 10.1002/adfm.201100297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen CS, Alonso JL, Ostuni E, Whitesides GM, Ingber DE. Cell Shape Provides Global Control of Focal Adhesion Assembly. Biochem. Biophys. Res. Commun. 2003;307:355–361. doi: 10.1016/s0006-291x(03)01165-3. [DOI] [PubMed] [Google Scholar]
- 32.Miller JS, Béthencourt MI, Hahn M, Lee TR, West JL. Laser-Scanning Lithography (LSL) for the Soft Lithographic Patterning of Cell-Adhesive Self-Assembled Monolayers. Biotechnol. Bioeng. 2006;93:1060–1068. doi: 10.1002/bit.20809. [DOI] [PubMed] [Google Scholar]
- 33.Hahn MS, Miller JS, West JL. Laser Scanning Lithography for Surface Micropatterning on Hydrogels. Adv. Mater. 2005;17:2939–2942. [Google Scholar]
- 34.Nagayama M, Uchida T, Gohara K. Temporal and Spatial Variations of Lipid Droplets During Adipocyte Division and Differentiation. J. Lipid Res. 2007;48:9–18. doi: 10.1194/jlr.M600155-JLR200. [DOI] [PubMed] [Google Scholar]
- 35.Peng R, Yao X, Cao B, Tang J, Ding J. The Effect of Culture Conditions on the Adipogenic and Osteogenic Inductions of Mesenchymal Stem Cells on Micropatterned Surfaces. Biomaterials. 2012;33:6008–6019. doi: 10.1016/j.biomaterials.2012.05.010. [DOI] [PubMed] [Google Scholar]
- 36.Oakes PW, Banerjee S, Marchetti MC, Gardel ML. Geometry Regulates Traction Stresses in Adherent Cells. Biophys. J. 2014;107:825–833. doi: 10.1016/j.bpj.2014.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rape AD, Guo W-h, Wang Y-l. The Regulation of Traction Force in Relation to Cell Shape and Focal Adhesions. Biomaterials. 2011;32:2043–2051. doi: 10.1016/j.biomaterials.2010.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xenogiannopoulou E, Iliopoulos K, Couris S, Karakouz T, Vaskevich A, Rubinstein I. Third-Order Nonlinear Optical Response of Gold-Island Films. Adv. Funct. Mater. 2008;18:1281–1289. [Google Scholar]
- 39.Gupta G, Tanaka D, Ito Y, Shibata D, Shimojo M, Furuya K, Mitsui K, Kajikawa K. Absorption Spectroscopy of Gold Nanoisland Films: Optical and Structural Characterization. Nano-technology. 2009;20:025703. doi: 10.1088/0957-4484/20/2/025703. [DOI] [PubMed] [Google Scholar]
- 40.Suzuki M, Shinohara Y, Ohsaki Y, Fujimoto T. Lipid Droplets: Size Matters. Microscopy. 2011;60:S101–S116. doi: 10.1093/jmicro/dfr016. [DOI] [PubMed] [Google Scholar]
- 41.Chawla A, Schwarz E, Dimaculangan D, Lazar M. Peroxisome Proliferator-Activated Receptor (PPAR) Gamma: Adipose-Predominant Expression and Induction Early in Adipocyte Differentiation. Endocrinology. 1994;135:798–800. doi: 10.1210/endo.135.2.8033830. [DOI] [PubMed] [Google Scholar]
- 42.Rosen ED, Sarraf P, Troy AE, Bradwin G, Moore K, Milstone DS, Spiegelman BM, Mortensen RM. PPARγ Is Required for the Differentiation of Adipose Tissue In Vivo and In Vitro. Mol. Cell. 1999;4:611–617. doi: 10.1016/s1097-2765(00)80211-7. [DOI] [PubMed] [Google Scholar]
- 43.James AW. Review of Signaling Pathways Governing MSC Osteogenic and Adipogenic Differentiation. Scientifica. 2013;2013:684736. doi: 10.1155/2013/684736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Allison M. Hemacord Approval May Foreshadow Regulatory Creep for HSC Therapies. Nat. Biotechnol. 2012;30:304–304. doi: 10.1038/nbt0412-304. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.