Abstract
Amyotrophic lateral sclerosis (ALS) is a rapidly progressing neurodegenerative disease characterized by motor neuron loss, leading to paralysis and death 2–5 years following disease onset1. Nearly all ALS patients contain aggregates of the RNA-binding protein TDP-43 in the brain and spinal cord2, and rare mutations in the gene encoding TDP-43 can cause ALS3. There are no effective TDP-43-directed therapies for ALS or related TDP-43 proteinopathies, such as frontotemporal dementia (FTD). Antisense oligonucleotides (ASOs) and RNA interference approaches are emerging as attractive therapeutic strategies in neurological diseases4. Indeed, treating a rodent model of inherited ALS (caused by a mutation in SOD1) with ASOs to SOD1 significantly slowed disease progression5. But since SOD1 mutations account for only ~2–5% of ALS cases, additional therapeutic strategies are needed. Silencing TDP-43 itself is probably not warranted given its critical cellular functions1,6 Here we present an unexpectedly powerful alternative therapeutic strategy for ALS, by targeting ataxin 2. Lowering ataxin 2 suppresses TDP-43 toxicity in yeast and flies7, and intermediate-length polyglutamine expansions in the ataxin 2 gene increase risk of ALS7,8. We used two independent approaches to test whether reducing ataxin 2 levels could mitigate disease in a mouse model of TDP-43 proteinopathy9. First, we crossed ataxin 2 knockout mice to TDP-43 transgenic mice. Lowering ataxin 2 reduced TDP-43 aggregation, had a dramatic effect on survival and improved motor function. Second, in a more therapeutically applicable approach, we administered ASOs targeting ataxin 2 to the central nervous system of TDP-43 mice. This single treatment markedly extended survival. Because TDP-43 aggregation is a component of nearly all ALS cases6, targeting ataxin 2 could represent a broadly effective therapeutic strategy.
To test the hypothesis that reducing ataxin 2 levels can rescue neurodegenerative phenotypes caused by TDP-43 accumulation, we first used a genetic approach. There are several transgenic mouse lines that express wild type or mutant TDP-43, using various strategies10. We selected a mouse line expressing human wild type (WT) TDP-43 under control of the Thy1 promoter, which drives pan-neuronal expression starting at around postnatal day seven (P7). We chose this mouse line because it presents robust and consistent phenotypes caused by abnormal TDP-43 accumulation. Whereas mice hemizygous for the TDP-43 transgene (TDP-43Tg/+) are viable, fertile, and grossly normal, mice harboring two copies of this transgene (TDP-43Tg/Tg) display profound motor dysfunction, resulting in an inability to walk around P21 and death around P249,11. In this study, we euthanized the animals when they were no longer able to right themselves. This humane euthanasia endpoint is a distinct advantage of this TDP-43 mouse model because it is directly dependent on motor dysfunction, an ALS-relevant phenotype. The degenerating neurons in brain and spinal cord of TDP-43Tg/Tg mice contain ubiquitinated and phosphorylated TDP-43 aggregates, the pathological hallmark of ALS patients2. This rapidly progressing phenotype provided a powerful readout of disease suppression to test potential therapeutic interventions. To lower ataxin 2 we used two independently generated lines of ataxin 2 knockout mice on different genetic backgrounds (see online methods). Heterozygous (Atxn2+/−) and homozygous (Atxn2−/−) mice are viable and fertile, with Atxn2−/− mice exhibiting some mid-life onset obesity phenotypes12,13. We crossed TDP-43Tg/+ mice with Atxn2+/− mice to produce TDP-43Tg/+Atxn2+/− offspring and then intercrossed these mice to produce TDP-43Tg/TgAtxn2+/+, TDP-43Tg/TgAtxn2+/−, TDP-43Tg/TgAtxn2−/−, and WT littermates (see online methods).
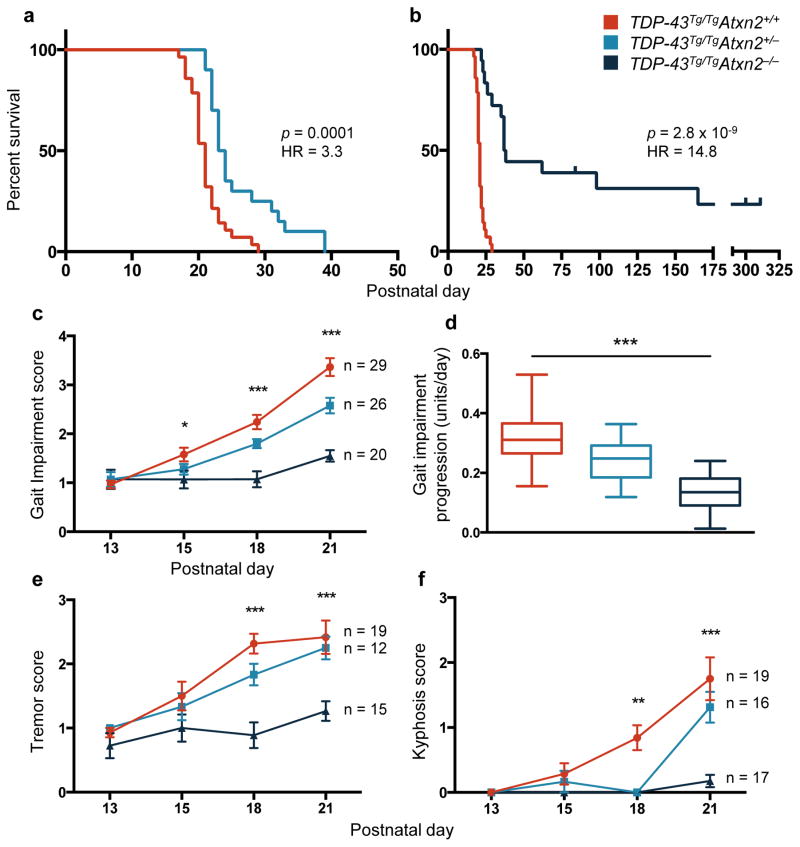
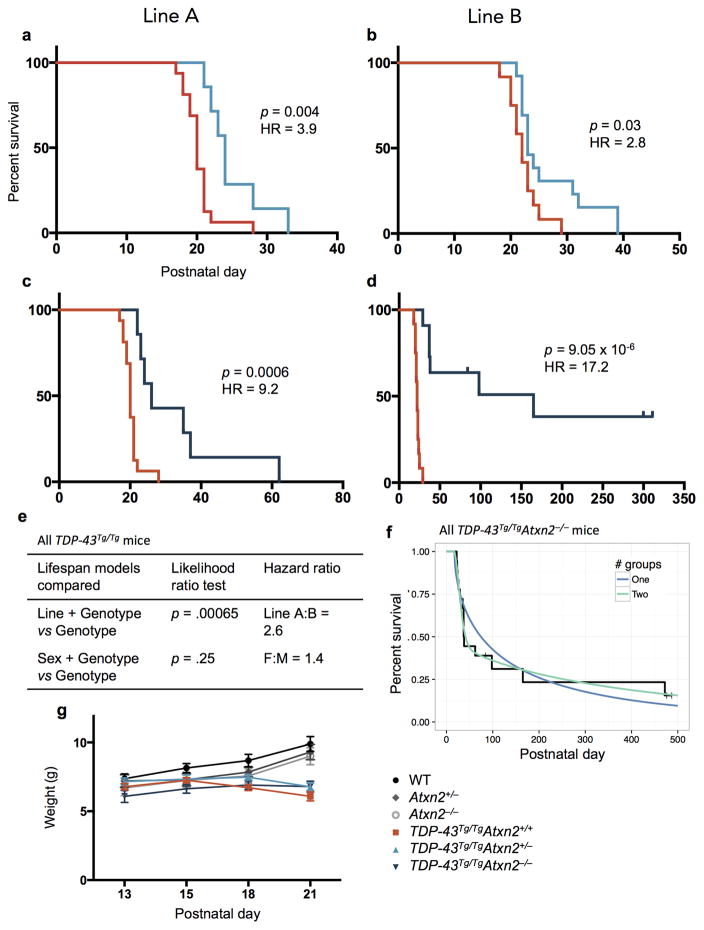
Decreased levels of ataxin 2 in TDP-43Tg/TgAtxn2+/− mice significantly improved lifespan compared to TDP-43Tg/TgAtxn2+/+ (Fig. 1a) and complete removal of ataxin 2 in TDP-43Tg/TgAtxn2−/− mice resulted in a dramatic 80% improvement in median lifespan (Fig. 1b), with several TDP-43Tg/TgAtxn2−/− mice surviving longer than 300 days. None of the TDP-43Tg/TgAtxn2+/+ mice survived longer than 29 days. We observed significant lifespan extension with ataxin 2 reduction in both mouse lines we created (Extended Data Fig. 1 a–d). Within the entire TDP-43Tg/TgAtxn2−/− population, we found evidence for two groups of responders (strong and weak), and the genetic background of the mice significantly contributed to this variability (Cox proportional hazards p = .002, line A:B HR = 6.8; Extended Data Fig. 1f).
Figure 1. Reducing levels of ataxin 2 extends lifespan, improves motor function, and slows the rate of disease progression in TDP-43 transgenic mice.
a,b) Kaplan-Meier survival curves comparing survival of TDP-43Tg/TgAtxn2+/+ (n = 28), TDP-43Tg/TgAtxn2+/− (n = 20) and TDP-43Tg/TgAtxn2−/− (n = 18) mice. Curves were compared by log rank test, and effect size was estimated by a Cox proportional hazards model (HR = hazard ratio). c) The gait impairment score of TDP-43Tg/Tg mice worsened with age, but this was attenuated by reducing ataxin 2. d) A line was fit to each mouse’s gait scores over time and the slope of that line was plotted, grouping animals by genotype. The error bars are min to max, and the three groups were compared by ordinary one-way ANOVA. e,f) Tremor and kyphosis scores were also decreased by lowering ataxin 2 in TDP-43Tg/Tg mice. c,e,f) At each age, the three genotypes were compared by ordinary one-way ANOVA. Means are plotted, and error bars indicate S.E.M. *p < .05, **p < .01, ***p < .001.
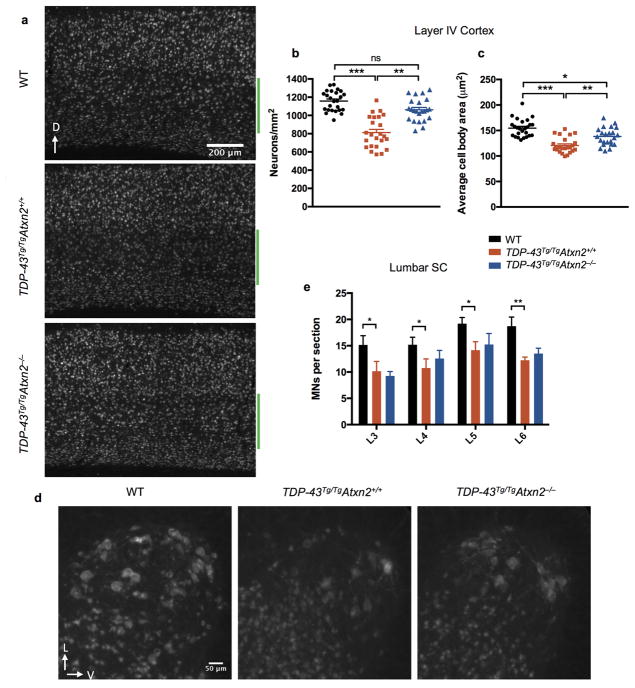
In addition to extending survival, reducing ataxin 2 slowed disease progression and improved motor function. All mice were able to walk efficiently by P13 (SI Video 1, 2). As previously described9, TDP-43Tg/Tg mice started to display a mild tremor and had some difficulty walking around P15, which steadily progressed to a severe tremor and complete lack of joint movement in the hindlimbs around P21 (SI Video 3). Reducing ataxin 2 levels in TDP-43Tg/TgAtxn2+/− and TDP-43Tg/TgAtxn2−/− animals significantly lowered gait impairment scores (see online methods) starting at P15 (Fig. 1c) and slowed the rate of progression of gait impairment (Fig. 1d). Lowering ataxin 2 also mitigated tremor (Fig. 1e) and kyphosis (hunched posture that can be caused by neurodegeneration; Fig. 1f). Even after an age at which all TDP-43Tg/TgAtxn2+/+ mice had reached the humane euthanasia endpoint (SI Video 4), several of the TDP-43Tg/TgAtxn2−/− mice had no overt motor dysfunction (SI Video 5). We confirmed TDP-43Tg/TgAtxn2+/+ mice have a ~30% and ~27% reduction in cortical layer V pyramidal neurons and lower motor neurons, respectively9. We found a significant rescue of layer V neurons and a trend towards lower motor neuron rescue in TDP-43Tg/TgAtxn2−/− mice (Extended Data Fig. 2).
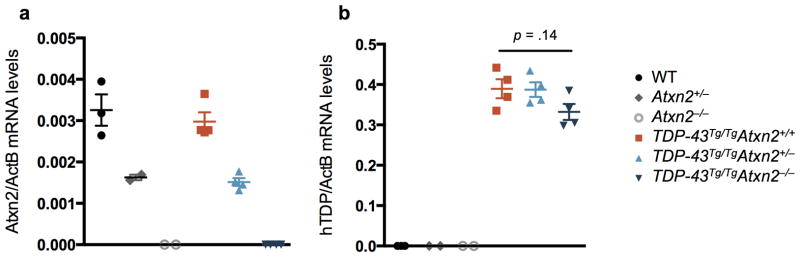
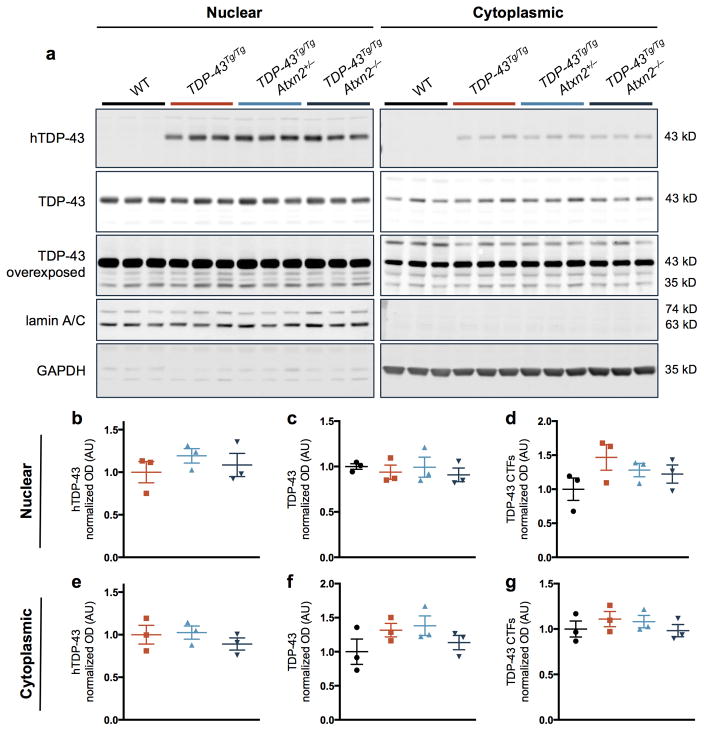
Reducing levels of ataxin 2 did not affect mRNA or protein levels of the human TDP-43 transgene in mouse brain tissue (Extended Data Fig. 3,4). Total TDP-43 (human and mouse) protein levels also were not significantly affected by ataxin 2 reduction (Extended Data Fig. 4). Thus, the beneficial effect of reducing ataxin 2 levels does not appear to act by reducing TDP-43 expression.
Ataxin 2 is an RNA-binding protein with multiple roles in RNA metabolism, including acting as a regulator of stress granule assembly14,15. Stress granules are highly dynamic, transient intracellular accumulations of RNA and protein that form when translation is stalled in response to cellular stress. We and others have proposed that concentrating aggregation-prone proteins like TDP-43 in stress granules can seed the formation of pathological protein aggregates in ALS16,17. Stress granule induction increases the levels of insoluble TDP-43, and TDP-43 inclusions can persist after stress granule dissolution18,19. Several lines of evidence suggest that TDP-43 inclusion formation may be directly involved in disease progression. ALS-associated mutations in TDP-43 increase the aggregation propensity and/or stability of the protein6,19,20. Additionally, TDP-43 inclusion pathology is correlated with neuron death in ALS patients21 and disease severity in TDP-43 mouse models9–11. Thus, we hypothesized that lowering ataxin 2 might reduce the propensity of TDP-43 to transit to stress granules and subsequently form pathological aggregates, and that this could prevent neurodegeneration.
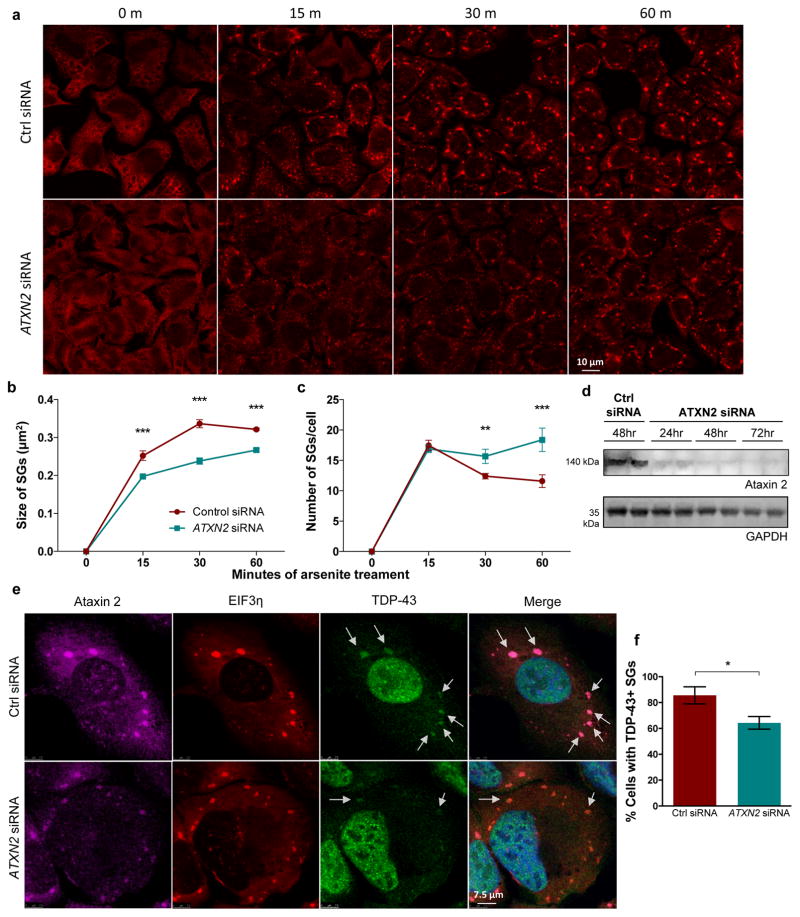
To investigate the mechanism by which ataxin 2 levels influence TDP-43 we first performed experiments with cultured human cells to visualize stress granules dynamics. Stress granules formed rapidly and, over time, underwent a maturation process in which they fused to form larger structures (Fig. 2a), consistent with their liquid-like material properties22. We knocked down ataxin 2 with siRNA and this markedly delayed the maturation of stress granules – at each time point, the stress granules were smaller and more numerous than in the control siRNA treated cells (Fig. 2a–c). Endogenous TDP-43 was recruited to stress granules, consistent with previous reports19, and was readily recognized by phosphorylation-specific and C-terminal epitope TDP-43 antibodies (Extended Data Fig. 5). Knocking down ataxin 2 significantly decreased the proportion of cells with stress granules containing endogenous TDP-43 (Fig. 2e,f). These data provide evidence that lowering ataxin 2 modulates stress granule dynamics and decreases the recruitment of TDP-43 to stress granules. We hypothesize that inhibiting the recruitment of TDP-43 into stress granules might reduce the propensity of TDP-43 to form pathological inclusions.
Figure 2. Knockdown of ataxin 2 delays the maturation of stress granules and decreases recruitment of TDP-43 to stress granules.
a) An EIF3η antibody was used to visualize stress granules (SGs) in U2OS cells. During continued stress, stress granules fuse to form larger structures. ATXN2 siRNA inhibited maturation of stress granules, resulting in smaller, more numerous stress granules at each time point (quantified in b–c). d) Western blot of ataxin 2 knockdown (source data in SI Fig. 1). e, f) Ataxin 2 siRNA treatment caused fewer cells to have TDP-43-positive stress granules at 60 minutes of arsenite exposure. b,c,f) The mean of 3 separate wells (116–146 cells per well) is plotted for each data point. Two-tailed t-tests were used to compare treatment groups. Means are plotted, and error bars represent SEM. *p < .05, **p < .01, ***p < .001.
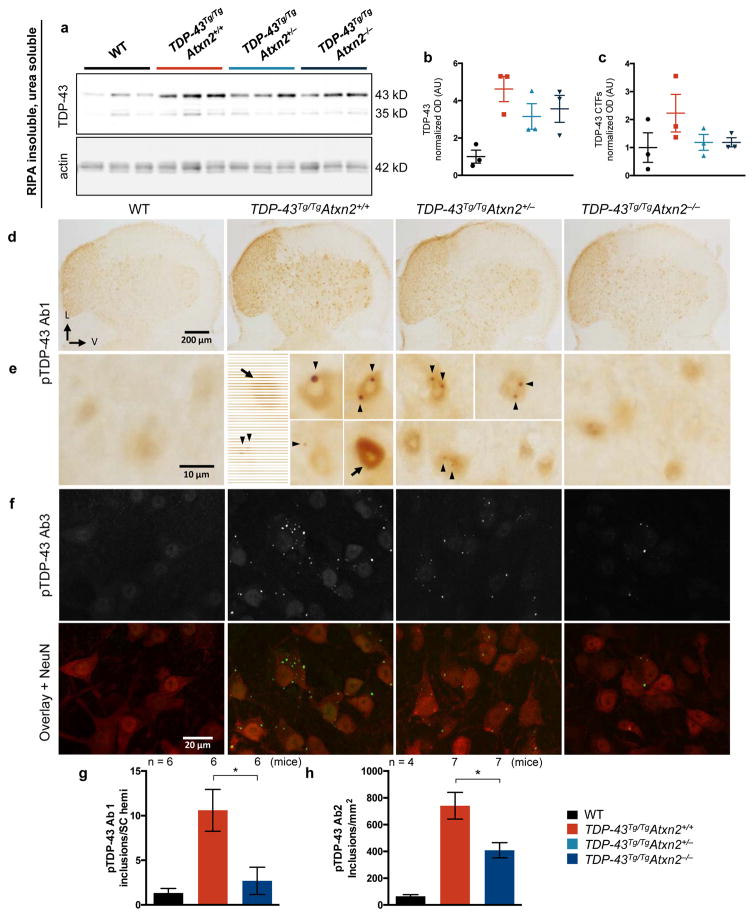
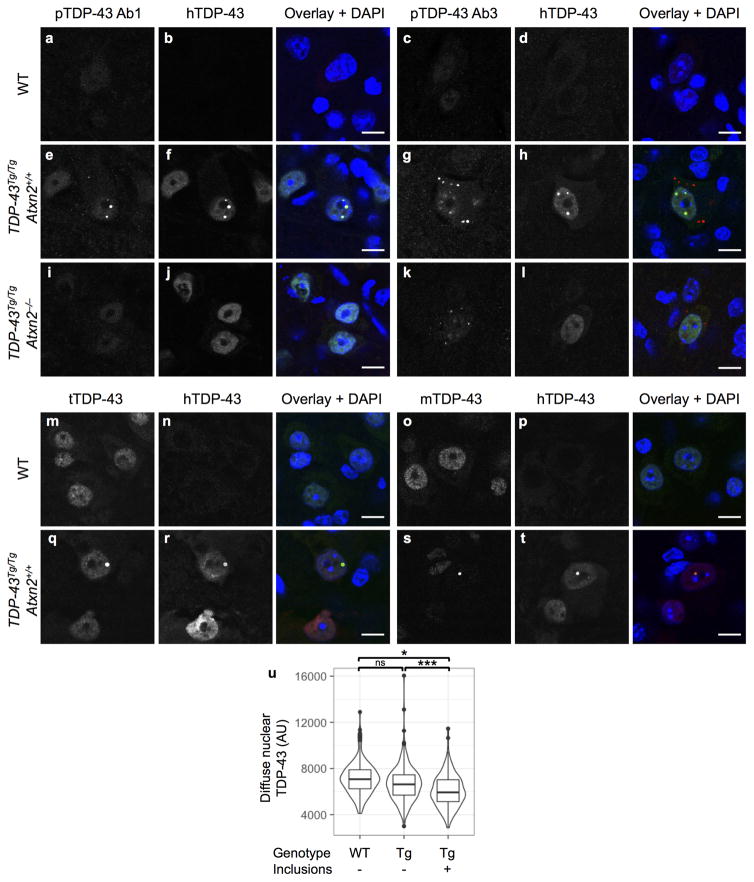
We next tested this hypothesis in vivo by defining the impact of lowering ataxin 2 levels on TDP-43 inclusion formation in TDP-43 transgenic mice. Aggregated TDP-43 in ALS patient neurons can become detergent insoluble, phosphorylated, and cleaved to form C-terminal fragments (CTFs). It is not clear what role these pathological alterations play in pathogenesis, but they are highly correlated with, and indicative of, pathological inclusion formation23,24. We found RIPA insoluble, urea soluble full-length TDP-43 was four times higher in TDP-43Tg/TgAtxn2+/+ mice than WT littermates (Fig. 3a,b). We also detected insoluble TDP-43 CTFs in TDP-43 transgenic mice (Fig. 3a,c). Reducing ataxin 2 moderately decreased levels of insoluble full-length and CTF TDP-43 (Fig. 3a–c). To visualize TDP-43 inclusions, we performed immunohistochemistry on spinal cord sections of TDP-43 transgenic mice. We used three different phospho-specific TDP-43 antibodies (pTDP-43 Ab1, Ab2, Ab3). We detected round, predominantly nuclear inclusions in TDP-43Tg/Tg mice when immunostaining with pTDP-43 Ab1 or with TDP-43 antibodies that are not phosphorylation specific (Extended Data Fig. 6e,f,q,s). The other two pTDP-43 antibodies tested (pTDP-43 Ab2 and Ab3) recognized nuclear and cytoplasmic inclusions that were much more numerous (Extended Data Fig. 6g). We did not detect inclusions in WT littermates with any of the TDP-43 antibodies tested (Extended Data Fig. 6a–d,m–p). Additionally, we found that cells with nuclear inclusions had lower levels of diffuse nuclear TDP-43 staining (Extended Data Fig. 6u). This suggests inclusion formation may sequester TDP-43 and cause a partial loss of TDP-43 function, which can be toxic to neurons in vivo25. To investigate whether ataxin 2 reduction had an impact on TDP-43 pathology, we quantified spinal cord inclusions and found that pTDP-43 Ab1-positive and pTDP-43 Ab2-positive were reduced by 75% and 45%, respectively, in TDP-43Tg/TgAtxn2−/− mice (Fig. 3d–h). These results support the hypothesis that lowering ataxin 2 mitigates TDP-43 toxicity by decreasing the propensity of TDP-43 to form pathological aggregates.
Figure 3. Lowering ataxin 2 levels decreases TDP-43 pathology.
a) RIPA-insoluble, urea soluble full-length and CTF TDP-43 are elevated in TDP-43Tg/TgAtxn2+/+ mice (quantified b–c), and appeared moderately reduced in TDP-43Tg/TgAtxn2−/− mice (source data in SI Fig. 1). d) Lumbar spinal cord hemispheres (SC hemi) stained with pTDP-43 Ab1. e) Magnification to the cellular level reveals weak, diffuse nuclear staining in WT mice, and pTDP-43 inclusions (arrow heads) and strong diffuse nuclear pTDP-43 (arrows) in TDP-43Tg/TgAtxn2+/+ mice. Fluorescent stains (Extended Data Fig. 6e) demonstrate that most pTDP-43 Ab1 inclusions are nuclear. f) pTDP-43 Ab3 reveals inclusions that are much more numerous and usually cytoplasmic (Extended Data Fig. 6g). Lowering ataxin 2 decreased pTDP-43 Ab1 inclusions at P21 (g), and pTDP-43 Ab2 inclusions at P23 (h) in lumbar spinal cord. Two-tailed t-tests were performed between groups of interest. Means are plotted, and error bars indicate S.E.M. *p < 0.05
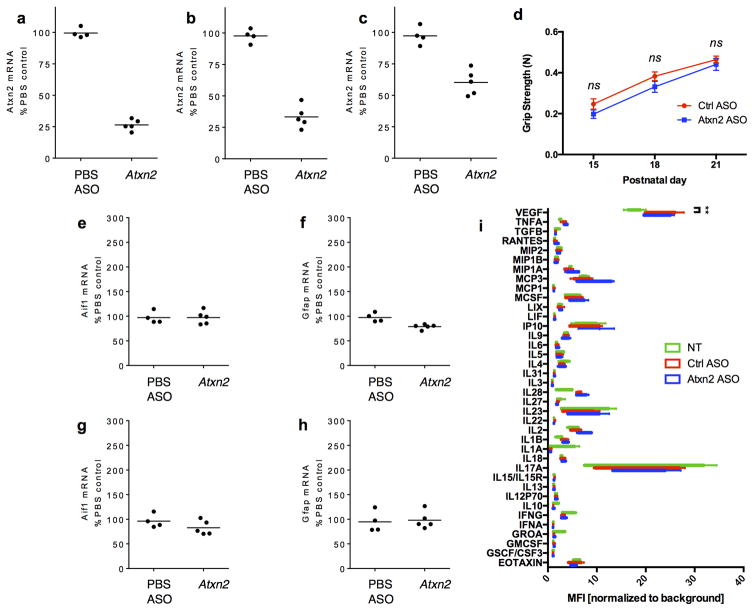
Given the striking amelioration of disease seen with ataxin 2 genetic reduction, we sought to further explore the translational potential of lowering ataxin 2 levels in a more therapeutically-applicable manner using antisense oligonucleotides (ASOs). ASOs are highly stable, synthetic nucleic acids that specifically hybridize with an mRNA target, triggering RNase H-mediated cleavage and degradation of the mRNA while leaving the ASO intact4. We screened a collection of ASOs for safety and efficacy (Extended Data Fig. 7 and 26), and used the most effective and well-tolerated ASO for further studies. Intracerebroventricular (ICV) injection of 4 μg of the Atxn2 ASO at P1 did not affect the home-cage behavior or grip strength of WT mice (Extended Data Fig. 7d). We also did not detect an increase in expression of genes associated with gliosis or inflammation (Extended Data Fig. 7e–i), suggesting this dose of ASO did not produce adverse side effects.
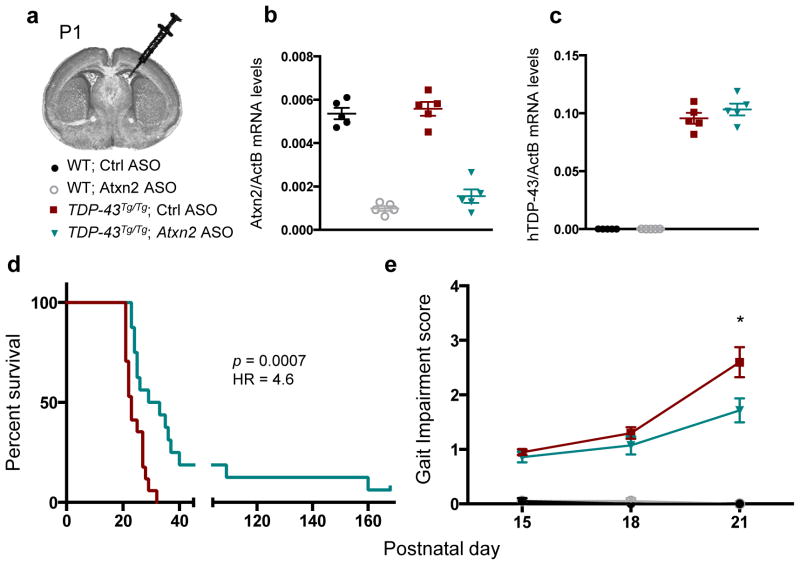
To test whether ASOs targeting Atxn2 could mitigate disease in TDP-43 transgenic mice, we administered TDP-43 transgenic mice and WT littermates with a one-time ICV injection of a non-targeting control ASO or Atxn2 ASO at P1 (Fig. 4a). Both ASOs were well tolerated by the mice at this dose, and the Atxn2 ASO successfully reduced levels of Atxn2 mRNA by 77% on average when assessed at P21 (Fig. 4b). Neither ASO affected expression levels of the human TDP-43 transgene (Fig. 4c). TDP-43 transgenic mice injected with the Atxn2 ASO had a striking 35% increase in median lifespan compared to control ASO injected littermates (Fig. 4d) and had significantly improved motor performance by P21 (Fig. 4e; SI Video 6). Two of the 16 TDP-43Tg/Tg mice treated with Atxn2 ASO survived over 120 days, whereas no control ASO treated TDP-43Tg/Tg mouse lived past 32 days. Therefore, a single administration of ASOs targeting Atxn2 into the central nervous system is sufficient to greatly prolong survival and improve motor performance of TDP-43 transgenic mice.
Figure 4. ASOs targeting ataxin 2 extend lifespan and improve motor performance in TDP-43 transgenic mice.
a) P1 mouse pups were treated with Ctrl or Atxn2 ASO via ICV injection. At P21, mRNA levels of Atxn2 (b) were decreased by 77% in the Atxn2 ASO-injected mouse brains without affecting mRNA levels of the human TDP-43 transgene (c). d) Lifespan was significantly extended by Atxn2 ASO treatment (n = 16) vs Ctrl ASO (n = 17). Curves were compared by log rank test, and effect size was estimated using a Cox proportional hazards model. e) Gait impairment score was also improved in Atxn2 ASO treated mice (n = 14) by P21 compared to Ctrl ASO (n = 20). Two-tailed t-tests were used to compare the two treatment groups at each age. Means are indicated, and error bars indicate S.E.M. *p < 0.05
We have shown by two independent approaches that lowering levels of ataxin 2 markedly increases lifespan and improves motor function in TDP-43 transgenic mice. Additionally, we have demonstrated that lowering ataxin 2 decreases the burden of TDP-43 inclusions, which provides mechanistic evidence that ataxin 2 may modulate toxicity by affecting the aggregation propensity of TDP-43. In support of this hypothesis, ataxin 2 and TDP-43 physically interact in an RNA-dependent manner and ALS-associated polyglutamine expansions in ataxin 2, which enhance stability7, can increase TDP-43 pathological modifications27. We demonstrate that lowering ataxin 2 impairs the maturation of stress granules, which are hypothesized to facilitate the pathological aggregation of TDP-4316,17. Indeed, reducing ataxin 2 in cultured cells decreases recruitment of endogenous TDP-43 to stress granules. Beyond ataxin 2, this general strategy of targeting factors required for the assembly of ribonucleoprotein granules per se could be pursued as a way to mitigate TDP-43 pathological aggregation and neurodegeneration.
Reduction of ataxin 2 has exciting therapeutic potential for ALS and FTD because TDP-43 pathology is a component of 97% of ALS cases and nearly 50% of FTD cases6. Although complete knockout of ataxin 2 seems well tolerated in mouse, further preclinical studies are required to assess the long-term safety and efficacy of ataxin 2 knockdown in the mammalian central nervous system. It is, perhaps, inspiring that there are currently clinical trials to test ASO therapies for Huntington disease (targeting HTT)28, familial ALS (targeting SOD1)29, and spinal muscular atrophy (targeting SMN2 splicing)30. Indeed, in patients with familial ALS caused by SOD1 mutations, ASOs targeting SOD1 administered as an intrathecal infusion were well tolerated29. Reducing ataxin 2 levels to treat TDP-43 proteinopathy is the first proposed ASO therapy for neurodegenerative disease designed to target a modifier gene that is not directly causative of the disease. This type of approach will likely be essential for treating sporadic ALS, which makes up over 90% of cases, because of the vital cellular roles of TDP-43.
Online Methods
Mouse crosses
TDP-43 transgenic mice were generated by Samir Kumar-Singh’s lab and have been previously described9,11. TDP-43Tg/+ mice were purchased from JAX (stock no: 012836, B6;SJL-Tg(Thy1-TARDBP)4Singh/J). TDP-43Tg/+ mice were maintained on a B6/SJL hybrid background by crossing with F1 hybrid mice from JAX to propagate the strain (stock no: 100012, B6SJLF1/J). Ataxin 2 knockout mouse lines have been previously described12,13. Atxn2+/− mice created by G.A.’s laboratory were backcrossed to C57/Bl6 mice for 10 generations and then maintained on the congenic C57/Bl6 background by crossing with WT non-littermates within the same line. Atxn2+/− mice created by S.M.P.’s laboratory were maintained on a B6129S hybrid background by crossing with Atxn2+/− males with F1 hybrid females from JAX (stock no: 101043, B6129SF1/J). Atxn2+/− mice from G.A. or S.M.P. were crossed with TDP-43Tg/+ mice in order to create lines A and B, respectively. Data for this project were collected from the offspring of TDP-43Tg/+Atxn2+/−, TDP-43Tg/+Atxn2+/+, and TDP-43Tg/+Atxn2−/− mice each crossed with other non-littermate mice (to avoid genetic drift) of the same genotype, respectively, within the same line.
We collected data for gait lifespan, gait impairment, and other phenotypes in two independent trials for each line. These trials were performed 4 months apart with different breeding pairs. After we performed a search of current literature analyzing lifespan in mouse models of neurodegenerative disease, we concluded that 20 animals total was likely a sufficient number in two separate trials to see an effect size worth additional consideration. The first trial of the study was performed with approximately 10 animals per genotype. We intended to do a power analysis using data from this cohort, but the lifespan and the other behavioral analysis data were already statistically significant. Therefore, standard power analysis methods were no longer applicable. We repeated the study in a second trial for a final sample size of 18–28 animals (Figure 1). All mice were genotyped through Transnetyx (Cordova, TN). Protein, RNA, and histological analyses were performed using mice from line B. Approximately equal numbers of males and females were used in all analyses.
ASO validation and administration
We screened a collection of ASOs designed to target various regions of the mouse Atxn2 mRNA. After screening Atxn2 ASOs for their ability to reduce Atxn2 levels in cultured mouse cells, and for toxicity in WT mice, we tested several ASOs by delivering them through a single intracerebroventricular (ICV) injection into the brain of P1 mice, and used the most effective and well-tolerated ASO for further studies. For the data presented in Fig. 4 and Extended Data Fig. 7d,i, P1 neonatal mice received an intracerebroventricular (ICV) injection of 3 μL of ASO in PBS (a total of 45 μg) were injected into the left ventricle using a NanoFil 10 μL syringe (World Precision Instruments) and a 33-gauge needle. The coordinates for injection were 2 mm anterior to the lambdoid suture, 1 mm lateral from the sagittal suture, and 2 mm deep. Within each litter, half of the pups (randomly chosen) were treated with a control ASO (CsCoToAoTAGGACTATCCAGGAA) and the other half were treated with an ASO targeting mouse Atxn2 (CTTCACATTTCGATCCAACA). Both ASOs were developed and synthesized by Ionis Pharmaceuticals. ASOs were synthesized as described 26 and were 20 bp in length, with five 2′-O-methoxyethyl (MOE) modified nucleotides at each end of the oligonucleotide, and ten DNA nucleotides in the center. The backbone of the ASOs consists of a mixture of phosphorothioate (PS) and phosphodiester (PO) linkages: 1-PS, 4-PO, 10-PS, 2-PO and 2-PS (5′ to 3′).
For the data presented in Extended Data Fig 7a–c, e–h, WT mice were injected via ICV at P1 with 45 μg in 3 μL of Atxn2 ASO or PBS control. Mice were sacrificed at P28 for RNA analysis. Quantitative PCR for Atxn2 mRNA was performed using forward primer CACTTCAGATTTCAACCCGAAC, reverse primer TGACTGGTAGCGAGAAGGT, and probe TAGTTAATGGAGGTGTTCCCTGGCC. In an independent study to determine the safety of repeated injections of the Atxn2 ASO, we injected WT mice at 5 weeks of age and again at 9 weeks then performed an observational study for 8 weeks. The animals moved normally within the cage, and did not have any overt tremor or gait impairment.
Care of TDP-43Tg/Tg mice
Humane experimental protocols were performed as approved by the Administrative Panel of Laboratory Animal Care (APLAC) of Stanford University, an institution accredited by the Association for the Assessment and Accreditation of Laboratory Animal Care (AAALAC). In crosses where impaired TDP-43Tg/Tg mice were possible offspring, high-fat breeder chow was placed in the cage from P16-21 to prepare the pups for prompt weaning. To avoid aggression towards the impaired TDP-43Tg/Tg mice by littermates and parents and to ensure the TDP-43Tg/Tg mice received adequate nutrition, all pups were weaned at P21. TDP-43Tg/Tg mice that had not reached the euthanasia endpoint by this time were weaned into cages separate from other littermates. All TDP-43Tg/Tg were given wet food (Clear2O diet gel 76A) in a cup on the floor on of the cage from P21 until death to ensure that the impaired mice could easily chew their food and readily access food and water. Even severely impaired mice were observed eating, defecating, and micturating. The mice were weighed every few days and no mice were found to weigh less than 75% of their maximum body weight.
Phenotype scoring and humane euthanasia endpoint determination
All phenotype scoring was performed blinded to the genotype or treatment group of the animal, and the order in which the animals were tested each day was random. Mice were placed on a textured plastic surface within an empty cage and observed as they moved around the cage. Gait impairment scoring was adapted from previously described methods31. The gait impairment score measures dysfunction, not overall motor proficiency, and is therefore translatable even to young mice. A score of 0, no impairment, was given if the mouse walked normally. A score of 1 was given if the mouse had a tremor or appeared to limp while walking. A score of 2 was given if the mouse had a severe tremor, severe limp, lowered pelvis, or feet pointing away from the body during locomotion (“duck feet”). A score of 3 was given if the mouse had difficulty moving forward, minimal joint movement, feet not being used to generate forward motion, difficulty staying upright, or its abdomen dragging on the ground. A score of 4 marked the euthanasia endpoint in which the mouse fell over and was unable to right itself within 30 s on all 3 of 3 trials. The endpoint was tested daily after a mouse had received a gait score of 3. Some of the severely impaired mice took 10–15 minutes to be fully roused, and therefore, mice were not tested for the euthanasia endpoint until they were moving around their cage. A humane euthanasia endpoint was used instead of natural death in survival analysis for the welfare of the animals, but it also served as a more precise endpoint that is directly dependent on motor dysfunction (an ALS-relevant phenotype) and less dependent on insufficient food and water intake. The mice did not appear emaciated or severely dehydrated prior to euthanasia, nor did they dip below 75% of their maximum body weight. Body weight did not differ among TDP-43Tg/TgAtxn2+/+, TDP-43Tg/TgAtxn2+/−, and TDP-43Tg/TgAtxn2−/− mice (Extended Data Fig. 1g).
For the kyphosis score, if the mouse was able to easily straighten its spine as it walked, and did not have persistent kyphosis, it received a score of 0. If the mouse exhibited mild kyphosis but was able to straighten its spine, it received a score of 1. If it was unable to straighten its spine completely and maintained persistent but mild kyphosis, it received a score of 2. If the mouse maintained pronounced kyphosis as it walked or while it sat, it was assigned a score of 3. Tremor was scored independently from 0 (not observed) to 3 (severe).
Mouse tissue collection
Anesthetized mice were perfused with PBS and the brain and spinal cord were carefully dissected and washed in chilled PBS. The brain was cut in half sagittally. The brain hemisphere used for immunohistochemistry, as well as the cervical and lumbar enlargements of the spinal cord, were placed in 4% PFA in PBS at 4°C for 48 hours then stored in 30% sucrose in PBS for at least 24 hours. The other brain hemisphere was cut into 8 predefined sections. These brain sections and the remaining spinal cord segments (upper cervical, lower thoracic/upper lumbar, and sacral) were flash frozen in liquid nitrogen for later RNA or protein collection and stored at −80°C. For all experiments using these samples (analyzing RNA or protein), the investigator was blinded to the genotype of the animal during the protocol and the samples were processed in a random order. Only biological replicates from individual mice are plotted in graphs.
RNA extraction and qPCR
RNA was extracted using the PureLink RNA Mini Kit (Life Technologies user guide publication MAN0000406), using TRIzol reagent and on-column PureLink DNase I treatment. Surfaces and equipment were cleaned with RNaseZap, and RNase-free pipette tips were used. In the first step of the protocol, flash-frozen tissue was kept on dry ice until 200 μL of TRIzol reagent was added. A Kontes motor pestle with a clean RNase-free disposable pestle were used to rapidly and thoroughly homogenize the sample (~45 strokes). The rest of the TRIzol (800 μL) was then added and inverted to mix. The remainder of the RNA isolation protocol was carried out as directed by the aforementioned user guide. The RNA was converted to cDNA using the High Capacity cDNA Reverse Transcription Kit. Standard TaqMan reagents, and the StepOnePlus Real-Time PCR System were used for qPCR with the following assays: human TDP-43 (Hs00606522_m1), mouse Atxn2 (Mm00485946_m1), and mouse ActB (Mm02619580_g1). Each reaction was performed in triplicate. The relative transcript levels were calculated in excel by the following formula: (POWER (2, − (CT Mean target)))/(POWER (2, − (CT Mean ActB))).
Protein extraction
Nucleocytoplasmic fractioning was performed in a manner similar to that described previously 32. All buffers were chilled and had Halt protease and phosphatase inhibitors, 1 mM DTT, and 1 mM PMSF added right before use. Snap-frozen tissue was kept frozen on dry ice and precisely weighed. All subsequent volumes are based on this initial weight. From the addition of hypotonic buffer on, samples were kept chilled on ice. All centrifugation steps were performed in a 4°C cold room. Briefly, tissue was homogenized by ~30 strokes of a Kontes motor pestle in 200 μL hypotonic buffer (10 mM Hepes, 10 mM NaCl, 1 mM KH2PO4, 5 mM NaHCO3, 5 mM EDTA, 1 mM CaCl2, 0.5 mM MgCl2). After homogenization, additional hypotonic buffer was added to a final volume of 10 μL/mg, and the tubes were inverted to mix. After 10 min on ice, 2.5 M sucrose (0.5 μL/mg) was added. The homogenate was pipetted up and down to resuspend and centrifuged at 6,300 g for 10 min. The supernatant was collected as the cytoplasmic fraction. The pellet was washed four times in chilled TSE buffer (10 μL/mg; 10 mM Tris, 300 mM sucrose, 1 mM EDTA, 0.1% IGEPAL, pH 7.5). For each wash, the homogenate was pipetted up and down to resuspend, the tube was inverted to clean sides of tube, and the sample was centrifuged at 1,000 × g for 5 min. The supernatant was clear by the final wash. The pellet of nuclei was resuspended in chilled RIPA buffer (5 μL/mg; Sigma). The sample was homogenized by pipetting up and down and incubated on ice for 10 min. During this incubation, each sample was passed through a 21G needle 20 times to shear viscous chromatin in the solution. The samples were spun at 21,000 g for 10 min, and the supernatant was saved as the nuclear fraction.
Sequential solubility fractioning was performed in a similar manner as was described previously33. Extraction was performed on ice using chilled buffers until urea buffer was added. Tissue was weighed. All subsequent volumes are based on this initial weight. RIPA buffer (5 μL/mg; Sigma) with Halt protease and phosphatase inhibitor and 5 mM EDTA added just prior to use, was added to tissue chunk and thoroughly homogenized with Kontes motor pestle for approximately 45 strokes. The homogenates were incubated on ice for 10 minutes and were transferred to polycarbonate centrifuge tubes. The samples were centrifuged (100,000g, 4C, 30 min) using a TLA 120.2 rotor. The supernatant was promptly and gently transferred to a new tube and stored at −80 °C. Pellet was washed by re-extraction in the same volume of RIPA buffer using the motor pestle for approximately 30 strokes. The samples were centrifuged again (100,000 rpm, 4C, 30 min) and the supernatant was discarded. The pellets were extracted in urea buffer (2 μL/mg; 7M urea, 2M thiourea, 4% CHAPS, 30 mM Tris, pH 8.5) prepared just prior the experiment, homogenized using a motor pestle for approximately 30 taps, and centrifuged (100,000 rpm, 22C, 30 min). Supernatants were saved as the urea fraction and stored at −80 °C in aliquots.
Western blotting
Nuclear and cytoplasmic fractions were heated at 70°C for 10 min with 1x LDS buffer. Proteins in LDS were run down 4–12% bis tris gels and transferred to nitrocellulose membranes. The membranes were blocked in Odyssey blocking buffer (LI-COR) at RT for 1 hour and treated over night at 4 °C with primary antibodies at the following dilutions: 1:5000 Rb TDP-43 C-term (Sigma-Aldrich T1580), 1:1000 Ms human-specific TDP-43 (Novus Biologicals H00023435-M01), 1:1000 Gt Lamin A/C (Santa Cruz 6215), 1:50,000 Ms GAPDH (Sigma-Aldrich G8795), 1:1000 Ms Ataxin 2 (BD 611378). Membranes were rinsed with PBST (0.1% Tween-20), washed x3 for 10 min, and treated with 1:20,000 Alexa fluor 680 or 790 conjugated IgG (H+L) raised in Donkey (Life Technologies) at RT for 1 hour, rinsed with PBST, and washed 3x for 10 min before being developed on a LI-COR Odyssey scanner at laser intensity well below saturation.
Luminex assay
This assay was performed in the Human Immune Monitoring Center at Stanford University. Mouse 38-plex kits were purchased from eBiosciences/Affymetrix and used according to the manufacturer’s recommendations with modifications as described below. Briefly: Beads were added to a 96 well plate and washed in a Biotek ELx405 washer. Samples were added to the plate containing the mixed antibody-linked beads and incubated at room temperature for 1 hour followed by overnight incubation at 4°C with shaking. Cold and Room temperature incubation steps were performed on an orbital shaker at 500–600 rpm. Following the overnight incubation plates were washed in a Biotek ELx405 washer and then biotinylated detection antibody added for 75 minutes at room temperature with shaking. Plate was washed as above and streptavidin-PE was added. After incubation for 30 minutes at room temperature wash was performed as above and reading buffer was added to the wells. Each sample was measured in duplicate. Plates were read using a Luminex 200 instrument with a lower bound of 50 beads per sample per cytokine. Custom assay Control beads by Radix Biosolutions are added to all wells.
This assay was performed with brain tissue from P21 WT mice untreated (n = 5) or given P1 ICV injection of the Atxn2 ASO (n = 4) or Ctrl ASO (n = 5). Each sample was processed in duplicate. The bead count and the coefficient of variation between duplicates were checked for each sample and epitope, and no cause was found to exclude values. Median fluorescence intensity (MFI) was averaged between the duplicates and normalized by dividing by the buffer background MFI for each respective epitope.
Mouse CNS Histology
Fixed sections were mounted in OCT and cut to a thickness of 40 μm using a Leica CM3050 S Cryostat. Sections were stored in cryoprotective media (0.01 M sodium phosphate, 30% glycerin, 30% ethylene glycol) at −20°C. Washes were done 4 times for 5 min each in TBST (0.1% Tween-20), unless otherwise noted, at RT. Washes and incubation steps were performed with gentle rotation using an orbital rotator. The following primary antibodies were used: 1:2000 Rb TDP-43 phospho 409/410 “pTDP-43 Ab1” (Cosmo Bio TIP-PTD-P01), 1:2000 Rb TDP-43 phospho 403/404 “pTDP-43 Ab2” (Cosmo Bio TIP-PTD-P05), 1:1000 Rb TDP-43 phospho 409/410 “pTDP-43 Ab3” (a gift from Leonard Petrucelli’s laboratory, affinity purified 3655), 1:1000 Ms TDP-43 human-specific (Novus Biologicals H00023435-M01), 1:1000 TDP-43 C-term (Sigma-Aldrich T1580), 1:500 Ms NeuN (Millipore MAB377), 1:1000 Rb NeuN antibody (abcam ab177487), and 0.04 μg/mL Rb TDP-43 mouse specific (a gift from Virginia Lee’s lab: 2341-aa379).
For DAB-amplified immunohistochemistry, free floating sections were placed in a 12 or 24 well plate with a rectangular specimen net insert, washed, pretreated for 20 min at RT in TBST with 0.1 % Triton-X and 0.6% hydrogen peroxide, washed, and blocked in 10% normal goat serum for 1 hour at RT. Primary antibody was diluted in TBST with 10% normal goat serum and incubated overnight at 4°C. The sections were then washed, treated with biotinylated goat anti rabbit IgG (H+L) secondary antibody (Vector) at 1:500 in TBST for 1 hour at RT, washed, treated with an ABC kit (Vectastain) for 1 hour at RT, and washed using 0.1 M Tris pH 7.4 for the final wash. DAB solution was prepared by dissolving a 10 mg DAB tablet (Sigma-Aldrich) in 20 mL of 0.1 M Tris pH 7.4. The solution was vortexed to dissolve, passed through a 0.45 μm filter unit, and 6 μL of 30% H2O2 was added right before use. All sections used in a single experiment were incubated simultaneously for 30 s to 2 min until the desired color change was reached. Sections were washed with 0.1 M Tris x4 for 5 min and mounted onto Superfrost Plus Microscope Slides (Fisher Scientific) in PB using a paintbrush. The slides were allowed to dry overnight and incubated in CitriSolv (Fisher Scientific) for a few minutes before immediate coverslipping with Entellan (Electron Microscopy Sciences). Sections were imaged using a Nikon Eclipse 50i microscope. pTDP-43 Ab1 inclusions per section were quantified by hand at 20x magnification blinded to the identity of the mice. Mice were quantified in a random order, but for technical reasons, all spinal cord hemispheres for a single mouse were quantified at the same time. For each mouse, 14–26 lumbar spinal cord hemispheres were quantified with 6 mice per genotype.
Alternatively, the sections were stained using immunofluorescence. The sections were washed then blocked/pre-treated in TBST with 0.1% triton-X and 10% normal goat or donkey serum (depending on secondary antibody host) for 1 hour at RT. Primary antibodies were diluted in TBST with 10% serum at 4°C overnight. The sections were then washed, treated with Alexa Fluor conjugated secondary antibodies (Life Technologies) diluted at 1:1000 in TBST with 10% serum for 3 hours, and washed again with Hoechst (1:5000) added to the final wash. Sections were mounted on frosted slides in PB using a paintbrush, and when the sections were no longer visibly wet, the slides were coverslipped using Prolong diamond antifade mountant (Molecular Probes). Slides were stored at 4°C for short term and −20°C for long term.
High quality representative images of Inclusions recognized by pTDP-43 Ab2 (Fig. 3f) were taken on a Zeiss LSM510 Meta Confocal microscope using a 10 μm z-stack with .37 μm steps (maximal intensity projections are shown). Inclusions recognized by pTDP-43 Ab2 were quantified using 44 μm-thick z-stacks at 10x magnification (1 spinal cord hemisphere per image) on a Leica DMI6000B fluorescent microscope. Maximum intensity projections were created using Fiji. Images were randomized and blinded by renaming each image with a random number using python. Photoshop CS6 soft round brushes were used to occlude white mater and the dorsal horn. The images were then run through the MetaXpress granularity module to automatically quantify the number of inclusions per image in an unbiased manner. This method detects inclusions of a predetermined size range based on changes in surrounding pixel intensity. The optimal parameters for this experiment were empirically determined to be an approximate size range of 1.3–5.2 μm in diameter and pixel intensity change of 2500 graylevels. The area of gray matter quantified in each image was measured using Fiji (the area quantified for each hemisphere was approximately 0.5 mm2). The number of inclusions per image was divided by the area of that image, and these values were averaged for each mouse (average of 19 images per mouse).
To quantify diffuse nuclear TDP-43, spinal cord sections were stained with 1:1000 TDP-43 C-term antibody, and imaged using a Leica DMI6000B fluorescent microscope. Using Fiji, a small circle was drawn in each nucleus (away from the inclusion, if an inclusion is present) to measure the diffuse TDP-43 signal. For TDP-43Tg/TgAtxn2+/+ mice, cells with an obvious inclusion(s) and 2 nearby cells in the same focal plane without inclusions were quantified. For WT mice, 5 cells were quantified per image. There was no way to blind the experimenter because the inclusions either obviously present or absent in each image. Three mice per genotype were analyzed, with an average of 18 images and 126 cells counted per mouse.
To quantify cortical layer V neurons, 40 uM-thick sagittal brain sections were processed for immunofluorescence, as detailed above, using a Rb NeuN antibody. Images were taken at 5x using a Leica DMI6000B fluorescent microscope. Images were then blinded and processed in a random order. Quantification was performed using Fiji. Layer V was outlined using the polygon selection tool, “measure” was used to find the area of the selection, a threshold was applied using the “triangle” algorithm, the area outside the selection was removed using “clear outside,” and “watershed” was used to separate overlapping nuclei. The “analyze particles” tool was used to count nuclei that were greater than 50 μm in area and had a circularity from 0.2–1.0. The number of nuclei for each image was divided by the layer V area. For each mouse, 3–8 sections were quantified (see “Statistical analyses”).
Cell culture
To knockdown ataxin 2, Dharmacon ON-TARGETplus SMARTpool siRNA was used with the following target sequences: CCAAAGAGUAGUUAAUGGA, AAAGGUAUAUCACCAGUUG, GACAAGCCCUUCUUUCUAC, CUAAACGCAUGUCUUCAGA. Stress granule experiments were performed using 0.5 mM sodium arsenite in U2OS cells. Counting was done on a Leica Widefield scope at 63x. Only cells with more than one EIF3η puncta and seemingly viable nuclei were considered. Cells with at least one TDP-43 puncta which co-localized with EIF3η were considered to be positive for TDP-43 granules, otherwise they were considered to be negative. Santa Cruz Biotechnology EIF3η (sc-16377) and Proteintech C-terminal TDP-43 antibody (12892-1-AP) were used for quantification. Phosphorylation-specific TDP-43 antibodies from CosmoBio (TIP-PTD-M01 and TIP-PTD-P05) and an ataxin 2 antibody (BD 611378) were also used to in stress granule images. TDP-43 positive stress granules were quantified at 60 min of sodium arsenite exposure. At this time point, almost all cells had stress granule formation, but the few without were excluded from quantification. 116–146 cells were counted up per well for 3 separate wells for each siRNA used. The U2OS cell line was purchased from ATCC (HTB-96), an official cell line repository organization, and has not been validated since purchase. We tested the cell line for mycoplasma contamination by PCR in 2012, and it was negative. Since then, we regularly check for mycoplasma by DAPI staining, a method routinely used in the field. Mycoplasma contaminated cells show DAPI-positive stain in the cytoplasm.
Statistical analyses
Analyses were performed using R version 3.1.3 and Prism 6 (GraphPad), and graphs were plotted using Prism 6. Survival curves were compared using the survdiff function in the survival R package, which performs a log-rank test, and effect sizes are reported as the hazard ratios given by the coxph function, which fits a Cox proportional hazards model. For comparisons among different genotypes or treatment groups, ordinary one-way ANOVA or two-tailed, unpaired t-test were used when comparing three groups or two groups, respectively. For TDP-43 inclusion quantification, the number of inclusions per spinal cord section or image field was averaged for each animal. The animals were grouped by genotype and comparisons of interest were made between two groups using a two-tailed, unpaired t-test. All data analyzed by ANOVA or t-test appeared to have a Gaussian distribution, and therefore parametric tests were used. If two groups compared by t-test appeared to have different standard deviations, an F-test was used to compare variances. We did not find any significant differences in variances among groups.
An ordinary two-way ANOVA was used to analyze Luminex 38-plex mouse cytokine assay data. Factor 1 was protein assayed; factor 2 was mouse treatment group. Pairwise comparisons among the three treatment groups were made for each protein. The Holm-Sidak method was used to compute multiplicity adjusted p-values. All 114 pairwise tests were considered within one family.
Linear mixed models (LMM, implemented using the lme4 R package) were used to determine if genotype has an effect on cortical layer V neuron density or average neuron cell body area. The LMM helps alleviate the pitfalls of statistically comparing groups with small sample sizes, while appropriately taking into account that multiple measurements came from the same mouse. A likelihood ratio test was used to compare the null hypothesis including only random effects terms for mouse and anatomical section to the alternative hypothesis including these terms as well as a fixed effect for genotype. A similar LMM was used to determine whether the presence of an inclusion had an effect on the level of diffuse TDP-43 in neuronal nuclei. A likelihood ratio test was used to compare the null hypothesis including only random effects terms for mouse and image to the alternative hypothesis including these terms as well as whether an inclusion is present.
Whether mouse genetic background (line) affects lifespan was assessed using a Cox proportional hazards likelihood ratio test, comparing the null model including only genotype to an alternative model including genotype and line. The effect of sex on lifespan was tested analogously.
We assessed the evidence supporting the presence of two groups of responders (weak responders and strong responders) within the TDP-43Tg/TgAtxn2−/− population using two approaches, one Bayesian and one frequentist, both using the RStan R package. Both analyses suggest modest but significant support for two groups. In both analyses we compared how well a single Weibull distribution fit the TDP-43Tg/TgAtxn2−/− life span data compared to a mixture of two Weibull distributions. The Weibull distribution is a commonly used parametric survival model which generalizes the exponential distribution, with hazard function λpt−1 where λ is the rate parameter, p is the shape parameter, and t is time.
In the Bayesian analysis, we estimated the Watanabe-Akaike information criterion (WAIC) 34 for the one and two component model using four chains of Hamiltonian Monte Carlo 35 with 2000 iterations each, the first 1000 of which were discarded as warmup. The difference in WAIC values was 3.3, suggesting positive support for the two component model. In the frequentist analysis we consider the one and two component models as null and alternative hypotheses respectively. We obtain maximum likelihood estimates of the component parameters (shapes and rates) for the two models, using LBFGS and marginalizing over mixture assignments. We use the likelihood ratio between these two models as a test statistic. We used a parametric bootstrap, sampling 1000 times from the empirical null (fit one component model) and refitting both models, to obtain the sampling distribution of the likelihood ratio. Under this distribution we obtain an empirical p-value of 0.02 and thereby reject the one component null hypothesis in favor of the alternative two component hypothesis.
Extended Data
Extended Data Figure 1. Reduction of ataxin 2 using two independent lines of Atxn2 knockout mice lines extends lifespan of TDP-43 transgenic mice.
Lines A and B were generated using Atxn2+/− mice from a congenic C57Bl/6 and a hybrid B6129S background, respectively. Within line A, TDP-43Tg/TgAtxn2+/− (n = 7) (a) and TDP-43Tg/TgAtxn2−/− (n = 7) (c) mice lived significantly longer than TDP-43Tg/TgAtxn2+/+ mice (n = 16). Within line B, TDP-43Tg/TgAtxn2+/− (n = 13) (b) and TDP-43Tg/TgAtxn2−/− (n = 11) (d) mice also lived significantly longer than TDP-43Tg/TgAtxn2+/+ mice (n = 12). Curves were compared by log rank test and effect size estimated by a Cox proportional hazards model (HR = hazard ratio). e) After taking genotype into account, the line that the mice came from, but not the sex of the mice, significantly affected lifespan. A Cox proportional hazards likelihood ratio test was used to compare the null model including only genotype to an alternative model including genotype and line or genotype and sex. f) We found evidence for two groups of responders (strong and weak) in the TDP-43Tg/TgAtxn2−/− population (parametric bootstrap p = 0.02, see online methods). The Kaplan-Meier curve of all TDP-43Tg/TgAtxn2−/− mice from both lines is plotted, and the one and two group models are graphed. g) Knockout of Atxn2 did not affect weight in non-transgenic or TDP-43Tg/Tg adolescents. Means are plotted, and error bars indicate S.E.M.
Extended Data Figure 2. Cortical layer V and lower motor neuron loss in TDP-43 transgenic mice.
a) Representative NeuN stains of a sagittal sections through cortex. Layer V is marked by a green bar. Layer V neurons were 30% less numerous (b) and had smaller cell bodies (c) in TDP-43Tg/TgAtxn2+/+ mice. These phenotypes were significantly ameliorated in TDP-43Tg/TgAtxn2−/− mice. Four mice were quantified per genotype, and the values for individual brain sections are plotted. Genotype groups were compared using linear mixed models with a random effect to appropriately account for the multiple measurements per mouse (see online methods). d) Representative NeuN stains of L5 lumbar ventral horn showing large lower motor neurons. e) Quantification of motor neuron cell bodies present in the ventral horn of the lumbar enlargement at levels L3-L6. There was an 27% decrease in motor neurons on average in TDP-43Tg/TgAtxn2+/+ mice compared to WT. Six P23 animals were used per genotype. Two-tailed t-tests were performed between groups of interest. Means are plotted, and error bars indicate S.E.M. *p < 0.05, **p < 0.01.
Extended Data Figure 3. Lowering ataxin 2 levels does not affect expression of the human TDP-43 transgene.
a) Atxn2 mRNA levels were decreased in Atxn2+/− mouse brain by ~50% and completely absent in Atxn2−/− mice. b) Among TDP-43Tg/Tg mice, Atxn2 reduction did not significantly affect levels of the human TDP-43 (hTDP-43) transgene by ordinary one-way ANOVA. Samples were collected at P21. Error bars indicate S.E.M.
Extended Data Figure 4. Protein levels of TDP-43 are not significantly affected by ataxin 2 reduction in TDP-43Tg/Tg mice.
a) Nucleocytoplasmic fractioning of mouse brain tissue segregated the nuclear marker lamin A/C from cytoplasmic marker GAPDH. Nuclear levels of human TDP-43 (b) or total full-length TDP-43 (c) were not altered among the genotypes, though nuclear TDP-43 C-terminal fragments (CTFs) were slightly elevated in TDP-43Tg/TgAtxn2+/+ mice (d). e–g) Cytoplasmic hTDP-43 (e) and total TDP-43 CTFs (g) were also unaltered among the genotypes, though cytoplasmic full-length TDP-43 (f) seemed slightly elevated in TDP-43Tg/TgAtxn2+/+ mice. Samples were collected at P21. Error bars indicate S.E.M. Gel source data in SI Fig 1.
Extended Data Figure 5. Stress granules contain phosphorylated TDP-43.
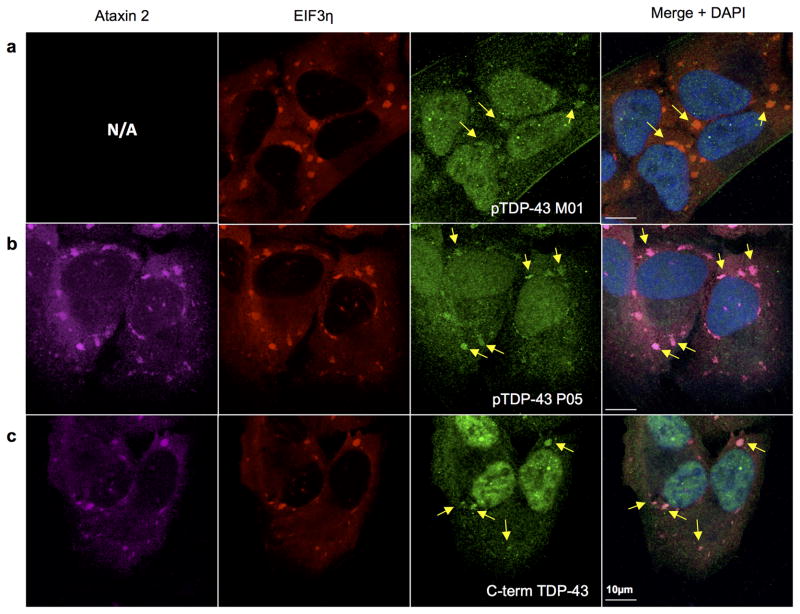
Two different phosphorylation-specific TDP-43 antibodies (a, b) and a C-terminal epitope TDP-43 antibody (c) readily stain stress granules, indicated by the stress granule markers EIF3η and ataxin 2.
Extended Data Figure 6. Two different types of inclusions are recognized in TDP-43Tg/Tg mice. a–d,m–p).
None of the TDP-43 antibodies tested recognized inclusions in WT mice. One of the three phosphorylated TDP-43 (pTDP-43) specific antibodies (e) and TDP-43 antibodies not phosphorylation-specific (f,h,q–t) recognized spherical, predominantly nuclear inclusions. The other two pTDP-43 specific antibodies (only one is shown) recognized smaller cytoplasmic and nuclear inclusions (g). i,j,l) The first type of inclusion was very rare in TDP-43Tg/TgAtxn2−/− mice. k) The second type of inclusion appeared smaller and reduced in number in TDP-43Tg/TgAtxn2−/− mice. q–t) Nuclear inclusions were effectively stained with total TDP-43 (tTDP-43), human-specific TDP-43 (hTDP-43), and mouse-specific TDP-43 (mTDP-43) antibodies. s) Diffuse mTDP-43 is greatly decreased in TDP-43Tg/Tg mice, an expected outcome of TDP-43 autoregulation. u) Levels of diffuse nuclear tTDP-43 were quantified by IF in WT neurons or TDP-43Tg/Tg neurons with or without inclusions. These 3 groups were compared in a pairwise fashion using linear mixed models with a term to appropriately account for multiple measurements per mouse (n = 3 mice per genotype, see online methods). Median and min to max are plotted. Images were taken in cervical spinal cord. Samples were collected at P21. Scale bars are 10 μm.
Extended Data Figure 7. An antisense oligonucleotide (ASO) targeting Atxn2 is able to successfully reduce mRNA levels throughout the central nervous system.
a) ICV injection at P1 of an ASO targeting Atxn2 was able to successfully reduce levels of Atxn2 mRNA in the spinal cord by ~75% when assessed at P28. Atxn2 reduction was also seen in the cortex (b) and cerebellum (c). d) Grip strength of WT mice was not effected by Ctrl or Atxn2 ASO injection (n = 16 per treatment). Mean and SEM plotted. Genetic markers of gliosis, Aif1 and Gfap, were not altered in the spinal cord (e, f) or cortex (g, h) after ASO injection. i) Using a Luminex 38-plex assay, we could not detect a significant difference in inflammatory markers among uninjected WT mice (n = 5) and WT mice treated with the Atxn2 (n = 4) or Ctrl (n = 5) ASOs (two-way ANOVA treatment group factor p = .32). However, the ASO treated animals had a small increase in 1 of the 38 markers, VEGF. Multiplicity-adjusted pairwise tests revealed that this difference was not significant for Atxn2 ASO treated mice (p = .17), but was for Ctrl ASO treated mice (p = .006). Min to max plotted. a–c,e–h) Biological replicates and means are plotted.
Supplementary Material
Supplemental Figure 1. Source data of gel images (uncropped Western blots).
This mouse was given a gait impairment score of 0.
This mouse was given a gait impairment score of 1.
This mouse was 21 days old and given a gait impairment score of 3 as it had very limited hindlimb joint movement and falls over.
This mouse was 22 days old and given a gait impairment score of 4 (humane euthanasia endpoint) because it was unable to right itself within 30 seconds of falling on its side on all 3 of 3 trials.
This TDP-43Tg/TgAtxn2−/− mouse was at 61 days old and still had no overt motor impairment. It was therefore given a gait impairment score of 0.
Examples of three P20 TDP-43Tg/Tg that received intracerebroventricular (ICV) administration of either the control ASO or the Atxn2 ASO at P1. The two severely impaired mice (gait impairment score of 4), which were unable to right themselves, received the control ASO whereas the one unimpaired mouse received the Atxn2 ASO.
Acknowledgments
This work was supported by NIH grants R01NS065317, R01NS09386501, R01NS073660 and R35NS097263 (A.D.G.), the National Science Foundation Graduate Research Fellowship (L.A.B.), the Robert Packard Center for ALS Research at Johns Hopkins (A.D.G.), Target ALS (A.D.G), the Glenn Foundation (A.D.G.), and the DFG grant AU96/13-1 (G.A.). We thank Leonard Petrucelli and Virginia Lee for sharing TDP-43 antibodies. We thank James Shorter and Leonard Petrucelli for comments on the manuscript and discussions. We thank Andrew Olsen and the Stanford Neuroscience Microscopy Service, supported by a grant from NIH (NS069375), for help with the confocal images, Yvonne Zuber (Stanford Veterinary Service Center) for mouse husbandry advice and support, Stanford’s Human Immune Monitoring Center (HIMC) for performing the Luminex assays.
Footnotes
P.J.-N., A.S. and F.R. are employed by Ionis Pharmaceuticals, a for-profit company that develops ASO therapies. The other authors declare no competing financial interest.
Author Contributions
L.A.B. and A.D.G. designed the experiments and wrote the paper. All authors reviewed and edited the manuscript. L.A.B. performed experiments and analyzed data. B.H. performed ASO injections and behavioral analyses on ASO-treated animals. G.B. helped with mouse dissections, and R.M. helped with mouse breeding and husbandry. D.A.K. helped perform statistical analyses. P.J.-N., A.S. and F.R. contributed ASOs, performed experiments to test ataxin 2 knockdown and immune response, and provided advice on designing experiments. J.M. and H.J.K. performed in vitro stress granule experiments. J.P.T. helped analyze stress granule experiments. G.A. and S.M.P. provided ataxin 2 knockout mice.
Data Availability Statement
The authors will make materials, data, code, and associated protocols promptly available to readers without undue qualifications. The ASOs used in this study are produced by IONIS pharmaceuticals, a for-profit company.
References
- 1.Taylor JP, Brown RH, Jr, Cleveland DW. Decoding ALS: from genes to mechanism. Nature. 2016;539:197–206. doi: 10.1038/nature20413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Neumann M, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 3.Lagier-Tourenne C, Cleveland DW. Rethinking ALS: the FUS about TDP-43. Cell. 2009;136:1001–1004. doi: 10.1016/j.cell.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Southwell AL, Skotte NH, Bennett CF, Hayden MR. Antisense oligonucleotide therapeutics for inherited neurodegenerative diseases. Trends Mol Med. 2012;18:634–643. doi: 10.1016/j.molmed.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 5.Smith RA, et al. Antisense oligonucleotide therapy for neurodegenerative disease. J Clin Invest. 2006;116:2290–2296. doi: 10.1172/JCI25424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ling SC, Polymenidou M, Cleveland DW. Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron. 2013;79:416–438. doi: 10.1016/j.neuron.2013.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elden AC, et al. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature. 2010;466:1069–1075. doi: 10.1038/nature09320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sproviero W, et al. ATXN2 trinucleotide repeat length correlates with risk of ALS. Neurobiol Aging. 2017;51:178e171–178e179. doi: 10.1016/j.neurobiolaging.2016.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wils H, et al. TDP-43 transgenic mice develop spastic paralysis and neuronal inclusions characteristic of ALS and frontotemporal lobar degeneration. Proc Natl Acad Sci U S A. 2010;107:3858–3863. doi: 10.1073/pnas.0912417107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McGoldrick P, Joyce PI, Fisher EM, Greensmith L. Rodent models of amyotrophic lateral sclerosis. Biochim Biophys Acta. 2013;1832:1421–1436. doi: 10.1016/j.bbadis.2013.03.012. [DOI] [PubMed] [Google Scholar]
- 11.Janssens J, et al. Overexpression of ALS-associated p.M337V human TDP-43 in mice worsens disease features compared to wild-type human TDP-43 mice. Mol Neurobiol. 2013;48:22–35. doi: 10.1007/s12035-013-8427-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kiehl TR, et al. Generation and characterization of Sca2 (ataxin-2) knockout mice. Biochem Biophys Res Commun. 2006;339:17–24. doi: 10.1016/j.bbrc.2005.10.186. [DOI] [PubMed] [Google Scholar]
- 13.Lastres-Becker I, et al. Insulin receptor and lipid metabolism pathology in ataxin-2 knock-out mice. Hum Mol Genet. 2008;17:1465–1481. doi: 10.1093/hmg/ddn035. [DOI] [PubMed] [Google Scholar]
- 14.Nonhoff U, et al. Ataxin-2 interacts with the DEAD/H-box RNA helicase DDX6 and interferes with P-bodies and stress granules. Mol Biol Cell. 2007;18:1385–1396. doi: 10.1091/mbc.E06-12-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaehler C, et al. Ataxin-2-like is a regulator of stress granules and processing bodies. PLoS One. 2012;7:e50134. doi: 10.1371/journal.pone.0050134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramaswami M, Taylor JP, Parker R. Altered ribostasis: RNA-protein granules in degenerative disorders. Cell. 2013;154:727–736. doi: 10.1016/j.cell.2013.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li YR, King OD, Shorter J, Gitler AD. Stress granules as crucibles of ALS pathogenesis. J Cell Biol. 2013;201:361–372. doi: 10.1083/jcb.201302044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parker SJ, et al. Endogenous TDP-43 localized to stress granules can subsequently form protein aggregates. Neurochem Int. 2012;60:415–424. doi: 10.1016/j.neuint.2012.01.019. [DOI] [PubMed] [Google Scholar]
- 19.Liu-Yesucevitz L, et al. Tar DNA Binding Protein-43 (TDP-43) Associates with Stress Granules: Analysis of Cultured Cells and Pathological Brain Tissue. PLoS One. 2010;5:e13250. doi: 10.1371/journal.pone.0013250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson BS, et al. TDP-43 is intrinsically aggregation-prone, and amyotrophic lateral sclerosis-linked mutations accelerate aggregation and increase toxicity. J Biol Chem. 2009;284:20329–20339. doi: 10.1074/jbc.M109.010264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geser F, et al. Evidence of multisystem disorder in whole-brain map of pathological TDP-43 in amyotrophic lateral sclerosis. Arch Neurol. 2008;65:636–641. doi: 10.1001/archneur.65.5.636. [DOI] [PubMed] [Google Scholar]
- 22.Molliex A, et al. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell. 2015;163:123–133. doi: 10.1016/j.cell.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neumann M, et al. Phosphorylation of S409/410 of TDP-43 is a consistent feature in all sporadic and familial forms of TDP-43 proteinopathies. Acta Neuropathol. 2009;117:137–149. doi: 10.1007/s00401-008-0477-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Igaz LM, et al. Expression of TDP-43 C-terminal Fragments in Vitro Recapitulates Pathological Features of TDP-43 Proteinopathies. J Biol Chem. 2009;284:8516–8524. doi: 10.1074/jbc.M809462200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang C, et al. Partial loss of TDP-43 function causes phenotypes of amyotrophic lateral sclerosis. Proc Natl Acad Sci U S A. 2014;111:E1121–1129. doi: 10.1073/pnas.1322641111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scoles D, et al. Antisense oligonucleotide therapy for spinocerebellar ataxia type 2. Nature. 2017 doi: 10.1038/nature22044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hart MP, Gitler AD. ALS-associated ataxin 2 polyQ expansions enhance stress-induced caspase 3 activation and increase TDP-43 pathological modifications. J Neurosci. 2012;32:9133–9142. doi: 10.1523/JNEUROSCI.0996-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kordasiewicz HB, et al. Sustained therapeutic reversal of Huntington’s disease by transient repression of huntingtin synthesis. Neuron. 2012;74:1031–1044. doi: 10.1016/j.neuron.2012.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller TM, et al. An antisense oligonucleotide against SOD1 delivered intrathecally for patients with SOD1 familial amyotrophic lateral sclerosis: a phase 1, randomised, first-in-man study. Lancet Neurol. 2013;12:435–442. doi: 10.1016/S1474-4422(13)70061-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Finkel RS, et al. Treatment of infantile-onset spinal muscular atrophy with nusinersen: a phase 2, open-label, dose-escalation study. Lancet. 2016;388:3017–3026. doi: 10.1016/S0140-6736(16)31408-8. [DOI] [PubMed] [Google Scholar]
- 31.Guyenet SJ, et al. A simple composite phenotype scoring system for evaluating mouse models of cerebellar ataxia. J Vis Exp. 2010 doi: 10.3791/1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mitchell JC, et al. Wild type human TDP-43 potentiates ALS-linked mutant TDP-43 driven progressive motor and cortical neuron degeneration with pathological features of ALS. Acta Neuropathol Commun. 2015;3:36. doi: 10.1186/s40478-015-0212-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walker AK, et al. Functional recovery in new mouse models of ALS/FTLD after clearance of pathological cytoplasmic TDP-43. Acta Neuropathol. 2015;130:643–660. doi: 10.1007/s00401-015-1460-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Watanabe S. Asymptotic equivalence of Bayes cross validation and widely applicable information criterion in singular learning theory. J Mach Learn Res. 2010;11:3571–3594. [Google Scholar]
- 35.Carpenter B, et al. Stan: A probabilistic programming language. J Stat Softw. 2016 doi: 10.18637/jss.v076.i01. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Source data of gel images (uncropped Western blots).
This mouse was given a gait impairment score of 0.
This mouse was given a gait impairment score of 1.
This mouse was 21 days old and given a gait impairment score of 3 as it had very limited hindlimb joint movement and falls over.
This mouse was 22 days old and given a gait impairment score of 4 (humane euthanasia endpoint) because it was unable to right itself within 30 seconds of falling on its side on all 3 of 3 trials.
This TDP-43Tg/TgAtxn2−/− mouse was at 61 days old and still had no overt motor impairment. It was therefore given a gait impairment score of 0.
Examples of three P20 TDP-43Tg/Tg that received intracerebroventricular (ICV) administration of either the control ASO or the Atxn2 ASO at P1. The two severely impaired mice (gait impairment score of 4), which were unable to right themselves, received the control ASO whereas the one unimpaired mouse received the Atxn2 ASO.