SUMMARY
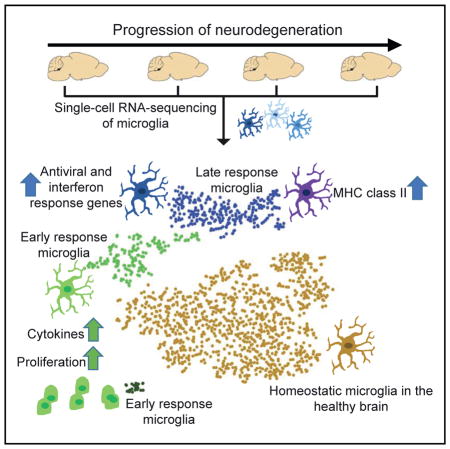
Microglia, the tissue-resident macrophages in the brain, are damage sensors that react to nearly any perturbation, including neurodegenerative diseases such as Alzheimer’s disease (AD). Here, using single-cell RNA sequencing, we determined the transcriptome of more than 1,600 individual microglia cells isolated from the hippocampus of a mouse model of severe neurodegeneration with AD-like phenotypes and of control mice at multiple time points during progression of neurodegeneration. In this neurodegeneration model, we discovered two molecularly distinct reactive microglia phenotypes that are typified by modules of co-regulated type I and type II interferon response genes, respectively. Furthermore, our work identified previously unobserved heterogeneity in the response of microglia to neurodegeneration, discovered disease stage-specific microglia cell states, revealed the trajectory of cellular reprogramming of microglia in response to neurodegeneration, and uncovered the underlying transcriptional programs.
In Brief
Mathys et al. use single-cell RNA sequencing to determine the phenotypic heterogeneity of microglia during the progression of neurodegeneration. They identify multiple disease stage-specific cell states, including two molecularly distinct reactive microglia phenotypes that are typified by modules of co-regulated type I and type II interferon response genes, respectively.
INTRODUCTION
Microglia are tissue-resident macrophages in the brain and spinal cord (Prinz and Priller, 2014). They clear apoptotic cells and are involved in both the elimination and maintenance of synapses for proper neural circuit wiring (Aguzzi et al., 2013). As the damage sensors for the CNS, microglia have been found to respond to nearly any CNS perturbation (Fourgeaud et al., 2016). Indeed, a growing body of evidence based on genome-wide association studies, transcriptomic, and epigenomic analyses, as well as experimental evidence in mouse models, implicates immunological mechanisms and their cellular component, microglia, in the pathogenesis of Alzheimer’s disease (AD) (Gjoneska et al., 2015; Mosher and Wyss-Coray, 2014; Neumann and Daly, 2013; Wang et al., 2015; Zhang et al., 2013). In brain tissue taken at autopsy from individuals with AD, microglia surround Aβ plaques, and their altered morphology indicates that these cells are responding to challenge (Bouvier et al., 2016; Heppner et al., 2015).
However, much remains to be learned about the molecular changes underlying the response of microglia in the AD brain. In particular, the signal that triggers the initial microglial response in the brain undergoing neurodegeneration remains to be determined. Genome-wide transcriptional profiling in microglia has revealed widespread changes in gene expression in mouse models of AD (Orre et al., 2014; Wang et al., 2015). However, ensemble-based approaches that measure gene expression from bulk populations of microglia cells in AD brains can only report population averages that may not reflect the response of individual cells or reveal cell subsets. Furthermore, these studies characterized the reactive microglia phenotype at only one time point, late in the progression of neurodegeneration. Therefore, it remains to be determined how the transcriptional programs in microglia cells change over time as they transition from their initial homeostatic state in the healthy brain to the reactive phenotypes seen in the neurodegenerating brain. Because the microglia phenotype may change drastically over the course of neurodegeneration, tracking with a fine temporal resolution is needed to capture the full spectrum of microglia cell states.
Here, we use single-cell RNA sequencing to examine the phenotypic heterogeneity of microglial cells in the healthy brain and in a mouse model of severe neurodegeneration with AD-like phenotypes. We identified multiple disease stage-specific microglia cell states that are almost exclusively observed in the diseased but not in the healthy brain. We delineate early- versus late-response gene modules and find that microglia proliferation is an early response to neuronal insult. We further identify two distinct reactive microglia phenotypes that arise at a later stage of neurodegeneration and are typified by modules of co-regulated type I and type II interferon response genes, respectively. Our work identifies previously unobserved heterogeneity in the response of microglia to neurodegeneration, discovers microglia cell states, reveals the trajectory of cellular reprogramming of microglia in response to neurodegeneration, and uncovers the underlying transcriptional programs.
RESULTS
RNA Sequencing Profiling of Single Microglial Cells Isolated from the Hippocampus
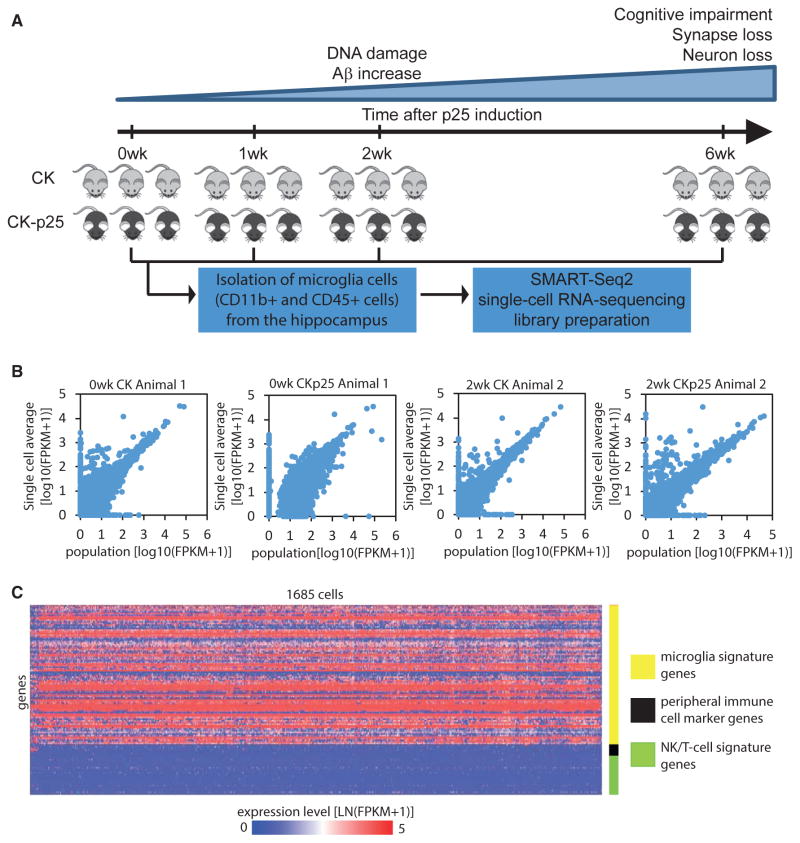
To determine the phenotypic heterogeneity and the transcriptional dynamics of microglia cells during the progression of neurodegeneration, we used the CK-p25 inducible mouse model of severe neurodegeneration. In CK-p25 mice, the expression of p25, the calpain cleavage product of Cdk5 activator p35, is under the control of the CamKII promoter and can be switched on by withdrawing doxycycline from the animal’s diet (Cruz et al., 2003; Cruz and Tsai, 2004; Fischer et al., 2005; Patrick et al., 1999). Although the CK-p25 model is not based on any genetic mutations associated with familial AD, it develops many aspects of AD-like pathology. A recent study reported that transcriptional profiles from CK-p25 mice and 5XFAD mice, a commonly used AD mouse model, show moderate but similar concordance with human AD brain signatures (Hargis and Blalock, 2017). Neurodegeneration in CK-p25 mice occurs in a temporally compressed but highly predictable manner (Cruz et al., 2003, 2006; Fischer et al., 2005). At 2 weeks after p25 induction, CK-p25 mice exhibit DNA damage and increased amyloid-β levels, followed by progressive neuronal and synaptic loss with cognitive impairment, which is severe by 6 weeks of p25 induction (Cruz et al., 2003, 2006; Fischer et al., 2005; Kim et al., 2008). Additional AD-like phenotypes found in 6-week induced CK-p25 mice include forebrain atrophy, astrogliosis, aberrant APP processing, and hyperphosphorylation of tau (Cruz et al., 2003, 2006). CK-p25 mice also display neurofibrillary tangle-like pathology after 27 weeks of p25 induction (Cruz et al., 2003). Importantly, as the p25 transgene is strictly expressed only in excitatory neurons (Figures S1A–S1D), this model permits us to examine the microglial response to neuronal cell death. We isolated cells expressing the microglia markers CD11b and CD45 from the hippocampus of three to four CK-p25 mice and three CK control littermates at each of the following four time points during the progression of neurodegeneration: before p25 induction and 1, 2, and 6 weeks after p25 induction (henceforth abbreviated as 0wk, 1wk, 2wk, and 6wk, respectively) (Figure 1A; Table S1). Individual CD11b-positive and CD45-positive microglial cells were sorted directly into RNA lysis buffer in 96-well plates using fluorescence-activated cell sorting (FACS) (Figure S2). To verify that our sorting protocol resulted in single cells in each well, we mixed microglia cells from two individually distinguishable mouse strains (wild-type and a Cx3cr1-knockout strain expressing TdTomato in microglia) and sorted single cells into the wells of a 96-well plate. We then measured the level of Cx3cr1 and TdTomato mRNA in each well using qPCR. In the large majority of wells, we detected either Cx3cr1 or TdTomato—but not both—mRNA species, confirming that the large majority of wells contained a single cell (Figures S1E–S1G). We prepared a total of 2,183 single-cell RNA sequencing libraries using a modified version of the Smart-Seq2 protocol (Picelli et al., 2014) and sequenced the libraries to a depth of 251,353 (median) mapped reads per cell. The average expression profile across single cells and the matching population profile were tightly correlated (Figures 1B and S1H), with an average Pearson product-moment correlation coefficient (r = 0.86) comparable with that observed in previous studies (Gaublomme et al., 2015; Shalek et al., 2014). We removed 498 cells on the basis of quality metrics (see Experimental Procedures), retaining 1,685 cells for further analyses. To verify that the cells isolated were indeed microglia cells, we compared the average expression profile of cells isolated from CK control mice with previously published expression profiles of specific brain cell types and with the expression profiles of monocytes and tissue-resident macrophages (Zhang et al., 2014; Lavin et al., 2014). We found that the average expression profile of cells isolated from CK control mice clustered with previously published microglia profiles (Figures S1I and S1J). Comparison with the set of 86 microglia signature genes defined by Butovsky et al. (2014), showed that microglial marker genes (including Csf1r, Tmem119, P2ry12, Hexb, and Sall1) were expressed at a relatively uniform level in the large majority of cells. In contrast, marker genes of peripheral immune cells (Cdc20, Ccr2, Cd163, and Ly6c1), and natural killer cell and T cell signature genes (including Zap70, Skap1, and Cd247) (Bezman et al., 2012) were expressed in only a small subset of the cells (Figures 1C and S1K).
Figure 1. Single-Cell RNA Sequencing of Microglia Cells Isolated from the Hippocampus of CK-p25 Mice and CK Control Littermates.
(A) Workflow diagram for single-cell RNA sequencing of microglia cells isolated from the hippocampus of CK-p25 mice and CK control littermates at four different time points after p25 induction.
(B) Quality of single-cell RNA sequencing. Scatterplots compare transcript expression (log10[FPKM+1]) between the average of 95 single-cells and a bulk population of 200 cells. The data from four representative animals are shown.
(C) Heatmap showing the expression level of 86 microglia signature genes (yellow), genes preferentially expressed in peripheral immune cells (black), and natural killer cell and T cell signature genes (green) across the 1,685 CD11b- and CD45-positive cells analyzed in this study.
Non-linear Dimensionality Reduction Reveals Multiple Distinct and Disease Stage-Specific Microglia Cell States
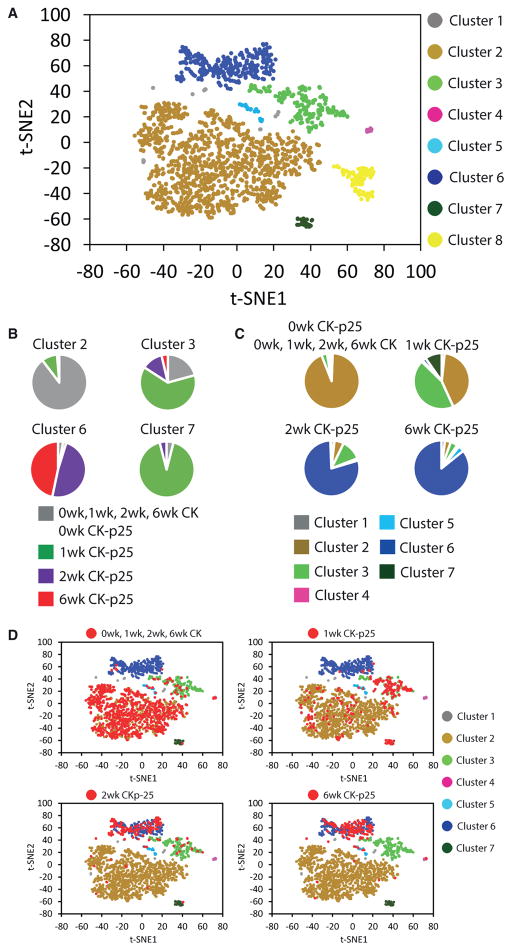
We analyzed 1,685 transcriptomes of single cells expressing the microglia markers CD11b and CD45, isolated from the hippocampus of 0wk, 1wk, 2wk, and 6wk CK-p25 and CK control littermates. Non-linear dimensionality reduction with t-distributed stochastic neighbor embedding (t-SNE) followed by density clustering revealed multiple distinct cell populations (Figure 2A) (van der Maaten and Hinton, 2008). Cluster 2 predominantly contained cells isolated from the CK control and 0wk CK-p25 (Figures 2B and 2C). Clusters 3 and 7 mainly contained cells isolated from 1wk CK-p25 mice, and cluster 6 was composed almost exclusively of cells isolated from 2wk and 6wk CK-p25 mice (Figures 2B and 2C). The large majority of cells isolated from the CK control mice at all four time points and from 0wk CK-p25 grouped together in cluster 2 (Figures 2B–2D). We obtained similar results after using a normalization technique to reduce the effects of confounders (Gaublomme et al., 2015) (Figures S3A and S3B), when we used a different RNA sequencing transcript quantification program, RSEM (Figures S3C and S3D), and when we used the GENECODE release M9 gene annotation (data not shown). We saw a similar disease stage-specific response of microglia to neurodegeneration using linear dimensionality reduction (data not shown) and found comparable results using the Single Cell Consensus Clustering (SC3) algorithm as an alternative approach to partitioning cellular subpopulations (Kiselev et al., 2016) (Figures S3E and S3F). Thus, our analysis revealed multiple microglia cell states in the progression of neurodegeneration that clustered separately from most of the cells isolated from the healthy brain (homeostatic microglia): an early-response state composed mainly of cells from 1wk CK-p25 mice and a late-response state composed of the large majority of cells isolated from 2wk and 6wk CK-p25 mice.
Figure 2. Non-linear Dimensionality Reduction Reveals Multiple Distinct and Disease Stage-Specific Microglia Cell States.
(A) Clustering of 1,685 CD11b and CD45 double-positive cells isolated from the hippocampus into eight populations. The t-SNE plot shows a two-dimensional representation of global gene expression profile relationships among 1,685 cells.
(B) Pie charts showing the composition of some of the clusters identified in (A).
(C) Pie charts showing the distribution of each group of cells indicated across the clusters identified in (A) (excluding cluster 8). Cells are grouped by genotype and time point.
(D) t-SNE plots as shown in (A). Cells isolated from CK control mice (at all four time points) and cells from CK-p25 mice 1, 2, and 6 weeks after p25 induction (0wk, 1wk, 2wk, and 6wk, respectively) are highlighted in red in individual panels.
The t-SNE analysis also revealed clusters 4 and 5 (Figure 2A), which contained only a small number of cells (9 and 21 cells, respectively) and expressed peripheral immune cell marker genes at higher levels relative to all other clusters, suggesting that they represent bone marrow-derived peripheral immune cells (Table S2). Cluster 8 (Figure 2A) contained cells that came exclusively from one out of three biological replicates (Table S3), indicating that it likely resulted from technical confounders and this cluster was therefore excluded from further analyses (see Experimental Procedures).
Single-Cell Differential Expression Analysis Reveals Hundreds of Genes Regulated during Cellular Reprogramming of Microglia in Response to Neurodegeneration
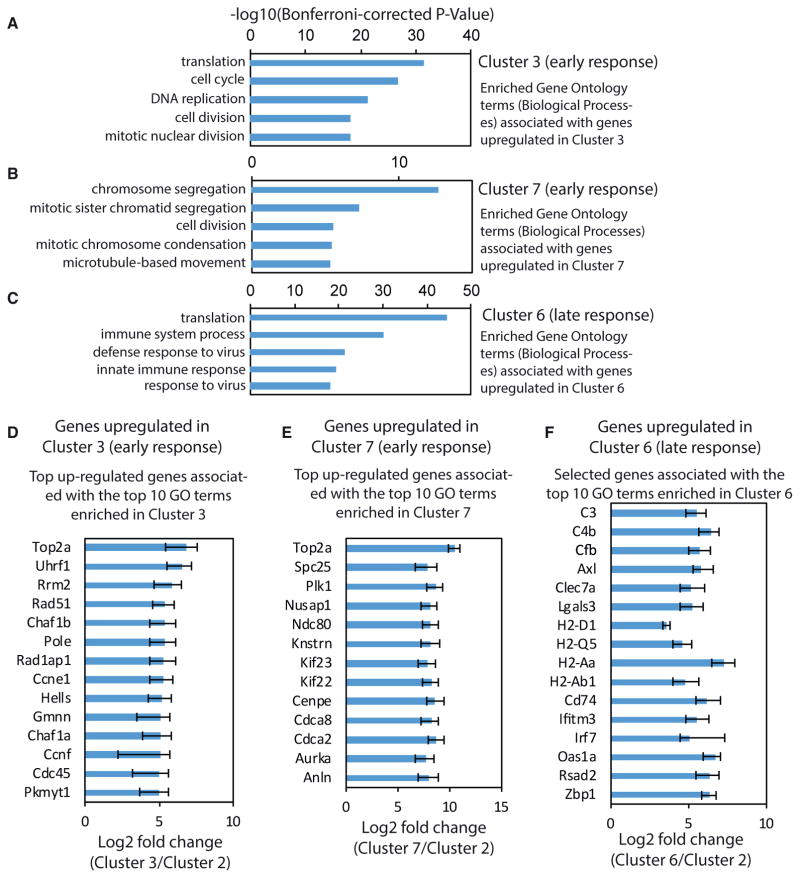
We next examined how microglia respond over time in the neurodegenerating brain environment by comparing homeostatic microglia (cluster 2) with early-response state (clusters 3 and 7) versus late-response state microglia (cluster 6). We used the single-cell differential expression (SCDE) software package (Kharchenko et al., 2014) to reveal hundreds of significantly up-and downregulated genes in the early and late-response microglia cell states, compared with homeostatic microglia (Figures S4A–S4C; Table S4). All future references to up- and downregulated gene expression refer to comparison with homeostatic microglia cluster 2. Gene Ontology (GO) term enrichment analysis revealed that cell cycle genes and genes involved in DNA replication and repair were over-represented among the genes upregulated in the early-response cluster 3 (Figures 3A and 3D). Cell cycle-related genes were also found to be upregulated in the early-response cluster 7. The top five GO terms enriched among the genes upregulated in cluster 7 were all related to cell division (Figures 3B and 3E). Plotting the average expression of G1/S and G2/M genes (Tirosh et al., 2016) revealed an approximate circle (Figure S4D), presumably reflecting progression along the cell cycle. Cells from Cluster 3 and Cluster 7 were separated by G1/S and G2/M phase scores, indicating they are in different phases of the cell cycle (Figure S4D). To measure the proliferative activity of microglia, we injected CK and CK-p25 mice with the thymidine analog, 5-ethynyl-2′-deoxyuridine (EdU), every second day during the first 2 weeks after p25 induction. Fluorescent labeling of EdU followed by flow cytometry analysis using the microglia markers CD11b and CD45 revealed that 3% of the microglia from CK control mice had incorporated EdU, in contrast to the significantly larger fraction (22%) seen in CK-p25 mice (Figure S4E). To test whether this increased proliferative activity is reflected in higher microglia density, we performed immunostaining of hippocampal sections and imaged across the major subregions in CK and CK-p25 mice. In 1wk CK-p25 mice, we observed that the density of Iba1-positive microglia was unchanged in the CA1 subregion (Figure S4F) but already significantly increased in CA3 (Figure S5G) and dentate gyrus (Figures S4H and S4K). In 2wk and 6wk CK-p25 mice, microglia density was significantly increased in all three hippocampal subregions examined (Figures S4F–S4K). These findings are consistent with our analysis of single-cell RNA sequencing (Figures 3A, 3B, 3D, and 3E) and EdU incorporation data (Figure S4E), indicating that hippocampal microglia show increased proliferation early in the response to neurodegeneration.
Figure 3. Single-Cell Differential Expression Analysis Reveals Hundreds of Genes Regulated during Cellular Reprogramming of Microglia in Response to Neurodegeneration.
(A–C) Bar plots showing the top 5 Gene Ontology (GO) terms (biological processes) associated with genes upregulated in (A) cluster 3 compared with cluster 2, (B) cluster 7 compared with cluster 2, and (C) cluster 6 compared with cluster 2.
(D and E) Bar graphs showing the fold change in gene expression of the top upregulated genes associated with the top 10 GO terms in cells of cluster 3 (D) and cluster 7 (E) compared with cells of cluster 2.
(F) Bar graph showing the fold change in gene expression of selected genes associated with the top 10 GO terms in cells of cluster 6 compared with cells of cluster 2.
For all panels, error bars show the 95% confidence interval.
Among the genes upregulated in the late-response cluster 6, we did not see cell cycle genes but rather saw over-representation of immune response-related genes instead (Figure 3C). The top five GO terms included immune system process, defense response to virus, and innate immune response (Figures 3C and 3F). The list of genes included those encoding major histo-compatibility complex (MHC) class 1 (H2-D1, H2-Q5) and class II (H2-Aa, H2-Ab1, Cd74) components. We also identified many interferon response genes (Ifitm3, Irf7) and genes associated with the GO term “defense response to virus” (OaS1a, Rsad2, Zbp1). Binding motifs of the interferon-regulatory factor (IRF) family were significantly enriched within a 1 kb window around the transcription start site of genes upregulated in cluster 6, providing further evidence for an enrichment of interferon response-related genes in late-response state microglia (Table S5). Overall, these data indicate that microglia mount a pronounced immune response during the later stages of neurodegeneration.
Temporally Distinct Transcriptional Dynamics among Immune Response-Related Genes during Microglia Activation
To further dissect the gene expression changes underlying cellular reprogramming of microglia, we analyzed the pairwise correlations between the expression levels of upregulated genes (see Experimental Procedures). Hierarchical clustering of the resulting correlation matrix of genes upregulated in the early-response cluster 3 cells revealed at least two major modules of co-regulated genes (Figure S5A). GO term enrichment analysis suggested that these two modules contained functionally distinct sets of genes. Whereas genes related to translation, metabolic processes, and immune response were over-represented in module 1, genes involved in cell cycle and DNA metabolic processes were over-represented in module 2 (Figure S5B). A similar separation into two functionally distinct modules was observed for the genes upregulated in the early-response cluster 7 (Figure S5C). Genes involved in cell cycle and mitosis were over-represented in module 1, while genes with a role in the immune response and metabolic processes were over-represented in module 2 (Figure S5D). Modules of co-regulated genes upregulated in the late-response cluster 6 also contained functionally distinct sets of genes (Figure S5E). Whereas genes involved in the immune response were over-represented in module 1, genes related to translation and metabolic processes were over-represented in module 2 (Figure S5F). Thus, this analysis identified functionally distinct modules of co-regulated genes in microglia responding to neurodegeneration. Interestingly, the GO terms enriched among module 1 genes upregulated in cluster 3 were also found to be the most highly enriched among the genes upregulated in cluster 6. Indeed, 199 of the 255 module 1 genes upregulated in cluster 3 were also found to be upregulated in cluster 6, indicating a partial overlap in the gene expression signatures that distinguish cells of the early and the late-response cluster from cells in the healthy brain (data not shown).
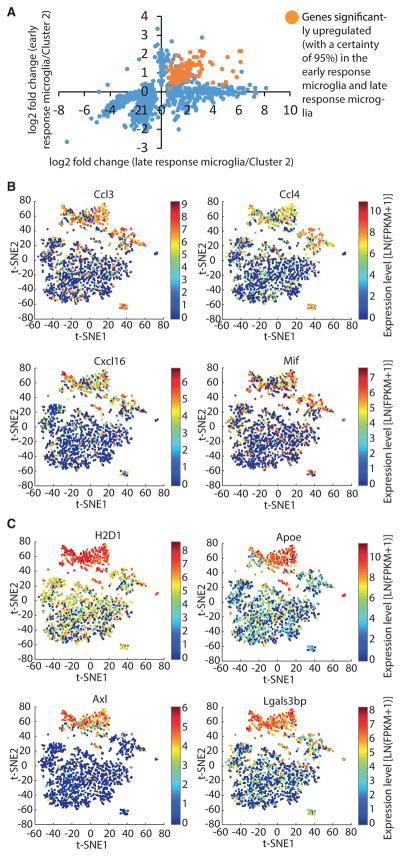
To more precisely examine the relationship between the early and late response of microglia cells, we next focused only on cluster 3 cells that were isolated from 1wk CK-p25 mice (Figure 2D). We found that a substantial fraction (191) of the genes significantly upregulated in late-response cluster 6 cells were also significantly upregulated (with a certainty of 95%) in CK-p25 microglia after only 1 week of p25 induction (Figure 4A). Of the two early-response clusters, we focused our analysis on cluster 3 because of relatively higher cell numbers, but a similar overlap in gene expression signatures could be seen in cluster 7, as 38 of the 57 module 2 genes upregulated in Cluster 7 were also found to be upregulated in cluster 6 (data not shown). When we plotted the fold change of gene expression in 1wk CK-p25 cluster 3 versus the fold change of gene expression in late-response cluster 6, we found roughly two groups of differentially expressed genes: one in which the early and late expression changes appeared to be correlated and a second group that was exclusively upregulated only in late-response microglia but not in early-response microglia (Figure 4A). Among the set of 191 early and consistently upregulated genes, 38 genes were significantly upregulated with a more stringent cutoff (p < 0.001) in cells of the early-response cluster 3 (data not shown). This latter set included genes encoding lysosomal cysteine proteases (Ctsb, Ctsz), the cysteine protease inhibitor Cst7, the inflammatory cytokine Mif, chemokines (Ccl12, Ccl3, Ccl4, Cxcl16), other immune response-related genes (Tlr2, Lilrb4), and genes involved in glycolysis (Pkm, Pgk1, Gapdh, Pgam1). These genes were also significantly upregulated (with a certainty of 95%) in the microglia of the early-response cluster 7 (Table S4). The set of genes that was exclusively upregulated during the later stages of neurodegeneration included many genes we had identified to be more than 10-fold upregulated in the cells of cluster 6, including complement components (C3, C4b, and Cfb), MHC class 1 (H2-D1, H2-Q4, H2-Q5) and class II (H2-Aa, H2-Ab1, Cd74) components, genes involved in the interferon response (Ifitm3, Irf7) and genes associated with the GO term “defense response to virus” (OaS1a, Rsad2, Zbp1). Overall, our analysis revealed a dichotomy in the transcriptional dynamics among genes significantly upregulated in late-response microglia and point to the possibility that the early-response cluster represents a transient, intermediate activation state of microglia.
Figure 4. Temporally Distinct Transcriptional Dynamics among Immune Response-Related Genes during Micro-glia Activation.
(A) Scatterplot comparing the fold change in gene expression in early- and late-response microglia for genes significantly differentially expressed. The genes significantly upregulated (with a certainty of 95%) in early- and late-response microglia are shown in orange. All other genes are shown in blue. Definition of early-response microglia: cells of cluster 3 that were isolated from CK-p25 mice 1 week after p25 induction (1wk CK-p25). Definition of late-response microglia: cells of cluster 6.
(B and C) t-SNE plots as shown in Figure 2A. Selected genes showing early and consistent upregulation (B) and selected genes exclusively upregulated in the late-response cluster 6 (C) are shown. Data points are colored by the expression levels of the genes indicated.
Next, we explored the transcriptional dynamics of these genes during the time course of neurodegeneration in individual microglia. This analysis provided evidence that a subset of immune response-related genes in late-response cluster 6, including chemokines Ccl3, Ccl4, and Cxcl16 and inflammatory cytokine Mif, were already upregulated in a subset of microglia 1 week after p25 induction (Figure 4B). In contrast, the expression of other immune-related genes, such as H2-D1, Axl, Apoe, and Lgals3bp, was increased in the majority of cells of the late-response cluster 6 but not in 1wk CK-p25 Cluster 3 (Figure 4C). In summary, our analysis of transcriptional dynamics during disease progression uncovered temporally distinct subsets of immune response-related genes during microglia activation in a neurodegenerative context.
Heterogeneous Late Response of Microglia to Neurodegeneration Is Typified by Different Modules of Co-regulated Genes
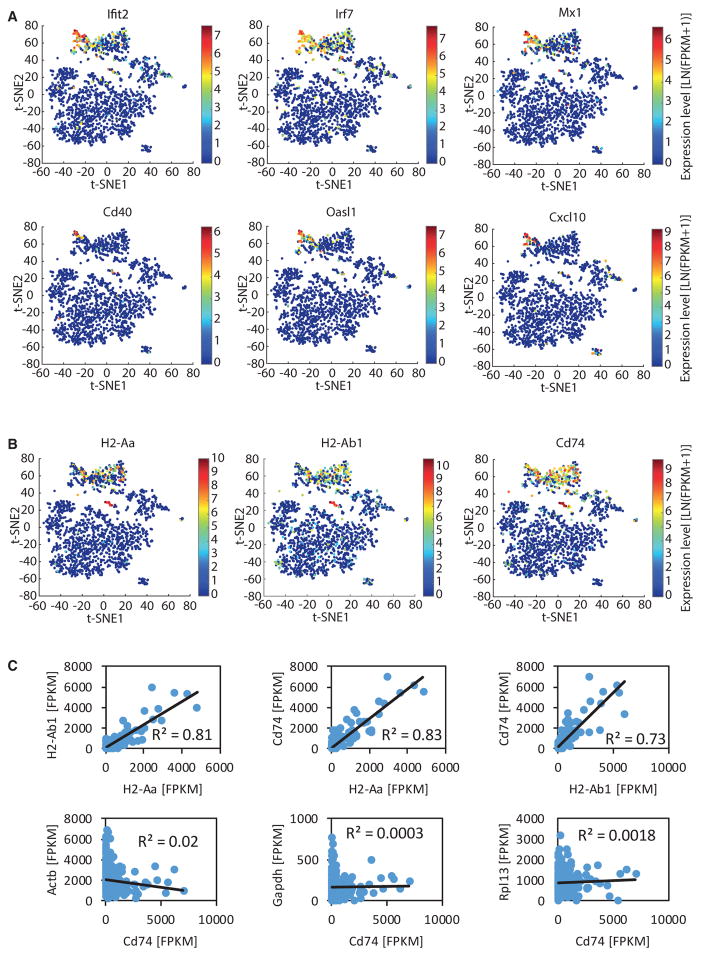
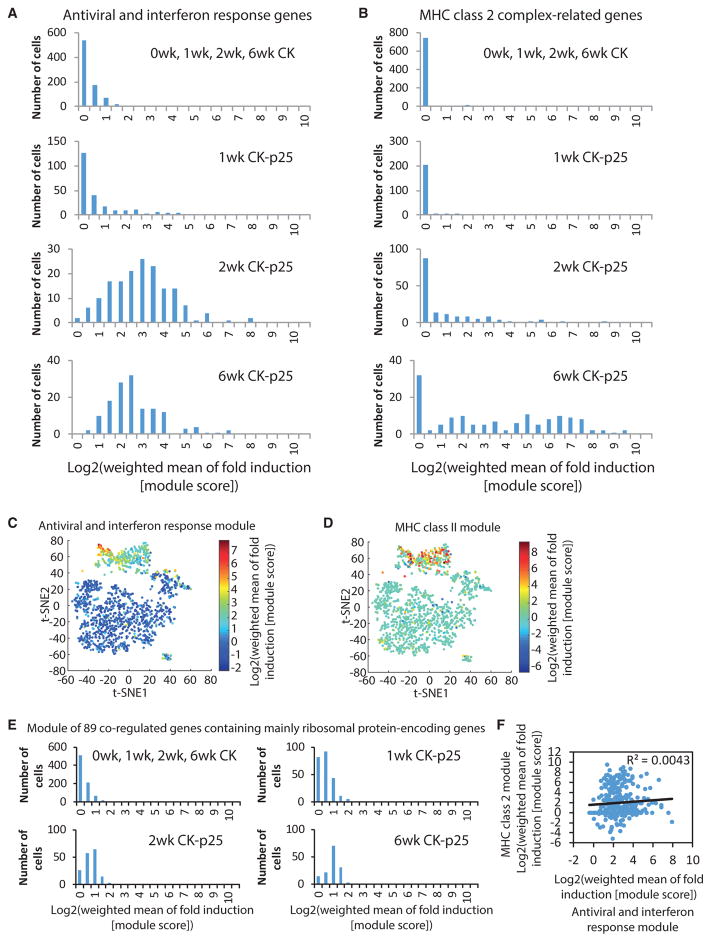
We next focused solely on the transcriptional dynamics in late-response microglia and discovered small subsets of single cells characterized by upregulation of specific gene sets, such as antiviral and interferon response genes (Figure 5A) and components of the MHC class II pathway (Figure 5B). To quantify the activation of these two gene sets within individual cells, we computed a module score as previously described (Shalek et al., 2014; see Experimental Procedures). We computed the module scores for a set of 132 co-regulated antiviral and interferon response genes. Most 2wk and 6wk CK-p25 microglia showed activation of the antiviral and interferon response gene expression module (Figures 6A and 6C). However, there was substantial variation across individual cells, with a small subset of cells exhibiting a fold induction module score that was at least an order of magnitude higher than the average (Figures 6A and 6C). We also computed the module scores for the four genes encoding components of MHC class II. Whereas some 2wk and 6wk CK-p25 microglia showed no detectable activation of the MHC class II module, other cells exhibited a fold induction value at least an order of magnitude higher than the average (Figures 6B and 6D). In contrast to the variability seen in antiviral and interferon response genes and MHC class II components, the induction scores of another module of co-regulated genes, containing mainly ribosomal protein-encoding genes exhibited a much narrower distribution (Figure 6E). Moreover, for the MHC class II components, although the expression of these genes was highly variable between cells, it was tightly correlated within individual cells and the expression of the MHC class II-related gene CD74 was not correlated with the expression of the housekeeping genes Actb, Gapdh, and Rpl13 (Figure 5C). These observations indicate that the extensive variability in the expression of MHC class II components measured across cells reflects true biological differences, rather than technical noise inherent to single-cell RNA sequencing experiments There was no correlation observed between the induction of the antiviral and interferon response module and the MHC class II module, indicating that the cells expressing high levels of antiviral and interferon response genes and the cells expressing high levels of MHC class II genes are not necessarily identical (Figure 6F). Thus, these data indicate that there are at least two distinct reactive microglia phenotypes in neurodegeneration.
Figure 5. Heterogeneous Late Response to Neurodegeneration of Microglia Is Typified by Different Modules of Co-regulated Genes.
t-SNE plots as shown in Figure 2A.
(A) Data points are colored by the expression level of selected antiviral and interferon response genes as indicated.
(B) Data points are colored by the expression level of the genes encoding MHC class II components as indicated.
(C) Scatterplots showing the correlation of the expression level of MHC class II related genes (H2-Aa, H2-Ab1, Cd74) and of Cd74 and the housekeeping genes Actb, Gapdh, and Rpl13 across the cells of cluster 6.
Figure 6. Two Distinct Reactive Microglia Phenotypes in Late Response to Neurodegeneration.
(A and B) Histograms showing the distribution of the weighted fold induction of a module of 132 antiviral and interferon response genes (A) and four MHC class II complex-related genes (B) across the cells of the groups indicated.
(C and D) t-SNE plots as shown in Figure 2A. Data points are colored by the weighted fold induction of (C) a module of 132 antiviral and interferon response genes and (D) a module of four MHC class II complex-related genes.
(E) Histograms showing the distribution of the weighted fold induction of a module of co-regulated genes mainly containing ribosomal protein-encoding genes across the cells of the groups indicated.
(F) Scatterplot showing the correlation between the induction of the antiviral and interferon response module and the MHC class II module across the cells isolated from CK-p25 mice 2 and 6 weeks after p25 induction.
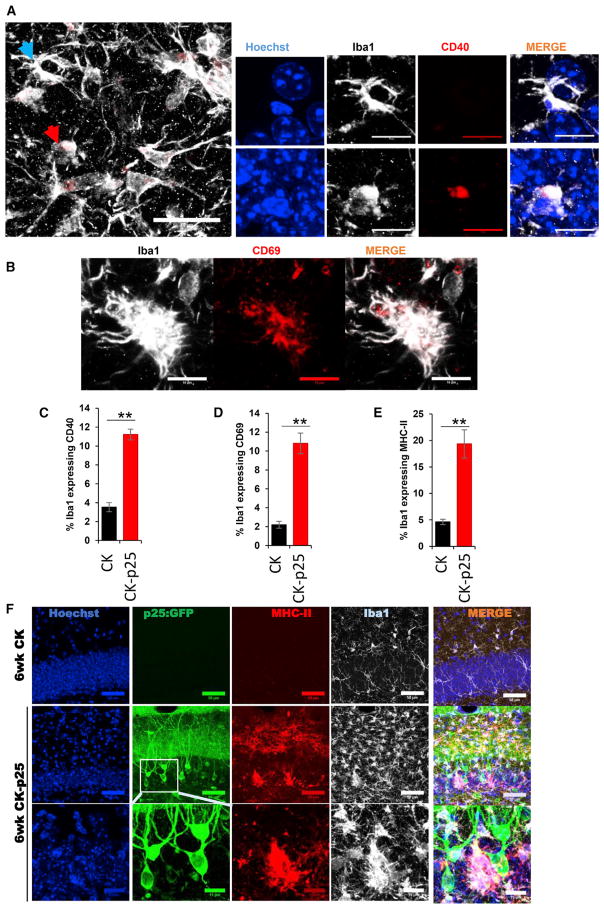
To corroborate the observed heterogeneity among microglia with an alternative approach, we performed immunostaining of hippocampal sections from CK and CK-p25 mice. To examine the expression of co-regulated antiviral and interferon response genes, we stained the sections with antibodies recognizing the protein products of Cd40 and Cd69 and for the microglia marker Iba1. We observed an increase in the number of CD40-expressing cells in all three hippocampal subregions of 6wk CK-p25 mice compared with CK control mice (dentate gyrus: Figures 7A, 7C, and S6A; CA1 and CA3: Figure S6B). Importantly, the immunostaining revealed that only a subset of the Iba1-positive cells detectably express CD40 (dentate gyrus: Figures 7A, 7C, and S6A; CA1 and CA3: Figure S6B). Similarly, we found an increased number of CD69-positive cells in all three hippocampal subregions of CK-p25 mice 6 weeks after p25 induction compared with CK control mice (dentate gyrus: Figures 7B, 7D, and S6C; CA1 and CA3: Figure S6D). However, again, only a subset of Iba1-positive cells detectably expressed CD69 in CK-p25 mice (dentate gyrus: Figures 7D and S6C; CA1 and CA3: Figure S6D). We next stained the sections with an antibody recognizing the MHC class II component CD74. In the hippocampus of CK control mice, only very few cells expressing the MHC class II component CD74 were detected (dentate gyrus: Figures 7E, 7F, and S6E; CA1 and CA3: Figure S6F). In contrast, the number of CD74-positive cells was dramatically increased in the hippocampus of CK-p25 mice 6 weeks after p25 induction (dentate gyrus: Figures 7E, 7F, and S6E; CA1 and CA3: Figure S6F). Importantly, the overall proportion of CD74-expressing microglia was limited to only a subset of cells, confirming the heterogeneity of the hippocampal microglia population in the response to neurodegeneration (dentate gyrus: Figures 7E, 7F, and S6E; CA1 and CA3: Figure S6F). Taken together, our immunostaining data show that the expression of two components of a module of co-regulated antiviral and interferon response genes (CD69 and CD40) as well as the MHC class II component CD74 are strongly upregulated in subsets of microglia in the hippocampus of CK-p25. Thus, our immunostaining data confirm a previously unobserved heterogeneity in the response of microglia to neurodegeneration.
Figure 7. Immunostaining Reveals Heterogeneous Late Response of Microglia to Neurodegeneration.
Immunostaining in the dentate gyrus of CK-p25 mice 6 weeks after p25 induction.
(A) Immunohistochemistry with anti-CD40 (red) and anti-Iba1 (white) antibodies. Cells indicated with blue and red arrows are shown at higher magnification on the right.
(B) Immunohistochemistry with anti-CD69 (red) and anti-Iba1 (white) antibodies.
(C–E) Quantification of the CD40 (C), CD69 (D), MHC2 (E), and Iba1 immunostaining. Values are percentages of Iba1-positive cells expressing CD40, CD69, and MHC2, respectively.
(F) Immunohistochemistry with anti-GFP (green), anti-CD74 (MHC2, red), and anti-Iba1 (white) antibodies. For all graphs, quantification is based on immunostaining in the dentate gyrus of seven CK mice and four CK-p25 mice, with two sections per animal.
Error bars show SEM. **p < 0.01; ***p < 0.001.
Gene Sets Induced in the CK-p25 Mouse Model of Neurodegeneration Also Tend to Be Upregulated in Microglia of Aged Human Individuals
Finally, we wondered whether the gene sets induced in the CK-p25 mouse model of neurodegeneration might also be relevant for the biology of human microglia. To this end, we compared our data with a recently published transcriptomic analysis of purified human cortical microglia (Galatro et al., 2017). Specifically, we asked whether the gene sets induced in the CK-p25 mouse model of neurodegeneration also tend to be upregulated in microglia of aged human individuals compared with microglia of younger individuals. Indeed, we observed that the expression of gene sets that are human orthologs of the genes significantly upregulated in clusters 3, 6, and 7 was positively correlated with age (Figures S7A, S7C, and S7E). In contrast, the expression of genes downregulated in clusters 3, 6, and 7 was not significantly correlated with age (Figures S7B, S7D, and S7F). We also found that the expression of human orthologs of the modules of co-regulated antiviral and interferon response genes and MHC class II components was positively correlated with age (Figures S7G and S7H). Specifically, the antiviral and interferon response genes Isg15, Oasl, Ifitm3, Irf7, and Ifi30 were found among the genes most significantly correlated with age in human microglia (Table S6). Thus, these data indicate that the gene sets induced in the CK-p25 mouse model of neurodegeneration also tend to be upregulated in microglia of aged human individuals.
DISCUSSION
In this study we comprehensively surveyed the transcriptome of microglia cells as they progressed from healthy to neurodegeneration states in the brain at single-cell resolution. We found a remarkable phenotypic heterogeneity of microglia: from early-response states, characterized by marked proliferation, to late-response states of mounting immune response. In this latter group, we discovered a further heterogeneity that was typified by functionally distinct modules of co-regulated genes.
Temporal Changes of Microglia Response to Neurodegeneration
We were interested to decipher the initial response of microglia to insult and to track the temporal changes of these cells over the course of neurodegeneration. We used the CK-p25 mouse model of severe neurodegeneration, as it displays key pathological hallmarks of AD in a temporally predictable manner, to study the response of microglia at fine- and single-cell resolution. Our single-cell RNA sequencing analysis identified two neurodegeneration-associated microglia cell states that are distinct from the microglia state in the healthy brain. Importantly, we found that microglia isolated at an early stage of neurodegeneration (early-response microglia cell state) are distinct from microglia isolated at a late stage of neurodegeneration (late-response microglia cell state). However, the early- and late-response microglia cell states are not completely unrelated, as we found that a substantial fraction of genes that are upregulated in late-response microglia are already increased in early-response microglia. Moreover, 1wk CK-p25 cells were distributed across homeostatic microglia cluster 2 and early-response cluster 3, and share characteristics with both microglia in the healthy brain and fully activated microglia at the later stages of neurodegeneration. These observations suggest that the early-response microglia cell state represents a transient intermediate activation state that constitutes part of the trajectory of cellular reprogramming of homeostatic microglia in response to neurodegeneration.
Although we cannot exclude the possibility that some of the cells analyzed are peripheral immune cells, our data suggest that the large majority of cells examined are microglia cells. First, the large majority of cells analyzed expressed microglia marker genes but not peripheral immune cells signature genes (Figures 1C and S1K). Second, only a very small fraction of cells expressed high levels of CD45, a characteristic of peripheral immune cells (Figure S2). Third, t-SNE analysis revealed two small clusters (clusters 4 and 5) that expressed peripheral immune cell marker genes, showing that our methods have sufficient power to detect the subtle differences in gene expression between mi-croglia and peripheral immune cells (Figure 2A).
Microglia Proliferate and Increase the Expression of a Small Set of Cytokines at an Early Stage of Neurodegeneration
Our previous study had revealed an increased level of transcripts encoding immune response-related genes in hippocampal tissue of CK-p25 mice (Gjoneska et al., 2015). However, it was unclear whether this increase was due to gene expression changes in microglia or to an expansion of the microglia population as a result of proliferation. Our data clearly demonstrate that microglia respond to neurodegeneration with both a dramatic reprogramming of their transcriptome and increased proliferation. We identified a small set of cytokines, including the pro-inflammatory cytokines MIF and TNF, that are upregulated in the late-response microglia cell state but already show upregulation 1 week after p25 induction. Interestingly, the pro-inflammatory cytokine TNF has recently been identified as one of the factors that are necessary and sufficient to induce a neurotoxic astrocyte cell state (Liddelow et al., 2017). This raises the possibility that early-response microglia may contribute to the initiation of a cascade that ultimately leads to neuronal death during the later stages of neurodegeneration. Given the pleiotropic effects of TNF reported (Probert, 2015), it is possible that increased expression of TNF in microglia serves multiple distinct functions that remain to be explored.
Two Distinct Reactive Microglia Phenotypes in Neurodegeneration
We discovered small subsets of microglia in the hippocampus of CK-p25 mice late in neurodegeneration typified by strong upregulated expression of selected gene sets, such as antiviral and interferon response genes and MHC class II components. Interestingly, these two subsets do not overlap entirely, indicating that not only are there at least two distinct reactive microglia phenotypes in the heterogeneous cell population but that there are likely multiple triggers of microglia activation. Interestingly, we detected many genes that are induced by type I interferon among the module of co-regulated antiviral and interferon response genes. In contrast, MHC class II genes have been shown to be induced by interferon-gamma (type II interferon) (O’Keefe et al., 2001). This raises the intriguing possibility that the two distinct subsets of reactive microglia we observed in CK-p25 mice could reflect exposure to type I interferon versus type II interferon.
What might be the trigger to induce expression of the co-regulated antiviral and interferon response gene module? We found that a subset of microglia with activated type I interferon pathway are in close proximity to neurons harboring DNA damage (Figures S6G and S6H). It is well established that DNA damage can lead to the induction of type 1 interferons (Härtlova et al., 2015). Thus, DNA damage might be one factor that contributes to the induction of antiviral and interferon response genes in microglia. Consistent with this idea, we found that the module of co-regulated antiviral and interferon response genes was significantly upregulated in microglia of the DNA-repair deficient mouse model Ercc1Δ/− compared with wild-type control mice (Figures S7I and S7J).
In the peripheral immune system, MHC class II molecules are responsible for presenting peptides derived from extracellular pathogens to T cells and are expressed by a subset of antigen-presenting cells, such as macrophages, dendritic cells, and B cells (Ting and Trowsdale, 2002). Whether the subset of microglia expressing high levels of MHC class II molecules indeed functions as antigen-presenting cells is an intriguing hypothesis that remains to be addressed. Alternatively, MHC class II molecules could serve a T cell-independent function in microglia. Supporting this idea, it has been shown that the presence of MHC class II molecules exacerbated both neurodegenerative symptoms and neuropathology in murine globoid leukodystrophy, most likely in a T cell-independent manner (Matsushima et al., 1994).
Recently, Keren-Shaul et al. (2017) reported the identification of a microglia type associated with neurodegenerative diseases, termed disease-associated microglia (DAM). The authors carried out single-cell RNA sequencing on microglia isolated from a mouse model of AD that expresses five human familial AD gene mutations (5XFAD) and from a mouse model of amyotrophic lateral sclerosis (mSOD1 [G93A] mice). The DAM microglia phenotype was observed only in the 5XFAD and mSOD1 (G93A) mice but not in the corresponding wild-type mice. Using a different mouse model of neurodegeneration (CK-p25), we identified a microglia cell state that was almost exclusively observed at the later stages of disease progression (late-response microglia). Interestingly, late-response microglia express increased levels of many genes that were also observed to be upregulated in DAM (e.g., Cd9, Itgax, Clec7a, Cd63, Spp1, Fth1, Axl, Lpl, Cst7, Ctsb, Apoe). Of the 278 genes significantly upregulated in DAM, 202 genes were also significantly upregulated in late-response microglia, suggesting a substantial similarity between the expression profiles of DAM and late-response microglia. This observation is consistent with the idea that the DAM program may be a primed set of genes that is expressed in response to varied conditions of altered homeostasis (Keren-Shaul et al., 2017). However, we found that many antiviral and interferon response genes were significantly upregulated in late-response microglia but not in DAM, suggesting that there may also be potential differences between these two microglia cell states. Keren-Shaul et al. (2017) proposed that DAM are generated through a two-step activation process. Homeostatic microglia first transition to an intermediate stage (stage 1 DAM) in a Trem2-independent manner, followed by a second, Trem2-dependent, transition to stage 2 DAM. We saw significant differences in the expression of both stage 1 DAM and stage 2 DAM characteristic genes in late-response microglia but much less so in early-response microglia (Figure S7K). This observation may indicate that early-response microglia are in an even more naive activation state than stage 1 DAM. Whether the induction of a DAM-like expression program in microglia of the CK-p25 model of neurodegeneration is protective, neutral, or deleterious remains to be determined.
In summary, our work identified previously unobserved heterogeneity in the response of microglia to neurodegeneration, including the discovery of microglia cell states, and uncovered transcriptional programs underlying the trajectory of cellular reprogramming of microglia in response to neurodegeneration. Our analysis also indicates that many of the gene expression regulatory events identified in our mouse model are conserved in aged human microglia. Thus, these insights into the molecular programs underlying microglia activation may pave the way for designing rational and efficient strategies to treat AD and other neurodegenerative diseases.
EXPERIMENTAL PROCEDURES
Animals
All animal work was approved by the Committee for Animal Care of the Division of Comparative Medicine at the Massachusetts Institute of Technology.
Isolation of Microglia from the Hippocampus
Hippocampal tissue was enzymatically digested using the Neural Tissue Dissociation Kit (P) (catalog number 130-092-628; Miltenyi Biotec) according to the manufacturer’s protocol, with minor modifications. The resulting cell suspension was then stained using allophycocyanin (APC)-conjugated CD11b mouse clone M1/70.15.11.5 (130-098-088; Miltenyi Biotec) and phycoerythrin (PE)-conjugated CD45 antibody (553081; BD PharMingen) according to the manufacturer’s (Miltenyi Biotec) recommendations. FACS was then used to purify CD11b and CD45 positive microglial cells.
Single-Cell RNA Sequencing Library Preparation
Single-cell RNA sequencing libraries were generated on the basis of the SMART-Seq2 protocol (Picelli et al., 2014) with minor modifications. Libraries were tagmented using the Nextera XT DNA Library Preparation Kit (catalog number FC-131-1096; Illumina) and the Nextera XT Index Kit version 2 Sets A, B, C, and D according to the manufacturer’s instructions with minor modifications. Specifically, reactions were run at one fourth the recommended volume, the tagmentation step was extended to 10 min, and the extension time during the PCR step was increased from 30 s to 60 s.
Click-iT Plus EdU Cell Proliferation Assay
Animals were intraperitoneally injected with 50 mg/kg EdU every second day during the first 2 weeks after p25 induction. Two weeks after p25 induction, the animals were transcardially perfused with ice-cold PBS, and microglia were isolated using the Neural Tissue Dissociation Kit (P) as described above. EdU incorporation was then detected using the Click-iT Plus EdU Pacific Blue Flow Cytometry Assay Kit (catalog number C10636; Thermo Fisher Scientfic) according to the manufacturer’s instructions.
Immunohistochemistry
Immunohistochemistry experiments were performed as described previously (Iaccarino et al., 2016).
Supplementary Material
Highlights.
Transcriptomes of 1,685 individual microglia cells from healthy and diseased brains
Identification of disease stage-specific microglia cell states
Trajectory of cellular reprogramming of microglia in response to neurodegeneration
Two distinct reactive microglia phenotypes in the neurodegenerating brain
Acknowledgments
We are grateful to E. Demmons and J.J. Trombetta for technical assistance. We thank members of the Tsai laboratory, J. Penney, H. Meharena, W. Ralvenius, and E. Gjoneska, for discussions and comments on the paper. We thank P. Wisniewski, C. Zollo, C. Johns, and P. Autissier of the Whitehead flow cytometry core facility for assistance with FACS. This work is supported in part by the JBP Foundation and NIH grant RF1 AG054321 to L.-H.T. H.M. was sup- ported by an Early Postdoc Mobility fellowship from the Swiss National Science Foundation (P2BSP3_151885).
Footnotes
AUTHOR CONTRIBUTIONS
H.M., C.A., P.L.D.J., R.M.R, A.R., and L.-H.T. designed experiments. H.M. performed the majority of experiments with help from E.M. C.A. performed the immunohistochemistry experiments. H.M. and F.G. performed the bioinformatics analysis with help from M.H. H.M., J.Z.Y., R.M.R., and L.-H.T. wrote the manuscript.
DATA AND SOFTWARE AVAILABILITY
The accession number for the RNA sequencing data reported in this paper is GEO: GSE103334.
Supplemental Information includes Supplemental Experimental Procedures, seven figures, and six tables and can be found with this article online at https://doi.org/10.1016/j.celrep.2017.09.039.
References
- Aguzzi A, Barres BA, Bennett ML. Microglia: scapegoat, saboteur, or something else? Science. 2013;339:156–161. doi: 10.1126/science.1227901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezman NA, Kim CC, Sun JC, Min-Oo G, Hendricks DW, Kamimura Y, Best JA, Goldrath AW, Lanier LL Immunological Genome Project Consortium. Molecular definition of the identity and activation of natural killer cells. Nat Immunol. 2012;13:1000–1009. doi: 10.1038/ni.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvier DS, Jones EV, Quesseveur G, Davoli MA, Ferreira AT, Quirion R, Mechawar N, Murai KK. High resolution dissection of reactive glial nets in Alzheimer’s disease. Sci Rep. 2016;6:24544. doi: 10.1038/srep24544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butovsky O, Jedrychowski MP, Moore CS, Cialic R, Lanser AJ, Gabriely G, Koeglsperger T, Dake B, Wu PM, Doykan CE, et al. Identification of a unique TGF-β-dependent molecular and functional signature in microglia. Nat Neurosci. 2014;17:131–143. doi: 10.1038/nn.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz JC, Tsai LH. A Jekyll and Hyde kinase: roles for Cdk5 in brain development and disease. Curr Opin Neurobiol. 2004;14:390–394. doi: 10.1016/j.conb.2004.05.002. [DOI] [PubMed] [Google Scholar]
- Cruz JC, Tseng HC, Goldman JA, Shih H, Tsai LH. Aberrant Cdk5 activation by p25 triggers pathological events leading to neurodegeneration and neurofibrillary tangles. Neuron. 2003;40:471–483. doi: 10.1016/s0896-6273(03)00627-5. [DOI] [PubMed] [Google Scholar]
- Cruz JC, Kim D, Moy LY, Dobbin MM, Sun X, Bronson RT, Tsai LH. p25/cyclin-dependent kinase 5 induces production and intra-neuronal accumulation of amyloid beta in vivo. J Neurosci. 2006;26:10536–10541. doi: 10.1523/JNEUROSCI.3133-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer A, Sananbenesi F, Pang PT, Lu B, Tsai LH. Opposing roles of transient and prolonged expression of p25 in synaptic plasticity and hippocampus-dependent memory. Neuron. 2005;48:825–838. doi: 10.1016/j.neuron.2005.10.033. [DOI] [PubMed] [Google Scholar]
- Fourgeaud L, Través PG, Tufail Y, Leal-Bailey H, Lew ED, Burrola PG, Callaway P, Zagórska A, Rothlin CV, Nimmerjahn A, Lemke G. TAM receptors regulate multiple features of microglial physiology. Nature. 2016;532:240–244. doi: 10.1038/nature17630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galatro TF, Holtman IR, Lerario AM, Vainchtein ID, Brouwer N, Sola PR, Veras MM, Pereira TF, Leite REP, Möller T, et al. Transcriptomic analysis of purified human cortical microglia reveals age-associated changes. Nat Neurosci. 2017;20:1162–1171. doi: 10.1038/nn.4597. [DOI] [PubMed] [Google Scholar]
- Gaublomme JT, Yosef N, Lee Y, Gertner RS, Yang LV, Wu C, Pandolfi PP, Mak T, Satija R, Shalek AK, et al. Single-cell genomics unveils critical regulators of Th17 cell pathogenicity. Cell. 2015;163:1400–1412. doi: 10.1016/j.cell.2015.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gjoneska E, Pfenning AR, Mathys H, Quon G, Kundaje A, Tsai LH, Kellis M. Conserved epigenomic signals in mice and humans reveal immune basis of Alzheimer’s disease. Nature. 2015;518:365–369. doi: 10.1038/nature14252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargis K, Blalock E. Transcriptional signatures of brain aging and Alzheimer’s disease: What are our rodent models telling us? Behav Brain Res. 2017;322:311–328. doi: 10.1016/j.bbr.2016.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Härtlova A, Erttmann SF, Raffi FA, Schmalz AM, Resch U, Anugula S, Lienenklaus S, Nilsson LM, Kröger A, Nilsson JA, et al. DNA damage primes the type I interferon system via the cytosolic DNA sensor STING to promote anti-microbial innate immunity. Immunity. 2015;42:332–343. doi: 10.1016/j.immuni.2015.01.012. [DOI] [PubMed] [Google Scholar]
- Heppner FL, Ransohoff RM, Becher B. Immune attack: the role of inflammation in Alzheimer disease. Nat Rev Neurosci. 2015;16:358–372. doi: 10.1038/nrn3880. [DOI] [PubMed] [Google Scholar]
- Iaccarino HF, Singer AC, Martorell AJ, Rudenko A, Gao F, Gillingham TZ, Mathys H, Seo J, Kritskiy O, Abdurrob F, et al. Gamma frequency entrainment attenuates amyloid load and modifies microglia. Nature. 2016;540:230–235. doi: 10.1038/nature20587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren-Shaul H, Spinrad A, Weiner A, Matcovitch-Natan O, Dvir-Szternfeld R, Ulland TK, David E, Baruch K, Lara-Astaiso D, Toth B, et al. A unique microglia type associated with restricting development of Alzheimer’s disease. Cell. 2017;169:1276–1290. e17. doi: 10.1016/j.cell.2017.05.018. [DOI] [PubMed] [Google Scholar]
- Kharchenko PV, Silberstein L, Scadden DT. Bayesian approach to single-cell differential expression analysis. Nat Methods. 2014;11:740–742. doi: 10.1038/nmeth.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Frank CL, Dobbin MM, Tsunemoto RK, Tu W, Peng PL, Guan JS, Lee BH, Moy LY, Giusti P, et al. Deregulation of HDAC1 by p25/Cdk5 in neurotoxicity. Neuron. 2008;60:803–817. doi: 10.1016/j.neuron.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiselev VY, Kirschner K, Schaub MT, Andrews T, Yiu A, Chandra T, Natarajan KN, Reik W, Barahona M, Green AR, Hemberg M, et al. SC3: consensus clustering of single-cell RNA-seq data. Nat Methods. 2016;14:483–486. doi: 10.1038/nmeth.4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavin Y, Winter D, Blecher-Gonen R, David E, Keren-Shaul H, Merad M, Jung S, Amit I. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell. 2014;159:1312–1326. doi: 10.1016/j.cell.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liddelow SA, Guttenplan KA, Clarke LE, Bennett FC, Bohlen CJ, Schirmer L, Bennett ML, Münch AE, Chung WS, Peterson TC, et al. Neurotoxic reactive astrocytes are induced by activated microglia. Nature. 2017;541:481–487. doi: 10.1038/nature21029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushima GK, Taniike M, Glimcher LH, Grusby MJ, Frelinger JA, Suzuki K, Ting JP. Absence of MHC class II molecules reduces CNS demyelination, microglial/macrophage infiltration, and twitching in murine globoid cell leukodystrophy. Cell. 1994;78:645–656. doi: 10.1016/0092-8674(94)90529-0. [DOI] [PubMed] [Google Scholar]
- Mosher KI, Wyss-Coray T. Microglial dysfunction in brain aging and Alzheimer’s disease. Biochem Pharmacol. 2014;88:594–604. doi: 10.1016/j.bcp.2014.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann H, Daly MJ. Variant TREM2 as risk factor for Alzheimer’s disease. N Engl J Med. 2013;368:182–184. doi: 10.1056/NEJMe1213157. [DOI] [PubMed] [Google Scholar]
- O’Keefe GM, Nguyen VT, Ping Tang LL, Benveniste EN. IFN-gamma regulation of class II transactivator promoter IV in macrophages and microglia: involvement of the suppressors of cytokine signaling-1 protein. J Immunol. 2001;166:2260–2269. doi: 10.4049/jimmunol.166.4.2260. [DOI] [PubMed] [Google Scholar]
- Orre M, Kamphuis W, Osborn LM, Jansen AH, Kooijman L, Bossers K, Hol EM. Isolation of glia from Alzheimer’s mice reveals inflammation and dysfunction. Neurobiol Aging. 2014;35:2746–2760. doi: 10.1016/j.neurobiolaging.2014.06.004. [DOI] [PubMed] [Google Scholar]
- Patrick GN, Zukerberg L, Nikolic M, de la Monte S, Dikkes P, Tsai LH. Conversion of p35 to p25 deregulates Cdk5 activity and promotes neurodegeneration. Nature. 1999;402:615–622. doi: 10.1038/45159. [DOI] [PubMed] [Google Scholar]
- Picelli S, Faridani OR, Björklund AK, Winberg G, Sagasser S, Sandberg R. Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc. 2014;9:171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- Prinz M, Priller J. Microglia and brain macrophages in the molecular age: from origin to neuropsychiatric disease. Nat Rev Neurosci. 2014;15:300–312. doi: 10.1038/nrn3722. [DOI] [PubMed] [Google Scholar]
- Probert L. TNF and its receptors in the CNS: the essential, the desirable and the deleterious effects. Neuroscience. 2015;302:2–22. doi: 10.1016/j.neuroscience.2015.06.038. [DOI] [PubMed] [Google Scholar]
- Shalek AK, Satija R, Shuga J, Trombetta JJ, Gennert D, Lu D, Chen P, Gertner RS, Gaublomme JT, Yosef N, et al. Single-cell RNA-seq reveals dynamic paracrine control of cellular variation. Nature. 2014;510:363–369. doi: 10.1038/nature13437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting JP-Y, Trowsdale J. Genetic control of MHC class II expression. Cell. 2002;109(Suppl):S21–S33. doi: 10.1016/s0092-8674(02)00696-7. [DOI] [PubMed] [Google Scholar]
- Tirosh I, Venteicher AS, Hebert C, Escalante LE, Patel AP, Yizhak K, Fisher JM, Rodman C, Mount C, Filbin MG, et al. Single-cell RNA-seq supports a developmental hierarchy in human oligodendroglioma. Nature. 2016;539:309–313. doi: 10.1038/nature20123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Maaten L, Hinton G. Visualizing data using t-SNE. J Mach Learn Res. 2008;9:2579–2605. [Google Scholar]
- Wang Y, Cella M, Mallinson K, Ulrich JD, Young KL, Robinette ML, Gilfillan S, Krishnan GM, Sudhakar S, Zinselmeyer BH, et al. TREM2 lipid sensing sustains the microglial response in an Alzheimer’s disease model. Cell. 2015;160:1061–1071. doi: 10.1016/j.cell.2015.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Gaiteri C, Bodea LG, Wang Z, McElwee J, Podtelezhnikov AA, Zhang C, Xie T, Tran L, Dobrin R, et al. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer’s disease. Cell. 2013;153:707–720. doi: 10.1016/j.cell.2013.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O’Keeffe S, Phatnani HP, Guarnieri P, Caneda C, Ruderisch N, et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci. 2014;34:11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.