Abstract
Proteins have the potential to replace synthetic small molecule drugs due to their specificity and selectivity in their activity. However, if administered in their native form, they can aggregate leading to immunogenicity or get cleared by renal and reticuloendothelial system which ultimately hampers their efficacy as therapeutic agents. Thus, the formulation in which proteins are administered plays a key role in retaining their biological activity and lowering immunogenicity while minimizing the dosage requirements. Enzyme-wrapping, using synthetic polymers, is a strategy employed to provide enzymes with lower immunogenicity, longer circulation times and better targeting capabilities. Protein-polymer complexation methods, involving covalent, non-covalent and electrostatic interactions, that can provide means to develop formulations for retaining enzyme stability are discussed in this chapter.
Keywords: nano-armoring, protein-polymer conjugates, polymer wrapping, protein encapsulation, emulsion polymerization, responsive nanogels, electrostatic complexation
1. Introduction
Enzymes utilize their selectivity in biochemical catalysis to play a vital role in most processes in living organisms. Therefore, a deviation from their optimal activity can lead to serious physiological abnormalities and sometimes be life threatening (Vellodi, 2005). While their exquisitely high specificity accounts for precisely controlling biochemical processes, this specifity also makes it very difficult to correct in case of their deficiency. Taking this into consideration, recombinant protein/enzymes technologies have been developed to “directly” supplement a deficient enzyme or to interfere with or augment an abnormal biological process (Leader, Baca & Golan, 2008). However, systemic administration of these therapeutic enzymes as such is often ineffective, as they lead to immunogenicity and cytotoxicity(Brange, Andersen, Laursen, Meyn & Rasmussen, 1997, Curatolo, Valsasina, Caccia, Raimondi, Orsini & Bianchetti, 1997, Hermeling, Crommelin, Schellekens & Jiskoot, 2004). Moreover, these proteins can also undergo clearance by renal and reticuloendothelial systems (Poznansky & Juliano, 1984), leaving only minimal fraction of the administered dose reaching the target site(Desnick & Schuchman, 2012).
To address these challenges, several formulation strategies have been laid out for delivering proteins more effectively into biological systems. These methods include PEG-ylation of proteins(Brocchini, Godwin, Balan, Choi, Zloh & Shaunak, 2008, Joralemon, McRae & Emrick, 2010, Nischan & Hackenberger, 2014, Pasut & Veronese, 2012), non-covalent and electrostatic methods to complex proteins with polymers,(Ayame, Morimoto & Akiyoshi, 2008, Ghosh, Yang, Arvizo, Zhu, Agasti, Mo, et al, 2010, González-Toro, Ryu, Chacko, Zhuang & Thayumanavan, 2012, Lee, Wang, Ye, Yoon, Chan & Yang, 2008, Mero, Ishino, Chaiken, Veronese & Pasut, 2011, Mueller, Capelle, Seyrek, Martel, Carrupt, Arvinte, et al, 2012) encapsulation of proteins/enzymes using inorganic networks,(Dyal, Loos, Noto, Chang, Spagnoli, Shafi, et al, 2003, Luckarift, Spain, Naik & Stone, 2004) encapsulation of enzymes in liposomal vesicles (Dai, Wang, Zhao, Li & Wang, 2006) and entrapment in hydrogels(Azagarsamy, Alge, Radhakrishnan, Tibbitt & Anseth, 2012, Cohen, Beaudette, Tseng, Bachelder, Mende, Engleman, et al, 2008, Molla, Marcinko, Prasad, Deming, Garman & Thayumanavan, 2014, Murthy, Xu, Schuck, Kunisawa, Shastri & Fréchet, 2003, Vermonden, Censi & Hennink, 2012).
In this chapter, some of these methods will be described in detail where enzymes are wrapped or conjugated with synthetic polymers(González-Toro et al, 2012, Matsumoto, González-Toro, Chacko, Maynard & Thayumanavan, 2013, Molla et al, 2014, Ventura, Eron, González-Toro, Raghupathi, Wang, Hardy, et al, 2015). This approach of modifying enzymes using polymers can be termed as nano-armoring of enzymes, since the resulting formulation of polymer –enzyme complex have nanoscale (10–100 nm) dimensions. The method of enzyme wrapping using polymers can be divided into three categories based on the nature of interaction between enzymes and polymers:
Covalent conjugation
Electrostatic complexation
Non-covalent entrapment
2. Covalent conjugation
This method involves the formation of a covalent bond between certain functional groups of the enzyme and the polymer(Heredia & Maynard, 2007). Although in some cases covalent modification of enzymes is associated with decreased biological activity,(Fishburn, 2008) it can be minimized either by site specific modification of enzymes(Nischan et al, 2014) where its active site is not affected or by using covalent modification which is reversible (Ventura et al, 2015). In this chapter, we will particularly focus on reversible covalent modification of enzyme using polymers, where the use of organic solvents is limited or completely avoided, because organic solvents have been implicated in the observed enzyme denaturation during the conjugation process(Babu, Moradian & Douglas, 2001, Knubovets, Osterhout & Klibanov, 1999).
Covalent conjugation between enzyme and polymer through a disulfide bond is an attractive option due to its good conjugating ability in aqueous conditions and its reversible nature (ability to cleave under the reducing conditions of cell cytosol). The reversible nature of this bond allows the enzyme to tracelessly dissociate from the polymer (i.e. without any remnant functional groups of the polymer in the protein), thereby retaining its original structure and function. However, it is to be noted that this approach is more suitable for enzymes that are targeted to the cytosol such as caspases, cytosolic protein kinases, cytochrome c, and RNAse.
The thiol reactive polymer that will be discussed here is an amphiphilic random co-polymer (PEGMA-PDSMA polymer) consisting of a hydrophilic polyethylene glycol-based and pyridyldisulfide-based monomers.(Ryu, Chacko, Jiwpanich, Bickerton, Babu & Thayumanavan, 2010, Ventura et al, 2015) This polymer is a suitable candidate for enzyme wrapping because of the following reasons: i) The polymer is water-dispersible ii) It is non-toxic and can potentially be a safe pharmaceutical excipient(Ryu et al, 2010) iii) The pyridyl disulfide group of the polymer can readily react with free cysteine thiols of the proteins.
The covalent conjugation between an enzyme and the polymer can be executed by utilizing the surface accessible cysteines on the enzyme and thiol-reactive pyridyl disulfide monomers of the polymer. One limitation to this approach is that, not all enzymes have surface accessible cysteine groups to react with the polymer. However, there exist some enzymes that have free cysteines at their active site(Chang & Yang, 2000); conjugation of polymer using those cysteines might affect their catalytic activity when conjugated. However, if the target delivery site is the cytosol, the enzyme can detach itself from the polymer through cleavage of the disulfide bond under the reducing conditions of the cytosol(Balendiran, Dabur & Fraser, 2004). This process should then restore the active site functionalities and thus the enzymatic function.
2.1 Synthesis of Thiol reactive (PEGMA:PDSMA) polymer
The monomer(Ghosh, Basu & Thayumanavan, 2006) and the polymer(Ventura et al, 2015) can be synthesized following literature protocols. An overview of the polymer synthesis is shown below:
Equipment and chemicals
Poly(ethylene glycol)monomethylether methacrylate (PEGMA; MW 475), 2,2′-dithiodipyridine, 2,2′-azobis-(2-methylpropionitrile) (AIBN), 4-cyano-4-(phenylcarbonothioylthio) pentanoic acid (chain transfer agent), and D,L-dithiothreitol (DTT) from Sigma-Aldrich, Pyridyl disulfide ethyl methacrylate (PDSMA) was prepared from a reported procedure(Ghosh et al, 2006)., Schlenk flask, magnetic stir bar, stirrer, hotplate, sand/oil bath, freeze pump thaw set up.
Procedure
- To synthesize a 1:1 PEGMA:PDSMA polymer, weigh 537 mg of PDSMA and 1.00 g of PEGMA into a 10 mL Schlenk flask with a magnetic stir bar.
- To synthesize a 3:7 PEGMA:PDSMA polymer, weigh 749 mg of PDSMA and 598 mg of PEGMA in a 10 mL Schlenk flask with a magnetic stir bar.
Add 1.2 mg of AIBN and 21 mg of 4-cyano-4-(phenylcarbonothioylthio)pentanoic acid in 3 mL THF
Close the Schlenk flask and perform freeze pump-thaw cycles (three cycles) to degas the reaction mixture with an argon inflow in the final cycle.
Seal the reaction vessel and place it in a preheated oil/sand bath at 70 °C with stirring at 350 rpm for 12 hours.
Pour the reaction mixture in 20 mL cold diethyl ether to obtain the polymer as precipitate.
2.2 Protein-polymer conjugation utilizing surface accessible cysteines of proteins
This approach is viable if the protein of interest has enough surface accessible cysteine groups in order to react with the polymer. As an example, a detailed method to conjugate caspase-3 enzyme with PEGMA-PDSMA polymer is described here.
2.2.1 Determination of surface accessible cysteine residues on the protein
Method overview
To quantify the amount of surface accessible cysteine residues on the protein, Ellman’s reagent (5,5′-dithiobis(2-nitrobenzoic acid)/ DTNB) was used. DTNB, upon reaction with thiols of proteins, generates a colored product 2-nitro-5-thiobenzoate (TNB) that absorbs at 412 nm. For every mole of thiol that reacts with DTNB, one mole of TNB is produced and by quantifying the amount of TNB, the number of cysteines per mole of protein can be estimated.
Equipment and chemicals
Ellman’s reagent (5,5′-dithiobis(2-nitrobenzoic acid)/ DTNB) from Sigma-Aldrich, Protein (caspase-3), 1.5 mL Eppendorf micro-centrifuge tubes, 20 mM Tris pH 8.0 buffer, UV-Visible spectrophotometer.
Procedure
Prepare a protein stock solution of 10 µM concentration in Tris pH 8.0 buffer.
Prepare Ellman’s reagent stock solution of 500 uM concentration in Tris pH 8.0 buffer.
Mix 50 µL of protein stock solution with 50 µL of Ellman’s reagent solution in a 1.5 mL micro-centrifuge tube and incubate for 30 minutes at room temperature.
Make the volume of this reaction mixture to 1 mL using Tris pH 8.0 buffer and measure the absorbance at 412 nm.
The number of reactive cysteines per protein can be calculated by fitting the absorption values into Beer-Lambert’s equation: A= ε.c.l where A= Absorbance, ε= molar extinction coefficient of TNB which is 14,150 M−1cm−1, c = concentration of TNB (which is equal to the concentration of cysteines of protein) and l is the absorption path length. The number of accessible cysteines per protein can be calculated by dividing the number of moles of cysteines (c) over number of moles of protein.
2.2.2) Reaction of enzyme with thiol reactive polymer & synthesis of nanogel around the protein
Once the number of surface accessible cysteines on the enzyme is estimated, we can calculate the amount of polymer to be used for conjugation. Typically, large excess of the polymer (molar excess of thiol reactive PDS functional groups on the polymer) should be used in order to completely engage the surface accessible cysteines on protein in conjugation with polymer.
Method overview
In this example, 0.06 mg of caspase-3 (0.002 micromoles of enzyme and 0.01 micromoles of total cysteines) is reacted with 3 mg of PEGMA:PDSMA polymer (containing 4 micromoles of PDS) to form a protein-polymer conjugate. To create a polymeric network around the enzyme, self-crosslinking ability of the polymer will be utilized.
Taking the above example in perspective, 3 mg of polymer has about 4 micromoles of PDS units. In order to achieve 20% crosslink density, i.e. to utilize 20 % of PDS in polymer for crosslinking, 10 mole percent DTT (0.4 micromoles) should be added. This is because each DTT can cleave two PDS units on the polymer to result in one disulfide linkage. This crosslinking process will result in a polymeric network around the enzyme creating a nano-armor. The additional unreacted PDS units on the polymer can be utilized to surface functionalize with thiol terminated PEG to provide better circulation ability or a thiol containing ligand for active targeting.
Equipment and chemicals
PBS buffer (pH 7.4), 3 mg/mL PEGMA:PDSMA polymer stock solution in PBS pH 7.4 buffer, stir plate, 7 mL glass vial and stir bar, caspase-3 stock solution 0.6 mg /mL PBS, 1mg/mL DTT stock solution in PBS (pH 7.4), PEG thiol of MW1000 Da from Lysan Bio inc (stock solution of 20 mg/mL in PBS), fridge/cold room with a magnetic stir plate at 4 °C.
Procedure
Take 1 mL of polymer stock solution in a 7 mL glass vial with an appropriate stir bar and place it on a magnetic stir plate.
Stir the polymer solution at 350 rpm at 20 °C for 15 minutes.
Add 100 µL (0.06 mg) of caspase-3 stock drop wise.
Stir the solution for 1 hour at 20 °C.
To this solution, add 62 µL of DTT stock solution (0.06 mg of DTT).
Stir the solution for an additional 1 hour at 20 °C.
Add 159 µL PEG-thiol stock solution dropwise and stir the solution for an additional 24 hours at 4 °C.
Tips on enzyme conjugation:
Allow gentle stirring and avoid air bubbles inside the reaction to avoid protein denaturation.
For the dropwise addition of the small volumes, use capillary pipette tips.
2.2.3 Separation of unreacted enzyme and byproducts from the enzyme-polymer complex
The disulfide exchange reaction to form protein-polymer conjugate, polymer self-crosslinking, and nanogel surface with PEG thiol will result in pyridothione and oxidized DTT byproducts which are removed by dialysis. The complete removal of pyridothione byproduct was evaluated by monitoring its corresponding absorption peak at 340 nm.
Equipment and chemicals
7000 Da MWCO snake skin dialysis membrane & Amicon Ultra 100,000 Da MWCO centrifugal filters from Fisher Scientific, ultracentrifuge, PBS (pH 7.4) buffer, 1 L beaker with a magnetic stir bar, fridge/cold room with a magnetic stir plate at 4 °C.
Procedure
Take the nanogel sample into 7000 Da MWCO snake skin dialysis membrane and dialyze against 1 L PBS (pH 7.4) buffer in a beaker for 24 hours by replacing PBS every 6 hours.
Collect the sample after dialysis and dialyze again using Amicon Ultra 100,000 Da MWCO centrifugal filters to remove unconjugated enzyme and to concentrate the sample to the desired volume.
2.2.4 Determination of enzyme-polymer complexation and enzyme release
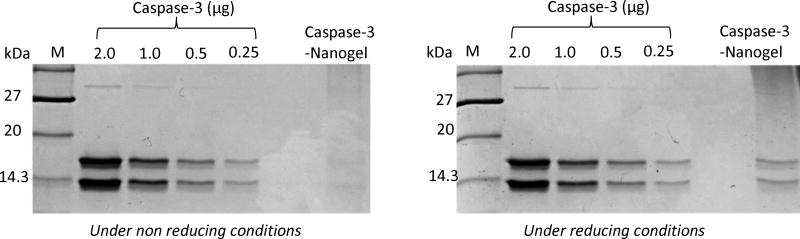
To verify the enzyme-polymer conjugation, SDS PAGE technique can be utilized. Under non-reducing conditions, the conjugate, being high molecular weight complex will have a lower mobility compared to just the protein by itself. However, the conjugate, upon incubation under reducing conditions, protein dissociates from the polymer and can be monitored by the appearance of a protein band.
Equipment and chemicals
SDS gel electrophoresis running buffer, DTT, 16% acrylamide gel, gel electrophoresis cell, enzyme-encapsulated nanogel solution (prepared in the previous steps).
Procedure
To test the enzyme encapsulation, take 50 µg of nanogel in a micro-centrifuge tube and heat it at 90 °C for 5 minutes (DTT which is normally used for protein denaturation should not be used).
To test the release of entrapped enzyme, take 50 µg of nanogel in a micro-centrifuge tube and incubate it in 100 mM DTT solution for one hour at 25 °C. Later, heat it at 90 °C for 5 minutes.
Analyze the samples by SDS-PAGE using 16% acrylamide gel and using coomassie blue for staining.
2.2.5 Enzyme activity study
Equipment and chemicals
Nanogel stock solution (with estimated 100 nM caspase-3 concentration based on protein feed during its preparation) , 100 nM caspase-3 enzyme stock solution in caspase-3 buffer, Amicon Ultra 3000 Da MWCO centrifugal filters from Fisher Scientific, ultra-centrifuge, caspase-3 buffer (20 mM HEPES pH 7.5, 5 mM CaCl2, 150 mM NaCl, and 10% PEG 400), 1 M and 50 mM DTT stock solutions in caspase-3 buffer, fluorogenic substrate (N-acetyl Asp-GLu-Val-Asp-7-amino-4-methylcoumarin) from Enzo Lifesciences (prepare 1 mM stock solution in caspase-3 buffer), DTT, temperature adjustable fluorescent plate reader.
Procedure
Take 90 µL of nanogel stock solution in a micro-centrifuge tube and add 10 µL of 1 M DTT (to make a final DTT concentration of 100 mM).
As a negative control, take 90 µL of nanogel stock solution and add 10 µL of 50 mM DTT (to make a final DTT concentration of 0.5 mM).
Take 90 µL of 100 nM enzyme stock solution in two separate tubes and add 10 µL of 1 M DTT and 50 mM DTT as a reference for 100 % enzyme activity.
Incubate the samples at 25 °C for 1 hour.
Take 90 µL of the each of the samples in a 96 black well plate
Add 10 µL of 1 mM Ac-DEVD-AMC substrate solution
Record the kinetic fluorescence by using λEx: 365 nm/ λEm: 495 nm for 7 minutes at 37 °C and plot the slope of the kinetic curve over time to measure the enzyme activity.
2.3 Appending a reactive thiol to protein
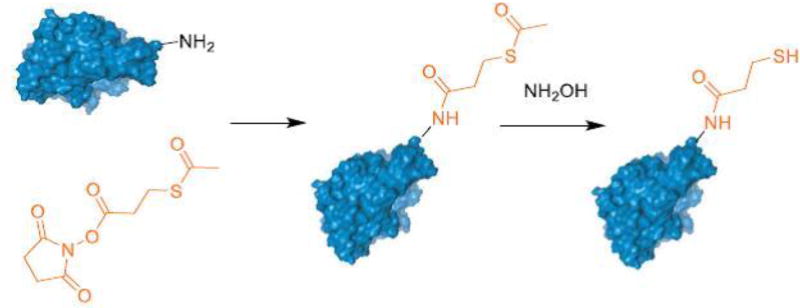
The success of the above methodology to conjugate enzyme with polymers using cysteines usually depends on the number of surface accessible cysteine units on the enzyme and their reactivity. However, this method is not very efficient with proteins having less accessible cysteine units. To circumvent this, an alternative approach can be used where proteins are modified to append a thiol functional group on their surface(Matsumoto et al, 2013). This can be achieved by following well-established protocols such as using SATP (N-succinimidyl-S-acetylthio propionate) or SATA (N-succinimidyl S-acetylthioacetate) to modify the lysine-based primary amines present on the protein surface.
Equipment and chemicals
SATP from Fisher Scientific (stock solution of 16 mg of SATP in 1 mL DMF, 50 mM sodium phosphate (pH 7.5) buffer solution with 1 mM EDTA), 0.5 M hydroxylamine hydrochloride in 50 mM sodium phosphate ( pH 7.5) buffer solution with 25 mM EDTA, Amicon Ultra 3000 Da MWCO centrifugal filters from Fisher Scientific, FPLC set up with superose 6, 10/300 column with temperature set up at 4 °C, Elution solvent for FPLC (50 mM sodium phosphate (pH 7.5) buffer containing 15 mM NaCl, UV visible spectrophotometer.
Procedure
Weigh 5 mg of protein of interest and dissolve it in 0.5 mL of pH 7.5 50 mM sodium phosphate solution with 1mM EDTA in a micro-centrifuge tube.
To that solution, add 5 µL of SATP stock solution, mix it gently and incubate for one hour at 20 °C.
Remove the unreacted SATP by ultrafiltration using Amicon Ultra centrifugal filters with suitable molecular weight cutoff (lower than the molecular weight of protein).
Resuspend the concentrated protein in the ultracentrifuge tube in 0.5 mL of pH 7.5 PBS.
For the deprotection of acetylated thiols, add 50 µL of hydroxylamine hydrochloride stock solution to the protein solution and incubate for 2 hours at 20 °C.
Use FPLC to purify the protein and concentrate it to the desired volume using ultrafiltration.
Calculate the amount of free thiol using Ellman’s assay described in Section 2.2.1.
Once the thiol moiety is generated on the protein, the method described earlier (Section 2.2.2 −2.2.3) should be followed for polymer conjugation and nanogel synthesis.
3. Electrostatic complexation
As discussed earlier, the disulfide mode of covalent conjugation would be an ideal choice if the target site to deliver the enzyme is cell cytosol. This is because of the cytosolic reducing environment, which can detach the polymer from the enzyme. However, if the enzyme’s target site of delivery is non-cytosolic (such as lysosomes) where no reducing environment is present, to cleave the polymer form the enzyme, disulfide conjugation strategy may not be a suitable choice. For example, β-galactosidase is an enzyme that is present in the lysosomes. The deficiency of this enzyme is associated with lysosomal storage diseases such as gangliosidosis, Morquio syndrome B, and galactosialidosis. Enzyme replacement therapies have been considered to supplement these deficient enzymes into the lysosomes(Desnick et al, 2012). Since lysosomes do not possess a reducing environment necessary to shed the disulfide based polymer wrapping around the enzyme, alternate methods of enzyme wrapping such as electrostatic complexation and entrapment of protein inside a polymeric matrix by non-covalent means could be of great use.
Proteins possess either a positive or a negative surface charge depending on their amino acid composition and pH of the solution in which they are present. Electrostatic complexation method involves utilizing the surface charge of a protein to complex with a polymer having a complementary charge. The example of electrostatic complexation that will be described here is based on enzyme β-galactosidase (β-gal) and polymeric nanogel derived from PEGMA:PDSMA amphiphilic random polymers(González-Toro et al, 2012). The isoelectric point (pI) of β-gal is 4.8, which suggests that at physiological pH (pH 7.4) the enzyme will have a net negative surface charge and therefore it can be complexed with a nanogel having a net positive charge. The protein complexation with nanogel involves three steps as follows:
Synthesis of nanogels
Surface charge generation on the nanogels
Electrostatic complexation of the protein with nanogels
3.1 Synthesis of polymer and nanogels
To synthesize PEGMA:PDSMA polymer 3:7 ratio, the method described in Section 2.1 can be adopted. The polymer, when dissolved in an aqueous solution, forms nano-micellar assemblies and has an ability to encapsulate hydrophobic guest molecules. By using a calculated amount of reducing agent (DTT), these micellar assemblies are crosslinked to form nanogels.
3.1.1 Encapsulation of hydrophobic guests by polymeric micelle
Hydrophobic guest encapsulation by the amphiphilic polymer was achieved by co-solvent method. A suitable choice of organic solvent should be made after considering solubility and stability of the hydrophobic molecules. Here we describe the encapsulation procedure using hydrophobic DiI dye that is soluble in acetone as an example.
Equipment and chemicals
DiI dye (1,1'-dioctadecyl-3,3,3',3'-tetramethylindocarbocyanine perchlorate (‘DiI’; DiIC18(3))) purchased from Fisher Scientific, acetone, PEGMA :PDSMA (3:7) polymer stock solution in PBS (pH 7.4) buffer, stir plate, 7 mL glass vial and stir bar, 1mg/mL DTT stock solution in PBS (pH 7.4).
Procedure
Prepare a 2 mg/mL DiI stock solution in acetone (depending on the solubility of the hydrophobic molecule an organic solvent must be chosen which is miscible in water and has low boiling point).
Take 1 mL of polymer stock solution in a 7 mL vial with a magnetic stir bar and stir the solution at 350 rpm.
To the polymeric solution, add 100 µL of hydrophobic stock solution dropwise under constant stirring. Care should be taken to ensure that the volume ratio of organic to aqueous solution is kept to a minimum (around 0.1 or less).
Allow this mixture to stir in open air for 6 hours to evaporate the organic solvent to form a polymeric micelle with guest molecules localized in its core.
The polymeric micelle solution should be passed through a 0.45 micron nylon filter to remove the un-encapsulated hydrophobic molecules.
3.1.2 Crosslinking of the polymeric micelle
Calculating the amount of DTT for a desired crosslinking density
1 mole of DTT can react with 2 moles of PDS functionalities in the polymer by a disulfide exchange reaction to generate one mole disulfide crosslinks. Therefore, to attain 10% crosslink density (utilize 10 mole % of PDS in polymer) 5 mole % of DTT relative to the amount of PDS functionalities should be used.
In the current example which has PEGMA:PDSMA in 3:7 ratio, every 10 mg of polymer contains 0.009 mmoles of PEGMA monomer (MW 473 g/mol) and 0.021 mmoles of PDSMA monomer (MW 255 g/mol). To make a 10% crosslink nanogel, the amount of DTT that is required will be 5% relative to the PDS, which is 0.001 mmoles.
Equipment and chemicals
10 mg/mL polymeric micelle stock solution prepared in the previous step, sodium phosphate buffer (5 mM pH 7.4), stir plate, 7 mL glass vial and stir bar, 1mg/mL DTT stock solution in phosphate buffer 5 mM (pH 7.4), 3000 Da MWCO snake skin dialysis membrane (purchased from Fisher Scientific), fridge/cold room with a magnetic stir plate at 4 °C.
Procedure
Take 1 mL of polymeric micelle solution which was prepared previously (10 mg/mL polymer concentration) in a vial with a magnetic stir bar.
Stir the solution at 350 rpm and to it add 154 µL of DTT stock solution dropwise.
Allow the reaction to stir for at least 1 hour at 25 °C.
Monitor the progress of reaction by measuring the absorption peak at 340 nm every 30 minutes (resulting from the pyridothione byproduct).
The crosslinking reaction is complete if there is no further increase in the absorbance at 340 nm over time.
Take the reaction mixture into 3000 Da MWCO snake skin dialysis membrane and dialyze it against 1 L sodium phosphate buffer (5 mM pH 7.4) solution for 24 hours at 4 °C by replacing the dialysis buffer every 6 hours to remove pyridothione byproduct produced in the crosslinking reaction.
3.2 Modification of surface charge on nanogels
The nanogels prepared so far using the above method do not possess any surface charge. However, the PDS functional groups present on these crosslinked nanogels provide an opportunity to modify their surface using a charged ligand. Depending on the surface charge of the protein, a complementary surface charge on the nanogel can be easily generated using this method.
In this example, β-galactosidase possesses a negative surface charge at neutral pH. To facilitate its electrostatic complexation with nanogels, a positive surface charge should be incorporated on the nanogels. In case if the enzyme possesses a positive surface charge, a negative surface charge needs to be generated on the nanogel.
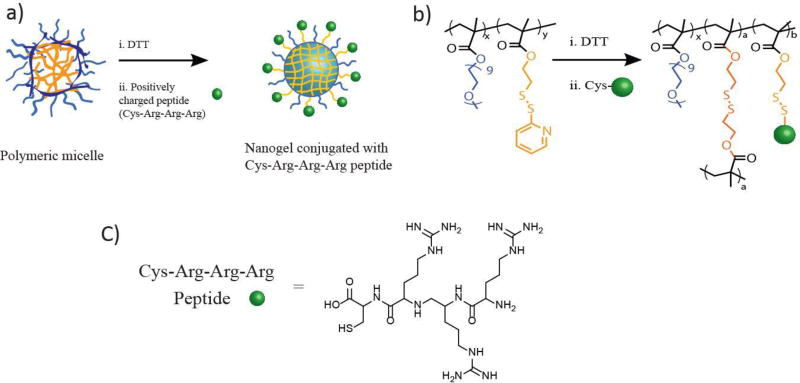
To generate a positive surface charge on the nanogel, a ligand such as triarginine peptide with a cysteine on the C-terminus (CRRR) can be used (González-Toro, Ryu, Chacko, Zhuang & Thayumanavan, 2012). The thiol group on cysteine will react with the reactive PDS functional groups of the nanogel thus, appending the positive triarginine group on the surface of the nanogel. The conjugation between nanogel and CRRR can be monitored by the evolution of the pyridothione group via its absorption spectrum, in addition to the surface charge measurements.
3.2.1 Surface modification of nanogel using CRRR peptide
The acceptable amount of ligand that can be conjugated to the nanogels depends on the available PDS functional groups on the nanogel. Therefore, while calculating the amount of ligand that can be conjugated to the nanogel, the crosslink density of nanogel (amount of PDS groups utilized from the polymer to make a nanogel) must be taken into account.
In this example, 10 mg of polymer (which contains 0.021 mmol of PDS monomer) was used to make a nanogel with 10% crosslink density (consumed 0.0021 mmol of PDS on polymer during crosslinking) as described in Section 3.1.2. Therefore, the number of available PDS groups on the resulting nanogel can be calculated by subtracting the moles of PDS groups consumed in crosslinking from the moles of PDS in the polymer which would be 0.021–0.0021=0.0189 mmol. Thus, the amount of thiol containing ligands that can be added for decorating nanogels must not exceed 0.0189 mmol.
Conjugation of peptide to nanogel
Equipment and chemicals
CRRR peptide (synthesized using a reported procedure(González-Toro et al, 2012) or available in commercial sources such as GenScript Corporation), Amicon Ultra 10,000 Da MWCO centrifugal filters, ultracentrifuge, stir plate, 7 mL glass vial and stir bar, nanogel stock solution (prepared in the previous step), PBS (pH 7.4) buffer.
Procedure
Take 1 mL of 10 mg /mL nanogel solution in a 7 mL glass vial with a magnetic stir bar placed on a stirrer with stirring speed of 350 rpm.
Add 5 mg of CRRR peptide (0.0084 mmoles) to the nanogel solution and allow it to stir for 12 hours at 25 °C.
Transfer the reaction mixture into Amicon Ultra 10k Da MWCO and dialyze the solution to remove unreacted CRRR peptide and pyridothione byproduct.
Resuspend the peptide conjugated nanogel solution in phosphate buffer to make it to the volume of desired concentration.
3.3 Electrostatic complexation of protein with nanogels and its characterization
The complementary surface charge of protein and nanogel can be utilized to achieve electrostatic complexation to result in the formation of nanogel-protein aggregates. The complexation ratio between the nanogel and protein needs to be optimized by carefully monitoring the properties of the resulting nano-aggregates. A spectroscopic method based on FRET can be used to optimize this complexation ratio. This can be achieved when nanogel and protein are labeled with two different dyes which are FRET partners. Fluorescein and DiI dyes make one such FRET pair, where the absorption spectrum of the DiI (acceptor) overlaps with the emission spectrum of fluorescein (donor).
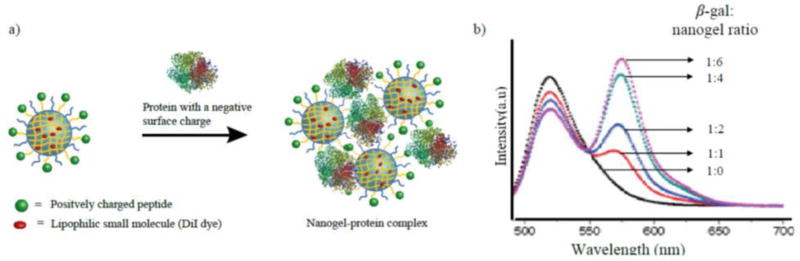
In this example, DiI dye (FRET acceptor) is encapsulated inside the nanogel and fluoroscein dye (FRET donor) is conjugated to β-galactosidase by labeling with FITC. When nanogel and β-gal form a complex, the associated proximity of these dyes lead to FRET observation, which is characterized by a decrease in the emission intensity of donor and increase in intensity of the acceptor. By keeping the amount of one complexing partner constant which is β-gal in this case (FRET donor), the nanogel (FRET acceptor) is slowly added in increments. The emission intensity of the donor (fluorescein) will continue to decrease until a stable complex is formed and further addition of nanogel will not result in any more decrease in the emission intensity of donor (Figure 9).
Figure 9.
a) Pictorial representation of protein complexing with nanogel b) FRET study to optimize the complexation ratio between nanogel and protein(González-Toro et al, 2012).
Equipment and chemicals
Fluoroscein isothiocyanate isomer I from Sigma-Aldrich, stock solution in DMSO solvent (4 mg/mL), 0.1 M sodium bicarbonate solution (pH 9.0), β-galactosidase protein, size exclusion chromatography system with sephadex G-25 as stationary phase and phosphate buffer (5 mM, pH 7.4) as mobile phase, UV-visible spectrophotometer, stir plate, 7 mL glass vial and stir bar.
Procedure to label the enzyme with dye
Take 2.5 mg of β-gal in 900 µL of 0.1 M sodium bicarbonate solution in a 7 mL glass vial with a magnetic stir bar.
Add 250 uL of FITC stock solution.
Wrap the vial with aluminum foil to protect it from light and allow the mixture for gentle stirring for 2 hours.
Purify the protein using SEC and concentrate using ultrafiltration using Amicon Ultra 30,000 Da MWCO tubes.
Calculate the protein concentration using UV-vis absorption spectroscopy and adjust the concentration of protein to the desired amount using sodium phosphate buffer (5 mM pH 7.4).
Procedure to complex protein with nanogel
Prepare 2 mg/mL stock solution of nanogel-CRRR using sodium phosphate buffer (5 mM pH 7.4).
Prepare 1 mg/mL stock solution of fluorescein labelled β-gal solution in sodium phosphate buffer (5 mM pH 7.4).
Prepare a range of complex solutions by using volume ratios between β-gal:nanogel-CRRR of 1:1 – 1:6 and incubate them for 1 hour at 25 °C.
Monitor emission spectra of the formed complex solutions by using excitation wavelength of 480 nm to identify the stable complexation ratio.
Identify the stable complexation ratio such that, increase in the amount of NG-CRRR addition does not result in decrease in the emission of fluorescein.
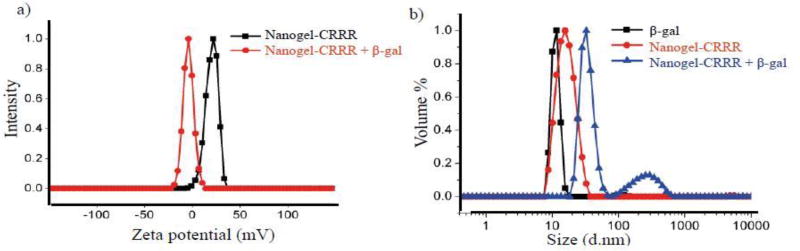
The success of this electrostatic complexation can also be verified by studying the surface charge and the size of the aggregates (Fig 10). Since the nanogel surface is positively charged, upon formation of nanoaggregate, a drift in the surface charge from positive to negative zeta potential can be observed. Similarly, complexes have a larger size compared to nanogel and protein by itself.
Figure 10.
Evaluation of complexation between nanogel and protein with a) change in surface charge b) increase in size.(González-Toro et al, 2012)
4. Non-covalent entrapment
Both the covalent conjugation and electrostatic complexation methods for nano-armoring of an enzyme rely on the physical and chemical properties of the protein surface. In the case of covalent conjugation, the density of thiol functional groups on the protein surface, which can participate in covalent bond formation, determines the success of the method. However, in electrostatic complexation, the extent of surface charge of the protein will ultimately decide the stability of the complex formed with the polymer.
Depending on the structural and physical properties of a particular protein, one of these methods can be more promising over the other. However, for proteins with less accessible functional groups on their surface and with less overall surface charge, non-covalent entrapment method would be a favorable choice. This method is based on inverse emulsion polymerization through which a hydrogel matrix can be synthesized around the protein to provide a nano-armor.
This methodology of protein entrapment in nanoscale hydrogels involves the following steps:
Preparation of inverse emulsion solution and nanogel synthesis
Nanogel extraction
Protein release and activity
4.1 Preparation of inverse emulsion solution and nanogel synthesis
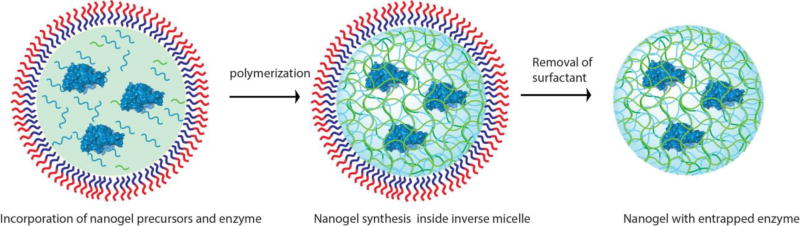
In this method of inverse emulsion polymerization, heptane and Brij L4 were chosen as the continuous phase and surfactant respectively. Addition of a small amount of water relative to the amount of organic solvent to this mixture will result in an inverse mini-emulsion which contains nanoscale water droplets stabilized by the surfactant in bulk organic phase.
The aqueous solution used in this process contains all the necessary starting materials to synthesize a polymeric nanogel matrix such as monomers, crosslinkers, free radical initiators along with the protein cargo that needs to be encapsulated inside the nanogel. The monomer is usually an acrylate or acrylamide derivative which results in linear chains upon polymerization, while the crosslinker based on bisacrylate or bisacrylamide derivatives can form a polymeric network resulting in a nanogel while trapping the protein in its network.
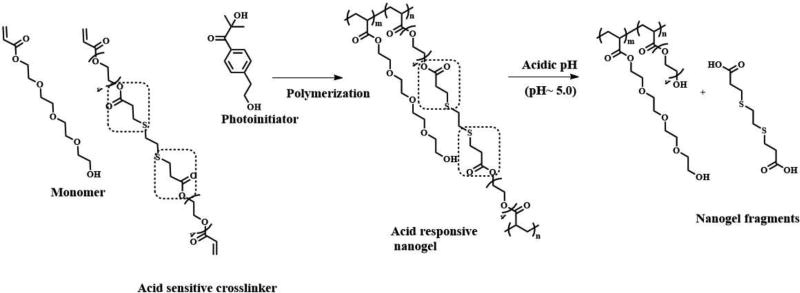
The ability of nanogel to release the entrapped protein cargo will depend on the responsive behavior of crosslinker used. Literature precedents show that the choice of crosslinkers depend on the desired stimuli-responsive features required on the nanogels such as pH(Standley, Kwon, Murthy, Kunisawa, Shastri, Guillaudeu, et al, 2004) or redox(Ashwinkumar, Maya & Jayakumar, 2014), light(Azagarsamy et al, 2012). In this example nanogel synthesis using a pH-responsive crosslinker, which is a derivative of β-thiopropionate ester(Molla et al, 2014) will be described (Figure 11).
Figure 11.
Synthetic scheme involved in nanogel synthesis and its responsive behavior towards acidic pH(Molla et al, 2014)
The feed molar ratio between the monomer and the crosslinker will determine the extent of crosslinked network within the nanogel. This ratio can be varied to fine tune the controlled release of the encapsulated cargo (protein) from the nanogel. In this example, the molar ratio between monomer and crosslinker was about 90:10. This is sufficient to make stable nanogels, even though there are some other examples in the literature where 97.5:2.5 was used to make nanogels which demonstrate that nanogel formation requires only a small amount of crosslinker relative to the amount of monomer(Azagarsamy et al, 2012).
To carry out the polymerization, there are different choices of free radical initiators that can be used, the criterion for which is that it must be soluble in aqueous solution along with monomer and crosslinker. For example, a photoinitiator such as Irgacure (2-hydroxy-4′-(2-hydroxyethoxy)-2-methylpropiophenone) can be used such that the nanogel formation can be triggered simply by exposure to UV light. Alternatively, ammonium persulphate initiator can be used where TEMED (tetramethylethylenediamine) catalyst is used to trigger polymerization reaction.
Equipment and chemicals
Brij L4 surfactant (Sigma-Aldrich), heptane, n-butanol, tetraethylene glycol acrylate monomer and β-thioester diacrylate crosslinker synthesized as per reported procedure (Molla et al, 2014), α-glucosidase enzyme, 5.5mg/mL stock solution of Irgacure (2-hydroxy-4′-(2-hydroxyethoxy)-2-methylpropiophenone) from Sigma-Aldrich, stir plate, 20 mL vial with magnetic stir bar, vortex mixer, bath sonicator, argon gas line with 18 G needle, UV chamber equipped with stirring plate.
Procedure
Take 0.6 g of Brij L4 surfactant and 5 mL of heptane in a 20 mL glass vial with a magnetic stir bar and mix thoroughly until a clear solution is obtained.
In a micro-centrifuge tube take 150 mg of tetraethylene glycol acrylate monomer (0.604 mmol), 18 mg of pH responsive β-thioester diacrylate crosslinker (0.06 mmol), 100 µL of initiator stock solution, 2 mg of α glucosidase and mix them thoroughly using vortex.
Mix the polymer precursors and protein (step-2) into the surfactant in heptane (step-1) and vortex for 5 minutes followed by sonication for an additional 5 minutes to form a mini-emulsion.
Under constant stirring at 500 rpm, purge the mini-emulsion solution with argon for about 10 min to remove oxygen and then close the cap of the vial.
Place the vial in a UV chamber with 365 nm light source for 20 minutes under constant stirring at 500 rpm to form nanogel.
4.2 Nanogel extraction
The next step in this procedure involves extraction of nanogels from the bulk organic solvent, which can be carried out using two different methods: solvent extraction method and gravimetric extraction method.
4.2.1 Solvent extraction method
Equipment and chemicals
PBS (pH 7.4 buffer), n-butanol, 50 mL centrifuge tubes made of polypropylene material, centrifuge, Pasteur pipettes, vortex mixer.
Procedure
In a 50 mL centrifuge tube measure 16 mL of PBS (pH 7.4) buffer.
To it, add the nanogel solution prepared in the previous step and mix it thoroughly using vortex for about a minute.
To it add 2 mL of n-butanol and centrifuge for 15 minutes at 3000 × g.
Discard the organic layer using a Pasteur pipette
Perform steps 3 and 4 twice to remove any surfactant present in the aqueous solution
Take small amount of aqueous solution (about 300 µL) and lyophilize it to calculate the concentration of nanogel using the same volume of PBS buffer as blank
Tip
For enzymes that are fragile, the amount of time involved in vortexing and sonication must be minimized as it could damage their structure to result in a loss of its enzymatic activity.
4.2.2 Gravimetric extraction procedure
In this method, the organic solvent and the surfactant from the nanogel reaction mixture are removed by taking advantage of the density of nanogel, which is associated with aqueous solvent. Since the nanogel is associated with trace amount of water, its density is more compared to that of bulk organic phase, which is heptane in this case. This difference in density can be utilized to pellet the hydrogel from the bulk organic phase by using a high centrifugal force.
Equipment and chemicals
PBS (pH 7.4 buffer), 10 mL centrifuge tubes made of polypropylene material, centrifuge.
Procedure
Take the nanogel solution formed after UV irradiation in a centrifuge tube.
Centrifuge for 30 minutes at 15,000 × g at 4 °C to pellet the nanogel.
Decant the supernatant and then wash the pellet with 5 mL of heptane.
Repeat steps 2 and 3 twice and resuspend the nanogel pellet in PBS solution or a suitable buffer of choice.
To calculate the concentration of nanogel take small amount of aqueous solution and lyophilize it to calculate the concentration of nanogel using the same volume of buffer as blank.
4.3 Activity assay for the released enzyme
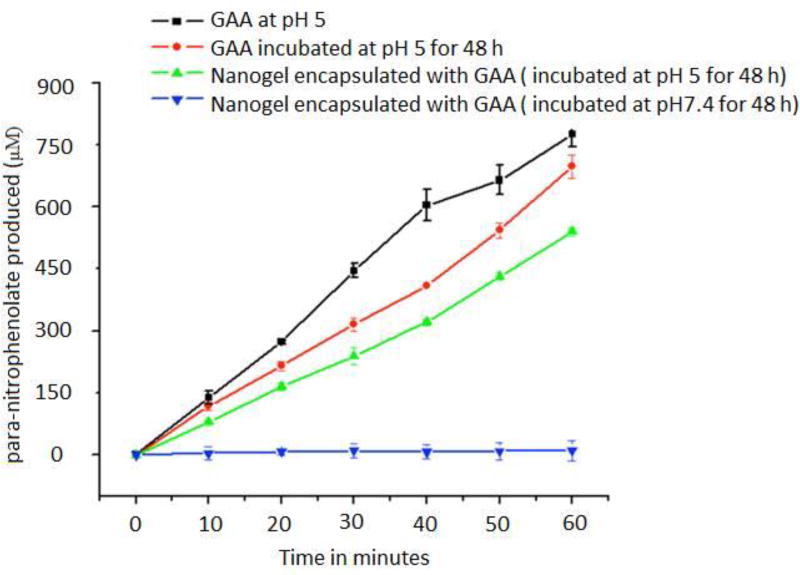
Protein quantification assay such as bicinconic acid assay (Pierce™ BCA Protein Assay protocol) can be utilized to estimate the amount of protein in the nanogel sample. In order to determine the activity of enzyme entrapped in the nanogel GAA, activity assay can be performed using para-nitrophenol-α-D-glucopyranoside as a substrate (Figure 13). The active enzyme will cleave the substrate to produce para-nitrophenolate product which has an absorbance at 400 nm.
Figure 13.
Activity recovery of enzyme after release from the nanogel (Molla et al, 2014)
Equipment and chemicals
100 mM citrate buffer (pH 5), 200 mM sodium borate buffer (pH 10), para-nitrophenol-α-D-glucopyranoside (Sigma-Aldrich), nanogel solution with an estimated concentration of encapsulated protein, 96 well clear flat-bottom plates, plate reader for measuring absorbance.
Procedure
Make a nanogel sample stock solutions in i) 100 mM citrate (pH 5) buffer and ii) PBS (pH 7.4) having protein concentration 0.34 µM and incubate it for 48 hours at 4 °C .
Prepare a α-glucosidase stock solution in 100 mM Citrate (pH 5) buffer and incubate it for 48 hours at 4 °C.
Take 50 µL of substrate solution in an ultra-centrifuge tube and add 50 µL of nanogel/ protein stock solution to it which was prepared in previous steps and mix gently.
For activity measurement, take 10 µL of the reaction mixture (step-2) and add it to 290 µL of 200 mM sodium borate buffer (pH 10) and measure the absorbance at 400 nm.
Take the activity measurement every 10 minutes for an hour to get the activity profile.
5. Conclusion
Nano-armoring enzymes using polymers is a useful technique to formulate therapeutic proteins for systemic administration. While armoring enzymes using polymers serve its purpose of providing a protective shield from aggregation and degradation, it also offers an additional feature of surface functionalization thus, providing targeting capabilities to the enzymes. In this chapter, we have discussed three different methods to formulate enzyme-polymer complexes based on the type of interactions between them. Depending on the physical and chemical properties of the enzyme of interest and the therapeutic target, a suitable formulation method can be applied to provide a nano-armor for the enzyme. Covalent means of enzyme-polymer complexes will be a suitable technique for proteins that have surface accessible and reactive functional groups. Alternatively, electrostatic complexation methods can be employed for enzymes having a significant overall surface charge at physiological pH. Non-covalent enzyme entrapment methods, on the other hand, would serve better if the enzyme of interest lacks surface accessible reactive functional groups or if using covalent means is detrimental to maintaining the activity of the protein cargo.
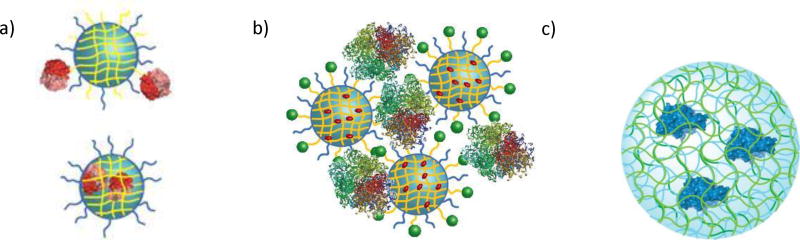
Figure 1.
Cartoon representing different types of enzyme wrapping using polymers. a) Covalent conjugation. b) Electrostatic complexation. C) Non-covalent entrapment in hydrogel network.
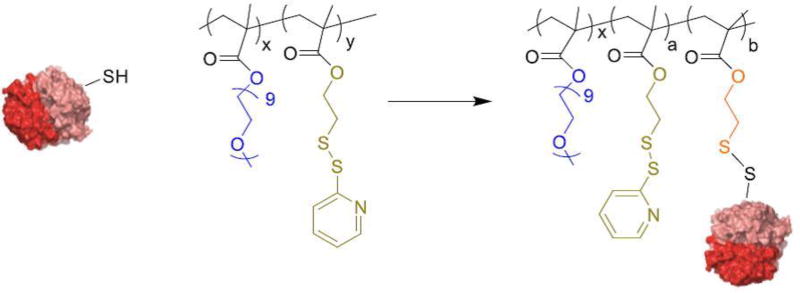
Figure 2.
Scheme involving PEDMA-PDSMA polymer conjugating with protein containing a thiol by a disulfide exchange reaction
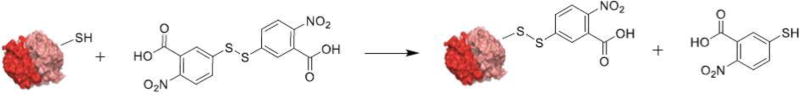
Figure 3.
Reaction scheme involved in quantifying the surface thiol groups on protein using Ellman’s reagent.
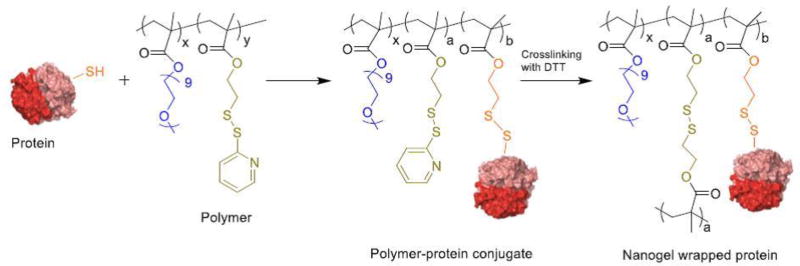
Figure 4.
Reaction scheme involving protein conjugation with polymer and self-crosslinking of polymer to result in a nanogel(Ventura et al, 2015)
Figure 5.
SDS gel electrophoresis evaluation of protein polymer complex under reducing and non-reducing conditions(Ventura et al, 2015).
Figure 6.
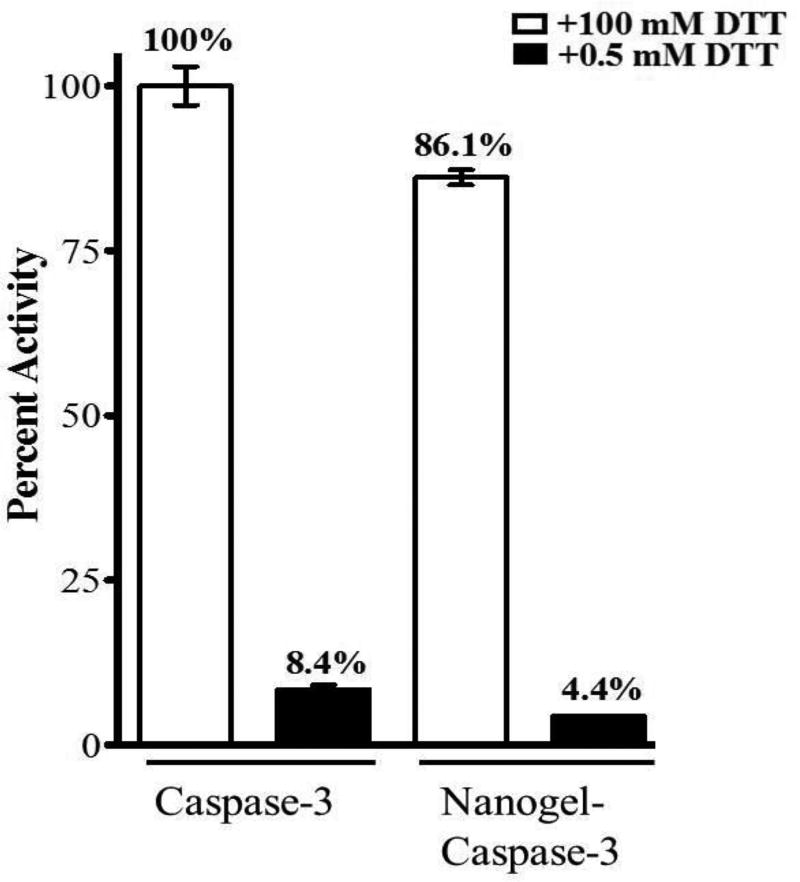
Activity recovery of enzyme after release from the nanogel (Ventura et al, 2015).
Figure 7.
Reaction scheme involving modification of amines on the protein to thiols using a SATP reagent
Figure 8.
a) Cartoon and b) Structural representation of nanogel synthesis and its surface modification with c) triarginine peptide for generating a positive charge on nanogel(González-Toro et al, 2012)
Figure 12.
Scheme involving nanogel synthesis using inverse emulsion polymerization
Acknowledgments
We thank the Army Research Office (W911NF-15-1-0568) and the National Institutes of Health (GM-065255 and CA-169140) for partial support of this research.
References
- 1.Ashwinkumar N, Maya S, Jayakumar R. Redox-responsive cystamine conjugated chitin-hyaluronic acid composite nanogels. RSC Advances. 2014;4:49547–49555. [Google Scholar]
- 2.Ayame H, Morimoto N, Akiyoshi K. Self-assembled cationic nanogels for intracellular protein delivery. Bioconjugate chemistry. 2008;19:882–890. doi: 10.1021/bc700422s. [DOI] [PubMed] [Google Scholar]
- 3.Azagarsamy MA, Alge DL, Radhakrishnan SJ, Tibbitt MW, Anseth KS. Photocontrolled nanoparticles for on-demand release of proteins. Biomacromolecules. 2012;13:2219–2224. doi: 10.1021/bm300646q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Babu KR, Moradian A, Douglas D. The methanol-induced conformational transitions of β-lactoglobulin, cytochrome c, and ubiquitin at low pH: A study by electrospray ionization mass spectrometry. Journal of the American Society for Mass Spectrometry. 2001;12:317–328. doi: 10.1016/s1044-0305(00)00226-9. [DOI] [PubMed] [Google Scholar]
- 5.Balendiran GK, Dabur R, Fraser D. The role of glutathione in cancer. Cell Biochemistry and Function. 2004;22:343–352. doi: 10.1002/cbf.1149. [DOI] [PubMed] [Google Scholar]
- 6.Brange J, Andersen L, Laursen ED, Meyn G, Rasmussen E. Toward understanding insulin fibrillation. Journal of Pharmaceutical Sciences. 1997;86:517–525. doi: 10.1021/js960297s. [DOI] [PubMed] [Google Scholar]
- 7.Brocchini S, Godwin A, Balan S, Choi J-w, Zloh M, Shaunak S. Disulfide bridge based PEGylation of proteins. Advanced drug delivery reviews. 2008;60:3–12. doi: 10.1016/j.addr.2007.06.014. [DOI] [PubMed] [Google Scholar]
- 8.Chang HY, Yang X. Proteases for cell suicide: functions and regulation of caspases. Microbiology and molecular biology reviews. 2000;64:821–846. doi: 10.1128/mmbr.64.4.821-846.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cohen JA, Beaudette TT, Tseng WW, Bachelder EM, Mende I, Engleman EG, Fréchet JM. T-cell activation by antigen-loaded pH-sensitive hydrogel particles in vivo: the effect of particle size. Bioconjugate chemistry. 2008;20:111–119. doi: 10.1021/bc800338n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Curatolo L, Valsasina B, Caccia C, Raimondi GL, Orsini G, Bianchetti A. Recombinant human IL-2 is cytotoxic to oligodendrocytes after invitro self aggregation. Cytokine. 1997;9:734–739. doi: 10.1006/cyto.1997.0228. [DOI] [PubMed] [Google Scholar]
- 11.Dai C, Wang B, Zhao H, Li B, Wang J. Preparation and characterization of liposomes-in-alginate (LIA) for protein delivery system. Colloids and surfaces B: Biointerfaces. 2006;47:205–210. doi: 10.1016/j.colsurfb.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 12.Desnick RJ, Schuchman EH. Enzyme Replacement Therapy for Lysosomal Diseases: Lessons from 20 Years of Experience and Remaining Challenges. Annual Review of Genomics and Human Genetics. 2012;13:307–335. doi: 10.1146/annurev-genom-090711-163739. [DOI] [PubMed] [Google Scholar]
- 13.Dyal A, Loos K, Noto M, Chang SW, Spagnoli C, Shafi KV, Ulman A, Cowman M, Gross RA. Activity of Candida rugosa lipase immobilized on γ-Fe2O3 magnetic nanoparticles. Journal of the American Chemical Society. 2003;125:1684–1685. doi: 10.1021/ja021223n. [DOI] [PubMed] [Google Scholar]
- 14.Fishburn CS. The Pharmacology of PEGylation: Balancing PD with PK to Generate Novel Therapeutics. Journal of Pharmaceutical Sciences. 2008;97:4167–4183. doi: 10.1002/jps.21278. [DOI] [PubMed] [Google Scholar]
- 15.Ghosh P, Yang X, Arvizo R, Zhu Z-J, Agasti SS, Mo Z, Rotello VM. Intracellular delivery of a membrane-impermeable enzyme in active form using functionalized gold nanoparticles. Journal of the American Chemical Society. 2010;132:2642–2645. doi: 10.1021/ja907887z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghosh S, Basu S, Thayumanavan S. Simultaneous and reversible functionalization of copolymers for biological applications. Macromolecules. 2006;39:5595–5597. [Google Scholar]
- 17.González-Toro DC, Ryu J-H, Chacko RT, Zhuang J, Thayumanavan S. Concurrent Binding and Delivery of Proteins and Lipophilic Small Molecules Using Polymeric Nanogels. Journal of the American Chemical Society. 2012;134:6964–6967. doi: 10.1021/ja3019143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.González-Toro DC, Ryu J-H, Chacko RT, Zhuang J, Thayumanavan S. Concurrent binding and delivery of proteins and lipophilic small molecules using polymeric nanogels. Journal of the American Chemical Society. 2012;134:6964–6967. doi: 10.1021/ja3019143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heredia KL, Maynard HD. Synthesis of protein-polymer conjugates. Organic & Biomolecular Chemistry. 2007;5:45–53. doi: 10.1039/b612355d. [DOI] [PubMed] [Google Scholar]
- 20.Hermeling S, Crommelin DJA, Schellekens H, Jiskoot W. Structure-Immunogenicity Relationships of Therapeutic Proteins. Pharmaceutical Research. 2004;21:897–903. doi: 10.1023/b:pham.0000029275.41323.a6. [DOI] [PubMed] [Google Scholar]
- 21.Joralemon MJ, McRae S, Emrick T. PEGylated polymers for medicine: from conjugation to self-assembled systems. Chemical Communications. 2010;46:1377–1393. doi: 10.1039/b920570p. [DOI] [PubMed] [Google Scholar]
- 22.Knubovets T, Osterhout JJ, Klibanov AM. Structure of lysozyme dissolved in neat organic solvents as assessed by NMR and CD spectroscopies. Biotechnology and bioengineering. 1999;63:242–248. [PubMed] [Google Scholar]
- 23.Leader B, Baca QJ, Golan DE. Protein therapeutics: a summary and pharmacological classification. Nature Reviews Drug Discovery. 2008;7:21–39. doi: 10.1038/nrd2399. [DOI] [PubMed] [Google Scholar]
- 24.Lee AL, Wang Y, Ye W-H, Yoon HS, Chan SY, Yang Y-Y. Efficient intracellular delivery of functional proteins using cationic polymer core/shell nanoparticles. Biomaterials. 2008;29:1224–1232. doi: 10.1016/j.biomaterials.2007.11.021. [DOI] [PubMed] [Google Scholar]
- 25.Luckarift HR, Spain JC, Naik RR, Stone MO. Enzyme immobilization in a biomimetic silica support. Nature biotechnology. 2004;22:211–213. doi: 10.1038/nbt931. [DOI] [PubMed] [Google Scholar]
- 26.Matsumoto NM, González-Toro DC, Chacko RT, Maynard HD, Thayumanavan S. Synthesis of nanogel-protein conjugates. Polymer chemistry. 2013;4:2464–2469. doi: 10.1039/C3PY00085K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mero A, Ishino T, Chaiken I, Veronese FM, Pasut G. Multivalent and Flexible PEG-Nitrilotriacetic Acid Derivatives for Non-covalent Protein Pegylation. Pharmaceutical Research. 2011;28:2412. doi: 10.1007/s11095-011-0468-8. [DOI] [PubMed] [Google Scholar]
- 28.Molla MR, Marcinko T, Prasad P, Deming D, Garman SC, Thayumanavan S. Unlocking a Caged Lysosomal Protein from a Polymeric Nanogel with a pH Trigger. Biomacromolecules. 2014;15:4046–4053. doi: 10.1021/bm501091p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mueller C, Capelle MAH, Seyrek E, Martel S, Carrupt P-A, Arvinte T, Borchard G. Noncovalent PEGylation: Different effects of dansyl-, l-tryptophan-, phenylbutylamino-, benzyl- and cholesteryl-PEGs on the aggregation of salmon calcitonin and lysozyme. Journal of Pharmaceutical Sciences. 2012;101:1995–2008. doi: 10.1002/jps.23110. [DOI] [PubMed] [Google Scholar]
- 30.Murthy N, Xu M, Schuck S, Kunisawa J, Shastri N, Fréchet JM. A macromolecular delivery vehicle for protein-based vaccines: acid-degradable protein-loaded microgels. Proceedings of the National Academy of Sciences. 2003;100:4995–5000. doi: 10.1073/pnas.0930644100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nischan N, Hackenberger CPR. Site-specific PEGylation of Proteins: Recent Developments. The Journal of Organic Chemistry. 2014;79:10727–10733. doi: 10.1021/jo502136n. [DOI] [PubMed] [Google Scholar]
- 32.Pasut G, Veronese FM. State of the art in PEGylation: The great versatility achieved after forty years of research. Journal of Controlled Release. 2012;161:461–472. doi: 10.1016/j.jconrel.2011.10.037. [DOI] [PubMed] [Google Scholar]
- 33.Poznansky MJ, Juliano RL. Biological approaches to the controlled delivery of drugs: a critical review. Pharmacological Reviews. 1984;36:277–336. [PubMed] [Google Scholar]
- 34.Ryu J-H, Chacko RT, Jiwpanich S, Bickerton S, Babu RP, Thayumanavan S. Self-Cross-Linked Polymer Nanogels: A Versatile Nanoscopic Drug Delivery Platform. Journal of the American Chemical Society. 2010;132:17227–17235. doi: 10.1021/ja1069932. [DOI] [PubMed] [Google Scholar]
- 35.Standley SM, Kwon YJ, Murthy N, Kunisawa J, Shastri N, Guillaudeu SJ, Lau L, Fréchet JMJ. Acid-Degradable Particles for Protein-Based Vaccines: Enhanced Survival Rate for Tumor-Challenged Mice Using Ovalbumin Model. Bioconjugate Chemistry. 2004;15:1281–1288. doi: 10.1021/bc049956f. [DOI] [PubMed] [Google Scholar]
- 36.Vellodi A. Lysosomal storage disorders. British Journal of Haematology. 2005;128:413–431. doi: 10.1111/j.1365-2141.2004.05293.x. [DOI] [PubMed] [Google Scholar]
- 37.Ventura J, Eron SJ, González-Toro DC, Raghupathi K, Wang F, Hardy JA, Thayumanavan S. Reactive self-assembly of polymers and proteins to reversibly silence a killer protein. Biomacromolecules. 2015;16:3161–3171. doi: 10.1021/acs.biomac.5b00779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vermonden T, Censi R, Hennink WE. Hydrogels for protein delivery. Chemical Reviews. 2012;112:2853–2888. doi: 10.1021/cr200157d. [DOI] [PubMed] [Google Scholar]