Abstract
Clinical phenotypes of congenital myasthenic syndromes and primary mitochondrial disorders share significant overlap in their clinical presentations, leading to challenges in making the correct diagnosis. Next generation sequencing is transforming molecular diagnosis of inherited neuromuscular disorders by identifying novel disease genes and by identifying previously known genes in undiagnosed patients. This is evident in two patients who were initially suspected to have a mitochondrial myopathy, but in whom a clear diagnosis of congenital myasthenic syndromes was made through whole exome sequencing. In patient 1, whole exome sequencing revealed compound heterozygous mutations c.1228C>T (p.Arg410Trp) and c.679C>T (p.Arg227*) in collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase (COLQ). In patient 2, in whom a deletion of exon 52 in Dystrophin gene was previously detected by multiplex ligation-dependent probe amplification, Sanger sequencing revealed an additional homozygous mutation c.1511_1513delCTT (p.Pro504Argfs*183) in docking protein7 (DOK7). These case reports highlight the need for careful diagnosis of clinically heterogeneous syndromes like congenital myasthenic syndromes, which are treatable, and for which delayed diagnosis is likely to have implications for patient health. The report also demonstrates that whole exome sequencing is an effective diagnostic tool in providing molecular diagnosis to patients with complex phenotypes.
Keywords: Congenital myasthenia, mitochondrial respiratory chain, mutation, DOK7, COLQ, Duchenne muscular dystrophy, whole exome sequencing
1. Introduction
Congenital myasthenic syndromes (CMS) are a family of inherited neuromuscular disorders characterized by fatigable muscle weakness, ptosis, ophthalmoplegia, respiratory and skeletal muscle weakness [1]. This clinical profile overlaps considerably with other myopathies and can lead to diagnostic challenges [2]. Electromyography and muscle histology may provide nonspecific results, resulting in patients being misdiagnosed and untreated [2–4]. Establishing the specific molecular diagnosis in a family allows treatment to be individualized as drug responses vary widely between different forms of CMS [5, 6].
The molecular diagnosis of genetically heterogeneous disorders like CMS and mitochondrial respiratory chain (MRC) disorders can be challenging, as traditional Sanger sequencing is often time consuming and expensive. Next generation sequencing (NGS) technology is now being applied as an efficient and cost-effective strategy for the diagnosis of Mendelian disorders, not only for screening of well characterised genes, but also for identification of novel disease genes and prenatal diagnosis [7, 8].
We report two patients with mutations in genes associated with CMS in whom there was no molecular diagnosis but a suspicion of mitochondrial myopathy was initially considered, based on mitochondrial enzyme studies which showed complex I deficiency. The report illustrates the diagnostic difficulty in differentiating between these disorders that share phenotypic similarities.
2. Case Reports
Patient 1
This boy’s clinical phenotype in early childhood and the abnormality on repetitive nerve stimulation (RNS) were initially presumed to represent a form of congenital myasthenia and were previously reported [9]. He was the youngest of three children (Figure 1A(i); II-3) to non-consanguineous parents of Caucasian and Filipino descent. From six months of age, he had frequent episodes of muscle weakness and respiratory failure often associated with febrile illness. At 20 months of age, when admitted with acute respiratory failure, there was abnormal decrement on RNS and excessive jitter with blocking on stimulated single fibre EMG (SFEMG). Genetic screening revealed no mutations in CHAT, CHRNA1, CHRNB, CHRNE and RAPSN. Because of a lack of response to pyridostigmine, absence of mutations in the common CMS genes, presence of limb-girdle weakness and absence of ptosis, a decision to do a muscle biopsy was taken to investigate the possibility of a congenital or mitochondrial myopathy. Muscle biopsy from neck extensors at 41 months of age showed mild variation in fibre size and marked (90%) type I predominance. Oxidative enzyme stains (NADH, SDH and COX/SDH) showed a normal pattern of staining. Diagnostic analysis of MRC enzymes showed significant reduction in complex I activity (and borderline reduction in complex III and complex IV) relative to complex II and citrate synthase, consistent with complex I deficiency (Table 1). Serum lactate was normal.
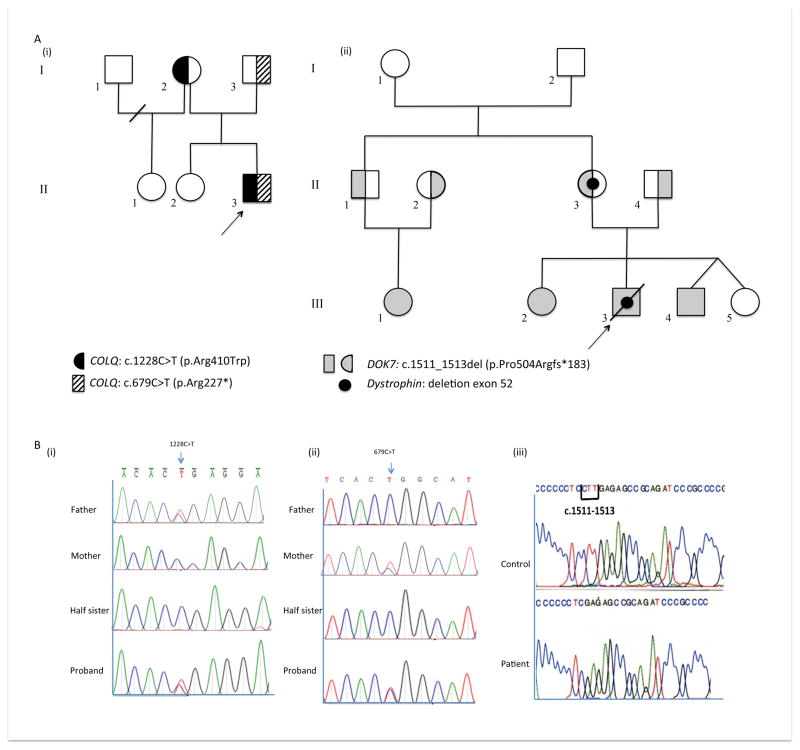
Figure 1.
(A) Pedigrees for family 1 (i) and 2 (ii). The proband is denoted by an arrow. (B) DNA sequence profile COLQ mutations for patient 1, parents and unaffected sister (i) c.1228C>T (p.Arg410Trp) mutation heterozygous in the father and proband (ii) c.679C>T (p.Arg227*) mutation heterozygous in the mother and proband (iii) Sanger sequencing of DOK7 near the end of exon 7 in a control and in patient 2 showing a homozygous c.1511_1513delCTT deletion in patient.
Table 1. Skeletal muscle respiratory chain enzymes for Patients 1 and 2.
Enzyme Activities are expressed as * nmol/min/mg or $/min/mg. Bolded values are below the reference range.
% Activity and % Ratio are expressed relative to protein or relative to marker enzymes as % of normal control mean. Bolded values are <20% or 21–30% of the normal control mean and represent Major or Minor enzyme criteria, respectively, in the Bernier diagnostic criteria [1]. Values in parentheses are expressed as % of the lowest normal control value, with values <80% of the lowest normal control regarded as deficient in NCMD diagnostic criteria [2].
| Ref Range Activity (nmol/min/mg)* or (/min/mg)$ | Patient 1 | Patient 2 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Activity (nmol/min/mg)* or (/min/mg)$ | % activity | % CS Ratio | % CII ratio | Activity | % activity | % CS Ratio | % CII ratio | ||
| Complex I* | 19–72 | 5 | 12 (26%) | 15 (21%) | 15 (23%) | 13 | 28 (60%) | 38 (54%) | 19 (29%) |
| Complex II* | 26–63 | 37 | 82 | 100 | - | 76 | 176 | 202 | - |
| Complex II+III* | 30–76 | 12 | 26 | 33 | 32 | 30 | 130 | 80 | 39 |
| Complex III$ | 12.8–50.9 | 11.1 | 38 | 45 | 45 | 32.7 | 183 | 130 | 65 |
| Complex IV$ | 3.3–9.1 | 3.1 | 46 | 57 | 57 | 5.0 | 116 | 92 | 46 |
| Citrate Synthase* | 85–179 | 105 | 81 | - | - | 107 | - | - | - |
On examination at the age of 4 years, the patient had proximal weakness including neck flexors and extensors. There was no ptosis, ophthalmoplegia or facial weakness. With disease progression he developed a scoliosis, lost ambulation (7 years) and required nocturnal Bilevel Positive Airway Pressure (BiPAP) (8 years). Treatment with a cocktail of carnitine, thiamine, riboflavin, menadione and Coenzyme Q provided no discernible benefit.
WES was performed on DNA samples from patient 1, his parents and half-sister. Agilent SureSelect Human All Exon kit (Agilent Technologies, Santa Clara, CA, USA) was used to capture human genomic DNA in exonic target regions, following the manufacturer’s protocols. WES data analysis (Supplementary Methods & Supplementary Table 1) in patient 1 revealed compound heterozygous c.1228C>T (p.Arg410Trp) and c.679C>T (p.Arg227*) mutations in acetylcholinesterase (AChE) collagen-like tail subunit gene (COLQ) (OMIM 603033), a gene linked to endplate acetylcholinesterase deficiency [10]. Mutations were confirmed by Sanger sequencing of the entire family of Patient 1 (Figure 1B(i) and (ii)). Both COLQ mutations have been previously associated with congenital myasthenic syndrome [10, 11]. No variants in nuclear genes relevant to MRC structure and function or known to cause complex I disorders were identified (Supplementary Table 2 & 3). In addition, mitochondrial genome sequencing (Western Australian Institute of Medical Research) using DNA from the muscle sample of the proband revealed no mtDNA encoded mutations or deletions as a cause of the complex I defect.
Repeat RNS studies at 5 Hz of Patient 1 showed a decremental response of 19% in the right abductor digiti minimi and abnormal jitter on single fibre EMG in the right orbicularis oculi. Treatment with salbutamol (2 mg TDS) resulted in significant improvement and he was able to walk short distances independently after 2 months of therapy.
Patient 2
The proband (Figure 1A(ii); III-3) was the second of four children of non-consanguineous parents of South Indian (Tamil) origin. Stridor and recurrent apnoea were noticed soon after birth. He had no ptosis, ophthalmoplegia or facial weakness. A nasojejunal tube was required for feeding. A muscle biopsy at 1 month of age showed dystrophic features with absence of dystrophin by immunohistochemical criteria. In addition, there was a normal fibre size distribution, and modified Gomori trichome, acid phosphatase, succinic dehydrogenase, ATPase, and reversed ATPase stains were normal.
Dystrophin gene analysis by MLPA showed a deletion of exon 52 inherited from his mother. The patient required continuous BiPAP from 5 months of age and died following an episode of aspiration. As Duchenne muscular dystrophy is not associated with neonatal onset and severe early phenotype, the muscle biopsy underwent MRC enzyme analysis and showed reduction in complex I activity relative to complex II, consistent with complex I deficiency (Table 1). Subsequently, his siblings (Figure 1A(ii) ; III-2 and III-4) and a maternal first cousin (Figure 1A(ii); III-1) presented with ptosis, ophthalmoplegia, stridor and mild proximal muscle weakness. Prenatal testing had shown that the male sibling (Figure 1B; III-4) did not carry the dystrophin deletion. Sanger sequencing in Patient 2’s cousin (Figure 1B; III-1) revealed she had a homozygous DOK7 (OMIM 610285) mutation (c.1511_1513delCTT; p.Pro504Argfs*183) (Figure 1B(iii) ) that has been previously reported as pathogenic, leading to CMS [12]. Cascade testing showed that her parents (Figure 1A(ii) ; II-1 and II-2) were heterozygous for the DOK7 deletion while patient 2 and his affected siblings (Figure 1A(ii); III-2 and III-4) were homozygous for this mutation.
3. Discussion
The congenital myasthenic syndromes are a group of rare genetically heterogeneous disorders of the neuromuscular junction. They are important to diagnose as they can cause significant disability and can be fatal if untreated. However, diagnosis can be difficult as they may have a variable phenotype, require specialized neurophysiological tests and often multiple genetic tests for diagnosis, and may give negative responses to cholinergic agonist challenge testing.
The presence of fatigable weakness, ptosis and ophthalmoplegia, though hallmarks of disorders of the neuromuscular junction, are not universally present in CMS [1], and may be part of the phenotype of mitochondrial disorders and congenital myopathies [13, 14]. Ptosis and ophthalmoplegia were absent in both probands and fatigable weakness was difficult to identify due to the chronic myopathy in patient 1 and very young age in patient 2.
In patient 1 the correct diagnosis was reached when pathogenic variants causing CMS were identified using NGS technology. There have been other reports where WES in patients thought to have a primary mitochondrial disorder eventually unexpectedly revealed a genetic diagnosis not directly related to mitochondrial respiratory chain structure or function [15–17]. Patient 2, who died in infancy, was diagnosed only when another affected family member who presented with more typical symptoms of CMS had DOK7 mutation testing using Sanger sequencing.
Clinical diagnosis for many neuromuscular disorders still depends on histological and enzymological examination of a muscle biopsy. In our probands the diagnostic process was complicated by finding probable complex I defects on MRC enzyme testing. Even though normal muscle morphology and immunohistochemical staining was seen in our patients, a suspicion of mitochondrial myopathy could not be ruled out, as in paediatric cases normal muscle histopathology is not always indicative of the absence of the disorder [18]. Moreover, blood lactate may be normal in patients with a mitochondrial myopathy, and CSF lactate, rarely performed in patients who have a neuromuscular phenotype, is a more sensitive marker [19].
MRC enzyme analysis typically has greater sensitivity than histology and lactate measurement for diagnosing MRC disorders in children [19]. However, it is not 100% specific as secondary MRC enzyme defects can be found in various neuromuscular, neurological and metabolic disorders [20–25]. A clear understanding as to why these defects occur is lacking [22]. Muscle complex I activity does not show a clear cut-off between patients with primary and secondary MRC disorders, which prompted us to develop diagnostic criteria for MRC disorders that rely on evidence from at least two relatively independent types of investigation [26]. Using these “Bernier” criteria, both patients had major criteria for an MRC enzyme defect (<20% residual activity relative to marker enzymes), but neither had sufficiently specific evidence from other sources to achieve a definite diagnosis, hence we defined them as having probable MRC disorders. An alternative set of diagnostic criteria, the NCMD criteria, defines MRC enzymes as being deficient if less than 80% of the lowest normal control value. The enzyme results would also have been interpreted as deficient by these criteria (Table 1), but again would not result in a definitive diagnosis due to the non-specific clinical and metabolic features [27]. Low complex I activity can be an artefact of improper sample handling or storage but this seemed unlikely since complex II activities were normal (Table 1). Our experience is that complex II is typically more labile than complex I in samples with delays in processing or improper storage [20].
Patient 1 was reclassified as having a mitochondrial myopathy as he showed no improvement with pyridostigmine treatment, and no mutations were identified in the common CMS genes. Neuromuscular transmission abnormalities have been reported in mitochondrial myopathies [28, 29]. Mitochondria located near the synapse have been implicated in playing a regulatory role in neurotransmitter release and disruption of this process could affect neurotransmission [30]. Mutations in nuclear and mitochondrial DNA encoded genes relevant to MRC structure and function were not found in either of our patients, suggesting that the complex I deficiency is likely to be secondary to the primary COLQ and DOK7 abnormalities.
Patient 2 also presented with the uncommon co-occurrence of a severe out-of-frame dystrophinopathy (X-linked) and an autosomal recessive neuromuscular disorder. He had a course that was more severe and rapidly progressive than in other family members. Dystrophinopathies, in isolation, do not present with such a severe infantile course. The antemortem muscle biopsy, and post-mortem diaphragm and intercostal muscle biopsy, showed severe dystrophic changes, and the dystrophinopathy may have contributed to his more severe phenotype.
When patients present with typical symptoms, as was the case of Patient 2’s cousin, Sanger sequencing is still a useful tool for diagnosis. However, it can be time consuming and expensive when patient’s present with a complex phenotype and a mutation is not evident in the common genes. Innovative technologies like targeted gene panels, whole exome and whole genome sequencing hold promise as a time and cost effective diagnostic tool in distinguishing between phenotypically similar but genetically heterogeneous syndromes like CMS, and should be considered during the clinical workup of patients. Establishing the correct diagnosis in patient 1 enabled appropriate therapy for his CMS with significant improvement. In conclusion, secondary complex I deficiency may be observed in patients with CMS, and could potentially be misleading. Therefore, where there is a continued suspicion of a CMS, access to next generation sequencing may help with establishing the correct genetic diagnosis.
Supplementary Material
Synopsis.
We report how the use of the modern next generation sequencing technology and traditional Sanger Sequencing, lead to the redefinition of the diagnosis in two patients suspected of having a mitochondrial respiratory chain disorder, and translated to an effective therapy for one of the patients.
Highlights.
Next generation sequencing has transformed the approach to genetic disease diagnosis
Mitochondrial dysfunction may confound the diagnosis of congenital myasthenic syndromes
delayed diagnosis of congenital myasthenic syndromes has therapeutic implications
Acknowledgments
We thank Simone Tregoning and Wendy Salter, Murdoch Childrens Research Institute, Melbourne, for enzyme assays and Rachael Duff from the West Australian Institute for Medical Research, Perth, for undertaking the mtDNA sequencing in Patient 1. This research was supported by Australian NHMRC grant 1026891 to J.C., an NHMRC Principal Research Fellowship to D.R.T and an Australian Mitochondrial Disease Foundation (AMDF) PhD Scholarship to M.J.M, an NIH Research Grant NS6277 to A.G.E, and a Research Grant from the Shenzhen Municipal Government of China (NO.CXZZ20130517144604091) to X.X. We also gratefully acknowledge donations to J.C. by the Crane and Perkins families.
Footnotes
Compliance with Ethics Guidelines
All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1975, as revised in 2000 (5). Informed consent was obtained from all patients for being included in the study.
Conflicts of Interest
Minal Menezes, Yiran Guo, Manoj Menezes, Jinlong Liang, Dong Li, Lisa G Riley, Nigel F Clarke, P. Ian.Andrews, Lifeng Tian, Richard Webster, Fengxiang Wang, Xuanzhu Liu, Yulan Chen, David R Thorburn, Brendan J Keating, Andrew Engel, Hakon Hakonarson, John Christodoulou and Xun Xu declare that they have no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lorenzoni PJ, Scola RH, Kay CS, Werneck LC. Congenital myasthenic syndrome: a brief review. Pediatric neurology. 2012;46:141–148. doi: 10.1016/j.pediatrneurol.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 2.Kinali M, Beeson D, Pitt MC, Jungbluth H, Simonds AK, Aloysius A, Cockerill H, Davis T, Palace J, Manzur AY, Jimenez-Mallebrera C, Sewry C, Muntoni F, Robb SA. Congenital myasthenic syndromes in childhood: diagnostic and management challenges. Journal of neuroimmunology. 2008;201–202:6–12. doi: 10.1016/j.jneuroim.2008.06.026. [DOI] [PubMed] [Google Scholar]
- 3.Gurnett CA, Bodnar JA, Neil J, Connolly AM. Congenital myasthenic syndrome: presentation, electrodiagnosis, and muscle biopsy. Journal of child neurology. 2004;19:175–182. [PubMed] [Google Scholar]
- 4.Pitt M. Neurophysiological strategies for the diagnosis of disorders of the neuromuscular junction in children. Developmental medicine and child neurology. 2008;50:328–333. doi: 10.1111/j.1469-8749.2008.02038.x. [DOI] [PubMed] [Google Scholar]
- 5.Engel AG. The therapy of congenital myasthenic syndromes. Neurotherapeutics: the journal of the American Society for Experimental Neuro Therapeutics. 2007;4:252–257. doi: 10.1016/j.nurt.2007.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liewluck T, Selcen D, Engel AG. Beneficial effects of albuterol in congenital endplate acetylcholinesterase deficiency and Dok-7 myasthenia. Muscle & nerve. 2011;44:789–794. doi: 10.1002/mus.22176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calvo SE, Compton AG, Hershman SG, Lim SC, Lieber DS, Tucker EJ, Laskowski A, Garone C, Liu S, Jaffe DB, Christodoulou J, Fletcher JM, Bruno DL, Goldblatt J, Dimauro S, Thorburn DR, Mootha VK. Molecular diagnosis of infantile mitochondrial disease with targeted next-generation sequencing. Science translational medicine. 2012;4:118ra110. doi: 10.1126/scitranslmed.3003310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wong L-JC. Translation to Clinical Diagnostics. 2013. Next Generation Sequencing. [Google Scholar]
- 9.Nogajski JH, Kiernan MC, Ouvrier RA, Andrews PI. Congenital myasthenic syndromes. Journal of clinical neuroscience: official journal of the Neurosurgical Society of Australasia. 2009;16:1–11. doi: 10.1016/j.jocn.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 10.Mihaylova V, Muller JS, Vilchez JJ, Salih MA, Kabiraj MM, D’Amico A, Bertini E, Wolfle J, Schreiner F, Kurlemann G, Rasic VM, Siskova D, Colomer J, Herczegfalvi A, Fabriciova K, Weschke B, Scola R, Hoellen F, Schara U, Abicht A, Lochmuller H. Clinical and molecular genetic findings in COLQ-mutant congenital myasthenic syndromes. Brain: a journal of neurology. 2008;131:747–759. doi: 10.1093/brain/awm325. [DOI] [PubMed] [Google Scholar]
- 11.Wargon I, Richard P, Kuntzer T, Sternberg D, Nafissi S, Gaudon K, Lebail A, Bauche S, Hantai D, Fournier E, Eymard B, Stojkovic T. Long-term follow-up of patients with congenital myasthenic syndrome caused by COLQ mutations. Neuromuscular disorders. 2012;22:318–324. doi: 10.1016/j.nmd.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 12.Cossins J, Liu WW, Belaya K, Maxwell S, Oldridge M, Lester T, Robb S, Beeson D. The spectrum of mutations that underlie the neuromuscular junction synaptopathy in DOK7 congenital myasthenic syndrome. Human molecular genetics. 2012;21:3765–3775. doi: 10.1093/hmg/dds198. [DOI] [PubMed] [Google Scholar]
- 13.Haas RH, Parikh S, Falk MJ, Saneto RP, Wolf NI, Darin N, Cohen BH. Mitochondrial disease: a practical approach for primary care physicians. Pediatrics. 2007;120:1326–1333. doi: 10.1542/peds.2007-0391. [DOI] [PubMed] [Google Scholar]
- 14.Menezes MP, North KN. Inherited neuromuscular disorders: pathway to diagnosis. Journal of paediatrics and child health. 2012;48:458–465. doi: 10.1111/j.1440-1754.2011.02210.x. [DOI] [PubMed] [Google Scholar]
- 15.Lieber DS, Calvo SE, Shanahan K, Slate NG, Liu S, Hershman SG, Gold NB, Chapman BA, Thorburn DR, Berry GT, Schmahmann JD, Borowsky ML, Mueller DM, Sims KB, Mootha VK. Targeted exome sequencing of suspected mitochondrial disorders. Neurology. 2013;80:1762–1770. doi: 10.1212/WNL.0b013e3182918c40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Craigen WJ, Graham BH, Wong LJ, Scaglia F, Lewis RA, Bonnen PE. Exome sequencing of a patient with suspected mitochondrial disease reveals a likely multigenic etiology. BMC medical genetics. 2013;14:83. doi: 10.1186/1471-2350-14-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dare JT, Vasta V, Penn J, Tran NT, Hahn SH. Targeted exome sequencing for mitochondrial disorders reveals high genetic heterogeneity. BMC medical genetics. 2013;14:118. doi: 10.1186/1471-2350-14-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koenig MK. Presentation and diagnosis of mitochondrial disorders in children. Pediatric neurology. 2008;38:305–313. doi: 10.1016/j.pediatrneurol.2007.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kirby DM, Crawford M, Cleary MA, Dahl HH, Dennett X, Thorburn DR. Respiratory chain complex I deficiency: an underdiagnosed energy generation disorder. Neurology. 1999;52:1255–1264. doi: 10.1212/wnl.52.6.1255. [DOI] [PubMed] [Google Scholar]
- 20.Thorburn DR, Chow CW, Kirby DM. Respiratory chain enzyme analysis in muscle and liver. Mitochondrion. 2004;4:363–375. doi: 10.1016/j.mito.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 21.Hui J, Kirby DM, Thorburn DR, Boneh A. Decreased activities of mitochondrial respiratory chain complexes in non-mitochondrial respiratory chain diseases. Developmental medicine and child neurology. 2006;48:132–136. doi: 10.1017/S0012162206000284. [DOI] [PubMed] [Google Scholar]
- 22.Schapira AH. Human complex I defects in neurodegenerative diseases. Biochimica et biophysica acta. 1998;1364:261–270. doi: 10.1016/s0005-2728(98)00032-2. [DOI] [PubMed] [Google Scholar]
- 23.Schapira AH, Cooper JM, Dexter D, Clark JB, Jenner P, Marsden CD. Mitochondrial complex I deficiency in Parkinson’s disease. Journal of neurochemistry. 1990;54:823–827. doi: 10.1111/j.1471-4159.1990.tb02325.x. [DOI] [PubMed] [Google Scholar]
- 24.Tabrizi SJ, Cooper JM, Schapira AH. Mitochondrial DNA in focal dystonia: a cybrid analysis. Annals of neurology. 1998;44:258–261. doi: 10.1002/ana.410440218. [DOI] [PubMed] [Google Scholar]
- 25.Parker WD, Jr, Parks J, Filley CM, Kleinschmidt-DeMasters BK. Electron transport chain defects in Alzheimer’s disease brain. Neurology. 1994;44:1090–1096. doi: 10.1212/wnl.44.6.1090. [DOI] [PubMed] [Google Scholar]
- 26.Bernier FP, Boneh A, Dennett X, Chow CW, Cleary MA, Thorburn DR. Diagnostic criteria for respiratory chain disorders in adults and children. Neurology. 2002;59:1406–1411. doi: 10.1212/01.wnl.0000033795.17156.00. [DOI] [PubMed] [Google Scholar]
- 27.Morava E, van den Heuvel L, Hol F, de Vries MC, Hogeveen M, Rodenburg RJ, Smeitink JA. Mitochondrial disease criteria: diagnostic applications in children. Neurology. 2006;67:1823–1826. doi: 10.1212/01.wnl.0000244435.27645.54. [DOI] [PubMed] [Google Scholar]
- 28.Emeryk B, Rowinska-Marcinska K, Nowak-Michalska T, Sawicka E. Muscular fatigability in mitochondrial myopathies. An electrophysiological study. Electromyography and clinical neurophysiology. 1992;32:235–245. [PubMed] [Google Scholar]
- 29.Ukachoke C, Ashby P, Basinski A, Sharpe JA. Usefulness of single fiber EMG for distinguishing neuromuscular from other causes of ocular muscle weakness. The Canadian journal of neurological sciences Le journal canadien des sciences neurologiques. 1994;21:125–128. doi: 10.1017/s0317167100049040. [DOI] [PubMed] [Google Scholar]
- 30.Ly CV, Verstreken P. Mitochondria at the synapse. The Neuroscientist: a review journal bringing neurobiology neurology and psychiatry. 2006;12:291–299. doi: 10.1177/1073858406287661. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.