Abstract
Crohn’s disease is associated with an altered innate immune response of pathogenic importance. This altered response can be associated to loss-of-function polymorphisms in the NOD2 (nucleotide-binding oligomerization domain-containing protein 2) gene, but also changes in transcriptional and post-transcriptional regulatory layers, including microRNA activity. Here, we characterized the link between NOD2 genotype and inflammatory-mediated changes in innate signaling by studying transcriptional and post-transcriptional activity in response to NOD2-agonist muramyl dipeptide in monocytes from healthy controls, and Crohn’s disease patients with and without NOD2 loss-of-function polymorphisms. We measured the expression of genes and microRNAs in monocytes from these subjects after stimulation with muramyl dipeptide. Gene expression profiles mainly distinguished the actual muramyl dipeptide response, but not the genotype. A hyper-responsive phenotype was found in Crohn’s disease patients without NOD2 mutations, characterized by upregulated cytokine receptors and general downregulation of microRNA expression. Conversely, microRNA expression could identify genotype-specific differences between subject groups but exhibited little change upon muramyl dipeptide treatment. Only two microRNAs showed muramyl dipeptide-induced response, including miR-155, which was found to regulate multiple genes and whose host gene was one of the highest muramyl dipeptide responders. miR-155 was upregulated in Crohn’s disease patients with NOD2 mutations following lipopolysaccharide and Escherichia coli treatment, but the upregulation was substantially reduced upon muramyl dipeptide treatment. While Crohn’s disease patients with NOD2 mutations on average showed a reduced muramyl dipeptide response, the cohort exhibited large individual variance: a small subset had inflammatory responses almost comparable to wild-type patients on both gene and miR-155 regulatory levels.
Crohn’s disease: Genetics impacts molecular regulation of innate immunity on miRNA, but not gene level
The genetics of people with Crohn’s disease affects the molecular drivers of their dysregulated immune responses. Some individuals with Crohn’s harbor mutations in the NOD2 gene, which encodes a pathogen recognition receptor that binds to a molecule called muramyl dipeptide (MDP). To better understand how alternations in NOD2 can lead to increased susceptibility to gut inflammation, Yun Chen, Mohammad Salem and colleagues from the University of Copenhagen and Herlev Hospital, Denmark, analyzed the expression patterns of both genes and small, regulatory microRNAs in blood cells from healthy controls and from Crohn’s patients with and without NOD2 mutations. They exposed the cells to MDP, and saw that although gene acticity changed dramatically as a response, there was little difference between subjects, regardless of genetics. Conversely, microRNA expression showed genotype-specific differences that weren not impacted by MDP treatment. The findings underscore the importance of microRNAs in Crohn’s disease.
Introduction
Crohn’s disease (CD)1 constitutes together with ulcerative colitis (UC)2 the two most prevailing disorders under the umbrella term inflammatory bowel disease (IBD).3 CD can affect any part of the gastrointestinal tract but most often the ileocecal region. General clinical manifestations of intermitting bowel inflammation are associated with abdominal pain, fever, prolonged diarrhea, and/or weight loss.4 In CD, genetic variations and environmental factors such as altered microbiota interact to produce the inflammatory background of the disease.1, 5 Along with changes in the adoptive immune system, an impaired innate immunity is believed to play a crucial role in the immunopathogenesis of CD.
Epidemiologic and linkage studies suggest that genetic factors play a clinical role in determining CD susceptibility.6, 7 One of the key genes identified through these studies is NOD2 (nucleotide-binding oligomerization domain-containing protein 2),8, 9 encoding a cytosolic pathogen recognition receptor that specifically binds to muramyl dipeptide (MDP), the smallest bioactive component of peptidoglycans present in most bacteria.10 Activation of NOD2 leads to induction of key immune signaling pathways involved in CD pathogenesis, including the upregulation of various pro-inflammatory cytokines.11 In addition, continuous MDP stimulation inhibits the development of experimental colitis in animal models,12 suggesting that normally functioning NOD2 signaling pathways might be directly involved in protecting and maintaining the intestinal homeostasis. Furthermore, innate signaling is altered even in NOD2 wild-type (WT) CD patients,13 implying post-transcriptional modifications of NOD2 signaling pathways could be involved in re-establishment of normal innate responses, and this mechanism could be a future therapeutic target aiming at restoring microbial responses in CD. In this context, recent studies have linked NOD2 to crucial post-transcriptional changes, including regulation by microRNAs (miRNAs) in different intestinal epithelial cell lines.14–16 miRNAs are short structured RNAs that play important roles in the regulation of gene transcription and translation in development and disease progression, including CD.17, 18 One of the most well-known miRNA regulators associated with inflammation and innate immunity is miR-155.19–22 Previous studies have shown an elevated level of miR-155 in colonic mucosa of CD and UC,23, 24 and depletion of miR-155 protects from experimental colitis in mice.25 Different pathogenic stimuli induce miR-155 in vitro, including viral infection20, 26, 27 and lipopolysaccharide (LPS).28 However, the role of miR-155 in immune cells in the context of CD and NOD2 polymorphisms is not well established.
There are three major single-nucleotide polymorphisms (SNPs) of the NOD2 gene (SNP8, SNP12, and SNP13) that are strongly associated to CD and cause a loss-of-function phenotype with a reduced response to MDP.11 SNP8 and SNP12 are missense mutations, whereas SNP13 is a frameshift mutation with the most drastic loss-of-function phenotype.29 The link between SNPs in NOD2 and CD-associated inflammatory responses might have implications for the disease not only because an innate immune dysfunction is important for the pathogenesis of CD, but also since normalization of innate signaling alleviates colitis.12, 30 However, controversies exist on how alterations in NOD2 signaling can lead to an increased susceptibility to inflammation, and what the role of post-transcriptional regulation in patients with/without disease-associated NOD2 variants are for the resulting pathogenic innate immune response.11 Therefore, transcriptional profiles of NOD2 mutant cells responding to MDP or bacterial stimulation have been analyzed using coverage-limited microarrays and focused either on mRNAs or miRNAs. The miRNA–mRNA regulation studies were either a single-target study31 or done in non-primary cells or immune cells derived from non-CD subjects.15, 32 Thus, it is important to investigate the interactions between miRNAs and genes genome-wide in patient-derived cells, and study how this is affected in innate immune response as function of different NOD2 genotypes. To do this, it is necessary to investigate both RNA species from the same cells upon inflammation induction in cells with and without NOD2 loss-of-function SNPs.
To this end, in this study we measured the expression of genes and miRNAs in human blood monocytes before and after MDP stimulation using genome-wide RNA and small RNA sequencing in an exploratory cohort consisting of healthy control subjects and CD patients with/without NOD2 loss-of-function SNPs (N = 7 in total), and subsequently validated our major findings in larger patient groups (N = 29, in total) as well as all identified miRNA-gene interaction candidates in the THP-1 cell line (N = 8). Our data suggest that gene expression effectively identifies pathological MDP/NOD2-associated immune gene responses in blood monocytes from subjects without NOD2 loss-of-function mutations. A pre-inflammatory, hyper-responsive phenotype was found in CD patients without NOD2 loss-of-function SNPs. Conversely, the miRNA regulatory layer was surprisingly static before and after MDP stimulation, but could predict subject and genotype groups regardless of MDP treatment. Only two miRNAs, miR-190A and miR-155, responded to MDP. CD patients with NOD2 loss-of-function SNPs were, as expected, on average less responsive to MDP compared to other subjects at both gene and miR-155 levels, but these patients displayed a surprisingly large variance in their responses ranging from a total loss to a nearly intact inflammatory response. Although NOD2 loss-of-function SNP patients displayed reduced miR-155 induction after MDP treatment, miR-155 induction was unchanged when treating with LPS or intact bacteria treatment, suggesting that NOD2 mutations only affect NOD2-pathway-elicited response and not other inflammatory pathways.
Results
Differential expression profiles between CDWT, CDNOD2, and control subjects
We first analyzed the miRNA and gene expression signatures between the three subject groups: CDWT, CDNOD2, and controls using small RNA-Seq and RNA-Seq as defined in the “Methods” section before MDP treatment (green arrows, Fig. 1 and top of Fig. 2a,b) and after MDP treatment (yellow arrows, Fig. 1 and top of Fig. 2c,d).
Fig. 1.
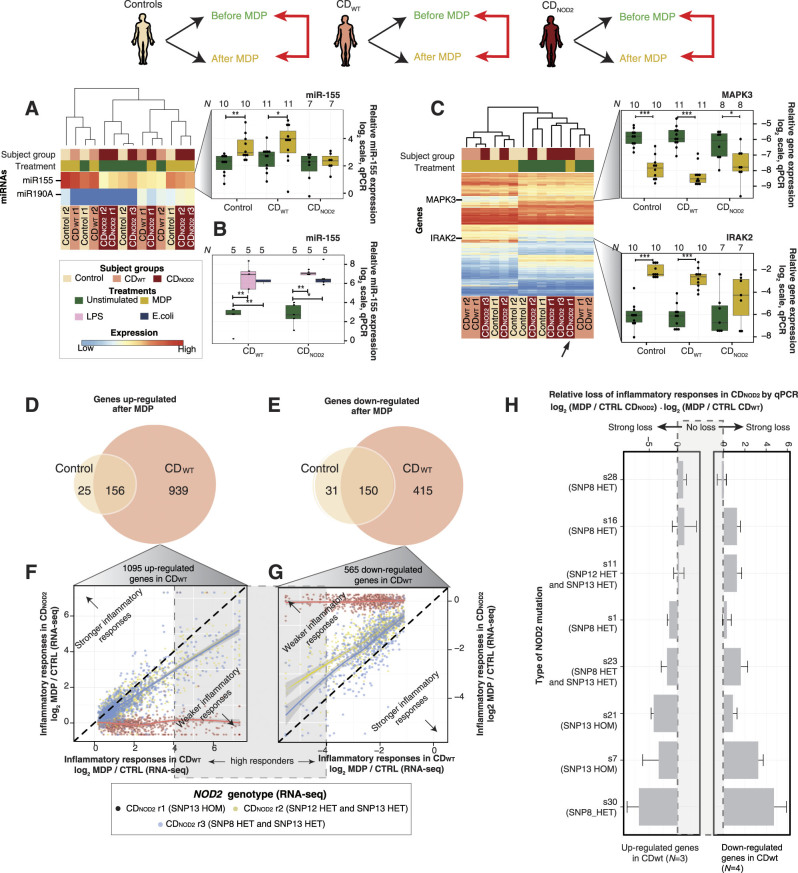
Overview of cohorts and differential expression analyses. Monocytes were derived from three subject groups: healthy controls, CD patients with WT NOD2 genotypes (CDWT), and CD patients with loss-of-function NOD2 genotypes (CDNOD2). Monocytes were analyzed by RNA-Seq and small RNA-Seq (black body outlines, number of tested individuals is indicated), and selected genes and miRNAs were validated by qPCR in a validation cohort (blue body outlines, number of tested individuals is indicated). We analyzed the difference between subject groups, before and after MDP stimulation (green and yellow arrows, shown in Figs. 2,3), and the response to MDP stimulation within each group (red arrows, shown in Fig. 4)
Fig. 2.
miRNA and gene expression patterns between subject groups. a Heat map showing the expression pattern of differentially expressed miRNAs between subject groups before MDP stimulation (green arrows in Fig. 1, also shown as schematics on top). Expression values are broken into permilles and assigned to a color gradient ranging from red (high expression) to blue (low expression). Rows indicate individual miRNAs, while columns show individual subjects. Rows and columns are ordered by hierarchical clustering using Euclidian distance, where the top dendrogram indicate similarity between subjects. Subject groups and MDP treatment are indicated by color above the heat maps. Note the clear separation between CDWT, CDNOD2, and controls. b As in a, but showing differentially expressed genes (by RNA-Seq) between subject groups before MDP treatment. c As in a, but showing differentially expressed miRNAs between subject groups after MDP stimulation. Note the clear separation between CDWT, CDNOD2, and controls. d As in a, but showing differentially expressed genes between subject groups after MDP treatment. e Left: Venn diagram showing the overlap between differentially expressed miRNAs between subject groups before and after MDP treatment. Right: expression-based clustering of the five shared miRNAs as in a, but only showing subjects, treatments, and the resulting dendrogram (color codes as in a–d). Note that subject groups are clearly separated, while MDP treatment status is not. f As in e, but for differentially expressed genes. Note that the shared gene set is not as efficient at distinguishing subject groups as the miRNA expression
Intriguingly, the miRNA expression signature could separate the three subject groups (controls, CDNOD2, and CDWT) both before and after MDP treatment. In both states, control subjects and CDNOD2 patients were more similar to each other than CDWT patients, where many miRNAs were more lowly expressed compared to the other two groups (Fig. 2a,c). Thus, the overall miRNA population was downregulated in CDWT patients, suggesting an overall lower impact of miRNA-mediated gene repression in this group.
In contrast, the differences of gene expression could not distinguish subject groups as clearly as the miRNA expression, because the differences between subject groups were much smaller than that observed by miRNA expression (see dendrogram in Fig. 2b,d, and Supplementary Fig. 1a, b).
We next analyzed the differentially expressed miRNAs and genes identified in these two analyses. Nearly half of miRNAs (42%, 5/12) that were differentially expressed between subject groups before MDP treatment were also differentially expressed between subject groups after MDP treatment (Fig. 2e, Venn diagram in the left panel). The expression levels of these shared miRNAs were highly consistent before and after MDP treatment (Supplementary Fig. 2a); thus, the overall regulatory layer of miRNAs seemed to be independent of MDP signaling between groups, and could at the same time separate the subject groups. Indeed, the five miRNAs that were differentially expressed between groups regardless of treatment could alone distinguish subject groups (but not MDP treatment) efficiently (Fig. 2e, dendrogram in the right panel).
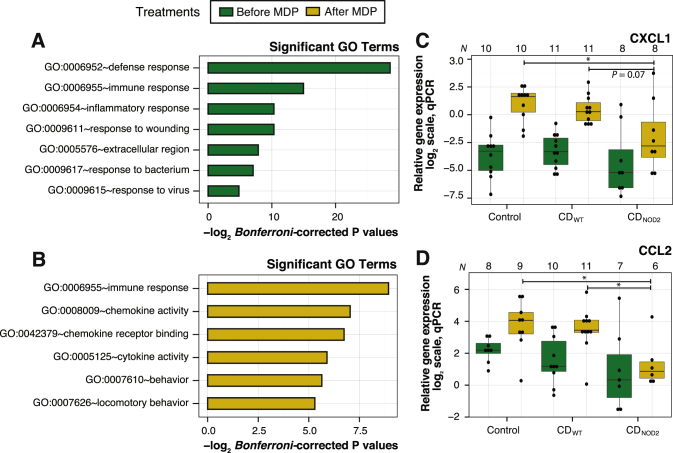
In contrast, only around 10% (13/108, Fig. 2f, also see Supplementary Fig. 1c) of genes that were significantly different between subject groups before MDP treatment were also significantly different between patient groups after MDP treatment. These shared genes could not separate the subject groups (Fig. 2f, dendrogram in the left panel). Consistently, these genes displayed a wide expression difference between subjects before and after MDP (Supplementary Fig. 2b). Genes that were differentially expressed between subject groups before MDP stimulation showed an enrichment of gene ontology (GO) terms relating to defense response, inflammatory response, response to wounding, and response to virus (Fig. 3a and Supplementary Fig. 3a). In contrast, genes differentially expressed between patient genotype groups after MDP treatment were enriched in chemokine and cytokine-related functions (Fig. 3b and Supplementary Fig. 3b). Quantitative PCR (qPCR) analysis for two selected chemokines, CXCL1 and CCL2, in a larger cohort confirmed upregulation after MDP treatment in healthy controls and CDWT, but not in CDNOD2 patients (Fig. 3c,d; Supplementary Fig. 3c, d show corresponding RNA-Seq data). This is consistent with the established link between NOD2 and cytokine induction through the NF-κB pathway.11
Fig. 3.
Functional annotation of differentially expressed genes between subject groups. a Enriched significant GO terms (Bonferroni-corrected P-values ≤ 0.05) for differential expressed genes between subject groups before MDP treatment. X-axis shows –log2(Bonferroni-corrected P-values) and Y-axis shows GO terms. b As in a, but showing significant GO terms for differential expressed genes between subject groups after MDP treatment. c qPCR validation for a selected chemokine, CXCL1, which exhibits a large difference between the subject groups after MDP treatment. X-axis shows subject groups before (dark green) and after MDP treatment (yellow). Numbers on top of the plot indicate the number of biological replicates passing the qPCR thresholds. Each subject is visualized as black dots. Y-axis shows the relative gene expression in log2 scale. Asterisks indicate level of significance (two-sided t-test): * indicates P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001. A single comparison with a P-value close to the significance threshold is indicated. d qPCR validation for a selected chemokine, CCL2, following the conventions of c
miRNA and gene expression response to MDP
In order to investigate cell response to MDP, we compared the difference in miRNA and gene expression before vs. after MDP treatment within each patient group (red arrows in Fig. 1b and top of Fig. 4). Consistent with the analysis above, only two miRNAs (miR-155 and miR-190A) were significantly changed as a response to MDP in any pairwise comparison. Because miR-155 was much higher expressed than miR-190A (Fig. 4a, left panel), and was previously established as a key inflammatory regulator,21, 22, 33 we validated its expression change by qPCR in the larger validation cohort. The results showed that miR-155 was significantly upregulated in controls and CDWT, but not in CDNOD2 after MDP treatment (P ≤ 0.05, paired two-sided t-test) (Fig. 4a, right panel; Supplementary Fig. 4a shows corresponding small RNA-Seq data). Notably, there was no difference in miR-155 upregulation between CDWT and CDNOD2 patients when cells were treated with either LPS or intact intestinal Escherichia coli bacteria (Fig. 4b). Thus, miR-155 could be upregulated by multiple inflammatory stimuli but only the MDP-triggered upregulation was dependent on the NOD2 genotype. This makes sense, as NOD2 is a specific sensor for MDP, while LPS and E. coli activate other NOD2-independent inflammatory pathways.
Fig. 4.
MDP-dependent miRNA and gene expression response within subject groups. a Left: Heat map as in Fig. 2a showing the two miRNAs that were differential expressed as a response to MDP within a subject group (schematic on top shows the analyzed comparisons). Right: qPCR validations for miR-155 are following the conventions of Fig. 3c. Asterisks indicate level of significance (two-sided paired t-test): * indicates P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001. b qPCR analysis of miRNA-155 expression LPS or intact E. coli in monocytes from CD patients, broken up by NOD2 genotype and treatment. Y-axis shows the log2 normalized fold change of mir-155 vs. expression control (see Supplementary Methods for normalization): dots strand for individual patients. Asterisks indicate level of significance (two-sided paired t-test): * indicates P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001. c Left: As in left of a, but showing genes that were differential expressed as a response to MDP within a subject group. The black arrow highlights the outlier CDNOD2 subject that shows no substantial change in response to MDP. Right: qPCR validations of IRAK2 and MAPK3 genes are shown in the right panel, organized as in the right of panel a. d Venn diagrams showing the overlap of upregulated genes in response to MDP within subject groups. e Venn diagrams showing the overlap of downregulated genes in response to MDP within subject groups. f Degree of inflammatory gene response of individual CDNOD2 patients compared to the average response of CDWT patients from the exploratory cohort. The inflammatory response is measured in a set of 1095 genes that were upregulated in CDWT. X-axis shows log2 fold change of the 1095 genes in CDWT patients after MDP. Y-axis shows corresponding fold changes in individual CDNOD2 patients. Each dot corresponds to one gene, colored by individual CDNOD2 patients. Solid lines represent fitted linear models for each CDNOD2 patient using the loess approach; 95% confidence intervals are indicated as gray shadows. The dashed diagonal in black corresponds to equivalent inflammatory response between CDNOD2 and CDWT patients. Genes located above the diagonal will correspond to stronger inflammatory response in a CDNOD2 patient than the average CDWT response. Conversely, genes located below the diagonal will correspond to decreased inflammatory response in CDNOD2 vs. CDWT patients. Genes with high log2 fold changes (>4) are considered high MDP responders (see main text). g As in e, but for the 565 downregulated genes in CDWT patients following MDP stimulation. The two parts of the plot indicating stronger or weaker inflammatory response in a CDNOD2 are now swapped to the opposite. Genes with low log2 fold changes (<−4) are considered high MDP responders (see main text). h Relative loss of inflammatory response in individual CDNOD2 patients compared to average CDWT patients in the validation cohort by qPCR. Each pair of horizontal bar plots shows the average degree of inflammatory response loss for a CDNOD2 patient, expressed as the average difference vs. CDWT patients for three upregulated genes in CDWT patients (CXCL1, CCL2, and IRAK2, left panel) and four downregulated genes in CDWT patients (MAPK3, PALD1, MAVS, and FLI1, right panel). Error bars indicate the standard error of the mean
Conversely, the gene expression profiles were drastically different after MDP treatment (Fig. 4c, left panel and Supplementary Fig. 4b) and could distinguish treated vs. non-treated samples with the exception of one CDNOD2 sample (see black arrows in Fig. 4c and Supplementary Fig. 4b). qPCR analysis in the larger validation cohort confirmed the changes in selected differential expressed genes (Fig. 4c, right panel and Supplementary Fig. 4c–f). Notably, CDWT patients had more than six times the number of upregulated genes than healthy controls (1095 vs. 181, Fig. 4d; also see Supplementary Fig. 4g). Moreover, the large majority of upregulated genes in healthy controls (86%, 156/181) were also upregulated in CDWT patients. To test whether these 156 shared upregulated genes had the same magnitude of MDP response, we plotted the average gene expression fold change after MDP in the healthy control group vs. corresponding fold change in the CDWT group (Supplementary Fig. 4h). In general, genes with high upregulation in the healthy control group had even higher upregulation in CDWT patients. This was also true for the host gene of miR-155, consistent with the higher upregulation of mature miR-155 in CDWT (Fig. 4a), and indicating an important role for miR-155 in MDP response in CD conditions.
Conversely, in the CDNOD2 patients, no genes were significantly upregulated or downregulated as a response to MDP (only ten genes were induced using the alternative significance calculation method, Supplementary Fig. 4g).
Together, these observations indicated that only subjects with WT NOD2 (healthy controls and CDWT patients) had substantial MDP-induced responses and CDWT patients had a stronger and wider MDP response. To investigate the responses only found in CDWT patients further, we mapped the shared upregulated genes between CDWT and control subjects and the genes only upregulated in CDWT subjects onto cytokine-receptor KEGG pathways.34 Strikingly, the receptors for upregulated cytokines were primarily upregulated in CDWT patients but not in healthy controls (Supplementary Fig. 5a, b), suggesting that the downstream inflammatory response in these pathways was blocked or diminished in healthy controls even if the relevant cytokines were present. These results strongly imply that CDWT patients had a wider inflammatory response including substantial activation of chemokine receptors and their downstream signaling pathways. Thus, these patients could be characterized as a phenotype with pre-inflammatory hyper-reactive innate response. An investigation of MDP-dependent downregulated genes showed that although the number of downregulated genes was smaller than upregulated genes, the results were consistent to the pattern observed above: on gene level, CDWT patients had a stronger and wider MDP-mediated inflammatory response than healthy controls, while CDNOD2 patients typically lost the majority of the inflammatory response to MDP (Fig. 4e and Supplementary Fig. 4g, h).
While we observed no significant gene changes upon MDP stimulation in CDNOD2 patients, the MDP states of two CDNOD2 subjects could be distinguished using gene expression profiles (see dendrogram in Fig. 4c, left panel). Thus, we reasoned that subtle expression changes might exist, and that the type of NOD2 mutation might influence the degree of expression change. To investigate this further, we focused on the genes found upregulated and downregulated upon MDP treatment in CDWT (1095 and 565 genes, respectively). For each CDNOD2 sample we plotted the RNA-Seq gene expression fold change after MDP treatment vs. the average gene expression fold change after MDP in the CDWT group (Fig. 4f,g, and Supplementary Fig. 4g, right panels). While this confirmed the overall lower inflammatory response in CDNOD2 samples vs. CDWT, it also showed clear differences in inflammatory response across the individual CDNOD2 samples. This was consistent between upregulated and downregulated genes and was particularly clear for genes with high fold changes. This indicated the loss of inflammatory responses in CDNOD2 patients was not binary but a gradient across individuals. To verify our observation in a larger cohort, we measured the expression of seven validated MDP-responsive genes in CDWT (three upregulated genes: CXCL1, CCL2, and IRAK2; four downregulated genes: MAPK3, MAVS, PALD1, and FLI1) in eight CDNOD2 and eleven CDWT samples before and after MDP treatment by qPCR (Fig. 4h). From this analysis, the large variance in inflammatory response was evident: some CDNOD2 patients showed virtually no inflammatory response, while some had inflammatory responses comparable to CDWT subjects. Overall, in our set, CDNOD2 patients carrying SNP13 HOM had the lowest responses to inflammation induced by MDP (Fig. 4f–h and Supplementary Fig. 4g).
miRNA-gene associations in MDP response
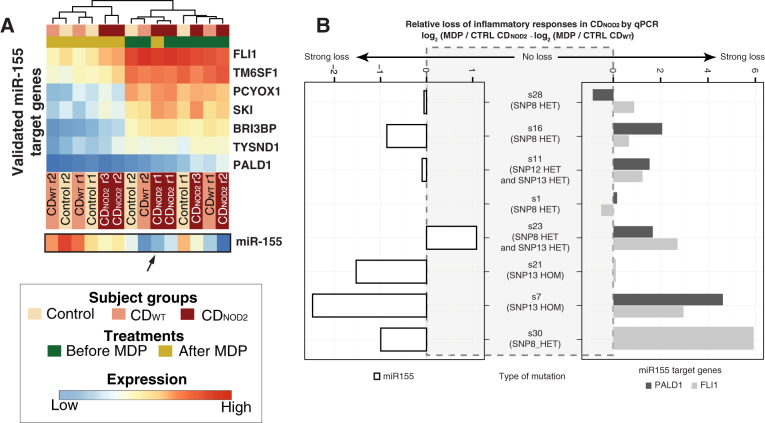
The analyses above did not take miRNA-elicited regulation of gene expression into account. To identify likely regulatory miRNA-gene associations, we used experimentally verified miRNA targets from mirTarBase 4.3,35 and required that the miRNA and gene pair in question had anti-correlated expression patterns (e.g., upregulation of miRNA and downregulation of gene, or vice versa) in any of the previously made pairwise expression analyses (any arrow in Fig. 1). When including all the differentially expressed miRNAs and genes, only 16 putative miRNA-gene pairs centered on miR-155 were identified (Fig. 5a). To further consolidate the regulatory role of miR-155, we transfected pre-miR-155 into human monocytic THP-1 cells and measured the expression change of the 16 candidate target genes with qPCR (Fig. 5a). Of these, PALD1, FLI1, PCYOX1, and BRI3BP were significantly repressed upon miR-155 transfection (P ≤ 0.05, two-sided t-test, Fig. 5b–e), while TYSND1, TM6SF1, and SKI were borderline significant (Fig. 5f–h, for details see “Methods”). It is important to consider that this type of experiment may include indirect effects; however, several of the genes had additional external evidence in the form of gene reporter assays (external evidence is summarized in Fig. 5i).
Fig. 5.
Identification of miR-155-gene associations. a miRNA-gene association network showing miR-155-gene interactions identified. Line color summarizes the qPCR-based transfection analysis results in the exploratory cohort (shown in b–h). Strong validations (bold lines, corresponding to b–e) and weak validations (gray lines, corresponding to f–h), and interactions that failed validations are shown as dotted lines. Strong and weak criteria are described in “Methods” (b–h). qPCR results of miR-155 transfected THP1 cells. Y-axis shows relative qPCR expression of the gene indicated on top of the plot. X-axis shows boxplots for cells that were not transfected, miR-155-transfected cells, and control-transfected cells. Dots indicate individual samples. Asterisks indicate level of significance (two-sided t-test): * indicates P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001. P-values for tests that are borderline significant are shown (see Methods). b–e show strong validation cases, f–h shows weak validation cases. i Summary of evidence for miR-155 regulation of the selected genes, from mirTarBase and this study. Black boxes indicate the evidence from the indicated type of experiments from mirTarBase. “Strong” and “weak” categories refer to the classification of validations in b–h
We found that this limited set of seven validated miR-155 targeting genes alone could distinguish MDP-treated cells from non-treated cells except for the same CDNOD2 subject that could not be correctly classified using the whole gene set (Fig. 6a). Because we identified a large variance across CDNOD2 patients on gene expression level following MDP stimulation, we hypothesized that a similar variation existed on miR-155 regulation level. In order to test this, we focused on miR-155 and the seven target genes identified above, and compared their MDP response between individual CDNOD2 patients and pooled CDWT patients (Supplementary Fig. 6), similarly to our previous gene expression comparison in Fig. 4f,g. As expected, the CDNOD2 subject that showed the smallest inflammatory responses at gene level also displayed the most drastic loss of miR-155 upregulation and downregulation of its seven validated targets (Supplementary Fig. 6). This variance across patients was confirmed by qPCR analysis in the larger cohort. This showed that the loss of miR-155 induction following MDP treatment varied across subjects, and the corresponding level of downregulation of the miR-155 target genes generally followed the miR-155 levels (Fig. 6b). In general, CDNOD2 subjects displaying a reduced MDP response of gene level also showed this on miR-155 level (Figs. 4h, 6b).
Fig. 6.
Classification power and variability of miR-155 and its targets across CDNOD2 patients. a Heat map as in Fig. 2a showing the expression pattern of the seven genes interacting with miR-155 indicated in a. The expression of miR-155 in corresponding libraries is indicated below the heat map. The black arrow highlights the outlier CDNOD2 subject that shows no substantial response to MDP. b Degree of response changes of individual CDNOD2 patients compared to the average response of CDWT patients from the exploratory cohort on miR-155 level (left panel) and two selected miR-155 target genes (PALD and FLI1, right panel). The plot follows the same conventions as Fig. 4h
Discussion
In this study we explored gene and miRNA expression in an initial set of human blood monocytes using genome-wide high-throughput sequencing techniques and verified our major conclusions by applying qPCR analyses in a larger validation cohort or THP-1 cells. All the samples were split by MDP treatment and were broken up by subject groups, including patients with NOD2 genotypes.
The results from pairwise differential expression studies between the subjects demonstrated that the MDP treatment resulted in considerable changes in gene expression. In particular, CDWT patients showed a much stronger and wider inflammation response compared to healthy controls. This is in line with a report investigating the regulation of specific inflammatory pathways upon innate engagement of monocytes in CD,36 but we here show that these changes are more general, involving activating cytokine receptors and their downstream signaling pathways. In addition, it is noteworthy that while the major inflammatory responses in the gene layer of CDWT patients corresponded to upregulation, miRNA expression was generally downregulated in these patients. This suggests that the innate hyper-responses upon inflammation in CDWT patients might be also mediated by generally reduced activity of the miRNA regulatory layer. Such a NOD2-dependent phenotype of a pre-inflammatory innate hyper-responsiveness state could be essential in shaping the inflammatory processes found in CD. In line with this, there is evidence of an increased risk of onset of CD in patients after infections with intestinal pathogens, e.g., Salmonella spp. or Campylobacter spp. 37, 38
In contrast to mRNA levels, miRNA expression signatures showed large differences between subject groups, but only two miRNAs changed significantly with MDP treatment including a key inflammatory regulator, miR-155. Studies investigating NOD2-specific miR-155 inductions have shown MDP stimuli alone has limited impact on miR-155 expression in human dendritic cells,15 mouse macrophages,27 and embryonic fibroblasts.39 Our results demonstrated that MDP could induce miR-155 and its host gene in human monocytes expressing WT NOD2, and this induction was particularly strong in monocytes from CD patients, suggesting the NOD2-based induction on miR-155 might differ across both cell and disease types.
Previous studies have shown that the NOD2 loss-of-function SNPs lead to decreased immune response,40, 41 but the differences between individual subjects and type of mutations were not addressed. Here, we showed that the effect of NOD2 loss-of-function SNPs is diverse: some individuals from our CDNOD2 cohort lost the MDP response, while some could mount an inflammatory response similar to that of WT NOD2 subjects. This observation was reflected on both general gene expression profiles, on miR-155 levels and on its target genes. Patients carrying the SNP13 HOM appeared to have the largest response loss, although to investigate the detailed effect of respective genotype, expression quantitative trait loci analyses across a much larger population will be necessary.
As we have not induced or rescued NOD2 mutations, these results are based on correlations between genotype and phenotype. However, as reduced induction of miR-155 in the CDNOD2 group occurred only when treating with MDP (a NOD2 agonist) and not when treating with LPS or intact bacteria, it is highly likely that the NOD2 mutations are the main drivers for the phenotypes observed and by extension, that inflammatory pathways not involving NOD2 are likely intact in these patients. Additional mutations carried by these patients may modulate the MDP response and may explain the wide MDP-response repertoire in CDNOD2 patients. Such mutations may not only affect protein-coding genes but also regulatory sites, including the disruption of miRNA-binding sites, which have been reported to occur in immune-associated genes, including NOD2 and IRGM.42, 43 However, to analyze such cases, genome-wide genotyping is necessary.
In summary, our study identified substantial and novel changes in NOD2 signaling of CDNOD2 and CDWT patients. The first group had on average dampened MDP responses, but the global response was highly varied across individuals, on both gene and miR-155 regulation levels. The CDWT group was characterized by a pre-inflammatory innate hyper-responsive phenotype and thus responds much stronger to MDP than cells from healthy controls. This hyper-responsive phenotype was characterized by activation a wide range of cytokine receptors and potentially also by a general downregulation of miRNA expression in these patients. Both types of dysregulated innate responses could, in different ways, increase susceptibility to intestinal inflammation in response to commensal bacteria. Moreover, we have shed light on an underappreciated role of miRNAs in identifying disease-specific and genotype-specific alterations in CD.
Methods
Study population and genotyping
The patient population in this study was derived from a total of 57 healthy controls (i.e., without any known GI disorders) and 236 patients with well-established CD attending the IBD Clinic at Department of Gastroenterology, Medical Section, Herlev Hospital. The diagnostic criteria for CD were based on established guidelines44 applying clinical, radiologic, endoscopic, and histologic criteria. Screening for the common NOD2 polymorphisms (i.e., SNP8 [Arg702Trp], SNP12 [Gly908Arg], and SNP13 [Leu1007fsinsC]) was performed as previously described.45 All participants in this study were WT in respect to autophagy-related 16-like 1 (ATG16L1) genotype. Patients included in the sequencing and validation studies were in complete clinical remission for at least 4 weeks prior to inclusion (Harvey-Bradshaw index ≤5),46 and did not receive glucocorticoids or biologics for at least 3 preceding months. Long-term treatment with thiopurines was, however, allowed if the dosing had been stable for more than 2 months. For RNA-Seq and small RNA-Seq, monocytes were isolated from peripheral blood samples from three subject groups (N = 7 in total): (I) healthy controls with WT NOD2 (N = 2), (II) CD patients with WT NOD2 (CDWT) (N = 2), and (III) CD patients with NOD2 SNPs (CDNOD2; one SNP13 homozygote—replicate 1, one SNP13 heterozygote and SNP12 heterozygote—replicate 2, and one SNP13 heterozygote and SNP8 heterozygote—replicate 3) (N = 3). Subsequently, miRNA/mRNA patterns identified in the RNA-Seq exploratory experiments were tested in a larger cohort (N = 29 in total) where monocytes were obtained from (I) healthy controls with WT NOD2 (N = 10), (II) CDWT (N = 11), (III) CDNOD2 (mixed NOD2 polymorphisms) (N = 8). The monocytes isolated from each participant were grown with or without MDP stimulation (see Supplementary Methods) for sequencing and qPCR analysis. Supplementary Table 1 shows additional subject data. Validations of miRNA-gene interactions were done with eight biological replicates in TPH-1 cells (see Supplementary Materials and Methods).
Real-time qPCR validation of mRNAs and miRNAs
RNA was purified from monocytes and THP-1 cells, and converted to cDNA as described in Supplementary Methods. Primer sequences are shown in Supplementary Table 2. For each primer pair, only the samples that passed qPCR thresholds were used in respective analyses (see “Methods”). Numbers of samples passing thresholds are shown on top of corresponding figures. Two-sided t-tests qPCR values were used as default statistical tests between two groups. Paired tests were conducted for comparisons within subject groups shown in Fig. 4 and Supplementary Fig. 4. P ≤ 0.05 was used as significance cutoff. For miRNA transfection experiments in THP-1 cells, validations were considered “strong” if miR-155-transfected samples vs. no transfected samples and miR-155-transfected samples vs. control-transfected samples were both significant. Validations were considered “weak” either if P < 0.06 in both comparisons, or if P ≤ 0.05 in one of the two comparisons. Validations were considered “negative” if none of the above applied.
RNA-Seq and small RNA-Seq preparation, mapping and quantification
Total RNA and short RNAs were purified using the Nucleospin RNA II isolation kit (Macherey-Nagel) as recommended by the manufacturer. Quantity and purity were determined on a NanoDrop1000, all samples had a 260/280 nm absorbance >1.9. Illumina libraries were constructed using TruSeq Stranded mRNA Sample Prep Kit and TruSeq Small RNA Library Preparation Kit (Illumina) following the manufacturer’s protocols. The libraries were sequenced on an Illumina Genome Analyser IIx running sequencing 76 cycles, single read, for the mRNA libraries, and 36 cycles, single read, for the short RNA libraries. Adapters were trimmed from reads, which were then, filtered by quality and mapped to the hg19 assembly (see Supplementary Methods).
Differential expression analysis
Since our samples were derived from three subject groups and two MDP conditions, many types of pair-wise comparisons between groups were feasible, which would answer different types of questions (Fig. 1). Three group comparisons were performed as shown in Fig. 1: (1) between patient groups “before MDP treatment” (green arrows), (2) between subject groups “after MDP treatment” (yellow arrows), and (3) between expression profiles of the same subject group before and after treatment “MDP response” (red arrows).
For differential expression of genes in the above pair-wise comparisons, we conducted a combined analysis using DESeq47 and Cuffdiff,48 where we required an adjusted P-value ≤ 0.05 (Benjamini–Hochberg False Discovery Rate, FDR) from DESeq combined with a non-adjusted P-value ≤ 0.05 from Cuffdiff (shown in main figures). The reciprocal analysis requiring FDR ≤ 0.05 from Cuffdiff combined with a non-adjusted P-value ≤ 0.05 from DESeq did not give substantially different results (shown in Supplementary Figs. 1, 3–5). Differential expression analysis for miRNAs was performed by DESeq47 requiring a FDR ≤ 0.05. All differentially expressed genes analyzed with both methods and miRNAs are listed in Supplementary Table 3.
Code availability
Aside from the programs mentioned above, computational analyses were performed in R. Code is available by request.
Ethical approval
The study was approved by the Scientific Ethics Committee of the Copenhagen Capital Region. Written informed consents from all included subjects were obtained prior to participation, and the project fulfilled the Helsinki V Declaration.
Electronic supplementary material
Acknowledgements
This work was supported by grants from Axel Muusfeldts Foundation, Holger og Ruth Hesses Mindefond, Else og Mogens Wedell-Wedellsborgs Foundation, Direktør Kurt Bønnelycke og hustru fru Grethe Bønnelyckes Foundation, P. A. Messerchmidt og Hustrus Foundation, the Foundation of Aase and Ejnar Danielsen, the Danish Council for Independent Research, the Lundbeck Foundation, and the Novo Nordisk Foundation. We thank Hanne Fuglsang, Nine Scherling, Noor Irmam, and Tine Helene Soendergaard for excellent technical assistance. We thank Kim Jensen for access to PCR instruments.
Data sharing
Small RNA and RNA-Seq data are deposited in the GEO database, ID: GSE69446.
Author contributions
Y.C. and M.S. are shared first authors. M.B. and J.B. are shared second authors. A.S., J.B.S., and O.H.N. designed the study and supervised the project. Y.C. analyzed the data. Y.C. and A.S. made the figures. M.S. made the sequencing libraries. M.S., M.B., J.B., Y.L., and M.C. made qPCR and miRNA validations. J.B.S. and O.H.N. did patient allocation. Y.C., M.S., J.B.S., O.H.N., and A.S. wrote the manuscript.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary Information accompanies the paper on the npj Genomic Medicine website (doi:10.1038/s41525-016-0001-4).
Contributor Information
Jakob Benedict Seidelin, Phone: +45 38 68 60 54, Email: jseidelin@dadlnet.dk.
Albin Sandelin, Phone: +45 22 45 66 68, Email: albin@binf.ku.dk.
Ole Haagen Nielsen, Phone: +45 38 68 36 21, Email: ole.haagen.nielsen@regionh.dk.
References
- 1.Baumgart DC, Sandborn WJ. Crohn’s disease. Lancet. 2012;380:1590–1605. doi: 10.1016/S0140-6736(12)60026-9. [DOI] [PubMed] [Google Scholar]
- 2.Danese S, Fiocchi C. Ulcerative colitis. N. Engl. J. Med. 2011;365:1713–1725. doi: 10.1056/NEJMra1102942. [DOI] [PubMed] [Google Scholar]
- 3.Abraham C, Cho JH. Inflammatory bowel disease. N. Engl. J. Med. 2009;361:2066–2078. doi: 10.1056/NEJMra0804647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kalla R, Ventham NT, Satsangi J, Arnott IDR. Crohn’s disease. Br. Med. J. 2014;349:g6670–g6670. doi: 10.1136/bmj.g6670. [DOI] [PubMed] [Google Scholar]
- 5.Jostins L, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McGovern DPB, Kugathasan S, Cho JH. Genetics of inflammatory bowel diseases. Gastroenterology. 2015;149:e2. doi: 10.1053/j.gastro.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braat H, Peppelenbosch MP, Hommes DW. Immunology of Crohn’s disease. Ann. N. Y. Acad. Sci. 2006;1072:135–154. doi: 10.1196/annals.1326.039. [DOI] [PubMed] [Google Scholar]
- 8.Hugot JP, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 9.Ogura Y, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature. 2001;411:603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 10.Mo J, et al. Pathogen sensing by nucleotide-binding oligomerization domain-containing protein 2 (NOD2) is mediated by direct binding to muramyl dipeptide and ATP. J. Biol. Chem. 2012;287:23057–23067. doi: 10.1074/jbc.M112.344283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salem M, Seidelin JB, Rogler G, Nielsen OH. Muramyl dipeptide responsive pathways in Crohn’s disease: from NOD2 and beyond. Cell. Mol. Life Sci. 2013;70:3391–3404. doi: 10.1007/s00018-012-1246-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Watanabe T, et al. Muramyl dipeptide activation of nucleotide-binding oligomerization domain 2 protects mice from experimental colitis. J. Clin. Invest. 2008;118:545–559. doi: 10.1172/JCI33145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seidelin JB, Broom OJ, Olsen J, Nielsen OH. Evidence for impaired CARD15 signalling in Crohn’s disease without disease linked variants. PLoS ONE. 2009;4:e7794. doi: 10.1371/journal.pone.0007794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Y, et al. miR-122 targets NOD2 to decrease intestinal epithelial cell injury in Crohn’s disease. Biochem. Biophys. Res. Commun. 2013;438:133–139. doi: 10.1016/j.bbrc.2013.07.040. [DOI] [PubMed] [Google Scholar]
- 15.Brain O, et al. The intracellular sensor NOD2 induces microRNA-29 expression in human dendritic cells to limit IL-23 release. Immunity. 2013;39:521–536. doi: 10.1016/j.immuni.2013.08.035. [DOI] [PubMed] [Google Scholar]
- 16.Chuang AY, Chuang JC, Zhai Z, Wu F, Kwon JH. NOD2 expression is regulated by microRNAs in colonic epithelial HCT116 cells. Inflamm. Bowel. Dis. 2014;20:126–135. doi: 10.1097/01.MIB.0000436954.70596.9b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kalla R, et al. microRNAs: new players in IBD. Gut. 2015;64:504–517. doi: 10.1136/gutjnl-2014-307891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sayed D, Abdellatif M. microRNAs in development and disease. Physiol. Rev. 2011;91:827–887. doi: 10.1152/physrev.00006.2010. [DOI] [PubMed] [Google Scholar]
- 19.Bai J, et al. Integrating analysis reveals microRNA-mediated pathway crosstalk among Crohn’s disease, ulcerative colitis and colorectal cancer. Mol. Biosyst. 2014;10:2317–2328. doi: 10.1039/C4MB00169A. [DOI] [PubMed] [Google Scholar]
- 20.O’Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. microRNA-155 is induced during the macrophage inflammatory response. Proc. Natl. Acad. Sci. U.S.A. 2007;104:1604–1609. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jablonski KA, Gaudet AD, Amici SA, Popovich PG, Guerau-de-Arellano M. Control of the inflammatory macrophage transcriptional signature by miR-155. PLoS ONE. 2016;11:e0159724. doi: 10.1371/journal.pone.0159724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seddiki N, Brezar V, Ruffin N, Lévy Y, Swaminathan S. Role of miR-155 in the regulation of lymphocyte immune function and disease. Immunology. 2014;142:32–38. doi: 10.1111/imm.12227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van der Goten J, et al. Integrated miRNA and mRNA expression profiling in inflamed colon of patients with ulcerative colitis. PLoS ONE. 2013;9:e116117–e116117. doi: 10.1371/journal.pone.0116117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fasseu M, et al. Identification of restricted subsets of mature microRNA abnormally expressed in inactive colonic mucosa of patients with inflammatory bowel disease. PLoS ONE. 2010;5:e13160. doi: 10.1371/journal.pone.0013160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singh UP, et al. miR-155 deficiency protects mice from experimental colitis by reducing T helper type 1/type 17 responses. Immunology. 2014;143:478–489. doi: 10.1111/imm.12328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang P, et al. Inducible microRNA-155 feedback promotes type I IFN signaling in antiviral innate immunity by targeting suppressor of cytokine signaling 1. J. Immunol. 2010;185:6226–6233. doi: 10.4049/jimmunol.1000491. [DOI] [PubMed] [Google Scholar]
- 27.Koch M, Mollenkopf H-J, Klemm U, Meyer TF. Induction of microRNA-155 is TLR- and type IV secretion system-dependent in macrophages and inhibits DNA-damage induced apoptosis. Proc. Natl. Acad. Sci. U. S. A. 2012;109:E1153–E1162. doi: 10.1073/pnas.1116125109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tili E, et al. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-alpha stimulation and their possible roles in regulating the response to endotoxin shock. J. Immunol. 2007;179:5082–5089. doi: 10.4049/jimmunol.179.8.5082. [DOI] [PubMed] [Google Scholar]
- 29.Economou M, Trikalinos TA, Loizou KT, Tsianos EV, Ioannidis JPA. Differential effects of NOD2 variants on Crohn’s disease risk and phenotype in diverse populations: a metaanalysis. Am. J. Gastroenterol. 2004;99:2393–2404. doi: 10.1111/j.1572-0241.2004.40304.x. [DOI] [PubMed] [Google Scholar]
- 30.Strober W, Asano N, Fuss I, Kitani A, Watanabe T. Cellular and molecular mechanisms underlying NOD2 risk-associated polymorphisms in Crohn’s disease. Immunol. Rev. 2014;260:249–260. doi: 10.1111/imr.12193. [DOI] [PubMed] [Google Scholar]
- 31.Pathak S, et al. miR-155 modulates the inflammatory phenotype of intestinal myofibroblasts by targeting SOCS1 in ulcerative colitis. Exp. Mol. Med. 2015;47:e164. doi: 10.1038/emm.2015.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu G, et al. Transcriptome and targetome analysis in MIR155 expressing cells using RNA-seq. RNA. 2010;16:1610–1622. doi: 10.1261/rna.2194910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O’Connell RM, Rao DS, Baltimore D. microRNA regulation of inflammatory responses. Annu. Rev. Immunol. 2012;30:295–312. doi: 10.1146/annurev-immunol-020711-075013. [DOI] [PubMed] [Google Scholar]
- 34.Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44:D457–D462. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hsu S-D, et al. miRTarBase update 2014: an information resource for experimentally validated miRNA-target interactions. Nucleic Acids Res. 2013;42:D78–D85. doi: 10.1093/nar/gkt1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Santaolalla R, Fukata M, Abreu MT. Innate immunity in the small intestine. Curr. Opin. Gastroenterol. 2011;27:125–131. doi: 10.1097/MOG.0b013e3283438dea. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gradel KO, et al. Increased short- and long-term risk of inflammatory bowel disease after salmonella or campylobacter gastroenteritis. Gastroenterology. 2009;137:495–501. doi: 10.1053/j.gastro.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 38.García Rodríguez LA, Ruigómez A, Panés J. Acute gastroenteritis is followed by an increased risk of inflammatory bowel disease. Gastroenterology. 2006;130:1588–1594. doi: 10.1053/j.gastro.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 39.Schulte LN, Westermann AJ, Vogel J. Differential activation and functional specialization of miR-146 and miR-155 in innate immune sensing. Nucleic Acids Res. 2013;41:542–553. doi: 10.1093/nar/gks1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li J, et al. Regulation of IL-8 and IL-1beta expression in Crohn’s disease associated NOD2/CARD15 mutations. Hum. Mol. Genet. 2004;13:1715–1725. doi: 10.1093/hmg/ddh182. [DOI] [PubMed] [Google Scholar]
- 41.Moreira LO, et al. The TLR2-MyD88-NOD2-RIPK2 signalling axis regulates a balanced pro-inflammatory and IL-10-mediated anti-inflammatory cytokine response to gram-positive cell walls. Cell. Microbiol. 2008;10:2067–2077. doi: 10.1111/j.1462-5822.2008.01189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Landi D, et al. Polymorphisms within micro-RNA-binding sites and risk of sporadic colorectal cancer. Carcinogenesis. 2008;29:579–584. doi: 10.1093/carcin/bgm304. [DOI] [PubMed] [Google Scholar]
- 43.Brest P, et al. A synonymous variant in IRGM alters a binding site for miR-196 and causes deregulation of IRGM-dependent xenophagy in Crohn’s disease. Nat. Genet. 2011;43:242–245. doi: 10.1038/ng.762. [DOI] [PubMed] [Google Scholar]
- 44.Van Assche G, et al. The second European evidence-based consensus on the diagnosis and management of Crohn’s disease: definitions and diagnosis. J. Crohns Colitis. 2010;4:7–27. doi: 10.1016/j.crohns.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 45.Yazdanyar S, Nordestgaard BG. NOD2/CARD15 genotype and common gastrointestinal diseases in 43,600 individuals. J. Intern. Med. 2010;267:228–236. doi: 10.1111/j.1365-2796.2009.02137.x. [DOI] [PubMed] [Google Scholar]
- 46.Harvey RF, Bradshaw JM. A simple index of Crohn’s-disease activity. Lancet. 1980;1:514. doi: 10.1016/S0140-6736(80)92767-1. [DOI] [PubMed] [Google Scholar]
- 47.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trapnell, C. et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc.7, 562–578, doi:10.1038/nprot.2012.016 (2012). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.