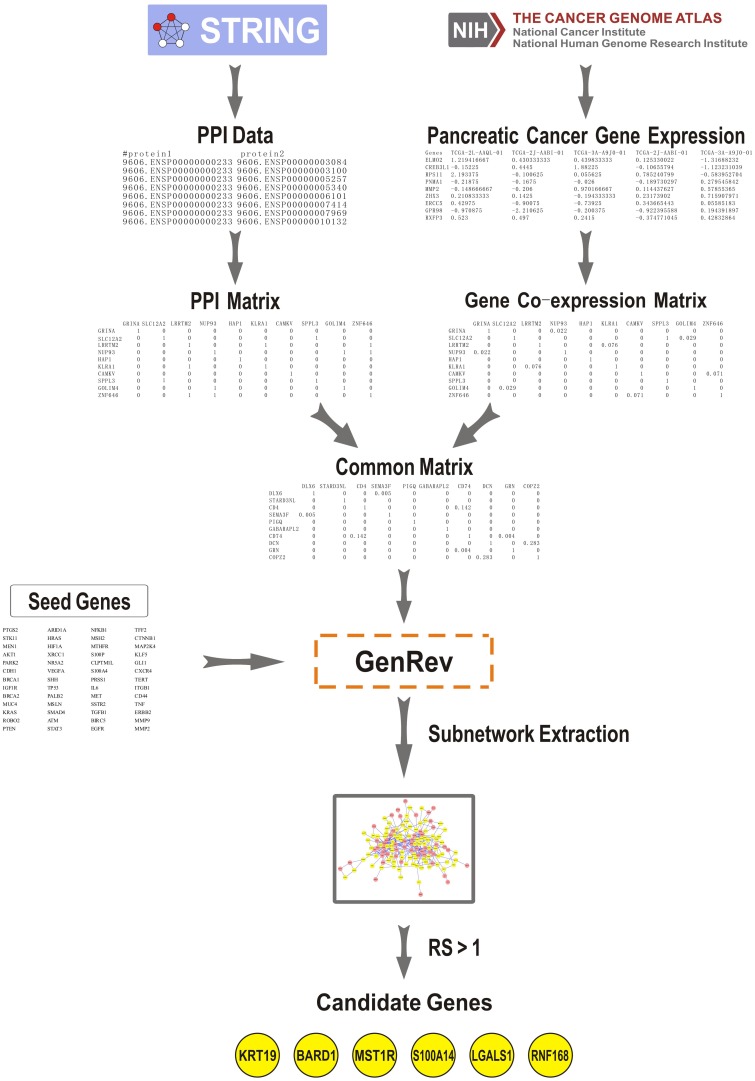
Figure 1. Summary workflow for prediction of pancreatic cancer candidate genes.
The approach was based on three steps: (1) We constructed a PPI network and a co-expression network using data derived from STRING (Search Tool for the Retrieval of Interacting Genes/Proteins) and The Cancer Genome Atlas (TCGA), respectively. A common network was obtained through comparing the two networks. (2) Seed genes and the common network were imported into GenRev which was used to extract a subnetwork with the limited k-walks algorithm. (3) 6 key candidate genes were identified among the subnetwork using an evaluation indicator denoted as ranking score (RS).