Significance
A well-investigated mechanism regulating synapse protein turnover and remodeling is the classical ubiquitin–proteasome system by which polyubiquitin (polyUb) chains linked through ubiquitin lysine 48 (K48) tag substrates for proteasomal degradation. Little is known about the role of nondegradable polyUb linkages, e.g., K63-linked chains, in the synapse, although these chains represent the second most abundant polyUb linkage in mammalian brain. We identified a bona fide K63-polyUb substrate in PSD-95, a major postsynaptic scaffold. K63-polyUb conjugation markedly augments PSD-95’s scaffolding capability and promotes its synaptic targeting and anchorage. We identify a K63-linkage–specific E3 ligase–deubiquitinase complex that controls activity-dependent assembly and disassembly of K63-polyUb chains on PSD-95. We show that this unconventional polyUb linkage promotes synapse maturation, efficacy, and plasticity.
Keywords: lysine 63-linked ubiquitination, PSD-95, TRAF6, CYLD, long-term depression
Abstract
Ubiquitination-directed proteasomal degradation of synaptic proteins, presumably mediated by lysine 48 (K48) of ubiquitin, is a key mechanism in synapse and neural circuit remodeling. However, more than half of polyubiquitin (polyUb) species in the mammalian brain are estimated to be non-K48; among them, the most abundant is Lys 63 (K63)-linked polyUb chains that do not tag substrates for degradation but rather modify their properties and activity. Virtually nothing is known about the role of these nonproteolytic polyUb chains at the synapse. Here we report that K63-polyUb chains play a significant role in postsynaptic protein scaffolding and synaptic strength and plasticity. We found that the postsynaptic scaffold PSD-95 (postsynaptic density protein 95) undergoes K63 polyubiquitination, which markedly modifies PSD-95’s scaffolding potentials, enables its synaptic targeting, and promotes synapse maturation and efficacy. TNF receptor-associated factor 6 (TRAF6) is identified as a direct E3 ligase for PSD-95, which, together with the E2 complex Ubc13/Uev1a, assembles K63-chains on PSD-95. In contrast, CYLD (cylindromatosis tumor-suppressor protein), a K63-specific deubiquitinase enriched in postsynaptic densities, cleaves K63-chains from PSD-95. We found that neuronal activity exerts potent control of global and synaptic K63-polyUb levels and, through NMDA receptors, drives rapid, CYLD-mediated PSD-95 deubiquitination, mobilizing and depleting PSD-95 from synapses. Silencing CYLD in hippocampal neurons abolishes NMDA-induced chemical long-term depression. Our results unveil a previously unsuspected role for nonproteolytic polyUb chains in the synapse and illustrate a mechanism by which a PSD-associated K63-linkage–specific ubiquitin machinery acts on a major postsynaptic scaffold to regulate synapse organization, function, and plasticity.
The diverse functions of ubiquitination are largely determined by structurally and functionally distinct polyubiquitin (polyUb) chain types (1). Conventional chains linked through lysine 48 (K48) of ubiquitin are the classical signal that targets substrates to the 26S proteasome for degradation, whereas K63-linked chains have emerged as a predominant unconventional linkage regulating protein interactions, trafficking, and kinase activation (2, 3). Ubiquitination is reversed by deubiquitinases (DUBs), a large group of proteases that cleave the isopeptide linkage between ubiquitin moieties (4). K63 polyubiquitination plays essential roles in immune and inflammatory pathways leading to NF-κB activation, mediated by the TNF receptor-associated factor (TRAF) family E3 ubiquitin ligases (2, 5). The DUB cylindromatosis tumor-suppressor protein (CYLD), which when mutated causes familial cylindromatosis (6), serves as a negative regulator of NF-κB signaling by deubiquitination (4).
The strength of excitatory synapses depends on the abundance and proper assembly of postsynaptic receptors, channels, and signaling complexes, anchored by molecular scaffolds in the postsynaptic density (PSD) (7). Regulated degradation of postsynaptic proteins by the ubiquitin–proteasome system (UPS) has emerged as a major mechanism for structural and functional modifications of synapses (8–11). UPS-mediated, activity-dependent remodeling of PSD proteins (12), including several synaptic scaffold “master organizers” (12–15), is believed to play a crucial role in organization of the PSD and maintenance of the synapse (15–17). Despite their potential to significantly impact protein network assembly, almost nothing is known about proteasome-independent K63-polyUb chains at the synapse. Intriguingly, ubiquitome analysis estimates that K63-polyUb chains are the second most abundant ubiquitin linkage in the rat brain, behind conventional K48-linkage (18), and the K63-specific CYLD is highly enriched in the PSD (19).
PSD-95 is a major postsynaptic scaffold in the membrane-associated guanylate kinase (MAGUK) family (7). Because of its modular structure and abundance in the PSD, PSD-95 binds numerous PSD proteins and serves as a core stabilizer of the molecular architecture of the PSD (20). PSD-95 associates with glutamate receptors and signaling complexes in the PSD, thus controlling synaptic strength (21) and regulating activity-dependent synaptic plasticity (22–26). PSD-95 also promotes dendritic spine morphogenesis and synapse maturation (27), likely by associating with proteins that regulate the spine cytoskeleton, such as SPAR (28) and GKAP/SAPAP (29, 30). The abundance of PSD-95 is regulated by activity through the UPS (13), and its targeting to synapse membranes is regulated by palmitoylation (31, 32), phosphorylation (33, 34), and neddylation (35). Consistent with a central role for PSD-95 in synaptic function and plasticity, mice lacking PSD-95 display impaired learning and reward-related behaviors (36, 37).
In this study, we show that K63-polyUb is a prominent ubiquitin linkage type in the PSD, identify a bona fide K63-polyUb substrate and its E2/E3/DUB complex in the synapse, and illustrate a mechanism by which this concept contributes to synapse assembly, function, and plasticity.
Results
K63-PolyUb Chains Are Present at the Synapse.
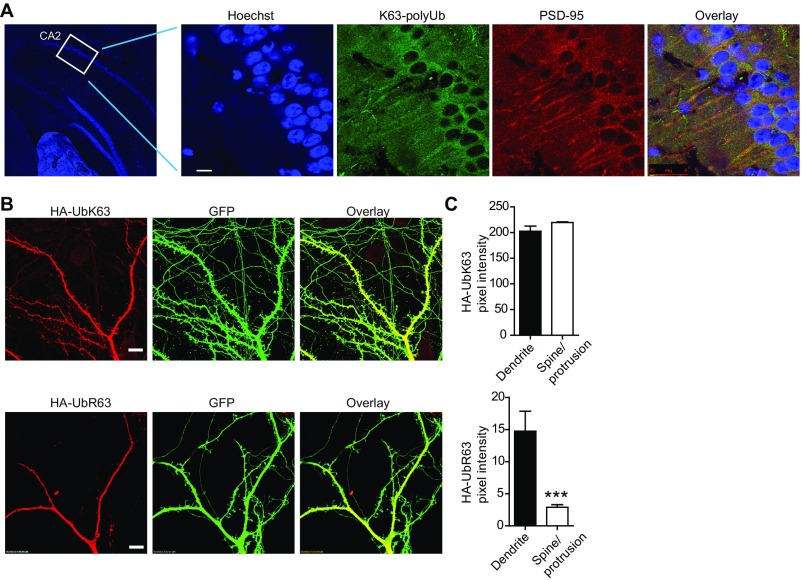
We first investigated whether K63-linked ubiquitination occurs at the synapse. Western blotting of forebrain lysates from mice at different postnatal ages using anti-UbK63, a linkage-specific antibody that selectively recognizes K63-linked polyUb (≥2) chains (38), detected high molecular weight K63-polyUb smear bands, which appeared at postnatal day 5 (P5) and became increasingly abundant thereafter (Fig. 1A). Pan-ubiquitin and K63-polyUb profiles in ubiquitin immunoprecipitates prepared from mouse forebrains and cultured rat hippocampal neurons revealed strikingly similar patterns (Fig. 1B). Immunofluorescence microscopy revealed abundant K63-polyUb staining in the mouse hippocampus (Fig. S1A), and K63-polyUb puncta partially colocalized with dendritic PSD-95 clusters in cultured hippocampal neurons (Fig. 1 C and D). To further confirm the presence of K63-polyUb at synapses, we cotransfected GFP (to label spines) and ubiquitin mutants HA-UbK63 and HA-UbR63 (Fig. 1G) into cultured neurons. HA-UbK63, but not HA-UbR63 (abolishing K63-polyUb synthesis), was abundant in spines (Fig. S1 B and C). These data indicate that K63-polyUb conjugates are present at synapses.
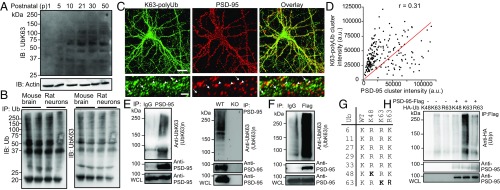
Fig. 1.
K63-polyUb is present at excitatory synapses and targets PSD-95 as a substrate. (A) Immunoblot (IB) analysis of K63-polyUb species in forebrain lysates prepared from mice at different postnatal stages. (B) Total ubiquitin and K63-polyUb (after stripping and reprobing) species in mouse forebrains (P30) and cultured rat hippocampal neurons at 21 d in vitro (DIV21) after immunoprecipitation (IP) with an anti-ubiquitin antibody. (C) Immunostaining of K63-polyUb and PSD-95 in cultured rat hippocampal neurons. [Scale bars, 20 μm (Upper) and 5 μm (Lower).] Arrowheads denote colocalized PSD-95 and K63-polyUb clusters, and arrows indicate K63-polyUb clusters lacking PSD-95 costaining. (D) Correlation plot of PSD-95 and K63-polyUb cluster intensities in C. Pearson’s r is shown. (E) K63-polyubiquitination of endogenous PSD-95 in mouse brain. WCL, whole-cell lysate. (F) K63-polyubiquitination of PSD-95–Flag in transfected HEK293FT cells. (G) Schematic showing ubiquitin lysine mutants used in this study. (H) Effects of HA-ubiquitin and mutants in promoting K63 ubiquitination of PSD-95–Flag in HEK293FT cells. Experiments were performed in the absence of proteasome inhibitors.
Fig. S1.
Additional characterizations of K63-polyUb presence in neurons and synapses. (A) Immunostaining of K63-polyUb and PSD-95 on mouse hippocampal sections. Hoechst 3342 was used to label neuronal nucleus. (Scale bar, 10 μm.) (B) Spine localization for HA-UbK63, but significantly less for HA-UbR63, in cotransfected cultured rat hippocampal neurons. (Scale bars, 10 μm.) (C) Quantification of HA fluorescence pixel intensities for dendritic spines or protrusions and shafts in B. n = 16 shafts and 534 spines for HA-UbK63; n = 11 shafts and 285 spines for HA-UbR63. ***P < 0.001; unpaired t tests.
PSD-95 Is a Substrate of K63 Ubiquitination.
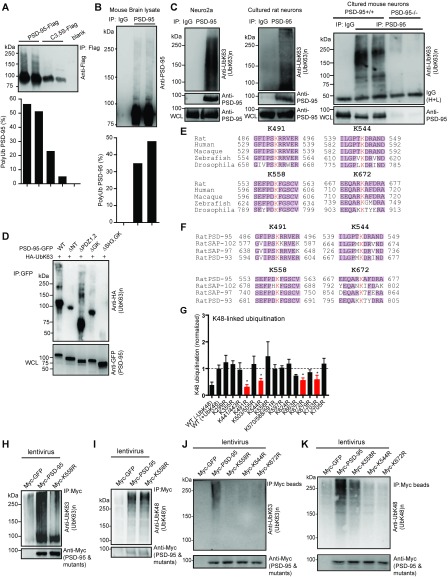
We next set to identify substrates that undergo K63-linked ubiquitination in the synapse. We chose PSD-95 because previous studies (13) had shown that PSD-95 is conjugated by polyUb, although the linkage type was unknown. Both ectopically expressed and endogenous PSD-95 appeared polyubiquitinated under denaturing conditions: We estimated that up to 45% and 36% of total PSD-95 were likely ubiquitinated in HEK293FT cells (Fig. S2A) and neurons (Fig. S2B), respectively, in the absence of proteasome inhibitors, suggesting strong PSD-95 ubiquitination via nonproteolytic linkages, e.g., K63. Indeed, we detected robust K63 polyubiquitination of endogenous PSD-95 in Neuro2a cells, rat hippocampal cultures, and brains of WT but not PSD-95–KO mice (Fig. 1E and Fig. S2C). Consistently, PSD-95–Flag was K63-polyubiquitinated when expressed in HEK293FT cells (Fig. 1F). In addition, polyubiquitination of PSD-95 was enhanced by coexpressed HA-UbK63 but not by HA-UbK48 or HA-UbR63 mutants (Fig. 1 G and H). Thus, PSD-95 is a bona fide substrate of K63 polyubiquitination.
Fig. S2.
Additional characterizations of PSD-95 polyubiquitination. (A and B) Simultaneous detection of both nonubiquitinated (∼95 kDa band) and ubiquitinated PSD-95 (higher molecular weight smears) in transfected HEK293FT cell (A) and mouse brain (B) lysates with anti-Flag (A) or anti–PSD-95 (B) following immunoprecipitation with anti-Flag (A) or anti–PSD-95 (B). The percentage of ubiquitinated PSD-95 compared with the total PSD-95 in the immunoprecipitate, defined as [high molecular weight smear intensity/(high molecular weight smear intensity + 95 kDa band intensity)] × 100, is shown at the bottom. Experiments were performed on denatured lysates in the absence of proteasome inhibitors. (C) K63-linked polyubiquitination of endogenous PSD-95 in Neuro2a cells, cultured rat hippocampal neurons, and cultured WT or PSD-95–KO mouse neurons. (D) K63 polyubiquitination of PSD-95 and mutants, analyzed by immunoprecipitation and immunoblotting using lysed HEK293FT cells transfected with the indicated plasmids. (E and F) Conservation of identified lysine sites in PSD-95 SH3 and GK domains across species (E) and in the MAGUK family (F). K491, K544, K558, and K672 (red) represent the lysine residue locations in rat PSD-95. (G) Quantification of K48 polyubiquitination in HEK293FT cells cotransfected with Flag-tagged WT PSD-95 or mutants and HA-UbK48. Cell lysates were immunoprecipitated with anti-Flag and immunoblotted with anti-HA for K48-linked polyubiquitination. Data are based on gel density analysis of high molecular weight ubiquitin smears for each K-to-R mutant and normalized to WT PSD-95 (+UbK48). Substantially impaired mutants are shown in red. n = 3–11 experiments. *P < 0.05; unpaired t tests vs. WT(+UbK48) control. (H) Severely impaired K63 polyubiquitination of K558R in rat hippocampal neurons. Rat hippocampal cultures were infected at DIV4 with lentiviruses expressing Myc-GFP, Myc–PSD-95, or Myc-K558R and were analyzed by immunoprecipitation with anti–PSD-95 and immunoblotting with anti-UbK63 antibody to asses K63 polyubiquitination. (I) The K558R mutation does not affect K48-linked polyubiquitination in neurons. The experiment was similar to that in H except that immunoblotting was done with anti-UbK48 antibody to assess K48 polyubiquitination. MG132 (10 μM) was used in this experiment. (J and K) Myc-544R, Myc-558R, and Myc-672R all nearly abolished K63 polyubiquitination of PSD-95, whereas only Myc-544R and Myc-672R severely impaired K48 polyubiquitination of PSD-95 in cultured rat hippocampal neurons infected with the indicated lentiviruses. Immunoprecipitation was done with Myc-conjugated agarose beads.
Mapping Ubiquitination Sites on PSD-95.
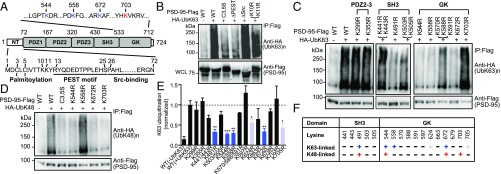
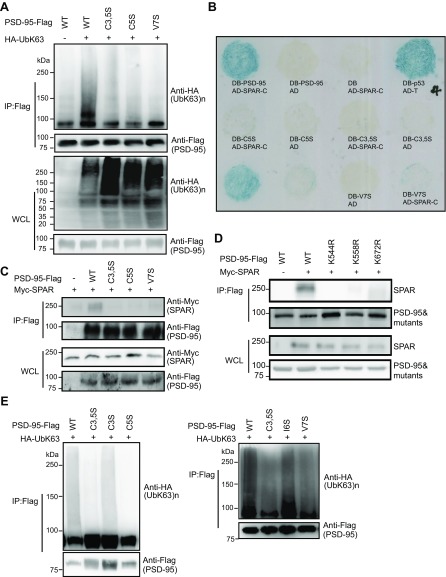
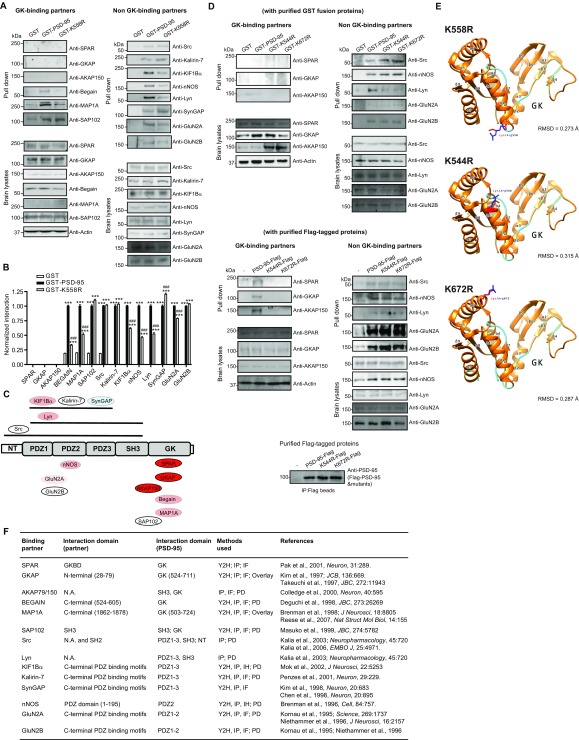
We next systematically mapped the K63 polyubiquitination sites on PSD-95 using site-directed mutagenesis. PSD-95 contains a unique N terminus (NT) followed by conserved protein-interaction domains (Fig. 2A). Deleting the NT abolished K63 ubiquitination (Fig. S2D). However, replacing both lysine residues (K10 and K11) within the NT with arginine did not substantially affect PSD-95 ubiquitination (Fig. 2B), indicating that they are not actual K63-polyUb conjugation sites. More likely, the NT is required for K63 ubiquitination of PSD-95 by mediating ubiquitin enzyme binding (see below and Fig. S3E). Supporting this, mutating cysteines 3 and 5 to serine (C3,5S) or deleting residues from proline 26 through asparagine 72 (ΔSrc) impacted PSD-95 K63 ubiquitination (Fig. 2B). C3,5S and ΔSrc mutants have been implicated in maintaining the structural integrity of PSD-95 to properly interact with the signaling enzymes palmitoyl transferases (39) and the tyrosine kinase Src (40). Interestingly, deleting the PEST sequence (R13–S25), a motif associated with UPS-mediated protein turnover (13), also substantially reduced K63 polyubiquitination of PSD-95 (Fig. 2B). Additional screening suggested that the primary K63 ubiquitination sites are localized in Src-homology 3–guanylate kinase (SH3-GK) domains (Fig. 2C and Fig. S2D). Of all 17 lysines within SH3-GK, mutation of four residues (K491 in SH3 and K544, K558, and K672 in GK) most substantially diminished K63 ubiquitination of PSD-95 (Fig. 2 C and E). These residues are conserved from Drosophila to humans and among MAGUK members (Fig. S2 E and F), suggesting evolutionary significance. PSD-95 was also conjugated by K48–polyUb, and all but one (K558R) of the mutants also drastically reduced K48-linked ubiquitination of PSD-95 (Fig. 2D and Fig. S2G). Another mutant, K703R, selectively impaired K48 while sparing K63 ubiquitination (Fig. 2 C and D). Importantly, the three GK lysine residues mediate PSD-95 ubiquitination similarly in neurons: K558R selectively impaired K63, whereas K544R and K672R impaired both K63 and K48 ubiquitination of PSD-95 (Fig. S2 H–K). Our results demonstrate that PSD-95 is dually modified by K48 and K63 linkages and identify linkage-specific sites that mediate PSD-95 ubiquitination (Fig. 2F).
Fig. 2.
Mapping ubiquitination sites on PSD-95. (A) Modular structure of PSD-95. GK, guanylate kinase-like; PDZ, PSD-95, Dlg, ZO-1; SH3, Src-homology 3. Within the NT, cysteine palmitoylation residues (C3, C5), the PEST motif, and the Src-binding region are shown. Within GK, four key lysine residues identified and used in this study are indicated. (B–D) K63- or K48-linked ubiquitination of PSD-95–Flag and mutants in HEK293FT cells. WCL, whole-cell lysate. (E) Quantification of K63-polyUb levels normalized to WT PSD-95 (+UbK63). n = 3–13 experiments. ***P < 0.001, **P < 0.01, *P < 0.05; unpaired t tests vs. WT(+UbK63). (F) Mutagenesis summary; “+” in dark blue or red indicates an essential contribution; “+” in light blue indicates a modest contribution; “−” indicates little or no contribution.
Fig. S3.
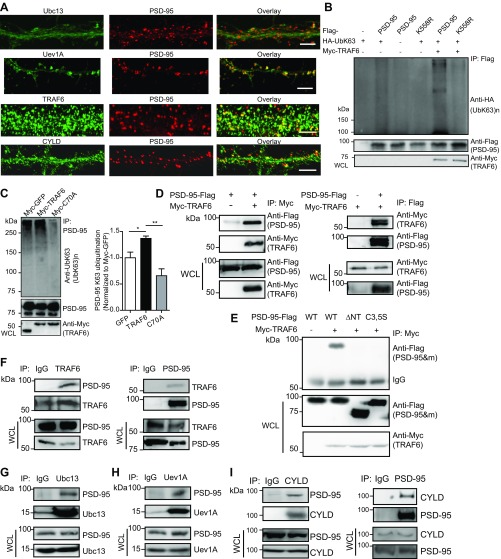
Additional experiments characterizing the E2/E3/DUB complex for K63 polyubiquitination of PSD-95. (A) Dendritic distributions of Ubc13, Uev1A, TRAF6, and CYLD in cultured hippocampal neurons and their localizations relative to PSD-95. (Scale bars, 5 μm.) (B) TRAF6 promotes K63-linked polyubiquitination of WT PSD-95 but not K558R. Lysates from HEK293FT cells transfected with the indicated plasmids were analyzed by immunoprecipitation and immunoblotting. (C) Effects of TRAF6 and C70A overexpression on PSD-95 K63 ubiquitination in cultured rat hippocampal neurons. Neurons were infected with lentiviruses expressing the indicated proteins, followed by immunoprecipitation and immunoblotting assays. Quantification is shown to the right. n = 3 independent experiments. *P < 0.05, **P < 0.01; unpaired t tests. (D) Interaction between TRAF6 and PSD-95 in HEK293FT cells. Cells were transfected with plasmids encoding PSD-95–Flag or Myc-TRAF6 or both, followed by immunoprecipitation and immunoblotting assays. (E) Lack of interaction between TRAF6 and PSD-95 mutants PSD-95ΔNT and C3,C5S in HEK293FT cells. Cells transfected with the indicated plasmids were analyzed by immunoprecipitation and immunoblotting. (F–I) Association of TRAF6 (F), Ubc13 (G), Uev1A (H), and CYLD (I) with PSD-95 in the mouse brain, analyzed by coimmunoprecipitation. Forebrain tissues were homogenated and used in immunoprecipitation and immunoblotting assays. IgG was used as a control in immunoprecipitation experiments.
A K63-Specific E2/E3/DUB Complex in the PSD.
We next searched for the molecular machinery of K63 ubiquitination in the synapse. The substrate specificity and linkage topology for ubiquitination are determined to a certain extent by limited ubiquitin-conjugating enzymes (E2s) and to a greater extent by ubiquitin ligases (E3s), a large and diverse family of proteins. To date, the only known K63-selective E2s are the Ubc13/Uev1A complex. We found that Ubc13 is present and Uev1A is enriched in the PSD (Fig. 3A), and both colocalized with PSD-95 in dendritic clusters (Fig. S3A).
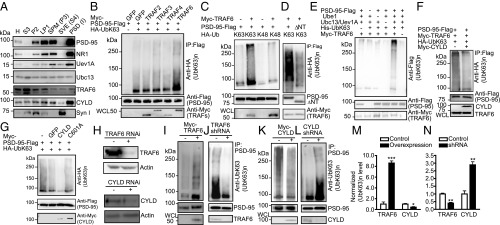
Fig. 3.
Identification of an E2/E3/DUB complex for PSD-95. (A) Immunoblots showing the presence of indicated proteins in subcellular fractions of the rat forebrain. H, homogenates; LP1, synaptosome fraction; P2, crude synaptosomal membranes; PSD (I), PSD fraction after one Triton X-100 extraction; S3, cytosol; SPM, synaptic plasma membrane (P3); SVE, synaptic vesicle enriched (S4). (B) Effects of TRAFs in promoting K63 ubiquitination of PSD-95 in HEK293FT cells. (C) TRAF6 promotes K63 but not K48 ubiquitination of PSD-95. (D) Impaired K63 ubiquitination of PSD-95ΔNT. (E) In vitro ubiquitination assay. (F) Immunoblots showing inhibition of TRAF6-promoted PSD-95 K63 ubiquitination by CYLD in HEK293FT cells. (G) In vitro deubiquitination assay. (H) Efficiency of TRAF6 and CYLD knockdown in cultured hippocampal neurons. (I–L) Effects of TRAF6 and CYLD overexpression (I and K) or knockdown (J and L) on PSD-95 K63 ubiquitination in neurons. (M and N) Quantification of I–L. n = 3 or 4 experiments; ***P < 0.001, **P < 0.01, *P < 0.05; unpaired t tests vs. controls.
To seek the E3 ligase responsible for K63 ubiquitination of PSD-95, we surveyed published PSD MS databases for likely candidates. The only relevant lead identified was TRAF3, a member of the TRAF cytoplasmic adaptor protein family (41). Structurally, TRAFs, including TRAF3 and six other members, are K63-specific E3 ligases involved in transducing interleukin and TNF signaling in the NF-κB pathway (2, 5). Surprisingly, ectopically expressed TRAF3 did not promote K63 ubiquitination of PSD-95; rather, screening a panel of additional TRAFs revealed that only TRAF6 promoted K63 ubiquitination of PSD-95 (Fig. 3B). TRAF6 is present in rat brain PSD (Fig. 3A), and in HEK293FT cells the overexpression of TRAF6 but not other TRAFs potently promoted K63, but not K48, ubiquitination of PSD-95 (Fig. 3C). TRAF6-facilitated K63 ubiquitination was diminished in K558R (Fig. S3B) and PSD-95ΔNT (Fig. 3D) mutants. In vitro ubiquitination assays using purified proteins showed that TRAF6, in conjunction with Ubc13/Uev1A, effectively delivered the ubiquitin moieties to PSD-95, which did not occur when TRAF6, UbK63, Ubc13, Uev1A, or PSD-95 was omitted from the reaction mix (Fig. 3E). TRAF6 interacted with PSD-95 but not with PSD-95ΔNT or -C3,5S in HEK293FT cells (Fig. S3 D and E), which explains their nearly abolished K63 ubiquitination (Fig. 2B and Fig. S2 A and D). Together, these results identify TRAF6 as a direct K63-specific E3 ligase for PSD-95.
A DUB that selectively cleaves K63 chains is CYLD (4). CYLD also is detected in PSD MS studies (42, 43) and, interestingly, is enriched in the PSD (19). We detected a markedly higher amount of CYLD in PSD compared with other fractions (Fig. 3A). In HEK293FT cells, CYLD inhibited TRAF6-induced K63 ubiquitination of PSD-95 (Fig. 3F). In vitro deubiquitination assays showed that WT CYLD, but not the C601A mutant lacking the DUB enzymatic activity, substantially diminished K63 ubiquitination of PSD-95 (Fig. 3G). These results suggest that CYLD is a PSD-enriched DUB that removes K63 chains on PSD-95.
Additional experiments confirm that TRAF6 and CYLD are E3 ligases and DUBs for PSD-95, respectively, in neurons. Both TRAF6 and CYLD colocalized with PSD-95 on dendrites of cultured hippocampal neurons (Fig. S3A). Interactions between endogenous PSD-95 and TRAF6, Ubc13, Uev1A, or CYLD in mouse forebrain were also detected (Fig. S3 F–I). Importantly, overexpression of TRAF6, but not the E3 ligase-deficient mutant C70A, increased, but knockdown of TRAF6 decreased, K63 ubiquitination of PSD-95 in hippocampal neurons (Fig. 3 I, J, M, and N and Fig. S3C). In contrast, CYLD overexpression decreased, but knockdown increased, K63 ubiquitination of PSD-95 (Fig. 3 K–N). Thus, TRAF6-Ubc13/Uev1A-CYLD is a neuronal E2–E3–DUB complex regulating K63 ubiquitination of PSD-95.
K63-PolyUb Chains Modify PSD-95 Scaffolding.
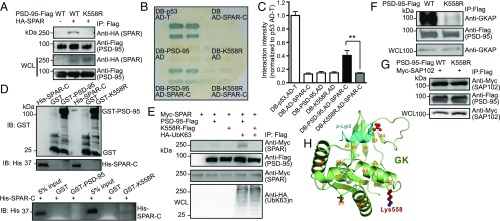
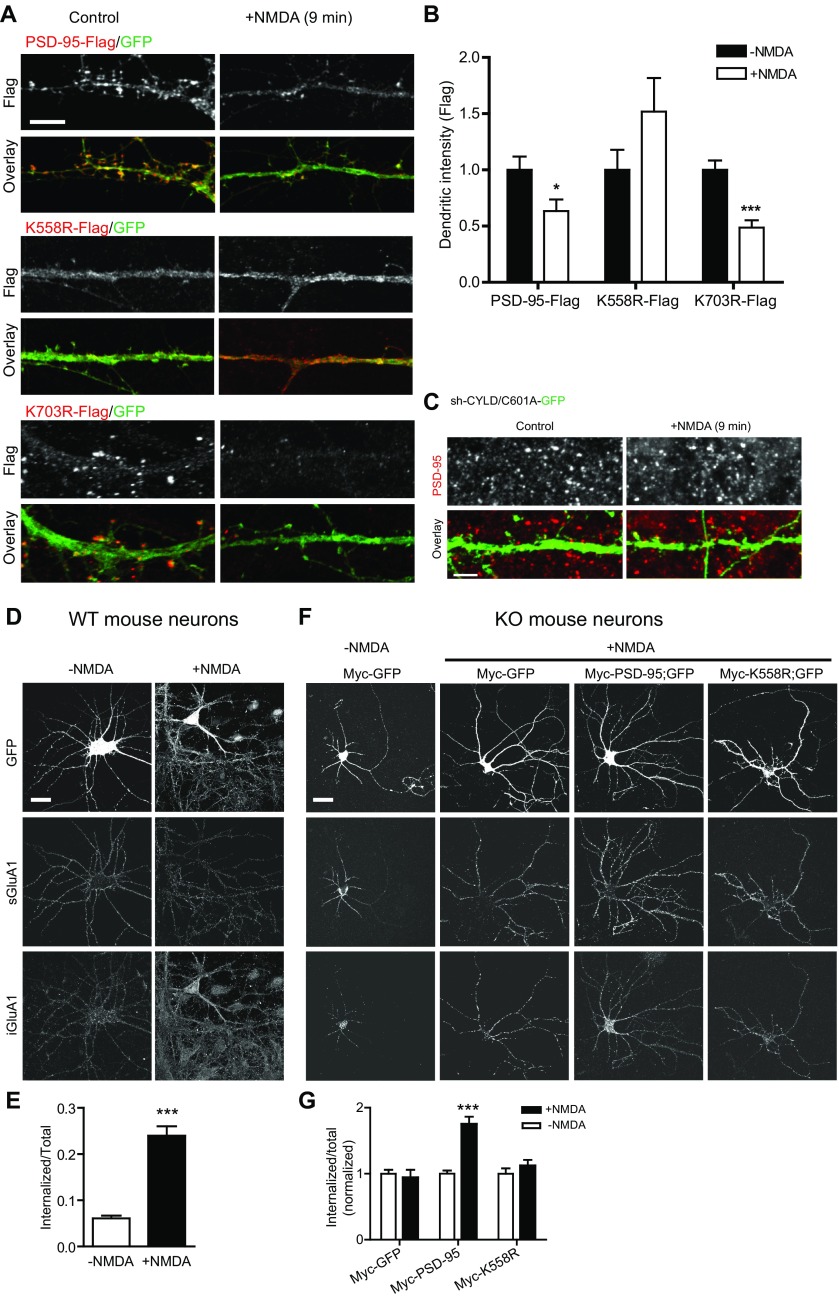
A hallmark role of K63-polyUb chains is modifying substrate properties. We thus examined the ability of K63-deficient mutants to interact with known PSD-95–binding proteins. WT PSD-95, but not K558R, coprecipitated in HEK293FT cells with SPAR (Fig. 4A), a primary GK-binding partner (28). Yeast two-hybrid assays confirmed that PSD-95 interacted with the SPAR C terminus, but this interaction was abolished by K558R in yeast (Fig. 4 B and C). We detected no interaction between His–SPAR-C and GST–PSD-95 or GST-K558R purified from Escherichia coli (Fig. 4D), where the ubiquitination machinery is absent, further supporting the notion that SPAR binds K63-ubiquitinated PSD-95 but not its unconjugated form. Importantly, the PSD-95–SPAR but not the K558R–SPAR interaction was “rescued” in an in vitro reconstitution assay using purified ubiquitinated PSD-95 expressed in HEK293FT cells (Fig. 4E). In HeLa cells, K558R also markedly weakened PSD-95 interaction with GKAP/SAPAP (Fig. 4F), another GK-binding scaffold enriched in the PSD that links PSD-95–NMDA receptor (NMDAR) to Shank–Homer complexes (29, 30). However, K558R did not affect PSD-95 interaction with SAP102, a GK-interacting MAGUK (44), in HEK293FT cells (Fig. 4G).
Fig. 4.
K63-polyUb regulates GK-target interactions. (A) Immunoblots showing lack of interaction between K558R and SPAR in HEK293FT cells. (B) Severely impaired interaction between K558R and SPAR-C in yeasts. (C) Quantification of B. n = 3 experiments; **P < 0.01; one-way ANOVA with post hoc Tukey’s test. (D, Upper) Immunoblots confirming expression of purified GST, GST–PSD-95, GST-K558R, and His–SPAR-C from E. coli. (Lower) GST pulldown showing that both purified GST–PSD-95 and GST-K558R do not interact with His–SPAR-C. (E) In vitro reconstitution of PSD-95–SPAR interaction. (F) Immunoblots showing markedly weakened interaction between transfected K558R-Flag and endogenous GKAP in HeLa cells. (G) Immunoblots showing unaltered K558–SAP102 interaction in cotransfected HEK293FT cells. (H) A ribbon diagram showing the structure of PSD-95 GK in complex with a phosphorylated target peptide derived from Lgl2. Lys558 (shown in the spherical model) is located in the α1/β2 loop opposite the GK target-binding groove.
The nearly abolished interactions between K558R and SPAR or GKAP were unlikely to be due to K–R substitution-related structural alterations to GK. First, structural analysis indicates that K558 is located in the loop connecting α1 and β2 of GK at the opposite side of the target-binding groove of GK, and its sidechain is also fully exposed (Fig. 4H). Thus, a mere K–R substitution is highly unlikely to alter the GK structure and its binding to targets, including SPAR, GKAP, and Lethal giant larvae 2 (Lgl2) (Fig. 5H) (45, 46). Second, NT mutants C3,5S, C5S, and V7S severely impaired K63 ubiquitination of PSD-95 (likely by interfering with E3 binding to the NT, see above) (Fig. S4A) and also disrupted PSD-95–SPAR interaction in yeast (Fig. S4B) and HEK293FT cells (Fig. S4C). Because these residues are located more than 500 aa from GK, it is unlikely that these mutations affect the GK structure.
Fig. 5.
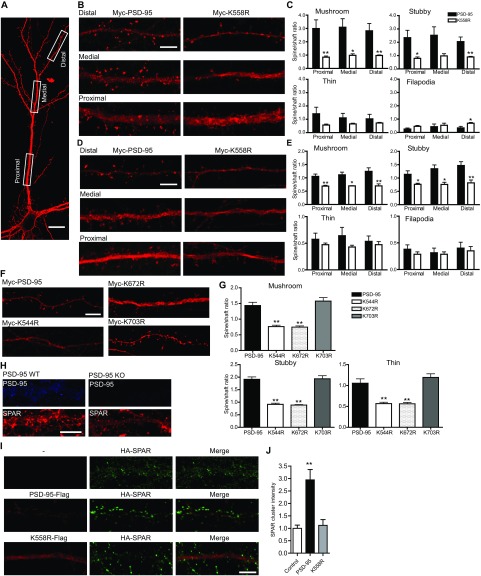
K63-polyUb regulates PSD-95 targeting to and clustering at dendritic spines. (A) PSD-95–KO mouse neurons transfected with Myc–PSD-95 or Myc-K558R and immunostained with anti-Myc. Arrowheads indicate PSD-95 and K558R clusters along dendrites. [Scale bars, 20 μm (Left) and 5 μm (Right).] (B) Spines at higher resolution. (Scale bars, 1 μm.) (C, Left) Quantification of spine/shaft Myc fluorescence ratio. n = 32–41 cells; ***P < 0.001, *P < 0.05; unpaired t tests. (Right) Cumulative probability of spine/shaft fluorescence ratios for all spines from each group. (D) Effects of TRAF6 or CYLD knockdown on PSD-95 clustering. (E) Quantification of PSD-95 clustering intensity and spine/shaft ratio. n = 22–34 cells. **P < 0.01, ***P < 0.001, unpaired t tests.
Fig. S4.
Additional experiments using expanded N-terminal and GK-domain mutants to show that K63 ubiquitination is important for PSD-95–SPAR interaction and is independent of PSD-95 palmitoylation. (A) Impaired K63 polyubiquitination of C3,5S-Flag, C5S-Flag, and V7S-Flag mutants, analyzed by immunoprecipitation and immunoblotting using lysed HEK293FT cells transfected with the indicated plasmids. (B) Severely impaired interactions between C3,5S, C5S, or V7S and SPAR-C in the yeast two-hybrid system. Yeast strain MaV203 was cotransformed with plasmids encoding DB–PSD-95, DB-C3,5S, DB-C5S, or DB-V7S and AD–SPAR-C. Protein–protein interaction was assayed by growing the yeast on selective medium lacking leucine and tryptophan. DB-p53 and AD-T were cotransformed as a positive control. DB and AD–SPAR-C, DB–PSD-95 and AD, DB-C3,5S, DB-C5S, or DB-V7S and AD were cotransformed as negative controls. Unlabeled yeast colonies at the lower left corner were unrelated experiments. (C) Severely impaired interactions between C3,5S-Flag, C5S-Flag, or V7S-Flag and Myc-SPAR in HEK293FT cells. Cells were transfected with indicated plasmids and analyzed by immunoprecipitation and immunoblotting. (D) Severely impaired interactions between K544R-Flag, K558R-Flag, or K672R-Flag and Myc-SPAR in HEK293FT cells. Cells were transfected with the indicated plasmids and analyzed by immunoprecipitation and immunoblotting. (E) Effects of several palmitoylation-regulating mutants on K63 polyubiquitination of PSD-95 analyzed by immunoprecipitation and immunoblotting using lysed HEK293FT cells transfected with the indicated plasmids.
To gain more insights into K63 regulation of PSD-95–target binding, we probed 14 PSD-95 interactions using GST pulldown on mouse brain lysates (Fig. S5 A–C). Consistent with the above results, neither GST-K558R nor GST–PSD-95 pulled down detectable amounts of brain SPAR and GKAP. Similar results were obtained for AKAP79/150, a GK- and SH3-binding synaptic scaffold for certain protein kinases and phosphatases (7); thus K63-polyUb may strongly influence this interaction as well. GST–PSD-95 recovered substantial amounts, but GST-K558 recovered significantly less of the GK-binding proteins Begain and MAP1A, suggesting that K63-polyUb facilitates these interactions. GST–PSD-95 and GST-K558R recovered similar amounts of SAP102; thus this GK interaction is unaffected by K63-polyUb. Finally, pulldown profiles of eight additional proteins interacting with non-GK domains of PSD-95 indicate that K63 ubiquitination had rather modest influence on these interactions.
Fig. S5.
Additional data showing that K63-polyUb regulates PSD-95–target interactions in a GK domain-preferred, target-dependent manner, using several K63-polyUb-deficient mutants. (A) GST pulldown analysis of 14 known PSD-95–binding proteins on mouse brain homogenates using purified GST, GST–PSD-95, and GST-K558R. Western blot analysis of each protein in total lysates is shown at the bottom. (B) Quantification of GST pulldown assays. Gel densitometric analysis was performed for individual proteins, and protein–protein interaction strength was presented as normalized levels. For all proteins except SPAR, GKAP, and AKAP, data were normalized to respective GST–PSD-95 controls. Protein bands for SPAR, GKAP, and AKAP were undetectable and not quantifiable and thus were assigned the value zero and included in the graph to indicate the lack of interaction between them and PSD-95 or K558R. The absence of these bands even for the WT full-length PSD-95 protein is consistent with our hypothesis that K63-linked ubiquitination of PSD-95, which does not occur for bacteria-expressed GST fusion proteins, is essential for PSD-95 to interact with SPAR, GKAP, and AKAP. n = 3 experiments; ***P < 0.001 vs. GST, ###P < 0.001 vs. GST–PSD-95; one-way ANOVA with Tukey’s post hoc tests. (C) Schematic summary of PSD-95–interacting proteins examined in A and B. The module(s) on PSD-95 to which each protein binds are depicted. The relative importance of K63-linked polyubiquitination on each interaction is color-coded: red indicates essential importance, light red indicates nonessential but facilitating, light blue indicates slightly inhibitory, and white indicates no effect. (D, Upper) GST pulldown analysis of three GK-binding and five non–GK-binding proteins on mouse brain homogenates, using purified GST, GST–PSD-95, GST-K544R, or GST-K672R. Western blot analysis of each protein in total lysates is shown at the bottom. (Lower) Pulldown analysis of interactions between K544R or K672R and three GK-binding and five non–GK-binding proteins on mouse brain homogenates using purified K544R-Flag or K672R-Flag from denatured HEK293FT cell lysates. Western blot analysis of each protein in total lysates is shown at the bottom. (E) Computational modeling shows that the K–R substitution at K558, K544, or K672 is not expected to significantly impact the GK structure. The orange model displays the X-ray crystallography model (Protein Data Bank ID code: 1KJW). The cyan model displays the mutant model as predicted by Phyre. Labels indicate positions of α-helices and β-sheets within each domain. Red/blue residues indicate lysine/arginine, respectively. The rmsd scores are under the X-ray model resolution of 1.8 Å, indicating highly similar structures (66). (F) Summary of PSD-95–target interaction literature details. IF, immunofluorescence; IH, immunohistochemistry; N.A., not available; PD, GST pulldown assay; Y2H, yeast two-hybrid assay.
We further extended the above analysis to additional GK domain K63 mutants, K544R and K672R (although both also impair K48 linkage). Both mutants nearly abolished the interaction with SPAR in HEK293FT cells (Fig. S4D). Similar to GST-K558R or GST–PSD-95, neither GST-K544R nor GST-K672R pulled down detectable SPAR, GKAP, or AKAP from brain lysates (Fig. S5D). In comparison, although PSD-95–Flag purified from HEK293FT cells expectedly pulled down significant amounts of SPAR, GKAP, and AKAP from brain lysates, purified K544R and K672R did not (Fig. S5D). Notably, non–GK-dependent interactions were not affected. Since neither mutation impacts the GK structure or its target binding property (Fig. S5E), we concluded that K63-polyUb assembled on GK modifies the scaffolding capability of PSD-95 in a GK-preferred and target-specific manner.
K63-PolyUb Targets PSD-95 to Synapses and Promotes Synapse Remodeling and Strength.
PSD-95 association with other PSD proteins is essential for its clustering at the synapse. When introduced into PSD-95–KO neurons (where the lack of endogenous PSD-95 prevented potential interference), Myc–PSD-95, but not the K63-specific mutant Myc-K558R, was preferentially concentrated in heads of mushroom, stubby, and thin spines (Fig. 5 A and B) and displayed a markedly higher spine/shaft ratio than Myc-K558R (Fig. 5C). The lack of K558R accumulation in spines was observed across the entire dendritic tree of neurons and also, albeit to a lesser degree, in transfected rat neurons in which endogenous PSD-95 was present (Fig. S6 A–E). The dual-linkage mutants Myc-K544R and Myc-K672R also showed drastically impaired spine-head accumulation, whereas the K48-specific mutant Myc-K703R was accumulated normally (Fig. S6 F and G). Knockdown of endogenous TRAF6 markedly decreased PSD-95 enrichment at spines and the spine/shaft ratio, whereas CYLD knockdown increased these parameters (Fig. 5 D and E). Finally, SPAR clustering was markedly reduced in cultured PSD-95–KO neurons, and PSD-95–Flag, but not K558-Flag, promoted synaptic clustering of HA-SPAR (Fig. S6 H–J). These results support the crucial role of K63-polyUb in the targeting and clustering of PSD-95 and its associated proteins at synapses.
Fig. S6.
Additional experiments showing that PSD-95 K63 ubiquitination is required for dendritic spine targeting of PSD-95 and SPAR. (A) A mouse hippocampal neuron transfected with Myc–PSD-95 (red) illustrating the selection strategy of proximal, medial, and distal dendritic segments. (Scale bar, 40 μm.) Proximal, medial, and distal dendrites were selected based on their distance from the soma or branching points. Specifically, proximal segments were within 100 μm of soma on the primary dendrites; medial segments were within 100 μm of the first branching points on the secondary dendrites; distal segments were on a tertiary or higher-order dendrite. (B) Impaired spine targeting of K558R in cultured PSD-95–KO hippocampal mouse neurons transfected with Myc–PSD-95 or Myc-K558R. (Scale bar, 5 μm.) (C) Quantification of the spine/shaft fluorescence ratio in B. n = 6–23 cells; **P < 0.01, *P < 0.05; unpaired t tests vs. respective controls. (D) Impaired spine targeting of K558R in cultured rat hippocampal neurons transfected with Myc–PSD-95 or Myc-K558R. (Scale bar, 5 μm.) (E) Quantification of the spine/shaft fluorescence ratio in D. n = 3–8 cells; **P < 0.01, *P < 0.05; unpaired t tests vs. respective controls. (F) Impaired spine targeting of K544R and K672R but not K703R in cultured rat hippocampal neurons transfected with Myc–PSD-95, Myc-K544R, Myc-K672R, or Myc-K703R. (Scale bar, 5 μm.) (G) Quantification of the spine/shaft fluorescence ratio in F. n = 10–16 cells; **P < 0.01; post hoc Dunnett’s tests following one-way ANOVA. (H) SPAR clustering is markedly diminished in cultured PSD-95–KO hippocampal mouse neurons, revealed by SPAR immunofluorescence. (Scale bar, 5 μm.) (I) PSD-95–Flag, but not K558R-Flag, significantly restored clustering of HA-SPAR in cotransfected cultured PSD-95–KO hippocampal mouse neurons. (Scale bar, 5 μm.) (J) Quantification of SPAR clustering intensity in I. n = 12–14 cells; **P < 0.01; post hoc Dunnett’s tests following one-way ANOVA.
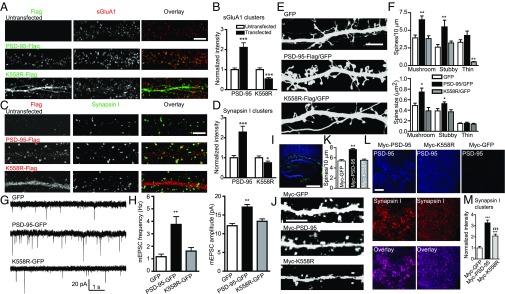
Synaptically localized PSD-95 serves as a “slot scaffold” to control synaptic AMPA receptor (AMPAR) content (21, 47). Consistent with ref. 27, overexpression of PSD-95–Flag in rat hippocampal neurons enhanced clustering of surface GluA1 (sGluA1) receptors (Fig. 6 A and B) as well as the presynaptic synapsin I (Fig. 6 C and D), supporting the notion that PSD-95 promotes synapse maturation (48). K558R-Flag overexpression did not enhance, and in fact reduced, sGluA1 and synapsin I clustering (Fig. 6 A–D). PSD-95–Flag, but not K558R-Flag, also significantly increased the density and head size of mushroom and stubby spines (Fig. 6 E and F). Functionally, miniature excitatory postsynaptic currents (mEPSCs) recorded from PSD-95–GFP transfected pyramidal neurons exhibited significantly higher frequency and amplitude compared with GFP-transfected controls, whereas K558R-GFP overexpression did not affect either mEPSC frequency or amplitude (Fig. 6 G and H). Similar to K558R, K544R and K672R lost the ability to enhance sGluR1 clustering and mushroom spine morphogenesis in neurons (Fig. S7).
Fig. 6.
K63-polyUb facilitates synapse maturation and strengthening. (A) sGluA1 clusters (red) from untransfected neurons or neurons transfected with PSD-95–Flag or K558R-Flag (green). (Scale bar, 5 μm.) (B) Quantification of sGluA1 cluster intensity, normalized to untransfected neurons. n = 28–29 cells; ***P < 0.001. (C) Synapsin I clusters (green) from untransfected and PSD-95–Flag or K558R-Flag transfected (red) neurons. (Scale bar, 5 μm.) (D) Quantification of synapsin I cluster intensity, normalized to untransfected neurons. n = 14–26 cells; ***P < 0.001, *P < 0.05. (E) GFP images of dendrites and spines from neurons (co)transfected with the indicated plasmids. (Scale bar, 5 µm.) (F) Quantification of spine density (Upper) and size (Lower) in E. n = 7–12 cells. **P < 0.01, *P < 0.05. (G) Representative mEPSCs from neurons transfected with indicated plasmids. (H) Mean mEPSC frequency (Left) and amplitude (Right). n = 27–38 cells. **P < 0.01. (I) Lentivirus expression of Myc–PSD-95 or Myc-K558R in the hippocampus in vivo. Viruses expressing Myc–PSD-95, Myc-K558R, or Myc-GFP and GFP within the same dual promoter vector (Fig. S7C) were injected into P20 PSD-95–KO brains. Nuclei are stained with Hoechst (blue). (Scale bar, 500 µm.) (J) Examples of apical dendrites (GFP) of infected pyramidal neurons. (Scale bar, 5 µm.) (K) Quantification of spine density in J. n = 28–31 cells. **P < 0.01. (L) Example PSD-95 and synapsin I puncta in infected striatum radiatum. (Scale bar, 5 μm.) (M) Quantification of synapsin I cluster intensity in L. n = 16–18 cells. ***P < 0.001 vs. Myc-GFP; ###P < 0.001 vs. Myc–PSD-95; unpaired t tests in B and D, one-way ANOVA with Dunnett’s’ test vs. GFP in F, H, and K, and one-way ANOVA with Tukey’s test in M.
Fig. S7.
Additional data on K544R and K672R supporting the notion that K63 ubiquitination of PSD-95 promotes dendritic spine formation and maturation in cultured rat hippocampal neurons. (A) sGluA1 clusters (red) from untransfected neurons or from neurons transfected with PSD-95–Flag, K544R-Flag, or K672R-Flag (green). (Scale bar, 5 μm.) (B) Quantification of sGluA1 cluster intensity, normalized to untransfected neurons. n = 9–29 cells; ***P < 0.001, **P < 0.01. PSD-95–Flag transfected and untransfected data are replotted from Fig. 6B for direct comparisons. (C) Plasmids containing dual-promoter expression cassettes. Syn, synapsin I promoter. (D) Representative GFP images of dendrites and spines from DIV21 neurons transfected with the indicated plasmids. (Scale bar, 5 µm.) (E) Quantification of spine density in D. n = 15–25 cells; **P < 0.01, *P < 0.05; one-way ANOVA followed by Dunnett’s test vs. GFP.
We next investigated the role of K63-polyUb in synapse remodeling in vivo. Lentiviruses expressing Myc–PSD-95/GFP, Myc-K558R/GFP, or Myc-GFP were injected into hippocampi of P20 PSD-95–KO mice (Fig. 6I), and dendritic spines and synapsin I clustering were analyzed at P34 (Fig. 6 J–M). Myc–PSD-95– but not Myc-K558R–expressing CA1 pyramidal neurons displayed a significant increase in spine density on apical dendrites over GFP control (Fig. 6 J and K), and synapsin I clustering was significantly more enhanced in Myc–PSD-95–injected than in K558R-injected mice (Fig. 6 L and M). In summary, our experiments in vitro and in vivo support the notion that K63-polyUb promotes synapse formation, maturation, and strength.
Regulation of K63-PolyUb Conjugation by Activity.
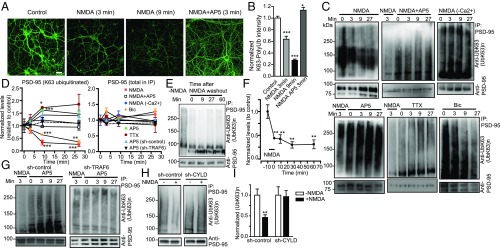
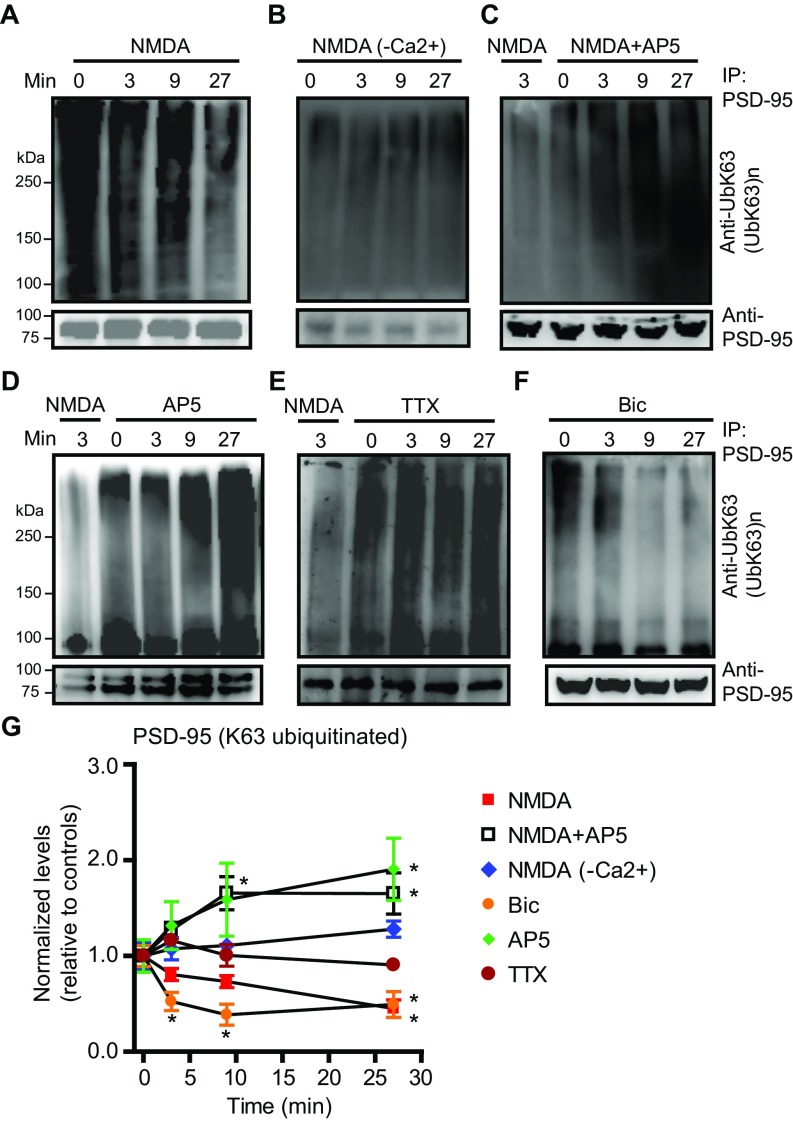
We next examined whether K63 ubiquitination is regulated by neuronal activity. Exposure to NMDA, a treatment used to induce chemical long-term depression (cLTD) in cultured neurons and slices (33, 49), resulted in a rapid (within minutes) and potent (∼73%) decrease of K63-polyUb conjugate level (Fig. 7 A and B). This effect was completely reversed by the NMDAR antagonist AP5. The profound loss of K63-polyUb staining was observed throughout the neuron, indicating a global disassembly of K63-polyUb chains, likely by deubiquitination of substrates.
Fig. 7.
Activity-dependent regulation of K63-polyUb conjugation in cultured rat hippocampal neurons. (A) Rapid and global loss of K63-polyUb signals following NMDA application. Neurons were treated with NMDA (30 μM), NMDA + AP5 (50 μM), or control solution for 3 or 9 min before immunostaining for K63-polyUb. (Scale bar, 40 μm.) (B) Quantification of A. n = 19–36 cells. ***P < 0.001, *P < 0.05; unpaired t tests. (C) Immunoblots showing levels of K63 polyubiquitinated PSD-95 at various time points after treatment with NMDA (30 μM), NMDA+AP5 (50 μM), NMDA in Ca2+-free artificial cerebrospinal (ACSF) fluid, AP5, TTX (2 μM), or bicuculline (Bic; 40 μM). (D) Quantification of C and G. K63-ubiquitinated and total PSD-95 in immunoprecipitation eluates following treatments were normalized to respective untreated controls. n = 3 or 4 experiments; ***P < 0.001, **P < 0.01, *P < 0.05; unpaired t tests vs. controls. (E and F) Immunoblots (E) and summary (F) showing prolonged loss of K63-polyUb from PSD-95 induced by NMDA. n = 3; **P < 0.01, one-way ANOVA with Tukey’s post hoc tests. (G) Immunoblots showing effects of AP5 on PSD-95 K63 ubiquitination in neurons infected with sh-control or sh-TRAF6 lentiviruses. (H) Immunoblots showing CYLD knockdown abolished NMDA-induced PSD-95 deubiquitination. (I) Summary of H. n = 6–8 experiments; **P < 0.01; paired t tests vs. controls.
We directly investigated activity regulation of PSD-95 ubiquitination. NMDA induced rapid loss of K63-polyUb from PSD-95, which was prevented in a Ca2+-free solution or by AP5 (Fig. 7 C and D). This NMDA-triggered deubiquitination was long lasting even after NMDA removal (Fig. 7 E and F), resembling that of cLTD (49). AP5 alone elicited a gradual increase of PSD-95 ubiquitination that reached a plateau by 10 min after treatment (Fig. 7 C and D), suggesting that PSD-95 is constitutively conjugated by K63-polyUb under resting conditions but simultaneously undergoes deubiquitination driven by spontaneous NMDAR activation. Consistently, blocking action potential-dependent synaptic activity with tetrodotoxin (TTX) or enhancing synaptic activity with the GABAA receptor blocker bicuculline respectively increased and decreased K63 ubiquitination of PSD-95 (Fig. 7 C and D). Knocking down TRAF6 prevented the AP5-elicited increase of PSD-95 ubiquitination (Fig. 7G), supporting the notion that TRAF6 mediates constitutive K63 ubiquitination of PSD-95. In contrast, CYLD knockdown abolished NMDA-induced loss of PSD-95 ubiquitination (Fig. 7 H and I), suggesting that CYLD is required for this activity-dependent deubiquitination. Similar results were also observed in acute mouse prefrontal slices (Fig. S8), suggesting that these regulations occur in native circuits. Together these results indicate that K63-polyUb conjugation to PSD-95 is controlled by activity through TRAF6 and CYLD.
Fig. S8.
Activity-dependent regulation of K63 ubiquitination of PSD-95 in mouse brain slices. (A–F) Immunoblots of K63 polyubiquitination of PSD-95 in mouse prefrontal cortical slices following various stimulations. Acutely cut slices (containing the anterior cingulated cortex and the prelimbic cortex) were treated with the specified agonists or antagonists for the time indicated, lysed, and analyzed by immunoprecipitation and immunoblotting assays. In C–E, a positive control (NMDA treatment for 3 min) was also included. Drugs were included in ACSF at the following concentrations: NMDA (30 μM), AP5 (50 μM), TTX (2 μM), and bicuculline (40 μM). Each experiment was performed using pooled slices from an independent PSD-95 WT mouse. (G) Quantification of A–F. Ubiquitinated and total PSD-95 in immunoprecipitation eluates following treatments were normalized to respective untreated controls. n = 3–5 experiments; *P < 0.05; unpaired t tests vs. controls.
K63-PolyUb Regulates Activity-Dependent PSD-95 Clustering.
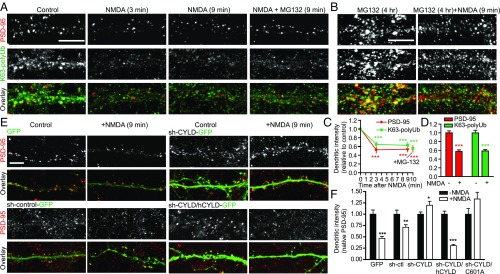
Given that K63-polyUb targets PSD-95 to synapses and that NMDA stimulation deubiquitinates PSD-95, we tested the hypothesis that K63-polyUb regulates activity-dependent PSD-95 clustering at synapses. NMDA induced a rapid decrease of endogenous PSD-95 staining intensity on dendrites, concomitant with a loss of dendritic K63-polyUb immunostaining (Fig. 8A). This NMDA-triggered PSD-95 declustering was not due to synaptic PSD-95 degradation, because including the proteasome inhibitor MG132 during stimulation did not prevent PSD-95 declustering (Fig. 8 A–D). Furthermore, when transfected into neurons, the K48-deficient K703R-Flag was quickly lost from spines following NMDA stimulation, whereas K558R-Flag was resistant to NMDA and maintained its diffuse dendritic localization (Fig. S9 A and B). This result confirmed that the rapid (within minutes) PSD-95 declustering following NMDAR activation was primarily mediated by translocation of PSD-95 away from synapses independent of proteasomal turnover. Finally, knocking down endogenous CYLD prevented NMDA-induced PSD-95 declustering which, importantly, was restored by cotransduction of an RNAi-resistant CYLD cDNA but not the DUB-dead mutant C601A (Fig. 8 E and F and Fig. S9C). These results demonstrate a critical role for K63-polyUb in activity-dependent regulation of PSD-95 clustering at synapses.
Fig. 8.
CYLD mediates NMDA-induced rapid PSD-95 declustering from spines. (A and B) NMDA-induced, proteasome-independent PSD-95 declustering on dendrites. (A) Neurons were treated with NMDA (30 μM), NMDA + MG132 (20 μM), or control solution for 3 or 9 min before immunostaining for PSD-95 or K63-polyUb. (B) Neurons were incubated with MG132 for 4 h before 9-min NMDA stimulation. (Scale bars, 5 μm.) (C) Quantification of dendritic PSD-95 and K63-polyUb fluorescence intensities in A. n = 22–24 cells; ***P < 0.001; unpaired t tests vs. no-treatment control. (D) Quantification of B. n = 20 cells; ***P < 0.001, unpaired t tests. (E) Effects of CYLD knockdown on NMDA-induced endogenous PSD-95 declustering. Neurons infected with the indicated vectors were treated with NMDA or control solution for 9 min before immunostaining for PSD-95. (Scale bar, 5 μm.) (F) Quantifications of E. n = 10–34 cells; ***P < 0.001, **P < 0.01, *P < 0.05; unpaired t tests.
Fig. S9.
Additional data on NMDA-triggered PSD-95 declustering and sGluA1 internalization in cultured rat (A–C) and mouse (D–G) hippocampal neurons. (A) Declustering of K703R-Flag, but not K558R-Flag, following NMDA stimulation. Neurons transfected with the indicated vectors were treated with NMDA or control solution for 9 min before immunostaining for Flag. Cotransfected GFP was used to identify positive cells. (Scale bar, 5 μm.) (B) Quantification of dendritic PSD-95 (Flag) fluorescence intensities in A. n = 13–22 cells; ***P < 0.001, *P < 0.05; unpaired t tests vs. respective no-treatment controls. (C) Representative images showing the coinfected CYLD-601A mutant failed to rescue the sh-CYLD abolishment of NMDA-triggered PSD-95 declustering. Neurons infected with sh-CYLD and C601A-GFP were treated with NMDA or control solution for 9 min before immunostaining for PSD-95. This experiment was run as a part of the experiments shown in Fig. 8 E and F. (Scale bar, 5 μm.) (D) NMDA-triggered AMPAR internalization assay in PSD-95 WT neurons. Representative images show surface and internalized GluA1 (iGluA1) receptors following NMDA (50 μM, 9 min) or control treatment. (E) Quantified sGluA1 internalization ratio in D. n = 20–22 cells; ***P < 0.001; unpaired t test. (F) NMDA-triggered sGluA1 internalization in PSD-95–KO neurons infected with Myc-GFP, Myc–PSD-95 (and GFP), or Myc-K558 (and GFP) (expressed with dual-Syn promoter vectors). (G) Quantification of NMDA-triggered sGluA1 internalization in F normalized to respective no-NMDA controls. n = 26–46 cells; ***P < 0.001; unpaired t tests.
K63 Deubiquitination and CYLD Are Required for cLTD.
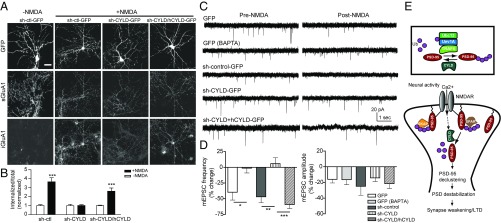
The rapid PSD-95 declustering associated with PSD-95 deubiquitination by NMDA suggests that K63 linkage has a role in LTD. We tested this hypothesis by first examining NMDAR-dependent AMPAR endocytosis in cultured hippocampal neurons. In agreement with earlier studies (26, 50), a brief application of NMDA induced robust internalization of sGluA1 receptors throughout the cell (Fig. S9 D and E). This NMDAR-triggered sGluA1 internalization was severely impaired in PSD-95–KO neurons (Fig. S9 F and G), as is consistent with an important role for PSD-95 in LTD (13, 25, 26, 33). Reintroducing WT PSD-95, but not K558R (which reached synapses, albeit to a lesser degree than WT), to PSD-95–KO neurons restored the NMDA-induced sGluA1 internalization (Fig. S9 F and G). Supporting the notion that deubiquitination is necessary for this process, CYLD knockdown in rat hippocampal neurons abolished NMDAR-triggered sGluA1 internalization, and, importantly, this effect was rescued by coexpressing the RNAi-resistant CYLD (Fig. 9 A and B). These experiments suggest important roles for K63 deubiquitination and for CYLD in cLTD.
Fig. 9.
CYLD is required for cLTD. (A) NMDA (50 μM, 9 min)-triggered sGluA1 internalization in cultured rat hippocampal neurons infected with indicated plasmids. (Scale bar, 30 μm.) (B) Quantification of A, normalized to respective no-NMDA controls. n = 13–19 cells; ***P < 0.001; unpaired t tests. (C) NMDA-triggered depression of mEPSCs in cultured rat hippocampal neurons transfected with indicated plasmids. Intracellular BAPTA: 15 mM. (D) Quantification of mEPSC frequency and amplitude. n = 6–8 cells; ***P < 0.001, **P < 0.01, *P < 0.05; one-way ANOVA with post hoc Tukey’s test. (E) Model for roles of nonproteolytic K63 linkage in the synapse using PSD-95 as a proof of principle. (Upper) An E2/E3/DUB complex controls the balance of K63 ubiquitination/deubiquitination of PSD-95. (Lower) K63-polyUb plays an essential role in synaptic maintenance and orderly organization of PSD-95 and LTD. PSD-95 is constitutively conjugated by K63-polyUb chains (by Ubc13/Uev1A–TRAF6), which maintains and compartmentalizes the protein (perhaps combined with other posttranslational modifications) to specific subdomains within the PSD. Synaptic activity opens NMDARs, allowing Ca2+ influx, and recruits/activates CYLD (dashed arrow), which subsequently removes K63-polyUb from PSD-95. Deubiquitinated PSD-95 translocates away from PSD, destabilizing the PSD and weakening the synapse. Palmitoylated PSD-95 is targeted to the synapse membrane but may not be incorporated in a particular scaffolding/signaling complex that depends on PSD-95 K63 ubiquitination.
We next examined NMDA-induced depression of mEPSCs, a functional form of cLTD sharing similar mechanisms with NMDAR-triggered AMPAR internalization (26, 50). In control neurons, bath-applied NMDA produced a significant decrease in mEPSC frequency (but only a small, insignificant decrease in amplitude), consistent with findings in ref. 50, which was blocked by loading the Ca2+ chelator BAPTA (Fig. 9 C and D). CYLD knockdown abolished this NMDAR-triggered mEPSC frequency depression, and this effect was rescued by coexpressing RNAi-resistant CYLD. Together, these results indicate that CYLD mediates cLTD, likely through deubiquitination and synaptic decumulation of PSD-95.
Discussion
The UPS has emerged as a major mechanism that deconstructs synapses and remodels neural circuits during development and behavior learning (8–11). We illustrate a UPS-independent mechanism by which ubiquitination has an unexpected, constructive role in synapse organization, function, and plasticity (Fig. 9E). Among eight polyUb chain types, K63 chains are considered the primary nondegradable linkage (51). The extended open conformations adopted by these chains are recognized not by the proteasome but by diverse ubiquitin-interacting proteins involved in endocytosis, kinase activation, and protein trafficking (1). These chains can also remodel the surface of substrate proteins, akin to phosphorylation, to directly regulate protein–protein interactions (3). The regulation of PSD-95 interactions with structurally distinct partners by K63-polyUb demonstrated by several K63-deficient mutants, suggests that K63 chains modify the conformation of PSD-95, particularly the GK domain, through surface remodeling. However, recognizing the high diversity and complexity of ubiquitin-binding domains (1), we cannot exclude the possibility that unidentified ubiquitin-binding domains may be embedded in these proteins that can serve as primary or secondary interaction sites. Regardless of the mechanism, our results suggest that a key role of K63 chains at the synapse is to regulate the orderly organization of the PSD protein network.
What are the endogenous K63-polyUb conjugation site(s) on brain PSD-95? While standard and well-accepted in ubiquitination site mapping and often required for validating high-throughput data, our mutagenesis results do not prove a lysine is actually conjugated by polyUb chains. A more direct approach is MS, which became available for rodent brain PSD-95 during the preparation and revision of this article (52). This evolving database has uncovered several potential ubiquitination sites on SH-GK of PSD-95, and, interestingly, only K558 independently converges with our low-throughput biochemical assay. Combined, these results show it is reasonable to conclude that K558 is an authentic ubiquitination site for brain PSD-95 that accounts for a substantial amount of total PSD-95 ubiquitination. Conversely, K491, K544, and K672, which we found to markedly influence PSD-95 ubiquitination, were not picked up by MS and could be false negatives due to resolution and/or sensitivity issues often associated with high-throughput experiments. Alternatively, some of these residues may not be true ubiquitin sites in native tissues but undergo other types of modifications that can critically regulate the ubiquitination of the cognate site(s). Indeed, based on the MS database (52), K544 and K672 may be acetylated. In either case, it is interesting to note that a K–R mutation at any of the four lysine residues we identified in SH3-GK severely impairs the total PSD-95–K63 ubiquitination to a similar extent, suggesting critical interdependence among these sites in regulating ubiquitination. This could be through complex but poorly understood spatiotemporal mechanisms underlying ubiquitination (53) or via crosstalk between ubiquitin network and other modifications (54). Regardless of the mechanistic details, given the established specificity of polyUb chain antibodies (38), our study (combined with the MS resource) provides little doubt that PSD-95 undergoes K63 ubiquitination and identifies a direct K63-polyUb site in K558 and several other potential sites that regulate this modification on a key synaptic scaffold.
Synaptic targeting of PSD-95 is regulated by protein palmitoylation and phosphorylation. Palmitoylation at C3 and C5 residues recruits PSD-95 to synapse membranes (31), and phosphorylation of S295 enhances synaptic accumulation of PSD-95 (33). However, palmitoylation does not seem to be absolutely required for K63 ubiquitination, as mutants impairing this modification were still ubiquitinated (C3S and I6S in Fig. S4E). Whether K63-polyUb influences palmitoylation and phosphorylation is unknown. Given the abundance of PSD-95 in PSD [∼300 copies (55)], its many binding partners (7), and that only a fraction of the protein undergoes ubiquitination at a given time, we suggest that, depending on activity patterns and surrounding molecular architectures, separate pools of differentially modified PSD-95 molecules simultaneously exist in the PSD. The different modification mechanisms may act in concert to target and compartmentalize PSD-95 to appropriate subdomains in PSD (Fig. 9E) for it to participate in various synaptic processes. Thus, K63 ubiquitination represents a mechanism joining phosphorylation and palmitoylation in synaptic targeting, assembly, and signaling.
Previous studies have shown that NMDA stimulation induces K48–polyUb accumulation and proteasome recruitment in dendritic spines (56) as well as presumable K48-linked ubiquitination and degradation of PSD-95 (13). In contrast, we found NMDA triggered rapid and widespread K63-polyUb loss and PSD-95 deubiquitination (K63). A careful comparison suggests that processes involving the two different linkages might take place at different time points: K63 deubiquitination occurs within minutes, whereas proteasomal ubiquitination and degradation of PSD-95 occur at or 5–10 min after treatment (13, 56). PSD-95 is constitutively conjugated by K63-polyUb chains, consistent with the lack of proteasome-associated ubiquitination events for PSD-95 at resting state (12, 57). These studies suggest tightly controlled modifications of PSD-95 by different ubiquitin topologies. One potential mechanism is ubiquitin chain editing, a process through which the removal of K63-polyUb chains from a substrate is followed by the addition of K48–polyUb chains (38, 53). Thus, PSD-95 represents a prime candidate for ubiquitin editing, and the K48/K63 dual sites we have identified may participate in the editing. This will await future investigations.
The activity-dependent assembly/disassembly of K63-polyUb chains may involve dynamic regulation of the E2–E3–substrate–DUB multiprotein complex we have identified in the PSD. The Ubc13/Uev1A dimer remains the only known K63-selective E2 that, together with the RING-domain family E3 ligase TRAF6, catalyzes the assembly of K63-polyUb chains on substrates. Our results suggest that the Ubc13/Uev1A–TRAF6 complex binds the PSD-95 NT and delivers ubiquitin moieties to cognate lysine sites. It remains to be determined how the E2–E3 enzymes sense neuron activity; however, CYLD appears to be recruited to the PSD following depolarization (19) or NMDA stimulation (58) in cultured neurons, which may contribute to activity-dependent regulations of K63-polyUb conjugation at synapses described here.
The K63 linkage represents an important mechanism regulating synapse development, function, and plasticity. First, by conjugating to a major synaptic scaffold, K63-polyUb chains promote synapse formation, maturation, and strength, as indicated by several K63-deficient mutants in vitro and in vivo. Second, we demonstrate a critical role for K63 linkage and CYLD in cLTD. Our data support the notion that cLTD-inducing NMDAR activation triggers a rapid cleavage of K63-polyUb chains from PSD-95 by CYLD, leading to mobilization of deubiquitinated PSD-95 away from the core of PSD and subsequent loss of synaptic AMPARs, a model (Fig. 9E) consistent with the slot hypothesis of synaptic strength control by PSD-95 (21, 47). We note that the immediate (within minutes) synaptic PSD-95 depletion by NMDA is due to PSD-95 translocation from, but not degradation at, the synapse. This finding is consistent with the observation that some PSD proteins undergo UPS-independent trafficking to or away from synapses (12). This view, however, is different from a previous report proposing NMDA-induced UPS-dependent degradation of PSD-95 at the synapse (13). The basis for the apparent inconsistency is unclear but may simply reflect different faces at different time points of the same process (e.g., ubiquitin editing; see above). Finally, K63 linkage may play a role in homeostatic plasticity. TRAF6 and CYLD are, respectively, potent positive and negative regulators of the TNF receptor and IL-1/Toll-like receptor signaling (2, 5). Several proinflammatory cytokines, including glia-released TNFα, known to control synaptic strength and scaling (59, 60), are present in the brain. This raises the possibility that K63 ubiquitination and synaptically localized TRAF6/CYLD may contribute to homeostatic maintenance of synaptic connectivity in response to changes in glial activity during normal adaptive processes or pathological conditions. Finally, mutations to the CYLD gene cause familial cylindromatosis (6), and patients often develop psychological problems. Our findings thus open up avenues in the study of neuro–immune and neuro–glial interactions involved in brain circuit development, dysfunction, injury, and repair.
There likely are other K63 substrates at the synapse. In Caenorhabditis elegans neurons, K63 ubiquitination has been implicated in mechanisms that regulate glutamate receptor trafficking (61). A substantial portion of K63-polyUb clusters do not colocalize with PSD-95 (Figs. 1C and 8A), supporting the presence of additional synaptic substrates. The abundance of CYLD in the PSD suggests that it may be a primary DUB for multiple proteins. Identification and characterization of these proteins can provide insights into the mechanisms of synapse development, function, and plasticity as well as related pathologies.
Materials and Methods
All procedures involving animals were approved by Harvard Medical School or State University of New York Upstate Medical University Institutional Animal Care and Use Committees. Methods details on molecular biology, biochemistry, virus construction and packaging, stereotaxic injection, immunohistochemistry, confocal microscopy and imaging analysis, and electrophysiology, as well as a full list of reagents, are included in SI Materials and Methods. All data are expressed as mean ± SEM. Comparisons were made with two-sided t tests or one-factor ANOVA with appropriate post hoc tests. Significance level was set at 0.05.
SI Materials and Methods
Animals.
The PSD-95–KO mouse strain (on an outbred MF-1 background) (37) and C57BL/6J mice were bred in-house in standard laboratory conditions (12-h light/dark cycle) with food and water provided ad libitum. Mouse breeding was carried out by housing one male and two or three female heterozygous mice in the same cage. Timed pregnant rats were purchased from Charles River Laboratories.
Cell Culture and Transfection.
HEK293T, HEK293FT, and Neuro2a cells were cultured in DMEM supplemented with 10% FBS (GIBCO). Cells were plated on 10-cm dishes and grown to 80% confluency before transfection using calcium phosphate. Low- (4 × 104), mid- (4 × 105), and high-density (1 × 106) hippocampal neurons were prepared from E18 rat embryos or mouse PSD-95 WT or KO embryos or P1 pups and were maintained in Neurobasal medium supplemented with B-27 and 2 mM l-glutamine (Invitrogen) for up to 21 DIV. Neurons were transfected with calcium phosphate at DIV7. For cotransfection of two plasmids, equal weights of plasmids were thoroughly mixed in CaCl2 solution and added to HBSS drop by drop while vortexing. The mixtures were then applied to neurons. One hundred percent efficiency of cotransfection is routinely obtained.
Antibodies.
The following antibodies were used: anti-Flag (Sigma, F7425), anti-Flag (Sigma, F3165), anti-Flag M2 Affinity Gel (Sigma, A2220), anti-HA (12CA5) (Roche, 11583816001), anti-HA-Peroxidase (Roche, 11667475001), anti–c-Myc (9E10) (Santa Cruz, sc-40), anti–c-Myc Agarose (Pierce/Thermo scientific, 20168), anti-GST (Santa Cruz, sc-138), anti-His (Qiagen, 34660), anti–K63-PolyUb (Millipore, 05-1308), anti–K48-PolyUb (Millipore, 05-1307), anti–PSD-95 (Cell Signaling, 2507), anti–PSD-95 (Santa Cruz, sc-32290), anti-GluN1 (Millipore, 05-432), anti-GluN2A (Millipore, 07-632), anti-GluN2B (Millipore, 06-600), anti–GluA1-NT (Millipore, MAB2263), anti-Synapsin I (Millipore, AB1543P), anti-TRAF6 (Abcam, ab33915), anti-CYLD (Abcam, ab60266), anti-CYLD (Cell Signaling, 8462), anti-GKAP (Abcam, ab65020), anti-SPAR (Millipore, ABN485), anti-AKAP150 (Millipore, 07-210), anti-Begain (Abcam, ab125461), anti-MAP1A (Abcam, ab101224), anti-SAP102 (StressGen, VAM-PS006), anti-Src (Cell Signaling, 2109), anti-Kalirin-7 (Abcam, ab52012), anti-KIF1B (Abcam, ab69614), anti-nNOS (BD, 610308), anti-Lyn (Millipore, 04-268), anti-SynGAP (Sigma, S-5437), anti-Ubc13 (ProSci, 3373), anti-Uev1A (ProSci, 3375), anti-actin (Millipore, MAB1501R), Alexa Fluor goat anti-mouse IgG 488 (Invitrogen, A-1100), Alexa Fluor goat anti-mouse IgG 568 (Invitrogen, A-11004), Alexa Fluor goat anti-mouse IgG 647 (Invitrogen, A21236), Alexa Fluor goat anti-rabbit IgG 488 (Invitrogen, A-11008), Alexa Fluor goat anti-rabbit IgG 568 (Invitrogen, A-11036), horseradish peroxidase-conjugated goat anti-mouse (Pierce, 31430), and horseradish peroxidase-conjugated goat anti-rabbit (Pierce, 31460).
Plasmids.
Enhanced GFP-tagged mammalian expression plasmids PSD-95∆NT–GFP, PSD-95∆PDZ1,2–GFP, PSD-95∆GK–GFP, PSD-95∆SH3–GK–GFP, and K558R-GFP were generated by site-directed mutagenesis using pEGFP–N2–PSD-95 as the template (62). Flag-tagged expression plasmids C3S-Flag, C5S-Flag, C3,5S-Flag, I6S-Flag, V7S-Flag, ∆PEST-Flag, ∆Src-Flag, K10R/K11R-Flag, K299R-Flag, K355R-Flag, K441R/K443R-Flag, K491R-Flag, K503R/K505R-Flag, K544R-Flag, K558R-Flag, K570R/K588R/K591R-Flag, K597R-Flag, K624R-Flag, K663R-Flag, K672R-Flag, K679R-Flag, K703R-Flag, and K705R-Flag were derived using pcDNA–PSD-95–Flag as the template (62). Myc-tagged expression plasmids Myc-TRAF2, Myc-TRAF3, Myc-TRAF4, Myc-TRAF6, Myc-CYLD, Myc-CYLD (C601A), and Myc-GFP were constructed using the pCMV-Myc vector (Clontech). Standard molecular biology techniques were used. Complete coding sequences of PSD-95, K544R, K558R, K672R, C5S, C3,5S, and V7S were cloned into pGEX-5x-1 vector for expression and purification of proteins in E. coli BL21Gold, and into pDBleu vector for yeast two-hybrid assays. DNA fragments corresponding to the SPAR C terminus were cloned into pET28a vector for protein expression and purification in E. coli BL21Gold and into pAD-Trp vector for yeast two-hybrid assays. pDB-p53 and pAD-T were control plasmids included in the Two-Hybrid System Kit (Invitrogen). Lentivirus transfer plasmids syn-Myc-GFP-syn-GFP, syn–Myc–PSD-95–syn–GFP, syn-Myc-K544R-syn-GFP, syn-Myc-K558R-syn-GFP, syn-Myc-K672R-syn-GFP, syn-Myc-K703R-syn-GFP, syn-Myc-TRAF6-syn-GFP, syn-Myc-TRAF6 C70A-syn-GFP, syn-Myc-hCYLD-syn-GFP (a human CYLD resistant to CYLD RNAi), and syn-Myc-hCYLD C601A-syn-GFP were derived from a custom-constructed dual synapsin promoter vector. HA-tagged ubiquitin (HA-Ub) and mutants were kindly provided by Zhijian James Chen, UT Southwestern Medical Center, Dallas. HA-SPAR and Myc-SPAR were kindly provided by Daniel T. S. Pak, Georgetown University, Washington, DC. Lentivirus vectors FUGW [kindly provided by David Baltimore, Caltech, Pasadena, CA) and pLLox3.7 [kindly provided by Roger Nicoll, University of California, San Francisco (UCSF)] were used as templates to generate TRAF6 and CYLD shRNA plasmids. All constructs were confirmed by DNA sequencing and protein expression.
Immunoprecipitation and Immunoblot Analysis.
HEK293FT cells were transfected with the indicated plasmids with calcium phosphate (Clontech) following the manufacturer’s instructions. When necessary (e.g., for the K48 ubiquitination assays in Fig. 2D), cells were treated with MG132 (Sigma) for 8 h beginning 40 h after transfection. At 48 h cells were collected and lysed in ice-cold radioimmunoprecipitation assay (RIPA) buffer [50 mM Tris⋅HCl (pH 7.4), 1% Nonidet P-40, 0.1% SDS, 0.25% deoxycholic acid, 150 mM NaCl, 1 mM EDTA, 1 mM PMSF, and protease inhibitor mixture (Roche)]. Neuro2a cells, mouse brain, and infected neuron cultures were similarly lysed/homogenized and cleared in ice-cold RIPA buffer. After preclearing with protein G beads (Amersham) at 4 °C for 1 h, lysates were incubated with primary antibodies at 4 °C for 2 h. After the addition of protein A/G beads (Santa Cruz) and incubation at 4 °C overnight, immunocomplexes were collected and washed with ice-cold RIPA buffer and eluted with SDS loading buffer by boiling for 5 min. The supernatant was collected by centrifugation and used in immunoblotting experiments. For immunoblotting, proteins were resolved by SDS/PAGE and transferred to PVDF membrane (Bio-Rad). After blocking and washing, PVDF membranes were incubated with primary antibodies at 4 °C overnight followed by incubation with secondary antibodies at room temperature for 1 h. Immunoreactive signals were detected with the SuperSignalWest Femto Maximum Sensitivity Substrate Kit (Pierce) and an LAS-3000 image system (Fujifilm). Densitometric analysis of discrete protein bands or of high molecular weight smears of polyubiquitinated protein species was carried out within linear range using ImageJ (NIH).
Ubiquitination and Deubiquitination Assays in Vivo and in Vitro.
At 48 h after transfection, HEK293T or HEK293FT cells were collected and washed in ice-cold PBS and lysed by denaturing 1% SDS RIPA buffer including 1 mM PMSF and protease inhibitor mixture. Cell lysates were heated at 100 °C for 5 min and diluted with SDS-free RIPA buffer (final SDS concentration, 0.1%). Diluted lysates were homogenized and cleared by centrifugation (at >12,000 rpm) at 4 °C for 20 min. Total proteins in suspensions were quantified using the Bio-Rad Dc Protein Assay Kit. Lysates were further normalized against PSD-95 (or PSD-95–Flag) by Western blot before immunoprecipitation and immunoblotting analysis of ubiquitinated species of PSD-95. Immunoprecipitation was done with anti–PSD-95, anti-Flag, anti-Myc, anti-GFP, or, in some experiments (Fig. S2 J and K), anti-Myc–conjugated agarose beads. Immunoblotting was done with anti-UbK63 or anti-HA for K63 polyubiquitinated PSD-95 and in some cases with anti-UbK48 for K48 polyubiquitinated PSD-95. In some experiments, membranes were stripped and reprobed with another antibody. Images were taken using an exposure time that allowed a high molecular weight smear to be clearly seen. Cultured neurons and mouse brain slices were processed similarly.
For the in vitro ubiquitination assay, PSD-95 was purified from denatured lysates of HEK293FT cells overexpressing PSD-95–Flag by immunoprecipitation with anti-Flag. TRAF6 and GFP were purified from lysates of HEK293FT cells overexpressing Myc-TRAF6 or Myc-GFP by immunoprecipitation with anti-Myc. Ubiquitination reactions were done in a 40-µL total volume containing 50 mM Tris⋅HCl (pH 7.5), 2.5 mM MgCl2, 2 mM ATP, 1 µg PSD-95–Flag, 10 µg His-UbK63 (Boston Biochem), 200 ng ubiquitin-activating enzyme Ube1 (Boston Biochem), 800 ng ubiquitin-conjugating enzymes Ubc13/Uev1A (Boston Biochem), and ∼50 ng Myc-TRAF6 or Myc-GFP. After incubation at 37 °C for 2 h, reactions were stopped by the addition of 3× protein loading buffer [190 mM Tris (pH 6.8), 6% SDS, 30% glycerol, 15% β-mercaptoethanol, and 0.003% bromophenol blue dye]. Samples were boiled for 5 min and resolved by 10% SDS/PAGE. Anti-Flag and anti-Myc antibodies were used for Western blotting. For the in vitro deubiquitination assay, PSD-95 was purified from denatured lysates of HEK293FT cells coexpressing PSD-95–Flag and HA-UbK63 by immunoprecipitation with anti-Flag. CYLD and CYLD (C601A) were purified from lysates of HEK293FT cells overexpressing Myc-CYLD or Myc-CYLD (C601A) by immunoprecipitation with anti-Myc. Deubiquitination reactions were performed in a 30-µL total volume containing 50 mM Tris⋅HCl (pH 7.5), 150 mM NaCl, 2 mM EDTA (pH 8.0), 2 mM DTT, Protein A/G beads bound with UbK63–PSD-95–Flag (5 µg), and ∼50 ng Myc-CYLD or Myc-CYLD (C601A). After incubation at 37 °C for 9 h, reactions were stopped by the addition of 3× protein loading buffer. Samples were boiled for 5 min and resolved by 10% SDS/PAGE. Anti-HA, anti-Flag, and anti-Myc antibodies were used for Western blotting.
Reconstitution of Protein Interactions in Vitro.
Denatured HEK293FT cell lysates were prepared as detailed in the ubiquitination assay. PSD-95–Flag and K558R-Flag were purified from lysates of HEK293FT cells coexpressing PSD-95–Flag or K558R-Flag and HA-GFP or HA-UbK63 by immunoprecipitation with anti-Flag. Myc-SPAR was purified from lysates of HEK293FT cells overexpressing Myc-SPAR by immunoprecipitation with anti-Myc. Bound proteins were eluted from Protein A/G beads using IgG elution buffer, and elution samples were neutralized with 1 M Tris⋅HCl and concentrated by a Superfilter Column (Millipore). PBS was used to exchange buffers during the concentration. Small volumes of samples (∼50 µL) were recovered and stored at −80 °C after quantification using the Bio-Rad DC Protein Assay Kit and immunoblotting. Reconstitution reactions were done in 500 µL PBS buffer including 1 mM PMSF, protease inhibitor mixture, 1 µg PSD-95–Flag or K558R-Flag, and ∼500 ng Myc-SPAR. After rotation at 4 °C for 2 h, 5 µg anti-Flag antibody was added to the solution mixture and rotated at 4 °C for 2 h; then protein A/G beads were added and rotated at 4 °C overnight. Immunoprecipitation and immunoblotting were done as described above with anti-Flag and anti-Myc antibodies.
Pulldown Assays.
GST, GST–PSD-95, GST-K544R, GST-K558R, GST-K672R, and His–SPAR-C were expressed and purified from E. coli BL21Gold. For protein (except Src) pulldown using brain lysates, mouse brain proteins were extracted with Nonidet P-40 buffer [50 mM Hepes (pH 7.4), 150 mM NaCl, 1 mM EDTA, 1% (vol/vol) Nonidet P-40] supplemented with a complete protease inhibitor mixture (Roche). For Src pulldown, mouse brain proteins were extracted with PBS supplemented with 1 mM PMSF and protease inhibitor mixture by sonication. After preclearing with Sepharose beads at 4 °C for 1 h, lysates were mixed with GST, GST–PSD-95, GST-K544R, GST-K558R, or GST-K672R bound to glutathione-conjugated Sepharose beads, and the binding reaction was incubated at 4 °C for 4 h. For pulldown of His–SPAR-C with GST–PSD-95 or GST-K558R, the His–SPAR-C eluate was mixed with GST, GST–PSD-95, or GST-K558R bound to glutathione-conjugated Sepharose beads in NETN buffer [100 mM NaCl, 20 mM Tris⋅Cl (pH 8.0), 1 mM EDTA, 0.5% (vol/vol) Nonidet P-40], and incubated at 4 °C for 4 h. Precipitates were washed extensively with lysis buffer. Proteins bound to glutathione beads were eluted and separated on SDS/PAGE 4–12% gel and subjected to Coomassie blue staining or immunoblot analysis. For pulldown experiments using purified Flag-tagged PSD-95 or mutants (Fig. S5D) on mouse brain lysates, PSD-95–Flag, K544R-Flag, or K672R-Flag were purified from denatured lysates of HEK293FT cells overexpressing PSD-95–Flag, K544R-Flag, or K672R-Flag by immunoprecipitation with anti-Flag M2 Affinity Gel. The remaining procedures were the same as for GST pulldown detailed above.
Subcellular Fractionation.
Subcellular fractionation of rat forebrain was performed as previously described (2) with minor modifications. Homogenates (H; the supernatant after the initial low-speed centrifugation), cytosol (S3), crude synaptosomal membranes (P2; containing synaptosomes and mitochondria), the synaptosome fraction (LP1), synaptic plasma membrane (SPM or P3), synaptic vesicle enriched fraction (S4), and PSD (I) (the PSD fraction after one Triton X-100 extraction) were obtained and resolved by SDS/PAGE and analyzed by Western blotting.
Yeast Two-Hybrid Assay.
Yeast strain MaV203 was transformed with plasmids encoding the GAL4 DNA activation domain (AD) fusion proteins (pAD-trp, pAD–trp–SPAR-C, and pAD-T), and GAL4 DNA-binding (DB) domain fusion proteins (pDB–leu–PSD-95, pDB-leu-C3,5S, pDB-leu-C5S, pDB-leu-V7S, pDB-leu-K558R, and pDB-p53). Protein–protein interaction was assayed by growing the yeast on selective medium lacking leucine and tryptophan, following the manufacturer’s manual (Invitrogen). Interaction strength was quantified by X-Gal color-densitometric analysis using ImageJ.
RNAi.
shRNA-expression cassettes targeting TRAF6 (5′-gagaacagatgcctaatca-3′) or CYLD (5′-ggctgtgcgcgcagacttc-3′) were cloned into FUGW-DsRed and pLLox3.7-EGFP vectors, respectively. A scrambled sequence was used as negative control for sh-TRAF6 (in FUGW-DsRed), and a shRNA sequence targeting luciferase (in pLLox3.7-EGFP) was used as an irrelevant control for sh-CYLD. These constructs were used to package lentivirus along with helper vectors or for transfection, where indicated.
Virus Preparation and Infection in Vitro.
For lentivirus production, HEK293FT cells were cotransfected by calcium phosphate with an indicated transfer plasmid (syn-Myc-GFP-syn-GFP, syn–Myc–PSD-95–syn–GFP, syn-Myc-K544R-syn-GFP, syn-Myc-K558R-syn-GFP, syn-Myc-K672R-syn-GFP, syn-Myc-TRAF6-syn-GFP, syn-Myc-TRAF6 C70A-syn-GFP, syn-Myc-CYLD-syn-GFP, syn-Myc-CYLD C601A-syn-GFP, sh-TRAF6, or sh-CYLD or sh-controls) plus two helper vectors, PSHX2 and PMDG2. Supernatants were collected 48 h after transfection and centrifuged to concentrate the virus. For neuron infection, concentrated virus solutions were added onto DIV4–7 hippocampal neurons and grown to DIV13–21 before Western blotting, immunostaining and imaging, or electrophysiology analyses. For coinfections, equal-titer concentrated viruses were thoroughly mixed and added onto DIV7 neurons. One hundred percent efficiency of coinfection into the same neurons is usually obtained under our experimental conditions.
Immunostaining, Confocal Microscopy, and Image Analysis.
Cultured neurons were fixed at DIV15 (or DIV21 in Figs. 1C and 6E and Figs. S1B and S3A) in 4% paraformaldehyde and 5% sucrose in PBS (or methanol in Figs. 1C and 8 A and B and Fig. S3A) for 10–15 min. Permeabilization of cells was done with 0.3% Tween-20 or 0.1% Triton X-100 in PBS. Cells were blocked with 1% goat serum for 1 h. Cells were incubated with primary antibodies at 4 °C for 3 h or overnight followed by incubation with secondary antibodies conjugated with appropriate Alexa dyes for 1 h. For labeling of sGluA1 receptors, cells were fed with anti–GluA1-NT antibody for 30 min before fixation and permeabilization. For AMPAR internalization, sGluA1 receptors were labeled in live neurons by incubation (37 °C, 20 min) with a mouse antibody against the N terminus of GluA1 [1:200 in the conditioned medium, Eagle's minimal essential medium (MEM) + 0.5 mM glutamine]. After the antibody was washed out, cells were treated with control medium (20 μM AMPAR antagonist DNQX and 100 μM mGluR antagonist LY341495 in MEM) for 5 min (26), followed by the application of NMDA (50 μM) for 9 min. NMDA was then washed out, and neurons were incubated in the control medium for an additional 10 min before fixation. Neurons were fixed, and sGluA1 receptors were labeled without permeabilization with a saturating amount of goat anti-mouse Alexa Fluor 568 secondary antibody, followed by permeabilization with 0.1% Triton X-100 for 30 min at room temperature and staining of internalized GluA1 with a goat anti-mouse Alexa Fluor 647 secondary antibody. Cells were mounted on glass slides.
Brain sections were prepared from 4-mo-old C57BL/6J mice. Mice were transcardially perfused with 4% paraformaldehyde in PBS. The dissected whole brains were postfixed in 4% paraformaldehyde and 5% sucrose in PBS at 4 °C overnight. Coronal sections (∼40 µm) were cut using a Microtome HM450 (Thermo Scientific), permeabilized with 0.3% Triton X-100 in PBS, and blocked with 1% BSA for 1 h. They were then incubated with primary antibodies at 4 °C overnight followed by incubation with secondary antibodies conjugated with appropriate Alexa dyes for 1 h. Sections were mounted on glass slides.
For confocal microscopy, image stacks along the z axis were acquired using a Leica TCS SP5 Spectral Confocal Microscope (Leica Microsystems) fitted with a 60× objective (NA 1.4) at pixel resolutions of 512 × 512 or 1,024 × 1,024. Image acquisition settings were kept the same for all scans when fluorescence intensity was compared. Maximum projection images were analyzed using ImageJ and NeuronStudio software. For quantification of the spine/shaft ratio, outlines of dendritic segments and spines or filopodia from neurons transfected with Myc–PSD-95, Myc-K544R, Myc-K558R, Myc-K672R, or Myc-K703R or infected with sh-control, sh-TRAF6, or sh-CYLD were carefully traced. Fluorescence intensity (per pixel) was then determined for the traced areas. The spine/shaft ratio for each spine was defined as the ratio of Myc or endogenous PSD-95 fluorescence intensity of the spine divided by the respective fluorescence intensity of the parent shaft. Spine types and filopodia were visually identified and manually classified following established guidelines. Briefly, spines with a medium-large head (≥0.5 μm in diameter) and a neck 0.3–2.0 μm in length were classified as mushroom; spines with a small-medium head (∼0.4–1.0 μm) and a long neck (>2.0 μm) were classified thin. Spines with a distinct head but without a neck were classified stubby. Protrusions without a distinct head and longer than 0.7 μm were classified as filopodia. For quantification of sGluA1 clustering, the fluorescence intensity of carefully traced sGluA1 clusters on randomly selected dendrites was measured for neurons (DIV15) transfected with PSD-95–Flag, K544R-Flag, K558-Flag, or K672R-Flag; sGluA1 clusters from neighboring untransfected neurons on the same culture dish were used as control. Similar quantification was done for synapsin I clustering, except that the fluorescence intensity of these presynaptic clusters opposing postsynaptic sites of PSD-95–Flag or K558R-Flag transfected neurons and untransfected neurons were compared. Cluster intensities for other native (e.g., PSD-95, K63-polyUb, and SPAR) and exogenous (e.g., HA-SPAR) proteins were measured similarly. For quantification of dendritic K63-polyUb, PSD-95, PSD-95–Flag, HA-UbK63 and HA-UbR63, spine HA-UbK63 and HA-UbR63, or whole-cell K63-polyUb fluorescence intensity, outlines of selected dendrites, spines, or cells were carefully traced, and the fluorescence signal was measured for the traced areas. For quantification of dendritic spine density (the number of total spines per 10 μm), GFP-expressing dendrites randomly selected from DIV21 neurons transfected or cotransfected with specified plasmids were measured. Inclusion of separately expressed “free” GFP was necessary to fill and morphologically define dendritic shafts and protrusions. For quantification of spine size, GFP-filled spine heads of randomly selected mushroom, stubby, and thin spines on selected dendritic branches were carefully traced, and traced areas were measured. For quantification of NMDA-triggered sGluA1 internalization, all dendrites of each randomly selected GFP-expressing pyramidal neuron were traced, and the fluorescence intensity in the red (surface) and blue (internalized) channels in the traced areas were measured. Dendritic sGluA1 internalization was defined as the ratio of endocytosed (red) to total (red + blue) receptors. NMDA-induced sGluA1 internalization was expressed as the level normalized to constitutive sGluA1 internalization in unstimulated control cells run simultaneously.
For data processing, we traced the entire dendritic tree of a neuron when measuring sGluA1 internalization. For others, one dendritic segment (60 μm or longer) per neuron was collected. Individual cluster or spine measurements were first grouped and averaged per neuron. Means from multiple neurons in the same group were then averaged to obtain a population mean and presented as mean ± SEM. Background was subtracted before fluorescence intensity measurement as necessary. Fluorescence intensities were measured as pixel intensities, defined as integrated pixel intensities (i.e., total eight-bit intensity values across all pixels contained in traced areas) in specific channels divided by the total number of pixels. All groups to be compared were run simultaneously using cells from the same culture preparations and transfection conditions. At least three batches of separately grown cultures were used to obtain the total cells imaged and quantified in each figure. For presentation, representative images were processed using Adobe Photoshop by adjusting brightness and contrast using the same parameters for all conditions in each experiment to enhance visual display.
Stereotaxic Virus Injection, Histology, and Analysis.
PSD-95–KO mice (at P20) were deeply anesthetized and placed in a stereotaxic frame. Lentivirus (1 μL) expressing Myc-GFP, Myc–PSD-95 (and GFP), or Myc-K558R (and GFP) (driven by separate synapsin promoters within the same vectors; see Plasmids section above) were injected bilaterally into the hippocampus. The coordinates of injections were (from bregma): anteroposterior, −0.27; mediolateral, ±0.26; dorsoventral, −0.16 mm for young brains. Two weeks after surgery, mice (P34) were killed, and their brains were collected and fixed. Coronal sections (80–100 µm) were cut for spine morphology analysis, and 40-µm sections were cut for immunostaining with anti-synapsin I and anti–PSD-95. Sections were mounted on glass slides. For spine analysis, confocal image stacks of ∼10 μm were acquired using a Leica SP5 confocal microscope fitted with a 40× oil objective at pixel resolutions of 512 × 512. Numbers of total spines (identified by GFP-filled protrusions) per 10-μm apical dendrites (striatum radiatum) were measured from GFP-expressing CA1 pyramidal neurons infected with Myc–PSD-95, Myc-K558R, or Myc-GFP. For synapsin I puncta analysis, image stacks of 5.024 μm along the z axis were acquired using the 40× oil objective on the SP5 microscope at pixel resolutions of 512 × 512 from GFP-labeled striatum radiatum in CA1. Puncta containing both synapsin I and Myc–PSD-95 or Myc-K558R were traced, and fluorescence intensity was measured. Images analysis was done using ImageJ as above.
Protein Structure and Modeling.
The ribbon diagram of the PSD-95 GK domain in complex with a phosphor-Lgl2 peptide is drawn based on our recently published work (46). For GK modeling, models were generated using Phyre2 (www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index), a homology modeling server which uses profile–profile matching algorithms to generate models based on sequence (63). Images were generated using UCSF Chimera (64). 3D superimposition rmsd was calculated in Chimera using the match-align tool (65).
Electrophysiology.
Whole-cell patch-clamp recordings were performed on cultured rat hippocampal pyramidal neurons at DIV13–15 after transfection or infection at DIV7. Pyramidal neurons were identified by their morphology. Cells were superfused at room temperature with an oxygenated (95% O2 and 5% CO2) extracellular solution containing (in mM) 126 NaCl, 25 NaHCO3, 11 glucose, 2.5 KCl, 1.2 NaH2PO4, 1.2 MgSO4, and 2.5 CaCl2. Recording pipettes (4.5–5.5 MΩ) were filled with an intracellular solution containing the following (in mM): 142 Cs-gluconate, 8 NaCl, 10 Hepes, 0.4 EGTA, 2.5 QX-314 [N-(2,6-dimethylphenylcarbamoylmethyl)triethylammonium bromide], 2 Mg-ATP, and 0.25 GTP-Tris (pH 7.25) (with CsOH). mEPSCs were recorded at −60 mV with picrotoxin (50 μM) and TTX (1 μM) present in the superfusion medium. Recordings of NMDA-triggered depression of mEPSCs were performed as previously described (50) with minor modifications. Specifically, TTX in the bath was replaced with lidocaine (200 μM). After 5–8 min baseline mEPSC recording at −60 mV, cells were stimulated by perfusing with a solution containing NMDA (50 μM) and glycine (20 μM), but not lidocaine, for 5 min. NMDA was then washed out by replacing the stimulation solution with the original lidocaine-containing solution for 7–9 min. During NMDA treatment and washout, cells were allowed at their native membrane potential without voltage clamping. After washout, cells were clamped again at −60 mV for another 7- to 15-min mEPSC recording. cLTD was calculated as the percentage change of mEPSC frequency after NMDA treatment over that of baseline.
All electrophysiological data were acquired using an Axon Multiclamp 700B amplifier (Molecular Devices) with Digidata 1440A and pClamp software (version 10; Molecular Devices). Signals were filtered at 1 kHz and digitized at 10–20 kHz. mEPSCs were analyzed by Mini Analysis 6 (Synaptosoft). All recordings were performed with the electrophysiologist blinded to the plasmids that had been transfected into the recorded neurons.
Statistical Analysis.
All data are expressed as mean ± SEM. Comparisons involving two groups were performed with two-sided unpaired t tests. One-factor ANOVA was performed for three or more data groups, followed by post hoc Tukey's multiple comparison tests or Dunnett’s tests for comparison with a specific control, as indicated. The significance level was set at 0.05.
Acknowledgments
We thank R. A. Nicoll, Z. J. Chen, D. Baltimore, and D. T. Pak for providing reagents and Eric Zajicek for helping with protein modeling. This study was supported by NIH Grants DA032283, MH106489, and RR026761 (to W.-D.Y.) and RR000168 (to the New England Primate Research Center/Harvard Medical School). M.Z. is supported by Research Grants Council of Hong Kong Grant AoE-M09-12.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1620153114/-/DCSupplemental.
References
- 1.Husnjak K, Dikic I. Ubiquitin-binding proteins: Decoders of ubiquitin-mediated cellular functions. Annu Rev Biochem. 2012;81:291–322. doi: 10.1146/annurev-biochem-051810-094654. [DOI] [PubMed] [Google Scholar]
- 2.Chen ZJ, Sun LJ. Nonproteolytic functions of ubiquitin in cell signaling. Mol Cell. 2009;33:275–286. doi: 10.1016/j.molcel.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 3.Mukhopadhyay D, Riezman H. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science. 2007;315:201–205. doi: 10.1126/science.1127085. [DOI] [PubMed] [Google Scholar]
- 4.Komander D, Clague MJ, Urbé S. Breaking the chains: Structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10:550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- 5.Chung JY, Park YC, Ye H, Wu H. All TRAFs are not created equal: Common and distinct molecular mechanisms of TRAF-mediated signal transduction. J Cell Sci. 2002;115:679–688. doi: 10.1242/jcs.115.4.679. [DOI] [PubMed] [Google Scholar]
- 6.Bignell GR, et al. Identification of the familial cylindromatosis tumour-suppressor gene. Nat Genet. 2000;25:160–165. doi: 10.1038/76006. [DOI] [PubMed] [Google Scholar]
- 7.Kim E, Sheng M. PDZ domain proteins of synapses. Nat Rev Neurosci. 2004;5:771–781. doi: 10.1038/nrn1517. [DOI] [PubMed] [Google Scholar]
- 8.DiAntonio A, Hicke L. Ubiquitin-dependent regulation of the synapse. Annu Rev Neurosci. 2004;27:223–246. doi: 10.1146/annurev.neuro.27.070203.144317. [DOI] [PubMed] [Google Scholar]
- 9.Tai HC, Schuman EM. Ubiquitin, the proteasome and protein degradation in neuronal function and dysfunction. Nat Rev Neurosci. 2008;9:826–838. doi: 10.1038/nrn2499. [DOI] [PubMed] [Google Scholar]
- 10.Mabb AM, Ehlers MD. Ubiquitination in postsynaptic function and plasticity. Annu Rev Cell Dev Biol. 2010;26:179–210. doi: 10.1146/annurev-cellbio-100109-104129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bingol B, Sheng M. Deconstruction for reconstruction: The role of proteolysis in neural plasticity and disease. Neuron. 2011;69:22–32. doi: 10.1016/j.neuron.2010.11.006. [DOI] [PubMed] [Google Scholar]
- 12.Ehlers MD. Activity level controls postsynaptic composition and signaling via the ubiquitin-proteasome system. Nat Neurosci. 2003;6:231–242. doi: 10.1038/nn1013. [DOI] [PubMed] [Google Scholar]
- 13.Colledge M, et al. Ubiquitination regulates PSD-95 degradation and AMPA receptor surface expression. Neuron. 2003;40:595–607. doi: 10.1016/s0896-6273(03)00687-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pak DT, Sheng M. Targeted protein degradation and synapse remodeling by an inducible protein kinase. Science. 2003;302:1368–1373. doi: 10.1126/science.1082475. [DOI] [PubMed] [Google Scholar]
- 15.Shin SM, et al. GKAP orchestrates activity-dependent postsynaptic protein remodeling and homeostatic scaling. Nat Neurosci. 2012;15:1655–1666. doi: 10.1038/nn.3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ehrlich I, Klein M, Rumpel S, Malinow R. PSD-95 is required for activity-driven synapse stabilization. Proc Natl Acad Sci USA. 2007;104:4176–4181. doi: 10.1073/pnas.0609307104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsai NP, et al. Multiple autism-linked genes mediate synapse elimination via proteasomal degradation of a synaptic scaffold PSD-95. Cell. 2012;151:1581–1594. doi: 10.1016/j.cell.2012.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Na CH, et al. Synaptic protein ubiquitination in rat brain revealed by antibody-based ubiquitome analysis. J Proteome Res. 2012;11:4722–4732. doi: 10.1021/pr300536k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dosemeci A, Thein S, Yang Y, Reese TS, Tao-Cheng JH. CYLD, a deubiquitinase specific for lysine63-linked polyubiquitins, accumulates at the postsynaptic density in an activity-dependent manner. Biochem Biophys Res Commun. 2013;430:245–249. doi: 10.1016/j.bbrc.2012.10.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen X, et al. PSD-95 is required to sustain the molecular organization of the postsynaptic density. J Neurosci. 2011;31:6329–6338. doi: 10.1523/JNEUROSCI.5968-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elias GM, Nicoll RA. Synaptic trafficking of glutamate receptors by MAGUK scaffolding proteins. Trends Cell Biol. 2007;17:343–352. doi: 10.1016/j.tcb.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 22.Béïque JC, Andrade R. PSD-95 regulates synaptic transmission and plasticity in rat cerebral cortex. J Physiol. 2003;546:859–867. doi: 10.1113/jphysiol.2002.031369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stein V, House DR, Bredt DS, Nicoll RA. Postsynaptic density-95 mimics and occludes hippocampal long-term potentiation and enhances long-term depression. J Neurosci. 2003;23:5503–5506. doi: 10.1523/JNEUROSCI.23-13-05503.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ehrlich I, Malinow R. Postsynaptic density 95 controls AMPA receptor incorporation during long-term potentiation and experience-driven synaptic plasticity. J Neurosci. 2004;24:916–927. doi: 10.1523/JNEUROSCI.4733-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu W, et al. Molecular dissociation of the role of PSD-95 in regulating synaptic strength and LTD. Neuron. 2008;57:248–262. doi: 10.1016/j.neuron.2007.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bhattacharyya S, Biou V, Xu W, Schlüter O, Malenka RC. A critical role for PSD-95/AKAP interactions in endocytosis of synaptic AMPA receptors. Nat Neurosci. 2009;12:172–181. doi: 10.1038/nn.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.El-Husseini AE, Schnell E, Chetkovich DM, Nicoll RA, Bredt DS. PSD-95 involvement in maturation of excitatory synapses. Science. 2000;290:1364–1368. [PubMed] [Google Scholar]
- 28.Pak DT, Yang S, Rudolph-Correia S, Kim E, Sheng M. Regulation of dendritic spine morphology by SPAR, a PSD-95-associated RapGAP. Neuron. 2001;31:289–303. doi: 10.1016/s0896-6273(01)00355-5. [DOI] [PubMed] [Google Scholar]
- 29.Kim E, et al. GKAP, a novel synaptic protein that interacts with the guanylate kinase-like domain of the PSD-95/SAP90 family of channel clustering molecules. J Cell Biol. 1997;136:669–678. doi: 10.1083/jcb.136.3.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takeuchi M, et al. SAPAPs. A family of PSD-95/SAP90-associated proteins localized at postsynaptic density. J Biol Chem. 1997;272:11943–11951. doi: 10.1074/jbc.272.18.11943. [DOI] [PubMed] [Google Scholar]
- 31.Craven SE, El-Husseini AE, Bredt DS. Synaptic targeting of the postsynaptic density protein PSD-95 mediated by lipid and protein motifs. Neuron. 1999;22:497–509. doi: 10.1016/s0896-6273(00)80705-9. [DOI] [PubMed] [Google Scholar]
- 32.Ho GP, et al. S-nitrosylation and S-palmitoylation reciprocally regulate synaptic targeting of PSD-95. Neuron. 2011;71:131–141. doi: 10.1016/j.neuron.2011.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim MJ, et al. Synaptic accumulation of PSD-95 and synaptic function regulated by phosphorylation of serine-295 of PSD-95. Neuron. 2007;56:488–502. doi: 10.1016/j.neuron.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 34.Steiner P, et al. Destabilization of the postsynaptic density by PSD-95 serine 73 phosphorylation inhibits spine growth and synaptic plasticity. Neuron. 2008;60:788–802. doi: 10.1016/j.neuron.2008.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vogl AM, et al. Neddylation inhibition impairs spine development, destabilizes synapses and deteriorates cognition. Nat Neurosci. 2015;18:239–251. doi: 10.1038/nn.3912. [DOI] [PubMed] [Google Scholar]
- 36.Migaud M, et al. Enhanced long-term potentiation and impaired learning in mice with mutant postsynaptic density-95 protein. Nature. 1998;396:433–439. doi: 10.1038/24790. [DOI] [PubMed] [Google Scholar]
- 37.Yao WD, et al. Identification of PSD-95 as a regulator of dopamine-mediated synaptic and behavioral plasticity. Neuron. 2004;41:625–638. doi: 10.1016/s0896-6273(04)00048-0. [DOI] [PubMed] [Google Scholar]
- 38.Newton K, et al. Ubiquitin chain editing revealed by polyubiquitin linkage-specific antibodies. Cell. 2008;134:668–678. doi: 10.1016/j.cell.2008.07.039. [DOI] [PubMed] [Google Scholar]
- 39.El-Husseini AE, et al. Dual palmitoylation of PSD-95 mediates its vesiculotubular sorting, postsynaptic targeting, and ion channel clustering. J Cell Biol. 2000;148:159–172. doi: 10.1083/jcb.148.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kalia LV, Pitcher GM, Pelkey KA, Salter MW. PSD-95 is a negative regulator of the tyrosine kinase Src in the NMDA receptor complex. EMBO J. 2006;25:4971–4982. doi: 10.1038/sj.emboj.7601342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fernández E, et al. Targeted tandem affinity purification of PSD-95 recovers core postsynaptic complexes and schizophrenia susceptibility proteins. Mol Syst Biol. 2009;5:269. doi: 10.1038/msb.2009.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peng J, et al. Semiquantitative proteomic analysis of rat forebrain postsynaptic density fractions by mass spectrometry. J Biol Chem. 2004;279:21003–21011. doi: 10.1074/jbc.M400103200. [DOI] [PubMed] [Google Scholar]
- 43.Jordan BA, et al. Identification and verification of novel rodent postsynaptic density proteins. Mol Cell Proteomics. 2004;3:857–871. doi: 10.1074/mcp.M400045-MCP200. [DOI] [PubMed] [Google Scholar]
- 44.Masuko N, et al. Interaction of NE-dlg/SAP102, a neuronal and endocrine tissue-specific membrane-associated guanylate kinase protein, with calmodulin and PSD-95/SAP90. A possible regulatory role in molecular clustering at synaptic sites. J Biol Chem. 1999;274:5782–5790. doi: 10.1074/jbc.274.9.5782. [DOI] [PubMed] [Google Scholar]
- 45.Zhu J, et al. Guanylate kinase domains of the MAGUK family scaffold proteins as specific phospho-protein-binding modules. EMBO J. 2011;30:4986–4997. doi: 10.1038/emboj.2011.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu J, et al. Phosphorylation-dependent interaction between tumor suppressors Dlg and Lgl. Cell Res. 2014;24:451–463. doi: 10.1038/cr.2014.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Opazo P, Sainlos M, Choquet D. Regulation of AMPA receptor surface diffusion by PSD-95 slots. Curr Opin Neurobiol. 2012;22:453–460. doi: 10.1016/j.conb.2011.10.010. [DOI] [PubMed] [Google Scholar]
- 48.Futai K, et al. Retrograde modulation of presynaptic release probability through signaling mediated by PSD-95-neuroligin. Nat Neurosci. 2007;10:186–195. doi: 10.1038/nn1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee HK, Kameyama K, Huganir RL, Bear MF. NMDA induces long-term synaptic depression and dephosphorylation of the GluR1 subunit of AMPA receptors in hippocampus. Neuron. 1998;21:1151–1162. doi: 10.1016/s0896-6273(00)80632-7. [DOI] [PubMed] [Google Scholar]
- 50.Beattie EC, et al. Regulation of AMPA receptor endocytosis by a signaling mechanism shared with LTD. Nat Neurosci. 2000;3:1291–1300. doi: 10.1038/81823. [DOI] [PubMed] [Google Scholar]
- 51.Xu P, et al. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 2009;137:133–145. doi: 10.1016/j.cell.2009.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hornbeck PV, et al. PhosphoSitePlus, 2014: Mutations, PTMs and recalibrations. Nucleic Acids Res. 2015;43:D512–D520. doi: 10.1093/nar/gku1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grabbe C, Husnjak K, Dikic I. The spatial and temporal organization of ubiquitin networks. Nat Rev Mol Cell Biol. 2011;12:295–307. doi: 10.1038/nrm3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hunter T. The age of crosstalk: Phosphorylation, ubiquitination, and beyond. Mol Cell. 2007;28:730–738. doi: 10.1016/j.molcel.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 55.Chen X, et al. Mass of the postsynaptic density and enumeration of three key molecules. Proc Natl Acad Sci USA. 2005;102:11551–11556. doi: 10.1073/pnas.0505359102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bingol B, et al. Autophosphorylated CaMKIIalpha acts as a scaffold to recruit proteasomes to dendritic spines. Cell. 2010;140:567–578. doi: 10.1016/j.cell.2010.01.024. [DOI] [PubMed] [Google Scholar]
- 57.Bingol B, Schuman EM. A proteasome-sensitive connection between PSD-95 and GluR1 endocytosis. Neuropharmacology. 2004;47:755–763. doi: 10.1016/j.neuropharm.2004.07.028. [DOI] [PubMed] [Google Scholar]
- 58.Thein S, et al. CaMKII mediates recruitment and activation of the deubiquitinase CYLD at the postsynaptic density. PLoS One. 2014;9:e91312. doi: 10.1371/journal.pone.0091312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Beattie EC, et al. Control of synaptic strength by glial TNFalpha. Science. 2002;295:2282–2285. doi: 10.1126/science.1067859. [DOI] [PubMed] [Google Scholar]
- 60.Stellwagen D, Malenka RC. Synaptic scaling mediated by glial TNF-alpha. Nature. 2006;440:1054–1059. doi: 10.1038/nature04671. [DOI] [PubMed] [Google Scholar]
- 61.Kramer LB, et al. UEV-1 is an ubiquitin-conjugating enzyme variant that regulates glutamate receptor trafficking in C. elegans neurons. PLoS One. 2010;5:e14291. doi: 10.1371/journal.pone.0014291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang J, et al. Inhibition of the dopamine D1 receptor signaling by PSD-95. J Biol Chem. 2007;282:15778–15789. doi: 10.1074/jbc.M611485200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pettersen EF, et al. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 65.Meng EC, Pettersen EF, Couch GS, Huang CC, Ferrin TE. Tools for integrated sequence-structure analysis with UCSF Chimera. BMC Bioinformatics. 2006;7:339. doi: 10.1186/1471-2105-7-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McGee AW, et al. Structure of the SH3-guanylate kinase module from PSD-95 suggests a mechanism for regulated assembly of MAGUK scaffolding proteins. Mol Cell. 2001;8:1291–1301. doi: 10.1016/s1097-2765(01)00411-7. [DOI] [PubMed] [Google Scholar]