Significance
Invasion is a major cause of cancer mortality, as exemplified by metastatic spread of peripheral malignancies or local intracranial invasion of glioblastoma. While individual mediators of invasion are identified, functional or structural interactions between these mediators remain undefined. We identified a structural cross-activating c-Met/β1 integrin complex that promotes breast cancer metastases and invasive resistance of glioblastoma to the antiangiogenic therapy bevacizumab. We show that tumor cells adapt to their microenvironmental stressors by usurping c-Met and β1 integrin, with c-Met displacing α5 integrin from β1 integrin to form a c-Met/β1 complex with far greater fibronectin affinity than α5β1 integrin. These findings challenge conventional thinking about integrin–ligand interactions and define a molecular target for disrupting metastases or invasive oncologic resistance.
Keywords: resistance, glioblastoma, angiogenesis, hypoxia, invasion
Abstract
The molecular underpinnings of invasion, a hallmark of cancer, have been defined in terms of individual mediators but crucial interactions between these mediators remain undefined. In xenograft models and patient specimens, we identified a c-Met/β1 integrin complex that formed during significant invasive oncologic processes: breast cancer metastases and glioblastoma invasive resistance to antiangiogenic VEGF neutralizing antibody, bevacizumab. Inducing c-Met/β1 complex formation through an engineered inducible heterodimerization system promoted features crucial to overcoming stressors during metastases or antiangiogenic therapy: migration in the primary site, survival under hypoxia, and extravasation out of circulation. c-Met/β1 complex formation was up-regulated by hypoxia, while VEGF binding VEGFR2 sequestered c-Met and β1 integrin, preventing their binding. Complex formation promoted ligand-independent receptor activation, with integrin-linked kinase phosphorylating c-Met and crystallography revealing the c-Met/β1 complex to maintain the high-affinity β1 integrin conformation. Site-directed mutagenesis verified the necessity for c-Met/β1 binding of amino acids predicted by crystallography to mediate their extracellular interaction. Far-Western blotting and sequential immunoprecipitation revealed that c-Met displaced α5 integrin from β1 integrin, creating a complex with much greater affinity for fibronectin (FN) than α5β1. Thus, tumor cells adapt to microenvironmental stressors induced by metastases or bevacizumab by coopting receptors, which normally promote both cell migration modes: chemotaxis, movement toward concentrations of environmental chemoattractants, and haptotaxis, movement controlled by the relative strengths of peripheral adhesions. Tumor cells then redirect these receptors away from their conventional binding partners, forming a powerful structural c-Met/β1 complex whose ligand-independent cross-activation and robust affinity for FN drive invasive oncologic processes.
Cooperation between integrins and receptor tyrosine kinases (RTKs) contributes to migration in normal and cancer cells. Previous studies have demonstrated colocalization of RTK c-Met with α5 or β1 integrin (1, 2) but the mechanisms and biologic consequences of these interactions remain undetermined. Invasion of tumor cells is directed by haptotaxis, cellular locomotion in response to concentration gradients of adhesive molecules in the extracellular matrix (ECM), and chemotaxis, cellular locomotion in response to concentration gradients of diffusible factors. Given the ability of integrins and c-Met to mediate haptotaxis and chemotaxis, respectively, these complementary functions suggest that interaction between c-Met and integrins could have a significant biologic impact on tumor cell migration.
While chemotactic c-Met and haptotactic integrins like α5 or β1 each contribute individually to tumor cell migration, the mechanism through which they interact to regulate migration and the specific aspects of cancer cell biology that rely on their interaction remain uncertain. Here, we demonstrate that c-Met and β1, not α5, integrin interact physically and functionally during two invasive cancer processes: metastases, the cause of 90% of cancer deaths (3), and glioblastoma (GBM) resistance to VEGF neutralizing antibody bevacizumab, a common occurrence after short-lived response to this agent (4–9).
Results
Physical Interaction Between c-Met and β1 Integrin Increases During Metastases.
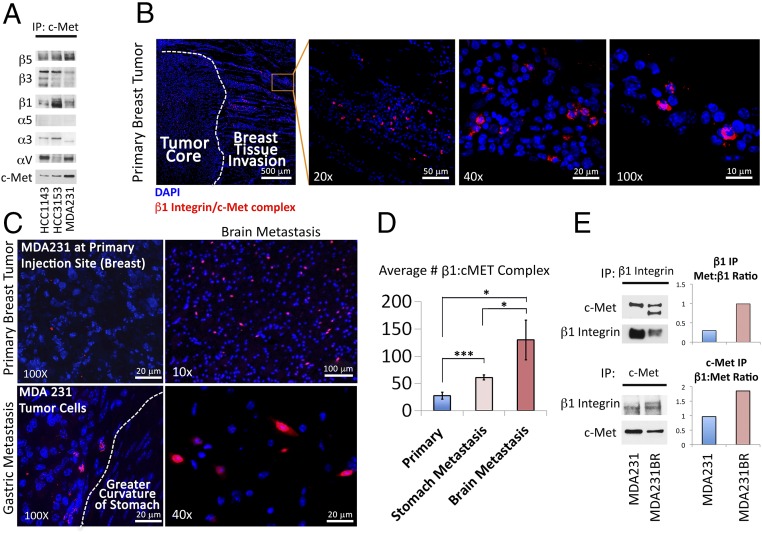
To determine which integrins interacted with c-Met in breast cancer cells, we performed a Western blot in cultured HCC1143, HCC3153, and MDA-MB-231 human breast adenocarcinoma cells, which revealed that c-Met was expressed in all three lines, while, of a panel of screened integrins, αV, α3, α5, β1, β3, and β5 integrins were expressed in all three lines (SI Appendix, Figs. S1 and S2). Immunoprecipitation then revealed that, of these six integrins, β1 integrin was the most oncologically pertinent one (5, 10) that bound c-Met in all three lines (Fig. 1A and SI Appendix, Fig. S3). To determine if this c-Met/β1 integrin interaction increased during metastases, we implanted MDA-MB-231 cells into the mammary fat pads of immunodeficient mice (n = 5), after which spontaneous metastases arose in one mouse. Using proximity ligation assays (PLAs), we detected c-Met/β1 integrin complex formation at the invasive front in all four primary tumors, particularly in the primary tumor with gross metastases (Fig. 1B and SI Appendix, Fig. S4). PLA revealed more signal (red dots indicative of c-Met/β1 integrin complex) in gastric (Fig. 1C and SI Appendix, Fig. S5) and brain (Fig. 1C and SI Appendix, Fig. S6) metastases relative to the primary tumor that metastasized, with quantification revealing more than double the amount of PLA signal in the gastric metastasis (P = 0.0006) and nearly fivefold more PLA signal in the brain metastasis (P = 0.01) relative to the primary tumor (Fig. 1D). Similarly, immunoprecipitation (IP) of lysates from MDA-MB-231-BR, an MDA-MB-231 derived cell line selected through serial propagation in vivo for its propensity for brain metastases (11), exhibited greater c-Met/β1 integrin complex in culture than MDA-MB-231 cells (Fig. 1E), suggesting durability of the complex within the metastatic phenotype.
Fig. 1.
A c-Met/β1 integrin complex forms in breast cancer metastases in a xenograft model. (A) c-Met IP of HCC1143, HCC3153, and MDA-MB-231 breast cancer cells to assess binding of ubiquitously expressed integrins (see SI Appendix, Fig. S2 for Western blot of full integrin panel in all three lines) to c-Met in each cell line. (B) PLA revealed c-Met/β1 integrin complex formation at the invasive edge of a MDA-MB-231 mammary pad xenograft. (C) Representative PLA images of c-Met/β1 complexes in the primary mammary pad tumor (Upper Left), gastric metastasis (Lower Left), and brain metastasis (Upper and Lower Right). (D) Quantification revealed over double the number of PLA signals per 100× field in the gastric metastasis (P = 0.0006) and nearly fivefold more signal in the brain metastasis (P = 0.01) versus primary tumor (n = 4 fields per tumor). (E) c-Met and β1 integrin IPs each revealed more c-Met/β1 integrin complex in cultured MDA-MB-231-BR brain-seeking cells versus MDA-MB-231 cells. *P < 0.05; ***P < 0.001.
The c-Met/β1 Complex Promotes Breast Cancer Cell Migration.
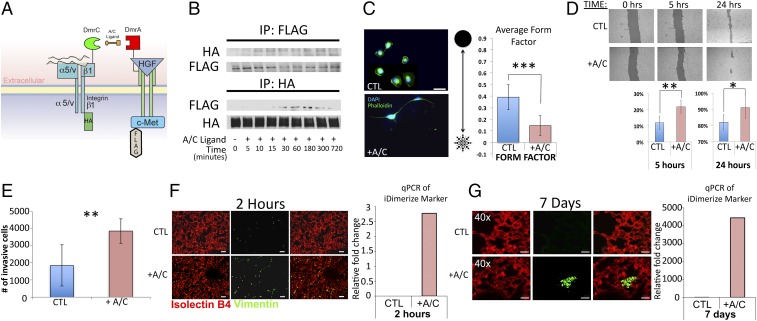
To determine if the c-Met/β1 complex detected in metastases promotes tumor cell motility, we engineered MDA-MB-231 cells to express β1 integrin and c-Met fused to FRB (DmrC) and FKBP (DmrA), respectively, creating MDA-MB-231-iDimerize-c-Met-β1 cells, which enabled us to increase c-Met/β1 complex formation using AP21967 (A/C ligand heterodimerizer), a derivative of rapamycin (12) (Fig. 2 A and B and SI Appendix, Figs. S7 and S8). AP21967 treatment changed morphology of MDA-MB-231-iDimerize-c-Met-β1 cells by giving rise to extensive protrusions emnating from the cytoplasm, pushing the cells toward the stellate morphology that is seen in invading mesenchymal cells (13), as evidenced by decreased circularity shape factor (P = 6 × 10−7) (Fig. 2C). Consistent with altered morphology being conducive to increased cell migration, AP21967 increased motility of MDA-MB-231-iDimerize-c-Met-β1 cells in scratch assays (P = 0.001 at 5 h, P = 0.02 at 24 h) (Fig. 2D) and increased invasion in Matrigel chambers (P = 0.007) (Fig. 2E and SI Appendix, Fig. S9).
Fig. 2.
Inducing c-Met/β1 integrin binding in breast cancer cells increases tumor cell migration and extravasation out of circulation. (A) MDA-MB-231 cells were engineered to express HA-tagged β1 integrin and FLAG-tagged c-Met fused to FRB (DmrC) and FKBP (DmrA), respectively, and treated with AP21967 (A/C heterodimerizer) to induce c-Met/β1 binding. (B) MDA-MB-231-iDimerize-c-Met-β1 cells were treated with AP21967 or vehicle (control), after which HA and FLAG IP confirmed that AP21967 induces c-Met/β1 binding. (C) MDA-MB-231-iDimerize-c-Met-β1 cells were assessed for morphology via a form factor plugin on ImageJ, revealing decreased circularity shape factor after AP21967 treatment (P = 6 × 10−7; n = 12 cells per group). (D) MDA-MB-231-iDimerize-c-Met-β1 cells were scratched at time = 0 and assessed after 5 and 24 h via mosaic imaging and analyzed using T-Scratch software to confirm that AP21967 induced migration (P = 0.001 at 5 h, P = 0.02 at 24 h) (n = 8 per group). (E) AP21967 also increased MDA-MB-231-iDimerize-c-Met-β1 invasion in Matrigel (n = 6 per group; P = 0.007). (F) MDA-MB-231-iDimerize-c-Met-β1 cells were pretreated for 2 h with or without AP21967, followed by tail vein injection into NSG mice (n = 6 mice per group). After 2 h, mice receiving AP21967-pretreated cells exhibited more human vimentin staining by immunohistochemistry versus those pretreated with vehicle, as shown in representative images here and quantified in SI Appendix, Fig. S9 (P = 0.02), with qPCR revealing the FKBP–c-Met fusion sequence in lungs of all mice receiving AP21967-pretreated cells, with the lack of PCR signal in all vehicle-treated mice precluding statistical comparison. (G) Analysis of lungs from the same experiment as F at 7 d postinjection revealed further increased human vimentin staining after tail vein injection of cells pretreated with AP21967 compared with no staining when injecting vehicle-pretreated cells (lack of staining in control group prevented statistical comparison). Similarly, qPCR at 7 d revealed increased tumor-specific FKBP–c-Met fusion sequence in mice receiving AP21967-treated cells, with lack of signal in most mice receiving vehicle-treated cells precluding statistical comparison. *P < 0.05; **P < 0.01; ***P < 0.001.
We then determined if inducing c-Met/β1 integrin binding promoted extravasation of MDA-MB-231 cells out of the circulation, a crucial step of the metastatic cascade. MDA-MB-231-iDimerize-c-Met-β1 cells were preincubated with AP21967 for 2 h and then injected i.v. via tail vein injection. Animals were killed 2 h and 7 d after injection, with increased human MDA-MB-231-iDimerize-c-Met-β1 cells detected in the lungs of mice receiving cells that were pretreated with AP21967 by two independent methods: immunostaining the lungs for human vimentin and PCR for the FKBP-c-Met fusion sequences, which would be unique to the implanted tumor cells. Two hours after tail vein injection of tumor cells, lung immunostaining revealed increased human vimentin staining when injecting cells pretreated with AP21967 versus those pretreated with vehicle (P = 0.02; Fig. 2F and SI Appendix, Fig. S10) and PCR for the FKBP-c-Met fusion sequences revealed no detectable signal in all mice receiving cells pretreated with vehicle, compared with consistent signal in all mice receiving cells pretreated with AP21967 (Fig. 2F). Seven days after tail vein injection of cells, there was increased lung human vimentin staining relative to 2 h when injecting cells pretreated with AP21967 compared with no detectable staining when injecting cells pretreated with vehicle (Fig. 2G and SI Appendix, Fig. S11). PCR at this 7-d time point revealed dramatic increase in the FKBP-c-Met fusion sequence transcript in all mice 7 d after receiving cells pretreated with AP21967 while only two of five mice receiving cells pretreated with vehicle had detectable transcript at this time point (Fig. 2G). Thus, while baseline MDA-MB-231 cells failed to establish metastases after initial circulatory passage through the lungs, inducing c-Met/β1 integrin complex formation in these cells caused them to reliably establish pulmonary metastases.
c-Met/β1 Complex Formation Increases During Invasive Resistance in Glioblastoma.
Each organ has unique extracellular matrix milieus that could be variably receptive to invasion by tumor cells with c-Met/β1 integrin complex formation. Due to the particularly significant levels of c-Met/β1 integrin complex we noted in brain metastases (Fig. 1 C and D and SI Appendix, Fig. S6), we investigated the role of the c-Met/β1 complex in the invasiveness of GBM, the most common malignant primary brain tumor. Unlike other aggressive cancers, GBM does not metastasize but rather invades locally. Because resistance to the antiangiogenic agent bevacizumab is a common driving event promoting transformation of GBM to a highly invasive phenotype, we investigated the role of the c-Met/β1 complex in bevacizumab resistance in GBM.
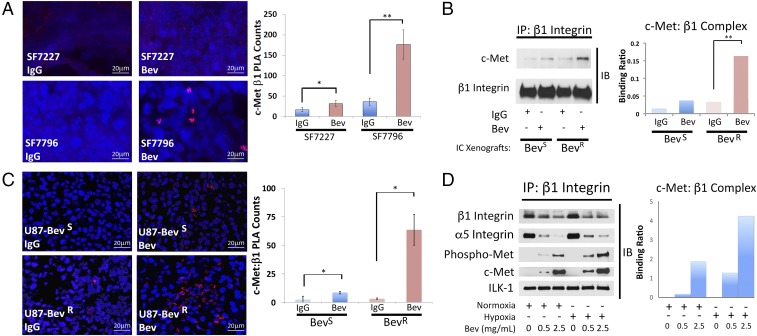
We analyzed c-Met/β1 complex formation in two GBM xenograft models of bevacizumab resistance created by our group. Our first model transfers the effects of prolonged antiangiogenic therapy directly from the patient to the mouse in the form of patient-derived xenografts (PDXs) that maintain the sensitivity or resistance to bevacizumab found in the patient tumor they derive from, while our second model recapitulates prolonged antiangiogenic therapy in mice. In the first model, bevacizumab caused a 5-fold increase in c-Met/β1 complex formation by PLA in intracranial resistant PDXs (SF7796) (P = 0.003), while bevacizumab also increased c-Met/β1 complex formation in intracranial responsive PDXs (SF7227 or SF7300) (P = 0.004–0.02), but by a lesser 2- to 3-fold factor (Fig. 3A and SI Appendix, Fig. S12). The second model involved U87-BevR and U87-BevS, isogeneic models of invasive bevacizumab resistance and sensitivity, respectively, that we have described (5, 6). Similar to the PDX findings, immunoprecipitated lysates of intracranial xenografts revealed robust physical c-Met/β1 integrin interactions in resistant U87-BevR xenografts treated with bevacizumab, compared with the sensitive bevacizumab-treated U87-BevS xenografts, and to control IgG-treated U87-BevR or U87-BevS xenografts (Fig. 3B). We confirmed this complex formation using PLAs, which revealed that bevacizumab increased c-Met/β1 complex formation in U87-BevS (P = 0.02) and U87-BevR (P = 0.02) xenografts, but the 19-fold increase in U87-BevR xenografts was far greater than the 3-fold increase in U87-BevS xenografts (Fig. 3C). Formation of the c-Met/β1 complex therefore increased with bevacizumab resistance in both in vivo models. The models also offered insight into the time course of complex formation, as the smaller increases noted in responsive xenografts treated with bevacizumab until progressing could represent a precursor to entrenched resistance, while continued treatment of resistant xenografts led to a much more robust increase in complex formation.
Fig. 3.
A c-Met/β1 complex forms in tumors that have evolved resistance to antiangiogenic therapy. (A) PLAs of GBM PDXs revealed more c-Met/β1 complex with bevacizumab treatment of resistant PDX SF7796 (P = 0.003), with a lesser increase occurring with bevacizumab treatment of intracranial responsive PDXs (SF7227) (P = 0.02). IP (B) and PLA (C) of U87-BevS and U87-BevR intracranial xenografts treated with IgG or bevacizumab revealed increased c-Met/β1 complex with bevacizumab treatment, particularly in resistant xenografts (P = 0.003 U87-BevR IP; P = 0.02 PLA for U87-BevS and U87-BevR). (D) U87 cells were cultured with increasing bevacizumab concentrations in normoxia and hypoxia for 48 h (Upper row). Cell lysates were immunoprecipitated with β1 and blotted for total and phosphorylated c-Met to assess their binding to β1 (Left). The ratio of c-Met binding to immunoprecipitated β1 based on band intensities is in the graph. *P < 0.05; **P < 0.01.
The c-Met/β1 Integrin Complex Is Driven by the Tumor Microenvironment.
Bevacizumab depletes VEGF, resulting in decreased vascularity and hypoxia in resistant tumors (8). Thus, we analyzed the impact of VEGF depletion and hypoxia on c-Met/β1 integrin complex formation. Complex formation was increased in U87 GBM cells in a dose-dependent manner by bevacizumab-induced depletion of all VEGF isoforms in culture (Fig. 3D and SI Appendix, Fig. S13) and in vivo (SI Appendix, Fig. S14) and by hypoxia in culture (Fig. 3D and SI Appendix, Fig. S13), with VEGF depletion and hypoxia in combination cooperating to increase c-Met/β1 complex formation in cultured cells even further than either factor did individually (Fig. 3D).
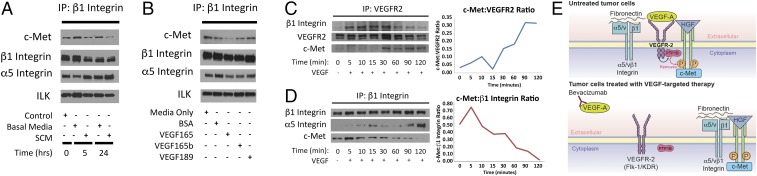
We then investigated the impact of vascular factors that would be altered during antiangiogenic therapy on complex formation. We assessed the impact of conditioned medium (CM) from endothelial cells, which decrease in abundance after VEGF-targeted antiangiogenic therapy, on complex formation in tumor cells. CM from human umbilical vein endothelial cells (HUVECs) decreased complex formation in cultured U87 cells (Fig. 4A and SI Appendix, Fig. S15). We then looked at how VEGF isoforms affect complex formation. While VEGF165, the predominant VEGF isoform, reduced complex formation, complex formation was not affected by VEGF189, an isoform that binds neuropilin-1 not VEGFR2 (14), and VEGF165b, an isoform that binds VEGFR2 without activating downstream pathways (15) (Fig. 4B and SI Appendix, Figs. S16 and S17). Binding of integrin-linked kinase (ILK), a protein associated with the integrin cytoplasmic domain, and α5 integrin, the predominant β1 heterodimer partner, to β1 did not change under conditions in Fig. 4 A and B, indicating that not all β1 binding partners were affected by these conditions. Formation of this complex thus occurs due to interactions among therapeutic stressors in the microenvironment, rather than by passive binding of up-regulated individual components.
Fig. 4.
VEGF binding to VEGFR2 sequesters c-Met and β1 integrin, preventing c-Met/β1 integrin complex formation. (A and B) U87 cells were cultured in (A) conditioned media (CM) from HUVEC cells and (B) with 100 ng/mL of VEGF isoforms 165, 165b, and 189. c-Met/β1 complex formation was then analyzed via IP. (C and D) U87 cells were treated with 100 ng/mL VEGF and (C) VEGFR2 binding to β1 and c-Met and (D) β1 integrin binding to c-Met were analyzed by IP at various time points. (E) Proposed mechanism by which VEGF binding to VEGFR2 sequesters c-Met and β1 integrin, preventing c-Met/β1 complex formation.
VEGF Suppresses the c-Met/β1 Complex Through VEGFR2-Mediated Sequestration.
To define the mechanism by which VEGF suppressed the formation of the c-Met/β1 integrin complex, we investigated the association of VEGFR2 with c-Met and β1 integrin. In cultured U87 cells, increasing duration of exposure to VEGF165 led to increased binding of VEGFR2 to β1 integrin and c-Met (Fig. 4C and SI Appendix, Fig. S18) and reduced formation of the c-Met/β1 complex (Fig. 4D and SI Appendix, Fig. S18). These findings suggest that VEGF binding to VEGFR2 sequesters c-Met and β1 integrin, preventing them from binding to each other, while bevacizumab-induced VEGF depletion liberates c-Met and β1 integrin from VEGFR2, allowing them to form a complex (Fig. 4E).
The c-Met/β1 Integrin Complex Promotes GBM Cell Migration.
To determine if the c-Met/β1 complex influences the GBM cell migration that defines resistance to antiangiogenic therapy, we utilized the same technology we employed in Fig. 2A for engineering cells with inducible c-Met/β1 integrin heterodimerization. As described above, by expressing β1 and c-Met fused to FRB (DmrC) and FKBP (DmrA), respectively, in U87 and U251 GBM cells, we were able to induce c-Met/β1 complex formation using AP21967 (12) (SI Appendix, Figs. S7 and S19–S21A). Complex induction with AP21967 led to increased c-Met phosphorylation, demonstrating that c-Met/β1 complex formation alone was sufficient for c-Met activation (SI Appendix, Fig. S21B). As with MDA-MB-231 cells, complex induction with AP21967 affected U87 and U251 GBM cell morphology by decreasing circularity shape factor (P = 1.6 × 10−6, 2.1 × 10−7) (SI Appendix, Fig. S22A), giving rise to the stellate morphology (13) seen in invading mesenchymal cells and bevacizumab-resistant GBM cells (5). Furthermore, complex induction with AP21967 increased migration (P = 0.006 at 5 and 25 h; SI Appendix, Fig. S22B) and invasion (P = 0.004–0.007; SI Appendix, Fig. S22C) but not proliferation (P = 0.2; SI Appendix, Fig. S23A) in GBM cells. Complex induction with AP21967 also promoted tumor cell survival in the hypoxia, but not the nutrient deprivation, associated with antiangiogenic therapy (three way ANOVA: P = 0.008 for AP21967–hypoxia interaction, P = 0.4 for AP21967-nutrient deprivation interaction; SI Appendix, Fig. S23B). These findings suggest that the c-Met/β1 complex contributes to migration and adaptation to the stress of devascularization in tumors that become resistant to antiangiogenic therapy.
c-Met/β1 Integrin Binding Drives Ligand-Independent Cross-Activation.
Having shown that c-Met/β1 integrin complex formation drives these two distinct invasive oncologic processes, we next sought to determine if c-Met-β1 integrin binding occurs exclusively between their extra- or intracellular domains. To do so in a manner free of the confounding influence of cells, we performed far-Western blotting, a technique derived from Western blotting to detect protein–protein interaction in vitro (16). Unlike Western blots, in which an antibody binds its target on a membrane, far-Western blotting utilizes either a tagged, or antibody-detectable protein (bait) to bind and detect a target protein (prey) that is denatured and renatured to conform to its 3D structure on the membrane (16). Thus, whereas Western blotting detects proteins, far-Western blotting detects protein:protein interactions. By far-Western blotting, binding of c-Met and β1 integrin occurred exclusively in their extracellular domains (Fig. 5 A and B), and β1 integrin in lysates from U87-BevR xenografts exhibited more robust binding to the c-Met extracellular, not intracellular, domain, compared with U87-BevS lysates (SI Appendix, Fig. S24).
Fig. 5.
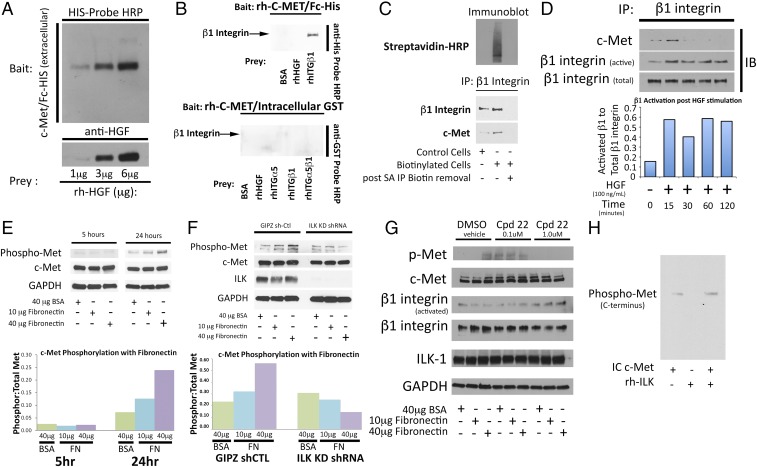
Localizing c-Met/β1 binding and identifying its consequences. (A) Far-Western blot of control bait (recombinant human extracellular His-tagged c-Met = rh-cMet) and prey (recombinant human HGF) with known interaction: HGF bound rh-cMET in a dose-dependent manner. (B) Far-Western blot of extracellular (EC) vs. intracellular (IC) c-Met to assess β1 binding. (Upper) His-tagged EC rh-cMet was the bait and preys were BSA (negative control), HGF (positive control), or recombinant β1. Band density reflects binding to EC rh-cMet, with binding of β1 to EC c-Met found. (Lower) Using BSA (negative control), HGF, recombinant α5, β1, and α5β1 as prey with GST-tagged IC rh-cMet as bait, lack of binding of IC c-Met to β1 was found. (C) To determine if c-Met/β1 binding occurred intracellularly, extracellular U87 proteins were selectively biotinylated, after which cell lysates were incubated with avidin-conjugated beads and eluted. (Upper) Nonbiotinylated proteins in the eluant and biotinylated proteins bound to beads were blotted with HRP-conjugated streptavidin, revealing signal only in proteins bound to beads, confirming biotinylation effectiveness. (Lower) Nonbiotinylated proteins in the eluant and nonbiotinylated whole cell lysates underwent β1 IP followed by blotting of the precipitant for β1 and c-Met, revealing c-Met/β1 complex only in whole cell lysates. Proteins bound to beads could not undergo IP as the acidic eluting solution disrupted protein–protein bonds. (D) U87 cells treated with HGF exhibited increased β1 activation and increased c-Met/β1 integrin complex formation. (E) U87 cells were plated on BSA or increasing FN concentrations for 5 and 24 h. Cells were blotted for total and phosphorylated c-Met. Bar graph represents ratio of phosphorylated c-Met to total c-Met for each condition. (F) U87 cells transduced with ILK shRNA or vector control were stimulated with FN. While cells with vector control maintained increased c-Met phosphorylation in response to increasing FN concentrations, cells with ILK shRNA lost this response. (G) U87 cells were treated with ILK inhibitor Cpd 22, which eliminated the ability of FN to drive c-Met phosphorylation. (H) Recombinant ILK was incubated with IC c-Met and adenosine 5′-O-(3-thiotriphosphate) (ATPγS), with c-Met phosphorylation detected by Western blot. While c-Met autophorphorylation occurred without ILK, c-Met phosphorylation was increased with ILK.
To determine if this binding of c-Met to β1 integrin first occurred before or after protein translocation to the cell membrane, we biotinylated U87 cellular proteins, allowing specific labeling of the extracellular portions of all cell membrane proteins, followed by column purification and elution of biotinylated proteins. IP revealed that the c-Met/β1 complex was not detected in the nonbiotinylated proteins, suggesting that complex formation occurred after protein translocation to the cell membrane (Fig. 5C).
We then investigated whether this extracellular binding of c-Met and β1 integrin allowed ligand-independent cross-activation. Increasing HGF concentrations increased c-Met/β1 integrin complex formation and increased β1 activation in U87 cells, indicating ligand-induced complex formation and ligand-independent cross-activation of one complex member by the other (Fig. 5D). Corresponding to this ability of HGF to activate β1, increasing concentrations of HGF increased adhesion of U87 cells to fibronectin (FN) (P = 0.02; SI Appendix, Fig. S25), without altering β1 levels (SI Appendix, Fig. S26), suggesting increased β1 functional activity.
We confirmed that the converse ligand-independent cross-activating relationship also existed by showing that β1 activation by its ligand fibronectin dramatically increased ligand-independent phosphorylation of c-Met in U87 cells (Fig. 5E). We then investigated the role of ILK, a mechanosensor which binds integrins for structural support (17), in this cross-activation of c-Met by activated β1. We confirmed by far-Western blotting that ILK binds the c-Met cytoplasmic domain (SI Appendix, Fig. S27), and showed that shRNA depletion of ILK (Fig. 5F) or treatment with Cpd 22, an ILK inhibitor (Fig. 5G), blocked ligand-independent phosphorylation of c-Met in the presence of fibronectin in U87 cells. We then investigated whether the necessity of ILK for ligand-independent phosphorylation of c-Met in the presence of fibronectin reflected true intrinsic kinase activity of ILK, a subject of debate in the literature (17), or if it reflected other functions of ILK, such as our finding that Cpd22 changed the morphology of U87 cells to more circular and reduced adhesion to fibronectin (SI Appendix, Fig. S28), consistent with the capacity described by others of ILK to act as a mechanosensor (17). We found that coincubating recombinant ILK with recombinant intracellular c-Met led to c-Met phosphorylation (Fig. 5H), a likely mechanism of the fibronectin-induced c-Met phosphorylation we observed. Further evidence supporting kinase activity of ILK and a mechanism by which this kinase could influence the c-Met signaling cascade was identified when we demonstrated that ILK phosphorylates AKT, a shared downstream mediator of c-Met and β1 integrin (SI Appendix, Fig. S29). Collectively, data in Fig. 5 indicates reciprocal ligand-independent cross-activation between c-Met and β1 integrin, demonstrating the power of their complex.
c-Met/β1 Complex Excludes α5 Integrin and Has Greater Fibronectin Affinity than α5β1 Integrin.
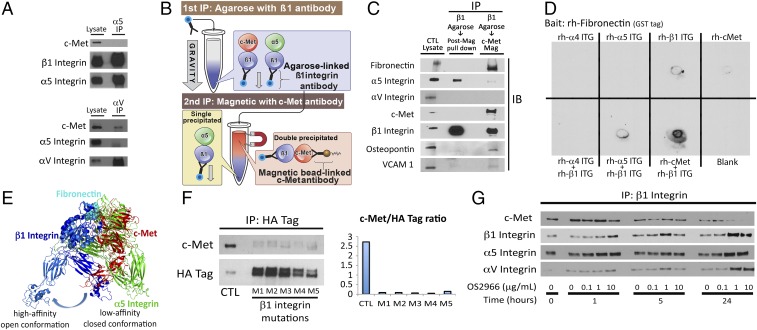
We then determined the relative affinities of β1 integrin for c-Met versus its predominant natural heterodimer partners α5 or αV integrin. In U87 GBM cells, c-Met could bind αV but not α5 integrin (Fig. 6A), similar to what we observed in breast cancer cells (Fig. 1A). We next devised a unique technique to isolate the c-Met/β1 complex to assess proteins specifically bound within the complex and to eliminate carried over protein bound to each protein when utilizing traditional IP techniques, a protocol we termed “sequential immunoprecipitation.” In this protocol, we immunoprecipitated agarose beads bound to β1 integrin antibody followed by IP of c-Met integrin antibody bound to magnetic beads as demonstrated in the schematic of Fig. 6B. This technique revealed almost no α5 or αV integrin in the β1+c-Met+ double immunoprecipitate (Fig. 6C), suggesting displacement of α5 and αV integrin by c-Met during c-Met/β1 integrin complex formation. Importantly, nearly all fibronectin, VCAM, and osteopontin, ECM proteins known to bind α5β1 and αvβ1, were bound to the β1+c-Met+ double IP, containing the c-Met/β1 complex, rather than the β1+c-Met− portion of the β1 integrin immunoprecipitate that failed to be pulled down in the second c-Met IP and therefore contained α5β1 and αvβ1 (Fig. 6C). This finding suggests that the c-Met/β1 complex binds fibronectin, the primary substrate along which cancer invasion occurs, along with VCAM and osteopontin, with greater affinity than α5β1 or αvβ1, the natural receptors for these ECM proteins. Consistent with these data, using dot blotting, we found that coincubated c-Met and β1 integrin bound recombinant fibronectin more efficiently than coincubated α5 and β1 integrin or coincubated α4 and β1 integrin (Fig. 6D).
Fig. 6.
c-Met/β1 integrin complex affinities and modeling. (A) IP revealed that αV but not α5 integrin bound c-Met in U87. (B) Sequential IP schematic: isolating the c-Met/β1 complex by binding β1 via agarose beads, followed by magnetics beads binding c-Met in complex with β1. (C) Sequential IP of β1 followed by c-Met revealed that a c-Met/β1 double IP contained far more fibronectin (FN), VCAM, and osteopontin and far less α5 than a β1 integrin single IP, with αV in neither, suggesting that c-Met displaces α5 from its β1 binding site and that c-Met/β1 integrin binds FN better than α5/β1 integrin. (D) Dot blotting revealed that coincubated c-Met+β1 bound FN far better than c-Met, α4, α5, or β1 alone; α5+β1; or α4+β1. (E) PyMOL analysis defined steric constraints causing c-Met to preferentially bind β1 in its high-affinity open conformation. (F) Five amino acids in β1 integrin predicted by ALA scanning as crucial to c-Met/β1 binding were changed to alanine by mutagenesis of a β1–HA fusion protein. HA IP of HEK cells expressing these constructs revealed decreased c-Met binding to each of five mutants versus wild-type β1–HA fusion protein. (G) Treating cultured U87 cells with variable concentrations of β1 neutralizing antibody OS2966 for 1, 5, and 24 h decreased c-Met/β1 complex formation as assessed by β1 IP.
To define c-Met/β1 integrin binding sites, we performed PyMOL modeling of c-Met/β1 integrin binding based on the crystal structures of each protein (18, 19). Both c-Met and α5 integrin contain seven-bladed β-propeller domains, with sequence identity of 15% between them (Cα rmsd 2.9 Å). α5 integrin interacts with β1 integrin through these β-propeller domains. Therefore, we modeled c-Met/β1 interaction based on the α5/β1 structure (Protein Data Bank 4wk4) using structural alignment. This analysis suggested that c-Met and α5 integrin share a common binding site on β1 integrin (SI Appendix, Fig. S30 A and B). The resulting complex was consistent with membrane orientation, as the C-terminal ends of c-Met and β1 integrin point in the same direction. These findings reveal that β1 integrin forms a complex with α5 or c-Met but never both at the same time, as both bind the same region of β1 integrin. This is further demonstrated by the superimposition of α5 integrin and c-Met in SI Appendix, Fig. S30C to demonstrate the similarity between the binding regions of these two proteins, allowing for β1 integrin’s exclusivity for a single partner in exchange for the other. Fibronectin, the primary ligand for α5/β1, binds α5/β1 through an Arg-Gly-Asp motif (20). Most of the contact with fibronectin is through β1 integrin, although there is limited contribution from α5. When c-Met replaces α5, PyMOL modeling predicts better contact with fibronectin (SI Appendix, Fig. S31). An explanation for this improved contact derived from our modeling, which revealed that the closed conformation of β1, known to have low affinity for fibronectin, has steric clash with the first IPT domain of c-Met (Fig. 6E). To resolve this clash, PyMOL modeling suggests a mechanism whereby c-Met overcomes this clash by keeping β1 in its high-affinity open conformation, providing a structural foundation for our sequential IP results and a functional explanation for why cancer cells benefit by replacing α5 with c-Met as a β1 binding partner.
Genetic or Pharmacologic Targeting of c-Met/β1 Integrin Binding.
Based on this PyMOL data, we investigated genetic and pharmacologic approaches to disrupt c-Met/β1 interaction. The ability of the smaller β1 molecule to bind c-Met proved readily targetable by site-directed mutagenesis of five individual amino acids identified as crucial for binding from our PyMOL modeling using the Rosetta ALA scanning method (21) (Fig. 6F and SI Appendix, Figs. S32 and S33 and Table S1) and OS2966, a therapeutic humanized β1 integrin neutralizing antibody (Fig. 6G). When targeting the ability of c-Met to bind β1 via site-directed mutagenesis of four regions of amino acids in the loops on the top of the c-Met propeller structure, regions suggested by PyMOL modeling to be important for c-Met/β1 binding, some reduction in complex formation occurred with three of the four analyzed mutations (SI Appendix, Figs. S34 and S35 and Table S1), but less robustly than seen with single amino acid changes in β1 integrin. When targeting c-Met pharmacologically via therapeutic artificial ligand NK4, coadministering NK4 and HGF reduced c-Met phosphorylation and c-Met/β1 complex formation (SI Appendix, Figs. S36 and S37). When targeting c-Met via onartuzamab, a neutralizing therapeutic antibody blocking four amino acids in the extracellular β-propeller Sema domain of c-Met (SI Appendix, Table S1), this antibody reduced the ability of some β1 antibodies to detect their epitopes, which may have been obscured by onartuzamab (SI Appendix, Fig. S38 and Table S1).
c-Met/β1 Complex Formation Increases in Patient Metastases and Bevacizumab-Resistant GBMs.
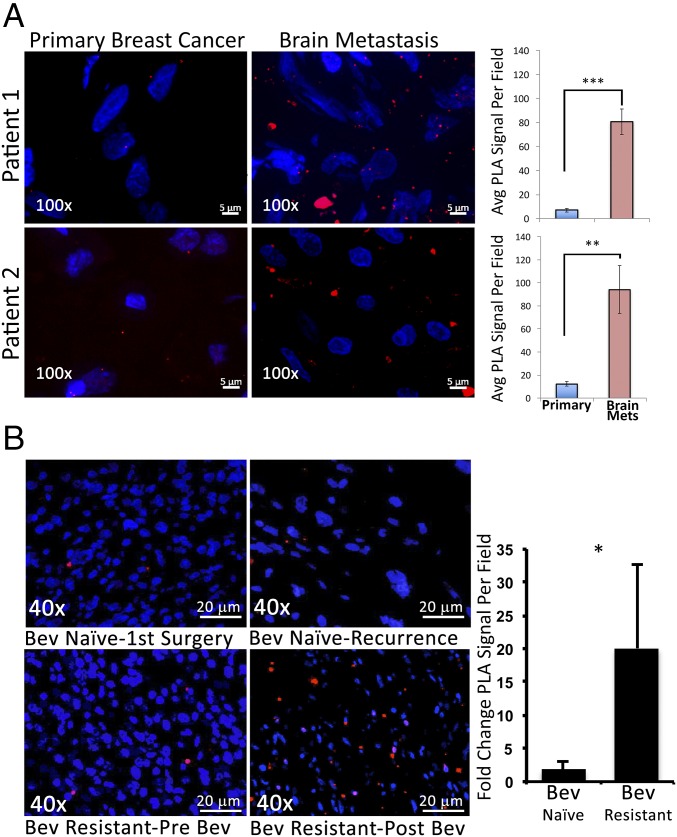
We then determined if formation of the c-Met/β1 complex is relevant to metastases and invasive resistance in patient samples. Using PLA, we found increased c-Met/β1 complex formation in patient breast cancer metastases to the brain compared with paired primary breast tumors from the same patients (P = 0.0006, 0.004) (Fig. 7A). This finding was in contrast to the fact that neither c-Met (P = 0.3) nor β1 integrin (P = 0.4) expression individually increased in archived microarray analysis (22) of paired primary breast tumors and metastases from eight patients (SI Appendix, Fig. S39). We then analyzed c-Met/β1 integrin complex formation in nine bevacizumab-resistant GBMs (5–7). PLAs detected 20-fold more c-Met/β1 complexes in bevacizumab-resistant GBMs compared with prebevacizumab (P < 0.05; Fig. 7B), greater than the 2-fold increase in recurrent GBMs not treated with bevacizumab (Fig. 7B). As with metastases, the increased c-Met/β1 complex formation in bevacizumab-resistant GBMs was far more than the 2- to 3-fold individual increase in phosphorylated c-Met or β1 integrin we previously reported in these tumors (5–7). These findings indicate that increased formation of the complex is a more specific mechanistic biomarker of metastases or bevacizumab resistance than up-regulation or activation of each individual factor.
Fig. 7.
In patients, the c-Met/β1 integrin complex is associated with breast cancer metastases and bevacizumab-resistant GBM. (A) Increased c-Met/β1 complex levels by PLA noted in brain metastases versus primary breast tumors from the same patients (P = 0.0006 patient 1; P = 0.004 patient 2; n = 5 fields per tumor). (B) c-Met/β1 integrin complex levels in nine GBM patients developing bevacizumab resistance and nine control GBM patients naïve to bevacizumab at diagnoses and recurrence were assessed with PLA. Representative PLA images from “Bev-naïve” and “Bev-resistant” patients with fold-change in PLA complex numbers after versus before tumor recurrences graphed (P < 0.05 Bev-resistant versus Bev-naïve). *P < 0.05; **P < 0.01; ***P < 0.001.
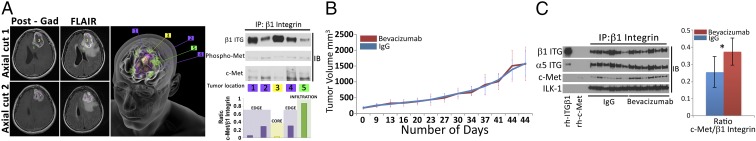
To define the regional variability in c-Met/β1 complex formation in patients, we obtained site-directed biopsies from a bevacizumab-resistant GBM (Fig. 8A). This technique allows the successful sampling of various regions of tumor, capturing the spectrum of hypoxia and nutrient deprivation occurring in GBM after evolving antiangiogenic therapy resistance. The c-Met/β1 integrin complex was detected in five regional biopsies from bevacizumab-resistant GBM. Increased complex formation occurred in invasive cells beyond the enhancing tumor edge (Fig. 8A and SI Appendix, Fig. S40), with the complex also harboring phosphorylated c-Met (Fig. 8A and SI Appendix, Fig. S41). Complex formation and bevacizumab resistance proved durable, as both were maintained in PDXs derived from this bevacizumab-resistant GBM, with the complex increasing further when the PDXs were treated with bevacizumab, to which they showed resistance (Fig. 8 B and C and SI Appendix, Fig. S42).
Fig. 8.
Regional variation and durability of c-Met/β1 complex in bevacizumab-resistant GBM. (A, Left) Locations on axial MRIs of five site-directed biopsies from a bevacizumab-resistant GBM. First column is T1 postgadolinium, and second is FLAIR imaging. Top row is axial cut 1, with biopsies 3 (tumor core), 4 (edge), and 5 (infiltration) shown. Bottom row is axial cut 2 with biopsies 1 and 2 (both edge) shown. (A, Middle) Artist’s 3D rendition of locations of the five site-directed biopsies taken from the bevacizumab-resistant GBM. (A, Right) Lysates from each location were used for IP with β1 integrin and immunoblotted for c-Met and phospho-Met. Quantified c-Met/β1 complex levels from each location graphed in the Lower Right categorized as tumor core, tumor edge, or infiltration. (B) The tumor from the patient in A was passaged into 10 SCID mice and treated with IgG (n = 5) or bevacizumab (n = 5). Tumor volume (in cubic millimeters) is plotted versus time. PDXs remained bevacizumab resistant and (C) c-Met/β1 integrin complex formation increased following treatment as shown by the β1 integrin IP of PDXs with blotting for c-Met. *P < 0.05.
We did not observe complex formation in a patient GBM treated with low-dose bevacizumab (5 rather than 10 mg/kg) (SI Appendix, Fig. S43), confirming the dose-dependent increase in c-Met/β1 integrin associated with bevacizumab treatment of cultured cells (Fig. 3D) and xenografts (SI Appendix, Fig. S14). The presence of small quantities of c-Met/β1 integrin complex in bevacizumab-naïve GBMs seen via PLA before the robust increase occurring with bevacizumab resistance led us to analyze c-Met/β1 integrin complex levels in GBMs at diagnosis to determine its impact on patient survival. The percent of β1 integrin bound to c-Met via IP ranged from 0 to 17% in lysates from GBMs taken at diagnosis. This percent correlated inversely with survival (P < 0.05; SI Appendix, Figs. S44 and S45). Thus, the c-Met/β1 integrin complex associated with bevacizumab resistance portended the poor prognosis that these patients experience (23).
Discussion
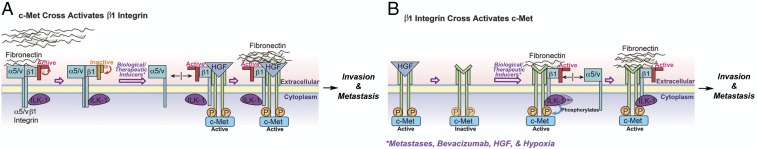
We describe a complex between c-Met and β1 integrin that forms during cancer metastases and the evolution of invasive resistance to antiangiogenic therapy. These two invasive processes are examples of aggressive cancer biology contributing to severely worsened patient prognosis (7, 23) whose mechanisms have yet to be fully defined. The power of the c-Met/β1 integrin complex we identified in driving these invasive oncologic processes lies in its robust affinity for fibronectin, a crucial component of the extracellular matrix that guides tumor cell invasion, and its ability to integrate and promote ligand-independent cross-activation of two receptors mediating chemotaxis and haptotaxis (Fig. 9).
Fig. 9.
Stimuli driving c-Met/β1 complex formation promote ligand-independent cross-activation of each receptor and enhance cancer cell invasion. (A) Biologic and therapeutic stimuli (metastases, bevacizumab resistance, HGF, and hypoxia) promote c-Met/β1 integrin complex formation, which maintains β1 integrin in its activate state even without fibronectin (FN), with the c-Met/β1 complex exhibiting greater FN affinity than β1 bound to its conventional heterodimer partners αV or α5. (B) Stimuli described in A promote c-Met/β1 complex formation, with the c-Met/β1 complex exhibiting greater affinity for FN than β1 bound to its α-heterodimer partners and FN promoting ILK-mediated c-Met phosphorylation even in the absence of HGF.
While c-Met and β1 integrin are each known to individually contribute to metastases (24, 25) and invasive bevacizumab resistance in GBM (5, 6, 26), the mechanisms through which these drive metastases or invasive resistance remain uncertain, as their high baseline expression levels do not change tremendously during acquisition of metastases (24, 25) or invasive resistance (5–7). The increase in c-Met/β1 integrin complex formation that we observed in metastases and bevacizumab-resistant GBM was more dramatic than the increase in the levels of c-Met and β1 integrin individually, suggesting that the complex drives these invasive processes more than the biology of the individual factors.
Integrin-Receptor Tyrosine Kinase Interactions.
Integrin-mediated adhesion and motility may be modulated by growth factors (27), while growth factor signaling can be modulated indirectly by integrins (28). Previous studies have demonstrated colocalization of c-Met and β1 integrin by immunostaining (1) and interaction of c-Met and α5 or β1 integrin by immunoprecipitation (1, 2). Our work demonstrates a true interaction between c-Met and β1 integrin with the utilization of far-Western blotting of recombinant proteins free of other cellular components, overcoming the limitation of colocalization studies, which provide indirect evidence, or immunoprecipitation, which sometimes artificially detects binding that can occur with micelles in immunoprecipitation of cell lysates. Furthermore, our work also expands the implications of c-Met binding β1 integrin beyond those described for other integrin–RTK interactions by demonstrating that the RTK c-Met displaces the α-heterodimer partner of β1 integrin to form a complex with β1 integrin, and that the resulting c-Met/β1 integrin complex is significantly more efficient at binding β1 integrin’s natural ligand fibronectin, a major substrate along which cancer invasion occurs. This switching of the β1 integrin binding partner from α5 integrin to RTK c-Met to increase fibronectin affinity is an efficient means of increasing tumor cell migration, which depends more on turnover of integrins in focal adhesions and affinity for extracellular matrix than cell surface expression of integrins (29). We also added molecular insight by defining residues critical for c-Met/β1 integrin binding in the loops on top of the c-Met propeller structure and the βI domain of β1 integrin. Finally, in showing the formation of this complex during metastases and therapeutic resistance, its regulation by factors in the microenvironment associated with metastases and resistance, and the ability of the complex to drive invasion and survival during hypoxia, we lend therapeutic implications to RTK–integrin interactions.
Role of VEGF in Complex Formation.
We integrated our finding that VEGF165 inhibits c-Met/β1 complex formation with the previous finding that binding of VEGF to VEGFR2 leads to formation of a VEGFR2/c-Met complex in which VEGFR2 inhibits c-Met phosphorylation (26). We found that VEGF bound to VEGFR2 independently sequesters c-Met and β1 integrin, with c-Met and β1 integrin free to form their complex in the absence of VEGF, as occurs during prolonged bevacizumab treatment. The resulting c-Met/β1 complex drives invasive resistance through the ligand independent cross-activation and more efficient fibronectin binding we demonstrated. Thus, c-Met may have context-dependent binding partners whose effects need to be balanced when administering bevacizumab.
c-Met/β1 Complex Formation Drives Ligand-Independent Activation of both Receptors.
Constitutive ligand-independent activation of RTKs such as c-Met (30) and EGFR (31) has been described in cancers through genetic alterations. Here, we identified an alternative mode of ligand-independent c-Met activation, as c-Met/β1 integrin complex formation allowed fibronectin binding to β1 integrin to drive c-Met phosphorylation. Indeed, PyMOL analysis revealed c-Met to sterically keep β1 integrin in its high-affinity open conformation, allowing the converse effect of c-Met driving ligand-independent β1 activation to occur as well.
ILK: Functionally Linking β1 Integrin and c-Met.
Given that integrins lack enzymatic activity, their signaling depends on recruiting adaptor proteins (32). One of the best described of these proteins is ILK, which binds some β integrin cytoplasmic domains. ILK contributes to actin rearrangement, cell polarization, migration, proliferation, and survival (33). Some have suggested that the kinase-like domain of ILK lacks catalytic activity (34), while others have identified kinase activity and reported that, while the atypical protein kinase domain of ILK lacks some amino acids considered essential for phosphotransferase activity, similar deficiencies are present in the catalytic domains of other known kinases (35). Our investigation supported kinase activity of ILK that phosphorylated c-Met, allowing the ligand-independent activation we found to occur in the complex, and AKT, a downstream mediator of c-Met. These findings link β1 signaling via ILK to c-Met and its associated downstream pathways.
Therapeutic Implications.
There are numerous therapeutic implications of our validation of the c-Met/β1 complex as a mechanism of metastases and invasive resistance. ILK inhibitors have been investigated as anticancer therapies (36) and our demonstration of their ability to decouple the receptor cross-talk in the c-Met/β1 complex may be an added advantage of this approach.
Furthermore, in the case of resistance to antiangiogenic therapy, the tendency for the c-Met/β1 complex to form at higher doses of these therapies suggests a role for investigating the impact of antiangiogenic therapy’s dosage on efficacy and resistance evolution. Such studies could determine that the notion that the maximum tolerable dose is most effective, while applicable to conventional chemotherapy targeting tumor cells, may not be applicable for antiangiogenic therapy targeting the tumor microenvironment. Instead, for antiangiogenic treatments, we may find that “less is more” (37).
The power of the c-Met/β1 complex could also be harnessed therapeutically by screening for small-molecule inhibitors that disrupt the extracellular binding of the residues we identified in the loops on top of the c-Met propeller and the βI domain of β1 integrin. Since complex up-regulation occurs more robustly in metastases and resistant tumors than increased activation of either individual factor, such an approach may prove more effective at targeting metastases or invasive resistance than c-Met or β1 integrin inhibition alone, which have produced concerns about toxicity or failed to produce meaningful results when used alone or in the setting of antiangiogenic therapy resistance (38, 39).
Methods
Study Approval.
Human tissue research was approved by the University of California San Francisco (UCSF) institutional review board (approval 11-06160). Animal experiments were approved by the UCSF institutional animal care and use committee (approval AN105170-02).
Cell Culture.
U87-MG (HTB-14, ATCC) and U251 (09063001, Sigma) human GBM cells and MDA-MB-231, HCC1143, and HCC3153 human breast cancer cells were passaged for fewer than 6 mo, verified by short tandem repeat (STR) profiling, and confirmed to be Mycoplasma-free. MDA-MB-231-BR brain-seeking cells were generated as described (11) and provided by S. K. Srivastava, Texas Tech University, Lubbock, TX. Cells were cultured in DMEM/F-12 supplemented with 10% FBS and 1% P/S and maintained at 37 °C. Hypoxia studies used an incubator that reduced oxygen from 20% (normoxia) to 1% (hypoxia).
Western Blot.
Human tissue samples and cell preparations were harvested in 1× radio immunoprecipitation buffer (10× RIPA; 9806, Cell Signaling) and one tablet each of PhoStop and Complete Mini (04906845001 and 04693124001, Roche). Insoluble materials were removed by centrifugation at 300 × g for 20 min at 4 °C. Protein concentration was determined using the bicinchronic acid (BCA) assay (23225, Thermo Scientific). Samples were prepared with 10–30 μg of protein in RIPA buffer with 4× LDS loading buffer (LP0001, Life Technologies). Samples were electrophoresed on SDS/PAGE gels, transferred to PVDF membranes, and probed with primary antibodies (SI Appendix, Table S2) overnight at 4 °C. Membranes were detected using HRP-conjugated secondary antibodies and imaged using radiographic film.
Immunoprecipitation.
Samples were prepared in 500 μL RIPA buffer containing 150–500 μg of protein. For β1 IP, 50 μL of protein A magnetic bead slurry (8740, Cell Signaling) were aliquoted per sample and washed with 500 μL of cell lysis buffer (9803, Cell Signaling). Beads were incubated with mouse monoclonal anti-β1 antibody (1:50; ab7168, Abcam) in 200 μL of 0.1% Triton-X 100 PBS for 15 min at room temperature (RT). Beads were magnetically precipitated and resuspended with sample lysate for incubation on a rotator (4 °C overnight). Antibody-bound beads were magnetically separated from the lysate supernatant and washed three times in 500 μL cell lysis buffer. Samples were eluted by precipitating beads on a magnetic rack and resuspending in 40 μL of 3× blue sample buffer 30× reducing agent: 1.25 M DTT (1:30; 7722s, Cell Signaling). Resulting samples were heated at 95 °C (5 min), centrifuged at 300 × g at RT, and magnetically precipitated. The supernatant was used for SDS/PAGE electrophoresis. Blots were probed with primary and secondary antibodies (SI Appendix, Table S2).
Engineering Cells with Inducible β1 Integrin and c-Met Interaction.
Plasmid information is in SI Appendix, Table S3. Our inducible protein–protein interaction system was the Lenti-X iDimerize heterodimer system (Clontech), which uses FKBP12 and FKBP rapamycin-binding domains altered to specifically bind the A/C heterodimerizer (AP21967) ligand. ITGB1 cDNA was amplified from pRK5 β1 vector (31786, Addgene) using Pfx50 high-fidelity Taq (Invitrogen) targeted for insertion into the 3′ multiple cloning site of the pLVX-Het1-DmrC vector. The PCR product was produced using B1-NotI-FW and B1-BamHI-HA RV primers (SI Appendix, Table S4), subcloned into pCR4-Blunt-TOPO, and inserted into pLVX-Het1-DmrC. Clones were screened with restriction digest and sequencing. The cMET coding region was amplified from p-ENTR-cMET (16042, Addgene) with primers in SI Appendix, Table S4. The PCR fragment was cloned into pCR4-Blunt-TOPO. The resulting pCR4-Blunt-TOPO-cMET(HIS) vector was digested with NotI and BglI and run on 0.75% low melting point agarose. The insert was ligated into NotI linearized pLVX-Het2. Lentiviral particles harboring constructs with an N-terminally DmrC-fused ITGB1 and N-terminally DmrA-fused Met coding sequences were generated using HEK293T cells and a third generation packaging system, with supernatants used to transduce MDA-MB-231, U87, or U251 cells for 24 h, followed by antibiotic selection.
PLA.
We utilized the Duolink Detection System (Sigma) on (i) cells with and without the iDimerize system on chamber slides and (ii) paraffin-embedded human tumor tissue and frozen tissue from intracranial U87-BevS, U87-BevR, SF7227, SF7300, or SF7796 xenografts, established as described (5, 40). Primary antibodies were mouse monoclonal anti-integrin β1 (1:50; ab52971, Abcam) and rabbit monoclonal anti–c-Met (1:250; ab51067, Abcam). Secondary antibodies were anti-mouse and anti-rabbit IgG Duolink in situ PLA probes (DUO92002/4, Sigma), respectively, per manufacturer instructions. PLA signal was assessed using Duolink in situ detection reagents (DUO92008, Sigma) and imaged using a Zeiss AxioObserver Z1 system equipped with an AxioCam MRm CCD and AxioVision software (Release 4.8). Images were quantified by counting dimerizations (red dots) per cell for chamber slides or per 100× field for tissue from four representative selections using the ImageJ cell counter plugin.
Assessment of c-Met and β1 Interaction in Cultured Cells.
U87 or MDA-MB-231 cells were cultured, in bevacizumab (Genentech), HGF (294-HGN-005/CF, R&D Systems), hypoxia, conditioned HUVEC media, or serum-free media containing VEGF165 (584501, Biolegend), VEGF165b (3045VE025CF, R&D Systems), or VEGF189 (8147VE025CF, R&D Systems) and complex formation was assessed via IP. c-Met activation through β1 signaling was assessed by plating U87 cells on increasing fibronectin amounts (sc-29011, Santa Cruz Biotechnology) and blotting for phosphorylated c-Met.
Migration and Proliferation Assays.
Transduced MDA-MB-231 or U87 cells were plated in six-well plates at 90% confluency. Cell monolayers were wounded using p200 pipette tips scratching the plastic surface from 12 to 6 o’clock. The wound was divided into four quadrants and imaged at 0, 5, and 24 h. Images were collated and analyzed using TScratch software (41). In parallel, the same cells were plated into black 96-well clear-bottom plates and analyzed using CyQuant cell proliferation assay kits (c7026, Thermo Scientific).
Animal Work.
For mammary xenografts, 1 × 105 MDA-MD-231 cells were injected unilaterally into the fourth mammary fat pad in 10 μL. For tail vein injections, 106 MDA-MB-231-iDimerize-c-Met-β1 cells in 100 μL were injected via tail veins. Some tumor cells were pretreated with 500 nM AP219667 in complete DMEM 2 h before tail vein injection. For intracranial xenografts, we intracranially implanted cells from previously described (5, 6) bevacizumab-resistant or sensitive xenografts into severe combined immunodeficient (SCID) nude mice. Animals were treated intraperitonially with IgG (I4506, Sigma) or Avastin (bevacizumab, Genentech) at designated doses. Resulting tumors were harvested and lysed into RIPA buffer and prepared for IP as described above. The s.c. tumors were established by injecting 5 × 105 cells s.c. into the flanks of SCID nude mice. Tumor volume was V = (Y × X2)/2, where Y is the longest diameter and X is the diameter perpendicular to that.
Sequential Immunoprecipitation.
U87-BevR xenograft lysates were immunodepleted for β1 integrin using anti-β1 integrin agarose-bound complex under nondenaturing conditions in cell lysis buffer on a rotator overnight at 4 °C (Cell Signaling). The resulting β1 integrin complex was harvested from solution by centrifugation at 300 × g for 1 min and eluted from agarose beads with Pierce IgG elution buffer (21004, Thermo Scientific). The eluent was mixed with magnetic beads coupled to anti-cMet antibody and incubated on a rotator overnight at 4 °C. We then used magnetic precipitation to produce the sequentially precipitated fraction. The immune-complexed β1 integrin/c-Met moieties were then eluted under denaturing conditions in 187.5 mM Tris⋅HCl (pH 6.8 at 25 °C), 6% (wt/vol) SDS, 30% glycerol and 0.03% (wt/vol) bromophenol blue with 0.125 M DTT and resolved by standard SDS/PAGE.
Modeling c-Met and β1 Integrin Interaction.
c-Met and β1 integrin extracellular crystal structures were available at resolutions of 2.80 Å (PDB 2uzx) and 1.85 Å (PDB 4wk4), respectively. β1 integrin was cocrystallized with integrin α5 (PDB 4wk4; SI Appendix, Fig. S30A). In this structure, α5 interacts with β1 integrin through the seven-bladed β-propeller. The extracellular domain of c-Met also contains a seven-bladed β-propeller domain. Therefore, we rationalized that c-Met may interact with β1 integrin through a β-propeller domain interface similar to the one between α5 and β1 integrin (SI Appendix, Fig. S30B). The two propellers, with 15% sequence identity, were superimposed using MultiProt software (42) with rmsd of 2.9 Å for the 292 aligned Cα positions (SI Appendix, Fig. S30C). The docking method was template based, which, when template is available, is considered more accurate (43). Template was from the known α5β1 integrin structure due to the structural similarities of c-Met and α5 integrin. To model the complex, c-Met was structurally aligned on α5 using MultiProt software. As a result of template-based modeling, only one docking model was obtained. We further repacked the interface to optimize side-chain positions and remove steric clashes using FireDock web server.
For study approval; morphology; immunofluorescence; qPCR; far-Western blotting; adhesion, kinase and survival assays; human tissue; microarray; and statistics, see SI Appendix, SI Methods.
Supplementary Material
Acknowledgments
Work was supported by grants to M.K.A.’s laboratory from American Brain Tumor Association, James S. McDonnell Foundation, American Cancer Society, NIH (5K02NS64167 and 1 R01 NS079697), UCSF Brain Tumor Specialized Program of Research Excellence (SPORE) Grant CA097257, and UCSF Resource Allocation Program. A.J. was a Howard Hughes Medical Institute fellow and is supported by NIH Grant 1F31CA203372-01.
Footnotes
Conflict of interest statement: M.K.A. is a member of the Scientific Advisory Board for Oncosynergy, Inc.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1701821114/-/DCSupplemental.
References
- 1.Barrow-McGee R, et al. Beta 1-integrin-c-Met cooperation reveals an inside-in survival signalling on autophagy-related endomembranes. Nat Commun. 2016;7:11942. doi: 10.1038/ncomms11942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mitra AK, et al. Ligand-independent activation of c-Met by fibronectin and α(5)β(1)-integrin regulates ovarian cancer invasion and metastasis. Oncogene. 2011;30:1566–1576. doi: 10.1038/onc.2010.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gupta GP, Massagué J. Cancer metastasis: Building a framework. Cell. 2006;127:679–695. doi: 10.1016/j.cell.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 4.Bergers G, Hanahan D. Modes of resistance to anti-angiogenic therapy. Nat Rev Cancer. 2008;8:592–603. doi: 10.1038/nrc2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carbonell WS, DeLay M, Jahangiri A, Park CC, Aghi MK. β1 integrin targeting potentiates antiangiogenic therapy and inhibits the growth of bevacizumab-resistant glioblastoma. Cancer Res. 2013;73:3145–3154. doi: 10.1158/0008-5472.CAN-13-0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jahangiri A, et al. Gene expression profile identifies tyrosine kinase c-Met as a targetable mediator of anti-angiogenic therapy resistance. Clin Cancer Res. 2013;19:1773–1783. doi: 10.1158/1078-0432.CCR-12-1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeLay M, et al. Microarray analysis verifies two distinct phenotypes of glioblastomas resistant to antiangiogenic therapy. Clin Cancer Res. 2012;18:2930–2942. doi: 10.1158/1078-0432.CCR-11-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pàez-Ribes M, et al. Antiangiogenic therapy elicits malignant progression of tumors to increased local invasion and distant metastasis. Cancer Cell. 2009;15:220–231. doi: 10.1016/j.ccr.2009.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kreisl TN, et al. Phase II trial of single-agent bevacizumab followed by bevacizumab plus irinotecan at tumor progression in recurrent glioblastoma. J Clin Oncol. 2009;27:740–745. doi: 10.1200/JCO.2008.16.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park CC, Zhang HJ, Yao ES, Park CJ, Bissell MJ. Beta1 integrin inhibition dramatically enhances radiotherapy efficacy in human breast cancer xenografts. Cancer Res. 2008;68:4398–4405. doi: 10.1158/0008-5472.CAN-07-6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gupta P, Adkins C, Lockman P, Srivastava SK. Metastasis of breast tumor cells to brain is suppressed by phenethyl isothiocyanate in a novel in vivo metastasis model. PLoS One. 2013;8:e67278. doi: 10.1371/journal.pone.0067278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muthuswamy SK, Gilman M, Brugge JS. Controlled dimerization of ErbB receptors provides evidence for differential signaling by homo- and heterodimers. Mol Cell Biol. 1999;19:6845–6857. doi: 10.1128/mcb.19.10.6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levi-Schaffer F, Slovik D, Armetti L, Pickholtz D, Touitou E. Activation and inhibition of mast cells degranulation affect their morphometric parameters. Life Sci. 2000;66:PL283–PL290. doi: 10.1016/s0024-3205(00)00538-5. [DOI] [PubMed] [Google Scholar]
- 14.Hervé MA, et al. Overexpression of vascular endothelial growth factor 189 in breast cancer cells leads to delayed tumor uptake with dilated intratumoral vessels. Am J Pathol. 2008;172:167–178. doi: 10.2353/ajpath.2008.070181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Woolard J, et al. VEGF165b, an inhibitory vascular endothelial growth factor splice variant: Mechanism of action, in vivo effect on angiogenesis and endogenous protein expression. Cancer Res. 2004;64:7822–7835. doi: 10.1158/0008-5472.CAN-04-0934. [DOI] [PubMed] [Google Scholar]
- 16.Wu Y, Li Q, Chen XZ. Detecting protein-protein interactions by far western blotting. Nat Protoc. 2007;2:3278–3284. doi: 10.1038/nprot.2007.459. [DOI] [PubMed] [Google Scholar]
- 17.Fukuda K, Knight JD, Piszczek G, Kothary R, Qin J. Biochemical, proteomic, structural, and thermodynamic characterizations of integrin-linked kinase (ILK): Cross-validation of the pseudokinase. J Biol Chem. 2011;286:21886–21895. doi: 10.1074/jbc.M111.240093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nagae M, et al. Crystal structure of α5β1 integrin ectodomain: Atomic details of the fibronectin receptor. J Cell Biol. 2012;197:131–140. doi: 10.1083/jcb.201111077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rickert KW, et al. Structural basis for selective small molecule kinase inhibition of activated c-Met. J Biol Chem. 2011;286:11218–11225. doi: 10.1074/jbc.M110.204404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xia W, Springer TA. Metal ion and ligand binding of integrin α5β1. Proc Natl Acad Sci USA. 2014;111:17863–17868. doi: 10.1073/pnas.1420645111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kortemme T, Kim DE, Baker D. Computational alanine scanning of protein-protein interfaces. Sci STKE. 2004;2004:pl2. doi: 10.1126/stke.2192004pl2. [DOI] [PubMed] [Google Scholar]
- 22.Manso L, et al. Analysis of paired primary-metastatic hormone-receptor positive breast tumors (HRPBC) uncovers potential novel drivers of hormonal resistance. PLoS One. 2016;11:e0155840. doi: 10.1371/journal.pone.0155840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clark AJ, et al. Neurosurgical management and prognosis of patients with glioblastoma that progresses during bevacizumab treatment. Neurosurgery. 2012;70:361–370. doi: 10.1227/NEU.0b013e3182314f9d. [DOI] [PubMed] [Google Scholar]
- 24.Takeuchi H, et al. c-MET expression level in primary colon cancer: A predictor of tumor invasion and lymph node metastases. Clin Cancer Res. 2003;9:1480–1488. [PubMed] [Google Scholar]
- 25.Carbonell WS, Ansorge O, Sibson N, Muschel R. The vascular basement membrane as “soil” in brain metastasis. PLoS One. 2009;4:e5857. doi: 10.1371/journal.pone.0005857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu KV, et al. VEGF inhibits tumor cell invasion and mesenchymal transition through a MET/VEGFR2 complex. Cancer Cell. 2012;22:21–35. doi: 10.1016/j.ccr.2012.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klemke RL, Yebra M, Bayna EM, Cheresh DA. Receptor tyrosine kinase signaling required for integrin alpha v beta 5-directed cell motility but not adhesion on vitronectin. J Cell Biol. 1994;127:859–866. doi: 10.1083/jcb.127.3.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Howe A, Aplin AE, Alahari SK, Juliano RL. Integrin signaling and cell growth control. Curr Opin Cell Biol. 1998;10:220–231. doi: 10.1016/s0955-0674(98)80144-0. [DOI] [PubMed] [Google Scholar]
- 29.Nagano M, Hoshino D, Koshikawa N, Akizawa T, Seiki M. Turnover of focal adhesions and cancer cell migration. Int J Cell Biol. 2012;2012:310616. doi: 10.1155/2012/310616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dai Y, Siemann DW. Constitutively active c-Met kinase in PC-3 cells is autocrine-independent and can be blocked by the Met kinase inhibitor BMS-777607. BMC Cancer. 2012;12:198. doi: 10.1186/1471-2407-12-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pines G, Köstler WJ, Yarden Y. Oncogenic mutant forms of EGFR: Lessons in signal transduction and targets for cancer therapy. FEBS Lett. 2010;584:2699–2706. doi: 10.1016/j.febslet.2010.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schiller HB, Friedel CC, Boulegue C, Fässler R. Quantitative proteomics of the integrin adhesome show a myosin II-dependent recruitment of LIM domain proteins. EMBO Rep. 2011;12:259–266. doi: 10.1038/embor.2011.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Legate KR, Montañez E, Kudlacek O, Fässler R. ILK, PINCH and parvin: The tIPP of integrin signalling. Nat Rev Mol Cell Biol. 2006;7:20–31. doi: 10.1038/nrm1789. [DOI] [PubMed] [Google Scholar]
- 34.Wickström SA, et al. Integrin-linked kinase controls microtubule dynamics required for plasma membrane targeting of caveolae. Dev Cell. 2010;19:574–588. doi: 10.1016/j.devcel.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hannigan GE, McDonald PC, Walsh MP, Dedhar S. Integrin-linked kinase: Not so ‘pseudo’ after all. Oncogene. 2011;30:4375–4385. doi: 10.1038/onc.2011.177. [DOI] [PubMed] [Google Scholar]
- 36.Hannigan G, Troussard AA, Dedhar S. Integrin-linked kinase: A cancer therapeutic target unique among its ILK. Nat Rev Cancer. 2005;5:51–63. doi: 10.1038/nrc1524. [DOI] [PubMed] [Google Scholar]
- 37.de Groot JF. High-dose antiangiogenic therapy for glioblastoma: Less may be more? Clin Cancer Res. 2011;17:6109–6111. doi: 10.1158/1078-0432.CCR-11-1853. [DOI] [PubMed] [Google Scholar]
- 38.Wen PY, et al. Society for Neuro-Oncology Annual Meeting. Oxford Press; New York: 2009. A phase 2 study of XL184, an inhibitor of Met, VEGFR2, and RET, in patients with progressive glioblastoma multforme. [Google Scholar]
- 39.Stupp R, et al. European Organisation for Research and Treatment of Cancer (EORTC) Canadian Brain Tumor Consortium CENTRIC study team Cilengitide combined with standard treatment for patients with newly diagnosed glioblastoma with methylated MGMT promoter (CENTRIC EORTC 26071-22072 study): A multicentre, randomised, open-label, phase 3 trial. Lancet Oncol. 2014;15:1100–1108. doi: 10.1016/S1470-2045(14)70379-1. [DOI] [PubMed] [Google Scholar]
- 40.Jahangiri A, et al. Gene expression profile identifies tyrosine kinase c-Met as a targetable mediator of antiangiogenic therapy resistance. Clin Cancer Res. 2013;19:1773–1783. doi: 10.1158/1078-0432.CCR-12-1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liang CC, Park AY, Guan JL. In vitro scratch assay: A convenient and inexpensive method for analysis of cell migration in vitro. Nat Protoc. 2007;2:329–333. doi: 10.1038/nprot.2007.30. [DOI] [PubMed] [Google Scholar]
- 42.Shatsky M, Nussinov R, Wolfson HJ. A method for simultaneous alignment of multiple protein structures. Proteins. 2004;56:143–156. doi: 10.1002/prot.10628. [DOI] [PubMed] [Google Scholar]
- 43.Gront D, et al. Assessing the accuracy of template-based structure prediction metaservers by comparison with structural genomics structures. J Struct Funct Genomics. 2012;13:213–225. doi: 10.1007/s10969-012-9146-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.