Significance
Phototrophic bacteria have provided fundamental insight into the biological mechanism of solar energy conversion. Early events in photosynthesis are carried out by the antenna apparatus for light-harvesting (LH) and the reaction center (RC) for charge separation. Here we describe a system for expressing a chimeric LH1-RC complex from two phylogenetically distant phototrophic purple bacteria: the LH1 from Thermochromatium tepidum and the RC from Rhodobacter sphaeroides. This system is exploited to definitively localize Ca2+ within the Tch. tepidum LH1 complex. The hybrid photocomplexes also provide powerful new tools for probing photosynthetic energy transfer, identifying intrinsic LH1–RC interactions, monitoring altered behavior of carotenoids in a nonnative environment, and linking specific amino acid residues to the specific spectroscopic properties of different phototrophic organisms.
Keywords: photosynthesis, light harvesting, Ca binding, Qy transition, Thermochromatium tepidum
Abstract
The native core light-harvesting complex (LH1) from the thermophilic purple phototrophic bacterium Thermochromatium tepidum requires Ca2+ for its thermal stability and characteristic absorption maximum at 915 nm. To explore the role of specific amino acid residues of the LH1 polypeptides in Ca-binding behavior, we constructed a genetic system for heterologously expressing the Tch. tepidum LH1 complex in an engineered Rhodobacter sphaeroides mutant strain. This system contained a chimeric pufBALM gene cluster (pufBA from Tch. tepidum and pufLM from Rba. sphaeroides) and was subsequently deployed for introducing site-directed mutations on the LH1 polypeptides. All mutant strains were capable of phototrophic (anoxic/light) growth. The heterologously expressed Tch. tepidum wild-type LH1 complex was isolated in a reaction center (RC)-associated form and displayed the characteristic absorption properties of this thermophilic phototroph. Spheroidene (the major carotenoid in Rba. sphaeroides) was incorporated into the Tch. tepidum LH1 complex in place of its native spirilloxanthins with one carotenoid molecule present per αβ-subunit. The hybrid LH1-RC complexes expressed in Rba. sphaeroides were characterized using absorption, fluorescence excitation, and resonance Raman spectroscopy. Site-specific mutagenesis combined with spectroscopic measurements revealed that α-D49, β-L46, and a deletion at position 43 of the α-polypeptide play critical roles in Ca binding in the Tch. tepidum LH1 complex; in contrast, α-N50 does not participate in Ca2+ coordination. These findings build on recent structural data obtained from a high-resolution crystallographic structure of the membrane integrated Tch. tepidum LH1-RC complex and have unambiguously identified the location of Ca2+ within this key antenna complex.
Structural and functional research on bacterial photosynthesis has paved the way for our understanding of the basic mechanism of solar energy conversion in natural systems. The early events in photosynthesis are carried out by two distinct components: the antenna apparatus for light energy collection and the reaction center (RC) for charge separation. In purple phototrophic bacteria, the antenna system consists of two large pigment-protein complexes: the core light-harvesting complex (LH1) that surrounds the RC and the peripheral light-harvesting complex (LH2) that surrounds the LH1. Both LH1 and LH2 are large oligomers formed from a basic structural subunit composed of two integral membrane polypeptides (α and β) that associate with bacteriochlorophyll (BChl) and carotenoid molecules. LH1 is present in all purple bacteria, whereas LH2 is absent in some species (1). In addition to light harvesting, LH1 is also involved in quinone transport between the RC and the quinone pool in the photosynthetic membrane (2).
The LH1 complex from the thermophilic purple sulfur bacterium Thermochromatium tepidum, first isolated from a Yellowstone hot spring by Madigan (3), is unique in its characteristic red-shifted Qy absorption band at 915 nm (4, 5). This contrasts with the LH1 complexes from most other purple bacteria, which show Qy absorbance of 875–890 nm. The Tch. tepidum LH1-RC core complex thus provides a unique model for studying “uphill” energy transfer from LH1 to the RC (6, 7). The unusual red shift of the Tch. tepidum LH1-Qy has been attributed to the binding of Ca2+ to the C-terminal domain of LH1 polypeptides (8, 9). A recent crystallographic structure of the Tch. tepidum LH1-RC revealed that LH1 forms concentric cylinders around the RC with the inner α- and outer β-polypeptide rings tightly connected by 16 Ca2+ on the periplasmic membrane surface (10). The 3.0-Å electron density map suggested that each Ca2+ in LH1 was coordinated by the main chain oxygen atom of α-Trp46, the side chains of α-Asp49 and α-Asn50, and the C-terminal carboxyl group of β-Leu46 in the adjacent subunit (10) (SI Appendix, Fig. S1). However, unambiguous positioning of Ca2+ within the Tch. tepidum LH1-RC could not be deduced from the crystal structure alone.
To positively identify the Ca2+ coordinating residues and to reveal their contributions to the LH1-Qy shift, we constructed a genetic system for heterologously expressing the Tch. tepidum LH1 complex in a deletion strain of Rhodobacter sphaeroides, a phylogenetically distant relative of Tch. tepidum that is readily amenable to genetic manipulation. By exploiting this system, we functionally expressed the Tch. tepidum LH1 complex in Rba. sphaeroides despite the two phototrophs sharing <40% sequence identity in their LH1 α- and β-polypeptides. In addition, the carotenoid composition of Tch. tepidum LH1 was replaced with that of Rba. sphaeroides. Site-directed mutants obtained from these constructions were then used to rigorously characterize the Tch. tepidum LH1 complex and unequivocally identify how Ca2+, a cation that influences both the thermal stability and unique spectral properties of Tch. tepidum LH1 (8, 11), is positioned within the precise structure of this important photocomplex. Although this system is shown to verify the Ca-binding site within the Tch. tepidum LH1 complex, the hybrid photocomplexes also provide powerful new research tools for probing photosynthetic energy transfer and quinone transport pathway, identifying intrinsic LH1–RC interactions, monitoring altered behavior of carotenoid molecules in a nonnative environment, linking specific amino acid residues to the specific spectroscopic properties of different phototrophic organisms, and advancing artificial photosynthesis.
Results
Construction of a Heterologous Expression System for Synthesizing the Tch. tepidum LH1 Complex and Production of Site-Directed Mutants.
A mutant of Rba. sphaeroides strain IL106 lacking the RC, LH1, and LH2 complexes (designated strain DPP1) was used to construct a genetic system for synthesizing the Tch. tepidum LH1 polypeptides (12) (SI Appendix, Materials and Methods). The constructed strain (designated strain TS2) contains a chimeric pufBALM gene cluster with pufBA from Tch. tepidum and pufLM from Rba. sphaeroides. Using this strain, site-specific mutations in Tch. tepidum LH1 were designed with the following rationale. An alanine residue was inserted at position 43 of the Tch. tepidum α-polypeptide (SI Appendix, Fig. S1), designated α-ins43A, to mimic the sequence of the LH1 complex (Qy at 889 nm) of Allochromatium vinosum, a mesophilic relative of Tch. tepidum, and to examine the effect of this deletion on the LH1-Qy transition. The amino acids α-Asp49, α-Asn50, and β-Leu46 in Tch. tepidum LH1 were replaced by alanines (designated α-D49A, α-N50A, and β-L46A, respectively), because these residues had been tentatively assigned as Ca-coordinating ligands in the LH1-RC crystal structure (10). Finally, to specifically assess the role of the C-terminal residue of the β-polypeptide in Ca2+ binding, a deletion mutant of the β-Leu46, designated β-L46del, was prepared.
Expression and Properties of the Chimeric LH1-RC Complexes.
All Rba. sphaeroides strains harboring the chimeric pufBALM gene clusters grew phototrophically at virtually similar growth rates as Rba. sphaeroides (SI Appendix, Fig. S2). The heterologously expressed Tch. tepidum LH1 polypeptides were assembled as functional pigment-protein complexes in all strains, as confirmed by the absorption spectra of intact cells and intracytoplasmic membrane (ICM) preparations (Fig. 1 and SI Appendix, Fig. S3). The identities of the expressed products were confirmed by TOF/MS measurements. No PufX polypeptide was detected in the chimeric LH1-RCs from both TOF/MS and HPLC analyses. The LH1 complex from Rba. sphaeroides strain TS2 contained the wild-type sequences of Tch. tepidum and showed a Qy transition at ∼915 nm identical to that of the native LH1 complex from Tch. tepidum (13). This result demonstrates that Rba. sphaeroides can effectively express LH1 complexes from a purple bacterium with significantly different LH1 polypeptides and spectroscopic properties from those of the native Rba. sphaeroides LH1. It also implies that the LH1-Qy position is primarily defined by the structures of the α- and β-polypeptides.
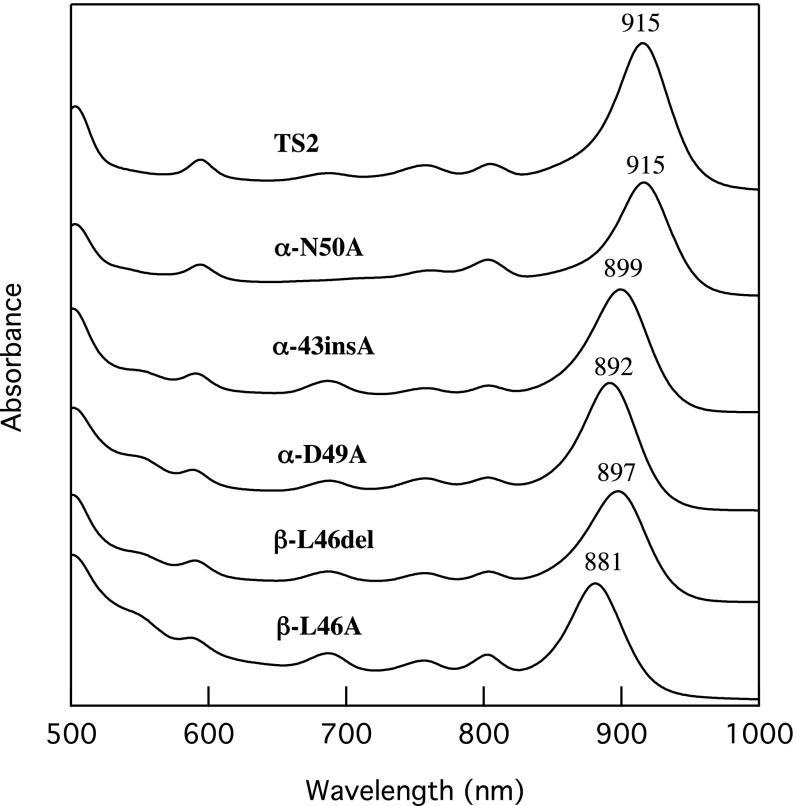
Fig. 1.
Absorption spectra of crude (strains TS2 and α-N50A) and detergent-treated (strains α-43insA, α-D49A, β-L46del and β-L46A) ICMs from the Rba. sphaeroides mutant strains. All membrane preparations were suspended in 20 mM Tris⋅HCl (pH 8.5) buffer and the spectra were recorded at room temperature.
The LH1 polypeptides in strains TS2 and α-N50A were well assembled with BChl a, whereas mutant strains α-43insA, α-D49A, β-L46A, and β-L46del contained variable levels of BChl a that were not properly incorporated within the LH1 polypeptides, as shown by their absorption characteristics between 750 and 800 nm (SI Appendix, Fig. S3). There was a tendency for strains expressing LH1 complexes with Qy bands more blue-shifted from that of the native Tch. tepidum LH1 to have higher proportions of misassembled complexes; this implies that the site-specific mutations that induce larger structural changes have more detrimental effects on pigment assembly. However, most of the misassembled components could be removed from the crude ICMs by mild detergent-treatment (Fig. 1). Unless stated otherwise, the detergent-treated ICMs of α-43insA, α-D49A, β-L46A, and β-L46del were used in subsequent measurements.
Introduction of different site-specific mutations into the Tch. tepidum LH1 polypeptides resulted in LH1 complexes with differing Qy transitions (Fig. 1). The LH1 complex of strain α-N50A showed a Qy at 915 nm, the same as that of the native Tch. tepidum LH1. This indicates that the asparagine residue, the 50th residue from the N terminus in the α-polypeptide (Asn50), does not participate in Ca binding in the Tch. tepidum LH1 complex, in agreement with preliminary results from a 1.9-Å structure of the Tch. tepidum LH1-RC (14). In contrast, the LH1 from strain α-43insA, which contains an Alc. vinosum-like α-polypeptide sequence (15), exhibited a blue-shifted Qy band at 899 nm. Comparison of this with the native Alc. vinosum LH1-Qy (889 nm) (16) suggests that the deletion at position 43 in the Tch. tepidum α-polypeptide contributes significantly to the Qy red shift. Mutation of the α-Asp49 to Ala (strain α-D49A) resulted in a blue-shifted Qy band at 892 nm. This result agrees with the assignment of this residue as a Ca-binding ligand in the crystal structure (10), and indicates that this residue also plays an important role in maintaining the red-shifted Qy of the Tch. tepidum LH1 complex. Both deletion and substitution of the β-Leu46 residue resulted in blue-shifted Qy transitions, highlighting the importance of this C-terminal residue. In fact, the β-L46A substitution yielded the most blue-shifted Qy transition of all mutations introduced in this study (Fig. 1).
Spectroscopic Characterization of the Strain TS2 Chimeric LH1-RC Complex.
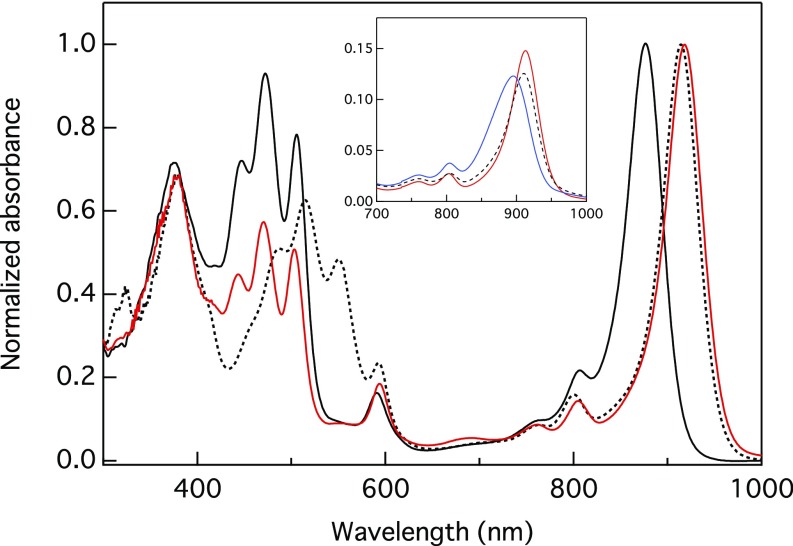
The LH1 complex from strain TS2 (Fig. 1) was sufficiently stable to be solubilized in an LH1-RC form and was obtained at high purity. Fig. 2 compares the spectra of purified LH1-RC complexes from strains TS2, DP2 (an LH2-deficient Rba. sphaeroides strain; SI Appendix, Materials and Methods), and native Tch. tepidum. Typically, the chimeric LH1-RC from strain TS2 displayed an LH1-Qy band in the range of 915–917 nm. A remarkable feature is the incorporation of spheroidene (the major carotenoid in Rba. sphaeroides) into the LH1 complex of strain TS2 in place of spirilloxanthin, which is present in the native Tch. tepidum LH1 complex (13). Notably, however, fewer spheroidenes per LH1 complex were incorporated than in wild-type Rba. sphaeroides LH1. Quantitative carotenoid analyses revealed the presence of 17.6 ± 1.0 carotenoid molecules per LH1-RC of strain TS2, indicating that each LH1 αβ pair incorporated only one carotenoid molecule. This contrasts with the two carotenoid molecules per αβ pair in the LH1 complex from strain DP2, the same as in wild-type Rba. sphaeroides (17).
Fig. 2.
Comparison of the absorption spectrum of the chimeric LH1-RC complex purified from strain TS2 (red curve) with the spectra of the LH1-RC complexes purified from strain DP2 (black curve) and from Tch. tepidum (dotted curve). All spectra were normalized at LH1-Qy bands. (Inset) Changes in the absorption spectra of the TS2 LH1-RC (red) on addition of EDTA to a final concentration of 5 mM (blue curve), followed by addition of CaCl2 to a final concentration of 175 mM (dashed curve).
Table 1 shows the carotenoid composition of purified LH1-RC complexes from the Rba. sphaeroides expression system. Spheroidene was the major component in all complexes, although there were variations in the composition of spheroidene derivatives among the complexes. The spheroidene-dominant carotenoids in the LH1 complex of strain TS2 exhibited absorption maxima at 443, 470, and 503 nm, slightly blue-shifted from those of the carotenoids in the LH1 complex of strain DP2 (447, 472, and 505 nm). This may reflect a difference in the protein environment between the two LH1 complexes (18).
Table 1.
Carotenoid composition (mol % of total carotenoids) in purified LH1-RC complexes from Rba. sphaeroides strains DP2, TS2, and α-N50A
| Carotenoid | DP2 | TS2 | α-N50A |
| OH-spheroidene | 23.8 | 6.9 | 7.5 |
| Spirilloxanthin | 0.9 | 7.7 | 6.3 |
| Demethylspheroidene | 9.9 | 9.4 | 15.6 |
| Spheroidenone | 3.0 | 1.8 | 1.1 |
| Spheroidene | 62.4 | 74.2 | 69.5 |
The existence of Ca-binding sites in the LH1 from strain TS2 was probed using EDTA titration experiments. Fig. 2, Inset shows a typical example in which on addition of EDTA, the LH1-Qy band blue-shifted from 915 nm to 895 nm and displayed a broadened band shape. The absorption maximum then red-shifted back to 912 nm following the addition of excess CaCl2. These data provide additional evidence that the Ca-binding sites in Tch tepidum LH1 are also present in the heterologously expressed LH1 complex.
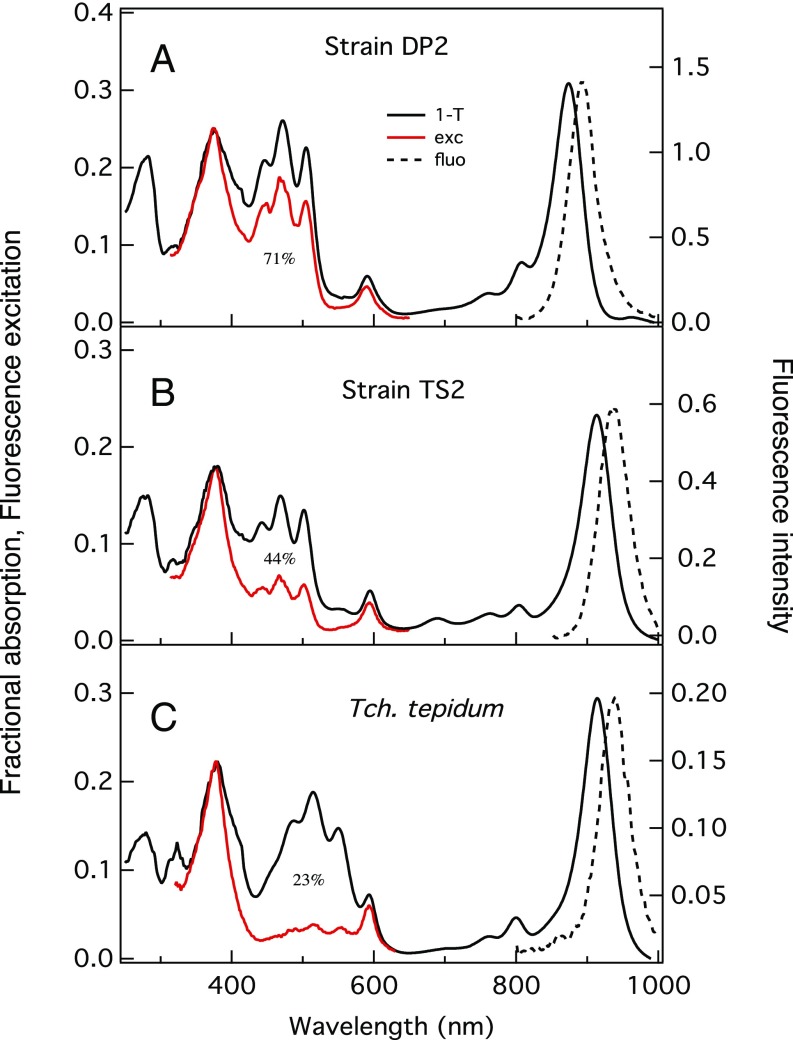
Fig. 3 compares the steady-state fluorescence excitation spectrum of the purified LH1-RC complex from strain TS2 with the spectra from strain DP2 and from Tch. tepidum. The overall efficiency of excitation energy transfer from the nonnative carotenoids to BChl a in the LH1 complex from strain TS2 was calculated as 44%. This value is significantly lower than that of the native Rba. sphaeroides LH1 complex from strain DP2 (71%) despite a similar carotenoid composition, but is much higher than that of the spirilloxanthin-containing Tch. tepidum LH1 complex (23%). This result suggests that energy transfer efficiency may be affected not only by the conjugation length of the carotenoid, but also by the protein environment, as well as the gap between the absorption maxima of the carotenoid and BChl a molecules and the number of carotenoids present in the LH1 complex. The efficiencies for the LH1 complexes from strain DP2 and native Tch. tepidum are compatible with those reported for other spheroidene- and spirilloxanthin-containing LH1 complexes, respectively (19–22).
Fig. 3.
Analysis of the efficiency of steady-state excitation energy transfer from carotenoid to BChl a in purified LH1-RC complexes from strains DP2 (A) and TS2 (B), and from Tch. tepidum (C). Fluorescence emission spectra (dashed lines) were plotted to match the maximum intensities to those of the LH1-Qy bands in fractional absorption spectra (1 − T, where T is transmittance) (solid black) on the same scale. Fluorescence excitation spectra (red) detected at fluorescence emission maxima were normalized at LH1 Soret bands in fractional absorption spectra.
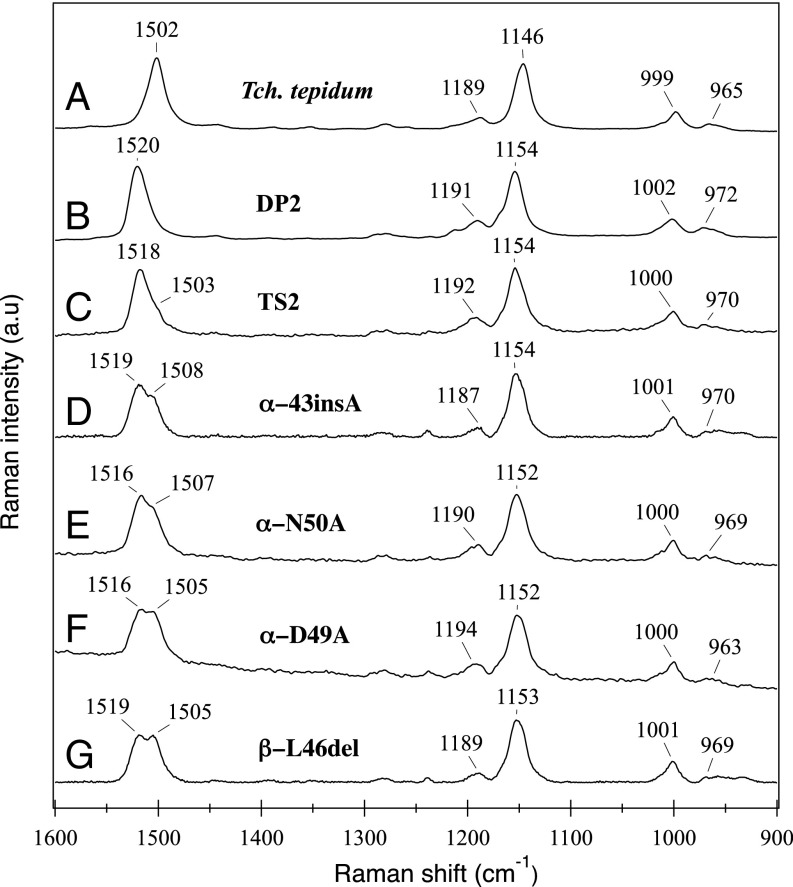
The characteristics of chimeric and native LH1-RC complexes were further examined by resonance Raman spectroscopy. On excitation at 532 nm in the absorption region of carotenoids, the LH1 from strain TS2 yielded a spectrum essentially identical to that from strain DP2 (Fig. 4B), a spectrum characteristic of all-trans spheroidene. The intense bands at 1,518 cm−1 and 1,154 cm−1, assigned to the νC=C and νC-C/δCH modes of all-trans spheroidene, respectively (23), were shifted by 16 cm−1 and 8 cm−1 from those of all-trans spirilloxanthin in Tch. tepidum LH1 (24) due to the relatively shorter CC bond lengths. The intense peak at 1,518 cm−1 in strain TS2 LH1 contained a shoulder around 1,505 cm−1, which became more apparent or split into two peaks in the spectra of mutant LH1 complexes (Fig. 4).
Fig. 4.
Resonance Raman spectra with excitation at 532 nm for the purified LH1-RC complexes from Tch. tepidum (A), strain DP2 (B), and ICM from strains TS2 (C), α-43insA (D), α-N50A (E), α-D49A (F), and β-L46del (G).
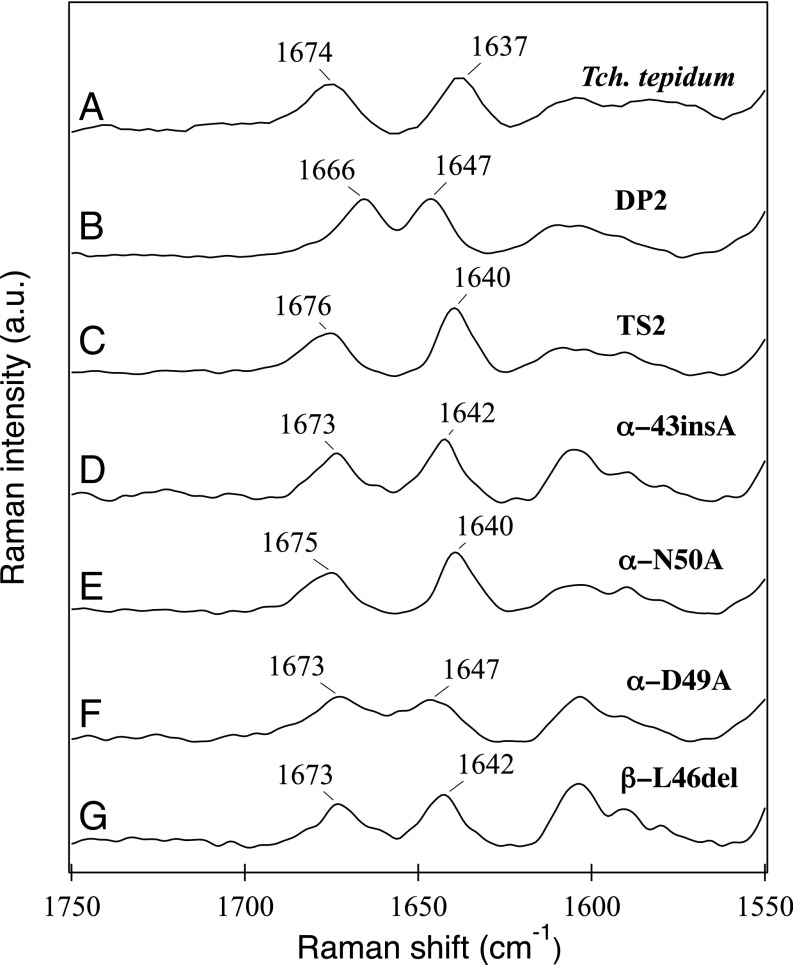
Interactions between BChl a and LH1 polypeptides were investigated by near-infrared resonance Raman spectroscopy (Fig. 5). The C3-acetyl C=O stretching band of the TS2 LH1 at 1,640 cm−1 was up-shifted by 3 cm−1 from that of native Tch. tepidum LH1 (24) but down-shifted 7 cm−1 from that of strain DP2 LH1. This result indicates that the interaction between the C3-acetyl group of BChl a and the LH1 polypeptides in strain TS2 is slightly weaker than that in the native Tch. tepidum LH1 complex but is stronger than that in the wild-type Rba. sphaeroides LH1 complex. As for the BChl a C13-keto group, the C=O stretching band at 1,676 cm−1 for strain TS2 LH1 is close to that of wild-type Tch. tepidum LH1, but significantly blue-shifted by 10 cm−1 from that of strain DP2 LH1. These data suggest a very weak (if any) hydrogen-bonding interaction between the C13-keto group and the LH1 polypeptides of strain TS2, whereas such hydrogen bonds likely form in the strain DP2 LH1 complex (25).
Fig. 5.
Near-infrared resonance Raman spectra with excitation at 1,064 nm for the purified LH1-RC complexes from Tch. tepidum (A) and strain DP2 (B), and ICM from strains TS2 (C), α-43insA (D), α-N50A (E), α-D49A (F), and β-L46del (G).
Characterization of Site-Specific Mutants of Tch. tepidum LH1 Complexes.
Because spheroidene was incorporated into the chimeric LH1-RC complexes as the major carotenoid, the resonance Raman spectra excited at 532 nm for the mutants were basically identical to those of wild-type Rba. sphaeroides LH1-RC. However, distinct differences were observed for the LH1 mutants in the C=C stretching region (Fig. 4), in which the intense bands around 1,518 cm−1 split into two bands, one around 1,505–1,508 cm−1 and the other around 1,516–1,519 cm−1. The relative intensity of the former to the latter increased in the order of β-L46del > α-D49A > α-N50A > α-ins43A > TS2. These results likely reflect a distorted conformation of the spheroidene molecules from a planar geometry along the C=C backbone in the mutant LH1 complexes (26). If this is so, then such distortion could influence the properties of BChl a in the LH1 complex and can be detected by resonance Raman spectroscopy with excitation at 1,064 nm. The results presented in Fig. 5 show that α-N50A with its LH1-Qy band at 915 nm gave a C3-acetyl C=O stretching band at 1,640 cm−1, whereas this band shifted to 1,642 cm−1 for α-ins43A and β-L46del and to 1,647 cm−1 for α-D49A.
A linear relationship exists between a downshift in the C3-acetyl stretching mode, which is sensitive to hydrogen bonding, and a red shift in the LH Qy maximum (25, 27) (SI Appendix, Fig. S4). This indicates that hydrogen-bonding interactions indeed occur in the LH1 complexes of the chimeric mutants. Moreover, the ratio of the splits in the carotenoid νC=C bands (∼1,505 cm−1/∼1,519 cm−1) also reveal a correlation with the LH1-Qy red shift. This finding indicates that conformational changes of carotenoid molecules have little if any effect on LH1 absorption properties, but do influence the LH1-Qy transition energy.
Discussion
The Tch. tepidum LH1 complex has been successfully expressed in Rba. sphaeroides mutant strains. In addition to their use in probing the coordination and function of Ca in Tch. tepidum LH1, these strains are valuable new tools for exploring several key primary processes in photosynthesis in greater detail. These include, but are not limited to (i) producing various hybrid LH1-RC complexes with vastly different biochemical and spectroscopic properties; (ii) investigating the intrinsic nature of the LH1–RC interactions; and (iii) verifying the roles of specific amino acids in energy transfer-coupled electron/proton transport processes. Because the LH1 complex surrounds the RC, it serves as both the entrance route of excitation energy to the RC and the exit route of reduced ubiquinones from the RC to the quinone pool and cytochrome bc1 complex (2). Therefore, the chimeric LH1-RC complexes constructed herein also provide excellent subjects for probing the dynamic processes of energy and electron transfers in photosynthesis by time-resolved spectroscopy.
At present, no genetic manipulation or expression system is available for Tch. tepidum, while several heterologous expression systems exist in other species and have been used to study various functions of the bacterial photosynthetic apparatus (28–33). However, our construction differs from other chimeric LH1-RCs in at least three major ways: (i) the complex contains components from distantly related purple bacteria, the purple sulfur bacterium Tch. tepidum (γ-Proteobacteria) and the purple nonsulfur bacterium Rba. sphaeroides (α-Proteobacteria); (ii) the LH1 transferred to Rba. sphaeroides differs significantly in primary structure from its native LH1 complex (37% sequence identity for both α- and β-polypeptides); and (iii) the Qy transition of the chimeric LH1 absorbs light of significantly longer wavelengths compared with that of the RC in the host strain (915 nm vs. 868 nm for the special pair).
All of the modified LH1 complexes produced in this study supported anaerobic, phototrophic growth of Rba. sphaeroides, and similar growth rates as seen in strain DP2 were observed for all mutant strains. These findings indicate that light energy absorbed by the modified LH1 complexes is indeed transferred to the RC for charge separation, even though these are energetically “uphill” processes (∼490 cm−1) occurring within a structurally nonnative LH1-RC complex. Phototrophic growth also demonstrates that the electron and proton transports necessary to support photophosphorylation are operational in each mutant derivative. Thus, in addition to the applications listed previously, the chimeric photocomplexes described here make available a powerful platform for experimentally dissecting the mechanism of uphill energy transfer and related early events in photosynthesis.
The characteristic feature of the LH1-Qy transition in native Tch. tepidum was reproduced in the heterologously expressed LH1 complex containing wild-type sequences, indicating that the spectroscopic properties of BChl a molecules in LH1 complexes are ultimately determined by the primary structures of α- and β-polypeptides. As revealed by resonance Raman spectroscopy (Fig. 5), the interaction between BChl a and LH1 polypeptides in the heterologously expressed LH1 complex containing wild-type sequences (strain TS2) was virtually identical to that seen in the native Tch. tepidum LH1 complex. The near-infrared Raman results show that the hydrogen-bonding interactions between the BChl a C3-acetyl group and LH1 polypeptides in the native Tch. tepidum (24) are also major factors regulating the Qy transition of the expressed LH1 complexes.
The PufX polypeptide in Rba. sphaeroides is known to have important roles in supporting photosynthetic growth, structural organization of the LH1-RC core complex, and cyclic electron transfer (34). In this work, the Rba. sphaeroides pufX gene was present and unaltered in the mutant derivatives (SI Appendix, Fig. S5); however, its expression product could not be detected in the chimeric LH1-RC complexes. While we are still attempting to detect PufX in the chimeric complexes, our results thus far suggest that the PufX polypeptide, if expressed, is not required for (or is unable to promote) assembly of the Tch. tepidum LH1 in Rba. sphaeroides, and thus is not incorporated in the LH1-RC complex.
Site-specific mutagenesis of this study provided an ideal tool for clarifying uncertainties in the assignments of the crystallographic electron density map for the Ca-binding residues in the Tch. tepidum LH1 complex. Both α-Asp49 and α-Asn50 were tentatively assigned as the Ca-binding residues in the 3.0-Å structure (10). Mutation of the α-Asp49 to Ala resulted in a blue shift of the LH1-Qy transition to 892 nm, whereas mutation of the α-Asn50 to Ala did not alter the Qy position. These results are completely consistent with recent findings from both high-resolution Ca-bound (14) and Sr(Ba)-substituted Tch. tepidum LH1-RC structures (35) that point to α-Asp49 as a key metal-binding residue; in contrast, it is now clear that α-Asn50 does not participate in Ca coordination. In addition, our results for the two β-L46 mutants highlight the importance and sensitivity of this C-terminal residue. Substitution of the C-terminal β-Leu46 residue resulted in a blue-shifted LH1-Qy transition (Fig. 1). This result would be difficult to interpret if the C-terminal carboxyl group serves as a ligand to Ca, as has been tentatively assigned in the 3.0-Å structure (10). Recently, it has become clear from a high-resolution structure of the Tch. tepidum LH1-RC (14) that β-Leu46 is not directly involved in Ca binding, but does participate in an extensive hydrogen-bonding network with its neighboring β-polypeptide and water molecules around the C-terminal end. Deletion of β-Leu46 also resulted in a blue-shifted LH1-Qy band to 897 nm, which can be attributed to a disruption of the hydrogen-bonding network and a conformational change in the C-terminal region of the β-polypeptide.
The major difference between the native and heterologously expressed Tch. tepidum LH1 complexes was their carotenoid compositions. As spheroidene was biosynthetically incorporated into the expressed LH1 complexes in place of the native spirilloxanthin, our heterologous LH1 expression system could also serve as a powerful tool for probing the behavior of different carotenoids in a nonnative environment, as evident from the resonance Raman results (Fig. 4). Splittings of the νC=C bands in the 1,500–1,520 cm−1 region were observed for carotenoids in the expressed LH1 complexes, indicating either that some of the spheroidenes in the expressed LH1 complexes differ slightly in their conformations from those in the native Rba. sphaeroides LH1 complex or that minor carotenoids such as demethylspheroidene and/or spirilloxanthin may contribute to the low-frequency component (SI Appendix, Results and Discussion).
A final benefit of our use of site-directed mutagenesis in this study is that it allowed direct verification of a long-standing hypothesis on the effect of a deletion in the Tch. tepidum LH1 α-polypeptide (8, 9, 11). Despite the high degree of sequence identities between Tch. tepidum and Alc. vinosum LH1 polypeptides (SI Appendix, Fig. S1), a deletion is present at position 43 in the Tch. tepidum LH1 α-polypeptide where an Ala exists in the corresponding Alc. vinosum LH1 polypeptide (15). Insertion of an Ala residue into the Tch. tepidum LH1 α-polypeptide at this position (strain α-43insA) resulted in a blue shift of its LH1-Qy band to 899 nm (Fig. 1). This indicates that the inserted Ala in the Tch. tepidum LH1 α-polypeptide prevents formation of a proper Ca-binding pocket and subsequent incorporation of Ca2+ into the complex; this is not required in the mesophilic Alc. vinosum, since Ca2+ has no apparent effect on the spectroscopic properties of its LH1-RC complex (36).
Materials and Methods
Construction of a Rba. sphaeroides mutant lacking genes encoding the LH1-RC (pufBALM) and LH2 (pucBA) polypeptides (designated DPP1) was performed using Rba. sphaeroides strain IL106 (12). A mutant lacking only LH2, designated DP2, was generated as well. The chimeric puf operon consisting of Tch. tepidum pufBA and Rba. sphaeroides pufLMX was cloned with a suicide vector pJP5603 and introduced into the Rba. sphaeroides mutant strain DPP1 by conjugal transfer from Escherichia coli S17-1λpir. The mutant recovering phototrophic growth by incorporation of the chimeric puf construct to the genomic DNA through a homologous recombination was named TS2. Site-specific mutagenesis and other genetic manipulations on the Tch. tepidum LH1 polypeptides were performed as described in SI Appendix, Materials and Methods. All expression strains were grown under phototrophic (anoxic/white light) conditions and, for certain experiments, with 850-nm LED illumination. The LH1-RC complexes from strains DP2, TS2, and α-N50A were purified by solubilizing the ICM with 0.23% lauryldimethylamine N-oxide in 20 mM Tris⋅HCl (pH 8.5), followed by anion-exchange chromatography (Toyopearl 650S; TOSOH) with 0.1% n-dodecyl β-d-maltopyranoside at 4 °C. Details of sample preparation and spectroscopic measurements are provided in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank Ayumi Imai and Megumi Kobayashi for their technical assistance in preparing the LH1-RC complex from strain DP2, and the Kao Corporation for providing lauryldimethylamine N-oxide. This work was supported by the Ministry of Education, Culture, Sports, Science, and Technology of Japan (MEXT; Grants-in-Aid for Scientific Research B 16H04174, to Z.-Y.W.-O. and C 16K07295, to Y.K.), Takeda Science Foundation, a Sumitomo Foundation Grant for Basic Science Research Projects, the Kurata Memorial Hitachi Science and Technology Foundation (Z.-Y.W.-O.), and a Japan Society for the Promotion of Science Research (JSPS) Fellowship (to T.K.). This work was supported in part by Grant-in-Aid for JSPS KAKENHI Grant 15K00642, MEXT KAKENHI Grant 24107004, and the Strategic Research Base Development Program for Private Universities from MEXT, Japan (to K.I.).K.V.P.N. was supported in part by grants from the Tokyo Ohka Foundation and the Precursory Research for Embryonic Science and Technology program of the Japan Science and Technology Agency.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1703584114/-/DCSupplemental.
References
- 1.Robert B, Cogdell RJ, van Grondelle R. The light-harvesting system of purple bacteria. In: Green BR, Parson WW, editors. Light-Harvesting Antennas in Photosynthesis. Kluwer; Dordrecht, The Netherlands: 2003. pp. 169–194. [Google Scholar]
- 2.Wang-Otomo Z-Y. Recent understanding on the photosystem of purple photosynthetic bacteria. In: Sugiyama M, Fujii S, Nakamura S, editors. Solar to Chemical Energy Conversion. Springer International; Cham, Switzerland: 2016. pp. 379–390. [Google Scholar]
- 3.Madigan MT. A novel photosynthetic purple bacterium isolated from a Yellowstone hot spring. Science. 1984;225:313–315. doi: 10.1126/science.225.4659.313. [DOI] [PubMed] [Google Scholar]
- 4.Garcia D, Parot P, Vermeglio A, Madigan MT. The light-harvesting complexes of a thermophilic purple sulfur photosynthetic bacterium Chromatium tepidum. Biochim Biophys Acta. 1986;850:390–395. [Google Scholar]
- 5.Nozawa T, Fukuda T, Hatano M, Madigan MT. Organization of intracytoplasmic membranes in a novel thermophilic purple photosynthetic bacterium as revealed by absorption, circular dichroism and emission spectra. Biochim Biophys Acta. 1986;852:191–197. [Google Scholar]
- 6.Ma F, Yu L-J, Wang-Otomo Z-Y, van Grondelle R. Temperature dependent LH1→RC energy transfer in purple bacteria Tch. tepidum with shiftable LH1-Qy band: A natural system to investigate thermally activated energy transfer in photosynthesis. Biochim Biophys Acta. 2016;1857:408–414. doi: 10.1016/j.bbabio.2015.12.006. [DOI] [PubMed] [Google Scholar]
- 7.Ma F, Yu L-J, Hendrikx R, Wang-Otomo Z-Y, van Grondelle R. Direct observation of energy detrapping in LH1-RC complex by two-dimensional electronic spectroscopy. J Am Chem Soc. 2017;139:591–594. doi: 10.1021/jacs.6b11017. [DOI] [PubMed] [Google Scholar]
- 8.Kimura Y, et al. Calcium ions are involved in the unusual red shift of the light-harvesting 1 Qy transition of the core complex in thermophilic purple sulfur bacterium Thermochromatium tepidum. J Biol Chem. 2008;283:13867–13873. doi: 10.1074/jbc.M800256200. [DOI] [PubMed] [Google Scholar]
- 9.Yu L-J, Kato S, Wang Z-Y. Examination of the putative Ca2+-binding site in the light-harvesting complex 1 of thermophilic purple sulfur bacterium Thermochromatium tepidum. Photosynth Res. 2010;106:215–220. doi: 10.1007/s11120-010-9596-y. [DOI] [PubMed] [Google Scholar]
- 10.Niwa S, et al. Structure of the LH1-RC complex from Thermochromatium tepidum at 3.0 Å. Nature. 2014;508:228–232. doi: 10.1038/nature13197. [DOI] [PubMed] [Google Scholar]
- 11.Kimura Y, Yu L-J, Hirano Y, Suzuki H, Wang Z-Y. Calcium ions are required for the enhanced thermal stability of the light-harvesting reaction center core complex from thermophilic purple sulfur bacterium Thermochromatium tepidum. J Biol Chem. 2009;284:93–99. doi: 10.1074/jbc.M806840200. [DOI] [PubMed] [Google Scholar]
- 12.Kondo M, et al. Photocurrent and electronic activities of oriented His-tagged photosynthetic light-harvesting/reaction center core complexes assembled onto a gold electrode. Biomacromolecules. 2012;13:432–438. doi: 10.1021/bm201457s. [DOI] [PubMed] [Google Scholar]
- 13.Suzuki H, et al. Purification, characterization and crystallization of the core complex from thermophilic purple sulfur bacterium Thermochromatium tepidum. Biochim Biophys Acta. 2007;1767:1057–1063. doi: 10.1016/j.bbabio.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 14.Yu L-J, Suga M, Wang-Otomo Z-Y, Shen J-R. Crystal structure of LH1-RC supercomplex from Thermochromatium tepidum at 1.9 Å resolution. The 17th International Congress on Photosynthesis Research. 2016 Available at schd.ws/hosted_files/ps2016/03/Abstract_Book_PS17.pdf. Accessed September 20, 2017.
- 15.Nagashima S, Shimada K, Matsuura K, Nagashima KVP. Transcription of three sets of genes coding for the core light-harvesting proteins in the purple sulfur bacterium, Allochromatium vinosum. Photosynth Res. 2002;74:269–280. doi: 10.1023/A:1021280104053. [DOI] [PubMed] [Google Scholar]
- 16.Wang Z-Y, Shimonaga M, Suzuki H, Kobayashi M, Nozawa T. Purification and characterization of the polypeptides of core light-harvesting complexes from purple sulfur bacteria. Photosynth Res. 2003;78:133–141. doi: 10.1023/B:PRES.0000004328.11219.79. [DOI] [PubMed] [Google Scholar]
- 17.Qian P, et al. Three-dimensional structure of the Rhodobacter sphaeroides RC-LH1-PufX complex: Dimerization and quinone channels promoted by PufX. Biochemistry. 2013;52:7575–7585. doi: 10.1021/bi4011946. [DOI] [PubMed] [Google Scholar]
- 18.Tonouchi N, Kosumi D, Sugisaki M, Nango M, Hashimoto H. How do surrounding environments influence the electronic and vibrational properties of spheroidene? Photosynth Res. 2015;124:77–86. doi: 10.1007/s11120-015-0095-z. [DOI] [PubMed] [Google Scholar]
- 19.Noguchi T, Hayashi H, Tasumi M. Factors controlling the efficiency of energy transfer from carotenoids to bacteriochlorophyll in purple photosynthetic bacteria. Biochim Biophys Acta. 1990;1017:280–290. [Google Scholar]
- 20.Cogdell RJ, Andersson PO, Gillbro T. Carotenoid singlet states and their involvement in photosynthetic light-harvesting pigments. J Photochem Photobiol B. 1992;15:105–112. [Google Scholar]
- 21.Davis CM, Bustamante PL, Loach PA. Reconstitution of the bacterial core light-harvesting complexes of Rhodobacter sphaeroides and Rhodospirillum rubrum with isolated α- and β-polypeptides, bacteriochlorophyll a, and carotenoid. J Biol Chem. 1995;270:5793–5804. doi: 10.1074/jbc.270.11.5793. [DOI] [PubMed] [Google Scholar]
- 22.Nakagawa K, et al. Probing the effect of the binding site on the electrostatic behavior of a series of carotenoids reconstituted into the light-harvesting 1 complex from purple photosynthetic bacterium Rhodospirillum rubrum detected by stark spectroscopy. J Phys Chem B. 2008;112:9467–9475. doi: 10.1021/jp801773j. [DOI] [PubMed] [Google Scholar]
- 23.Robert B. The electronic structure, stereochemistry and resonance Raman spectroscopy of carotenoids. In: Frank HA, Young AJ, Britton G, Cogdell RJ, editors. The Photochemistry of Carotenoids. Kluwer; Dordrecht, The Netherlands: 1999. pp. 189–201. [Google Scholar]
- 24.Kimura Y, et al. Metal cations modulate the bacteriochlorophyll-protein interaction in the light-harvesting 1 core complex from Thermochromatium tepidum. Biochim Biophys Acta. 2012;1817:1022–1029. doi: 10.1016/j.bbabio.2012.03.016. [DOI] [PubMed] [Google Scholar]
- 25.Sturgis JN, Olsen JD, Robert B, Hunter CN. Functions of conserved tryptophan residues of the core light-harvesting complex of Rhodobacter sphaeroides. Biochemistry. 1997;36:2772–2778. doi: 10.1021/bi962524a. [DOI] [PubMed] [Google Scholar]
- 26.Gall A, et al. Effects of mutagenesis on the detailed structure of spheroidenone in the Rhodobacter sphaeroides reaction centre examined by resonance Raman spectroscopy. Photosynth Res. 1999;59:223–230. [Google Scholar]
- 27.Sturgis JN, Robert B. Pigment binding-site and electronic properties in light-harvesting proteins of purple bacteria. J Phys Chem. 1997;101:7227–7231. [Google Scholar]
- 28.Zilsel J, Lilburn TG, Beatty JT. Formation of functional inter-species hybrid photosynthetic complexes in Rhodobacter capsulatus. FEBS Lett. 1989;253:247–252. doi: 10.1016/0014-5793(89)80969-x. [DOI] [PubMed] [Google Scholar]
- 29.Hunter CN. Genetic manipulation of the antenna complexes of purple bacteria. In: Blankenship RE, Madigan MT, Bauer CD, editors. Anoxygenic Photosynthetic Bacteria. Kluwer; Dordrecht, The Netherlands: 1995. pp. 473–501. [Google Scholar]
- 30.Kortlüke C, Breese K, Gad’on N, Labahn A, Drews G. Structure of the puf operon of the obligately aerobic, bacteriochlorophyll a-containing bacterium Roseobacter denitrificans OCh114 and its expression in a Rhodobacter capsulatus puf puc deletion mutant. J Bacteriol. 1997;179:5247–5258. doi: 10.1128/jb.179.17.5247-5258.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fulcher TK, Beatty JT, Jones MR. Demonstration of the key role played by the PufX protein in the functional and structural organization of native and hybrid bacterial photosynthetic core complexes. J Bacteriol. 1998;180:642–646. doi: 10.1128/jb.180.3.642-646.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crouch LI, Jones MR. Cross-species investigation of the functions of the Rhodobacter PufX polypeptide and the composition of the RC-LH1 core complex. Biochim Biophys Acta. 2012;1817:336–352. doi: 10.1016/j.bbabio.2011.10.009. [DOI] [PubMed] [Google Scholar]
- 33.Nagashima KVP, et al. Exchange and complementation of genes coding for photosynthetic reaction center core subunits among purple bacteria. J Mol Evol. 2014;79:52–62. doi: 10.1007/s00239-014-9634-z. [DOI] [PubMed] [Google Scholar]
- 34.Holden-Dye K, Crouch LI, Jones MR. Structure, function and interactions of the PufX protein. Biochim Biophys Acta. 2008;1777:613–630. doi: 10.1016/j.bbabio.2008.04.015. [DOI] [PubMed] [Google Scholar]
- 35.Yu L-J, Kawakami T, Kimura Y, Wang-Otomo Z-Y. Structural basis for the unusual Qy red-shift and enhanced thermostability of the LH1 complex from Thermochromatium tepidum. Biochemistry. 2016;55:6495–6504. doi: 10.1021/acs.biochem.6b00742. [DOI] [PubMed] [Google Scholar]
- 36.Kimura Y, et al. Effects of calcium ions on the thermostability and spectroscopic properties of the LH1-RC complex from a new thermophilic purple bacterium Allochromatium tepidum. J Phys Chem B. 2017;121:5025–5032. doi: 10.1021/acs.jpcb.7b03341. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.