Significance
CTCF interacts with the genome through thousands of sites and organizes the genome into topological domains, but whether CTCF has direct functions in the maintenance of genome stability is not known. Here, we report that CTCF depletion increases chromosomal instability and activates the DNA damage response. We show that CTCF is recruited to sites of DNA lesions in a process that depends on DNA damage signaling and the DNA-binding domain of CTCF, and that CTCF participates in homologous recombination repair of DNA double-strand breaks by interacting with Rad51 and promoting Rad51 repair foci formation. Thus, CTCF maintains genome stability by participating in DNA repair, highlighting a potential link between genome organization and genome stability.
Keywords: CTCF, chromatin, genome stability, DNA repair, homologous recombination
Abstract
CTCF is an essential epigenetic regulator mediating chromatin insulation, long-range regulatory interactions, and the organization of large topological domains in the nucleus. Phenotypes of CTCF haploinsufficient mutations in humans, knockout in mice, and depletion in cells are often consistent with impaired genome stability, but a role of CTCF in genome maintenance has not been fully investigated. Here, we report that CTCF maintains genome stability, is recruited to sites of DNA damage, and promotes homologous recombination repair of DNA double-strand breaks (DSBs). CTCF depletion increased chromosomal instability, marked by chromosome breakage and end fusions, elevated genotoxic stress-induced genomic DNA fragmentation, and activated the ataxia telangiectasia mutated (ATM) kinase. We show that CTCF could be recruited to drug-induced 53BP1 foci and known fragile sites, as well as to I-SceI endonuclease-induced DSBs. Laser irradiation analysis revealed that this recruitment depends on ATM, Nijmegen breakage syndrome (NBS), and the zinc finger DNA-binding domain of CTCF. We demonstrate that CTCF knockdown impaired homologous recombination (HR) repair of DSBs. Consistent with this, CTCF knockdown reduced the formation of γ-radiation–induced Rad51 foci, as well as the recruitment of Rad51 to laser-irradiated sites of DNA lesions and to I-SceI–induced DSBs. We further show that CTCF is associated with DNA HR repair factors MDC1 and AGO2, and directly interacts with Rad51 via its C terminus. These analyses establish a direct, functional role of CTCF in DNA repair and provide a potential link between genome organization and genome stability.
The insulator protein CTCF organizes the genome into local chromatin loops and large topological domains and carries out numerous diverse nuclear functions by binding to thousands of sites in the genome (1–3). CTCF mutations in humans leads to microcephaly and intellectual disability (4). Its conditional knockout in mice results in the loss of neuronal cells (5, 6), blocked differentiation and proliferation in lymphocytes, and massive apoptosis in the developing limb (7, 8), while mice with a CTCF hemizygous mutation are predisposed to cancer (9). In Drosophila, CTCF mutations lead to instability of the ribosomal RNA gene loci (10). Experiments in cultured cells have implicated CTCF in the protection against UV-induced apoptosis (11). While these complex phenotypes could result from the diverse reported nuclear functions of CTCF, they are also consistent with impaired genome stability. However, whether and, if so, how CTCF plays a role in genome maintenance have not been systematically investigated. Here we report that CTCF has a direct role in the maintenance of genome stability, and that its depletion results in chromosomal instability and activation of the DNA damage response (DDR). CTCF is recruited to sites of DNA damage in a process that depends on ataxia telangiectasia mutated (ATM) and Nijmegen breakage syndrome (NBS), and CTCF facilitates the homologous recombination directed DNA double-strand break (DSB) repair by interacting with Rad51 and promoting the formation of Rad51 repair foci.
Results
CTCF Prevents Genomic Instability.
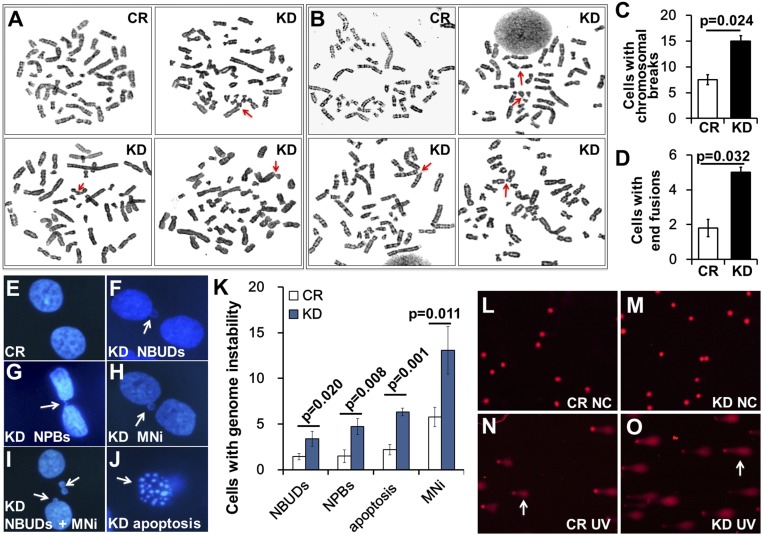
To directly test whether CTCF depletion increases genomic instability, we performed cytogenetic analysis of chromosomes in CTCF-depleted B-cell leukemia BALL-1 cells. We found a higher incidence of chromosomal abnormalities, including twofold increases in chromosomal breakage (Fig. 1 A and C) and end fusions (12, 13) (Fig. 1 B and D). As chromosomal instability usually manifests as nuclear buds (NBUDs), nucleoplasmic bridges (NPBs), micronuclei (MNi), and increased apoptosis, we evaluated the effect of CTCF depletion on these markers (14) and found significant increases in NBUD, NPB, and MNi formation, along with an elevated percentage of apoptotic cells (Fig. 1 E–K). These results suggest an increased level of genomic instability and DNA damage in CTCF-depleted cells.
Fig. 1.
CTCF knockdown led to chromosomal instability and aggravated drug-induced genome fragmentation. (A and C) Cytogenetic analysis of chromosomal breaks in control (CR) or CTCF knockdown (KD) BALL-1 cells (arrows). (B and D) Cytogenetic analysis for telomere fusions before and after CTCF depletion in BALL-1 cells. Arrows indicate fused chromosomal ends. (E–K) Representative images (arrows) of NBUDs, NPBs, and MNi, as well as apoptotic cells, in the CTCF-depleted BALL-1 cells. Statistical results from E–J were obtained from analysis of >100 mitotic cells each. (L–O) U2OS cells were transfected with siCR (CR) or siCTCF (KD) for 72 h, then plated in six-well plates for another 24 h before being treated with 60 J/cm2 of UV, followed by collection for comet analysis. Arrows point to genomic DNA migrated away from the original location in the nucleus.
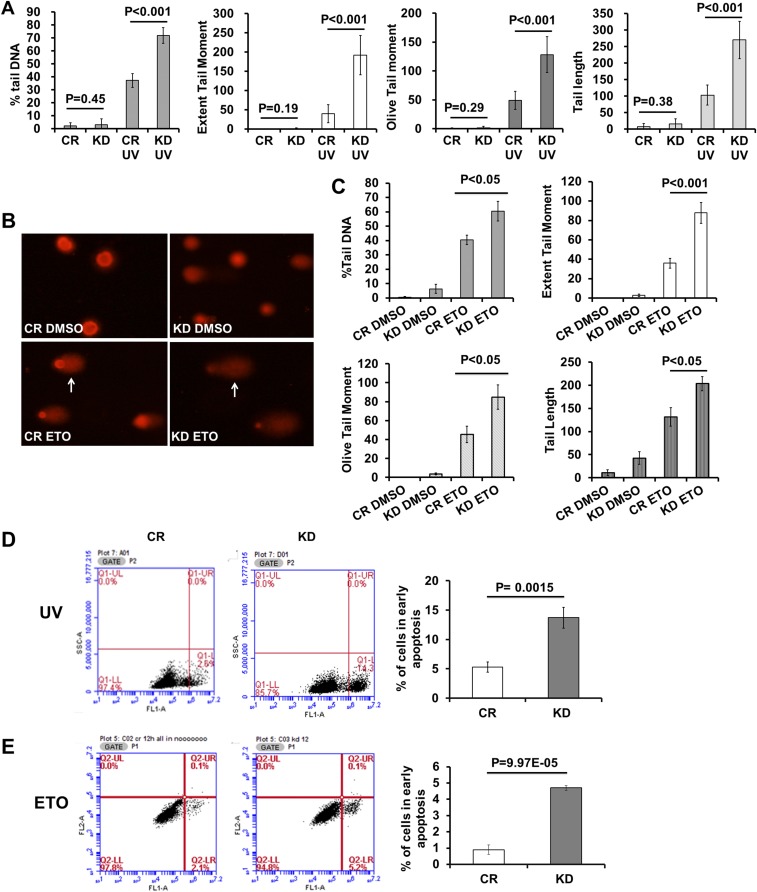
We next measured the effect of CTCF depletion on UV-induced genomic DNA fragmentation using the comet assay, and observed that the UV-treated cells produced significant amounts of fragmented genomic DNA that migrated away from the original locations, while untreated cells showed little or no migration after electrophoresis (Fig. 1 L and N). CTCF depletion increased the amount and migration distance of this fragmented genomic DNA (Fig. 1 M and O). Statistical analysis confirmed that the percentage of tail DNA and the extent of tail movement increased after CTCF knockdown, indicative of more DNA breaks (Fig. S1A). A similar effect of CTCF knockdown was seen in these cells treated with etoposide (ETO), a topoisomerase II inhibitor known to induce a large amount of DSBs (Fig. S1 B and C). Because genotoxic stress treatments will result in apoptosis in treated cells, we measured the effects of CTCF depletion on UV- and ETO-induced apoptosis, and found that the knockdown indeed increased apoptosis in these treated cells (Fig. S1 D and E). Taken together, these results demonstrate that CTCF prevents genomic instability.
Fig. S1.
CTCF depletion increased genotoxic stress-induced genome instability and apoptosis in U2OS cells. (A) U2OS cells were transfected with siCR (CR) or siCTCF (KD) for 72 h, then plated into six-well plates for another 24 h before being treated with 60 J/cm2 of UV. Then these cells were collected for comet analysis. Statistical analysis of the comet assay in more than 150 cells was done using casp-1.2.2 software. (B) U2OS cells were transfected with siCR (CR) and siCTCF (KD) for 72 h, plated in six-well plates for 24 h, and then treated with 25 μM etoposide for 1 h. Then cells were collected for comet analysis. Arrows points to genomic DNA migrated away from the nucleus. (C) Comet assay results of more than 150 cells were analyzed with casp-1.2.2 software. (D) U2OS cells were transfected with siCR and siCTCF for 72 h, plated in 24-well plates for 24 h, and then exposed to UV. Cells were allowed to recover in medium for 12 h before collection for apoptosis analysis. The apoptotic cells were counted based on Annexin V staining using Alexa Fluor 488-conjugated Annexin V (Invitrogen) according to the manufacturer’s instructions. Cell apoptosis was analyzed by flow cytometry; 10,000 cells per sample were counted. (E) U2OS cells were transfected with siCR and siCTCF for 72 h, plated in six-well plates for 24 h, and then incubated with 25 μM ETO for 30 min. Cells were allowed to recover in medium without etoposide for 12 h before collection for apoptosis analysis. The same method as in C was used to detect apoptosis.
CTCF Knockdown Activates the DDR.
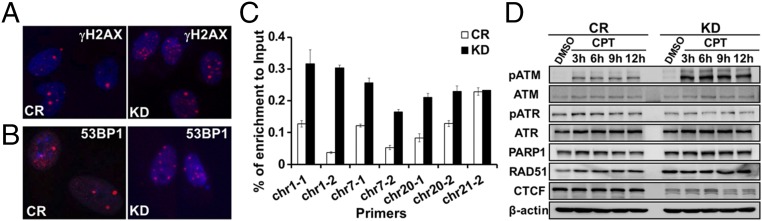
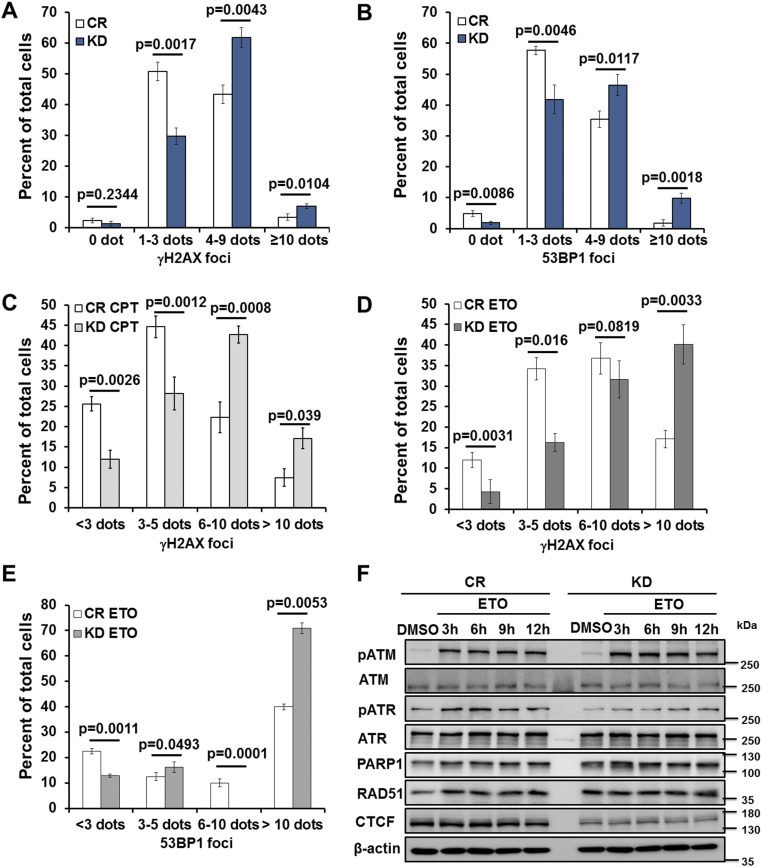
Defects in genome maintenance cause the accumulation of unrepaired genomic lesions, the increased expression of common fragile sites (CFS), and the activation of the DDR (15, 16). To determine whether CTCF depletion increases DDR activation, we examined the immunofluorescence signal of phosphorylated histone variant H2AX (γH2AX) in CTCF-depleted cells, as γH2AX is an indicative of ongoing DNA repair and unresolved DNA lesions (17). We found a strong increase in the number of γH2AX foci in CTCF knockdown cells compared with control cells (Fig. 2A and Fig. S2A). Consistent with this, CTCF depletion increased ETO and camptothecin (CPT; a topoisomerase I inhibitor inducing single-stranded DNA breaks) increased the number of γH2AX foci (Fig. S2 C and D). We also examined 53BP1 immunofluorescence signals in CTCF-depleted cells, as 53BP1 is a key factor in repairing DSBs generated by fragile site expression and other types of DNA damage (18, 19). We saw similar increases in 53BP1 foci number by CTCF knockdown in untreated cells (Fig. 2B and Fig. S2B) and in ETO-treated cells (Fig. S2E). To further test whether CTCF prevents genomic instability, we examined the effect of CTCF depletion on γH2AX occupancy at several known fragile sites by chromatin immunoprecipitation (ChIP) followed by quantitative PCR (20). Indeed, knockdown of CTCF in human primary fibroblast BJ cells led to increased γH2AX binding at all sites examined except site chr21-2 (Fig. 2C). These results suggest that CTCF maintains genome stability, and that its depletion activates DDR signaling.
Fig. 2.
CTCF knockdown activates the DNA damage response. (A and B) γH2AX (A) and 53BP1 (B) immunofluorescence in CR and CTCF knockdown (KD) BJ cells. (C) γH2AX ChIP experiment in CR and CTCF KD cells. The genomic regions amplified were as described previously (20) (P < 0.05, except for chr21-2). Primers sequences are listed in Table S1). (D) Western blot analysis in CR or CTCF KD U2OS cells treated with DMSO or CPT. β-actin was used as a loading control.
Fig. S2.
CTCF depletion results in genomic instability and increased sensitivity to genotoxic stress. (A and B) Control (CR) and CTCF knockdown (KD) BJ cells were subject to immunofluorescence using antibodies against γH2AX (A) and 53BP1 (B). Quantification is shown in the graphs. (C) After control or CTCF depletion for 48–72 h, BJ cells were plated in 24-well plates for 24 h and then incubated with 2 μM CPT for 0.5 h. Cells were allowed to recover in medium without camptothecin for 1 h before being fixed for immunofluorescence analysis. γH2AX foci were counted in more than 100 cells. (D and E) CTCF was depleted for 48–72 h in BJ cells, which were plated in 24-well plates for 24 h and then incubated with 25 μM ETO for 30 min. Cells were allowed to recover in medium without etoposide for 1 h before being fixed for immunofluorescence analysis using the indicated antibodies. γH2AX foci and 53BP1 foci were counted in more than 100 cells. (F) CTCF was depleted with siRNA in U2OS cells for 48–72 h, followed by treatment with DMSO and 25 μM ETO for 1 h. Cells were placed in fresh medium for the indicated times and then processed for Western blot analysis using antibodies against the indicated proteins. β-actin served as a loading control.
To directly measure the activation of DDR, we analyzed key DDR proteins by Western blot, and found that the phosphorylation level of ATM was strongly increased by CTCF knockdown in CPT- and ETO-treated cells (Fig. 2D and Fig. S2F), while total ATM expression remained unchanged. In addition, Rad51, a single-stranded DNA-binding protein responsible for HR repair (21, 22), showed modest increases in CTCF depleted cells. In contrast, the ATR kinase was only minimally affected by the knockdown (Fig. 2D and Fig. S2F), as was the level of PARP1, a protein that mediates single-stranded DNA repair. Thus, CTCF knockdown led to a stronger activation of the ATM kinase in drug-treated cells and thus an elevated level of DDR.
CTCF Is Recruited to Sites of DNA Damage.
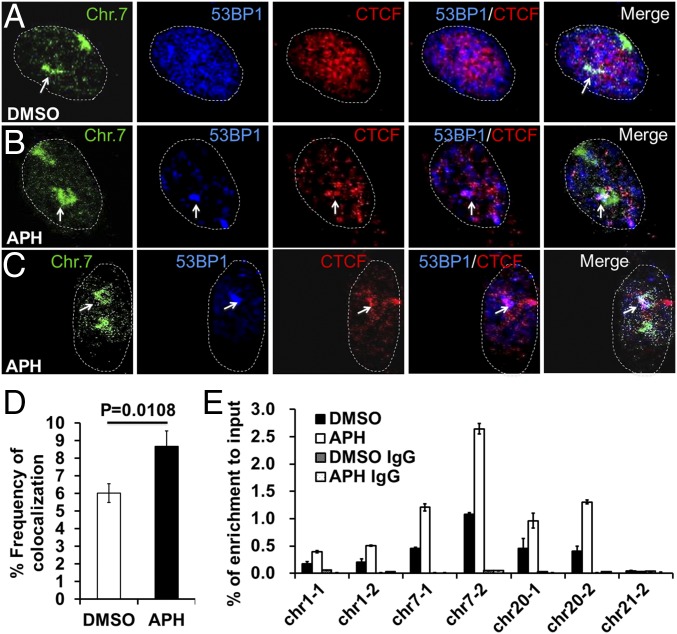
To determine whether CTCF is present at sites of DNA damage, we combined immunofluorescence with fluorescence in situ hybridization (immune-FISH) and observed increased colocalization of CTCF, 53BP1, and chr7 following fragile site expression-inducing aphidicolin (APH) treatment (Fig. 3 A–C). Statistical analysis showed that APH treatment produced a 45% increase in the colocalization frequency of CTCF, 53BP1, and chr7 (from 6.0% to 8.7%; Fig. 3D), suggesting that CTCF is recruited to expressed fragile sites. The localization of CTCF to sites of DNA damage is also supported by immunofluorescence analysis of either APH-treated (to induce DNA breaks at fragile sites in the genome) or ETO-treated (to induce DSBs) cells, where CTCF colocalized with most of the 53BP1 foci in both untreated and drug-treated cells, suggesting that CTCF was recruited to the induced DNA damage sites (Fig. 4 A and B).
Fig. 3.
Interaction of CTCF with fragile sites following APH treatment. (A–C) BJ cells were treated with DMSO (A) or 0.4 μM APH (B and C) for 24 h, followed by serum starvation for another 24 h. DNA immuno-FISH was performed with whole chromosome painting probes. The procedure was combined with immunofluorescence using antibodies against 53BP1 and CTCF. Representative images of colocalization of CTCF, 53BP1, and chr7 are shown. (D) Colocalization frequency of 53BP1 and CTCF immunofluorescence signals, and the FISH signals of chr7 as shown in A–C. (E) CTCF ChIP conducted in U2OS cells that had been incubated in DMSO or 0.4 μM APH for 24 h before being fixed with 1% formaldehyde. The genomic regions amplified were chosen according to previously published ChIP sequencing data (20) (P < 0.05, except for the control site chr21-2).
Fig. 4.
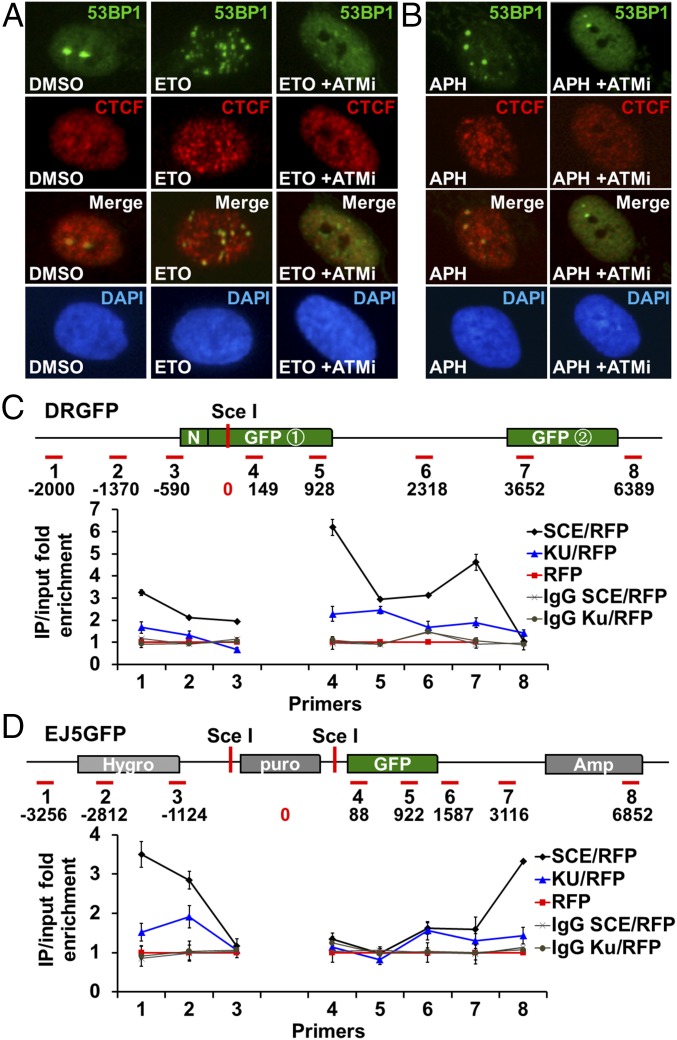
CTCF is recruited to sites of DNA damage. (A) Immunofluorescence using anti-53BP1 and anti-CTCF antibodies in BJ cells treated with DMSO and 25 μM ETO in the presence or absence of ATMi or KU55933 for 1 h. (B) CTCF and 53BP1 immunofluorescence in BJ cells treated with 0.4 μM APH for 24 h in either the presence or absence of ATMi for 3 h. (C and D) DR-GFP U2OS (C) and EJ5-GFP-U2OS (D) cells were transfected with either a blank plasmid plus an RFP plasmid or an I-SceI expression plasmid plus the RFP plasmid. For ATM inhibition, these cells were pretreated with ATMi (25 mmol/L) for 1 h and then incubated with the inhibitors for an additional 8 h during I-SceI transfection. ChIP was performed 24 h later with CTCF antibody. The ChIP DNA was then analyzed by quantitative PCR using primers designed for different regions on the DR-GFP and EJ5-GFP plasmids. The red bars and the coordinates represent distances from the I-SceI cutting sites. ChIP signals are presented here as fold of enrichment over control.
To test whether CTCF directly interacts with sites of DNA lesions, we measured CTCF occupancy at CFS 1q23.2, FRA7D, 20q13.33, and 21q22.3 by ChIP in U2OS cells, as these sites have been shown to be enriched with γH2AX in U2OS (20). We found significant of CTCF enrichment at these tested fragile sites compared with the control site, chr21-2, a site with little or no CTCF binding (Fig. 3E). Importantly, CTCF occupancy at these sites increased following APH treatment, suggesting that CTCF binding to these fragile sites was induced as a result of increased DDR signaling and fragile site expression. In particular, the induced CTCF binding to FRA 7D (tested by primer chr7-2; Fig. 3E) on chromatin 7 after APH treatment is consistent with the immuno-FISH experiments shown in Fig. 3 A–C. These results suggest that CTCF interacts with the DNA sequences near the sites of DNA lesions, and that the binding could be induced by the DDR.
To obtain more direct evidence supporting the interaction of CTCF at sites of DNA damage, we investigated whether CTCF interacts with the DNA near DSB ends by ChIP in a U2OS cell line carrying the DR-GFP reporter (23, 24) (Fig. S3C). Binding of CTCF to the DNA sequences around the I-Sce I endonuclease cutting site was minimal in the absence of the I-Sce I enzyme, but was strongly induced by introducing I-Sce I, as shown in Fig. 4C. A similar result was also using a second inducible DSB system in a U2OS cell line carrying the EJ5-GFP reporter (23), where CTCF is also recruited to most sites flanking the I-Sce I induced break point (Fig. 4D). These results confirm that CTCF was recruited to DSB ends, consistent with the increased recruitment of CTCF seen in drug- induced sites of DNA damage (Fig. 3). Of interest is the apparent absence of a CTCF-binding motif in DR-GFP and EJ5-GFP reporters, suggesting that the interaction between CTCF and sites of DNA damage is independent of a known DNA-binding consensus in the experiments described in Fig. 4 C and D, unlike most other reported CTCF-binding sites in the genome, where a consensus is usually present (1).
Fig. S3.
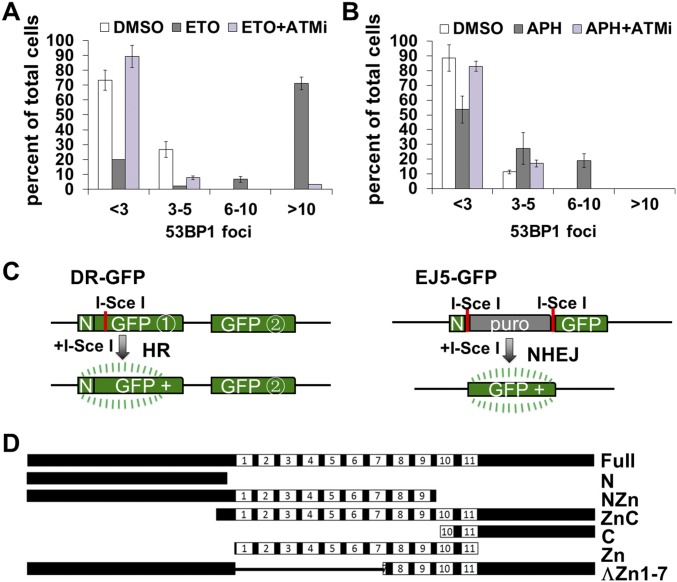
Statistical analysis of drug-induced 53BP1 foci and diagrams of HR and NHEJ reporters and CTCF segments. (A) Immunofluorescence using anti-53BP1 and anti-CTCF antibodies in BJ cells treated with DMSO and 25 μM ETO in the presence or absence of ATMi or KU55933 for 1 h. The 53BP1 foci were quantified. Control cells, n = 172; APH-treated cells, n = 135; ATMi cells, n = 130. (B) CTCF and 53BP1 immunofluorescence in BJ cells treated with 0.4 μM APH for 24 h in either presence or absence of ATMi for 3 h. The 53BP1 foci were quantified. Control cells, n = 140; APH-treated cells, n = 110; APH + ATMi cells, n = 141. (C) Schematic representation of the HR (DR-GFP) and NHEJ (EJ5-GFP) assay. (D) Diagram of CTCF and various regions fused to RFP.
CTCF Recruitment Depends on ATM, NBS, and the DNA-Binding Domain of CTCF.
To determine whether DDR signaling is necessary for CTCF recruitment to sites of DNA damage, we used an inhibitor of the ATM kinase (ATMi), KU55933, to block the DDR in ETO- and APH-treated cells. We observed a strong inhibition of CTCF recruitment to the 53BP1 foci, along with simultaneous reductions of both 53BP1 foci number and size (Fig. 4 A and B and Fig. S3 A and B), suggesting that CTCF recruitment to the drug-induced DNA damage sites is ATM-dependent. To confirm this, we examined CTCF binding to DNA sequences near DSB ends by ChIP in the presence of ATMi, and indeed observed significant reductions in CTCF occupancy near DSBs in both DR-GFP and EJ5-GFP reporters (Fig. 4 C and D). Thus, CTCF recruitment to DSBs requires DDR signaling initiated by ATM.
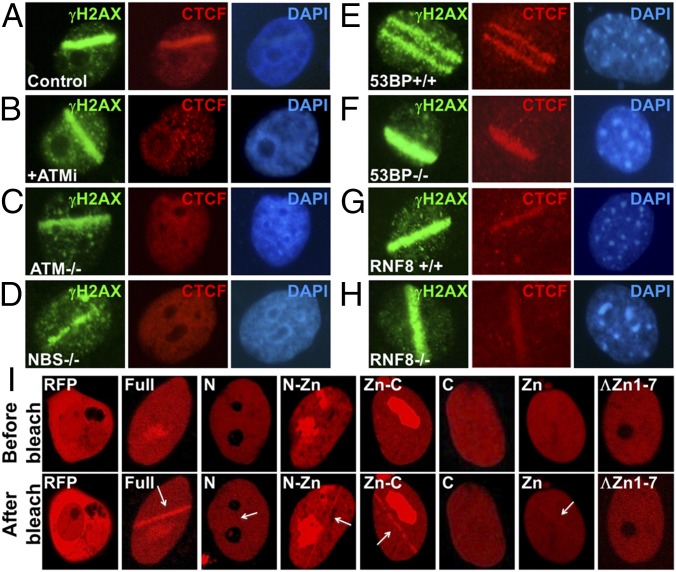
To further characterize CTCF recruitment to sites of DNA damage, we used a laser microbeam to induce DNA lesions in U2OS cells, followed by double-immunofluorescence staining with γH2AX and CTCF antibodies. Laser microirradiation resulted in colocalization of CTCF with the DSBs tracks in the irradiated nucleus (indicated by γH2AX signals; Fig. 5A), but this colocalization was absent in ATMi-treated and ATM-deficient cells (Fig. 5 B and C), confirming the observations in Fig. 4.
Fig. 5.
CTCF recruitment depends on ATM, NBS, and the DNA-binding domain of CTCF. (A–H) Laser irradiation analysis was done with an Olympus FluoView FV1000 confocal microscope in U2OS cells (A), U2OS cells in the presence of ATMi (B), human ATM-deficient cells (C), NBS1- deficient cells (D), wild-type MEF cells from a 53BP1 control mouse (E), 53BP1-deficient mouse MEF cells (F), wild-type MEF cells from an RNF8 control mouse (G), and RNF8-deficient MEF cells (H). These cells were scanned with a 405-nm laser and then fixed for immunofluorescence. γH2AX antibody was used to detect laser-induced sites of DNA lesions, while CTCF antibody was used to detect potential enrichment of CTCF at sites of damage. (I) RFP-tagged full-length CTCF and domains of CTCF were transfected into U2OS cells, followed by laser micro-irradiation and observation under the confocal microscope.
Using this method, we then examined CTCF recruitment in cell lines deficient for NBS1, 53BP1, or RNF8 (Fig. 5 D–H), and found that cells deficient in NBS1 also failed to recruit CTCF to the laser-irradiated regions (Fig. 5D), while 53BP1 or RNF8 deficiency did not affect this recruitment (Fig. 5 E–H). This result suggests that the Mre11-Rad50-Nbs1 (MRN) complex (25) is necessary for the recruitment of CTCF to sites of DNA damage.
To dissect the domains of CTCF responsible for its recruitment, we created fusion proteins between sections of CTCF and the RFP protein. The full-length CTCF molecule was divided into several sections, and RFP tags were added (26, 27) (Fig. S3D). The full- length N- and C-terminal truncations colocalized with laser-induced DNA damage regions, while the N and C termini alone showed no or minimal recruitment into the laser-induced regions of DNA damage (Fig. 5I). The zinc finger region showed weak but detectable recruitment, and when one to seven zinc finger domains were removed from CTCF (28), recruitment was essentially abolished (Fig. 5I). Thus, the zinc finger region is necessary for targeting CTCF to laser-induced sites of DNA damage. Taken together, the results from Figs. 4 and 5 demonstrate that CTCF recruitment to regions of DNA damage is ATM, NBS1, and CTCF DNA-binding domain dependent.
CTCF Promotes HR-Directed DNA DSB Repair.
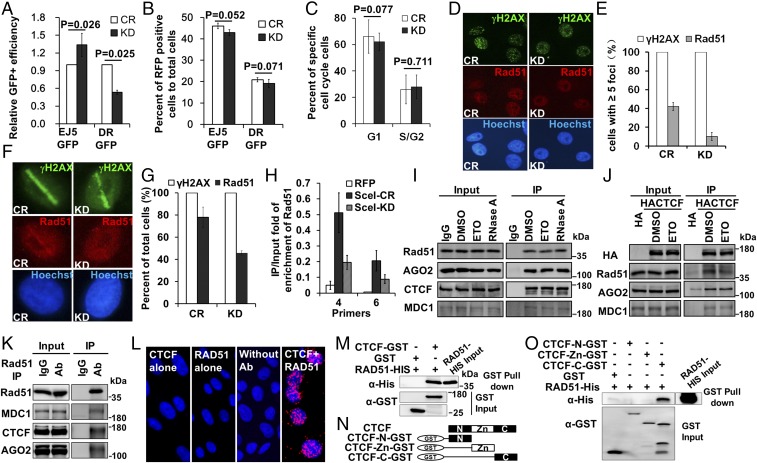
To test whether CTCF functions in DSB repair, we used two established assay systems, DR-GFP and EJ5-GFP (Fig. S3C), to measure the effect of CTCF depletion on HR and nonhomologous end-joining repair (NHEJ), respectively. We found that when CTCF level was reduced by siRNA, there was a 47% reduction in relative HR efficiency, from 1.0 to 0.53 (Fig. 6A), suggesting a function of CTCF in HR. In contrast, knockdown elevated the efficiency of NHEJ from 1.0 to 1.34, a 34% increase (Fig. 6A), indicating that CTCF is not necessary for NHEJ. Since NHEJ is a compensatory repair choice when HR is blocked, this result is also consistent with a role of CTCF in HR, a phenotype similar to the depletion of several repair factors in the HR pathway (29–31). In these experiments, an RFP expression plasmid was included to control for transfection efficiency, while cell cycle distributions were assessed before and after CTCF knockdown in the modified U2OS cell lines to control for cell cycle effects. We found no significant differences in transfection efficiency between control cells and knockdown cells (Fig. 6B), or any significant changes in the cell population at different stages of the cell cycle after CTCF knockdown (Fig. 6C). Thus, these experiments suggest that CTCF promotes an HR type of DNA repair.
Fig. 6.
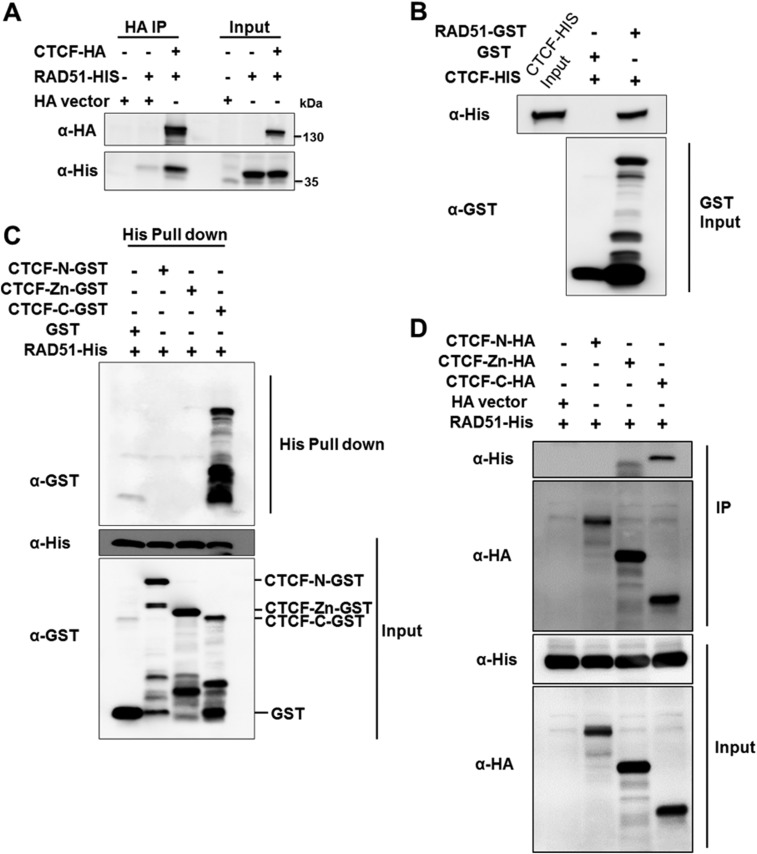
CTCF promotes HR-directed DSB repair. (A) After CTCF depletion for 48–72 h, DR-GFP U2OS or EJ5-GFP-U2OS cells were transfected with I-SceI plasmid. After another 48 h, GFP expression was detected with FACS. (B) Plasmids expressing RFP were also transfected into the aforementioned cells to monitor for transfection efficiency. (C) Propidium iodide (PI) was used to stain the DNA, and DNA content was quantified by FACS to monitor the cell cycle distribution of these cells. (D and E) CTCF was knocked down for 48–72 h in U2OS cells, followed by X-ray irradiation (with 5 Gy). After a 1-h recovery, the cells were fixed for immunostaining to detect Rad51 recruitment (P < 0.001). (F and G) U2OS cells were transfected with siCR (NC) and siCTCF (KD) for 72 h before be subjected to laser scanning and immunostaining to detect Rad51 recruitment (P < 0.05). (H) ChIP analysis of Rad51 recruitment at two locations of high CTCF occupancy (Fig. 4C) near I-SceI–induced breaks in the DR-GFP U2OS cell line before and after CTCF knockdown (P < 0.05). (I) U2OS cells were treated with DMSO or 25 μM ETO for 1 h, and the lysates were untreated or treated with 100 μg/mL RNase A before being subjected to immunoprecipitation using CTCF antibody- or IgG-preincubated Dynabeads. Immunoprecipitates (IP) and whole-cell extracts (Input) were immunoblotted with the indicated antibodies. (J) HA-CTCF expression plasmid or HA vector transfected in U2OS cells were treated with DMSO or 25 μM ETO for 1 h. The cell lysates were then subjected to immunoprecipitation using HA antibody-preincubated Dynabeads, and Western blot analysis was performed with the indicated antibodies. (K) U2OS cells lysates were subjected to immunoprecipitation using Rad51 antibody- or IgG-preincubated Dynabeads, followed by Western blot analysis with the indicated antibodies. (L) CTCF and Rad51 interactions in U2OS cells detected by PLA, shown as red spots. (M) GST or GST-CTCF was preincubated with GST-tagged protein purification resin for 2 h at 4 °C, followed by the addition of His-Rad51 and overnight incubation at 4 °C. Complexes were washed six times with binding buffer. Proteins were visualized by Western blot analysis using the indicated antibodies. (N) Diagram of GST-tagged regions of CTCF. (O) GST or GST-tagged truncations of CTCF were preincubated with GST-tagged protein purification resin, followed by the addition of His-Rad51 and incubation for pull-down analysis. Proteins were visualized by Western blot analysis using the indicated antibodies.
CTCF Interacts with Rad51 and Is Necessary for Formation of Rad51-Labeled DNA Repair Foci.
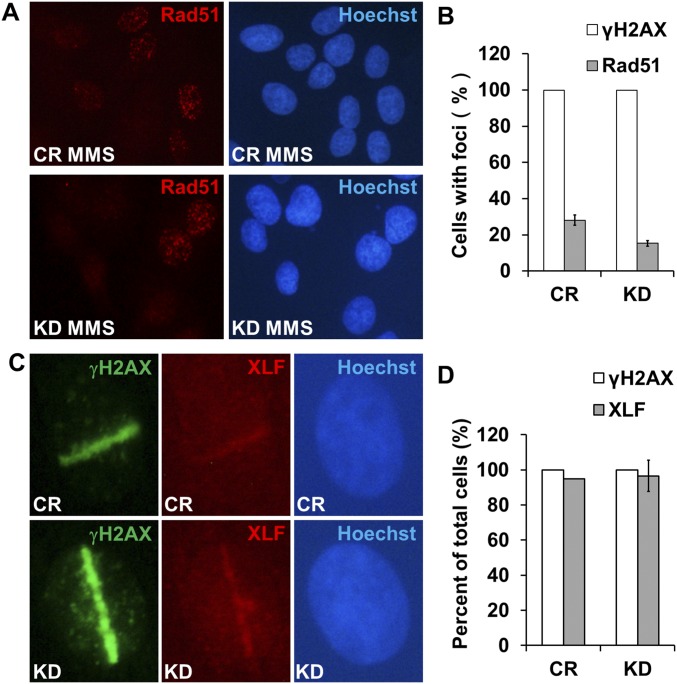
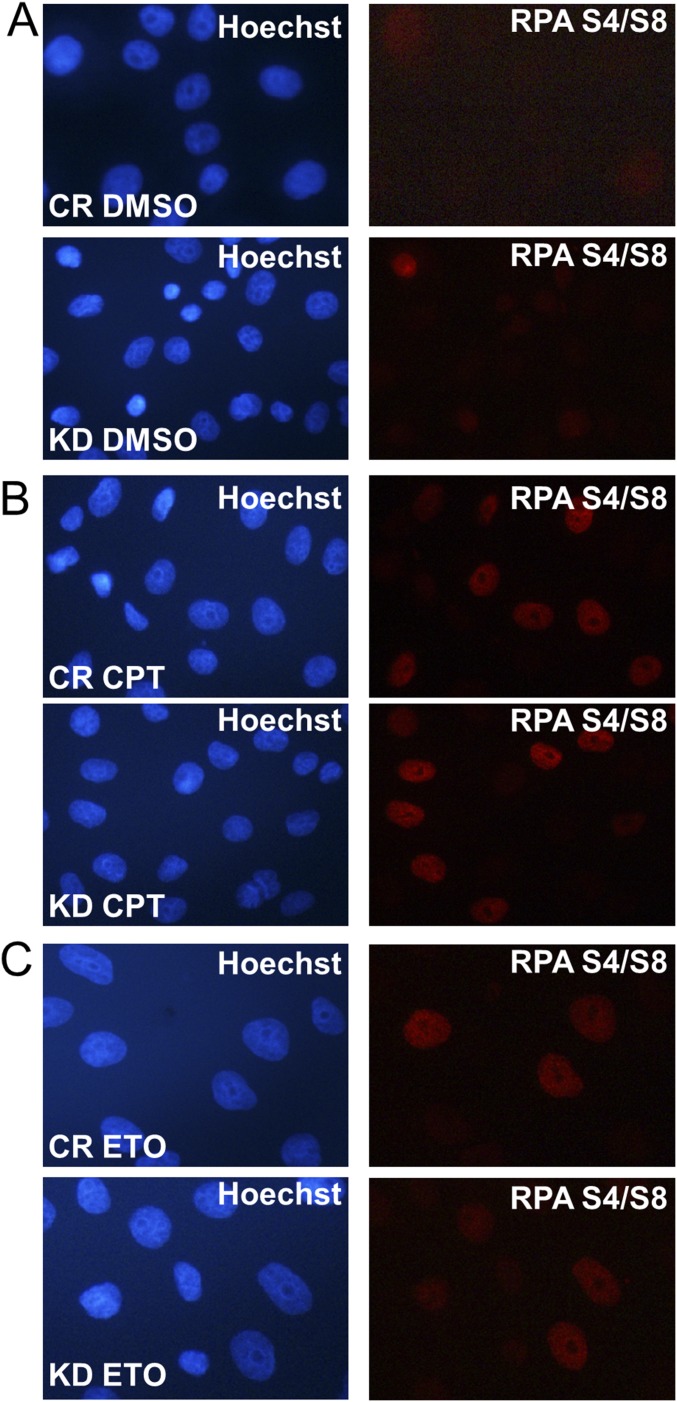
To determine how CTCF functions in the HR, we analyzed X-ray irradiation-induced Rad51 foci formation in the CTCF knockdown cells, since Rad51 is a key repair factor in HR (32–34), and aggregates into DNA repair foci after ionizing irradiation or DNA damage-inducing drug treatment (35, 36). We found that the knockdown decreased the percentage of cells with five or more X-ray–induced Rad51 foci decreased from 42.13% to <10.17% (Fig. 6 D and E). A similar result was obtained in cells treated with methyl methanesulphonate (MMS), a drug that can trigger single-stranded DNA breaks. Here CTCF depletion also led to a decrease in the percentage of Rad51 foci-positive cells (Fig. S4 A and B). We further tested the effect on Rad51 recruitment by CTCF depletion in the laser-induced DNA lesion experiments. Cells depleted of CTCF consistently showed deficient Rad51 recruitment to laser-induced DNA damage regions (Fig. 6 F and G). In contrast, XLF, a protein in the NHEJ repair pathway, was not affected by the knockdown (Fig. S4 C and D). To confirm the effect of CTCF depletion on the recruitment of Rad51, we performed ChIP using an Rad51 antibody, and observed decreased Rad51 binding in two tested sites (primers 4 and 6 in the DR-GFP reporter; Fig. 4C) after CTCF knockdown (Fig. 6H). We also examined the effect of CTCF knockdown on RPA S4/S8 foci formation, and found that these foci were unaffected (Fig. S5). Taken together, these results suggest that CTCF promotes HR by facilitating the recruitment of Rad51.
Fig. S4.
CTCF depletion impairs MMS-induced Rad51 foci formation, but has no effect on XLF recruitment to sites of laser-induced DNA lesions. (A and B) After CTCF was knocked down for 48–72 h using siRNA, U2OS cells were treated with 1 mM MMS for 1 h, then allowed to recover for 1 h before being fixed for immunostaining to detect Rad51 recruitment. The Rad51 foci-positive cells were quantified. Control cells, n = 196; CTCF knockdown cells, n = 222. (C and D) U2OS cells were transfected with siCR and siCTCF for 72 h before confocal microscopy laser scanning. The scanned cells were allowed to recover for 10 min before being fixed for immunostaining to detect XLF recruitment. The cells with XLF recruitment were quantified. Control cells, n = 38; CTCF knockdown cells, n = 58.
Fig. S5.
CTCF knockdown has no effect on single-stranded DNA production. CTCF was depleted for 48–72 h using siRNA in U2OS cells, followed by treatment with DMSO (A), 2 μM CPT (B), or 25 μM ETO (C) for 1 h. The cells were allowed to recover for 1 h in fresh medium before being fixed for immunostaining to detect RPA S4/S8 recruitment. Immunofluorescence was performed with anti-RPA S4/S8 phosphorylated antibody, and Hochest 33342 was used to indicate the cell nuclei.
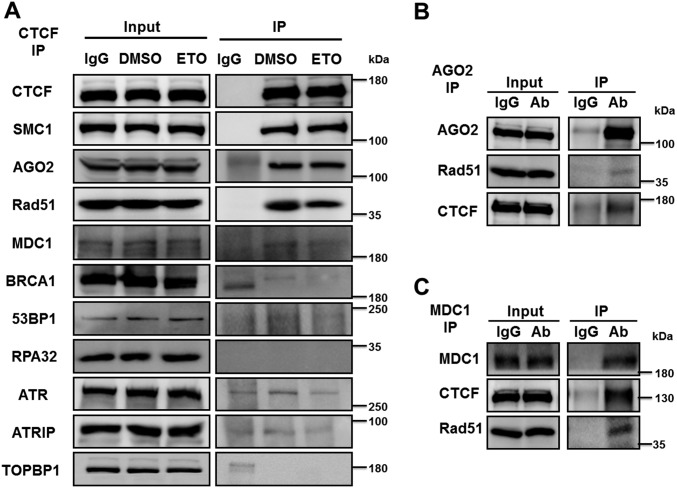
To determine whether CTCF physically interacts with any repair components, we conducted in vivo coimmunoprecipitation (co-IP) experiments and found that endogenous CTCF interacts with MDC1, Rad51, and AGO2 but not with other tested DDR components, including BRCA1, 53BP1, RPA32, ATR, ATRIP, and TOPBP1 (Fig. S6A). Analysis of anti-CTCF immunoprecipitates from U2OS cells incubated with antibody to Rad51, AGO2, and MDC1 revealed that these interactions were not affected by ETO or RNase A treatment, suggesting that the interactions are constitutive and independent of RNA (Fig. 6I). In agreement with interaction between endogenous CTCF and these DNA repair factors, the HA-tagged exogenous CTCF also exhibited interactions with Rad51, AGO2, and MDC1 (Fig. 6J). In the reciprocal experiments, anti-Rad51, -AGO2, and -MDC1 immunoprecipitates immunoblotted with antibody to CTCF also showed associations between CTCF and Rad51, AGO2, and MDC1 (Fig. 6K and Fig. S6 B and C). The CTCF–Rad51 interaction suggests that CTCF promotes HR repair foci formation by facilitating Rad51 recruitment.
Fig. S6.
CTCF interacts with MDC1, Rad51, and AGO2. (A) U2OS cells were treated with DMSO at a dilution of 1/1,000 or 25 μM ETO for 1 h. Then lysates were subjected to immunoprecipitation using CTCF or IgG-preincubated Dynabeads. Immunoprecipitates (IP) and whole-cell extracts (input) were immunoblotted with the indicated antibodies. (B) U2OS cell lysates were subjected to immunoprecipitation using AGO2 antibody or IgG-preincubated Dynabeads. The samples of IP and input were used for subsequent Western blot analysis with the indicated antibodies. (C) U2OS cells lysates were subjected to immunoprecipitation using MDC1 antibody or IgG-preincubated Dynabeads. The samples of IP and input were used for subsequent Western blot analysis with the indicated antibodies.
The in vivo CTCF–Rad51 interaction was further confirmed by co-IP using cotransfected HA-tagged CTCF and His-tagged Rad51 (Fig. S7A), and confirmed by a proximity ligation assay (PLA) (37), which allows direct visualization of in vivo protein–protein interactions (Fig. 6L). To determine whether the CTCF–Rad51 interaction is direct, we performed an in vitro GST pull-down analysis, and found that GST-CTCF can pull down His-Rad51 (Fig. 6M) and that GST-Rad51 can pull down His-CTCF (Fig. S7B). To determine the domain of CTCF responsible for the interaction, we examined the N and C termini and the zinc finger domain of CTCF, and found that the C terminus of CTCF directly interacts with Rad51 in both GST pull-down and co-IP analyses (Fig. 6 N and O and Fig. S7 C and D). These experiments suggest that CTCF may promote Rad51 repair foci formation by facilitating Rad51 recruitment.
Fig. S7.
CTCF directly interacts with Rad51. (A) HA-CTCF and His-Rad51 were coexpressed in HEK293T cells for 48 h. The cell lysates were then subjected to immunoprecipitation using HA antibody-preincubated Dynabeads, and Western blot analysis was performed with the indicated antibodies. (B) Purified recombinant GST-Rad51 interacts with His-CTCF. GST or GST-Rad51 was preincubated with GST-tagged purification resin for 2 h at 4 °C, followed by the addition of His-CTCF and overnight incubation at 4 °C. Complexes were washed six times with binding buffer, and then proteins were visualized by Western blot analysis using the indicated antibodies. (C) Purified recombinant His-Rad51 interacts the with GST-tagged C terminus of CTCF. His-Rad51 was preincubated His-tag purification resin for 2 h at 4 °C, followed by the addition of GST-CTCF-N, GST-CTCF-Zn, or GST-CTCF-C and overnight incubation at 4 °C. Complexes were washed six times with binding buffer, and proteins were visualized by Western blot analysis using the indicated antibodies. (D) CTCF-N-HA, CTCF-Zn-HA, CTCF-C-HA, and His-Rad51 were coexpressed in HEK293T cells for 48 h. The cell lysates were then subjected to immunoprecipitation using HA antibody-preincubated Dynabeads, and Western blot analysis was performed with the indicated antibodies.
Discussion
Here we have provided several lines of evidence supporting a previously undescribed function of CTCF in the maintenance of genome stability and DNA repair. CTCF knockdown increased genomic instability, marked by elevated levels of chromosomal breakage and fragile site expression. In addition, CTCF was physically recruited to sites of DNA lesions, with recruitment dependent on DNA damage signaling, the MRN complex, and the zinc finger DNA-binding domain of CTCF. Furthermore, CTCF participated in the HR type of DSB repair by promoting the recruitment and thus the formation of Rad51-dependent DNA repair foci. These activities suggest a potential link between genome organization and genome maintenance.
Considering its multiple reported nuclear activities, CTCF also could contribute to genome maintenance indirectly, for example, by regulating gene transcription. A recent study reported that CTCF maintains p53 expression and promotes p53 function in the DNA damage response through epigenetic regulation and binding to the antisense transcript originated from the p53 locus (27), suggesting that the RNA-binding properties of CTCF may play a role in DNA damage repair. Here we found that recruitment of CTCF to sites of DNA damage did not depend on the RNA-binding domain of CTCF, and that RNase A did not disrupt the association of CTCF and other DDR proteins (Fig. 6I), suggesting that the DNA repair activity of CTCF that we observed is separable from the effect of CTCF on p53.
The interaction of CTCF and sites of DNA damage exhibits different characteristics from most reported CTCF binding in the genome, in that it is induced by DNA damage and requires DNA damage signaling, but does not appear to require a known DNA-binding motif. This finding leads to the interesting possibility that CTCF binding to the genome could be far more dynamic and regulated than has been reported previously (2, 3, 38). Precisely how CTCF acts in DNA repair will require extensive further investigation. Since CTCF directly interacts with Rad51, and CTCF knockdown disrupted the formation of Rad51 repair foci but did not affect single-stranded DNA formation (RPA S4/S8 foci formation was unaffected; Fig. S5), we believe that CTCF acts downstream of RPA but upstream of Rad51 repair foci formation, possibly by facilitating Rad51 recruitment via the C terminus of CTCF.
We hypothesize that CTCF promotes DNA repair by recruiting Rad51, and that together with Rad51, CTCF stabilizes the DSB ends and/or homologous DNA sequences with the local chromosome architectural network, which could prevent the ends of DSBs from drifting apart and could facilitate the pairing of homologous sequences necessary for repair. Such an activity is supported by a “homing” phenomenon of P elements in Drosophila whereby transposable elements carrying genomic sequences containing CTCF-binding sites tend to integrate back to the very genomic regions from which these sequences derive (39).
Materials and Methods
BJ, BALL-1, and U2OS cells were obtained from American Type Culture Collection. DR-GFP-U2OS and EJ5-GFP-U2OS cell lines were obtained from Jeremy Stark, Beckman Research Institute of the City of Hope, Duarte, CA. ATM−/−, NBS1−/−, 53BP1−/−, and RNF8−/− cell lines were obtained from Matthew Weitzman, Children’s Hospital of Philadelphia, Philadelphia. Human ATM−/− cells were originally from Yosef Shiloh, Sackler School of Medicine, Tel Aviv University, Tel Aviv, human NBS−/− cells were originally from Patrick Concannon, Genetics Institute, University of Florida, Gainesville, FL, and 53BP1 and RNF8 MEFs were originally from Junjie Chen, The University of Texas MD Anderson Cancer Center, Houston. The sources of antibodies, plasmids, and reagents and detailed methodology for immunofluorescence, immuno-FISH, cytokinesis-block micronucleus cytome assay, laser irradiation analysis, CTCF deletion mutant cloning, siRNA- and shRNA-mediated knockdown of CTCF, cell cycle analysis, apoptosis assays, immunoprecipitation and Western blot analyses, ChIP, recombinant protein expression and purification, GST pull-down assays, and PLA are described in SI Materials and Methods.
SI Materials and Methods
Cells and Antibodies.
BJ, BALL-1, and U2OS cells were obtained from American Type Culture Collection. DR-GFP-U2OS and EJ5-GFP-U2OS cell lines were obtained from Jeremy Stark, Beckman Research Institute of the City of Hope, Duarte, CA. ATM−/−, NBS1−/−, 53BP1−/−, and RNF8−/− cell lines were obtained from Matthew Weitzman, Children’s Hospital of Philadelphia, Philadelphia. Human ATM−/− cells were originally from Yosef Shiloh, Sackler School of Medicine, Tel Aviv University, Tel Aviv, human NBS−/− cells were originally from Patrick Concannon, Genetics Institute, University of Florida, Gainesville, FL, and 53BP1 and RNF8 MEFs were originally from Junjie Chen, The University of Texas MD Anderson Cancer Center, Houston. Cells were grown in DMEM (Gibco) supplemented with 10% FBS and 1% penicillin-streptomycin in a humidified 5% CO2 atmosphere at 37 °C. CTCF polyclonal antibodies were purchased from Abcam and also custom made by GL Biochem; CTCF monoclonal antibodies were obtained from EMD Millipore. Antibodies against γH2AX, Rad51, BRCA1, ATM S1981, ATM, β-actin, and HA were purchased from Abcam. Alexa Fluor 594 goat anti-mouse IgG (H + L) and Alexa Fluor 488 goat anti-rabbit IgG (H + L) antibodies were obtained from Life Technologies.
Immunofluorescence.
BJ and U2OS cells were seeded on glass coverslips in 24-well plates at 1 d before the experiments. Cells were fixed with 4% paraformaldehyde at 4 °C for 60 min and then extracted with 0.2% Triton X-100 in PBS for 10 min. Nuclei were visualized by staining with Hoechst 33342. Images were acquired using a Nikon 80i or an Olympus FluoView FV1000 confocal microscope.
Immuno-FISH.
BJ cells were seeded on glass coverslips in 24-well plates at 1 d before treatment with 0.4 μM APH. After a 24-h serum starvation, the cells were fixed with 4% paraformaldehyde at room temperature for 30 min, extracted with 0.5% Triton X-100 in PBS for 10 min, blocked with 5% BSA in PBS for 1 h, and incubated with primary antibody and secondary antibody for 1 h. The cells were then deproteinized with 0.1 mol/L HCl for 7 min, digested with 20 μg/mL RNase A for 20 min, and incubated with probes in hybridization buffer at 95 °C for 4 min. Finally, the cells were washed with 0.4× SSC at 72 °C for 2 min and with 0.05% Tween-20 in 2× SSC at 37 °C for 30 s. Images were acquired using an Olympus FV1000 system.
Cytokinesis-Blocking Micronucleus Cytome Assay.
BALL-1 cells were seeded on glass coverslips in 24-well plates at 1 d before lentivirus infection. The cells were then infected with shCr or shCTCF lentivirus for 72 h and then treated with 3 μg/mL cytochalasin-B for 72 h. After a 24-h recovery, the cells were fixed with paraformaldehyde at room temperature for 30 min and then incubated with 1 μg/mL Hoechst 33342 for 1 min. Images were acquired with a Nikon Eclipse 80i microscope.
Laser Irradiation Analysis.
A 405-nm wavelength laser from an Olympus FluoView FV1000 confocal microscope was focused through a 60× objective lens. One scan of the laser light at full power delivers ∼1,600 nW. Cells were scanned 500 times with the 405-nm laser at full power as described previously (40). The cells were incubated with Opti-MEM (Gibco) in glass-bottom dishes placed in chambers to prevent evaporation on a 37 °C hotplate.
CTCF Deletion Mutant Cloning.
CTCF deletion mutants were made by PCR using the primers in Table S1. PCR products were subsequently cloned into RFP-tagged pmCherry-C1 vector.
Table S1.
Sequences of primers used for ChIP-RT PCR
| Primer | Sequence |
| Fragile site chr1-1 F | TGATGAGAGGGGTCTTCAGG |
| Fragile site chr1-1 R | TGTTCTCTCCTTGGGCCTTA |
| Fragile site chr1-2 F | AGGCCCAAGGAGAGAACAGT |
| Fragile site chr1-2 R | GCAGTCTGGGTTCCAGAGG |
| Fragile site chr7-1 F | TAACCGAAGTGCCTCCAGAC |
| Fragile site chr7-1 R | GGACAGCCTACTGCGTTAGC |
| Fragile site chr7-2 F | CAAAGGGTGGTGCTGTGA |
| Fragile site chr7-2 R | GCTCAGGGGGAGAAGGTG |
| Fragile site chr20-1 F | CCCTCCAGCAGAGTTCAGAC |
| Fragile site chr20-1 R | ATTGAAGGCAACCTGGTGAG |
| Fragile site chr20-2 F | TGTGATGTGTTGGCTGCAA |
| Fragile site chr20-2 R | ATTGAAGGCAACCTGGTGAG |
| Fragile site chr21-2 F | GGAGAGAACTCAGGCTCCAG |
| Fragile site chr21-2 R | CATTTCAGGGGGACTTGGT |
| U3 primer 1 F | TGACGTCAATGGGTGGAGTA |
| U3 primer 1 R | CCTCGACCCATGGTAATAGC |
| U3 primer 2 F | CGTTACTCCCACAGGTGAGC |
| U3 primer 2 R | CGTCATTAAACCAAGCGCTAA |
| U3 primer 3 F | TCCCCTTCTCCATCTCCAG |
| U3 primer 3 R | GGCATGAACATGGTTAGCAG |
| U3 primer 4 F | CATGCCCGAAGGCTACGT |
| U3 primer 4 R | CGGCGCGGGTCTTGTA |
| U3 primer 5 F | TGAGAATGAAGTGTATGTGGAACAG |
| U3 primer 5 R | TGTGGTTTCCAAATGTGTCAG |
| U3 primer 6 F | TACCCGCTTCCATTGCTC |
| U3 primer 6 R | AAGTAGCACGTCTCACTAGTCTCG |
| U3 primer 7 F | CTTCTTCAAGGACGACGGC |
| U3 primer 7 R | GTAGTTGTACTCCAGCTTGTG |
| U2 primer 1 F | GCTCGGTCTACTCTGGCATC |
| U2 primer 1 R | TAATGGCCACCTGGAAGAAG |
| U2 primer 2 F | GATGTAGGAGGGCGTGGATA |
| U2 primer 2 R | ATAGGTCAGGCTCTCGCTGA |
| U2 primer 3 F | ATGCCCTGGCTCACAAATAC |
| U2 primer 3 R | CCCATATGTCCTTCCGAGTG |
| U2 primer 4 F | GAGCTTCCCGGCATAGTACA |
| U2 primer 4 R | CAATCACGCCAGAAATGTTG |
| U2 primer 5 F | CTGGTAGGGACCCGAGTGTA |
| U2 primer 5 R | CACTTGGCCCTTGATGATCT |
| U2 primer 6 F | GCCTACATACCTCGCTCTGC |
| U2 primer 6 R | CAACCCGGTAAGACACGACT |
| Rad51 F | ATGGCAATGCAGATGCAG |
| Rad51 R | TCAGTCTTTGGCATCTCCC |
| CTCF F | ATGGAAGGTGATGCAGTCG |
| CTCF R | TCACCGGTCCATCATGCT |
| CTCF-N F | ATGGAAGGTGATGCAGTCG |
| CTCF-N R | TTACTGGAATGTCTTCTT |
| CTCF-Zn F | TGTGAGCTTTGCAGTTAC |
| CTCF-Zn R | TTAACAATTATCAGCATG |
| CTCF-C F | GCTGGCCCAGATGGCG |
| CTCF-C R | TTACCGGTCCATCATGC |
siRNA- or shRNA-Mediated Knockdown of CTCF.
siRNAs targeting GTAGAAGTCAGCAAATTAA of CTCF were transfected to U2OS cells using Lipofectamine 2000 (11668019; Life Technologies) according to the manufacturer’s instructions. Because BJ cells are very difficult to transfect, we used lentivirus to perform CTCF knockdown in BJ cells. pLKO.1 vector-based shRNA constructs for knocking down CTCF were a generous gift from Paul M. Lieberman (Wistar Institute). shControl in pLKO.1 vector (target sequence TTATCGCGCATATCACGCG) was designed to poorly target the Escherichia coli DNA polymerase and was confirmed to not affect human gene transcription. Packaging vectors pMDLg/pRRE, pRSV-Rev, and pCMV-VSVG were used for lentiviral production. In brief, 20 μg of shRNA constructs were cotransfected with 10 μg of each packaging vector into ∼107 293T cells. Viral supernatants were harvested with a 0.45-mm syringe filter at both 48 and 72 h posttransfection. During the CTCF knockdown of BJ, Polybrene (Sigma-Aldrich) was added to the medium at 8 μg/mL, followed by incubation with lentivirus for 12 h and then removal of the solution. Western blot analysis of CTCF protein levels was done at 72–96 h after lentivirus infection.
Cell Cycle Analysis.
U2OS cells were collected and fixed with 75% (vol/vol) ethanol at 4 °C overnight and then resuspended in PBS containing PI (20 μg/mL) and RNase A (10 μg/mL) at 37 °C in the dark for 30 min. Cell cycle distribution was analyzed by flow cytometry (LSR Fortessa; BD Biosciences). Data were collected from 10,000 cells per sample.
Apoptosis Assay.
Cells were collected, washed, and stained with the Annexin-V FITC Kit (Beyotime Biotechnology) according to the manufacturer’s protocol. In brief, cells were collected and washed twice with 1× binding buffer and then incubated in 100 μL of labeling solution containing 1 μL of annexin-V FITC conjugate and 10 μL of PI in the dark for 15 min at room temperature. Fluorescence of the samples was analyzed by flow cytometry using an LSR Fortessa cytometer. Data were collected from 10,000 cells per sample.
Immunoprecipitation and Western Blot Analyses.
U2OS cells (1 × 107) were harvested after two washes in ice-cold PBS and then lysed in 1 mL of RIPA buffer with added protease inhibitors and DNase I (Roche Diagnostics). Dynabeads Protein G (Invitrogen) were incubated with 1 μg of antibodies for immunoprecipitation. The cell lysates were incubated with Dynabeads Protein G-coupled antibody overnight at 4 °C with rotation. An aliquot was incubated with IgG (ab2410; Abcam) as a control to determine background binding. Western blot analysis was performed following standard protocols using antibodies at a 1:1,000 dilution. Detection was done using Super Signal West Pico Chemiluminescent Substrate (OH192607; Thermo Fisher Scientific).
ChIP.
ChIP assays were carried out with the Chromatin Immunoprecipitation Assay Kit (EMD Millipore) according to the manufacturer’s protocol with minor modifications. Cells were fixed with formaldehyde (final concentration 1% vol/vol; Sigma-Aldrich), followed by the addition of glycine (125 mM) to stop the reaction. Cells were washed three times with ice-cold PBS, then scraped from culture dishes into a microfuge tubes. Cells were collected by centrifugation at 5,000 × g for 10 min at 4 °C, and then lysed with lysis buffer with protease inhibitors and sonicated to yield 200- to 500-bp DNA fragments. The samples were clarified by centrifugation at 13,000 × g for 15 min at 4 °C. The supernatant was diluted 10-fold in IP dilution buffer with protease inhibitors. An aliquot (1/20) of each chromatin supernatant was reserved as the input sample. Dynabeads Protein G with a magnetic stand were used for immunoprecipitation. The chromatin supernatant was incubated with 5 μg of antibody overnight at 4 °C with rotation. An aliquot was incubated with IgG (ab2410; Abcam) as a control to determine background binding. The beads were washed for 5 min at 4 °C with rotation, twice with low-salt buffer, once with high-salt buffer, once with LiCl buffer, and twice with TE buffer. Immunocomplexes were eluted by adding 210 μL of elution buffer and incubating for 15 min at 65 °C. The beads were spun at 15,871 × g for 1 min, after which 200 μL of eluted solution was transferred to a new tube. Cross-links were reversed by incubation for 7 h at 65 °C, with a final concentration of 200 mM NaCl. The samples were then treated with RNase A and digested with proteinase K. DNA was purified with the QIAquick PCR Purification Kit (28104; Qiagen) and used as a template for real-time PCR.
Recombinant Protein Expression and Purification.
CTCF, Rad51, and CTCF deletion mutants were cloned from human primary fibroblast BJ cells using the primers listed in Table S1. PCR products were subsequently cloned into GST-fused expression vector pGEX-6p-1 or His-fused expression vector pET-28a as indicated. Recombinant proteins were purified from Rosetta (DE3), grown at 37 °C in LB medium supplemented with ampicillin or kanamycin. At OD600 = 0.6, 1 mM isopropyl β-D-1-thiogalactopyranoside was added to the culture, followed by incubation at 28 °C for 16 h. GST and His fusions were purified using GST-Tagged and His-Tagged Protein Purification Kits (Beyotime; P2262 and P2226) according to the manufacturer’s protocol.
Pull-Down Assay.
GST pull-down assays using GST alone or GST-tagged protein and His-tagged protein were performed in 1 mL of binding buffer (50 mM Tris⋅HCl pH 8.0, 150 mM NaCl, 1 mM EDTA, 1% Nonidet P-40, 1 mM DTT, 10 mM MgCl2 and protease inhibitors). GST or GST-tagged proteins were preincubated GST-tagged purification resin for 2 h at 4 °C, followed by the addition of His-tagged protein and overnight incubation at 4 °C. Complexes were washed six times with binding buffer, and proteins were visualized by Western blot analysis using the indicated antibodies.
PLA.
Interactions between CTCF and RAD51 in U2OS cells were detected by in situ PLA according to the manufacturer’s protocol (Sigma-Aldrich). Cells were first fixed with 4% PFA for 30 min at room temperature and then incubated with the primary antibodies at 4 °C overnight. PLA minus and PLA plus probes (containing the secondary antibodies conjugated to oligonucleotides) were added, followed by incubation for 1 h at 37 °C. More oligonucleotides were then added and allowed to hybridize to the PLA probes. Ligase was used to join the two hybridized oligonucleotides into a closed circle. The DNA was then amplified with rolling circle amplification. Nuclei were visualized by staining with Hoechst 33342. Images were acquired with a confocal microscope.
Acknowledgments
We thank Matthew Weitzman, Ping Zheng, and Yun Zhang for reagents and Jeremy Stark for reagents, insightful discussions, and a critical reading of the manuscript. This work was supported by Strategic Priority Research Program of the Chinese Academy of Sciences Grants XDB13020400 and KSZD-EW-Z-009; Chinese Academy of Sciences Grant KSCXZ-EW-BR-6; Yunnan Provincial Government Grants 2011HA005, 2013FA051, and 2016FB039; and National Natural Science Foundation of China Grants 81471966, 81672040, U1602226, 2016YFC1200404 (to J.Z.), and 31601157 (to F.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1704076114/-/DCSupplemental.
References
- 1.Kim TH, et al. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ong CT, Corces VG. CTCF: An architectural protein bridging genome topology and function. Nat Rev Genet. 2014;15:234–246. doi: 10.1038/nrg3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ghirlando R, Felsenfeld G. CTCF: Making the right connections. Genes Dev. 2016;30:881–891. doi: 10.1101/gad.277863.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gregor A, et al. De novo mutations in the genome organizer CTCF cause intellectual disability. Am J Hum Genet. 2013;93:124–131. doi: 10.1016/j.ajhg.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hirayama T, Tarusawa E, Yoshimura Y, Galjart N, Yagi T. CTCF is required for neural development and stochastic expression of clustered Pcdh genes in neurons. Cell Rep. 2012;2:345–357. doi: 10.1016/j.celrep.2012.06.014. [DOI] [PubMed] [Google Scholar]
- 6.Watson LA, et al. Dual effect of CTCF loss on neuroprogenitor differentiation and survival. J Neurosci. 2014;34:2860–2870. doi: 10.1523/JNEUROSCI.3769-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ribeiro de Almeida C, et al. Critical role for the transcription regulator CCCTC-binding factor in the control of Th2 cytokine expression. J Immunol. 2009;182:999–1010. doi: 10.4049/jimmunol.182.2.999. [DOI] [PubMed] [Google Scholar]
- 8.Soshnikova N, Montavon T, Leleu M, Galjart N, Duboule D. Functional analysis of CTCF during mammalian limb development. Dev Cell. 2010;19:819–830. doi: 10.1016/j.devcel.2010.11.009. [DOI] [PubMed] [Google Scholar]
- 9.Kemp CJ, et al. CTCF haploinsufficiency destabilizes DNA methylation and predisposes to cancer. Cell Rep. 2014;7:1020–1029. doi: 10.1016/j.celrep.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guerrero PA, Maggert KA. The CCCTC-binding factor (CTCF) of Drosophila contributes to the regulation of the ribosomal DNA and nucleolar stability. PLoS One. 2011;6:e16401. doi: 10.1371/journal.pone.0016401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li T, Lu L. Functional role of CCCTC binding factor (CTCF) in stress-induced apoptosis. Exp Cell Res. 2007;313:3057–3065. doi: 10.1016/j.yexcr.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deng Z, et al. A role for CTCF and cohesin in subtelomere chromatin organization, TERRA transcription, and telomere end protection. EMBO J. 2012;31:4165–4178. doi: 10.1038/emboj.2012.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O’Sullivan RJ, Karlseder J. Telomeres: Protecting chromosomes against genome instability. Nat Rev Mol Cell Biol. 2010;11:171–181. doi: 10.1038/nrm2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fenech M. Cytokinesis-block micronucleus cytome assay. Nat Protoc. 2007;2:1084–1104. doi: 10.1038/nprot.2007.77. [DOI] [PubMed] [Google Scholar]
- 15.Durkin SG, Glover TW. Chromosome fragile sites. Annu Rev Genet. 2007;41:169–192. doi: 10.1146/annurev.genet.41.042007.165900. [DOI] [PubMed] [Google Scholar]
- 16.Sancar A, Lindsey-Boltz LA, Unsal-Kaçmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 17.Bekker-Jensen S, Mailand N. Assembly and function of DNA double-strand break repair foci in mammalian cells. DNA Repair (Amst) 2010;9:1219–1228. doi: 10.1016/j.dnarep.2010.09.010. [DOI] [PubMed] [Google Scholar]
- 18.Lukas C, et al. 53BP1 nuclear bodies form around DNA lesions generated by mitotic transmission of chromosomes under replication stress. Nat Cell Biol. 2011;13:243–253. doi: 10.1038/ncb2201. [DOI] [PubMed] [Google Scholar]
- 19.Panier S, Boulton SJ. Double-strand break repair: 53BP1 comes into focus. Nat Rev Mol Cell Biol. 2014;15:7–18. doi: 10.1038/nrm3719. [DOI] [PubMed] [Google Scholar]
- 20.Harrigan JA, et al. Replication stress induces 53BP1-containing OPT domains in G1 cells. J Cell Biol. 2011;193:97–108. doi: 10.1083/jcb.201011083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Godin SK, Sullivan MR, Bernstein KA. Novel insights into RAD51 activity and regulation during homologous recombination and DNA replication. Biochem Cell Biol. 2016;94:407–418. doi: 10.1139/bcb-2016-0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawabata M, Kawabata T, Nishibori M. Role of recA/RAD51 family proteins in mammals. Acta Med Okayama. 2005;59:1–9. doi: 10.18926/AMO/31987. [DOI] [PubMed] [Google Scholar]
- 23.Gunn A, Bennardo N, Cheng A, Stark JM. Correct end use during end joining of multiple chromosomal double-strand breaks is influenced by repair protein RAD50, DNA-dependent protein kinase DNA-PKcs, and transcription context. J Biol Chem. 2011;286:42470–42482. doi: 10.1074/jbc.M111.309252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pierce AJ, Johnson RD, Thompson LH, Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13:2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Williams RS, Williams JS, Tainer JA. Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochem Cell Biol. 2007;85:509–520. doi: 10.1139/O07-069. [DOI] [PubMed] [Google Scholar]
- 26.Zlatanova J, Caiafa P. CTCF and its protein partners: Divide and rule? J Cell Sci. 2009;122:1275–1284. doi: 10.1242/jcs.039990. [DOI] [PubMed] [Google Scholar]
- 27.Saldaña-Meyer R, et al. CTCF regulates the human p53 gene through direct interaction with its natural antisense transcript, Wrap53. Genes Dev. 2014;28:723–734. doi: 10.1101/gad.236869.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Phillips JE, Corces VG. CTCF: Master weaver of the genome. Cell. 2009;137:1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bennardo N, Cheng A, Huang N, Stark JM. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet. 2008;4:e1000110. doi: 10.1371/journal.pgen.1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stark JM, Pierce AJ, Oh J, Pastink A, Jasin M. Genetic steps of mammalian homologous repair with distinct mutagenic consequences. Mol Cell Biol. 2004;24:9305–9316. doi: 10.1128/MCB.24.21.9305-9316.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yun MH, Hiom K. CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature. 2009;459:460–463. doi: 10.1038/nature07955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adamson B, Smogorzewska A, Sigoillot FD, King RW, Elledge SJ. A genome-wide homologous recombination screen identifies the RNA-binding protein RBMX as a component of the DNA-damage response. Nat Cell Biol. 2012;14:318–328. doi: 10.1038/ncb2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davies OR, Pellegrini L. Interaction with the BRCA2 C terminus protects RAD51-DNA filaments from disassembly by BRC repeats. Nat Struct Mol Biol. 2007;14:475–483. doi: 10.1038/nsmb1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang J, Ma Z, Treszezamsky A, Powell SN. MDC1 interacts with Rad51 and facilitates homologous recombination. Nat Struct Mol Biol. 2005;12:902–909. doi: 10.1038/nsmb991. [DOI] [PubMed] [Google Scholar]
- 35.Haaf T, Golub EI, Reddy G, Radding CM, Ward DC. Nuclear foci of mammalian Rad51 recombination protein in somatic cells after DNA damage and its localization in synaptonemal complexes. Proc Natl Acad Sci USA. 1995;92:2298–2302. doi: 10.1073/pnas.92.6.2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tarsounas M, Davies D, West SC. BRCA2-dependent and -independent formation of RAD51 nuclear foci. Oncogene. 2003;22:1115–1123. doi: 10.1038/sj.onc.1206263. [DOI] [PubMed] [Google Scholar]
- 37.Boussemart L, et al. eIF4F is a nexus of resistance to anti-BRAF and anti-MEK cancer therapies. Nature. 2014;513:105–109. doi: 10.1038/nature13572. [DOI] [PubMed] [Google Scholar]
- 38.Liu Z, Scannell DR, Eisen MB, Tjian R. Control of embryonic stem cell lineage commitment by core promoter factor, TAF3. Cell. 2011;146:720–731. doi: 10.1016/j.cell.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fujioka M, Wu X, Jaynes JB. A chromatin insulator mediates transgene homing and very long-range enhancer-promoter communication. Development. 2009;136:3077–3087. doi: 10.1242/dev.036467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lan L, et al. Accumulation of Werner protein at DNA double-strand breaks in human cells. J Cell Sci. 2005;118:4153–4162. doi: 10.1242/jcs.02544. [DOI] [PubMed] [Google Scholar]