Significance
Our experiments provide the analysis of lipid metabolite circadian oscillations in a cellular system synchronized in vitro, suggesting cell-autonomous diurnal changes in lipid profiles independent of feeding. Moreover, our work represents a comprehensive comparison between the lipid composition of human skeletal muscle derived from sedentary healthy adults, receiving hourly isocaloric solutions, and human primary skeletal myotubes cultured in vitro. A substantial number of lipid metabolites, in particular membrane lipids, exhibited oscillatory patterns in muscle tissue and in myotube cells, where they were blunted upon cell-autonomous clock disruption. As lipid oscillations in skeletal muscle membrane lipids may impact on insulin signaling and on the development of insulin resistance, studying the temporal lipid composition of human muscle is therefore of utmost importance.
Keywords: lipid metabolism, circadian clock, human skeletal muscle, human primary myotubes, lipidomics
Abstract
Circadian clocks play an important role in lipid homeostasis, with impact on various metabolic diseases. Due to the central role of skeletal muscle in whole-body metabolism, we aimed at studying muscle lipid profiles in a temporal manner. Moreover, it has not been shown whether lipid oscillations in peripheral tissues are driven by diurnal cycles of rest–activity and food intake or are able to persist in vitro in a cell-autonomous manner. To address this, we investigated lipid profiles over 24 h in human skeletal muscle in vivo and in primary human myotubes cultured in vitro. Glycerolipids, glycerophospholipids, and sphingolipids exhibited diurnal oscillations, suggesting a widespread circadian impact on muscle lipid metabolism. Notably, peak levels of lipid accumulation were in phase coherence with core clock gene expression in vivo and in vitro. The percentage of oscillating lipid metabolites was comparable between muscle tissue and cultured myotubes, and temporal lipid profiles correlated with transcript profiles of genes implicated in their biosynthesis. Lipids enriched in the outer leaflet of the plasma membrane oscillated in a highly coordinated manner in vivo and in vitro. Lipid metabolite oscillations were strongly attenuated upon siRNA-mediated clock disruption in human primary myotubes. Taken together, our data suggest an essential role for endogenous cell-autonomous human skeletal muscle oscillators in regulating lipid metabolism independent of external synchronizers, such as physical activity or food intake.
Circadian oscillations are daily cycles in behavior and physiology that are driven by the existence of underlying intrinsic biological clocks with near 24-h periods. This anticipatory mechanism has evolved to ensure that all aspects of behavior and physiology, including metabolic pathways, are temporally coordinated with daily cycles of rest–activity and feeding to provide the organism with an adaptive advantage (1). In mammals, circadian oscillations are driven by a master pacemaker, located in the suprachiasmatic nucleus (SCN) of the hypothalamus, which orchestrates subsidiary oscillators in peripheral organs via neuronal, endocrine, and metabolic signaling pathways (2, 3).
Large-scale gene expression datasets suggest that, in mammals, the vast majority of circadian-gene expression is highly organ-specific (4–6). Key metabolic functions in peripheral organs are subject to daily oscillations, such as carbohydrate and lipid metabolism by the liver, skeletal muscle, and endocrine pancreas (7).
Skeletal muscle is a major contributor of whole-body metabolism and is the main site of glucose uptake in the postprandial state (8). Therefore, perturbations in glucose sensing and metabolism in skeletal muscle are strongly associated with insulin resistance in type 2 diabetes (T2D) (9). Recent data support a fundamental role for the circadian muscle clock in the regulation of glucose uptake, with a significant reduction in Glut4 mRNA and protein levels in two muscle specific Bmal1 knockout models (10, 11). In addition, rhythmic genes involved in carbohydrate metabolism were down-regulated while those involved in lipid metabolic processes were up-regulated in mice bearing a muscle-specific Bmal1 KO (12). In line with these observations, it has been noted that disruption of circadian rhythmicity is associated with hyperlipidemia and hepatic steatosis in ClockΔ19 mice (13), with obesity in mice that bear an adipocyte-specific deletion of Bmal1 (14), and with reduced whole-body weight and reduction of total triglycerides and fatty acids in blood plasma of Per2-deficient mice (15).
Lipidomic analysis of liver from WT and clock-disrupted Per1/Per2 KO mice demonstrated a comparable fraction of oscillating lipids in the presence or absence of a functional clock. However, substantial differences in their composition and peak phase of oscillation were observed, suggesting that both endogenous clocks and fasting–feeding cycles contribute to the observed profile (16). Furthermore, quantitative characterization of the nuclear and mitochondrial lipidome of mouse liver uncovered daily lipid oscillations with distinct phases in these intracellular organelles (17).
In humans, diurnal variations in glucose tolerance have long been described (18), and rhythmic profiles of plasma glucose, insulin, and triglycerides have been detected (19, 20). Furthermore, a lipidomic analysis of human plasma has shown that ∼18% of all lipid profiles oscillate in a circadian manner independent of sleep and food intake (19). Similarly, metabolomics studies of plasma, saliva, and urine revealed that around 20% of all identified metabolites, among them many fatty acids, were oscillating (21–23).
Fasting–feeding cycles accompanying rest–activity rhythms represent major timing cues in the synchronization of peripheral clocks (24). However, whether lipids in peripheral tissues oscillate in a cell-autonomous manner has not yet been resolved. As the existence of self-sustained circadian oscillators operative in human primary skeletal myotubes established from muscle biopsies, cultured and synchronized in vitro, has been recently demonstrated (25–27), we sought to address this question. In this study, we explored the regulation of lipid metabolite oscillations in human skeletal muscle in vivo and in vitro, focusing on lipids involved in membrane organization (phospholipids and sphingolipids) and those involved in energy metabolism, such as triglycerides. Serial human muscle biopsies were analyzed, using a controlled protocol designed to minimize the effect of confounding factors, and were compared with primary myotubes established from human donor biopsies. Using a large-scale lipidomic approach, we monitored the time course of skeletal muscle lipid metabolites under physiological conditions and under siRNA-mediated cellular clock disruption, allowing us to dissect the impact of the cell-autonomous muscle clock from external factors.
Results
The Temporal Lipidome of Human Skeletal Muscle in Vivo.
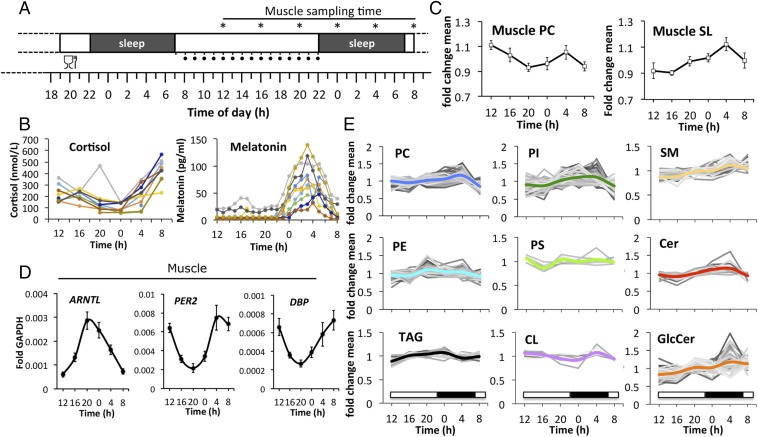
To investigate the time course of lipid metabolites in human skeletal muscle, we analyzed serial biopsies from the vastus lateralis muscle, taken every 4 h across 24 h (Fig. 1A). Samples were obtained from 10 healthy volunteers (see SI Appendix, Table S1 for donor characteristics), using a controlled protocol designed to minimize the effect of confounding factors (SI Appendix, SI Materials and Methods). Blood samples were drawn every hour, starting on the testing day over a span of 24 h. Plasma concentration profiles of melatonin and cortisol were highly synchronized across subjects, with the melatonin zenith reached at 0300 hours ± 38 min (SD) and the cortisol nadir at 0000 hours ± 40 min (SD) (Fig. 1B).
Fig. 1.
Temporal analysis of human skeletal muscle lipids in vivo. (A) Experimental design for the collection of muscle tissue samples (vastus lateralis) from healthy human subjects (n = 10, asterisks indicate muscle-sampling times). One standardized meal was provided on the evening before the testing day, as illustrated. The hourly intake of isocaloric solutions during the testing day is indicated by black dots. For protocol details, see Materials and Methods and SI Appendix, SI Materials and Methods. (B) Time course of plasma cortisol (Left) and melatonin (Right). (C) Temporal profiles of major PC species (Left; mean ± SEM, n = 10) and major SLs (Right, mean of total GlcCer, SM, Cer ± SD). (D) RT-qPCR results for ARNTL, PER2, and DBP on mRNA samples extracted from human skeletal muscle biopsies. Data were normalized to GAPDH (mean ± SEM). (E) Temporal profiles of lipid metabolites clustered by lipid class. Only lipid species that were detected in all participants are shown. Values were normalized to the lipid mean value per individual analysis and then averaged across all subjects (n = 10) for each time point. The thick colored line represents the average of each lipid class.
Using a targeted lipidomics-based approach, we next examined the diurnal profiles of triacylglycerides (TAGs), glycerophospholipids (PLs), and sphingolipids (SLs) in human skeletal muscle. Out of 1,058 lipid metabolites analyzed, 532 lipids were detected in all participants at all time points (32 TAGs, 181 SLs, and 319 PLs). Lipid raw data were normalized to the phosphorus content of each lipid extract to correct for differences in the amount of starting material. Each lipid value was then divided by its daily mean value, calculated individually for each biopsy. Examples of normalized and unnormalized lipid profiles are shown in SI Appendix, Fig. S1. Normalized lipid values were averaged for each time point across all donors (n = 10). The major phosphatidylcholine (PC) profile, representing the most abundant PC lipids, as well as the major SL profile, representing the most abundant sphingomyelins (SMs), ceramides (Cers), and glycosylceramides (GlcCers), showed enrichment at 0400 hours (Fig. 1C). The SL profile varied up to 20% in amplitude across 24 h, with the valley reached at 1600 hours and the peak reached at 0400 hours (Fig. 1C). In parallel to the lipidomic analysis, core clock transcript levels were monitored by quantitative reverse transcription PCR (RT-qPCR), with RNA extracted from the same human skeletal muscle biopsy material. As shown in Fig. 1D, core clock transcript expression was highly rhythmic, with ARNTL (BMAL1) exhibiting an oscillatory profile antiphasic to PER2 and DBP. The zenith of ARNTL expression was reached around 2000 hours, preceding the 0400 hours peak of PC and SL lipid accumulation by about 8 h.
Next, all lipid metabolite profiles, including low abundant lipids, were clustered by lipid class (Fig. 1E and Dataset S1). Phosphatidylinositol (PI), PC, SM, GlcCer, and Cer lipids exhibited peak levels around 0400 hours while phosphatidylethanolamine (PE), phosphatidylserine (PS), and TAG levels peaked at earlier time points, with zeniths reached on average at 2000 hours and 0000 hours, respectively (Fig. 1E and Dataset S1). Overall, lipid levels changed more than 20% over the course of 24 h, with variability observed among lipid classes (Fig. 1E and Dataset S1).
Lipidomes of Human Skeletal Muscle and Primary Myotubes Are Comparable, with Main Differences Seen for CL and PS.
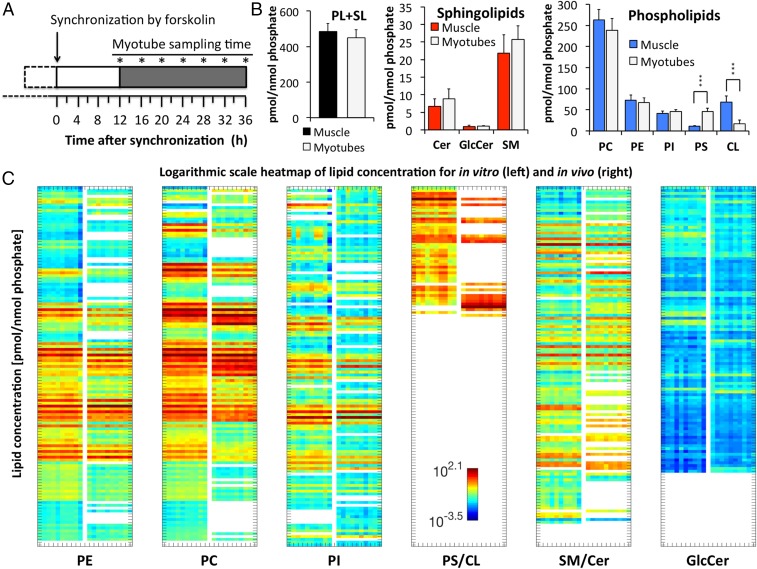
We next aimed to assess whether the in vivo lipid oscillations, observed in skeletal muscle biopsies, persist in vitro. To this end, human primary myoblasts were established from skeletal muscle biopsies (gluteus maximus) obtained from 10 healthy donors (see SI Appendix, Table S2 for donor characteristics). Cells were synchronized in vitro with a pulse of forskolin, and samples were collected every 4 h over 24 h, as shown in Fig. 2A.
Fig. 2.
Comparison between the lipidome of human skeletal muscle and primary myotubes. (A) Experimental design: Human primary myoblasts were differentiated into myotubes, synchronized with forskolin, and harvested at the indicated seven time points (asterisks). (B) Lipid concentrations of muscle tissue and myotubes; Sum of PL plus SL (Left), SL (Middle), PL (Right). Data represent the average across all time points. Mean ± SD; ***P < 0.001. (C) Logarithmic scale heat map of lipid concentration (pmol/nmol phosphate), shown for in vitro (n = 10) and in vivo (n = 10). Lipids were sorted by lipid class (PE, PC, PI, PS, CL, SM, Cer, and GlcCer) and lipid name, and separated for each lipid class into two columns (left columns, in vitro; right columns, in vivo). Data represent the mean concentration of each metabolite across all time points per individual participant. Colors indicate the log concentration of each metabolite.
Out of 1,058 analyzed lipid metabolites, 637 were detected in all myotube cultures at all time points (20 TAGs, 215 SLs, 402 PLs). When averaged across all time points, lipid levels were comparable between human skeletal muscle and primary myotubes. Combined PL and SL levels (Fig. 2 B, Left), as well as SL levels alone (Fig. 2 B, Middle), were not significantly different. However, a decrease in mitochondria-specific cardiolipin (CL) and an increase in PS were observed in human primary myotubes compared to muscle tissue (Fig. 2 B, Right). Overall, the lipid composition was highly homogenous among the 10 muscle and primary myotube samples but differed with regard to lipid chain length and degree of desaturation (Fig. 2C and Dataset S2). The strongest difference was observed for CLs, where the most abundant CL of skeletal muscle (CL72:8_C18:2) was not detected on average in primary myotubes (Fig. 2C, SI Appendix, Fig. S2, and Dataset S2). TAGs were not absolutely quantified but analyzed in a relative manner, and are therefore not included in Fig. 2 B and C.
Lipid Metabolite Oscillations Persist in Vitro in Synchronized Human Myotubes.
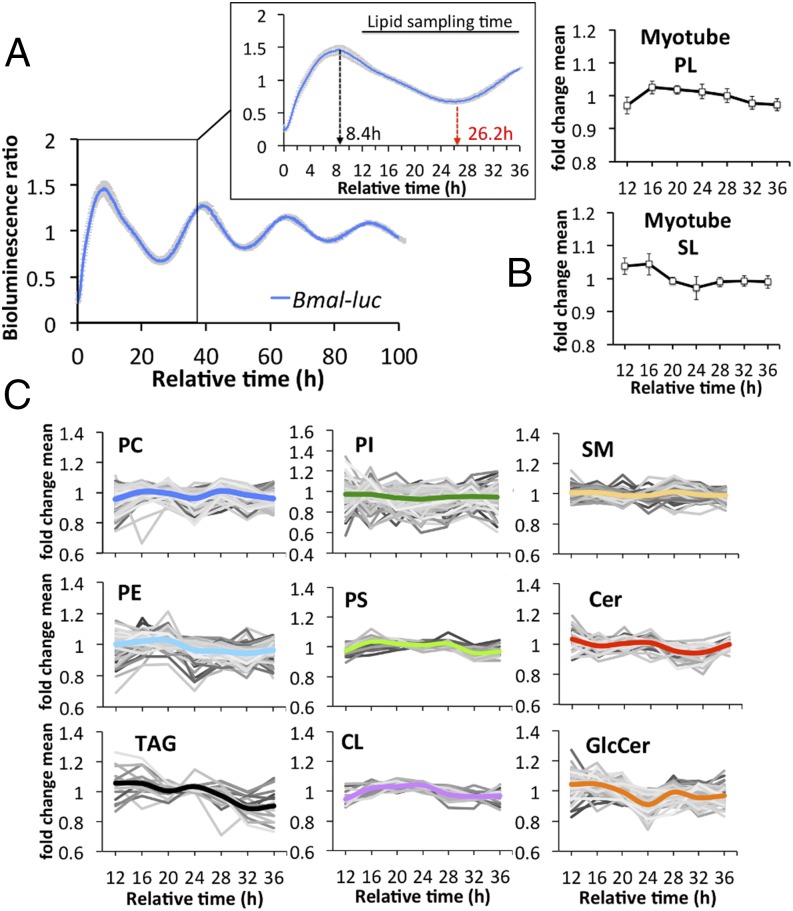
Human primary myotube lipids were analyzed as described for the in vivo analysis (for normalization, see SI Appendix, Fig. S3). To monitor core clock properties in synchronized human primary myotubes, circadian bioluminescence was continuously recorded for several days, using a lentiviral Bmal1-luc reporter (27). Bmal1-luc reporter profiles recorded from all 10 myotube cultures demonstrated a high degree of alignment (Fig. 3A). The first peak of Bmal1-luc reporter activity was observed about 8 h after forskolin synchronization (Fig. 3 A, Inset), and as previously demonstrated for endogenous BMAL1 in human primary myotubes (27). The Bmal1-luc reporter activity peak recorded 8 h after synchronization (Fig. 3B) was followed about 8 h later by a peak of the most abundant PL and SL lipids. This phase coherence was consistent with human skeletal muscle (Fig. 1 C and D). The most abundant PC lipids also exhibited enrichment at the 16-h time point; however, a second peak was observed 28 h following synchronization (SI Appendix, Fig. S4).
Fig. 3.
Lipid oscillations are preserved in human skeletal myotubes. (A) Average detrended Bmal1-luc bioluminescence profile of all human myotube cultures (n = 10). Mean ± SEM. (Inset) Bmal1-luc bioluminescence profile between 0 and 36 h. (B) Average temporal profile of major phospholipid PC, PE, PI, PS, and CL species (Upper; mean ± SD), and major sphingolipid SM, GlcCer, and Cer species (Lower, mean ± SD). (C) Temporal profiles of lipid metabolites clustered by lipid class. Only lipid species that were detected in all cultures are shown. Values were normalized to the lipid mean value per individual analysis and then averaged across all for each time point. The thick colored line represents the average of each lipid class.
When temporal profiles of lipid metabolites were averaged across all myotube cultures, individual lipid profiles were oscillating for each lipid class (Fig. 3C and Dataset S3). However, the amplitude of the mean (colored lines in Figs. 1E and 3C) was smaller in vitro than in vivo for some lipid classes (compare PC and PI in Figs. 1E and 3C and Dataset S3). This observed reduction in fold change might be in part attributed to the weaker effect of in vitro synchronization of primary myotubes by a single forskolin pulse, compared with the combination of central and peripheral synchronizing stimuli driving the skeletal muscle clock in vivo. The average PE, TAG, and CL profiles displayed a single peak 16 to 20 h following synchronization whereas PC and GlcCer profiles peaked twice within a 24-h period, at 16 h and 28 h, respectively (Fig. 3C and Dataset S3).
A Similar Percentage of Lipid Metabolites Exhibit Circadian Oscillations in Vivo and in Vitro.
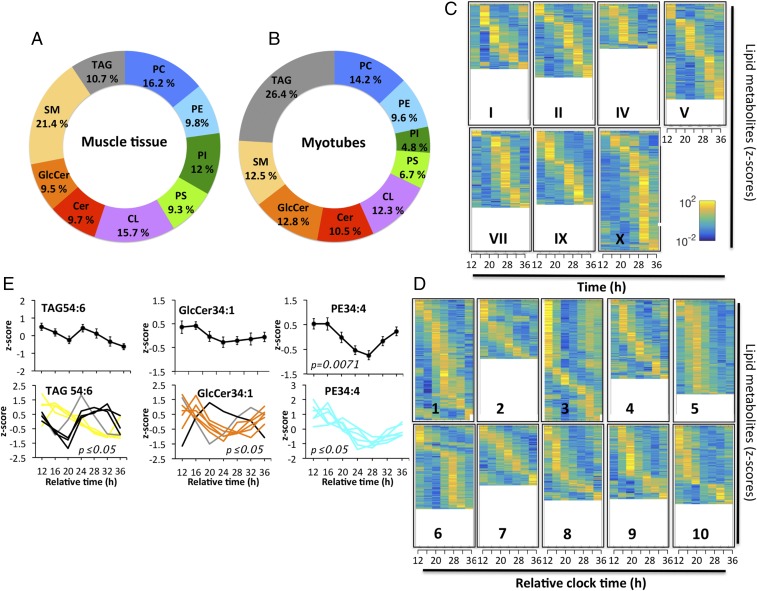
We next identified rhythmic lipid metabolites in human skeletal muscle and primary myotubes based on individually analyzed lipid profiles, using a widely used nonparametric algorithm, JTK_CYCLE (28). As the in vivo sample collection was incomplete (55 out of 60 samples were available), the calculation could only be performed on the basis of seven subjects for whom complete datasets were available. The percentage of circadian lipids per lipid class was determined as the number of cycling lipids vs. the total number of lipids detected per lipid class, averaged across the seven participants (Fig. 4A and Dataset S4). Rhythmic lipids were identified for each lipid class, suggesting a widespread circadian regulation of lipid metabolism (Fig. 4A and Dataset S4). A similar analysis was performed for human primary myotubes. Here, the percentage of rhythmic lipids per lipid class was calculated based on seven time points (Fig. 4B and Dataset S5). Strikingly, the average percentage of rhythmic lipids across the nine lipid classes was comparable between skeletal muscle and myotubes (12.7% vs. 12.4%, respectively). With regard to specific lipid classes, SM was the most rhythmic one in vivo whereas TAGs were the most rhythmic lipids in vitro (Fig. 4 A and B and Datasets S4 and S5).
Fig. 4.
Identification of rhythmic lipid metabolites in myotubes and skeletal muscle. (A) Percentage of diurnal lipid metabolites per lipid class in skeletal muscle (n = 7). (B) Percentage of circadian lipids per lipid class in primary myotubes (n = 10). The average across all lipid classes was 12.7% for skeletal muscle and 12.4% for primary myotubes. (C) Heat maps of rhythmic lipids in human skeletal muscle biopsies (n = 7). The average percentage of rhythmic lipids was 20.3% across all muscle donors based on the number of cycling lipids vs. detected lipids in each donor. (D) Heat maps of rhythmic lipid species in primary skeletal myotubes (n = 10). The average percentage of rhythmic lipids was 18.6%. Normalized z-scores of lipid metabolites are indicated in yellow (high) and blue (low). (E) Representative rhythmic lipids of skeletal myotube cultures: TAG54:6 (rhythmic in eight cultures), GlcCer34:1 (rhythmic in seven), and PE34:4 (rhythmic in six). (Upper) Average lipid profiles (mean ± SEM). (Lower) Individual lipid profiles, adjusted P value according to JTK_CYCLE.
If oscillating lipids were sorted according to their peak phase and plotted for each individual participant separately, the interindividual variability with regard to the total number of rhythmic metabolites and their phase preference became visible (Fig. 4 C and D and Datasets S4 and S5). Across all seven in vivo participants, the average percentage of rhythmic lipids in human skeletal muscle was 20.3% (based on the number of cycling lipids vs. detected lipids in each donor). For the in vitro study, the average percentage of rhythmic lipids was 18.6% (based on the number of cycling lipids vs. detected lipids per myotube culture).
The most rhythmic metabolites in vitro were TAG54:6 and GlcCer34:1, significantly oscillating in eight and seven myotube cultures, respectively (Fig. 4 E, Lower). However, if averaged across all 10 cultures, those lipids were not circadian (Fig. 4 E, Upper), due to phase differences between individual cultures (Fig. 4 E, Lower) and noncircadian oscillations in the remaining cultures. In contrast, PE lipids showed a higher degree of alignment, with PE34:4 being rhythmic on average across all myotube cultures and being one of the most rhythmic PE lipids on an individual level (Fig. 4E and Dataset S5). The most rhythmic lipids in human muscle and primary myotubes are listed in SI Appendix, Table S3.
Metabolic Clustering of Rhythmic Lipid Metabolites.
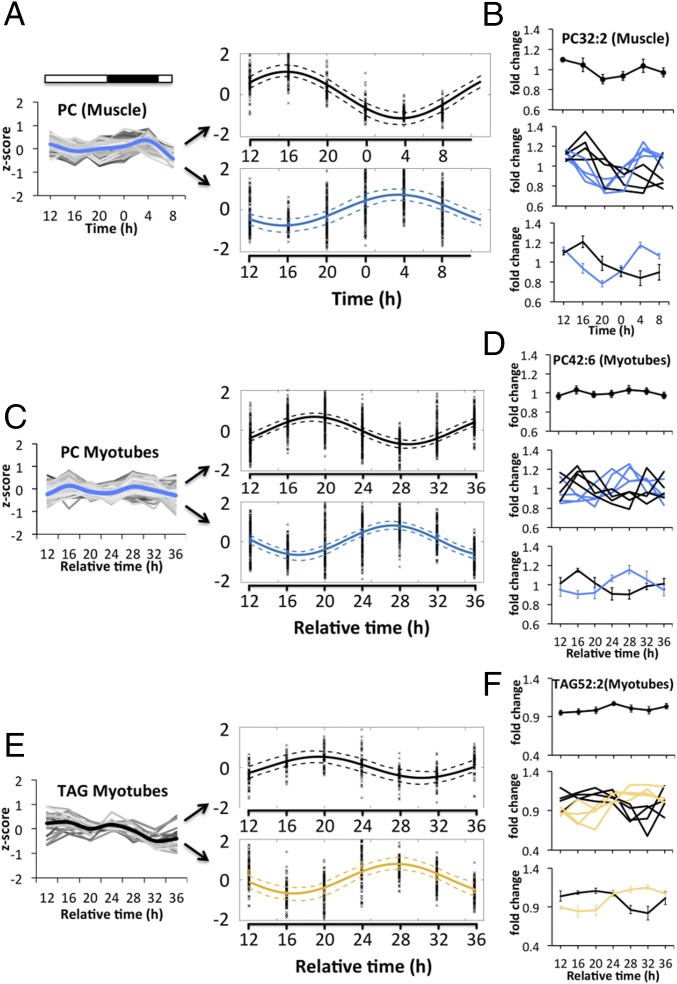
Based on the observed variance within the in vivo and in vitro donor cohorts (Fig. 4 C and D), we next investigated whether rhythmic lipids can be clustered into different subgroups based on their temporal profile. To assess this point, we applied nearest-neighbor clustering on the phase and amplitude of each profile to identify distinct lipid clusters within one lipid class both in vivo and in vitro. Following this approach, individual PC profiles of skeletal muscle (Fig. 5 A, Left and Dataset S6) could be separated into two clusters, with a single sinusoid fitted for all profiles in each cluster, exhibiting zenith levels at 1600 hours and 0400 hours, respectively (Fig. 5 A, Right). Lipid clusters were further dissected by investigating individual lipid metabolites (Fig. 5B and Dataset S6). PC32:2, which was identified as highly rhythmic in skeletal muscle upon JTK_CYCLE analysis (rhythmic in five out of seven muscle donors), was not significantly rhythmic if averaged across all participants, due to the phase variance between individual profiles (Fig. 5 B, Top and Middle and Dataset S6). However, individual profiles could be clustered into two major oscillatory subgroups with zeniths reached at 1600 hours and 0400 hours, respectively (Fig. 5 B, Middle and Bottom and Dataset S6).
Fig. 5.
Overlapping lipid clusters exist within different lipid classes in vivo and in vitro. (A) The total PC profile of skeletal muscle (Left) can be separated into two major PC clusters (Right) with zenith levels reached at 1600 hours and 0400 hours, respectively. The dashed line indicates the 95% confidence interval of the calculated fit. (B) PC32:2 is not rhythmic on average across all muscle donors (Top) but becomes rhythmic upon subcluster analysis (Middle and Bottom). (C) The total PC profile of human myotubes (Left) can be separated into two major PC clusters (Right) with zenith levels reached between 16 and 20 h and 28 h after synchronization, respectively. (D) PC42:6 is not rhythmic on average across all myotube cultures (Top) but becomes rhythmic upon subcluster analysis (Middle and Bottom). (E) The total TAG profile of myotubes (Left) can be separated into two major clusters (Right) with zenith levels reached at 20 h and 28 h, respectively. (F) TAG52:2 is not rhythmic on average across all myotube cultures (Top) but becomes rhythmic upon subcluster analysis (Middle and Bottom).
Similarly, when PC lipids derived from human primary myotubes (Fig. 5 C, Left and Dataset S6) were analyzed using this algorithm, two major subgroups emerged with zenith levels reached at 16 to 20 h and 28 h after synchronization (Fig. 5 C, Right). Such clustering was further validated on a single lipid level for the highly rhythmic PC42:6 (Fig. 5D and Dataset S6). PC32:2 was less rhythmic in primary myotubes (cycling in 3 out of 10 cultures) but could also be sorted according to phase into two subclusters (SI Appendix, Fig. S5A).
Overlapping lipid profiles were also observed within other lipid classes, including TAGs. Indeed, the total TAG profile of primary myotubes clustered into two groups, with zenith levels reached at 20 h and 28 h after synchronization (Fig. 5E and Dataset S6). TAG52:2 was not rhythmic across all myotube cultures; however, it became rhythmic upon subgroup analysis, with zeniths reached at 20 h and 28 to 32 h after synchronization (Fig. 5F and Dataset S6). Due to limitations of the in vivo TAG dataset (lower number of technical replicates), those could not be sorted using the here-described clustering algorithm; however, certain TAG lipids, such as TAG58:9, displayed subgroup oscillations upon individual analysis (SI Appendix, Fig. S5B).
Diurnal Lipid Metabolite Levels Correlate with Transcript Profiles Encoding for Key Enzymes Involved in Their Biosynthesis.
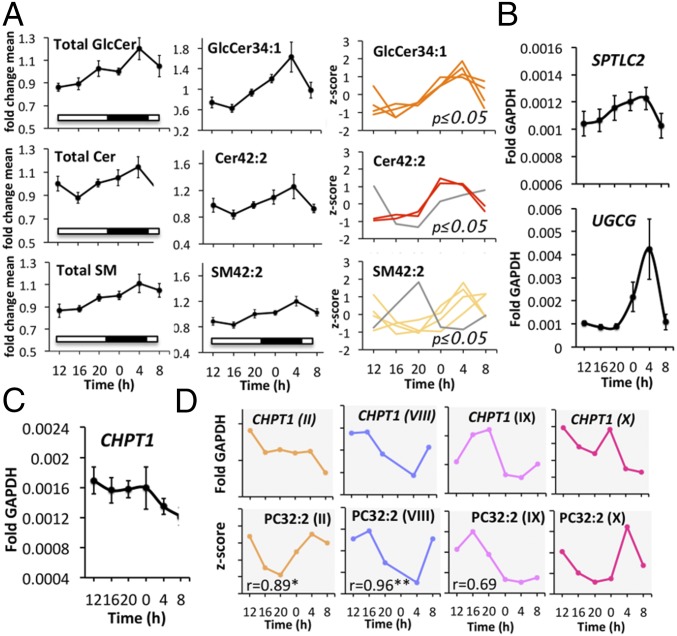
In human skeletal muscle, all three major SL classes (GlcCer, Cer, and SM) reached their zenith levels around 0400 hours (Figs. 1E and 6 A, Left). A similar accumulation pattern was found on a single metabolite level for GlcCer34:1, Cer42:2, and SM42:2 (Fig. 6 A, Middle and Right). Importantly, GlcCer34:1 was also one of the most rhythmic SLs in primary myotubes (Fig. 4E and SI Appendix, Table S3).
Fig. 6.
Temporal lipid profiles and mRNA profiles of genes implicated in their biosynthesis are correlated in vivo. (A) Total profiles of GlcCer, Cer, SM (Left, mean ± SEM); profiles of GlcCer34:1, Cer42:2, and SM42:2 (mean ± SEM; Middle); individual profiles of GlcCer34:1, Cer42:2, and SM42:2 that were identified as circadian (Right). (B) RT-qPCR gene expression profile of SPTLC2, and UGCG normalized to GAPDH (mean ± SEM). (C) RT-qPCR gene expression profile of CHPT1 normalized to GAPDH (mean ± SEM). (D) Individual gene expression profile of CHPT1 normalized to GAPDH in four subjects and temporal profile of PC32:2 in the same four subjects. The r numerical value indicates the corresponding Pearson correlation coefficient; *P < 0.05, **P < 0.01.
Next, we investigated the temporal gene expression profile of enzymes involved in de novo SL biosynthesis, using the same biopsy material used for the lipidomic analysis. Remarkably, the temporal expression profiles of SPTLC2 and UGCG, coding for subunit 2 of serine palmitoyltransferase and UDP-Glucose ceramide glucosyltransferase, respectively, correlated with the diurnal profiles of the three major SL classes (Fig. 6B). The transcripts of both key enzymes of de novo SL biosynthesis reached their peak levels between 0000 hours and 0400 hours, similar to the profile of their corresponding lipid metabolites (Fig. 6 A and B).
Moreover, we assessed whether a similar correlation could also be made for other lipid classes by analyzing the temporal mRNA profile of choline phosphotransferase 1 (CHPT1), involved in PC biosynthesis. While, on average, transcript levels of CHPT1 exhibited no clear oscillatory pattern (Fig. 6C), individually analyzed CHPT1 transcript levels were oscillating in a subset of muscle biopsy donors with distinct phases (Fig. 6 D, Upper). Strikingly, the oscillatory transcript expression pattern of CHPT1 and the diurnal profiles of the most rhythmic PC lipid (PC32:2) were highly similar in a subset of muscle donors (Fig. 6 D, Lower), with profile II and VIII being significantly correlated.
Chain Length, but Not the Degree of Desaturation, Influences the Circadian Profile of Lipid Metabolites.
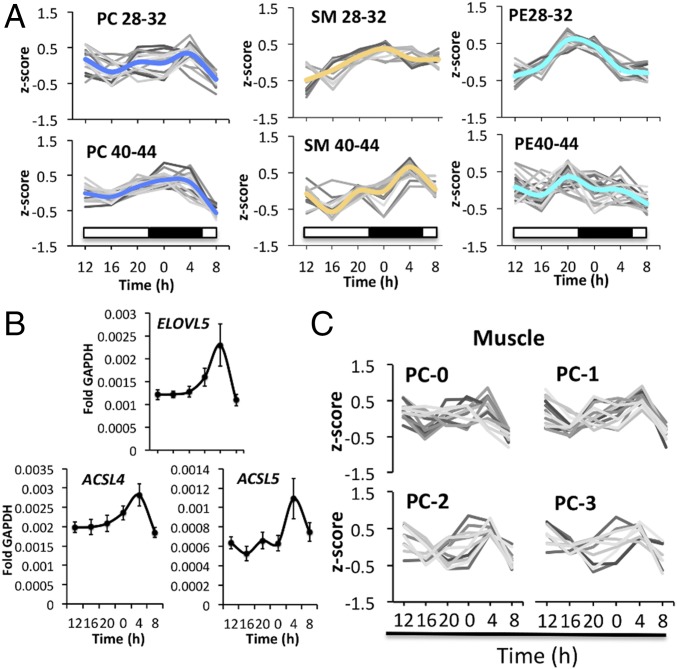
PC and SM lipid classes share the same head group, phosphocholine, and their biosynthesis pathways are interconnected. We noticed that temporal profiles of PC and SM showed similarities in vivo (Fig. 7A and Dataset S7) and in vitro (SI Appendix, Fig. S6A). When lipid profiles were clustered according to chain length, lipids that carried very long chain fatty acids (PC40-44, SM40-44) were oscillating in a synchronized manner, with zenith levels reached at 0400 hours (Fig. 7A and Dataset S7) compared with shorter chain PC an SM species. In contrast, PE lipids were not further synchronized upon lipid elongation. Shorter chain PEs (PE28-32) were well synchronized while very long chain PEs (PE40-44) were less synchronized and not enriched at the 0400 hours time point. Similar observations were made in vitro for PC, SM, and PE lipids of primary myotubes (SI Appendix, Fig. S6A).
Fig. 7.
Lipid chain length but not desaturation influences the circadian profile of PC and SM lipid metabolites. (A) Average temporal profile of PC, SM, and PE lipids, sorted by chain length in skeletal muscle (n = 10). PC, PE: 28-32 and 40-44 represents the sum of carbon atoms in both fatty acyl chains; SM lipids: 28-32 and 40-44 represents the sum of carbon atoms in the sphingoid base (C18) plus fatty acyl chain. (B) RT-qPCR results for ELOVL5, ACSL4, and ACSL5 on mRNA samples extracted from human skeletal muscle biopsies. Data were normalized to GAPDH (mean ± SEM). (C) Average PC lipid profiles (skeletal muscle) sorted by their degree of desaturation. The number of double bonds is indicated by the suffix 0, 1, 2, and 3.
Human long chain acyl-CoA synthetases activate fatty acids to form acyl-CoAs. The activated fatty acids can then be used for the biosynthesis of triglycerides and phospholipids, or for the generation of energy via β-oxidation. Moreover, these fatty acyl-CoAs can be elongated by fatty acid elongases (ELOVLs). We therefore investigated the temporal transcript levels of long chain fatty acyl-CoA synthetases (ACSL3-5), fatty acid elongase 5 (ELOVL5), and acyl-CoA dehydrogenases, responsible for the oxidation of long and very long chain fatty acids (ACAD and ACADVL). As shown in Fig. 7B, ELOVL5, ACSL4, and ACSL5 transcript levels oscillate with zenith levels reached at 0400 hours, coinciding with the peak accumulation of very long chain PC and SM lipids (Fig. 7A), whereas ACSL3, ACAD, and ACADVL did not oscillate (SI Appendix, Fig. S6B).
In contrast, the degree of desaturation did not have an effect on lipid rhythmicity, both in vivo and in vitro. Completely saturated lipids, or lipids bearing one or multiple double bonds in their fatty acyl chains, were equally aligned in skeletal muscle and in primary myotubes (Fig. 7C, SI Appendix, Fig. S6C, and Datasets S7 and S9).
Lipid Rhythms Are Driven by the Cell-Autonomous Molecular Circadian Clock.
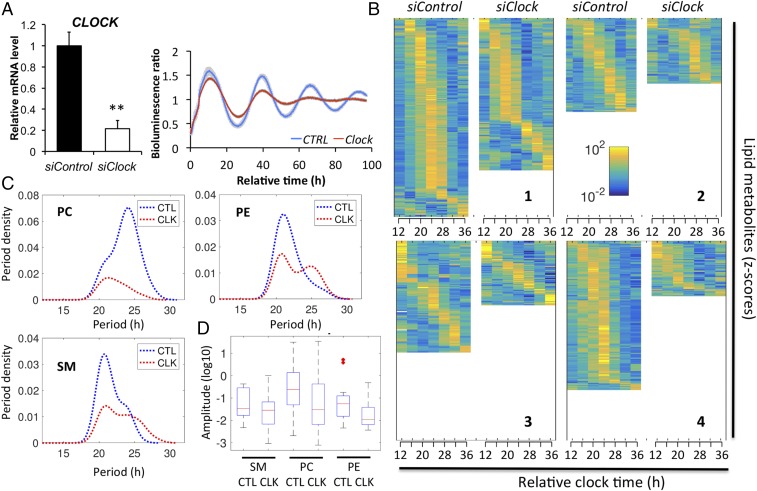
Finally, to investigate the effect of a functional molecular clock operative in human skeletal myotubes on lipid metabolism, we applied an efficient siClock transfection protocol, resulting in ∼80% knockdown of CLOCK transcript levels (Fig. 8 A, Left). Circadian amplitude of the Bmal1-luc reporter activity profile was strongly blunted in siClock-transfected myotubes, compared with cells transfected with nontargeting sequences (siControl) in four independent experiments, validating clock disruption (Fig. 8 A, Right). Following siRNA transfection, human primary myotubes were synchronized in vitro with a pulse of forskolin, and samples were collected every 4 h over 24 h in four independent experiments to monitor the effect of CLOCK depletion on lipid metabolite oscillations (see SI Appendix, Table S2 for donor characteristics). Importantly, the number of oscillating lipid metabolites was reduced on average for all four myotube cultures by 39.4 ± 7.8% (mean ± SEM) upon CLOCK depletion, and as individually shown by heat map analysis (Fig. 8B and Dataset S8). The analysis of lipidomic data from three major lipid classes (PC, PE, and SM) by ARSER algorithm (29) suggested a significant reduction in the frequency of oscillations with a period between 20 and 28 h upon siClock, as shown by period density plotted against the period length (Fig. 8C). Moreover, the mean amplitude of all circadian lipids was reduced in clock-disrupted myotubes, compared with their control counterparts for PC, PE, and SM (Fig. 8D).
Fig. 8.
siClock-mediated clock disruption attenuates circadian lipid oscillations in human skeletal myotubes. (A, Left) CLOCK mRNA was measured in human myotubes transfected with siControl or siClock by RT-qPCR and normalized to the mean of 9S and HPRT (mean ± SEM, n = 4; **P < 0.01). (Right) Average detrended Bmal1-luc bioluminescence profile (n = 4, mean ± SEM). Average Bmal1-luc oscillation profiles are shown for siControl (blue line) and siClock (red line) transfected myotubes. (B) Heat maps of rhythmic lipid species in primary skeletal myotubes transfected with either siControl or siClock in donors 1, 2, 3, and 4. Normalized z-scores of lipid metabolites are indicated in yellow (high) and blue (low). (C) Distribution of periodicity of oscillating PC, PE, and SM lipids for siControl (CTL) and siClock (CLK) (ARSER analysis, P value < 0.05). CLOCK knockdown induces a shift to include longer periods but mostly reduces the number of cycling lipids overall. (D) Box plot of amplitudes for PC, PE and SM raw lipid data for siControl and siClock of cycling lipids with a period between 22 and 26 (ARSER, P value < 0.05) after log transform. Knockdown lowers the amplitudes, with PC having the most pronounced effect.
Discussion
This study provides a systematic analysis of lipid metabolite oscillations in human skeletal muscle and in human primary myotubes. Moreover, it represents a comprehensive comparison between the lipid composition of human skeletal muscle in vivo and human primary myotube cells cultured in vitro. Importantly, rhythmic profiles of the lipid metabolites were blunted in myotubes upon clock perturbation, suggesting an essential role of the cell-autonomous oscillators for orchestrating circadian lipidomics.
Globally, lipid quantities were highly comparable between skeletal muscle and human primary myotubes (Fig. 2B), with the exception of CL and PS levels (Fig. 2 B, Right). Muscle biopsy donors were similar with regard to body mass index (BMI) but differed in age and muscle fiber type composition between the in vivo and in vitro cohorts (SI Appendix, Tables S1 and S2), as confirmed by myosin isoform analysis (SI Appendix, Fig. S7). While biopsies for the in vivo study originated from the vastus lateralis muscle (∼43% slow-twitch type I fibers) (30), samples for the in vitro study were established from the gluteus maximus that has a higher percentage of slow-twitch type I fibers (52 to 60%) (31).
CL, a lipid exclusively found in mitochondrial membranes, was reduced in human skeletal myotubes compared with muscle tissue (Fig. 2 B, Right), implying that the overall number of mitochondria is diminished in cell culture compared with muscle tissue, similar to mouse skeletal muscle and C2C12 myotubes (32). This effect might have been further enhanced by aging (33), due to the significant age difference between the older in vitro and younger in vivo cohorts in our study. Moreover, cells in culture rely mostly on glycolysis, leading to a further reduction in mitochondrial respiration (34). Additionally, a significant increase in PS was observed in primary myotubes compared with muscle tissue (Fig. 2 B, Right). Mammalian skeletal muscle is formed by the proliferation, differentiation, and fusion of myogenic precursor myoblast cells into multinucleated myotubes, and PS greatly enhances this fusion event (35, 36).
Overall, the lipid composition was more complex in primary myotubes (637 species) compared with skeletal muscle (532 metabolites), in particular for the lipid class of PCs (Fig. 2C). The fatty acid composition of skeletal muscle is reflected by the dietary intake of lipids in humans (37). The same holds true for the fatty acid composition of cells in culture where changes in serum influence the lipid composition of established cell lines. Of note, FBS is enriched in arachidonic (C20) and docosahexanoic (C22) acid (38) whereas a standard western diet mostly relies on C16- and C18-containing lipids. Consistently, very long chain PCs (PC40-44), containing C20 and C22 fatty acids in their fatty acyl chains, were highly abundant in primary myotubes, but less abundant or not detected in skeletal muscle (in Fig. 2C, compare the left and right columns of the PC heat map).
The data presented here demonstrate diurnal lipid oscillations of 20.3% in human skeletal muscle (Fig. 4). This value is similar to a study in human plasma, which showed 17.8% of oscillating lipid metabolites when analyzed on an individual basis (19). To unravel whether these metabolite oscillations are solely driven by diurnal cycles of rest–activity and food intake, or are controlled in a cell-autonomous manner, we conducted a similar temporal analysis in primary human skeletal myotubes synchronized in vitro. Recent studies conducted in other types of isolated human primary cells have demonstrated that peripheral clocks are functional in vitro, as shown for skin fibroblasts (39), thyrocytes (40), and pancreatic islets (41–43). Moreover, cell-autonomous oscillators of human primary skeletal myotubes have been recently assessed (25–27). Remarkably, we observed that lipid oscillations persisted in human primary myotubes synchronized in vitro, with a comparable percentage of oscillating lipid species (18.6%) (Fig. 4). However, as the in vivo biopsy collection was incomplete, with samples collected at six time points compared with seven time points in vitro, the percentage of oscillating metabolites in muscle tissue might be higher if analyzed in an identical manner. Furthermore, we cannot exclude that the results presented here are in part influenced by differences in fiber type composition between vastus lateralis and gluteus maximus, as demonstrated by myosin isoform analysis (SI Appendix, Fig. S7).
Globally, the percentage of lipid oscillations might even be underestimated as whole-cell lipid measurements can mask oscillations within subcellular compartments. Recent studies by Asher and coworkers identified that 17% of lipid metabolites oscillate when assessed in whole liver (16) whereas separate analyses of liver nuclei and mitochondria identified 34% and 31% of oscillating lipids, respectively, with distinct circadian phases between these organelles (17). However, it should be noted that participants in this study were confined to a semirecumbent position throughout the testing day while, in the mouse liver study, the data were collected from mice freely moving until the sampling point. Given that physical activity can modulate the level and oscillatory parameters of lipids, this may partly account for the observed differences.
The oscillation amplitude of individual lipid metabolite profiles was up to 30% across 24 h both in vivo and in vitro (Fig. 5), similar to previous analyses in mouse liver (17). Whereas such changes in amplitude are smaller than for diurnal transcript oscillations, they represent substantial differences in membrane homeostasis across 24 h.
How transcriptional rhythms are translated into lipid metabolites oscillations has not been addressed so far. Notably, peak levels of major PLs and SLs correlated with ARNTL (BMAL1) gene expression both in vivo (Fig. 1 C and D) and in vitro (Fig. 3 A and B), with phase coherence kept between core clock gene expression and lipid profiles indicating a potential role for the endogenous skeletal muscle clock in the regulation of lipid metabolism. In vivo, lipid levels peaked in the early morning hours, near the usual wake time, supporting a major role for the muscle clock in anticipating sleep-to-wake transition as has been proposed previously (44). To directly test the effect of cell-autonomous oscillators on lipid metabolic changes, we subsequently disrupted the endogenous clock, operative in human skeletal myotubes (27). Clock disruption had a strong impact on lipid metabolite oscillations. The number of oscillating lipid species was significantly reduced upon CLOCK depletion, as confirmed by JTK_CYCLE (Fig. 8B) and ARSER (Fig. 8C) analyses, with a strong impact on both PL and SL metabolites. Furthermore, amplitudes of the remaining oscillating lipid species were reduced (Fig. 8D), suggesting an overall blunting of the circadian rhythmicity in the absence of a functional cell-autonomous myotube clock.
Remarkably, zenith levels of enzymes involved in de novo SL biosynthesis (SPTLC2, UGCG) correlated well with the accumulation of SL species at the early morning hours in human skeletal muscle (Fig. 6). This correlation was not restricted to SLs alone as also the transcriptional regulation of PL biosynthesis showed enrichment at the early morning hours, with peak levels of ELOVL5, ACSL4, and ACSL5 transcripts correlating with peak levels of very long chain PC lipids (Fig. 7 A and B). Although the gene expression profile of choline phosphotransferase 1 (CHPT1), which catalyzes PC biosynthesis from CDP-choline, was not rhythmic on average, individual CHPT1 transcripts oscillated with profiles similar to the most rhythmic PC lipid metabolite in skeletal muscle (PC32:2) in a subset of biopsy donors (Fig. 6D and SI Appendix, Table S3).
Interestingly, we observed an effect of the circadian clock on lipid chain length but not on desaturation (Fig. 7 and SI Appendix, Fig. S6). This effect was characteristic for long and very long chain PC and SM lipids, which were highly synchronized in both skeletal muscle and primary myotubes, whereas other PLs like PE did not show this preference. Remarkably, a correlation based on chain length was also observed for long and very long chain PC and SM species in WT and Per1/2 KO mice whereas the contribution of desaturation was less prominent (17). Consistent with this observation is the rhythmic gene expression of ELOVL5, ACSL4, and ACSL5 in human skeletal muscle tissue, whose peak of expression correlated with peak levels of very long chain PC and SM lipids (Fig. 7 A and B). Importantly, all seven mammalian elongase family members are either rhythmically expressed or their expression is affected by core clock gene alterations in peripheral mouse tissues, with Elovl1, Elovl5, and Elovl6 being circadian in mouse skeletal muscle (5, 12, 45). Moreover, ACSL4 was shown to be rhythmic in mouse liver, and its rhythmicity was attenuated in ClockΔ19 mutant mice (46). We therefore hypothesize that enzymes involved in long chain fatty acid activation and elongation are important drivers of lipid oscillations. As very long chain PC and SM lipids make up a significant proportion of the outer leaflet of the plasma membrane (47, 48), these data suggest that the circadian clock might impact on plasma membrane function, with important consequences on receptor signaling and membrane glucose uptake.
Fasting–feeding and rest–activity cycles are major timing cues in the synchronization of peripheral clocks (24). The SCN molecular clock communicates with peripheral tissues, such as skeletal muscle, using neurohumoral, metabolic, and temperature signals (49–51). In our in vivo study, core clock alignment was demonstrated by plasma cortisol and melatonin profiles, as well as by muscle core clock gene expression (Fig. 1). In human primary myotubes, highly synchronized Bmal1-luc activity profiles showed alignment of the molecular clock also in the absence of the SCN (Fig. 3A). However, despite core clock alignment, we observed extensive variability among the individual participants with regard to lipid oscillations. This heterogeneity persisted in vitro, allowing us to identify oscillatory subgroups within different lipid classes (Fig. 5). Such variability with regard to lipid profiles has been previously observed in human plasma (19) and supports the existence of distinct diurnal metabolic phenotypes within the human population, possibly controlled on an epigenetic level.
Taken together, this work is a detailed characterization of the temporal human skeletal muscle lipidome, highlighting the essential role of the molecular oscillator in the circadian orchestration of muscle lipid metabolism. The observed substantial lipid oscillations further contribute to lipid complexity and could affect membrane properties and signaling. Given the current gap in understanding the enormous complexity of cellular lipids, circadian lipidomics may allow deciphering networks of coregulated lipids, due to their temporal regulation. Genetic animal models of clock disruption as well as nutritional studies have shown that the circadian clockwork is associated with altered lipid homeostasis, fatty liver, and obesity (52). Although various clock mutant mouse models exhibit altered lipid metabolism (53), it is challenging to dissect the contribution of the circadian clock on lipid metabolism from the impact of fasting–feeding cycles and rest–activity. In vitro synchronized primary cells may therefore pave the way for future studies that link circadian clock defects to insulin resistance, obesity, and T2D, and constitute a powerful model for studying peripheral oscillators in other human tissues. Of note, human primary myotubes established from donor muscle biopsies preserve their metabolic phenotype with regard to glucose metabolism (54) and insulin signaling (55, 56). As metabolic disorders, including obesity and T2D, are growing rapidly in modern societies (57), future studies aimed at deciphering clock functions in the regulation of lipid metabolism in context of these pathologies are therefore urgently warranted.
Materials and Methods
Human Skeletal Muscle Biopsies for the in Vivo Study.
Ten healthy volunteers were recruited for the in vivo study (see SI Appendix, Table S1 for donor characteristics). One week before the scheduled laboratory visit, participants had to adhere to a consistent daily feeding and sleeping routine.
Six 100-mg biopsy samples were acquired from the vastus lateralis muscle at 4-h intervals (1200, 1600, 2000, 0000, 0400, and 0800 hours) and immediately snap frozen under liquid nitrogen. The study was conducted in accordance with the Declaration of Helsinki and with the approval of the Health Research Authority [National Research Ethics Service (NRES) Committee South West; 14/SW/0123] and the Commission cantonale (Canton de Vaud) d’Éthique de la Recherche (Cer-VD). See SI Appendix, SI Materials and Methods for further details of study procedure.
Human Skeletal Muscle Biopsies for the in Vitro Study and Primary Myotube Cell Culture.
Muscle biopsies were derived from healthy donors with informed consent obtained from all participants (see SI Appendix, Table S2 for donor characteristics). The study conformed to the Declaration of Helsinki and the experimental protocol (“DIOMEDE”) was approved by the Ethical Committee SUD EST IV (Agreement 12/111) and performed according to the French legislation (Huriet’s law). All donors had HbAc1 levels inferior to 6.0% and fasting glycemia inferior to 7 mmol/L, were not diagnosed for T2D, neoplasia, or chronic inflammatory diseases, and were not doing shift work. Biopsies were taken from the gluteus maximus (n = 10) during planned surgeries. Human myoblasts were cultured in growth medium composed of HAM F-10 supplemented with 20% FBS, and 1% penicillin–streptomycin at 37 °C. After reaching confluence, myoblasts were differentiated into myotubes during 7 to 10 d in differentiation medium (DMEM supplemented with 2% FBS). Muscle differentiation was characterized by the fusion of myoblasts into polynucleated myotubes as previously described (27).
In Vitro Skeletal Myotube Synchronization, siRNA Transfection, and Real-Time Bioluminescence Recording.
Myotube cells were synchronized by 10 µM forskolin (Sigma) for 1 h once differentiation was achieved. Differentiated human myotubes were transfected with 20 nM siRNA targeting CLOCK (siClock) or with nontargeting siControl (Dharmacon; GE Healthcare), using HiPerFect transfection reagent (Qiagen) 24 h before synchronization, following the manufacturer’s protocol. Myotube bioluminescence recording was performed as described in ref. 27 and detailed in SI Appendix, SI Materials and Methods.
Quantitative RT-PCR.
RNA from human skeletal muscle samples (550 ng) was reverse-transcribed using a SuperScript VILO cDNA Synthesis Kit (Thermo Scientific). mRNA levels were determined by TaqMan real-time qPCR using a LightCycler 480 (Roche) following the manufacturer’s instructions. Copy numbers were normalized using GAPDH. The following TaqMan probes were used: PER2, Hs00256143_m1; ARNTL, Hs00253876_m1; DBP, Hs00609747_m1; CHPT1, Hs01012468_m1; ELOVL5, Hs01094711_m1; SPTLC2, Hs01027014_m1; UGCG, Hs00916612_m1; GAPDH, Hs02758991_m1; ACSL3, Hs00244853_m1; ACSL4, Hs00244871_m1; ACSL5, Hs01061754_m1; ACAD, Hs00155630_m1; ACADVL, Hs00825606_g1; MYH1, Hs00428600_m1; MYH2, Hs00430042_m1; MYH4, Hs00757977_m1; and MYH7, Hs01110632_m1.
RNA from human primary myotubes was reverse-transcribed, using SuperScript III reverse transcriptase (Thermo Scientific) and random hexamers, and then PCR-amplified on a LightCycler 480 (Roche). Values were normalized to the mean of two housekeeping genes (HPRT and 9S) as described previously (27).
Lipidomics Analysis.
The lipidomics analysis was performed as described in ref. 58 and detailed in SI Appendix, SI Materials and Methods.
Data Analysis.
Actimetrics LumiCycle analysis software (Actimetrics LTD) was used for bioluminescence data analysis. Normalized lipid values (fold change mean) were defined as the lipid value (phosphate normalized) for each time point divided by the lipid mean value, calculated separately for each donor. To identify circadian variations within the lipidomic dataset, phosphate values were z-scored within subjects and then further analyzed using the JTK_CYCLE (28) and ARSER (29) algorithms. The period width for the JTK_CYCLE analysis was set to fit a time frame of 20 to 28 h and a P value of ≤0.05 was considered statistically significant.
Statistical analyses concerning absolute lipid quantities were performed using a paired Student’s t test. Differences were considered significant for P ≤ 0.05 (*), P ≤ 0.01 (**), and P ≤ 0.001 (***) if not noted otherwise.
To determine the clustering, k-NN (nearest neighbors with k clusters) was applied to the phases and amplitudes in polar coordinates of all circadian signals for k = 1, 2, and 3 clusters. For further details, see SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Jacques Philippe and Anton Alekseev (University of Geneva) for constructive discussions, Dylan Thompson and Keith Stokes (University of Bath) for their assistance with data collection, and Isabelle Riezman (University of Geneva) for sample processing. This work was funded by Sinergia Swiss National Science Foundation Grant CRSII3-154405 (to H.R., C. Dibner, and E.L.); by Swiss National Science Foundation Grants 31003A-166700 (to C. Dibner) and 310030B_166686 (to H.R.); by SystemsX.ch (evaluated by the Swiss National Science Foundation) (H.R.); by the National Centre of Competence in Research Chemical Biology (H.R.); by the Fondation Romande pour la Recherche sur le Diabète; by the Fondation Ernst et Lucie Schmidheiny; by the Société Académique de Genève (C. Dibner); and by United Kingdom Biotechnology and Biological Sciences Research Council Grant BB/I008470/1 (to J.D.J.).
Footnotes
Conflict of interest statement: B.D.W. and F.G. are employees of Nestlé Institute of Health Sciences SA; L.G.K. is an employee of Nestec Ltd. J.A.B. has been a consultant for PepsiCo (Quaker) and Kellogg’s. The other authors have no conflict of interest to declare.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1705821114/-/DCSupplemental.
References
- 1.Asher G, Sassone-Corsi P. Time for food: The intimate interplay between nutrition, metabolism, and the circadian clock. Cell. 2015;161:84–92. doi: 10.1016/j.cell.2015.03.015. [DOI] [PubMed] [Google Scholar]
- 2.Albrecht U. Timing to perfection: The biology of central and peripheral circadian clocks. Neuron. 2012;74:246–260. doi: 10.1016/j.neuron.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 3.Dibner C, Schibler U, Albrecht U. The mammalian circadian timing system: Organization and coordination of central and peripheral clocks. Annu Rev Physiol. 2010;72:517–549. doi: 10.1146/annurev-physiol-021909-135821. [DOI] [PubMed] [Google Scholar]
- 4.Storch KF, et al. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417:78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- 5.Zhang R, Lahens NF, Ballance HI, Hughes ME, Hogenesch JB. A circadian gene expression atlas in mammals: Implications for biology and medicine. Proc Natl Acad Sci USA. 2014;111:16219–16224. doi: 10.1073/pnas.1408886111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Panda S, et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 7.Gachon F, Loizides-Mangold U, Petrenko V, Dibner C. Glucose homeostasis: Regulation by peripheral circadian clocks in rodents and humans. Endocrinology. 2017 doi: 10.1210/en.2017-00218. [DOI] [PubMed] [Google Scholar]
- 8.DeFronzo RA, et al. The effect of insulin on the disposal of intravenous glucose. Results from indirect calorimetry and hepatic and femoral venous catheterization. Diabetes. 1981;30:1000–1007. doi: 10.2337/diab.30.12.1000. [DOI] [PubMed] [Google Scholar]
- 9.Muoio DM, Newgard CB. Mechanisms of disease: Molecular and metabolic mechanisms of insulin resistance and beta-cell failure in type 2 diabetes. Nat Rev Mol Cell Biol. 2008;9:193–205. doi: 10.1038/nrm2327. [DOI] [PubMed] [Google Scholar]
- 10.Dyar KA, et al. Muscle insulin sensitivity and glucose metabolism are controlled by the intrinsic muscle clock. Mol Metab. 2013;3:29–41. doi: 10.1016/j.molmet.2013.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harfmann BD, et al. Muscle-specific loss of Bmal1 leads to disrupted tissue glucose metabolism and systemic glucose homeostasis. Skelet Muscle. 2016;6:12. doi: 10.1186/s13395-016-0082-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hodge BA, et al. The endogenous molecular clock orchestrates the temporal separation of substrate metabolism in skeletal muscle. Skelet Muscle. 2015;5:17. doi: 10.1186/s13395-015-0039-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Turek FW, et al. Obesity and metabolic syndrome in circadian clock mutant mice. Science. 2005;308:1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paschos GK, et al. Obesity in mice with adipocyte-specific deletion of clock component Arntl. Nat Med. 2012;18:1768–1777. doi: 10.1038/nm.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grimaldi B, et al. PER2 controls lipid metabolism by direct regulation of PPARγ. Cell Metab. 2010;12:509–520. doi: 10.1016/j.cmet.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adamovich Y, et al. Circadian clocks and feeding time regulate the oscillations and levels of hepatic triglycerides. Cell Metab. 2014;19:319–330. doi: 10.1016/j.cmet.2013.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aviram R, et al. Lipidomics analyses reveal temporal and spatial lipid organization and uncover daily oscillations in intracellular organelles. Mol Cell. 2016;62:636–648. doi: 10.1016/j.molcel.2016.04.002. [DOI] [PubMed] [Google Scholar]
- 18.Kalsbeek A, la Fleur S, Fliers E. Circadian control of glucose metabolism. Mol Metab. 2014;3:372–383. doi: 10.1016/j.molmet.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chua EC, et al. Extensive diversity in circadian regulation of plasma lipids and evidence for different circadian metabolic phenotypes in humans. Proc Natl Acad Sci USA. 2013;110:14468–14473. doi: 10.1073/pnas.1222647110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morgan L, Hampton S, Gibbs M, Arendt J. Circadian aspects of postprandial metabolism. Chronobiol Int. 2003;20:795–808. doi: 10.1081/cbi-120024218. [DOI] [PubMed] [Google Scholar]
- 21.Dallmann R, Viola AU, Tarokh L, Cajochen C, Brown SA. The human circadian metabolome. Proc Natl Acad Sci USA. 2012;109:2625–2629. doi: 10.1073/pnas.1114410109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davies SK, et al. Effect of sleep deprivation on the human metabolome. Proc Natl Acad Sci USA. 2014;111:10761–10766. doi: 10.1073/pnas.1402663111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giskeødegård GF, Davies SK, Revell VL, Keun H, Skene DJ. Diurnal rhythms in the human urine metabolome during sleep and total sleep deprivation. Sci Rep. 2015;5:14843. doi: 10.1038/srep14843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dibner C, Schibler U. Circadian timing of metabolism in animal models and humans. J Intern Med. 2015;277:513–527. doi: 10.1111/joim.12347. [DOI] [PubMed] [Google Scholar]
- 25.Hansen J, et al. Synchronized human skeletal myotubes of lean, obese and type 2 diabetic patients maintain circadian oscillation of clock genes. Sci Rep. 2016;6:35047. doi: 10.1038/srep35047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loizides-Mangold U, et al. Paraoxonase 1 (PON1) and pomegranate influence circadian gene expression and period length. Chronobiol Int. 2016;33:453–461. doi: 10.3109/07420528.2016.1154067. [DOI] [PubMed] [Google Scholar]
- 27.Perrin L, et al. Human skeletal myotubes display a cell-autonomous circadian clock implicated in basal myokine secretion. Mol Metab. 2015;4:834–845. doi: 10.1016/j.molmet.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hughes ME, Hogenesch JB, Kornacker K. JTK_CYCLE: An efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J Biol Rhythms. 2010;25:372–380. doi: 10.1177/0748730410379711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang R, Su Z. Analyzing circadian expression data by harmonic regression based on autoregressive spectral estimation. Bioinformatics. 2010;26:i168–i174. doi: 10.1093/bioinformatics/btq189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Staron RS, et al. Fiber type composition of the vastus lateralis muscle of young men and women. J Histochem Cytochem. 2000;48:623–629. doi: 10.1177/002215540004800506. [DOI] [PubMed] [Google Scholar]
- 31.Johnson MA, Polgar J, Weightman D, Appleton D. Data on the distribution of fibre types in thirty-six human muscles. An autopsy study. J Neurol Sci. 1973;18:111–129. doi: 10.1016/0022-510x(73)90023-3. [DOI] [PubMed] [Google Scholar]
- 32.Deshmukh AS, et al. Deep proteomics of mouse skeletal muscle enables quantitation of protein isoforms, metabolic pathways, and transcription factors. Mol Cell Proteomics. 2015;14:841–853. doi: 10.1074/mcp.M114.044222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murgia M, et al. Single muscle fiber proteomics reveals fibertype-specific features of human muscle aging. Cell Rep. 2017;19:2396–2409. doi: 10.1016/j.celrep.2017.05.054. [DOI] [PubMed] [Google Scholar]
- 34.Aguer C, et al. Galactose enhances oxidative metabolism and reveals mitochondrial dysfunction in human primary muscle cells. PLoS One. 2011;6:e28536. doi: 10.1371/journal.pone.0028536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jeong J, Conboy IM. Phosphatidylserine directly and positively regulates fusion of myoblasts into myotubes. Biochem Biophys Res Commun. 2011;414:9–13. doi: 10.1016/j.bbrc.2011.08.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van den Eijnde SM, et al. Transient expression of phosphatidylserine at cell-cell contact areas is required for myotube formation. J Cell Sci. 2001;114:3631–3642. doi: 10.1242/jcs.114.20.3631. [DOI] [PubMed] [Google Scholar]
- 37.Andersson A, Nälsén C, Tengblad S, Vessby B. Fatty acid composition of skeletal muscle reflects dietary fat composition in humans. Am J Clin Nutr. 2002;76:1222–1229. doi: 10.1093/ajcn/76.6.1222. [DOI] [PubMed] [Google Scholar]
- 38.Stoll LL, Spector AA. Changes in serum influence the fatty acid composition of established cell lines. In Vitro. 1984;20:732–738. doi: 10.1007/BF02618879. [DOI] [PubMed] [Google Scholar]
- 39.Brown SA, et al. The period length of fibroblast circadian gene expression varies widely among human individuals. PLoS Biol. 2005;3:e338. doi: 10.1371/journal.pbio.0030338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mannic T, et al. Circadian clock characteristics are altered in human thyroid malignant nodules. J Clin Endocrinol Metab. 2013;98:4446–4456. doi: 10.1210/jc.2013-2568. [DOI] [PubMed] [Google Scholar]
- 41.Perelis M, et al. Pancreatic β cell enhancers regulate rhythmic transcription of genes controlling insulin secretion. Science. 2015;350:aac4250. doi: 10.1126/science.aac4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pulimeno P, et al. Autonomous and self-sustained circadian oscillators displayed in human islet cells. Diabetologia. 2013;56:497–507. doi: 10.1007/s00125-012-2779-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saini C, et al. A functional circadian clock is required for proper insulin secretion by human pancreatic islet cells. Diabetes Obes Metab. 2016;18:355–365. doi: 10.1111/dom.12616. [DOI] [PubMed] [Google Scholar]
- 44.Schiaffino S, Blaauw B, Dyar KA. The functional significance of the skeletal muscle clock: Lessons from Bmal1 knockout models. Skelet Muscle. 2016;6:33. doi: 10.1186/s13395-016-0107-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dyar KA, et al. The calcineurin-NFAT pathway controls activity-dependent circadian gene expression in slow skeletal muscle. Mol Metab. 2015;4:823–833. doi: 10.1016/j.molmet.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kudo T, Tamagawa T, Kawashima M, Mito N, Shibata S. Attenuating effect of clock mutation on triglyceride contents in the ICR mouse liver under a high-fat diet. J Biol Rhythms. 2007;22:312–323. doi: 10.1177/0748730407302625. [DOI] [PubMed] [Google Scholar]
- 47.Li G, et al. Efficient replacement of plasma membrane outer leaflet phospholipids and sphingolipids in cells with exogenous lipids. Proc Natl Acad Sci USA. 2016;113:14025–14030. doi: 10.1073/pnas.1610705113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Meer G, Voelker DR, Feigenson GW. Membrane lipids: Where they are and how they behave. Nat Rev Mol Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Abraham U, et al. Coupling governs entrainment range of circadian clocks. Mol Syst Biol. 2010;6:438. doi: 10.1038/msb.2010.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Balsalobre A, Marcacci L, Schibler U. Multiple signaling pathways elicit circadian gene expression in cultured Rat-1 fibroblasts. Curr Biol. 2000;10:1291–1294. doi: 10.1016/s0960-9822(00)00758-2. [DOI] [PubMed] [Google Scholar]
- 51.Brown SA, Zumbrunn G, Fleury-Olela F, Preitner N, Schibler U. Rhythms of mammalian body temperature can sustain peripheral circadian clocks. Curr Biol. 2002;12:1574–1583. doi: 10.1016/s0960-9822(02)01145-4. [DOI] [PubMed] [Google Scholar]
- 52.Adamovich Y, Aviram R, Asher G. The emerging roles of lipids in circadian control. Biochim Biophys Acta. 2015;1851:1017–1025. doi: 10.1016/j.bbalip.2014.11.013. [DOI] [PubMed] [Google Scholar]
- 53.Panda S. Circadian physiology of metabolism. Science. 2016;354:1008–1015. doi: 10.1126/science.aah4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bourlier V, et al. Enhanced glucose metabolism is preserved in cultured primary myotubes from obese donors in response to exercise training. J Clin Endocrinol Metab. 2013;98:3739–3747. doi: 10.1210/jc.2013-1727. [DOI] [PubMed] [Google Scholar]
- 55.Bouzakri K, et al. Reduced activation of phosphatidylinositol-3 kinase and increased serine 636 phosphorylation of insulin receptor substrate-1 in primary culture of skeletal muscle cells from patients with type 2 diabetes. Diabetes. 2003;52:1319–1325. doi: 10.2337/diabetes.52.6.1319. [DOI] [PubMed] [Google Scholar]
- 56.Green CJ, Pedersen M, Pedersen BK, Scheele C. Elevated NF-κB activation is conserved in human myocytes cultured from obese type 2 diabetic patients and attenuated by AMP-activated protein kinase. Diabetes. 2011;60:2810–2819. doi: 10.2337/db11-0263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Krug EG. Trends in diabetes: Sounding the alarm. Lancet. 2016;387:1485–1486. doi: 10.1016/S0140-6736(16)30163-5. [DOI] [PubMed] [Google Scholar]
- 58.Loizides-Mangold U, David FP, Nesatyy VJ, Kinoshita T, Riezman H. Glycosylphosphatidylinositol anchors regulate glycosphingolipid levels. J Lipid Res. 2012;53:1522–1534. doi: 10.1194/jlr.M025692. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.