Significance
Monitoring neuronal activity in freely moving, behaving animals is a holy grail of neuroscience. Here we present a noninvasive tool that links the calcium profiles of specific fly neurons to real-time behavior. Optogenetic manipulation of two groups of circadian neurons indicates that they drive sleep or locomotor activity. The two calcium patterns are also distinct and couple well to the sleep/activity phases of fly behavior. The sleep-promoting neurons appear more active when the flies initiate daytime sleep, whereas the activity-promoting neurons appear to fire more strongly coincident with the evening locomotor activity peak. This new approach is complementary to electrophysiological recording and GCaMP imaging, especially for small organisms and behavorial paradigms for which these more traditional methods are not practical.
Keywords: circadian, sleep, Drosophila, optogenetics, calcium
Abstract
There are no general methods for reliably assessing the firing properties or even calcium profiles of specific neurons in freely moving flies. To this end, we adapted a GFP-based calcium reporter to luciferase that was expressed in small subsets of circadian neurons. This Tric-LUC reporter allowed a direct comparison of luciferase activity with locomotor activity, which was assayed in the same flies with video recording. The LUC profile from activity-promoting E cells paralleled evening locomotor activity, and the LUC profile from sleep-promoting glutamatergic DN1s (gDN1s) paralleled daytime sleep. Similar profiles were generated by novel reporters recently identified based on transcription factor activation. As E cell and gDN1 activity is necessary and sufficient for normal evening locomotor activity and daytime sleep profiles, respectively, we suggest that their luciferase profiles reflect their neuronal calcium and in some cases firing profiles in wake-behaving flies.
Locomotion and sleep are highly regulated biological processes in animals (1, 2). Under standard 12-h light:12-h dark (LD 12:12) laboratory conditions at a constant temperature of 25 °C, Drosophila locomotor activity is maximal in the morning around lights-on and in the evening around lights-off. These two daily activity bouts are often referred to as the morning (M) and evening (E) activity peaks (3–5). Flies generally take a midday siesta between these two activity peaks, and they experience consolidated nighttime sleep for most of the 12 h of darkness after lights-off (6).
This sleep–wake pattern is tightly regulated by the Drosophila circadian neuronal circuitry. This circuitry comprises approximately 75 pairs of neurons that express the core circadian proteins. Clock neurons can be further divided into several groups based on their anatomic position within the adult brain (6). The ventral lateral neurons (LNvs) consist of small and large cells (s-LNvs and l-LNvs, respectively). The PDF-positive s-LNvs are major fly pacemaker neurons (7–9). They also drive morning activity, although recent results show this relationship to be less exclusive (10). Another group consists of dorsal lateral neurons (LNds). Three of these LNds, along with the PDF-negative fifth s-LNv, are necessary for the timing of the E activity peak and thus are referred to collectively as E cells (4, 10). Their activity is also necessary and sufficient to generate a proper evening activity peak, and they are major locomotor activity-promoting neurons in a more general sense. For example, blocking neurotransmitter release from these cells diminishes baseline locomotor activity (10). In addition, there are three groups of dorsal neurons, the DN1s, DN2s, and DN3s (11–14). The DN1s in particular are most important from the locomotor activity-sleep standpoint, as they are crucial for promoting daytime sleep. They do this by inhibiting the E cells and the locomotor activity E peak that they cause (15).
The daily oscillations of clock proteins, such as period (PER) and timeless (TIM), are quite well synchronized in all circadian neuron groups (16, 17). However, their diverse functions suggest that their peak neuronal activities, when they achieve maximal firing rates, occur at different circadian times. For example, the DN1 peak firing rate might occur in the daytime when DN1s drive the siesta, whereas the E cell peak firing rate might occur coincident with lights-off, when these cells drive the evening locomotor activity peak.
Relevant to these possibilities are three recent studies that addressed the circadian neuronal activity and calcium patterns of the DN1s and E cells. One study assayed the DN1s electrophysiologically from dissected brains (18), while the other two studies used tethered flies and GCaMP6 to image calcium within the different circadian neuron groups (19, 20). All three studies indicated that the DN1s fire more in the morning and less in the evening. The imaging study also reported that the calcium activity within the E cells peaks several hours before the end of the subjective day in constant darkness (DD). As both studies were carried out under nonphysiological conditions, that is, not in normally waking, moving, and sleeping (freely moving) flies, the calcium or firing patterns might not reflect those patterns occurring under the standard assay conditions used in behavioral studies.
To address this issue, we previously developed a noninvasive in vivo calcium monitoring tool for recording neuronal activity in real-time in wake-behaving flies (15). That luciferase-based reporter used CaLexA, a transcription factor fusion protein dependent on calcium for nuclear import (21). We have now improved the calcium monitoring by switching to a Tric-LUC reporter. Tric (transcriptional reporter of intracellular Ca2+) uses a binary expression system that is also dependent on Ca2+. Importantly, TRIC-GFP has enhanced temporal resolution compared with the CaLexA-GFP in flies (22). In addition, we have compared Tric-LUC with newly developed neuronal activity-regulated transcriptional reporters, which, like the two calcium reporters, express luciferase in response to neuronal firing caused by optogenetic stimulation (23). Two of these three transcriptional reporters express in DN1s as well as E cells and show comparable sensitivity to Tric-LUC; thus, we used both systems to assay the activity patterns of the DN1s and the LNds under normal LD conditions.
We first used new, highly specific split GAL4 drivers. By requiring an overlap in the expression patterns of two promoters, these drivers are restricted to specific target neurons (24). They confirm that the gDN1s are sleep-promoting and that the E cells are activity-promoting. Importantly, the gDN1 calcium profile is roughly consistent with a previous electrophysiology assay of DN1 firing rate (18). The E cell calcium profile is very different; it peaks in the evening time and is minimized during the nighttime sleep period. Importantly, the gDN1 and E cell calcium patterns correlate closely with the actual daytime sleep and locomotor activity profiles, respectively, as determined by simultaneous real-time recording of behavior in freely moving flies. The profiles of the new transcriptional reporters were generally very close to the profile of the Tric-LUC calcium reporter and validate the functional relevance of the Tric-LUC patterns. Considered together with the importance of these two neuronal groups for the locomotor activity program, it is likely that the calcium and reporter patterns reflect neuronal activity, at least in part.
Results
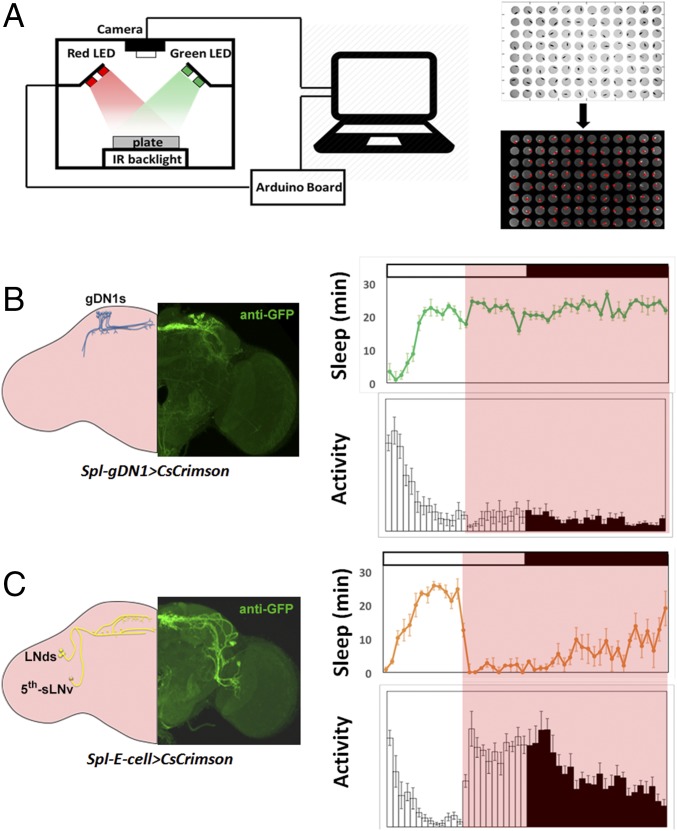
We previously described the effect of CsChrimson activation on a subset of circadian neurons, the five to six sleep-promoting gDN1s in each hemisphere. Here we repeated this experiment and also expressed CsChrimson in activity-promoting E cells (25, 26). In addition, two highly specific split-Gal4 (Spl) drivers were used to activate the three to four gDN1s and the four E cells present in each hemisphere (24, 27) (Fig. 1 B and C and Fig. S1 A and B). Behavior was monitored using Flybox in standard LD cycles (15) (Fig. 1A).
Fig. 1.
Identifying sleep-promoting and activity-promoting circadian neurons. (A) Schemes of Flybox, which has red and green LEDs to provide optogenetic activation and inhibition, and a camera that can record behavior of flies in a 96-well plate in real time. (Right) Raw and processed images recorded from Flybox. (B) Optogenetic activation of gDN1s promotes persistent sleep. (Left) Expression pattern of the split GAL4 line that labels gDN1s (Spl-gDN1) in one brain hemisphere. (Right) Sleep and activity profile of Spl-gDN1 > CsCrimson flies under optogenetic activation. Error bars represent SEM. n = 24 for each group. (C) Optogenetic activation of E cells results in persistent activity. (Left) Expression pattern of split GAL4 line that labels E cells (Spl-E) in one brain hemisphere. (Right) Sleep and activity pattern of Spl-E cell > CsCrimson flies under optogenetic activation. Error bars represent SEM. n = 23 for each group.
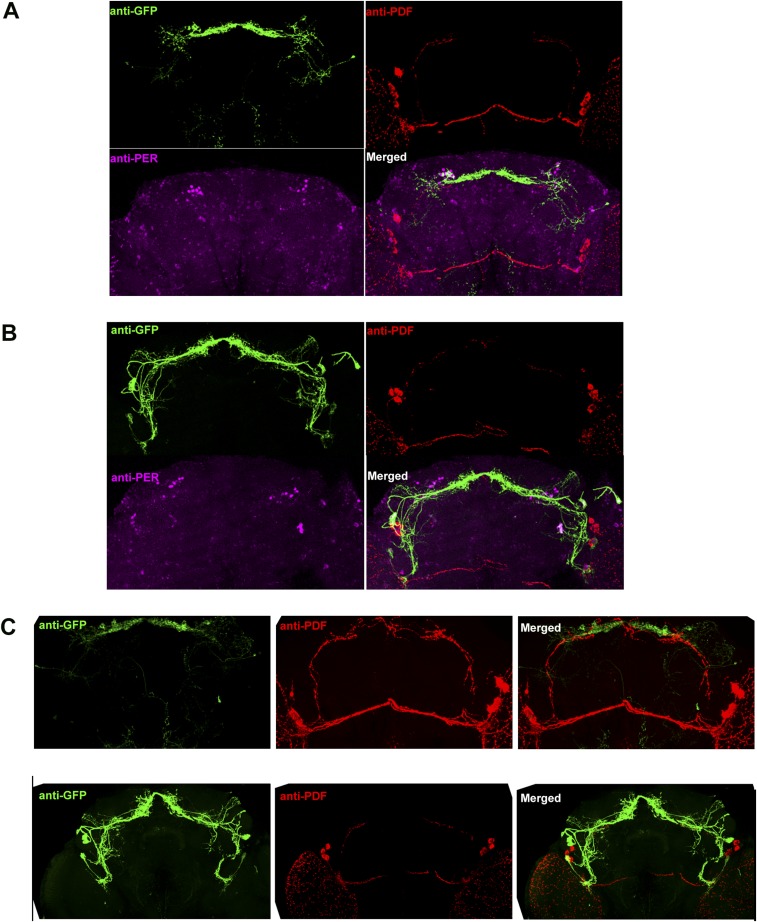
Fig. S1.
Anatomic characterization of Spl-E cell and Spl-gDN1 expression. (A) Representative GFP (green), PDF (red), and PER (magenta) costaining in a brain of Spl-gDN1 > UAS-CsCrimson fly. (B) Representative GFP (green), PDF (red), and PER (magenta) costaining in the brain of an Spl-E cell > UAS-CsCrimson fly. (C) GFP (green) and PDF (red) costaining of Spl-gDN1 > UAS-CsCrimson (Upper) and Spl-E cell > UAS-CsCrimson (Lower) brains. Spl-gDN1 GAL4 labels three to four DN1s in each hemisphere, and Spl-E cell GAL4 labels three LNds as well as the fifth s-LNv in each hemisphere.
As described previously, activation of the gDN1s rapidly puts the flies to sleep (15). In this experiment, activation began in the middle of the day and caused persistence of the midday siesta, which completely inhibited the E activity peak that would normally occur near lights-off (Fig. 1B).
In contrast, activation of E cells rapidly inhibited the siesta and promoted persistent activity (Fig. 1C). This caused a premature evening locomotor activity peak, which lasted for much longer after lights-off than would normally be the case, i.e., in the absence of continued activation. The different activation effects were also evident from the quantitative sleep parameters (Fig. S2A).
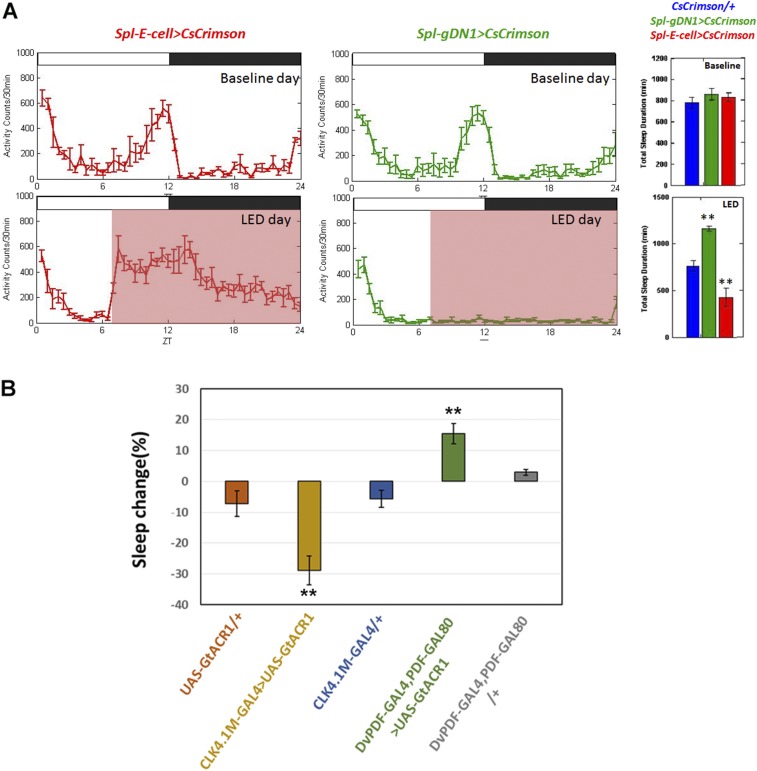
Fig. S2.
Effects of optogenetic activation and inhibition on activity and sleep profiles. (A) Locomotor activity profile of Spl-E cells > UAS-CsCrimson (Left, red) and Spl-gDN1 > UAS-CsCrimson (Middle, green) on the baseline day (Upper) and LED day (Lower). (Right) Quantification of 24-h sleep level on the baseline and LED days. The black boxes and white boxes indicate dark and light periods, respectively. The pink boxes denote the red light (637-nm) stimulation window. Genotypes of each group are labeled above the panel. Error bars correspond to SEM. n = 24 for each group. (B) Percentage of daytime sleep change during the LED day compared with the baseline day of E cell and gDN1-inhibited flies. Genotypes of each group are listed below the panel. n = 16 for Spl-gDN1 > UAS-GtACR1 and Spl-E cell > UAS-GtACR1 groups; n = 12 for three control groups. **P < 0.001, one-way ANOVA. Error bars represent SEM.
Given that the gDN1s are close to the dorsal surface, whereas the E cells are much more ventral (15, 28), successful activation of E cells indicates that sufficient light can penetrate into the brain to activate these cells. We also silenced gDN1s and E cells only in adults by expressing the inward rectifying K+ channel, UAS- Kir2.1 (29). Consistent with previous inhibition experiments (15), our data show that inhibition of gDN1s specifically reduces sleep and promotes locomotor activity during the middle of the day (Fig. 2A). Furthermore, suppression of E cell activity reduces evening locomotor activity and extends the siesta (Fig. 2A). We also observed similar behavioral results with optogenetic inhibition, by expressing the algal Guillardia theta anion channelrhodopsins (GtACRs) with strong DN1 and E cell drivers (30). The data indicate that the gDN1s are necessary and sufficient to inhibit activity and promote sleep, especially in the middle of the day. Moreover, the E cells are necessary for normal evening locomotor activity levels (Fig. 2B and Fig. S2B). This confirms our results from a previous study, which was less rigorous and used a less-specific driver (10). As silencing the four E cells predominantly affects the evening locomotor activity peak (see above), this is consistent with their firing at this time (see below).
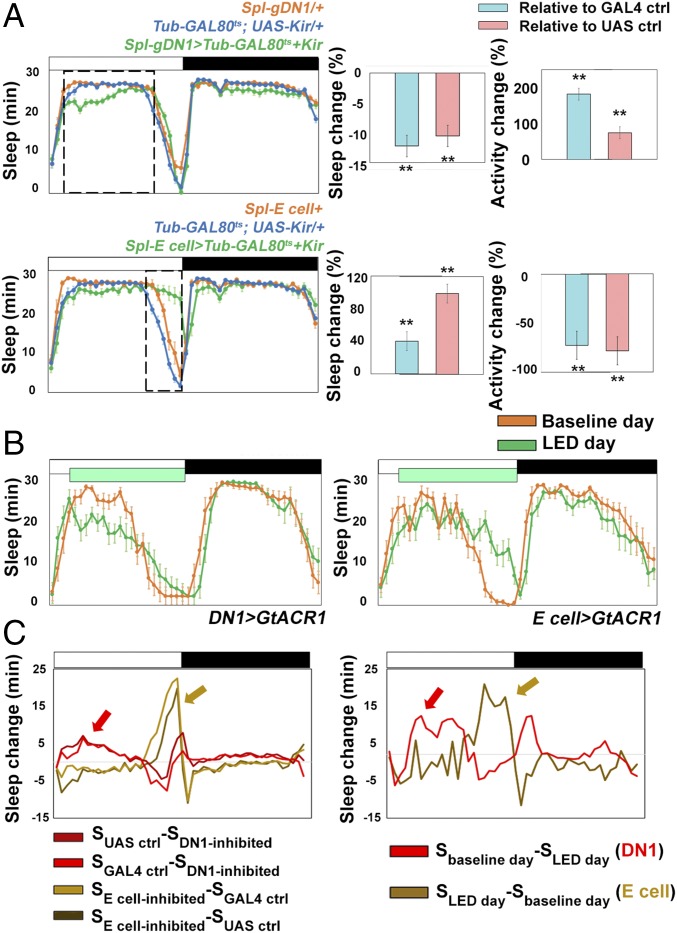
Fig. 2.
Effect of silencing E cells and gDN1s on sleep profiles. (A) Temperature-induced expression of Kir2.1 in different circadian neurons affects sleep and locomotor activity at different circadian times. (Upper) Sleep profiles of Spl-E cell > Tub-GAL80ts+UAS- Kir2.1 (green), Tub-GAL80ts/+; UAS- Kir2.1/+ (blue), and Spl-E cell/+ (orange) at 29 °C. (Lower) Sleep profiles of Spl-DN1 > Tub-GAL80ts+UAS- Kir2.1 (green), Tub-GAL80ts/+; UAS- Kir2.1/+ (blue), and Spl-DN1/+ (orange) at 29 °C. (Right) Quantification of relative sleep and activity change during the time window. The dashed box indicates the time window (ZT2-9 for Upper; ZT9-12 for Lower) within which the relative change of sleep and locomotor activity was calculated. n = 16 for each group. **P < 0.001, two-tailed Student’s t test. Error bars represent SEM. (B) Optogenetic inhibition of DN1s by expression of GtACR1 (driven by Clk4.1M-GAL4) reduces daytime sleep (Left), and optogenetic inhibition of E cells (driven by DvPDF-GAL4;PDF-GAL80) increases sleep during the evening (Right). The green box indicates the time window when the 530-nm green LEDs were turned on. n = 16 for each group. Error bars represent SEM. The quantification of experimental and control groups is shown in Fig. S2B. (C) Sleep change profiles of gDN1-inhibited flies and E cell-inhibited flies during LD. (Left) Profile calculated from the data in A. (Right) Profile calculated from the data in B. Red and brown arrows point to the putative peaks of DN1 and E cell neuronal activity in LD. The genotypes used to calculate the profiles are labeled below the panels.
To estimate the endogenous neuronal activity peak of gDN1s as well as E cells, we calculated the sleep-loss curve of gDN1-inhibited flies and the sleep-gain curve of E cell-inhibited flies during LD (Fig. 2C). Those curves predict that gDN1s fire during early daytime to promote sleep (red arrows in Fig. 2C). In contrast, E cells are predicted to have a specific activity peak during the evening to drive evening locomotor activity (brown arrows in Fig. 2C).
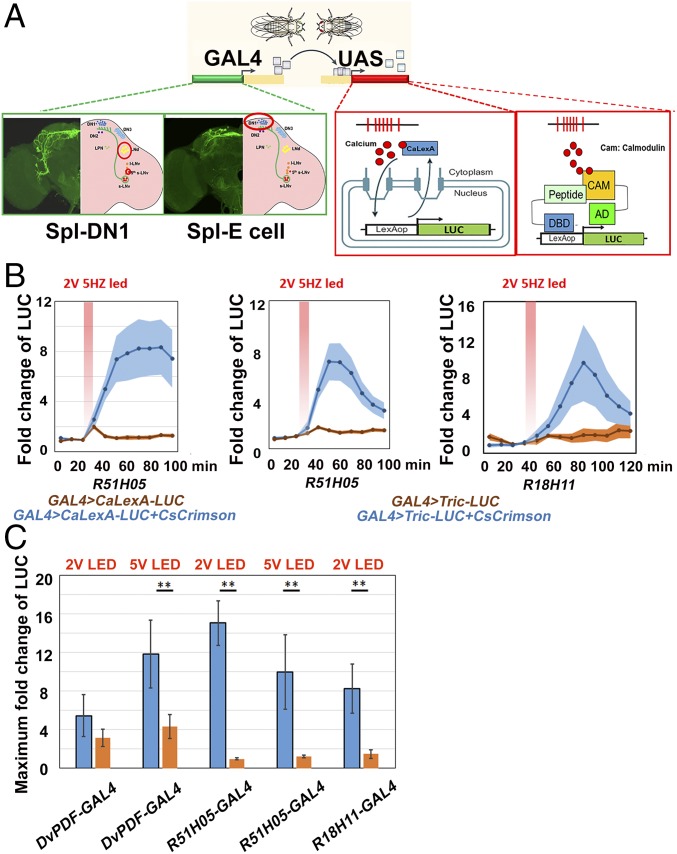
How do the experimentally determined calcium profiles correlate with the well-defined functional roles of gDN1s and E cells during the course of a normal LD cycle? To address this question, we turned to a strategy that we recently used, in vivo calcium monitoring with CaLexA-LUC in small numbers of discrete neurons in wake-behaving flies (15). Since calcium activity is an indicator of neuronal firing and nuclear entry of the transcription factor CaLexA is dependent on intracellular calcium levels, CaLexA is a surrogate reporter for neuronal activity. To assay in vivo calcium dynamics with an additional system, we developed the Tric-LUC reporter (Fig. 3A), which uses a calmodulin (CAM)–CAM-binding peptide interaction to achieve rapid calcium-dependent transcription activation (22).
Fig. 3.
Using in vivo calcium reporters to monitor temporal patterns within circadian pacemakers. (A) Schemes of CaLexA-LUC and Tric-LUC calcium reporters. (B) Comparing CaLexA-LUC and Tric-LUC responses to CsChrimson activation. The fold change of luminescence was calculated as the ratio of the luminescence level after CsChrimson activation to the baseline luminescence level. The red shaded box indicates the 2 V (0.59∼0.63 mW/mm2), 10-min, 5-Hz, 627-nm light pulse. The genotypes of each line are shown below. n = 8 for R18H11-GAL4 > Tric-LUC+CsCrimson; n = 15 for R18H11-GAL4 > Tric-LUC; n = 16 for other groups. Shading represents SEM. (C) Comparing the Tric-LUC response between different circadian neurons. The maximum fold changes of luminescence after optogenetic stimulation are plotted. The blue bars indicate GAL4 > Tric-LUC+CsCrimson, and the orange bars indicate GAL4 > Tric-LUC only. The GAL4 lines used in each group and the voltages of LED stimulation (2 V: 0.59∼0.63 mW/mm2; 5 V: 0.9∼1 mW/mm2) are shown below each histogram. n = 16 for each group. **P < 0.001, two-tailed Student’s t test. Error bars represent SEM.
To compare these two systems in freely moving flies, we expressed the transgenes along with CsChrimson with a previously characterized GAL4 driver line, R51H05, which is strongly expressed in the gDN1s (31, 32). The flies were exposed to a 10-min, 2 V (0.59∼0.63 mW/mm2), 5-Hz LED stimulation (Fig. 3B). There was a prominent increase in luciferase activity with both systems compared with the controls. This increase occurred rapidly and was evident by the first 10-min interval after the termination of LED exposure. There were important quantitative differences between the systems, however. Although CaLexA-LUC generated more luciferase activity than Tric-LUC, light stimulated the Tric-LUC system more rapidly. It reached half-maximal activity by approximately 20 min after termination of LED exposure, compared with 30 min or longer for CaLexA-LUC. Tric-LUC activity also decreased more rapidly and completely than CaLexA-LUC activity. The more dynamic nature of the Tric-LUC pattern compared with CaLexA-LUC is not unexpected, given the different way in which calcium regulates their activities (Discussion). We note that the Tric-LUC increase was slower with an additional gDN1 driver (R18H11) under the same conditions (Fig. 3B), which may reflect somewhat different driver strengths or cell-specificity between the two gDN1 drivers. Nonetheless, the more dynamic nature of the Tric-LUC caused us to favor it for comparisons of the different neurons across circadian time.
We next assayed the extent to which Tric-LUC responds to optogenetically induced firing in circadian neurons that lie deeper within the fly brain. These may be less amenable to Tric-LUC monitoring despite their activation with the red LED. Therefore, we coexpressed the CsChrimson and Tric-LUC reporters in the lateral circadian neurons with DvPDF-GAL4 and measured LUC activity after LED stimulation with different voltages. We compared the results with those obtained with drivers expressed in the gDN1s.
Although Tric-LUC expression in the lateral neurons responds to LED stimulation, 5 V (0.9∼1 mW/mm2) is required for a substantial response. In contrast, gDN1 expression of Tric-LUC requires only 2 V (0.59∼0.63 mW/mm2), which may even be superior to 5 V (Fig. 3C). It is uncertain whether this difference reflects more sensitive CsChrimson stimulation or better Tric-LUC monitoring from the more superficial dorsal neurons. Nonetheless, the data indicate that Tric-LUC is suitable for lateral neuron as well as dorsal neuron temporal monitoring of calcium. Therefore, we used Tric-LUC to compare the calcium/firing patterns between the sleep-promoting gDN1s and the activity-promoting E cells across a diurnal day under standard LD conditions.
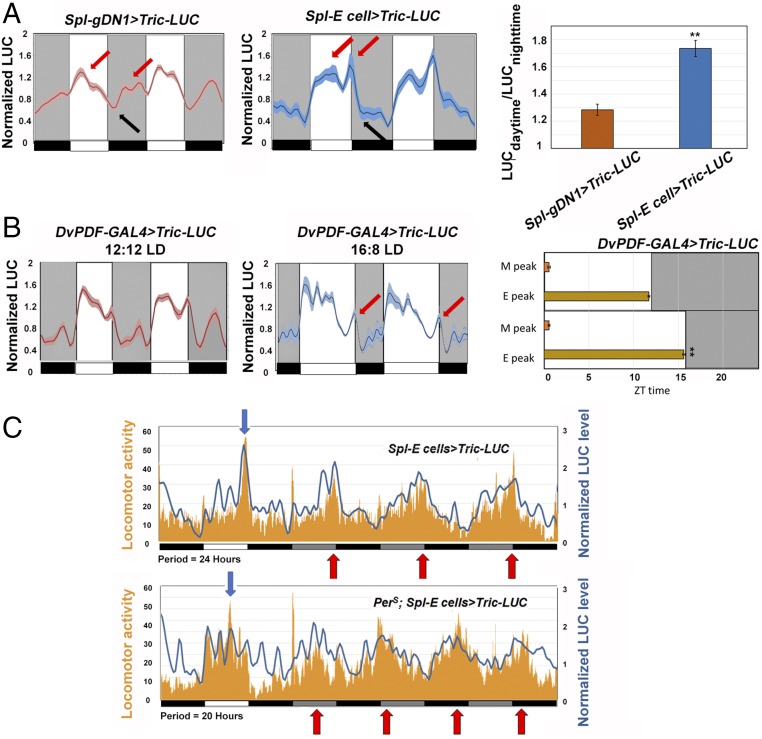
The gDN1 pattern exhibits a broad peak at about ZT0-4, shortly after lights-on, and a trough at ZT12-16, shortly after lights-off (Fig. 4A, Left; first red arrow and black arrow, respectively). A second peak occurs in the nighttime, which may just reflect the broad anticipation of the major peak (Fig. 4A, Left; second red arrow). Although this pattern does not precisely coincide with that expected for sleep- and siesta-promoting neurons (Discussion), it is very different from the E cell Tric-LUC pattern; it is high in the daytime, has a sharp peak coincident with lights-off, and then has a trough at nighttime (Fig. 4A, Middle; three arrows, left to right, respectively). This pattern is roughly consistent with the fly diurnal locomotor activity (Fig. 4A, Right; note the much higher daytime/nighttime LUC activity for E cells than for DN1s) and the activity-promoting role of the E cells (Discussion). Especially noteworthy is the prominent Tric-LUC peak centered at lights-off (Fig. 4A, Middle, second red arrow), which is coincident with the major evening locomotor activity peak. This relationship persists in constant darkness (see below).
Fig. 4.
Exploring the Tric-LUC patterns under different conditions. (A) The Tric-LUC pattern in the two circadian neuron groups. The normalized LUC levels of Spl-gDN1 > Tric-LUC (red curve, Left) and Spl-E cell > Tric-LUC (blue curve, Middle) from a 2-d recording are plotted. The light and dark periods are indicated by the white and gray backgrounds. The red arrows indicate peaks of LUC activity, and the black arrows point to the troughs of LUC activity. n = 24 for each group. Shading corresponds to SEM. (Right) The ratio of daytime LUC activity divided by nighttime LUC activity. **P < 0.001, two-tailed Student’s t test. Error bars represent SEM. (B) Environment input changes the calcium pattern of the circadian neurons. LUC activity from DvPDF-GAL4 > Tric-LUC flies were recorded in either 12:12 LD cycles (Left) or 16:8 LD cycles (Right). The red arrows point to the evening peak of DvPDF-GAL4 > Tric-LUC flies. n = 16 for each group. Shading corresponds to SEM. (Right) Peak times of the M and E LUC profiles in 12:12 LD (Upper) and 16:8 LD (Lower). **P < 0.001, two-tailed Student’s t test. Error bars correspond to SEM. (C) Real-time unfiltered raw data recordings of Spl-E-cell > Tric-LUC (Upper) and perS; Spl-E-cell > Tric-LUC flies (Lower). Mean LUC levels and locomotor activities from WT and perS groups are plotted. Tric-LUC levels (blue) and locomotor activities (orange) were recorded at the same time. The plate was placed into the TopCount NXT recording chamber every 30 min. The period of these flies is shown below the panel. Black boxes and white boxes indicate dark and light periods in LD, respectively. Gray boxes and black boxes indicate subjective day and night in DD, respectively. Red arrows point to the major locomotor activity peaks in DD, and the blue arrows point to the evening activity peaks in LD.
The gDN1 pattern is quite similar to that reported from a recent electrophysiological study that assayed firing rate and membrane potential temporal profiles of dissected brain DN1s (18). There are also recent GCaMP6 imaging studies of tethered flies (19, 20). These DN1 calcium patterns are 3- to 4-h advanced compared with the Tric-LUC and dissected brain patterns. In addition, these imaging studies assayed the E cell calcium patterns. Notably, the imaging E cell LD pattern is quite broad and missing a sharp peak coincident with lights-off (20), a feature of both the LD locomotor activity and the E cell Tric-LUC patterns (Fig. 4A, Middle).
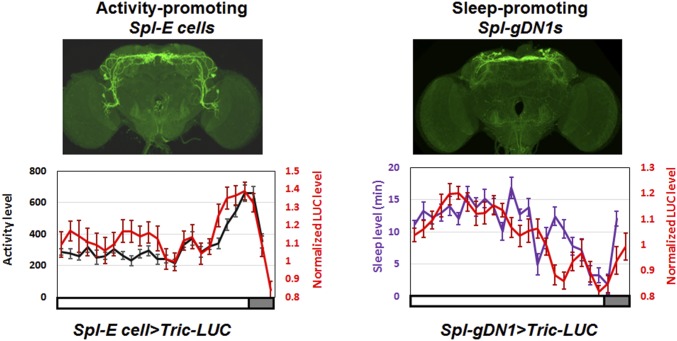
The close relationship between E cell Tric-LUC and locomotor activity is underscored by comparing the two patterns in the daytime at ZT0-12; they are nearly coincident (Fig. S3, Left). This is also the case for the gDN1 Tric-LUC and sleep profiles; they are both minimal at the end of the day, coincident with the E activity peak (Fig. S3, Right). Note that the locomotor activity and sleep profiles were measured by video monitoring at the same time and in the same flies as the Tric-LUC profiles.
Fig. S3.
Daytime activity/sleep patterns track calcium levels within the two different circadian neuron groups. The red curve represents the real-time Tric-LUC level monitored by the TopCount NXT plate reader at 30-min intervals, and the black and purple curves represent the activity and sleep levels, respectively, recorded simultaneously by a webcam in the 96-well plate format. (Upper) The GAL4 expression patterns used in the experiment. The white and gray boxes below the figures indicate dark and light periods, respectively. A full 12-h light period is shown, followed by the beginning of the dark period. n = 20 for the Spl-E cell > Tric-LUC group (Left); n = 16 for the Spl-gDN1 > Tric-LUC group (Right). Error bars represent SEM.
To further compare the E cell Tric-LUC patterns with E activity, we altered the environmental conditions or the fly genetic background. To express Tric-LUC in M as well as E cells, we used the DvPDF-GAL4 driver. Changing from LD 12:12 to LD 16:8 (summer time) shifted the evening Tric-LUC peak away from the main diurnal activity profile, but this evening Tric-LUC peak still remained coincident with lights-off and the evening peak of locomotor activity; the morning Tric-LUC peak, which appears after lights-on, was not shifted (Fig. 4B).
We assayed for an effect of genotype as well as light in the short period perS mutant strain and a control wild-type strain (Fig. 4C). As expected, perS flies have an advanced evening locomotor activity peak in LD compared with WT flies (Fig. 4C; blue arrows). Although these unfiltered raw data patterns are noisier and have some calcium peaks without obvious locomotor activity (especially the perS mutant strain, for unknown reasons), LD locomotor activity roughly corresponds to increased Tric-LUC activity (Fig. 4C, blue arrows), and perS affects both patterns. We also compared the E cell Tric-LUC activity and behavioral patterns from these grouped flies by extending the monitoring into DD (constant darkness; the red arrows in Fig. 4C indicate the major DD locomotor activity peaks.) The two profiles correlate well, especially for the WT strain, which suggests that these E cells are also a general source of locomotor activity drive in DD. This idea is consistent with previous work (10, 17). Taken together with E cell activation and inhibition experiments (10) (Figs. 1C and 2 A and B), these environmental and genetic perturbations further suggest that the E cell Tric-LUC evening peak reflects E cell firing.
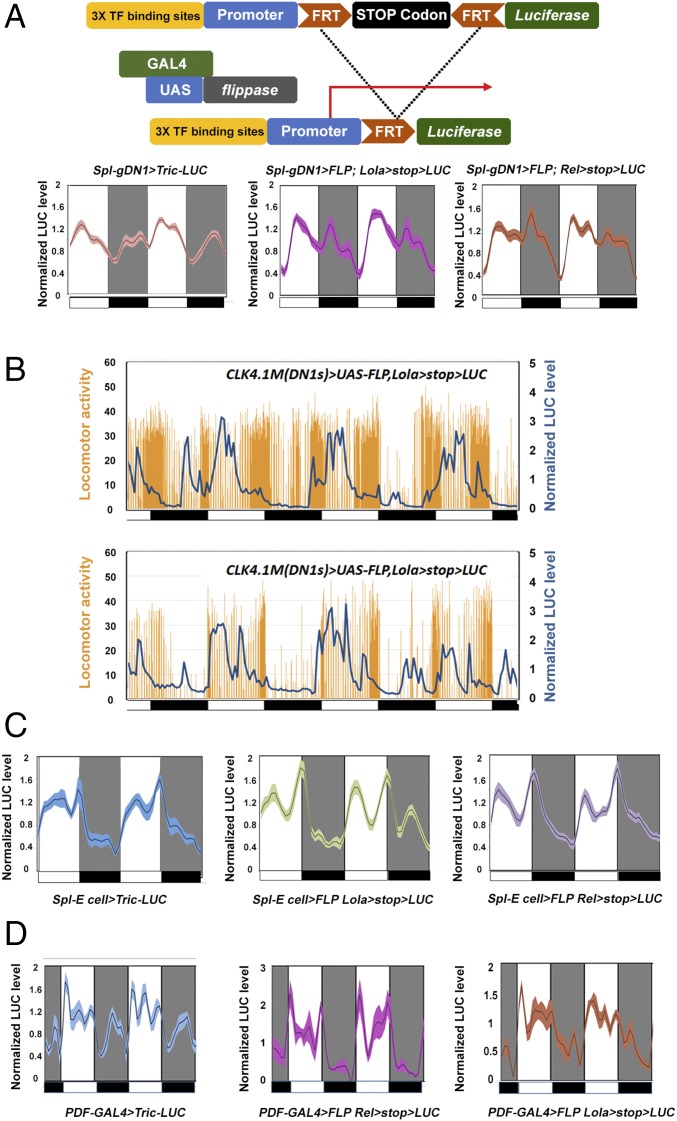
To complement the Tric-LUC monitoring strategy, we turned to recently identified luciferase reporters based on a different principle (Fig. 5A, Upper). These reporters contain multimerized transcription factor motifs, which were identified in a screen for Drosophila activity-regulated genes (ARGs) (23, 33). Of the three reporters that express and are activated by firing within generic neurons (Elav-GAL4), one (Eip78c) gave no signal with the circadian neuron drivers, but the two others were positive. These reporters, lola and relish, gave rise to essentially identical profiles in all cases.
Fig. 5.
Comparing neuronal activity reporters and calcium reporters within the circadian neurons. (A) The neuronal activity reporters have a different pattern than Tric-LUC in the gDN1s. (Upper) Schematic of the neuronal activity reporters. (Lower) Comparison of the Tric-LUC reporter and the Relish-LUC as well as Lola-LUC reporters in gDN1s. Light and dark periods are indicated by white and gray backgrounds, respectively. n = 24 for the Spl-gDN1 > Tric-LUC group; n = 16 for the other groups. Shading correspond to SEM. (B) Real-time recording of gDN1 LUC and locomotor activity of individual flies. The comparison shows that DN1 LUC activity is negatively associated with fly daytime activity level. The locomotor activity (orange histogram) and the LUC activity (blue curve) of individual CLK4.1m(DN1s) > UAS-FLP,Lola > stop > LUC flies were recorded simultaneously for 4 d in LD. (Upper) Recordings from individual flies with relatively high locomotor activity levels. (Lower) Recordings from individual flies with relatively low locomotor activity levels. Black boxes and white boxes indicate dark and light periods, respectively. (C) The neuronal activity reporter patterns are indistinguishable from the Tric-LUC pattern in E cells. The panels show normalized LUC activity from Spl-E cell > UAS-Tric-LUC, Spl-E cell > UAS-FLP Lola > stop > LUC, and Spl-E cell > UAS-FLP Relish > stop > LUC groups. Light and dark periods are indicated by white and gray backgrounds, respectively. n = 24 for the Spl-E cell > Tric-LUC group; n = 16 for the other groups. Shading corresponds to SEM. (D) The neuronal activity reporter patterns are also indistinguishable from the Tric-LUC pattern in the PDF neurons. The panels show normalized LUC activity from PDF-GAL4 > UAS-Tric-LUC, PDF-GAL4 > UAS-FLP Lola > stop > LUC, and PDF-GAL4 > UAS-FLP Relish > stop > LUC groups. Light and dark periods are indicated by the white and gray backgrounds, respectively. n = 16 for each group. Shading corresponds to SEM.
With the gDN1 driver, the ARG profiles also had a trough shortly after lights-off, but differed somewhat from the Tric-LUC profiles during the night (Fig. 5A, Lower). This suggests a modest difference in expression patterns within the gDN1s or perhaps different monitoring principles, e.g., calcium vs. firing (Discussion). Interestingly, DN1 reporter activity precisely tracks siesta onset immediately after the morning locomotor activity bout, which is best observed in different individual flies with different locomotor activity/siesta levels; reporter activity has fully decreased by the time that evening locomotor activity begins to appear (Fig. 5B). These profiles are fully consistent with the daytime sleep-promoting role of DN1s (Figs. 1B and 2 A and B). Notably, the E cell profiles of the two positive ARG reporters are essentially identical to the Tric-LUC profiles, making calcium monitoring likely for the ARG reporters (Fig. 5C; although see Discussion).
The quite complex ARG profiles of PDF-expressing circadian neurons are also nearly identical to their Tric-LUC profiles (Fig. 5D). We do not know the significance or functional correlates of these multiple luciferase peaks. One possible source of complexity is the different subgroups of PDF-expressing cells, such as the small and large LNvs (19), and another is ectopic expression. However, no comparable complexity is apparent in the tethered fly GCaMP6 imaging of PDF cells (19).
Discussion
To assay the calcium profiles of circadian neurons in wake-behaving flies, we expressed the Tric-LUC calcium reporter system as well as recently developed transcriptional reporters in different groups of circadian neurons. Notably, the major evening peak of locomotor activity always corresponds to a peak in the E cell Tric-LUC profile. As E cell neuronal activity is necessary and sufficient for evening locomotor activity, we suggest that this profile reflects, at least in part, E cell firing under wake-behaving conditions. A similar argument applies to the gDN1 profile, which has a peak that coincides with the major daytime sleep episode. The luciferase patterns from these two systems are similar but not precisely identical to previous GCaMP6 imaging results of circadian neurons in tethered flies and to electrophysiological recordings of circadian neurons in dissected brains.
We note that different neurons may have different Tric-LUC sensitivity. For example, they manifest different responses to 2 V (0.59∼0.63 mW/mm2) vs. 5 V (0.9∼1 mW/mm2) stimulation of CsChrimson (Fig. 3C). Although this could reflect light penetration (the E cells are deeper in the brain than the gDN1s), it could also reflect differences in the intrinsic properties of different circadian neurons, for example, a depolarization block in E cells. A differential response to firing strength or frequency would also explain why the gDN1s might respond differently to optogenetic stimulation vs. temperature stimulation with dTrpA1 (31); for example, this could reflect a differential release of neurotransmitters and neuropeptides in response to tonic vs. bursting stimulation. These considerations also raise the possibility that different neurons might respond differently to different modes of firing, or that the same neuron might even respond differently at different circadian times. In any case, it is likely that these LUC reporters are largely insensitive to important differences in individual firing patterns. More generally, luciferase profiling should be viewed as a strategy that is complementary to calcium imaging and electrophysiological recording and is especially relevant to biological problems that do not require millisecond resolution.
Nonetheless, the broad LUC patterns of gDN1s and E cells conform roughly to what one imagines their firing might be based on their known functional outputs. The gDN1s have a peak in the daytime and a second peak or shoulder at night, roughly corresponding to the daytime siesta and nighttime sleep (Fig. 4A). Inspection of individual flies strengthens the association of the DN1 pattern with daytime sleep, although the nighttime shoulder is less consistently present in the noisier individual fly records (Fig. 5B). The E cell pattern is different and notably diurnal, with a peak in the daytime and a trough at night (Fig. 4A). As noted above, this E cell daytime pattern is bifurcated; the second peak coincides with evening locomotor activity, and they shift together in response to environmental or genetic perturbations (Fig. 4 B and C). Inspection of individual fly records reinforces the association of E cell calcium with the evening activity peak in LD (Fig. 4C) and with overall locomotor activity in DD (Fig. S3). It would be interesting to assay the E cell pattern at 29 °C, which causes a more nocturnal activity pattern (34). Would the E cell and gDN1 Tric-LUC patterns also change?
The enhanced temporal resolution of Tric-LUC relative to CaLexA-LUC is necessary to visualize a discrete E cell LUC evening peak. This enhanced resolution is evident from three features of the LUC profiles after the pulse of optogenetic stimulation (Fig. 3A): Tric-LUC has lower activity levels, a more rapid increase, and then a more rapid decline from peak values. All of these features reflect more rapid inactivation relative to the more stable CaLexA. This makes sense from the reporter designs; Tric-mediated transcription should immediately decline along with calcium, whereas CaLexA incorporates no specific feature to decrease activity as calcium levels fall. On the other hand, one can imagine that the nuclear entry of CaLexA-LUC might be more sensitive to cytoplasmic calcium levels than Tric-LUC. This might be because the CaLexA design is based on calcineurin, which might require lower Ca2+ levels than the calmodulin design features of TRIC. Calcium imaging as well as CaLexA-LUC presumably measures mostly cytoplasmic calcium, which might more accurately reflect synaptic events than nuclear calcium. In this context, it is a somewhat mysterious how and why Tric-LUC and the transcriptional reporters appear to track neuronal activity so well. Perhaps nuclear and cytoplasmic calcium levels equalize on a time scale much faster than firing rate changes of circadian and sleep assays (hours). In addition, the close correspondence between transcriptional reporters and Tric-LUC suggests that the two positive transcriptional reporters may also be assaying calcium levels.
Nonetheless, a couple of interesting features emerge from the comparison of the two positive transcriptional reporters and the Tric-LUC reporter. (The negative reporter Eip78c may simply reflect little or no expression of that transcription factor in circadian neurons.) The somewhat different profiles of the transcriptional reporters and Tric-LUC in the gDN1s may indicate that the transcriptional reporters do not just assay calcium but also respond to a particular mode of firing, for example, bursting or tonic. That mode would differ from calcium levels in the gDN1s, but not in the E cells or PDF cells. This suggests that different transcriptional reporters could be screened for their responsiveness to different firing regimes and then used to explore those firing possibilities in wake-behaving flies. It also suggests that at least part of the ARG reporter profile reflects features of firing that differ from calcium, an argument that then impacts interpretation of the nearly identical ARG and E cell Tric-LUC profiles.
The correspondence between only some LUC peaks and behavior (e.g., Figs. 4C and 5B) suggests that the profiles may reflect firing at some times but calcium dynamics without firing at other times. Alternatively, the entire pattern may reflect firing changes but have no behavioral consequences at some times for any number of reasons, for example, because other neurons are firing at the same time with an opposite, canceling effect on behavior.
The E cell Tric-LUC evening peak occurs when the gDN1 Tric-LUC pattern is at a trough (black arrow in Fig. 4A). This might reflect low gDN1 sleep-promoting drive at this time, which would help the strong E cell firing and locomotor activity drive at the same time; however, the two patterns do not completely conform to predictions from previous results. For example, the gDN1s have been shown to promote sleep by inhibiting E cell activity, so we predicted that a peak in the DN1 Tric-LUC pattern would anticipate a trough in the E cell pattern; no such relationship between the two patterns is apparent. On the contrary, the DN1 peak appears to follow the E cell peak, both in the daytime and at night. As discussed previously (15), this discrepancy might reflect the fact that other circadian features—cycling transcriptional events, for example—might determine the timing of the peak inhibitory drive from the gDN1s. The discrepancy might also reflect some gDN1 heterogeneity, for example, in temporal firing patterns and/or calcium levels. gDN1 heterogeneity might also impact the differences between the gDN1 Tric-LUC pattern and the transcriptional reporter patterns; for example, DN1s may include activity-promoting subgroups as well as the sleep-promoting gDN1s (35). We see no similar differences between the E cell patterns or even the more complex PDF cell patterns between the two reporter systems, suggesting that the gDN1s may indeed be more heterogeneous than other circadian groups.
In any case, Tric-LUC and the transcriptional reporters provide informative calcium patterns from small discrete brain regions. The imaging assay of Greenspan et al. (36) achieves similar ends and may even have higher temporal resolution; however, that assay was limited to neurons close to the dorsal surface of the brain and also required a quite sophisticated mirror and imaging apparatus. Tric-LUC, in contrast, is relatively low tech and ideal for circadian, sleep, and other behavioral studies, where extremely rapid temporal resolution is probably unnecessary. Importantly, our approach works in different behavioral assays, some of which might not be amenable to imaging.
It is impossible to know whether GCaMP6 imaging is measuring neuronal firing or residual calcium rhythms. This consideration is particularly relevant to cells that are known to express molecular rhythms, and underscores the importance of knowing the function of these neurons as well as simultaneously profiling behavior. Our strategy of combining luciferase reporters with FlyBox video recording fulfills these requirements and is especially useful for researchers focused on broader patterns like those relevant to circadian and sleep-wake profiles.
Methods and Materials
Fly Strains and Rearing Conditions.
DvPDF-GAL4 and PDF-GAL4 was provided by J. H. Park, Clk4.1M-GAL4 was provided by Paul Hardin, UAS-CaLexA was provided by Jing Wang, UAS-GtACR1 was provided by Adam Claridge-Chang, Spl-DN1 (JRC_SS00781) and Spl-E cell (JRC_MB122B) GAL4s were provided by Gerald M. Rubin. LexAop-LUCattp40 was generated by Xiaojing Gao and Liqun Luo. The following lines were ordered from the Bloomington Stock Center: Pdfr (R18H11)-GAL4 (48832), Vglut (R51H05)-GAL4 (41275), UAS-CsChrimson (55136), UAS-flp (55806), UAS-Tric (62830), UAS-Kir2.1 (6595), and Tub -GAL80ts (7108). Flies were reared on standard cornmeal/agar medium supplemented with yeast. The adult flies were entrained in 12:12 LD cycles at 25 °C. The flies carrying Tub -GAL80ts and UAS-Kir2.1 were maintained at 21 °C.
Feeding of Retinal.
All trans-Retinal (Sigma-Aldrich) was dissolved in EtOH as a 40 mM stock solution. For CsChrimson and GtACRs experiments (Figs. 1, 2B, and 3), flies were transferred to 96-well plates loaded with 300 μL of food (5% sucrose and 2% agar) containing 400 μM all trans-Retinal (Sigma-Aldrich) for at least 3 d before any optogenetic experiments.
Setup of Optogenetics, Video Recording System, and Luciferase Assays.
The behavioral setup for the optogenetics and video recording system has been described previously (15). In brief, flies were loaded into white 96-well Microfluor 2 plates (Fisher) containing 5% sucrose and 2% agar food with or without 400 μM ATR as described above. We used 0.08∼0.1 mW/mm2 627-nm red- light pulses or 530-nm green-light pulses from eight LEDs to irradiate flies expressing CsChrimson or GtACRs within the DN1s and E cells. The LEDs were controlled by an Arduino board to modulate the voltage and frequency. Different LED intensities and frequencies were used (Fig. 3). Fly behavior was recorded by a web camera (Logistic C910) without an IR filter. The movement of flies was calculated with Pysolo software and transformed into a MATLAB (MathWorks)-readable file. Five pixels/s (50% of the full body length) was defined as a minimum movement threshold. The activity and sleep analyses were performed with a signal-processing toolbox implemented in MATLAB as described above.
For in vivo luciferase assays, flies were loaded onto white 96-well Microfluor 2 plates (Thermo Fisher Scientific). They were loaded with 5% sucrose and 2% agar food as described above but also containing 20 mM d-luciferin potassium salt (Gold Biotechnology) (15). Plates were loaded into the stacker of a TopCount NXT luminescence counter (PerkinElmer). Assays were carried out in an incubator under LD conditions. Luminescence counts were collected for 3–5 d in LD or DD at 25 °C. A nine plate mode in the stacker was used (Figs. 4 A and B and 5 A–C). The reading cycle is approximately 1 h for nine plates. For optogenetic stimulation with the luciferase assays (Fig. 3 B and C), 627-nm LEDs mounted to a pair of heat sinks were positioned symmetrically in the chamber to ensure uniform illumination of one 96-well plate. CsChrimson stimulation was provided with 0.59∼0.63 mW/mm2 (2 V) and 0.9∼1 mW/mm2 (5 V) (Fig. 3 B and C). The intensity of light varies with the distance of wells on the plate to the LED. Flies prefed with ATR for 2 d were loaded onto a plate. Single plates were kept in the LED chamber for 8 min and then automatically transferred to the TopCount NXT counter for a 2-min luminescence reading. To assay fly locomotor activity and the Tric-LUC or transcriptional luciferase reporters at the same time (Figs. 4C and 5B), a single plate was recorded with a web camera (Logistic C910) attached to the top of chamber. During each 30 min period, the plate sat in the video chamber for 28 min and then was automatically transferred to the TopCount NXT for a 2-min luminescence reading. The raw luminescence data were filtered, detrended, and normalized by MATLAB code (37) and plotted in Microsoft Excel. All experiments were repeated at least three times.
Locomotor and Sleep Experiments and Statistical Analysis.
Locomotor activity of individual male flies (aged 3–7 d) was measured with a video recording system under 12:12 LD conditions (15). The activity and sleep analysis was performed with pysolo (38) and a signal-processing toolbox implemented in MATLAB. Group activity was also generated and analyzed with MATLAB.
All statistical analyses were conducted using IBM SPSS software. The Wilks–Shapiro test was used to determine data normality. Normally distributed data were analyzed with the two-tailed unpaired Student t test and one-way ANOVA, followed by the Tukey–Kramer honest significant difference test as the post hoc test or two-way ANOVA with post hoc Bonferroni multiple comparisons. Data are presented as mean behavioral responses, and error bars represent the SEM. Differences between groups were considered significant at P < 0.05.
Fly Brain Immunocytochemistry.
Immunostaining was performed as described previously (39). Fly heads were removed and fixed in PBS with 4% paraformaldehyde and 0.008% Triton X-100 for 45–50 min at 4 °C. Fixed heads were washed in PBS with 0.5% Triton X-100 and then dissected in PBS. The brains were blocked in 10% goat serum (Jackson Immunoresearch) and then incubated with primary antibodies at 4 °C overnight or longer. For GFP, PDF, and PER costaining, chicken anti-GFP antibody (ab13970; 1:1,000; Abcam), mouse anti-PDF antibody (1:1,000; Developmental Studies Hybridoma Bank), and rabbit anti-PER antibody (1:200) were used as primary antibodies. After three washes with 0.5% PBST, the brains were incubated with Alexa Fluor 488-conjugated anti-chicken, Alexa Fluor 555-conjugated anti-rabbit, and Alexa Fluor 635-conjugated anti-mouse (Molecular Probes) at 1:500 dilution. The brains were washed three more times, followed by mounting in Vectashield Mounting Medium (Vector Laboratories) and viewed sequentially in 1.1-μm sections on a Leica SP5 confocal microscope. For comparison of the fluorescence signals from different conditions, the laser intensity and other settings were set at the same level during each experiment. Fluorescence signals were quantified by ImageJ as described previously.
Acknowledgments
We thank Meghana Holla, Nhi Nguyen, Hyung Jae Jung, Manuel J. Diaz, and Madelen Diaz for generous help; Dr. Katharine C. Abruzzi, Dr. Matthias Schlichting, Dr. Orie Shafer, and Dr. Leslie Griffith for helpful discussions and comments on early versions of this manuscript; and Dr. Liqun Luo, Dr. Adam Claridge-Chang, and Dr. Gerald Rubin for fly stocks. We are particularly indebted to Dr. Rubin and the Howard Hughes Medical Institute’s Janelia Research Campus for prepublication access to the unpublished Spl-DN1 (JRC_SS00781) and Spl-E cell (JRC_MB122B) GAL4 strains. More detailed characterization of these lines will be presented in Dionne, Rubin, and Nern (manuscript in preparation).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1706608114/-/DCSupplemental.
References
- 1.Allada R, Siegel JM. Unearthing the phylogenetic roots of sleep. Curr Biol. 2008;18:R670–R679. doi: 10.1016/j.cub.2008.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Campbell SS, Tobler I. Animal sleep: A review of sleep duration across phylogeny. Neurosci Biobehav Rev. 1984;8:269–300. doi: 10.1016/0149-7634(84)90054-x. [DOI] [PubMed] [Google Scholar]
- 3.Helfrich-Förster C. Differential control of morning and evening components in the activity rhythm of Drosophila melanogaster sex-specific differences suggest a different quality of activity. J Biol Rhythms. 2000;15:135–154. doi: 10.1177/074873040001500208. [DOI] [PubMed] [Google Scholar]
- 4.Grima B, Chélot E, Xia R, Rouyer F. Morning and evening peaks of activity rely on different clock neurons of the Drosophila brain. Nature. 2004;431:869–873. doi: 10.1038/nature02935. [DOI] [PubMed] [Google Scholar]
- 5.Stoleru D, Peng Y, Agosto J, Rosbash M. Coupled oscillators control morning and evening locomotor behaviour of Drosophila. Nature. 2004;431:862–868. doi: 10.1038/nature02926. [DOI] [PubMed] [Google Scholar]
- 6.Tataroglu O, Emery P. Studying circadian rhythms in Drosophila melanogaster. Methods. 2014;68:140–150. doi: 10.1016/j.ymeth.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Renn SC, Park JH, Rosbash M, Hall JC, Taghert PH. A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell. 1999;99:791–802. doi: 10.1016/s0092-8674(00)81676-1. [DOI] [PubMed] [Google Scholar]
- 8.Lear BC, Zhang L, Allada R. The neuropeptide PDF acts directly on evening pacemaker neurons to regulate multiple features of circadian behavior. PLoS Biol. 2009;7:e1000154. doi: 10.1371/journal.pbio.1000154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yao Z, Shafer OT. The Drosophila circadian clock is a variably coupled network of multiple peptidergic units. Science. 2014;343:1516–1520. doi: 10.1126/science.1251285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo F, Cerullo I, Chen X, Rosbash M. PDF neuron firing phase-shifts key circadian activity neurons in Drosophila. Elife. 2014;3:02780. doi: 10.7554/eLife.02780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y, Liu Y, Bilodeau-Wentworth D, Hardin PE, Emery P. Light and temperature control the contribution of specific DN1 neurons to Drosophila circadian behavior. Curr Biol. 2010;20:600–605. doi: 10.1016/j.cub.2010.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang L, et al. DN1(p) circadian neurons coordinate acute light and PDF inputs to produce robust daily behavior in Drosophila. Curr Biol. 2010;20:591–599. doi: 10.1016/j.cub.2010.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cavanaugh DJ, et al. Identification of a circadian output circuit for rest:activity rhythms in Drosophila. Cell. 2014;157:689–701. doi: 10.1016/j.cell.2014.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Im SH, Li W, Taghert PH. PDFR and CRY signaling converge in a subset of clock neurons to modulate the amplitude and phase of circadian behavior in Drosophila. PLoS One. 2011;6:e18974. doi: 10.1371/journal.pone.0018974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo F, et al. Circadian neuron feedback controls the Drosophila sleep–activity profile. Nature. 2016;536:292–297. doi: 10.1038/nature19097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yoshii T, Vanin S, Costa R, Helfrich-Förster C. Synergic entrainment of Drosophila’s circadian clock by light and temperature. J Biol Rhythms. 2009;24:452–464. doi: 10.1177/0748730409348551. [DOI] [PubMed] [Google Scholar]
- 17.Roberts L, et al. Light evokes rapid circadian network oscillator desynchrony followed by gradual phase retuning of synchrony. Curr Biol. 2015;25:858–867. doi: 10.1016/j.cub.2015.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flourakis M, et al. A conserved bicycle model for circadian clock control of membrane excitability. Cell. 2015;162:836–848. doi: 10.1016/j.cell.2015.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liang X, Holy TE, Taghert PH. Synchronous Drosophila circadian pacemakers display nonsynchronous Ca2+ rhythms in vivo. Science. 2016;351:976–981. doi: 10.1126/science.aad3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liang X, Holy TE, Taghert PH. A series of suppressive signals within the Drosophila circadian neural circuit generates sequential daily outputs. Neuron. 2017;94:1173–1189.e4. doi: 10.1016/j.neuron.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Masuyama K, Zhang Y, Rao Y, Wang JW. Mapping neural circuits with activity-dependent nuclear import of a transcription factor. J Neurogenet. 2012;26:89–102. doi: 10.3109/01677063.2011.642910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao XJ, et al. A transcriptional reporter of intracellular Ca(2+) in Drosophila. Nat Neurosci. 2015;18:917–925. doi: 10.1038/nn.4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen X, Rahman R, Guo F, Rosbash M. Genome-wide identification of neuronal activity-regulated genes in Drosophila. Elife. 2016;5:e19942. doi: 10.7554/eLife.19942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pfeiffer BD, et al. Refinement of tools for targeted gene expression in Drosophila. Genetics. 2010;186:735–755. doi: 10.1534/genetics.110.119917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Inagaki HK, et al. Optogenetic control of Drosophila using a red-shifted channelrhodopsin reveals experience-dependent influences on courtship. Nat Methods. 2014;11:325–332. doi: 10.1038/nmeth.2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klapoetke NC, et al. Independent optical excitation of distinct neural populations. Nat Methods. 2014;11:338–346. doi: 10.1038/nmeth.2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pfeiffer BD, et al. Tools for neuroanatomy and neurogenetics in Drosophila. Proc Natl Acad Sci USA. 2008;105:9715–9720. doi: 10.1073/pnas.0803697105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Helfrich-Förster C, et al. Development and morphology of the clock-gene-expressing lateral neurons of Drosophila melanogaster. J Comp Neurol. 2007;500:47–70. doi: 10.1002/cne.21146. [DOI] [PubMed] [Google Scholar]
- 29.Johns DC, Marx R, Mains RE, O’Rourke B, Marbán E. Inducible genetic suppression of neuronal excitability. J Neurosci. 1999;19:1691–1697. doi: 10.1523/JNEUROSCI.19-05-01691.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mohammad F, et al. Optogenetic inhibition of behavior with anion channelrhodopsins. Nat Methods. 2017;14:271–274. doi: 10.1038/nmeth.4148. [DOI] [PubMed] [Google Scholar]
- 31.Kunst M, et al. Calcitonin gene-related peptide neurons mediate sleep-specific circadian output in Drosophila. Curr Biol. 2014;24:2652–2664. doi: 10.1016/j.cub.2014.09.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jenett A, et al. A GAL4-driver line resource for Drosophila neurobiology. Cell Rep. 2012;2:991–1001. doi: 10.1016/j.celrep.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tanenhaus AK, Zhang J, Yin JC. In vivo circadian oscillation of dCREB2 and NF-κB activity in the Drosophila nervous system. PLoS One. 2012;7:e45130. doi: 10.1371/journal.pone.0045130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parisky KM, Agosto Rivera JL, Donelson NC, Kotecha S, Griffith LC. Reorganization of sleep by temperature in Drosophila requires light, the homeostat, and the circadian clock. Curr Biol. 2016;26:882–892. doi: 10.1016/j.cub.2016.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seluzicki A, et al. Dual PDF signaling pathways reset clocks via TIMELESS and acutely excite target neurons to control circadian behavior. PLoS Biol. 2014;12:e1001810. doi: 10.1371/journal.pbio.1001810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grover D, Katsuki T, Greenspan RJ. Flyception: Imaging brain activity in freely walking fruit flies. Nat Methods. 2016;13:569–572. doi: 10.1038/nmeth.3866. [DOI] [PubMed] [Google Scholar]
- 37.Levine JD, Funes P, Dowse HB, Hall JC. Signal analysis of behavioral and molecular cycles. BMC Neurosci. 2002;3:1. doi: 10.1186/1471-2202-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gilestro GF, Cirelli C. pySolo: A complete suite for sleep analysis in Drosophila. Bioinformatics. 2009;25:1466–1467. doi: 10.1093/bioinformatics/btp237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang CH, Hinteregger E, Shang Y, Rosbash M. Light-mediated TIM degradation within Drosophila pacemaker neurons (s-LNvs) is neither necessary nor sufficient for delay zone phase shifts. Neuron. 2010;66:378–385. doi: 10.1016/j.neuron.2010.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]