Significance
The Drosophila circadian clock controls daily rhythms in physiology, metabolism, and behavior via ∼24-h transcriptional feedback loops. CLOCK requires its heterodimeric partner CYCLE to initiate clock function in canonical groups of brain neurons and peripheral tissues, but Clock expression can also induce clocks in ectopic locations. Here, we show that CLOCK stabilizes CYCLE in canonical and ectopic clock cells, where CYCLE is normally rapidly degraded, and that ectopic clocks also require CRYPTOCHROME. This work defines the genetic architecture and molecular mechanisms required for clock initiation that are likely conserved in other eukaryotes and suggests that ectopic clocks can be generated by Clock, cycle, and cryptochrome expression in naive cells.
Keywords: circadian clock, Drosophila, protein stability, CYCLE, cryptochrome
Abstract
The Drosophila circadian clock keeps time via transcriptional feedback loops. These feedback loops are initiated by CLOCK-CYCLE (CLK-CYC) heterodimers, which activate transcription of genes encoding the feedback repressors PERIOD and TIMELESS. Circadian clocks normally operate in ∼150 brain pacemaker neurons and in many peripheral tissues in the head and body, but can also be induced by expressing CLK in nonclock cells. These ectopic clocks also require cyc, yet CYC expression is restricted to canonical clock cells despite evidence that cyc mRNA is widely expressed. Here we show that CLK binds to and stabilizes CYC in cell culture and in nonclock cells in vivo. Ectopic clocks also require the blue light photoreceptor CRYPTOCHROME (CRY), which is required for both light entrainment and clock function in peripheral tissues. These experiments define the genetic architecture required to initiate circadian clock function in Drosophila, reveal mechanisms governing circadian activator stability that are conserved in perhaps all eukaryotes, and suggest that Clk, cyc, and cry expression is sufficient to drive clock expression in naive cells.
Circadian clocks drive daily rhythms in metabolism, physiology, and behavior in a wide array of organisms. The identification of “clock genes” in Drosophila revealed that the circadian timekeeping mechanism is based on transcriptional feedback loops (1), which are used to keep time in most, if not all, eukaryotes. Despite this mechanistic conservation, the core components of animal, plant, and fungal feedback loops differ (2). In the Drosophila feedback loop, CLOCK-CYCLE (CLK-CYC) heterodimers activate period (per) and timeless (tim) transcription, PER-TIM complexes feed back to repress CLK-CYC transcription, and degradation of PER-TIM complexes release CLK-CYC to initiate the next cycle of transcription (1). These feedback loops operate in only ∼150 brain neurons and many, but not all, peripheral tissues in adults (reviewed in refs. 3 and 4).
Because CLK-CYC initiates clock function as a differentiated feature of most, if not all, brain pacemaker neurons that control activity rhythms (5), activation of these two genes is thought to determine which cells and tissues will contain circadian clocks. The activation of Clk has been well documented in brain pacemaker neurons (5, 6), but comparatively little is known about cyc expression. We recently showed that a fully functional GFP-cyc transgene expresses GFP-CYC protein exclusively in circadian pacemaker neurons (5), suggesting that CYC expression is limited to clock cells. However, the lack of enrichment of cyc mRNA in brain pacemaker neurons suggests that cyc is broadly expressed (7).
During fly development Clk is activated in all cells that will ultimately contain circadian clocks, but expressing Clk in cells that normally lack clock function can generate ectopic clocks (8). Like canonical clock cells, these ectopic clocks require cyc and show robust rhythms in per and tim mRNA and protein cycling in light-dark (LD) cycles that dampen in constant darkness (DD) (8, 9). This result is consistent with the possibility that cyc mRNA is broadly expressed, yet CYC is detected only in canonical clock cells (5). These observations suggest that Clk is required for CYC expression to initiate clock function, but how Clk promotes CYC accumulation and whether these clock components are sufficient to initiate clock function is not known.
Here we show that Clk controls CYC accumulation by stabilizing CYC in cultured Drosophila Schneider 2 (S2) cells. Likewise, CYC accumulates specifically in ectopic cells expressing Clk, indicating that CLK also stabilizes CYC in vivo. CLK and CYC, however, are not sufficient for ectopic clock function; cry is also required to entrain and/or maintain these clocks. This work reveals genes that initiate circadian clock function, defines conserved mechanisms underlying the accumulation of activator complexes in eukaryotes, and suggests that Clk, cyc, and cry expression are sufficient to program clock function in naive Drosophila cells.
Results
CYC Protein Is Stabilized by CLK.
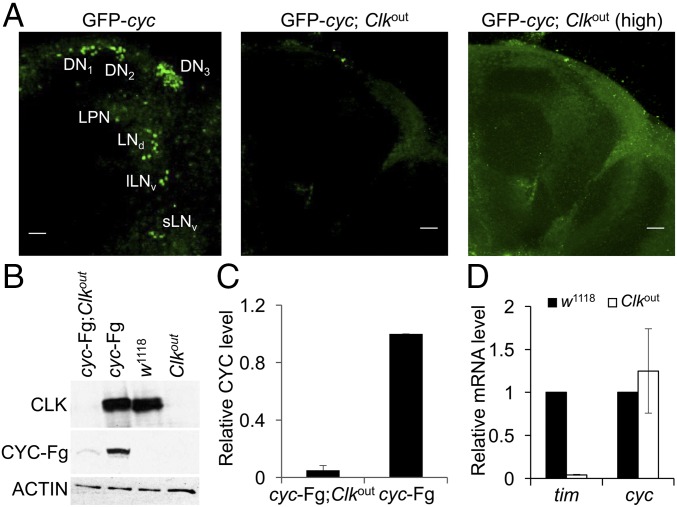
Previous work showing that cyc mRNA is not enriched in pacemaker neurons suggests that cyc is also expressed in nonclock cells (7). Broad cyc expression is consistent with the ability of Clk to generate clocks in nonclock brain neurons (8, 9), but contrasts with the pacemaker neuron-specific accumulation of GFP-CYC (5). To reconcile these data, we propose that cyc mRNA is broadly expressed and CYC accumulates only in cells that express Clk. If CYC accumulation is dependent on Clk, then loss of Clk in clock neurons should also eliminate CYC. Indeed, GFP-CYC was not detectable in pacemaker neurons from Clkout null mutant flies (10) (Fig. 1A). To determine if Clk is required for CYC accumulation in fly heads, where most clock gene expression emanates from retinal photoreceptors (11), we used a cyc-FLAG transgene that fully rescues clock function (12). The levels of CYC-FLAG in Clkout heads was reduced >10-fold compared with controls having intact clocks (Fig. 1 B and C). In contrast, cyc mRNA levels are the same in control (w1118) and Clkout heads (Fig. 1D), indicating that Clk is not required for cyc transcription. These results show that Clk promotes CYC accumulation at the posttranscriptional level.
Fig. 1.
CYC protein is expressed at low levels in Clkout flies. (A) GFP-CYC expression in brain pacemaker neurons was assessed in GFP-cyc and GFP-cyc; Clkout flies entrained in LD for at least 3 d and collected at ZT2. Immunostaining was performed on dissected brains using anti-GFP antibody and imaged by confocal microscopy as described (Materials and Methods). (Left) An 80-μm projected Z-series image of a right brain hemisphere from a GFP-cyc fly. (Middle) A 104-μm projected Z-series image of a right brain hemisphere from a GFP-cyc; Clkout fly. (Right) Same projected Z-series as Middle image with an increased laser power (high). Brains are oriented where lateral is to the right and dorsal is at the top. DN1, DN2, DN3, LPN, LNd, lLNv, and sLNv refer to pacemaker neuron groups as defined in the text. (Scale bar, 10 μm.) All images are representative of six or more brains. (B) Western blots containing proteins from the heads of control (w1118), cyc-FLAG (cyc-Fg), cyc-FLAG;Clkout (cyc-Fg;Clkout), and Clkout flies collected at ZT14 were probed with CLK, FLAG, and β-ACTIN antibodies to measure CLK, CYC-FLAG (CYC-Fg), and β-ACTIN (ACTIN), respectively. β-ACTIN was used as a loading control. (C) Relative CYC-FLAG levels on Western blots described in B were determined by measuring band intensities using Image J software (Materials and Methods). Values represent mean ± SEM from three independent experiments. (D) RT-qPCR analysis of tim and cyc mRNA levels in heads from control (w1118) and Clkout flies collected at ZT14. Values represent mean ± SEM from five independent experiments.
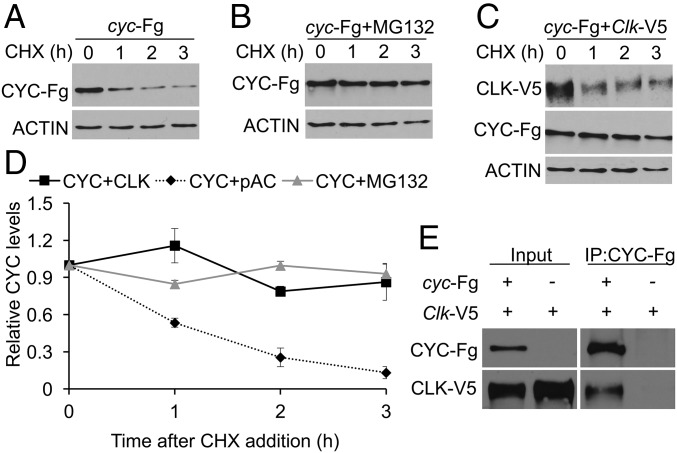
Reduced levels of CYC in Clkout flies could result from decreased synthesis or increased stability. Although there is evidence that transcription factors such as BMAL1 and HIF2α act in the cytoplasm to enhance translation (13, 14), we favor the possibility that CYC is stabilized by CLK as a product of heterodimer formation, which can stabilize other heterodimeric transcription factors (15, 16). To test whether CLK stabilizes CYC, we first determined the half-life of FLAG-tagged CYC in Drosophila S2 cells. S2 cells were transfected with pMK33-cyc-FLAG plasmid, CYC-FLAG expression was induced, translation was inhibited using cycloheximide (CHX), and samples were collected as described (Materials and Methods). The levels of CYC-FLAG declined rapidly after CHX addition, with a half-life of ∼1 h (Fig. 2 A and D). To identify the CYC degradation pathway, we measured the CYC-FLAG half-life after treatment with the 26S proteasome inhibitor MG132. CYC-FLAG levels were unchanged in the presence of MG132, indicating that CYC is degraded by proteasome (Fig. 2 B and D). To determine the impact of CLK on CYC protein stability, we measured CYC levels in the presence of V5-tagged CLK. CYC-FLAG was stabilized in the presence of CLK-V5 with a half-life of ∼9 h, demonstrating that CLK stabilizes CYC (Fig. 2 C and D). When CYC-FLAG and CLK-V5 were coexpressed in S2 cells, CLK-V5 was coimmunoprecipitated with CYC-FLAG, demonstrating that CLK and CYC are in the same complex (Fig. 2E). These results show that CLK stabilizes CYC by protecting CYC from proteasomal degradation.
Fig. 2.
CYC protein is stabilized when coexpressed with CLK. S2 cells transfected with pMK-cyc-FLAG (cyc-Fg) plasmid alone or in combination with pAc-Clk-V5 (Clk-V5) plasmid were incubated with CuSO4 for 1 h to induce cyc-Fg expression and then treated with CHX to inhibit translation. (A) S2 cells transfected with cyc-Fg. (B) S2 cells transfected with cyc-Fg and treated with MG132 at 0 h. (C) S2 cells cotransfected with cyc-Fg and Clk-V5. Proteins were extracted from cells harvested at the indicated times after CHX addition and used to prepare Western blots that were probed with anti-FLAG, anti-CLK, and anti–β-ACTIN antibodies. (D) Relative CYC-Fg levels were quantified using Image J software as described (Materials and Methods) and plotted as the mean value ± SEM from four independent experiments. (E) Protein extracts from cells transfected with Clk-V5 alone or Clk-V5 and cyc-Fg were subjected to immunoprecipitation using anti-FLAG antibody. Western blots containing cell extracts (Input) or immune complexes (IP) were probed with anti-FLAG and anti-CLK antibodies.
Clk Promotes CYC Accumulation in Ectopic Cells, but Is Not Sufficient for Clock Function in All Ectopic Cells.
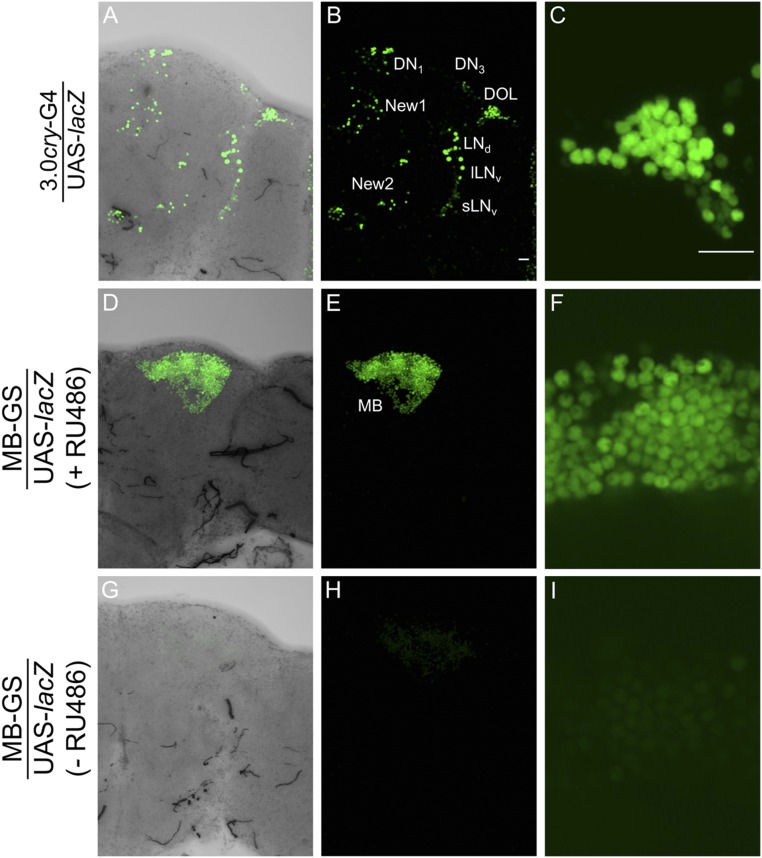
If CLK stabilizes CYC in vivo as it does in S2 cells, we predict that CYC will accumulate in cells that ectopically express CLK. To test this prediction, Clk was driven in cry-expressing clock and nonclock neurons using the 3.0cry-Gal4 driver (17) and in nonclock-expressing mushroom body neurons using the hormone-activated MB-GeneSwitch (MB-GS) driver (18). We first confirmed the spatial expression pattern of these drivers by using them to activate UAS-lacZnls, which expresses nuclear-localized β-galactosidase. As expected, the 3.0cry-Gal4 driver is expressed in a subset of pacemaker neurons including ∼8 DN1s, ∼2 DN3s, sLNvs, lLNvs, and ∼6 LNds and in nonclock cell groups including the new 1, new 2, and dorsal optic lope (DOL) neurons (Fig. S1 A–C). Likewise, the MB-GS driver was strongly expressed in mushroom body neurons in the presence, but not the absence, of RU486 inducer (Fig. S1 D–I). We then determined whether CLK stabilizes CYC in nonclock cells by generating flies that contain the 3.0cry-Gal4 or MB-GS driver, a UAS-Clk responder, and the GFP-cyc transgene, collecting these flies at Zeitgeber Time 2 (ZT2, where ZT0 is lights on and ZT12 is lights off) and immunostaining them with GFP to detect CYC and PER to mark CLK-CYC–dependent gene expression.
Fig. S1.
Localization of GAL4-dependent expression in brain. (A) UAS responder expressing nuclear-targeted lacZ (UAS-lacZ) was expressed under the control of the 3.0cry-Gal4 (3.0cry-G4) and MB-GS drivers. Immunostaining with LacZ antibody was performed on dissected adult fly brains and imaged by confocal microscopy. A projected Z-series image of the right brain hemisphere, where lateral is right and dorsal is top. Pacemaker neuron groups are as defined in the text. The 3.0cry-G4 drives expression in some pacemaker neurons plus new 1, new 2, and DOL cells, and MB-G4 drives expression in mushroom body neurons (MB). (A–C) A 136-μm projected Z-series image of the right brain hemisphere from a 3.0cry-G4/UAS-lacZ fly. (A) Image shown in B that includes the transmitted light view to show the outline of the brain hemisphere. (C) Magnified 58-μm projected Z-series image of the DOL region from B. (D–F) A 120-μm projected Z-series image of the right brain hemisphere from an MB-GS/UAS-lacZ induced with RU486. (D) Image shown in E that includes the transmitted light view to show the outline of the brain hemisphere. (F) Magnified 2-μm image of the MB region from E. (G–I) A 128-μm projected Z-series image of the right brain hemisphere from MB-GS/UAS-lacZ without RU486 induction. (G) Image shown in H that includes the transmitted light view to show the outline of the brain hemisphere. (I) Magnified 2-μm image of the MB region from H. (Scale bars, 10 μm.) The magnification of panels A, B, D, E, G, and H and panels C, F, and I are the same. All images are representative of six or more brains.
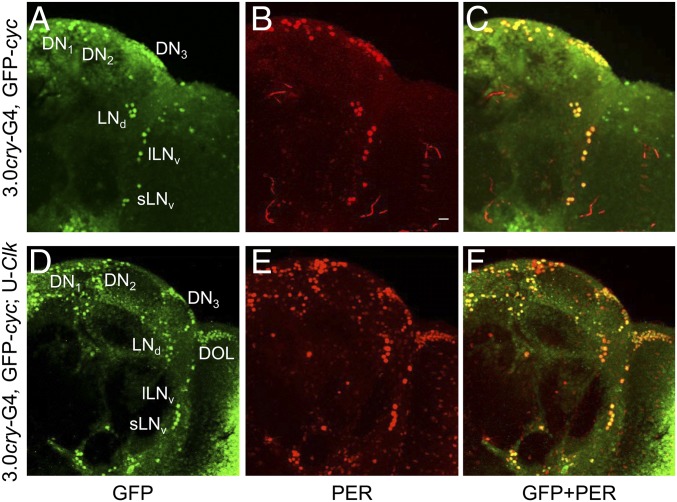
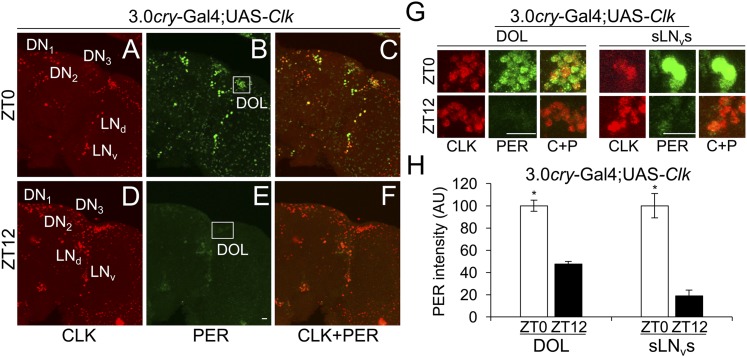
When the 3.0cry-Gal4 driver was used to express Clk, GFP-CYC expression was expanded to include all endogenous pacemaker neurons and 3.0cry-Gal4 driver-specific nonclock cells (Fig. 3 D–F). Among the different nonclock cell groups, we focused on DOL cells since they comprise ∼20 cells that are spatially segregated from pacemaker neurons and other 3.0cry-Gal4–expressing cells. GFP-CYC was detected in DOL cells in the presence, but not in the absence, of Clk expression (Fig. 3 A–C), demonstrating that CLK promotes CYC accumulation in vivo. Moreover, PER also accumulates in pacemaker neurons and DOL cells (Fig. 3E), indicating that CLK-CYC activates downstream target genes. Consistent with previous results (8, 9), PER levels cycle in DOL cells during 12 h light:12 h dark (LD) cycles (Fig. 4 B, E, G, and H), although PER cycling amplitude in DOL cells is less robust than in sLNvs (Fig. 4 A–H). These results show that Clk expression promotes CYC accumulation and PER cycling in DOL cells.
Fig. 3.
CLK expression in DOL cells promotes GFP-CYC accumulation. The 3.0cry-Gal4, GFP-cyc (3.0cry-G4, GFP-cyc), and 3.0cry-Gal4, GFP-cyc; UAS-Clk/+ (3.0cry-G4, GFP-cyc; U-Clk) flies were entrained in LD for at least 3 d and collected at ZT2. Immunostaining with GFP and PER antibodies was performed on dissected brains and imaged by confocal microscopy. Projected Z-series images of right brain hemispheres are shown, where lateral is right and dorsal is top. Pacemaker neuron groups are as defined in the text, and DOL cells are as defined in Fig. S1. Colocalization of GFP (green) and PER (red) is shown as yellow. (A–C) A 76-μm projected Z-series image of a 3.0cry-G4, GFP-cyc fly brain immunostained with GFP (A), PER (B), or GFP and PER (C). (D–F) An 86-μm projected Z-series image of a 3.0cry-G4, GFP-cyc; UAS-Clk fly brain immunostained with GFP (D), PER (E), or GFP and PER (F). GFP and PER immunostaining is detected in the indicated groups of pacemaker neurons and DOL cells, as well as in additional ectopic locations. (Scale bar, 10 μm.) All images are representative of six or more brains.
Fig. 4.
Clk expression in DOL cells is sufficient for PER cycling in LD. The 3.0cry-Gal4/+; UAS-Clk/+ (3.0cry-Gal4; UAS-Clk) flies were entrained in LD for 3 d and collected at ZT0 and ZT12. Immunostaining with CLK and PER antibodies was performed on dissected brains and imaged by confocal microscopy. Projected Z-series images of right brain hemispheres are shown, where lateral is right and dorsal is top. Pacemaker neuron groups are as defined in the text, and DOL cells are as defined in Fig. S1. Colocalization of CLK (red) and PER (green) is shown as yellow. (A–C) An 88-μm projected Z-series image of a brain from flies collected at ZT0 and immunostained with CLK (A), PER (B), or CLK and PER (C). (D–F) A 76-μm projected Z-series image from flies collected at ZT12 and immunostained with CLK (D), PER (E), or CLK + PER (F). (Scale bar, 10 μm). Panels A–F are the same magnification. (G) Magnified 24-μm projected Z-series images of DOL cells (Left) or magnified 12-μm projected Z-series images of sLNvs (Right) from flies collected at ZT0 in A–C or ZT12 in D–F. (Scale bar, 10 μm.) C+P, CLK + PER. All images are representative of six or more brains. (H) PER immunostaining intensity was quantified in DOL cells and sLNvs from flies collected at ZT0 and ZT12. AU, arbitrary units. Error bars indicate ±SEM. PER intensity was significantly (*P < 0.001) higher in both DOL cells and sLNvs at ZT0 than at ZT12 by two-tailed Student’s t test.
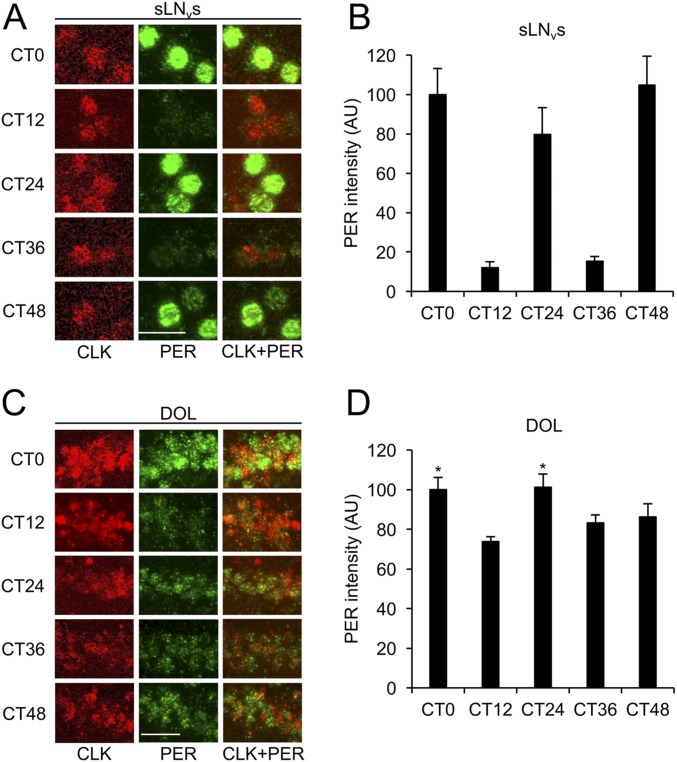
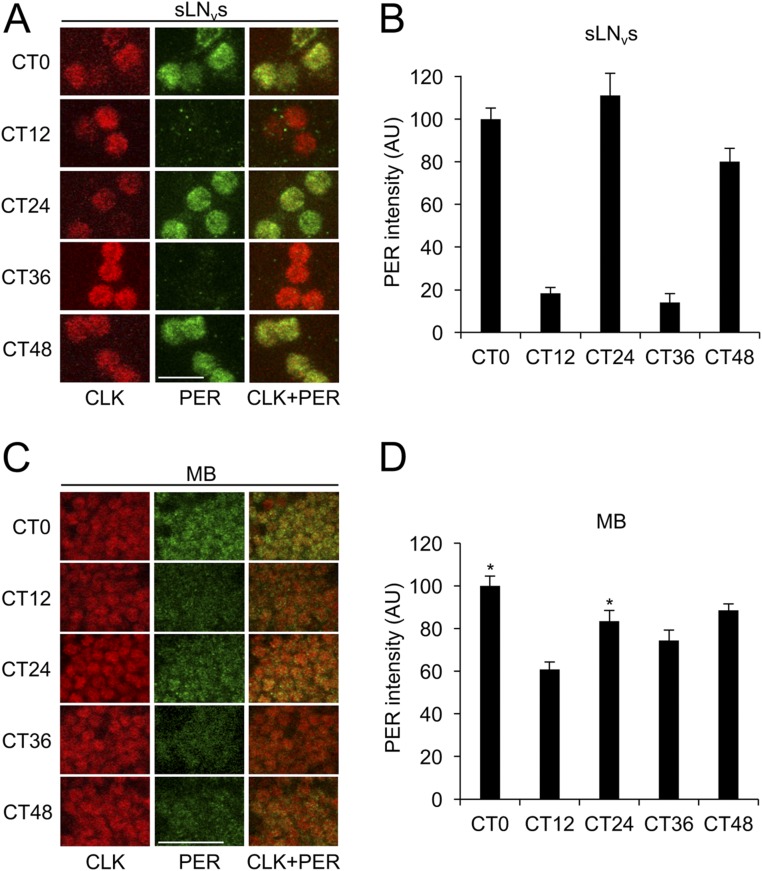
To determine if PER cycling in DOL cells is driven by LD cycles, we monitored PER rhythms in DOL cells and sLNvs during DD. Flies containing 3.0cry-Gal4 and UAS-Clk were entrained in LD for 3 d, transferred to DD, and collected every 12 h for 2 d starting at circadian time 0 (CT0), which corresponds to subjective lights on. In sLNvs, PER abundance showed significant (P < 0.05) circadian cycling with high levels at CT0, CT24, and CT48 and low levels at CT12 and CT36 (Fig. S2 A and B). In DOL cells, PER abundance was not significantly rhythmic, although PER levels at CT0 and CT24 were significantly (P < 0.01) higher than at CT12 (Fig. S2 C and D), indicative of a rapidly dampened rhythm.
Fig. S2.
Clk expression in DOL cells supports PER cycling that rapidly dampens in DD. The 3.0cry-Gal4/+; UAS-Clk/+ flies were entrained in LD cycles for 3 d, transferred to constant darkness, and collected at CT0, CT12, CT24, CT36, and CT48. Immunostaining with anti-PER and anti-CLK antibodies was performed on adult brains and imaged by confocal microscopy. Colocalization of CLK (red) and PER (green) is shown as yellow. (A) The 22-μm projected Z-series images of sLNvs from flies collected at the indicated times and immunostained with CLK (Left column), PER (Middle column), or CLK and PER (Right column). (B) PER immunostaining intensity from sLNvs was quantified in AU as described in Materials and Methods. Error bars indicate ±SEM. The overall effects of time of day were significant (P < 0.0001) by one-way ANOVA. Time-dependent cycling was significant (P < 0.05) by Tukey post hoc analysis. (C) The 26-μm projected Z-series images of DOL cells from flies collected at the indicated times and immunostained with CLK (Left column), PER (Middle column), or CLK and PER (Right column). (D) PER immunostaining intensity from DOL cells was quantified in AU as described above. Error bars indicate ±SEM. The overall effects of time of day were significant (P < 0.01) by one-way ANOVA. Time-dependent cycling was not significant by Tukey post hoc analysis. Asterisks denote significant (P < 0.05) increase in PER in DOL cells at CT0 and CT24 compared with CT12 by two-tailed Student’s t test. (Scale bars, 10 μm.) All images in panels A and C are the same magnification. All images are representative of six or more brains.
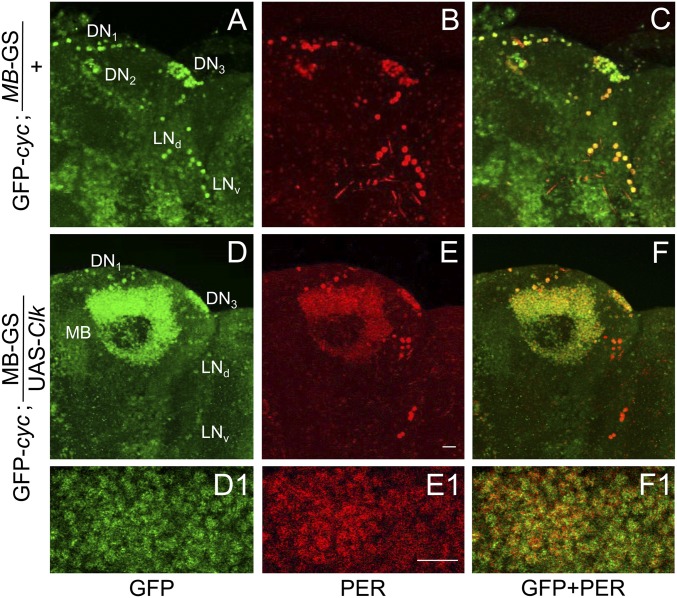
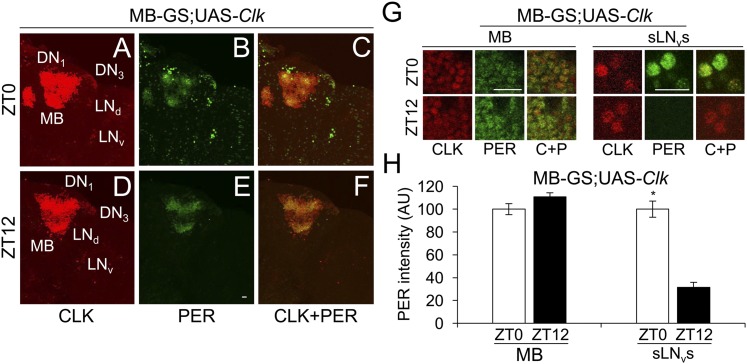
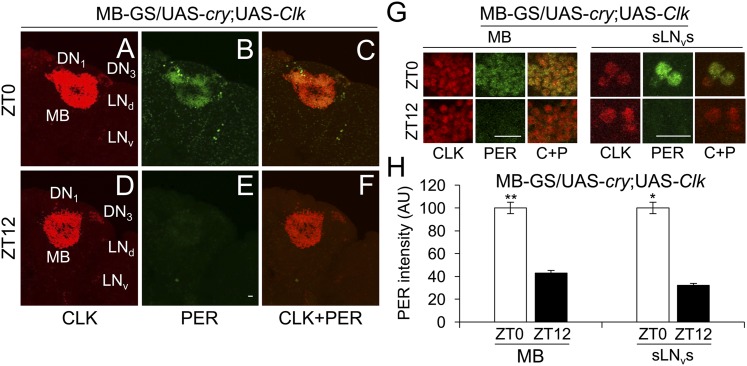
When MB-GS was used to drive Clk, GFP-CYC was detected in all endogenous pacemaker neurons plus MB neurons (Fig. 5 D, D1, F, and F1), but only in endogenous pacemaker neurons in controls lacking MB-GS–driven Clk (Fig. 5 A and C). As in DOL cells, Clk expression supports PER accumulation in MB neurons (Fig. 5 B, E, and E1), indicating that CLK engages CYC to drive target gene transcription. However, PER levels were constant in MB neurons at ZT0 and ZT12 (Fig. 6 B, E, G, and H), in contrast to the robust rhythms of PER staining intensity seen in pacemaker neurons (Fig. 6 A–H) or in DOL cells during LD (Fig. 4 B, E, G, and H). From these results, we conclude that, even though Clk expression in MB neurons promotes CYC accumulation, it is not sufficient to support clock function.
Fig. 5.
CLK expression in MB neurons promotes GFP-CYC accumulation. GFP-cyc; MB-GS and GFP-cyc; MB-GS/UAS-Clk flies induced with RU486 were entrained and collected as described in Fig. 3. Immunostaining with GFP and PER antibodies was performed on dissected brains and imaged by confocal microscopy. Projected Z-series images of right brain hemispheres are shown, where lateral is right and dorsal is top. Pacemaker neuron groups are as defined in the text, and MB neurons are as defined in Fig. S1. Colocalization of GFP (green) and PER (red) is shown as yellow. (A–C) A 106-μm projected Z-series image of a GFP-cyc; MB-GS fly brain immunostained with GFP (A), PER (B), or GFP and PER (C). (D–F) A 118-μm projected Z-series image of a GFP-cyc; MB-GS/UAS-Clk fly brain immunostained with GFP (D), PER (E), and GFP and PER (F). GFP and PER immunostaining are detected in the indicated groups of pacemaker neurons and in mushroom body neurons. (D1, E1, and F1) Magnified 2-μm image of MB neurons shown in D–F. (Scale bar, 10 μm.) All images are representative of six or more brains.
Fig. 6.
Clk expression in MB neurons does not support PER cycling in LD. MB-GS/+; UAS-Clk/+ (MB-GS; UAS-Clk) flies were induced with RU486, entrained and collected as described in Fig. 4. Immunostaining with CLK and PER antibodies was performed on dissected adult brains and imaged by confocal microscopy. Projected Z-series images of right brain hemispheres are shown, where lateral is right and dorsal is top. Pacemaker neuron groups are as defined in the text, and MB neurons are as defined in Fig. S1. Colocalization of CLK (red) and PER (green) is shown as yellow. (A–C) A 136-μm projected Z-series image of a brain from flies collected at ZT0 and immunostained with CLK (A), PER (B), or CLK and PER (C). (D–F) A 136-μm projected Z-series image of a brain from flies collected at ZT12 and immunostained with CLK (D), PER (E), or CLK and PER (F). (Scale bar, 10 μm). Panels A–F are the same magnification. (G) Magnified 2-μm images of MB neurons (Left) and magnified 18-μm projected Z-series images of sLNvs (Right) from flies collected at ZT0 in A–C or at ZT12 in D–F. (Scale bar, 10 μm.) C+P, CLK + PER. All images are representative of six or more brains. (H) PER immunostaining intensity was quantified in MB neurons and sLNvs from flies collected at ZT0 and ZT12. AU, arbitrary units. Error bars indicate ±SEM. PER intensity was significantly (*P < 0.01) higher in sLNvs at ZT0 than at ZT12 by two-tailed Student’s t test.
CRY Is Required for Ectopic Clock Function.
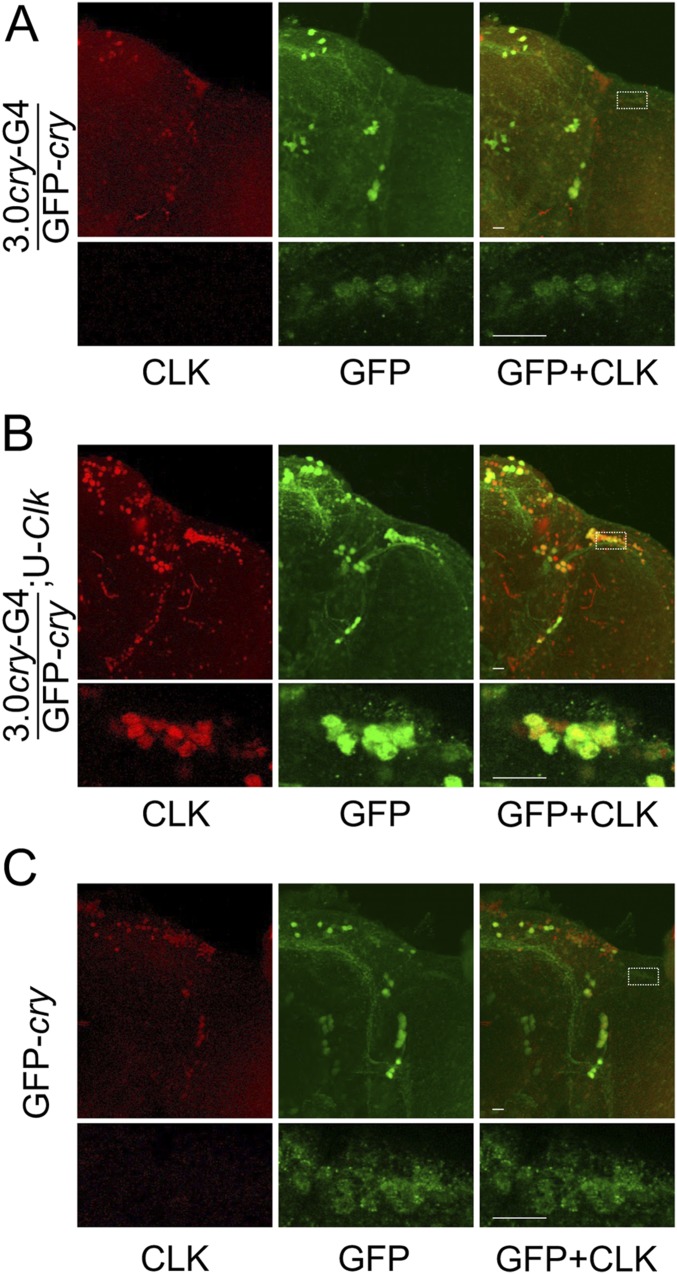
The ability of 3.0cry-Gal4–driven Clk, but not MB-GS–driven Clk, to generate ectopic clocks likely results from gene expression differences in these target cell populations. One obvious difference is that 3.0cry-Gal4 presumably drives expression only in CRY-positive cells, whereas no CRY is detected in MB neurons targeted by MB-GS (19–21). To confirm that CRY is expressed in DOL cells, we used a GFP-cry transgene to mark cells that endogenously express CRY with high sensitivity (19) and found that GFP-CRY is indeed expressed in DOL cells, albeit at lower levels than in pacemaker neurons (Fig. S3 A and C). Importantly, when 3.0cry-Gal4 was used to express UAS-Clk in GFP-cry flies, GFP-CRY levels increased substantially (Fig. S3B), suggesting that Clk-dependent factors enhance cry expression in DOL cells. Since cry expression is required for light entrainment and/or clock function in multiple peripheral tissues (22–24), cry may also be required for ectopic clock function. To test this possibility, we used MB-GS to drive both Clk and cry expression in the presence of RU486 and assessed PER levels in MB neurons at ZT0 and ZT12. PER levels cycled robustly in MB neurons upon Clk and cry coexpression (Fig. 7), demonstrating that cry is indeed necessary for clock function in MB neurons.
Fig. S3.
Endogenous CRY expression in DOL cells is enhanced by Clk expression. The 3.0cry-Gal4/GFP-cry (3.0cry-G4/GFP-cry), 3.0cry-Gal4/GFP-cry;UAS-Clk/+ (3.0cry-G4/GFP-cry;U-Clk), and GFP-cry flies were entrained in LD cycles for 3 d, transferred to constant darkness, and collected at CT24 on the first day of DD. Immunostaining with CLK and GFP (to detect GFP-CRY) antibodies was performed on adult brains and imaged by confocal microscopy. Colocalization of CLK (red) and GFP (green) is shown as yellow. (A) The 130-μm projected Z-series images of the right hemisphere from a 3.0cry-G4/GFP-cry brain (Top) and magnified 2-μm sections of the DOL region (Bottom) from the brain on Top immunostained with CLK (Left column), GFP (Middle column), or CLK and GFP (Right column). White rectangle, DOL region used for magnified image on Bottom. (B) The 140-μm projected Z-series images of the right hemisphere from a 3.0cry-G4/GFP-cry;U-Clk brain (Top) and magnified 2-μm sections of the DOL region (Bottom) from the brain on Top immunostained with CLK (Left column), GFP (Middle column), or CLK and GFP (Right column). White rectangle, DOL region used for magnified image on Bottom. (C) The 146-μm projected Z-series images of the right hemisphere from a GFP-cry brain (Top) and magnified 2-μm sections of the DOL region (Bottom) from the brain on Top immunostained with CLK (Left column), GFP (Middle column), or CLK and GFP (Right column). White rectangle, DOL region used for magnified image on Bottom. All images are representative of six or more brains. (Scale bars, 10 μm.) All top images and all bottom images in panels A–C are the same magnification.
Fig. 7.
Clk and cry expressions are required to support PER cycling in MB neurons during LD. MB-GS/UAS-cry; UAS-Clk/+ (MB-GS/UAS-cry; UAS-Clk) flies were induced with RU486, entrained and collected as described in Fig. 4. Immunostaining with CLK and PER antibodies was performed on dissected brains and imaged by confocal microscopy. Projected Z-series images of the right brain hemispheres are shown, where lateral is right and dorsal is top. Pacemaker neuron groups are as defined in the text, and MB neurons are as defined in Fig. S1. Colocalization of CLK (red) and PER (green) is shown as yellow. (A–C) A 122-μm projected Z-series image of a brain from flies collected at ZT0 and immunostained with CLK (A), PER (B), or CLK and PER (C). (D–F) A 122-μm projected Z-series image of a brain from flies collected at ZT12 and immunostained with CLK (D), PER (E), or CLK and PER (F). (Scale bar, 10 μm). Panels A–F are the same magnification. (G) Magnified 2-μm images of MB neurons (Left) and magnified 16-μm projected Z-series images of sLNvs from flies collected at ZT0 in A–C or at ZT12 in D–F. (Scale bar, 10 μm.) C+P, CLK + PER. All images are representative of six or more brains. (H) PER immunostaining intensity was quantified in MB neurons and sLNvs from flies collected at ZT0 and ZT12. AU, arbitrary units. Error bars indicate ±SEM. PER intensity was significantly (**P < 0.001) higher in MB neurons and significantly higher in sLNvs (*P < 0.01) at ZT0 than at ZT12 by two-tailed Student’s t test.
We then determined whether PER cycling in MB neurons coexpressing Clk and cry persisted in DD. Although PER levels in sLNvs showed significant (P < 0.05) circadian cycling with peaks at CT0, CT24, and CT48 and troughs at CT12 and CT36 (Fig. S4 A and B), the levels of PER in MB neurons did not show significant cycling (Fig. S4 C and D). However, PER levels in MB neurons at CT0 and CT24 were significantly (P < 0.01) higher than at CT12 (Fig. S4D), indicating that PER oscillations dampen rapidly in DD. Thus, ectopic clocks in MB neurons, like those in DOL cells, show a robust rhythm in PER cycling that quickly dampens in DD.
Fig. S4.
Clk and cry expression in MB neurons supports PER cycling that rapidly dampens in DD. MB-GS/UAS-cry; UAS-Clk/+ flies were induced with RU486, entrained in LD cycles for 3 d, transferred to DD, and collected at CT0, CT12, CT24, CT36, and CT48. Immunostaining with PER and CLK antibodies was performed on adult brains and imaged by confocal microscopy. Colocalization of CLK (red) and PER (green) is shown as yellow. (A) The 22-μm projected Z-series images of sLNvs from flies collected at the indicated times and immunostained with CLK (Left column), PER (Middle column), or CLK and PER (Right column). (B) PER immunostaining intensity from sLNvs was quantified in AU as described in Materials and Methods. Error bars indicate ±SEM. The overall effects of time of day were statistically significant (P < 0.0001) by one-way ANOVA. Time-dependent cycling was significant (P < 0.05) by Tukey post hoc analysis. (C) The 2-μm images of MB neurons from flies collected at indicated times and immunostained with CLK (Left column), PER (Middle column), or CLK and PER (Right column). (D) PER immunostaining intensity from MB neurons was quantified in AU as described above. Error bars indicate ±SEM. The overall effects of time of day were significant (P < 0.01) by one-way ANOVA. Time-dependent cycling was not significant by Tukey post hoc analysis. Asterisks denote a significant (P < 0.05) increase in PER in DOL cells at CT0 and CT24 compared with CT12 by two-tailed Student’s t test. All images are representative of six or more brains. (Scale bars, 10 μm.) All images in panels A and C are the same magnification.
Discussion
Here, we show that clock neuron-specific CYC accumulation in clock neurons and in Clk-dependent ectopic clocks in brain neurons is due to stabilization of CYC by CLK. In pacemaker neurons and whole fly heads, CLK is required for the accumulation of CYC (Fig. 1), demonstrating that Clk is required for CYC accumulation in clock cells. Experiments in S2 cells showed that CYC has a short ∼1-h half-life due to proteasomal degradation that is lengthened approximately ninefold when Clk is coexpressed (Fig. 2). Since CLK and CYC form complexes in S2 cells, the most parsimonious conclusion is that CLK-CYC heterodimerization stabilizes CYC via protection from proteasomal degradation.
Costabilization of heterodimeric transcription factors is not common, but two C/EBP family members, Ig/EBP and CHOP, are stabilized upon heterodimerization (16), and the Neurospora zinc-finger-PAS circadian activator White Collar 1 (WC1) is stabilized by White Collar 2 (WC2) upon WC1-WC2 heterodimerization (15). Our data account for CYC accumulation solely in Clk-expressing neurons and further define the first events required to initiate clock function in Drosophila. In mammals, Bmal1 mRNA is expressed at high levels, but BMAL1 levels are low in Clock−/− animals (25). Given that Clock and Bmal1 are orthologs of Clk and cyc, respectively (2), the stabilization of BMAL1 by CLOCK may be a conserved property of these proteins. Moreover, since WC2 stabilizes WC1 in Neurospora, stabilization of one circadian activator by its partner may be a conserved feature of eukaryotic clocks.
CLK likely binds CYC soon after CYC synthesis to produce stable CLK-CYC heterodimers. Since cyc mRNA does not cycle (26), CLK-CYC production is likely driven by rhythms in Clk mRNA, which peak near dawn (27, 28). Increased CLK-CYC production near dawn apparently offsets degradation due to CLK phosphorylation early in the day, resulting in constant CLK (and thus CLK-CYC) levels (29, 30). Just as CYC levels are low in the absence of CLK, CLK levels decrease in the absence of CYC despite high levels of Clk mRNA (30, 31). Consequently, the vast majority of CLK and CYC are present as stable CLK-CYC heterodimers, which apparently accumulate at levels determined by CLK abundance. If CLK levels fall, as in the Clkar mutant, then target gene cycling is diminished and rhythmic behavior is disrupted (32). Likewise, increased CLK activity, as seen in flies lacking Clk 3′UTR regulatory sequences, disrupts CLK-CYC target gene transcription and behavioral rhythms (33). The loss of Clk 3′UTR regulatory sequences causes ectopic Clk expression in the brain and production of additional PIGMENT DISPERSING FACTOR (PDF) neuropeptide-expressing neurons, which likely account for variable CLK-CYC target gene expression and arrhythmic behavior, respectively (33).
Because cyc mRNA expression is not restricted to pacemaker neurons in the brain (7), and ectopic clock generation by Clk is cyc-dependent (9), CLK is predicted to stabilize CYC in nonclock cells. Indeed, Clk expression promotes CYC accumulation in cry-expressing nonclock neurons and in MB neurons (Figs. 3 and 5). Although cyc mRNA can give rise to CYC when Clk is ectopically expressed, cyc mRNA function in nonclock cells is not known. CYC could be generated and rapidly degraded in many nonclock cell types, but protected from degradation by other binding partners that are induced, for example, in response to environmental stress. Alternatively, cyc mRNA may function directly, independent of producing CYC protein, in nonclock cells. Further studies are necessary to define cyc mRNA function in nonclock cells.
Once CYC is stabilized by CLK, CLK-CYC complexes can activate target gene transcription. In cry-expressing DOL cells, Clk expression induces ectopic clock function as measured by rhythms in PER accumulation that parallel those in pacemaker neurons during LD (Fig. 4). PER rhythms persist with a reduced amplitude on DD day 1 and are lost by DD day 2 (Fig. S2). This inability to maintain a robust rhythm is reminiscent of fly peripheral clocks that show lower amplitude rhythms than brain pacemaker neurons (34), which maintain high-amplitude rhythms via reinforcing neuronal signaling (34, 35). Nevertheless, Clk-induced ectopic clocks maintain high-amplitude PER rhythms under LD conditions (Fig. 4), indicative of a functional molecular clock. Although CLK-CYC activates feedback repressors to drive ectopic clock function, other clock components including posttranslational regulators (e.g., kinases, phosphatases) must be expressed in ectopic cells (1). These posttranslational clock regulators are likely widely expressed since they control many regulatory pathways, although some could be activated via ectopic Clk expression since they contain E-box regulatory elements bound by CLK-CYC (12).
Although Clk is sufficient to generate ectopic clocks in DOL cells, this was not the case in MB neurons, where Clk expression led to constant PER levels during LD (Fig. 6). One difference between DOL cells and MB neurons is that DOL cells express CRY (Fig. S3 A and C), while MB neurons lack CRY expression (20, 21). Since CRY mediates light entrainment in some pacemaker neurons and is necessary for light entrainment and clock function in peripheral tissues (19, 22, 23, 36–39), our inability to generate an ectopic MB clock may be due to the lack of CRY. Indeed, expressing both Clk and cry in MB neurons resulted in robust PER cycling in LD (Fig. 7), indicative of ectopic clock function. These Clk- and cry-induced PER rhythms in MB neurons mirror those in pacemaker neurons during LD, but rapidly dampen in DD (Fig. S4). The rapid dampening of PER rhythms in MB neurons is similar to that in DOL cells during DD (Fig. S2) and is faster than that in peripheral clocks using per-luciferase or tim-luciferase reporter assays (22, 23).
The inability of DOL cells and MB neurons to sustain clock function in DD likely stems from multiple factors including suboptimal or nonrhythmic expression of genes that contribute to timekeeping (e.g., Clk, cry, posttranscriptional regulators) and a lack of intercellular coupling that sustains robust rhythms in pacemaker neurons (34, 35). Although 3.0cry-Gal4–driven UAS-Clk enhances GFP-CRY expression in DOL cells (Fig. S3B), MB-GS–driven UAS-Clk is apparently unable to activate cry expression in MB neurons, suggesting that CLK can engage factors in DOL cells to increase cry expression, but cannot engage factors required to activate cry in MB neurons. These results suggest that the properties of molecular clocks in canonical and ectopic cells differ depending on the function and gene expression characteristics of the cell.
Our experiments demonstrate that cry, like Clk, is required for ectopic clock function. Since cyc is also necessary for ectopic clock function, it is possible that naive Drosophila cells can be programmed to express molecular clocks by expressing Clk, cyc, and cry. If such clock programming is possible, this work could lead to the development of Drosophila cell lines having clocks that operate in LD. The resulting cell lines would be analogous to monarch DpN1 cells, which possess a robust molecular clock that operates only in LD (40), yet represent a valuable tool for understanding the molecular machinery required for feedback loop function.
Materials and Methods
The following Drosophila strains were used in this study: w1118, w; Cyo/Sco; TM2/TM6B, cyc01 (26), Clkout (10), GFP-cyc; cyc01 (5), w; cyc-Flag (12), 3.0cry-Gal4 (17), MB-GeneSwitch (18), UAS-Clk (8), UAS-lacZ (#3955; Bloomington Drosophila Stock Center), UAS-cry (37), and GFP-cry (19). The pMK33-TAP-3XFLAG-6XHis expression vector (10) was used to generate the pMK33-TAP-3XFLAG-6XHis-dcyc (pMK33-cyc-Flag) plasmid for inducing cyc expressing in S2 cells. S2 cells maintained in Schneider’s Drosophila medium with 10% FBS and antibiotics were transfected, and gene expression was induced under conditions used to measure proteasomal degradation and protein half-life (10, 41). Western blots containing S2 cell and fly-head extracts and immunoprecipitates were prepared, probed with antisera, and quantified as described (10). RT-qPCR was carried out on fly heads as described (42). RU486 induction of MB-GS expression was carried out as described (18) with modifications. Antibody staining and imaging in adult brains was carried out as previously described (5, 43, 44). Immunostaining in clock cells was quantified from digital images of fly brains as described (9). For details concerning plasmid construction, S2 cell experiments, Western blot analysis, RT-qPCR analysis, RU486 induction, immunoprecipitations, and brain immunostaining, imaging, and quantification, please refer to SI Materials and Methods.
SI Materials and Methods
Plasmid Construction.
The pMK33-cyc-Flag plasmid was constructed by inserting the cyc ORF into the pMK33-TA.
P-3XFLAG-6XHis expression vector (10). The cyc ORF was amplified from genomic DNA using XhoI cyc forward (5′-ctattc CTC GAG ATG GAA GTT CAG GAG TTC TGC G-3′) and SpeI cyc reverse (5′-aattcc ACT AGT TTA TAA GAA CAC GGA ATT CTT GGC GA-3′) primers and subcloned into TA to form TA-cycORF (Invitrogen). The cyc ORF was removed from TA-cycORF with XhoI and SpeI and inserted into pMK33-TAP-3XFLAG-6XHis to form pMK33-cyc-Flag.
The S2 Cell Experiments.
S2 cells were maintained in Schneider’s Drosophila medium (Invitrogen) containing 10% FBS with (100 units/mL) penicillin and streptomycin (100 g/mL) (Invitrogen). The S2 cells at 40–80% confluence were transiently transfected with pMK33-cyc-Flag and pAc-Clk-V5 or pAc-V5 using Effectene Transfection Reagent (Qiagen) (6.4 μL of Enhancer, 10 μL of Transfection Reagent, 0.8 μg of total DNA), and 36 h after transfection cells were induced with copper sulfate (500 μM) as described (10). After a 1-h induction, cells from independent transfections were treated with CHX (10 μg/mL, C7698; Sigma) or CHX plus MG132 (50 μM; Sigma) and harvested 0, 1, 2, and 3 h later to measure proteasomal degradation and protein half-life (10, 41).
Western Blot Analysis.
The S2 cells and fly-head extracts were prepared using radioimmunoprecipitation assay (RIPA) buffer and used to prepare Western blots as described (10). Ten micrograms (for probing with CLK antibody) or 1 μg (for probing with FLAG and ACTIN antibodies) of S2 cell extract and 10 μg of fly-head extract were loaded on gels for Western blotting. Blots were probed with guinea pig anti-CLK (1:3,000; gp50), mouse anti-FLAG (1:30,000; Sigma), and mouse anti-actin (1:50,000; Abcam) antibodies. Horseradish peroxidase-conjugated secondary antibodies (Sigma) were diluted 1:1,000. Immunoblots were visualized using ECL plus reagent (GE Life Sciences). The ImageJ program was used to quantify protein abundance as described (10).
Quantitative PCR Analysis.
Quantitative PCR was carried out on fly heads as described (42). The following gene-specific primer pairs were used: rp49 forward primer (5′-TACAGGCCCAAGATCGTGAA-3′), rp49 reverse primer (5′-GCACTCTGTTGTCGATACCC-3′), tim forward primer (5′-GGTGGCATCTGTGTACGAAA-3′), tim reverse primer (5′-GATCTCGGTTCGCTCAAGTC-3′), cyc forward primer (5′-TGGACAATCACCCGAACATAC-3′), and cyc reverse primer (5-CTGAGGCAGGAAACCAATCA-3).
Immunoprecipitation.
Cell extracts were prepared using lysis buffer (50 mM Tris⋅HCl, pH 7.5, with 150 mM NaCl, 1 mM EDTA, and 1% Triton X-100), containing 0.5 mM PMSF, 1 mM Na3VO4, and 1 mM NaF and complete EDTA-free protease inhibitor mixture (Roche Applied Science). Proteins were immunoprecipitated using Anti-FLAG M2 Magnetic Beads according to manufacturer instructions (Sigma-Aldrich). One milligram of protein was added to Tris-buffered saline (pH 7.4) washed beads and incubated with agitation at 4 °C overnight. Beads were collected using an appropriate magnetic separator, and the supernatant was kept on the side. Beads were then washed three times with lysis buffer, transferred to a new tube, mixed with 20 μL of 2× sample buffer [125 mM Tris⋅HCl, pH 6.8, 4% SDS, 20% (vol/vol) glycerol, and 0.004% bromophenol blue, 10% 2-mercaptoethanol] and boiled for 5 min. The supernatant was used for Western blotting as described above.
RU486 Induction.
Late-stage pupae were transferred to an empty vial with one Kimwipe moistened with water before they emerged. Newly emerged flies were fed with either 5% sucrose and 1.25% agarose with or without 500 μg/mL RU486 for 5 d before collecting for dissection (18).
Immunostaining Adult Brains.
Antibody staining of adult brain was carried out as previously described (5). Adult brains were dissected and fixed with 3.7% formaldehyde. Samples were washed and incubated with primary and secondary antibodies in a solution containing 1× PBS, 5% BSA, 5% goat serum (Sigma) (5% donkey serum for primary antibodies raised in goat), 0.03% sodium deoxycholate, and 0.3% Triton X-100. The following primary antibodies were used: goat anti-CLK dC-17 (Santa Cruz Biotechnology) 1:100, goat anti-PER (Santa Cruz Biotechnology) 1:500, mouse anti–β-galactosidase (Promega) 1:200, rabbit anti-GFP ab6556 (Abcam) 1:500, preabsorbed rabbit anti-PER (gift from Michael Rosbash, Brandeis University, Waltham, MA) 1:15,000, and rabbit anti-GFP ab290 (Abcam) 1:2,000. The following secondary antibodies were used at a 1:200 dilution: goat anti-rabbit Alexa 647 (Molecular Probes), goat anti-rabbit Alexa 488 (Molecular Probes), donkey anti-rabbit Alexa 488 (Jackson ImmunoResearch Laboratories), donkey anti-mouse Alexa 488 (Jackson ImmunoResearch Laboratories), and donkey anti-goat Cy5 (ImmunoResearch Laboratories).
Imaging and Quantification.
Adult fly brains were imaged using an Olympus FV1000 confocal microscope as described previously (5). Confocal stacks were imaged using an Olympus FV1000 confocal microscope equipped with 20×/0.85 N.A. and 100× 1.40 N.A. oil-immersion objectives. For double-labeling experiments, sequential scans of the argon ion 488-nm and HeNe (633 nm for AlexaFluor 647 and Cy5) lasers were used to avoid bleed-through between channels. For imaging AlexaFluor 488 and either AlexaFluor 647 or Cy5, Argon 488 and HeNe 633-nm lasers were used, with the 488/543/633-nm dichroic mirror for excitation. Fluorescence signals were separated by a dichroic beam splitter (560 nm long-pass). A spectral detector set to 500–555 nm was used for AlexaFluor 488, and a detector with a 650-nm long-pass filter was used for AlexaFluor 647 or Cy5 signals. The Fluoview “Hi-Lo” look-up table was used to set the maximal signal below saturation and set the background to near zero using the high voltage and offset controls. Z-series images were obtained at a 2-μm step size, and Kalman averaging was not used. Original images were saved as 12-bit OIB format and processed using FV1000 confocal software to generate maximum intensity projections (Z-projections). Images were adjusted for brightness and contrast using Adobe Photoshop. For each genotype and time point, brain images were acquired using the same settings (power, gain, offset) at the same time.
Data of sLNvs and ectopic clocks were quantified from digital images using the Fiji version of ImageJ (44). CLK staining was used to mark the ectopic clock cell and sLNv nuclei, and PDF staining was used to identify sLNvs. For ectopic clock cells, the Fiji plugin 3D object counter was used to measure the total number of cells (43). The sum of the fluorescence intensity for all cells was measured, and the average intensity for each cell was calculated. An average was then calculated for each time point and scaled so that the typical peak, usually ZT0/CT0 for ectopic clocks, was 100 AU as described (9). Error Bar ±SEM. One-way ANOVA with post hoc Tukey test was used to identify time-dependent cycling (P < 0.05). A Student’s t-test was performed to compare data between two time points.
Acknowledgments
We thank the following people for providing fly stocks for this study: Patrick Emery for UAS-cry, Michael Rosbash for w; cyc-Flag and cyc01, Ravi Allada for UAS-Clk, and Gregg Roman for MB-GeneSwitch. Stocks obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537) were also used in this study. We also thank Michael Rosbash for the pAC-Clk-V5 plasmid and Stan Vitha from the Texas A&M Microscopy and Imaging Center for help with confocal microscopy. This work was supported by NIH Grant NS094807.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1707143114/-/DCSupplemental.
References
- 1.Tataroglu O, Emery P. The molecular ticks of the Drosophila circadian clock. Curr Opin Insect Sci. 2015;7:51–57. doi: 10.1016/j.cois.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bell-Pedersen D, et al. Circadian rhythms from multiple oscillators: Lessons from diverse organisms. Nat Rev Genet. 2005;6:544–556. doi: 10.1038/nrg1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Menet JS, Hardin PE. Circadian clocks: The tissue is the issue. Curr Biol. 2014;24:R25–R27. doi: 10.1016/j.cub.2013.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shafer OT, Yao Z. Pigment-dispersing factor signaling and circadian rhythms in insect locomotor activity. Curr Opin Insect Sci. 2014;1:73–80. doi: 10.1016/j.cois.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu T, Mahesh G, Houl JH, Hardin PE. Circadian activators are expressed days before they initiate clock function in late pacemaker neurons from Drosophila. J Neurosci. 2015;35:8662–8671. doi: 10.1523/JNEUROSCI.0250-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Houl JH, Ng F, Taylor P, Hardin PE. CLOCK expression identifies developing circadian oscillator neurons in the brains of Drosophila embryos. BMC Neurosci. 2008;9:119. doi: 10.1186/1471-2202-9-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nagoshi E, et al. Dissecting differential gene expression within the circadian neuronal circuit of Drosophila. Nat Neurosci. 2010;13:60–68. doi: 10.1038/nn.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao J, et al. Drosophila clock can generate ectopic circadian clocks. Cell. 2003;113:755–766. doi: 10.1016/s0092-8674(03)00400-8. [DOI] [PubMed] [Google Scholar]
- 9.Kilman VL, Allada R. Genetic analysis of ectopic circadian clock induction in Drosophila. J Biol Rhythms. 2009;24:368–378. doi: 10.1177/0748730409343761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mahesh G, et al. Phosphorylation of the transcription activator CLOCK regulates progression through a ∼24-h feedback loop to influence the circadian period in Drosophila. J Biol Chem. 2014;289:19681–19693. doi: 10.1074/jbc.M114.568493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glossop NR, Hardin PE. Central and peripheral circadian oscillator mechanisms in flies and mammals. J Cell Sci. 2002;115:3369–3377. doi: 10.1242/jcs.115.17.3369. [DOI] [PubMed] [Google Scholar]
- 12.Abruzzi KC, et al. Drosophila CLOCK target gene characterization: Implications for circadian tissue-specific gene expression. Genes Dev. 2011;25:2374–2386. doi: 10.1101/gad.178079.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lipton JO, et al. The circadian protein BMAL1 regulates translation in response to S6K1-mediated phosphorylation. Cell. 2015;161:1138–1151. doi: 10.1016/j.cell.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Uniacke J, et al. An oxygen-regulated switch in the protein synthesis machinery. Nature. 2012;486:126–129. doi: 10.1038/nature11055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng P, Yang Y, Gardner KH, Liu Y. PAS domain-mediated WC-1/WC-2 interaction is essential for maintaining the steady-state level of WC-1 and the function of both proteins in circadian clock and light responses of Neurospora. Mol Cell Biol. 2002;22:517–524. doi: 10.1128/MCB.22.2.517-524.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hattori T, Ohoka N, Inoue Y, Hayashi H, Onozaki K. C/EBP family transcription factors are degraded by the proteasome but stabilized by forming dimer. Oncogene. 2003;22:1273–1280. doi: 10.1038/sj.onc.1206204. [DOI] [PubMed] [Google Scholar]
- 17. Hao Zheng, Ng F, Yixiao Liu, Hardin PE (2008) Spatial and circadian regulation of cry in Drosophila. J Biol Rhythms 23:283–295. [DOI] [PMC free article] [PubMed]
- 18.Mao Z, Roman G, Zong L, Davis RL. Pharmacogenetic rescue in time and space of the rutabaga memory impairment by using gene-switch. Proc Natl Acad Sci USA. 2004;101:198–203. doi: 10.1073/pnas.0306128101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Agrawal P, et al. Drosophila CRY entrains clocks in body tissues to light and maintains passive membrane properties in a non-clock body tissue independent of light. Curr Biol. 2017;27:2431–2441.e3. doi: 10.1016/j.cub.2017.06.064. [DOI] [PubMed] [Google Scholar]
- 20.Benito J, Houl JH, Roman GW, Hardin PE. The blue-light photoreceptor CRYPTOCHROME is expressed in a subset of circadian oscillator neurons in the Drosophila CNS. J Biol Rhythms. 2008;23:296–307. doi: 10.1177/0748730408318588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoshii T, Todo T, Wülbeck C, Stanewsky R, Helfrich-Förster C. Cryptochrome is present in the compound eyes and a subset of Drosophila’s clock neurons. J Comp Neurol. 2008;508:952–966. doi: 10.1002/cne.21702. [DOI] [PubMed] [Google Scholar]
- 22.Ivanchenko M, Stanewsky R, Giebultowicz JM. Circadian photoreception in Drosophila: Functions of cryptochrome in peripheral and central clocks. J Biol Rhythms. 2001;16:205–215. doi: 10.1177/074873040101600303. [DOI] [PubMed] [Google Scholar]
- 23.Krishnan B, et al. A new role for cryptochrome in a Drosophila circadian oscillator. Nature. 2001;411:313–317. doi: 10.1038/35077094. [DOI] [PubMed] [Google Scholar]
- 24.Levine JD, Funes P, Dowse HB, Hall JC. Advanced analysis of a cryptochrome mutation’s effects on the robustness and phase of molecular cycles in isolated peripheral tissues of Drosophila. BMC Neurosci. 2002;3:5. doi: 10.1186/1471-2202-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Debruyne JP, et al. A clock shock: Mouse CLOCK is not required for circadian oscillator function. Neuron. 2006;50:465–477. doi: 10.1016/j.neuron.2006.03.041. [DOI] [PubMed] [Google Scholar]
- 26.Rutila JE, et al. CYCLE is a second bHLH-PAS clock protein essential for circadian rhythmicity and transcription of Drosophila period and timeless. Cell. 1998;93:805–814. doi: 10.1016/s0092-8674(00)81441-5. [DOI] [PubMed] [Google Scholar]
- 27.Bae K, Lee C, Sidote D, Chuang KY, Edery I. Circadian regulation of a Drosophila homolog of the mammalian clock gene: PER and TIM function as positive regulators. Mol Cell Biol. 1998;18:6142–6151. doi: 10.1128/mcb.18.10.6142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Darlington TK, et al. Closing the circadian loop: CLOCK-induced transcription of its own inhibitors per and tim. Science. 1998;280:1599–1603. doi: 10.1126/science.280.5369.1599. [DOI] [PubMed] [Google Scholar]
- 29.Kim EY, Edery I. Balance between DBT/CKIepsilon kinase and protein phosphatase activities regulate phosphorylation and stability of Drosophila CLOCK protein. Proc Natl Acad Sci USA. 2006;103:6178–6183. doi: 10.1073/pnas.0511215103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu W, Zheng H, Houl JH, Dauwalder B, Hardin PE. PER-dependent rhythms in CLK phosphorylation and E-box binding regulate circadian transcription. Genes Dev. 2006;20:723–733. doi: 10.1101/gad.1404406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Glossop NR, Lyons LC, Hardin PE. Interlocked feedback loops within the Drosophila circadian oscillator. Science. 1999;286:766–768. doi: 10.1126/science.286.5440.766. [DOI] [PubMed] [Google Scholar]
- 32.Allada R, Kadener S, Nandakumar N, Rosbash M. A recessive mutant of Drosophila clock reveals a role in circadian rhythm amplitude. EMBO J. 2003;22:3367–3375. doi: 10.1093/emboj/cdg318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lerner I, et al. Clk post-transcriptional control denoises circadian transcription both temporally and spatially. Nat Commun. 2015;6:7056. doi: 10.1038/ncomms8056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weiss R, Bartok O, Mezan S, Malka Y, Kadener S. Synergistic interactions between the molecular and neuronal circadian networks drive robust behavioral circadian rhythms in Drosophila melanogaster. PLoS Genet. 2014;10:e1004252. doi: 10.1371/journal.pgen.1004252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mezan S, Feuz JD, Deplancke B, Kadener S. PDF signaling is an integral part of the Drosophila circadian molecular oscillator. Cell Rep. 2016;17:708–719. doi: 10.1016/j.celrep.2016.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Egan ES, et al. An extraretinally expressed insect cryptochrome with similarity to the blue light photoreceptors of mammals and plants. J Neurosci. 1999;19:3665–3673. doi: 10.1523/JNEUROSCI.19-10-03665.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Emery P, So WV, Kaneko M, Hall JC, Rosbash M. CRY, a Drosophila clock and light-regulated cryptochrome, is a major contributor to circadian rhythm resetting and photosensitivity. Cell. 1998;95:669–679. doi: 10.1016/s0092-8674(00)81637-2. [DOI] [PubMed] [Google Scholar]
- 38.Emery P, et al. Drosophila CRY is a deep brain circadian photoreceptor. Neuron. 2000;26:493–504. doi: 10.1016/s0896-6273(00)81181-2. [DOI] [PubMed] [Google Scholar]
- 39.Stanewsky R, et al. The cryb mutation identifies cryptochrome as a circadian photoreceptor in Drosophila. Cell. 1998;95:681–692. doi: 10.1016/s0092-8674(00)81638-4. [DOI] [PubMed] [Google Scholar]
- 40.Zhu H, et al. Cryptochromes define a novel circadian clock mechanism in monarch butterflies that may underlie sun compass navigation. PLoS Biol. 2008;6:e4. doi: 10.1371/journal.pbio.0060004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu Y, Loros J, Dunlap JC. Phosphorylation of the Neurospora clock protein FREQUENCY determines its degradation rate and strongly influences the period length of the circadian clock. Proc Natl Acad Sci USA. 2000;97:234–239. doi: 10.1073/pnas.97.1.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu W, Zheng H, Price JL, Hardin PE. DOUBLETIME plays a noncatalytic role to mediate CLOCK phosphorylation and repress CLOCK-dependent transcription within the Drosophila circadian clock. Mol Cell Biol. 2009;29:1452–1458. doi: 10.1128/MCB.01777-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bolte S, Cordelières FP. A guided tour into subcellular colocalization analysis in light microscopy. J Microsc. 2006;224:213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- 44.Schindelin J, et al. Fiji: An open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]