Significance
Using single-molecule FRET imaging, we show that programmed base-pair mismatches between the peptidyl-tRNA anticodon and the mRNA codon dramatically prolong elongation factor G (EF-G)–catalyzed translocation. Mismatched peptidyl-tRNA–mRNA pairing within the pretranslocation complex specifically inhibits peptidyl-tRNA engagement of the small subunit P site, a rate-limiting process in translocation characterized by large-scale, intramolecular conformational changes within the EF-G(GDP)–bound ribosome complex. Consistent with the E site being vacant during this period, we find that elongation factor P (EF-P) can rescue this translocation defect. These findings reveal an unexpected relationship between tRNA decoding at the A site and translocation, and suggest an alternative mode of action for miscoding-inducing drugs as well as a novel function of EF-P in the cell to rescue ribosomes stalled by miscoding errors.
Keywords: ribosome, translocation, fidelity, EF-P, aminoglycosides
Abstract
Directional transit of the ribosome along the messenger RNA (mRNA) template is a key determinant of the rate and processivity of protein synthesis. Imaging of the multistep translocation mechanism using single-molecule FRET has led to the hypothesis that substrate movements relative to the ribosome resolve through relatively long-lived late intermediates wherein peptidyl-tRNA enters the P site of the small ribosomal subunit via reversible, swivel-like motions of the small subunit head domain within the elongation factor G (GDP)-bound ribosome complex. Consistent with translocation being rate-limited by recognition and productive engagement of peptidyl-tRNA within the P site, we now show that base-pairing mismatches between the peptidyl-tRNA anticodon and the mRNA codon dramatically delay this rate-limiting, intramolecular process. This unexpected relationship between aminoacyl-tRNA decoding and translocation suggests that miscoding antibiotics may impact protein synthesis by impairing the recognition of peptidyl-tRNA in the small subunit P site during EF-G–catalyzed translocation. Strikingly, we show that elongation factor P (EF-P), traditionally known to alleviate ribosome stalling at polyproline motifs, can efficiently rescue translocation defects arising from miscoding. These findings help reveal the nature and origin of the rate-limiting steps in substrate translocation on the bacterial ribosome and indicate that EF-P can aid in resuming translation elongation stalled by miscoding errors.
In bacteria, the process of directional substrate translocation is catalyzed by the five-domain GTPase, elongation factor G (EF-G) (Fig. 1A). EF-G, in complex with GTP [EF-G(GTP)], engages the pretranslocation (PRE) complex containing deacylated tRNA in the Peptidyl (P) site and peptidyl-tRNA in the Aminoacyl (A) site to facilitate the precise, directional movement of the messenger RNA (mRNA)-[tRNA]2 module (1–4) by one codon relative to the ribosome.
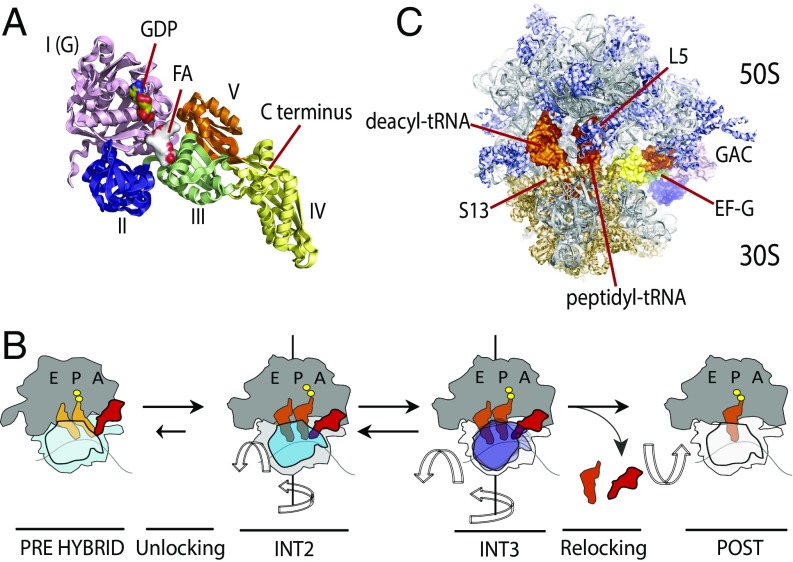
Fig. 1.
Structural models of the bacterial ribosome and EF-G and translocation scheme. (A) EF-G structural domains I–IV, GDP, FA, and the C terminus are indicated. The structural model is based on Protein Data Bank (PDB) ID code 4V7B (14). (B) Mechanistic framework of ribosome translocation (11) in which EF-G(GTP) binds preferentially to PRE complexes in which the small subunit (blue) has rotated with respect to the large subunit (dark gray) to induce “unlocking,” a process that enables the mRNA and tRNA substrates to begin to move with respect to the small subunit. Unlocking requires EF-G–catalyzed GTP hydrolysis. EF-G–induced unlocking promotes the formation of the INT2 complex, characterized by tRNA compaction, 18–20° head swivel (blue), and partial small subunit back-rotation (light gray). Reversible motions of the head domain away from the large subunit central protuberance (purple), together with complete small subunit back-rotation (white), lead to the INT3 complex. Recognition of peptidyl-tRNA within the small subunit P site triggers reverse swivel of the small subunit head domain, returning it to its classical position (white) from which EF-G(GDP) dissociates. (C) EF-G–bound, FA-stalled PRE complex showing compacted positions of deacylated (deacyl) and peptidyl-tRNAs (orange). The rRNA and large-subunit (50S) proteins are shown in gray and purple, respectively, and small-subunit (30S) proteins are shown in tan. The positions of ribosomal proteins S13 and L5 are indicated, as well as the GTPase-activating center (GAC) and EF-G. The structural model is based on PDB ID code 4W29 (14).
Ensemble measurements (5–9), together with pre–steady-state single-molecule FRET (smFRET) imaging (refs. 10, 11 and references therein) and structural studies (12–16), have revealed a comprehensive model of the translocation mechanism. In this framework, EF-G preferentially engages the PRE complex, in which the small subunit has rotated with respect to the large by ∼8–10° and the deacylated P-site tRNA has adopted a hybrid, P/E position within the P site (Fig. 1B). Upon engaging the A site, EF-G catalyzes GTP hydrolysis to enable rapid “unlocking” of the rotated small subunit to trigger large-scale structural rearrangements in both EF-G and the ribosome (5, 10, 17, 18). These early structural events facilitate the movements of both deacylated tRNA and peptidyl-tRNA through partially translocated, chimeric intersubunit hybrid positions (19–22). Such processes generate a relatively long-lived late intermediate in the translocation process, INT2, which can be efficiently trapped by the antibiotic fusidic acid (FA) (14, 23, 24) (Fig. 1C). Structures of the FA-stalled state have revealed that the formation of partially translocated tRNA positions correlates with reverse rotation of the small subunit body domain with respect to the large such that it is only partially rotated (∼2–4°) (6, 18, 19, 22), as well as a swivel-like motion of the small subunit head domain (∼18–20°) in the direction of translocation (5, 12, 14, 15, 24, 25).
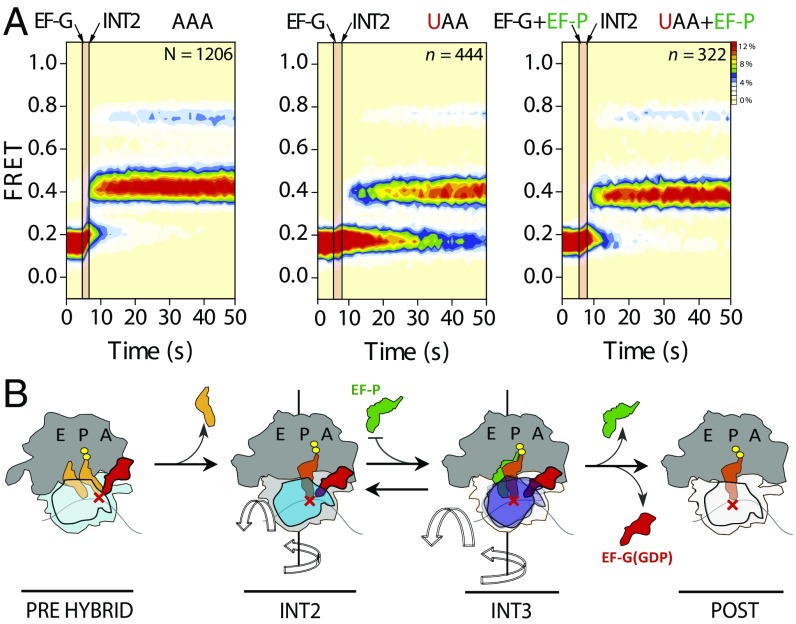
Complete translocation from the INT2 intermediate state entails rapid, reversible fluctuations into, and out of, a transient intermediate conformation (INT3) (11), defined by exaggerated movements of the small subunit head domain away from the large subunit central protuberance (Fig. 1B). Such motions, which may reflect tilting away from the subunit interface (26) and/or hyperswivel-like motion in the direction of translocation are rate-limiting to the translocation mechanism (11). Although their precise nature remains to be determined, reversible INT2 ↔ INT3 fluctuations have been hypothesized (11) to reflect conformational changes within the EF-G–bound ribosome that allow peptidyl-tRNA to fully engage the small subunit P site. Once the peptidyl-tRNA–mRNA complex has achieved a posttranslocation (POST)-like position within the small subunit P site, the head domain can then return to its unrotated, classical conformation, releasing EF-G(GDP) and completing the process of translocation (Fig. 1B).
To test the hypothesis that the peptidyl-tRNA engagement within the small subunit P site is rate-limiting to the EF-G–catalyzed translocation mechanism, we used a complementary set of FRET-based measurements (11, 20, 27–29) to investigate the impact of mismatches between the peptidyl-tRNA anticodon and the mRNA codon. Consistent with the newly established translocation framework (11), we observed that single mismatches in the codon-anticodon helix reduce the rate of translocation by up to nearly two orders of magnitude, where the defect principally manifests during the INT2 ↔ INT3 exchange (Fig. 1B). These findings suggest that the rate-limiting step in translocation specifically relates to the recognition and engagement of the peptidyl-tRNA anticodon/mRNA codon pair within the small subunit P site. This unexpected relationship between A-site decoding and translocation suggests that miscoding antibiotics may impact protein synthesis by impairing P-site recognition during EF-G–catalyzed translocation.
In accordance with improper positioning of peptidyl-tRNA within the P site, the translocation defects arising from mismatches in the codon-anticodon pair were found to be efficiently rescued by elongation factor P (EF-P), an abundant protein factor that binds the ribosome at a site that overlaps with Exit (E)-site tRNA (30). These observations imply that the impact of miscoding errors during tRNA selection at the A site may stem from defects in translocation that arise from poor recognition of the peptidyl-tRNA anticodon/mRNA codon pair at the P site. They further suggest a potentially novel EF-P function in the cell in which the protein binds the vacant E site at intermediate states of translocation to suppress defects arising from miscoding errors.
Results
To provide unique perspectives on the large-scale movements within reconstituted PRE complexes accompanying translocation, we used smFRET to image the relative movements of fluorophores site-specifically attached to S13 and L5 proteins within the small and large subunits, respectively; A- and P-site tRNAs; and EF-G (SI Materials and Methods). PRE complexes labeled in this fashion are fully functional in single-turnover and processive translation (10, 29, 31, 32).
To accommodate aminoacylated tRNA into the A site, where specific mismatches are programmed into the mRNA codon/peptidyl-tRNA anticodon pair, we incubated 70S initiation complexes with specific ternary complexes composed of aminoacyl-tRNA, GTP, elongation factor Tu, and elongation factor Ts (33) at elevated Mg2+ (15 mM) for 15 min at 25 °C in polymix buffer (34, 35). For consistency, control experiments were performed in the same manner using PRE complexes programmed with a cognate mRNA codon in the A site. Pre–steady-state smFRET measurements of translocation were subsequently performed in a low Mg2+ (5 mM) polymix buffer following previously established procedures (11).
Single-Mismatch, Near-Cognate Peptidyl-tRNAs Exhibit Defects in Translocation.
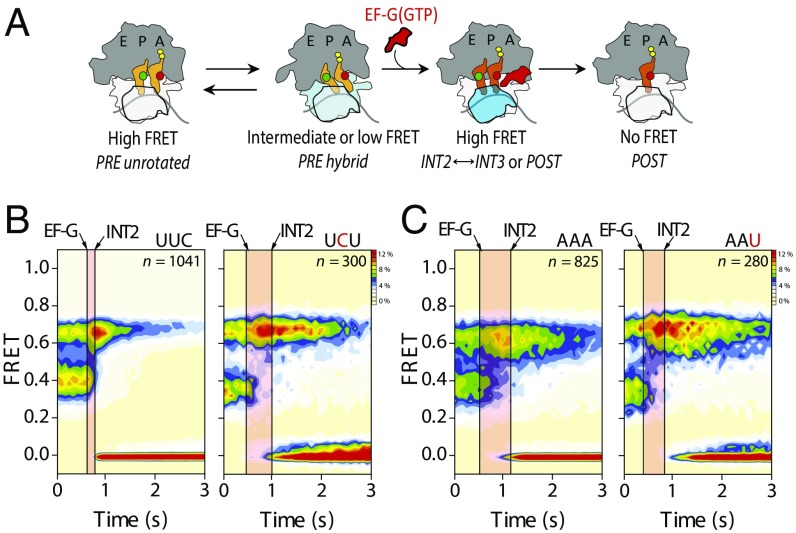
We first monitored the translocation of LD550-S13 (donor)– and LD650–peptidyl-tRNA (acceptor)–labeled PRE complexes bearing fMet-Lys-tRNALys in the A site (Fig. 2A), wherein the mRNA codon was either cognate (AAA) or programmed with a uridine mismatch at the first, second, or third position (Fig. 2B). As previously reported (11), PRE complexes labeled in this manner exhibited a mean FRET value of 0.13 ± 0.06, in line with the estimated distance between the N terminus of ribosomal protein S13 and peptidyl-tRNA in the PRE complex (Table S1). This FRET value represents both unrotated and rotated states of the PRE complex, which spontaneously exchange on a subsecond time scale (11, 28, 29).
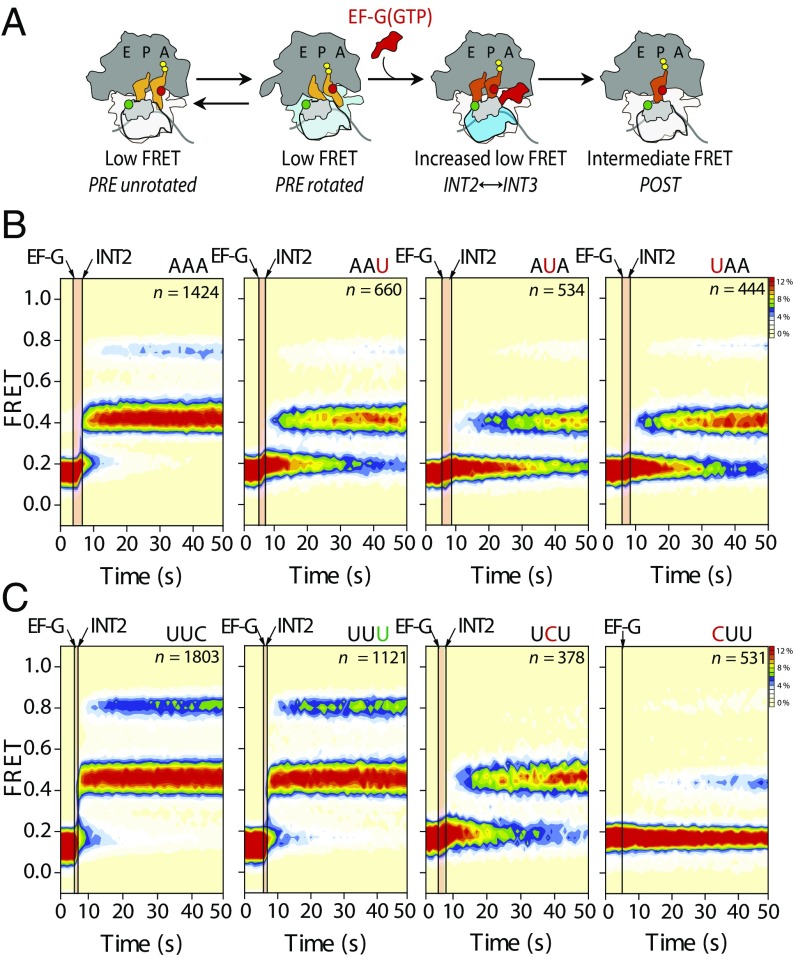
Fig. 2.
PRE complexes bearing miscoded peptidyl-tRNA within the A site display marked translocation defects. (A) Schematic of the translocation reaction coordinate monitored by an smFRET labeling strategy in which the N terminus of S13 (gray) and peptidyl-tRNA (orange) have been site-specifically labeled with donor (green) and acceptor (red) fluorophores, respectively (Materials and Methods). (B) Population FRET histograms showing pre–steady-state translocation of S13/tRNA-labeled PRE complexes bearing peptidyl-tRNALys programmed with cognate (AAA) and near-cognate (AAU, AUA, and UAA) mRNA codons. (C) Population FRET histograms showing pre–steady-state translocation of S13/tRNA-labeled PRE complexes bearing peptidyl-tRNAPhe programmed with cognate (UUC and UUU) and near-cognate (UCU and CUU) mRNA codons. Data were acquired at a 1-s time resolution. The number (n) of individual FRET trajectories in each histogram is indicated.
Table S1.
Approximate distances (Å) between sites of labeling within the ribosome
| Signal/structure | PRE (Classical) | PRE (Hybrid) | INT2 (FA) | POST |
| tRNA/tRNA | 35.7 (∼0.65 FRET) | 46.2 (0.2, 0.38 FRET) | 39.9 (∼0.65 FRET) | 39.9 (∼0.65 FRET/no FRET) |
| S13/tRNA (A-site acp3U47) | 74.7 (0.13 FRET) | 75.8 (0.13 FRET) | 71.3 (∼0.2 FRET) | 54.4 (0.42 FRET) |
| S13/L5 | 35.7 (0.75 FRET) | 44.3 (0.58 FRET) | 53.3 (∼0.50 FRET) | 37.1 (0.75 FRET) |
The interresidue distances between the various sites of labeling used in this study were determined from previously published structures along the translocation reaction coordinate: PRE (Classical; PDB ID code 4V6N) (14), PRE (Hybrid; PDB ID code 4V7D) (15), INT2 (FA-stabilized intermediate; PDB ID code 4V7B) (16), and POST (PDB ID code 4V5F) (17). The corresponding FRET states assigned to the structural pictures are indicated in parentheses for the specific complexes.
As expected (11), stopped-flow delivery of a saturating concentration (10 μM) of EF-G(GTP) to PRE complexes bearing a cognate codon-anticodon pair led to a rapid, transient increase in the mean FRET value (to ∼0.2 FRET), corresponding to INT2 formation, followed by a rate-limiting transition into intermediate-FRET (0.42 ± 0.06) or high-FRET (0.74 ± 0.06) POST conformations (Fig. 2B, Table 1, and Table S1). The “classical POST” state exhibits intermediate FRET, which reversibly fluctuates to a hybrid-like, high-FRET POST state, identified as such by puromycin release of the nascent peptide (Fig. S1). Strikingly, complexes programmed with mismatches in the codon-anticodon pair displayed dramatic (six- to 12-fold) defects in the overall translocation rate [kPOST; the rate of POST-state formation from the time of EFG-(GTP) injection] that could be specifically ascribed to the INT2-to-POST transition (Fig. 2B and Table 1). Even more dramatic reductions in translocation rates were observed between analogous cognate and mismatched PRE complexes bearing fMet-Phe-tRNAPhe in the A site (up to 80-fold defects; Fig. 2C, Table 1, and Fig. S1). For both systems, the observed translocation defects correlated with a reduction in rates of aminoacylated tRNA accommodation into the A site and peptidyl-tRNA dissociation after accommodation (SI Materials and Methods, Fig. S2, and Table S2). These findings suggest that mRNA codon/peptidyl-tRNA anticodon mismatches in the A site arising from errors during tRNA selection principally affect late steps in the process of translocation that occur after the INT2 complex is formed (Fig. 1B).
Table 1.
Rates of POST complex formation for PRE complexes programmed with matched and mismatched peptidyl-tRNA anticodon/mRNA codon pairs
| Codon | kPOST, s−1 | kPOST,cog/kPOST,ncog |
| AAA (COG-Lys) | 0.62 ± 0.01 | 1.0 ± 0.0 |
| AAU (NCOG-Lys) | 0.11 ± 0.01 | 5.8 ± 0.2 |
| AUA (NCOG-Lys) | 0.05 ± 0.01 | 12.3 ± 0.5 |
| UAA (NCOG-Lys) | 0.08 ± 0.02 | 7.6 ± 0.3 |
| UUC (COG-Phe) | 2.99 ± 0.24 | 1.0 ± 0.1 |
| UUU (COG-Phe) | 2.82 ± 0.34 | 1.1 ± 0.2 |
| UCU (NCOG-Phe) | 0.07 ± 0.01 | 40.4 ± 7.3 |
| CUU (NCOG-Phe) | 0.04 ± 0.01 | 74.8 ± 15.2 |
The ratios of kPOST [equivalent to krelocking described previously (11)] for cognate (COG) and near-cognate (NCOG) programmed PRE complexes are shown to indicate the relative translocation defect.
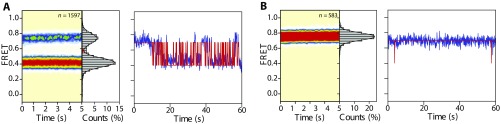
Fig. S1.
S13/tRNA-labeled POST complexes dynamically exchange between classical and hybrid states with peptidyl-tRNA bound within the P site. (A, Left) Population FRET histogram of S13/tRNA-labeled POST complexes bearing fMet-Phe-tRNAPhe programmed with a cognate (UUC) codon. (A, Right) Representative S13/tRNA FRET trace for the POST complex. (B, Left) Population FRET histogram of the POST complex shown in A following peptide release by incubation with puromycin (10 min, 25 °C, 2 mM puromycin). (B, Right) Representative S13/tRNA FRET trace for the puromycin-released POST complex. Comparing Fig.2 B and C, we observe that the stability of these hybrid-like peptidyl-tRNA configurations is sensitive to the nature of the nascent peptide. Data were acquired at a 40-ms time resolution. The number (n) of individual FRET trajectories in each histogram is indicated.
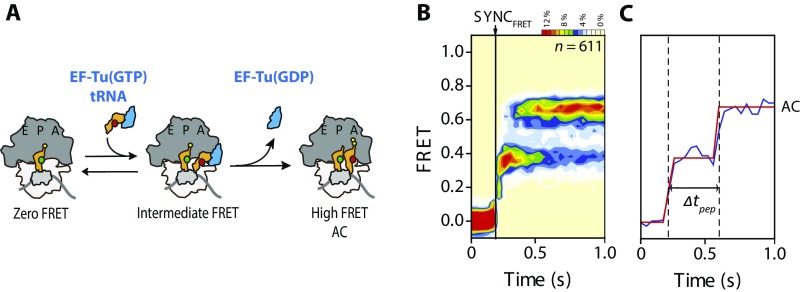
Fig. S2.
Quantitation of the rates of aminoacyl (aa)-tRNA accommodation for PRE complexes programmed with cognate and near-cognate peptidyl-tRNA anticodon/mRNA codon pairs in the A site. (A) Rate of aa-tRNA accommodation at high (15 mM) magnesium concentrations was calculated as described by tracking FRET between acceptor (red)-labeled EF-Tu(GTP)/aa-tRNA ternary complex and donor (green)-labeled 70S initiation complexes programmed with specific mRNA codons in the A site (Table S2). (B) Following delivery of EF-Tu(GTP)/aa-tRNA ternary complex (UUC in this histogram), traces are synchronized to the appearance of FRET to analyze the population behavior of tRNA selection. A fully formed PRE complex (one in which peptide bond formation has occurred after completion of tRNA selection) is defined as exhibiting a relatively stable (>120-ms duration) high-FRET (accommodated) state. (C) Accommodation time Δtpep was defined as the time between the first nonzero FRET dwell and the first stable (>120 ms) high-FRET–state dwell. kpep was calculated as the average accomodation rate based on exponential fits to the Δtpep distribution. Data were acquired at a 40-ms time resolution. The number (n) of individual FRET trajectories in each histogram is indicated.
Table S2.
Rate of aminoacyl-tRNA accommodation during tRNA selection at 15 mM Mg2+, rates of peptidyl-tRNA dissociation from the A site after accommodation at 5 mM Mg2+, and A-site tRNA occupancy at the time of translocation at 5 mM Mg2+ (relative to the occupancy measured after the formation of the PRE complex at 15 mM Mg2+)
| Codon | kpep, s−1 | kdiss, ×10−4⋅s−1 | O, % |
| AAA (COG-Lys) | 10.8 ± 2.1 | <5 ± 1 | >94 ± 1 |
| AAU (NCOG-Lys) | 3.8 ± 0.9 | 12 ± 3 | 87 ± 4 |
| AUA (NCOG-Lys) | 1.5 ± 0.4 | 179 ± 12 | 12 ± 2 |
| UAA (NCOG-Lys) | 3.9 ± 0.7 | 91 ± 15 | 33 ± 4 |
| UUC (COG-Phe) | 16.7 ± 1.8 | <5 ± 1 | >94 ± 1 |
| UUU (COG-Phe) | 15.3 ± 1.7 | <5 ± 1 | >94 ± 1 |
| UCU (NCOG-Phe) | 2.2 ± 0.4 | 209 ± 21 | 8 ± 2 |
| CUU (NCOG-Phe) | 1.2 ± 0.3 | 258 ± 17 | 5 ± 1 |
For the rates of accommodation of the tRNAs at 15 mM Mg2+, the formula kpep = [(koff/kon) + 1]/[(kpepnet)−1 − (kon)−1] was used, where koff, kon, and kpepnet are the rates of tRNA release from the A site, tRNA binding to the A site, and net tRNA accommodation from ternary complex delivery, respectively, according to the tRNA/tRNA signal (Fig. S2). The dissociation rate, kdiss, upon the shift to 5 mM Mg2+ was obtained by tracking the loss of LD650-labeled tRNA fluorescence over time by direct excitation. The corresponding A-site tRNA occupancy (O) at the moment of recording (2 min after the Mg2+ concentration shift) was calculated using kdiss. The rates of tRNA selection were calculated as described in Materials and Methods. COG, cognate; NCOG, near-cognate.
Early Translocation Steps Are only Modestly Altered by Codon-Anticodon Mismatches.
To specifically ascertain whether, and to what extent, steps preceding INT2 formation are affected by mismatches in the mRNA codon/peptidyl-tRNA anticodon pair, we monitored the translocation of donor- and acceptor-labeled deacylated and peptidyl-tRNAs (11, 32, 36) (Fig. 3A). To do so, PRE complexes were programmed with deacylated tRNAfMet (Cy3-s4U8-tRNAfMet) in the P site and either fMet-Phe-tRNAPhe or fMet-Lys-tRNALys (LD650-acp3U47-tRNAPhe/Lys) in the A site. In this experiment, we compared the translocation of PRE complexes programmed with cognate peptidyl-tRNAs in the A site with those with codon-anticodon mismatches in the A site most prone to miscoding (UCU, AAU; Table S2).
Fig. 3.
PRE complexes bearing matched and mismatched peptidyl-tRNA anticodon/mRNA codon pairs within the A site exhibit only modest differences in the rates of early translocation steps. (A) Schematic of the translocation reaction coordinate monitored in tRNA/tRNA-labeled PRE complexes, where the sites of donor (deacylated-tRNA) and acceptor (peptidyl-tRNA) fluorophore labeling are indicated with green and red circles, respectively. (B) Population FRET histograms showing pre–steady-state EF-G–catalyzed translocation of tRNA/tRNA-labeled PRE complexes bearing peptidyl-tRNAPhe in the A site programmed with either a cognate (Left, UUC) or near-cognate (Right, UCU) mRNA codon. The time delay between EF-G(GTP) addition and the average time to achieve the INT2 state is indicated. (C) Population FRET histograms showing pre–steady-state EF-G–catalyzed translocation of tRNA/tRNA-labeled PRE complexes bearing peptidyl-tRNALys in the A site programmed with either a cognate (Left, AAA) or near-cognate (Right, AAU) mRNA codon. Data were acquired at a 40-ms time resolution. The number (n) of individual FRET trajectories in each histogram is indicated.
As previously established (32, 36), both cognate (UUC, AAA) PRE complexes were highly dynamic, spontaneously transitioning between classical (A/A, P/P), hybrid-1 (H1; A/P, P/E), and hybrid-2 (H2; A/A, P/E) configurations on the subsecond time scale. These distinct ribosome conformations exhibited mean FRET values of ∼0.7 (high), ∼0.4 (intermediate), and ∼0.2 (low), respectively (Fig. 3 B and C and Table S3). As expected (11), EF-G(GTP)–catalyzed translocation of both complexes proceeded via the specific depletion of H1 and H2 hybrid states, rapidly enriching the high-FRET, tRNA-compacted INT2 configuration, followed by a loss of FRET upon deacylated-tRNA release (Fig. 3A).
Table S3.
FRET-state occupancies (%) for tRNA/tRNA-labeled PRE complexes
| Complex codon/FRET state | Low | Intermediate | High |
| UUC (COG) | 18 ± 2 | 38 ± 3 | 44 ± 5 |
| UCU (NCOG) | 11 ± 1 | 27 ± 1 | 62 ± 1 |
| AAA (COG) | 16 ± 1 | 31 ± 1 | 53 ± 2 |
| AAU (NCOG) | 23 ± 6 | 25 ± 3 | 52 ± 7 |
The FRET values corresponding to UUC were 0.67 ± 0.07 (high), 0.38 ± 0.1 (intermediate), and 0.20 ± 0.09 (low). For AAA, they were 0.64 ± 0.06 (high), 0.39 ± 0.13 (intermediate), and 0.20 ± 0.10 (low). For UCU, they were 0.69 ± 0.06 (high), 0.36 ± 0.13 (intermediate), and 0.19 ± 0.10 (low). For AAU, they were 0.63 ± 0.06 (high), 0.38 ± 0.13 (intermediate), and 0.19 ± 0.10 (low). COG, cognate; NCOG, near-cognate.
PRE complexes bearing mismatches in the A site also exhibited spontaneous fluctuations between classical and hybrid states (Table S3). As for PRE complexes bearing cognate peptidyl-tRNA, stopped-flow addition of EF-G(GTP) (10 μM) resulted in the specific depletion of hybrid configurations in both complexes (UCU and AAU), followed by rapid enrichment of a high-FRET, INT2-like configuration, and the loss of FRET (Fig. 3 B and C). For both systems, the rate of INT2 formation was either reduced or increased by approximately two- to fourfold, depending on the position of the codon-anticodon mismatch and peptidyl-tRNA identity (Fig. 3 B and C and Table S4). These results indicate that translocation steps preceding INT2 formation are only modestly affected by mRNA codon/peptidyl-tRNA anticodon mismatches in the A site (37). The observed rates of deacylated tRNA release from the E site parallel these modest defects (Table S4). Akin to FA-induced stalling of the translocation mechanism (11), the rates of deacylated tRNA release were nearly five- to 10-fold faster than formation of the POST complex (Table 1 and Table S4) and 10- to 50-fold faster than the rate of fluorophore photobleaching (∼0.06 s−1). We conclude from these findings that the most substantial translocation defect (∼98% for UCU, ∼95% for AAU) is subsequent to achieving the INT2 conformation.
Table S4.
Rates of INT2 formation and deacylated tRNA release from tRNA/tRNA-labeled PRE complexes (tRNA-tRNA FRET)
| Codon | kINT2, s−1 | kINT2, release, s−1 | krelease, s−1 | kPOST, s−1 |
| UUC (COG-Phe) | 8.5 ± 0.8 | 3.5 ± 0.3 | 2.4 ± 0.1 | 2.99 ± 0.24 |
| UCU (NCOG-Phe) | 2.3 ± 0.2 | 0.8 ± 0.3 | 0.6 ± 0.1 | 0.07 ± 0.01 |
| AAA (COG-Lys) | 1.5 ± 0.1 | 1.1 ± 0.3 | 0.6 ± 0.1 | 0.62 ± 0.01 |
| AAU (NCOG-Lys) | 3.6 ± 0.3 | 0.6 ± 0.2 | 0.5 ± 0.1 | 0.11 ± 0.01 |
The rate of INT2 formation (kINT2), the rate of deacylated tRNA release after INT2 formation (kINT2, release), and the rate of deacylated tRNA release from the E site from the moment of EF-G(GTP) injection (krelease), derived from tRNA/tRNA-labeled PRE complexes bearing peptidyl-tRNAPhe or peptidyl-tRNALys in the A site as indicated (Fig. 3), are shown. Rates of translocation (kPOST) derived from analogous measurements of S13/tRNA-labeled PRE complexes (Fig. 2A) are also shown. Rates were calculated as described in Materials and Methods. COG, cognate; NCOG, near-cognate.
mRNA Codon/Peptidyl-tRNA Anticodon Mismatches Inhibit Late Steps in Translocation.
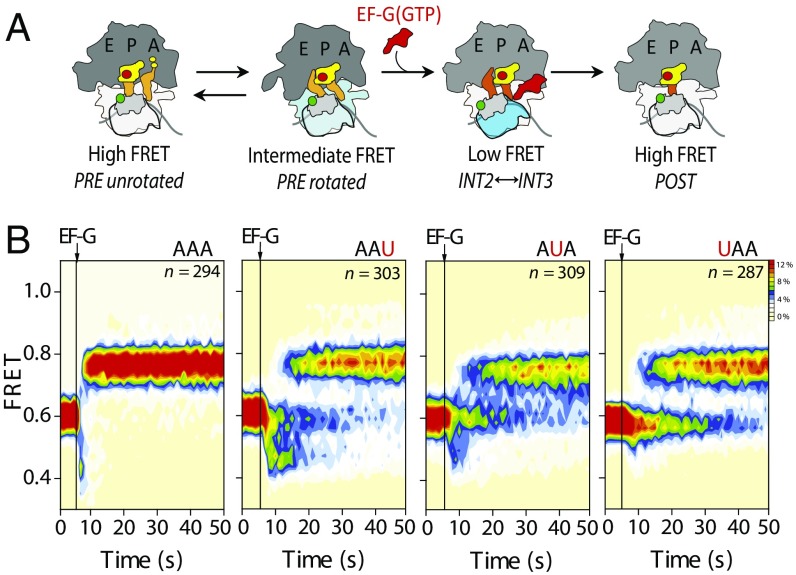
To probe the impact of codon-anticodon mismatches in the A site on small subunit head domain motions accompanying translocation, we evaluated PRE complexes labeled at ribosomal proteins S13 (donor, LD550) and L5 (acceptor, LD650) (11) (Fig. 4A). Complexes labeled in this manner bearing deacylated tRNAfMet in the P site and a cognate fMet-Lys-tRNALys in the A site displayed spontaneous fluctuations between intermediate- and high-FRET states. As previously established, these states reflect the interconversion between rotated (hybrid) and unrotated (classical) conformations of the PRE complex, respectively (11, 29).
Fig. 4.
PRE complexes bearing matched and mismatched peptidyl-tRNA anticodon/mRNA codon pairs within the A site exhibit marked differences in the rates of late translocation steps. (A) Schematic of the translocation reaction coordinate monitored in S13/L5-labeled PRE complexes, where the sites of donor (S13) and acceptor (L5) fluorophore labeling are indicated with green and red circles, respectively. (B) Population FRET histograms showing pre–steady-state EF-G–catalyzed translocation of S13/L5-labeled PRE complexes bearing peptidyl-tRNALys in the A site programmed with either a cognate (AAA) or near-cognate (AAU, AUA, or UAA) mRNA codon. Data were acquired at a 1-s time resolution. The number (n) of individual FRET trajectories in each histogram is indicated.
As expected (11), stopped-flow EF-G(GTP) addition (10 μM) to PRE complexes programmed with cognate peptidyl-tRNA (AAA) rapidly and efficiently converted the rotated population (0.58 ± 0.07 FRET) to an unrotated (0.75 ± 0.05 FRET) POST configuration, via transient, lower FRET (<0.5) conformations (Figs. 1B and 4B and Table S5). These lower FRET states principally reflect the transit between INT2 (∼0.5 FRET) and INT3 (∼0.4 FRET) conformations on the path to POST complex formation (11). In stark contrast, complexes bearing first-, second-, or third-position mismatches exhibited extended periods of delay before POST state formation in which lower FRET states were exhibited (Fig. 4B and Table S5). Similar delays were observed in phenylalanine-mismatched complexes (Fig. S3).
Table S5.
Translocation rates for S13/L5-labeled PRE complexes
| Codon/Rate | kPOST, s−1 |
| AAA (COG-Lys) | 0.52 ± 0.01 |
| AAU (NCOG-Lys) | 0.10 ± 0.02 |
| AUA (NCOG-Lys) | 0.06 ± 0.01 |
| UAA (NCOG-Lys) | 0.09 ± 0.01 |
| UUC (COG-Phe) | 2.36 ± 0.43 |
| UCU (NCOG-Phe) | 0.09 ± 0.02 |
Results are similar to those obtained for S13/tRNA-labeled PRE complexes (Table 1). Translocation rates were calculated as described in Materials and Methods. COG, cognate; NCOG, near-cognate.
Fig. S3.
EF-G–catalyzed translocation of S13/L5-labeled PRE complexes bearing peptidyl-tRNAPhe in the A site. (A) Translocation of PRE complexes programmed with the cognate (UUC) codon. Data were collected at a slow imaging frame rate (1 s−1) to capture the complete translocation reaction coordinate for both cognate and near-cognate PRE complexes. At this time resolution, cognate complexes transit INT2 and INT3 states in less than a single frame. Hence, the example FRET trace shown was obtained at 40 ms for the same complexes to illustrate the fluctuations between INT2 and INT3 states en route to POST complex formation (high FRET). (B) Translocation of PRE complexes programmed with the near-cognate (UCU) codon. A contour plot and a single-molecule trace show prolonged delays in the exchange between INT2 and INT3 states: population FRET histogram (Left, 1 s) and single-molecule trace (Right, 40 ms). The number (n) of individual FRET trajectories in each histogram is indicated. Rot., rotated.
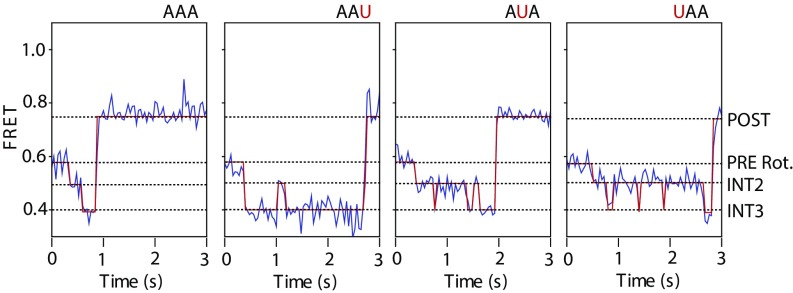
In line with these defects reflecting the engagement of the mRNA codon/peptidyl-tRNA anticodon pair at the P site, the position of the codon-anticodon mismatch had strong and distinct influences on the progression through late (lower FRET) translocation intermediates (Fig. 4B). Visual inspection of smFRET recordings revealed direct evidence of prolonged periods of reversible fluctuations between INT2 and INT3 FRET states before POST complex formation (Fig. 5 and Fig. S1B). These findings provide compelling evidence that P-site recognition, achieved via the INT3 conformation, is the rate-limiting step in substrate translocation on the bacterial ribosome.
Fig. 5.
PRE complexes bearing matched and mismatched peptidyl-tRNA anticodon/mRNA codon pairs within the A site exhibit extended periods of INT2-INT3 exchange on the path to the POST state. Representative S13/L5 translocation signal FRET traces for PRE complexes programmed with lysine cognate AAA and near-cognate AAU, AUA, and UAA codons. Data were acquired at a 40-ms time resolution. Rot., rotated.
EF-G Remains Bound to the Mismatch-Stalled Translocation Intermediates.
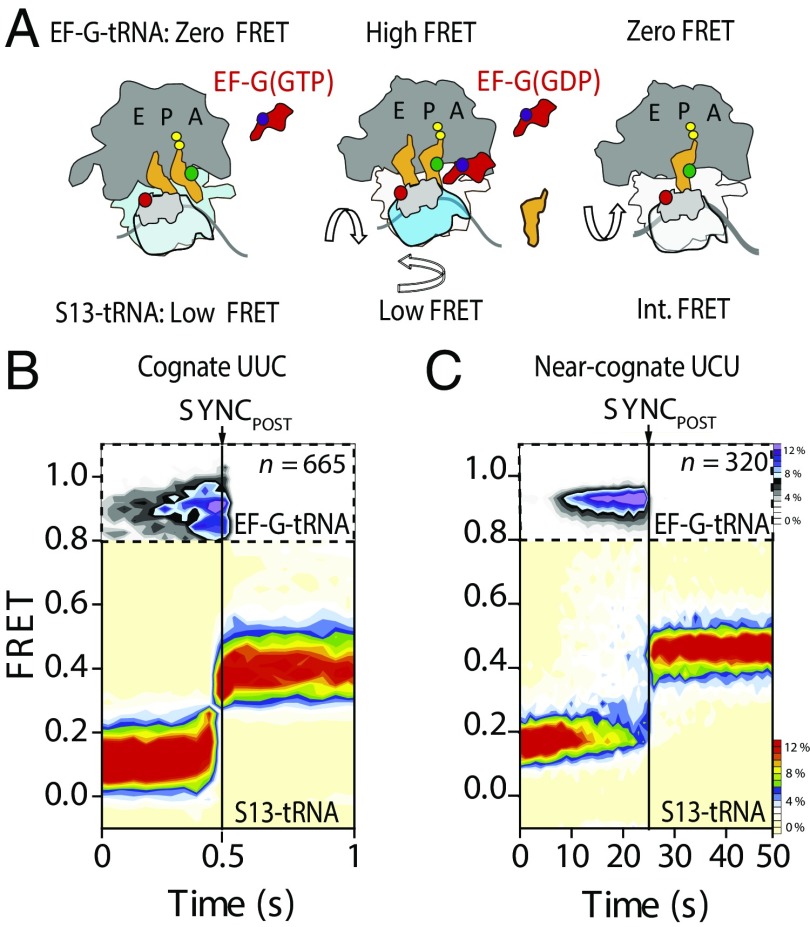
The translocation of cognate peptidyl-tRNA takes place while EF-G is bound to the ribosome (11). To establish whether EF-G remains affixed to the A site during the prolonged translocation of mismatched peptidyl-tRNAs, we performed three-color imaging to track the residence time of EF-G on the ribosome during translocation via FRET signals from donor-labeled (Cy3B) peptidyl-tRNA to acceptor-labeled (LD650) S13 and to acceptor-labeled (LD750) EF-G(GTP) (11) (Materials and Methods and Fig. 6A). For clarity, individual FRET recordings were synchronized to the loss of FRET between EF-G and peptidyl-tRNA.
Fig. 6.
EF-G resides on the ribosome through POST-state formation for PRE complexes bearing matched and mismatched peptidyl-tRNA anticodon/mRNA codon pairs within the A site. (A) Scheme of the three-color smFRET strategy to simultaneously image EF-G binding and translocation. The donor (green) fluorophore on the A-site tRNA has simultaneous FRET to an acceptor fluorophore (red) on protein S13 (light gray) in the small subunit head domain and to an acceptor fluorophore (purple) on EF-G (red). Int., intermediate. Population FRET histograms show the evolution of FRET between EF-G and peptidyl-tRNA (Top) and peptidyl-tRNA and ribosomal protein S13 (Bottom) in which both are postsynchronized (SyncPOST) to the loss of FRET between EF-G and tRNA for PRE complexes programmed with phenylalanine cognate (B, UUC) and near-cognate (C, UCU) codons. Data were acquired at 40-ms and 1-s time resolutions for cognate and near-cognate programmed PRE complexes, respectively. The number (n) of individual FRET trajectories in each histogram is indicated.
In line with the estimated rate of EF-G turnover in vitro under conditions of processive translation (∼1–3 s−1) (10, 11, 38, 39), the residence time of EF-G on cognate PRE complexes programmed with fMet-Phe-tRNAPhe in the A site was ∼250 ms (Fig. 6B). By contrast, a second position mismatch in the codon-anticodon pair increased the EF-G residence time 40-fold to 10 s (Fig. 6C). These data indicate that EF-G remains bound to the ribosome for the entire duration of the translocation stall until POST complex formation has been achieved (Table 1).
The Miscoding Translocation Defect in Aminoglycoside Action.
Aminoglycosides target the ribosome to affect the accuracy of aminoacyl-tRNA selection at the A site (40). Although known to increase miscoding rates, this effect alone is thought to be insufficient to explain their bactericidal activity (41, 42). Aminoglycosides have also been shown to bind to cognate PRE complexes to reduce the rates of EF-G–catalyzed translocation (43–47) and EF-G–catalyzed ribosome recycling (48). These perturbations to protein synthesis have been attributed to drug binding to the central bridge B2 region, including the helix 44 (h44) decoding site within the small subunit 16S rRNA and Helix 69 (H69) within the large subunit 23S rRNA (28, 49, 50). Aminoglycoside binding to h44 locally restructures the decoding center to induce extrahelical conformations of the universally conserved A1492 and A1493 residues (50, 51) to promote direct interactions with the codon–anticodon complex that promote miscoding (50, 52). Aminoglycoside binding to H69 impacts dynamic processes in the ribosome, including reversible subunit rotation (53).
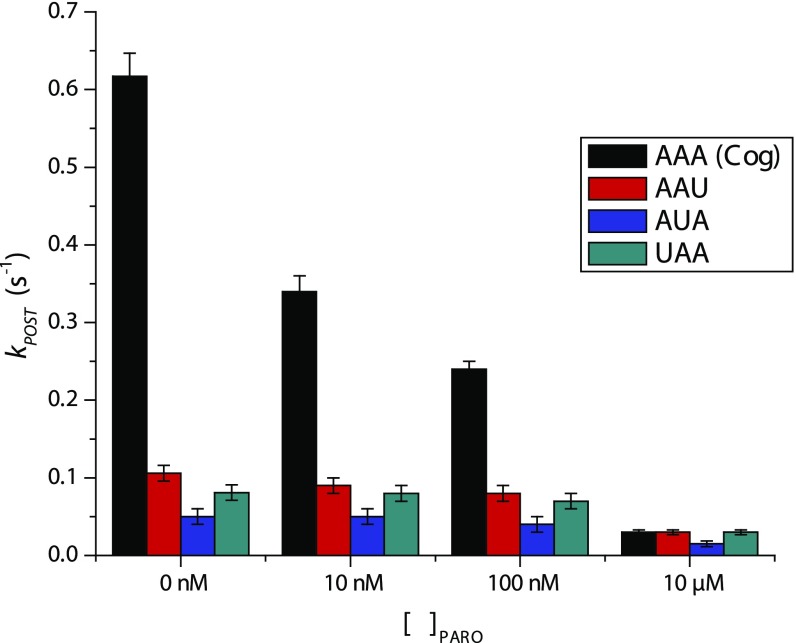
We examined the impacts of the 4,6-linked aminoglycoside paromomycin on the mechanism of translocation in the absence and presence of mismatches in the codon-anticodon pair. For these experiments, Lys-tRNALys was incorporated into the A site in the presence of 100 nM paromomycin, followed by either buffer exchange to remove the drug (29) or rinsing with buffer containing a specified paromomycin concentration to probe the drug’s additional impacts on the translocation mechanism. Consistent with efficient paromomycin removal from the ribosome after buffer exchange, cognate PRE complexes bearing fMet-Lys-tRNALys in the A site translocated at a rate indistinguishable from the uninhibited process (Table 1 and Fig. S4). In the presence of paromomycin (10 μM), the rate of translocation was reduced by up to 20-fold (Fig. S4).
Fig. S4.
Effect of paromomycin (PARO) on the translocation rates of PRE complexes programmed with cognate (Cog) and near-cognate peptidyl-tRNA anticodon/mRNA codon pairs in the A site. PRE complexes were prepared by incubating LD650-labeled Lys-tRNALys ternary complex with ribosomes bearing LD550-labeled S13 and programmed with the specified cognate or near-cognate codon in the A site. Cognate and near-cognate tRNA selection at the A site was performed in the presence of 100 nM paromomycin, followed by buffer exchange into Tris-polymix buffer containing 5 mM Mg2+ and the indicated paromomycin concentration. Translocation rates (kPOST) were calculated as described in Materials and Methods.
Analogous experiments performed on PRE complexes bearing mismatches in the mRNA codon/peptidyl-tRNA anticodon pair revealed that paromomycin only modestly reduced the rate of translocation beyond that caused by the mismatch alone (Fig. S4) up to a concentration of 100 nM. Notably, the concentrations of paromomycin required to substantially impact translocation rates appeared higher for near-cognate PRE complexes than for cognate PRE complexes.
EF-P Can Rescue Translocation Defects Arising from A-Site Miscoding.
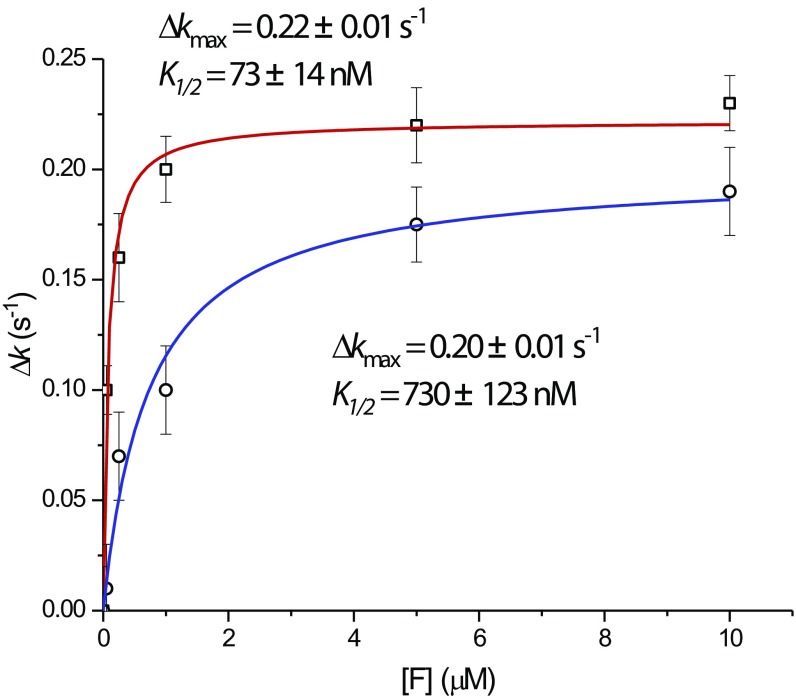
EF-P binds the ribosome between the ribosomal P and E sites (30). Interactions between domain I of EF-P and the CCA end of the P-site tRNA stimulate peptide bond formation in PRE complexes stalled by specific motifs, including polyproline sequences (54–57). Surprisingly, we find that saturating EF-P concentrations (10 μM) efficiently rescued miscoding-induced translocation defects (Fig. 7A and Table S6). These results reveal that ribosome complexes bearing mismatches within the peptidyl-tRNA anticodon/mRNA codon pair are specifically stalled at a stage before complete translocation after the release of deacylated tRNA from the E site (Fig. 3B and Table S4). Interestingly, EF-P lacking the β-lysinylation modification (K34A mutant; EF-Pm) that is essential to rescue of peptidyltransferase defects on the classically configured PRE complex (55, 56, 58, 59) also exhibited an ∼10-fold higher K1/2 (730 ± 123 nM, where K1/2 is the factor concentration at which the increment in the translocation rate is half its maximum) than the WT protein (73 ± 14 nM) for the translocation intermediate (Fig. S5 and Table S6).
Fig. 7.
EF-P partially rescues the translocation defect of PRE complexes bearing mismatched peptidyl-tRNA anticodon/mRNA codon pairs within the A site. (A) S13/tRNA translocation signal FRET histograms for PRE complexes programmed with lysine cognate (AAA) and near-cognate (UAA) codons, as well as the near-cognate codon-programmed complex with additional delivery of saturating EF-P (10 μM). Data were acquired at a 1-s time resolution. The number (n) of individual FRET trajectories in each histogram is indicated. (B) Model for EF-P–mediated rescue of stalled translocation of ribosomes with mismatched anticodon-codon pairs in the A site.
Table S6.
EF-P and its K34A mutant EF-Pm rescue PRE complexes stalled during translocation by mismatches between the peptidyl-tRNA anticodon and mRNA codon
| Codon | kPOST, s−1 | D = kPOST,cog/kPOST,ncog | kPOST, EF-P, s−1 | DEF-P = kPOST,cog/kPOST, EF-P | REF-P = D/DEF-P | kPOST, EF-Pm, s−1 | DEF-Pm = kPOST,cog/kPOST, EF-Pm | REF-Pm = D/DEF-Pm |
| AAA (COG) | 0.62 ± 0.01 | 1.0 ± 0.0 | 0.63 ± 0.02 | 0.98 ± 0.0 | 1.1 ± 0.0 | 0.55 ± 0.02 | 1.1 ± 0.0 | 0.9 ± 0.0 |
| AAU (NCOG) | 0.11 ± 0.01 | 5.8 ± 0.2 | 0.34 ± 0.04 | 1.8 ± 0.2 | 3.2 ± 0.4 | 0.30 ± 0.03 | 2.1 ± 0.2 | 2.8 ± 0.3 |
| AUA (NCOG) | 0.05 ± 0.01 | 12.3 ± 0.5 | 0.42 ± 0.04 | 1.5 ± 0.1 | 8.2 ± 0.6 | 0.35 ± 0.02 | 1.8 ± 0.1 | 6.8 ± 0.5 |
| UAA (NCOG) | 0.08 ± 0.02 | 7.6 ± 0.3 | 0.42 ± 0.03 | 1.5 ± 0.1 | 5.1 ± 0.4 | 0.52 ± 0.03 | 1.2 ± 0.1 | 6.3 ± 0.6 |
| UUC (COG) | 2.99 ± 0.24 | 1.0 ± 0.1 | 2.99 ± 0.71 | 1.0 ± 0.3 | 1.0 ± 0.3 | 2.82 ± 0.52 | 1.1 ± 0.2 | 0.9 ± 0.2 |
| UCU (NCOG) | 0.07 ± 0.01 | 42.7 ± 7.0 | 0.30 ± 0.05 | 10.0 ± 1.8 | 4.3 ± 1.0 | 0.32 ± 0.07 | 9.3 ± 2.2 | 4.6 ± 1.3 |
| CUU (NCOG) | 0.04 ± 0.01 | 74.8 ± 15.2 | 0.04 ± 0.01 | 74.8 ± 15.2 | 1.0 ± 0.3 | 0.05 ± 0.01 | 59.8 ± 12.9 | 1.3 ± 0.4 |
Shown are the rates of translocation (kPOST) for the various PRE complexes; the defect of the mismatched complexes translocation (D); the rate of translocation in the presence of 10 μM EF-P (kPOST, EF-P); the defect in translocation in the presence of EF-P (DEF-P); the EF-P “rescue factor” (REF-P; the fraction between D and DEF-P); the rates of translocation in the presence of 10 μM EF-Pm, the K34A mutant, the defect of translocation in the presence of EF-Pm (DEF-Pm), and the EF-Pm rescue factor (REF-Pm; the fraction between D and DEF-Pm). Translocation rates were calculated as described in Materials and Methods. COG, cognate; NCOG, near-cognate.
Fig. S5.
Fully modified (WT) and unmodified (K34A, EF-Pm) isoforms of EF-P have distinct capacities to rescue miscoding-stalled late intermediates of translocation. The plot shows the translocation rate difference of PRE complexes programmed with near-cognate AAU as a function of the concentration of the modified WT EF-P and mutant (K34A, EF-Pm). Δk = kEF-P − k0, where k0 = 0.11 s−1 (the rate of translocation observed in the absence of EF-P factors; Table 1). Data were well described by hyperbolic functions (Δk = Δkmax[F]/(K1/2 + [F]), where K1/2 is the factor concentration [F] at which the increment in the translocation rate is half its maximum) for both EF-P (□) and EF-Pm (○). Notably, the K1/2 of EF-Pm is 10.0 ± 2.6-fold higher than the K1/2 of WT EF-P. Translocation rates were calculated as described in Materials and Methods.
SI Materials and Methods
Materials.
GTP was purchased from Sigma and further purified on a Mono Q 5/50 GL ion exchange column (GE Healthcare Life Sciences). Kanamycin sulfate salt (Calbiochem), tetracycline hydrochloride (Sigma–Aldrich), ampicillin sodium salt (Amresco), paromomycin sulfate salt (Sigma-Aldrich), and FA sodium salt (Alexis Biochemicals) were used at the purity stated by the commercial suppliers (≥97%).
Preparation of Fluorescently Labeled Ribosomes, EF-G, and tRNAs.
Native 30S subunits enzymatically labeled on S13 (N-Sfp) with LD550 (Lumidyne Technologies), and 50S subunits labeled on L5 (N-Acp) with LD650 (Lumidyne Technologies) were prepared and purified as previously described (28, 29, 95). Cy3-fMet-tRNAfMet (s4U8) and Cy3B/LD650-Phe-tRNAPhe(acp3U47) were purified (98) and labeled (32) as previously described. The tRNALys was purchased (Chemical Block), purified by hydrophobic chromatography, and labeled with LD650 (acp3U47) as previously described (43).
Preparation of PRE Ribosome Complexes.
To prepare cognate and near-cognate complexes, surface-immobilized, empty A-site ribosomes were incubated in Tris-polymix buffer containing 15 mM Mg[OAc]2 with specific tRNA species (12 nM), together with elongation factor Tu (EF-Tu; 60 nM) and GTP (1 mM) for 15 min. Before EF-G–catalyzed translocation, PRE complexes were exchanged to Tris-polymix buffer containing 5 mM Mg[OAc]2 for 2 min. Pre–steady-state measurements of translocation were carried out by stopped-flow injection of EF-G (10 μM) together with GTP (1 mM) in Tris-polymix buffer containing 5 mM Mg[OAc]2 at 25 °C. The mixing time for such experiments was ∼100 ms.
Acquisition and Analysis of Two-Color smFRET Data.
Single-molecule fluorescence imaging was performed at 25 °C using a prism-based total internal reflection fluorescence microscope (32). Site-specifically labeled ribosome complexes programmed with 5′-biotinylated mRNA substrates were surface-immobilized within polyethylene glycol-passivated, streptavidin-coated, quartz microfluidic chambers (20, 32). Unless otherwise stated, all experiments were conducted in Tris-polymix buffer [50 mM Tris-OAc (pH 7.5), 100 mM KCl, 5 mM NH4OAc, 0.5 mM Ca(OAc)2, 0.1 mM EDTA, 5 mM putrescine, 1 mM spermidine, and 1.5 mM β-mercaptoethanol] with 5 mM Mg(OAc)2, a mixture of triplet-state quenchers (1 mM Trolox, 1 mM nitrobenzyl alcohol, and 1 mM cyclooctatetraene) (96), and an enzymatic oxygen scavenging system (97). All single-molecule imaging experiments were collected using a wide-field, prism-based total internal reflection fluorescence microscopy platform (99). Data were acquired using Hamamatsu ORCA-Flash 4.0(V2) sCMOS cameras controlled by home-made acquisition software written in LabVIEW. FRET measurements were performed by directly exciting the LD550/Cy3 donor fluorophore at 532 nm using a single-frequency laser (LaserQuantum), while simultaneously measuring fluorescence from both the LD550/Cy3 and LD650 fluorophores. Data were collected at a 40-ms or 1-s integration time. Using this approach, fluorescence and FRET trajectories could be obtained from thousands of surface-immobilized, fluorescently labeled ribosome complexes simultaneously, where an average of 500 photons were collected for each ribosome complex per image frame. Individual ribosome complexes were imaged before, during, and after stopped-flow injection of EF-G. The translocation of each complex was monitored at saturating EF-G concentration, and recorded single-molecule fluorescence and smFRET trajectories were extracted from the acquired images using the SPARTAN software package (99). Time-dependent donor and acceptor fluorescence intensities from individual ribosome complexes were transformed into FRET efficiency using the relationship, FRET = Iacceptor/(Idonor + Iacceptor). Traces were selected for analysis using the following criteria: a single catastrophic photobleaching event, at least a 8:1 signal-to-background noise ratio and 6:1 signal-to-signal noise ratio, less than four donor fluorophore blinking events, a correlation coefficient between donor and acceptor <0.5, and a lifetime of at least 50 frames in any nonzero FRET state ≥0.15. Reported FRET efficiency errors were obtained from the SD of normal distribution fits of compiled FRET histograms.
Individual FRET traces were analyzed by hidden Markov modeling-based idealization methods using the QuB software package, where kinetic analyses were conducted through idealization to specific models using the segmental k-means algorithm (32). In each model, transitions between all FRET states (including a photobleached/blinking state) were allowed. For the analysis of S13-tRNA FRET data, a two-state model was used with FRET values of 0.13, 0.42, and 0.74. For the analysis of tRNA-tRNA FRET data, a three-state model was used, with FRET values of 0.20, 0.38, and 0.66. For the analysis of S13-L5 FRET data, a two-state model was used with FRET values of 0.58 and 0.75.
Calculation of tRNA Accommodation and Translocation Rates.
The process of tRNA selection on the ribosome was monitored using smFRET by stopped-flow injection of Tris-polymix buffer containing ternary complex [60 nM EF-Tu, 12 nM Phe-tRNAPhe (LD650-acp3U47), and 1 mM GTP] to surface-immobilized ribosome complexes bearing deacylated tRNAfMet (Cy3-s4U8) in the P site, as previously described (31). Fluorescence and FRET traces were extracted and analyzed using automated analysis procedures implemented in SPARTAN (99) using the segmental k-means algorithm (100). Complete accommodation of Phe-tRNAPhe into the A site was marked by the first observation of a 120-ms dwell in a high (0.66 ± 0.07) FRET state, structurally assigned to the classical PRE complex configuration through previous investigations (31). This procedure was used to calculate the time from initial tRNA engagement of cognate and near-cognate tRNAs (codon recognition) to complete accommodation (Fig. S2).
Translocation rates, and the rates of forming specific translocation intermediates, were calculated as described by Wasserman et al. (11).
Monitoring FRET Between EF-G and the Ribosome.
To simultaneously monitor the translocation reaction coordinate and EF-G engagement with the A site, 150 nM LD750-labeled EF-G(GTP) was stopped-flow–delivered to surface-immobilized PRE complexes bearing LD650-labeled S13 and tRNAfMet in the P site and Cy3B-labeled fMet-Phe-tRNAPhe (acp3U47) in the A site. Cy3B was directly excited at 532 nm (LaserQuantum), while LD650 and LD750 intensities were simultaneously recorded. S13-tRNA FRET and EF-G–tRNA FRET, representing the relative changes in distance between acceptor 1 (S13) or acceptor 2 (EF-G) and the donor (A-site tRNA) during the translocation reaction coordinate, were calculated as follows: S13-tRNAFRET = ILD650/(ICy3B + ILD650) and EF-G–tRNAFRET = ILD750/(ICy3B + ILD750).
Translocating Traces Selection.
As previously described (11), traces undergoing translocation for S13-tRNA–labeled complexes were selected based on a shift from a low-FRET (PRE) state to a stable (1 s or longer dwell time) intermediate/high-FRET (POST) state. For PRE complexes in which both deacylated P-site and peptidyl A-site tRNA were labeled (tRNA-tRNA FRET), translocating traces were selected based on donor intensity loss (corresponding to labeled E-site tRNA loss) before acceptor intensity loss as well as high-FRET (intermediate/POST)–state occupation before FRET loss. E-site tRNA release rates were calculated by subtraction of the photobleaching rate from the net FRET loss rates. For the S13-L5–labeled complexes, translocating traces were selected based on FRET transition from the intermediate-FRET (rotated) state to the stable (1 s or longer dwell times) high-FRET (POST) state. The resulting times were compiled into histograms, transformed to cumulative distributions, and fit to exponential functions in Origin (OriginLab). Reported rate errors were obtained from the SDs of rates from three independent experiments.
Discussion
While the global rate and accuracy of translocation have been extensively investigated (37, 60–65), the specific impact of base-pairing mismatches in the peptidyl-tRNA anticodon/mRNA codon pair on the rate of peptidyl-tRNA translocation into the P site has yet to be directly examined in a pre–steady-state, single-turnover setting. The present investigations leverage the unique capacity of single-molecule methods to probe distinct subpopulations in a heterogeneous ensemble. By tracking the translocation reaction coordinate of the subpopulation of PRE complexes formed by miscoding at the A site from four distinct structural perspectives, we reveal direct evidence that mismatches in the mRNA codon/peptidyl-tRNA anticodon pair can reduce the rate of translocation by up to two orders of magnitude. Importantly, the translocation defects were observed to arise during late steps in the translocation mechanism. PRE complexes specifically displayed extended periods of fluctuation between the structurally defined INT2 state and the highly transient INT3 intermediate (Fig. 1B), whose structure is presently unknown.
The INT2 state (Fig. 1C) structure was trapped during the process of translocation using high concentrations of FA (11). This state is also referred to as the POST-like translocation intermediate (TI-POST) as both the mRNA and tRNA substrates are nearly fully translocated with respect to the small subunit body domain (12, 14, 15). The TI-POST is distinguished from the bona fide POST state in three structural respects. The small subunit head domain is swiveled by ∼18–20° in the direction of translocation, the small subunit body domain is rotated by ∼2–4° relative to the unrotated classical POST configuration, and peptidyl-tRNA has yet to fully engage the P site. As the movement of the peptidyl-tRNA codon-anticodon pair toward the P site entails a swivel-like motion of the small subunit head domain, together with reverse rotation of the small subunit body (13), we have hypothesized that INT2 ↔ INT3 exchange events reflect thermally driven, hyperswivel motions of the small subunit head domain within the EF-G(GDP)–bound ribosome. This model posits that the exaggerated movements of the head domain, together with reverse rotation of the body domain, enable peptidyl-tRNA to fully accommodate into the P site. This final step of mRNA and tRNA movement appears to rapidly trigger small subunit head domain relocking (reverse swivel) and POST complex formation from which EF-G(GDP) can swiftly release. Hence, the transit of peptidyl-tRNA from the A site to the P site (15) constitutes a P-site tRNA recognition process that is rate-limiting to the translocation mechanism on the bacterial ribosome and strongly dependent on proper pairing of the peptidyl-tRNA anticodon with the mRNA codon.
The precise nature of the miscoding-induced translocation defect requires further investigation. As has been observed for tRNA recognition at the A site (66), poorly matched mRNA codon-anticodon pairs may have impacts on shape-specific recognition at the P site as well as distortions in the peptidyl-tRNA body that are enforced by restructuring of the tRNA anticodon upon entrance and accommodation at the P site. The improper positioning of peptidyl-tRNA is, in turn, expected to affect EF-G interactions with the codon-anticodon pair as well as the ribosome. These alterations within the PRE complex increase the period in which fluctuations between INT2 and INT3 intermediates occur before the system resolves to the POST state (Fig. 5).
One of the most striking and important features of the observed translocation defect is that EF-G remains engaged with the ribosome until translocation has been completed (Fig. 6). Equally notable is that deacylated tRNA can release from the ribosome during the prolonged INT2 ↔ INT3 exchange (11) (Fig. 3). As the available INT2 (TI-POST) structures suggest considerable steric constraints on deacylated tRNA release from the E site (14, 15), we infer that tRNA dissociation occurs from the INT3 conformation, or a similar state, in which the E site is opened and solvent-accessible. Opening of the E site may include breaking of deacylated tRNA-L1 stalk interactions as well as exaggerated motions of the small subunit head domain. We conclude from these observations that EF-G contributes to maintaining the ribosome in a conformation where the small subunit P site exhibits low-affinity binding for improperly matched peptidyl-tRNA anticodon/mRNA codon pairs. The structural features of the INT3 complex that render the P site low-affinity and sensitive to the nature of the anticodon-codon pair may have important implications for ribosomal frameshifting (39, 67, 68), as well as the process of translation initiation, wherein the ribosomal P site must also exhibit low affinity for tRNA until the start site codon and initiator tRNA are properly matched (69).
The prolonging of translocation as a result of inappropriate aminoacyl-tRNA decoding at the A site is expected to have strong, negative impacts on cellular protein synthesis, particularly in the context of polysomes. The full repertoire of cellular mechanisms responsible for recognizing and resolving ribosome complexes stalled by miscoding errors is not presently known. In principle, such complexes could be dissociated through active mechanisms, where the release of peptidyl-tRNA would necessitate the actions of peptidyl-tRNA hydrolase (70, 71), or through rescue mechanisms that relieve the blockade. As the A site is occupied by EF-G, thus precluding mechanisms involving RelE, RelA, and tmRNA (26, 72–74), the recognition of such complexes may be most readily achieved through the vacant E site and/or the exaggerated positions of the small subunit head domain.
Consistent with this model, we find that EF-P can efficiently rescue translocation defects exhibited by miscoded complexes, albeit strongly dependent on the nature of the mismatch (Fig. 7A and Table S6). In the cell, EF-P’s impact on translocation is expected to hinge on the time delay between opening of the E site following deacylated tRNA release and peptidyl-tRNA engagement at the P site as well as EF-P’s concentration [estimated to range from 10–40 μM (5,000–20,000 copies per cell) (75, 76)]. These findings suggest that EF-P can potentially contribute to the cell’s tolerance of miscoding errors (42, 77, 78). Consistent with such a physiological role, EF-P has been implicated in the modulation of drug resistance (59, 79), including resistance to gentamicin, a 4,6-linked aminoglycoside that promotes miscoding during tRNA selection (80). As for EFP’s rescue of polyproline sequences (56), the translocation-rescuing activities of EF-P were also found to be strongly dependent on its conserved posttranslational modification located at the tip of domain 1 (Fig. S5). Further investigations will be needed to delineate whether the β-lysinylation modification simply increases EFP’s binding affinity to the vacant E site during the prolonged INT2 ↔ INT3 exchange or if the effect of the modification is to facilitate repositioning of peptidyl-tRNA within the large subunit P site to allosterically impact the P-site recognition process and translocation (Fig. 7B).
Given that stalled translocation complexes resulting from A-site miscoding errors bear only a single, chimeric, intersubunit, hybrid-state tRNA, such complexes may also be particularly prone to frameshifting or premature “drop off” from mRNA (42, 81). The successful translocation of mismatched mRNA codon/peptidyl-tRNA anticodon pairs may also result in proofreading through premature termination (82, 83). Such considerations suggest that the frequency of miscoding during cellular protein synthesis may be larger than functional estimates, as most of these depend on a fully translated reporter protein to produce a readout (84–86). Translocation stalling arising from miscoding may also influence cotranslational protein folding (87–90), and thus contribute to the aminoglycoside-induced cellular catastrophe that arises from the loss of cellular proton gradients at the outer membrane (91, 92). We therefore speculate that aminoglycoside-induced miscoding of aminoacyl-tRNA at the A site may contribute to bactericidal activity through the inhibition of the peptidyl-tRNA entry into the P site.
Further investigations will be needed to examine this proposed mode of aminoglycoside action and to query whether translocation-stalling phenomena are considerably different in distinct mRNA contexts or deeper into the protein ORF (93). In this context, it will be important to examine the physiological impacts of regulated changes in EF-P concentration, or that of its multifunctional eukaryotic homolog eIF5A (94).
Materials and Methods
Details are provided in SI Materials and Methods. Fluorescently labeled ribosomes, EF-G, and tRNAs were prepared as previously described (28, 29, 95). PRE complexes were generated on surface-immobilized ribosomes as described previously (32). PRE complex preparation differed in the 15-min incubation of the ternary complex solution at 15 mM Mg(OAc)2 to induce miscoding, followed by a wash with the original 5 mM Mg[OAc]2 polymix buffer. All experiments were conducted at 25 °C in Tris-polymix buffer [50 mM Tris-OAc (pH 7.5), 100 mM KCl, 5 mM NH4OAc, 0.5 mM Ca(OAc)2, 0.1 mM EDTA, 5 mM putrescine, 1 mM spermidine, and 1.5 mM β-mercaptoethanol] with 5 mM Mg(OAc)2, a mixture of triplet-state quenchers (1 mM Trolox, 1 mM nitrobenzyl alcohol, and 1 mM cyclooctatetraene) (96), and an enzymatic oxygen scavenging system (97). Acquisition and analysis of FRET data and selection of translocating traces were performed as previously described (11).
Acknowledgments
We thank R. Green (Johns Hopkins University) for providing the S13 knockout strain, P. Schultz (Scripps Research Institute) for providing the L5 knockout strain, and D. Wilson and P. Huter (Ludwig-Maximilian University Munich) for providing purified EF-P and its mutant counterpart. We also acknowledge helpful discussions, insights and cooperation provided by all members of the S.C.B. laboratory, especially Michael Wasserman, Roger Altman, Mikael Holm, and Randall Dass. This work was supported by the US NIH (Grant 5R01GM079238) and the National Science Foundation (Grant MCB-1412353).
Footnotes
Conflict of interest statement: S.C.B. has an equity interest in Lumidyne Technologies.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1707539114/-/DCSupplemental.
References
- 1.Holtkamp W, Wintermeyer W, Rodnina MV. Synchronous tRNA movements during translocation on the ribosome are orchestrated by elongation factor G and GTP hydrolysis. BioEssays. 2014;36:908–918. doi: 10.1002/bies.201400076. [DOI] [PubMed] [Google Scholar]
- 2.Munro JB, Sanbonmatsu KY, Spahn CM, Blanchard SC. Navigating the ribosome’s metastable energy landscape. Trends Biochem Sci. 2009;34:390–400. doi: 10.1016/j.tibs.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shoji S, Walker SE, Fredrick K. Ribosomal translocation: One step closer to the molecular mechanism. ACS Chem Biol. 2009;4:93–107. doi: 10.1021/cb8002946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen J, Tsai A, O’Leary SE, Petrov A, Puglisi JD. Unraveling the dynamics of ribosome translocation. Curr Opin Struct Biol. 2012;22:804–814. doi: 10.1016/j.sbi.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guo Z, Noller HF. Rotation of the head of the 30S ribosomal subunit during mRNA translocation. Proc Natl Acad Sci USA. 2012;109:20391–20394. doi: 10.1073/pnas.1218999109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ermolenko DN, Noller HF. mRNA translocation occurs during the second step of ribosomal intersubunit rotation. Nat Struct Mol Biol. 2011;18:457–462. doi: 10.1038/nsmb.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rodnina MV, Savelsbergh A, Katunin VI, Wintermeyer W. Hydrolysis of GTP by elongation factor G drives tRNA movement on the ribosome. Nature. 1997;385:37–41. doi: 10.1038/385037a0. [DOI] [PubMed] [Google Scholar]
- 8.Savelsbergh A, et al. An elongation factor G-induced ribosome rearrangement precedes tRNA-mRNA translocation. Mol Cell. 2003;11:1517–1523. doi: 10.1016/s1097-2765(03)00230-2. [DOI] [PubMed] [Google Scholar]
- 9.Pan D, Kirillov SV, Cooperman BS. Kinetically competent intermediates in the translocation step of protein synthesis. Mol Cell. 2007;25:519–529. doi: 10.1016/j.molcel.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Munro JB, Wasserman MR, Altman RB, Wang L, Blanchard SC. Correlated conformational events in EF-G and the ribosome regulate translocation. Nat Struct Mol Biol. 2010;17:1470–1477. doi: 10.1038/nsmb.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wasserman MR, Alejo JL, Altman RB, Blanchard SC. Multiperspective smFRET reveals rate-determining late intermediates of ribosomal translocation. Nat Struct Mol Biol. 2016;23:333–341. doi: 10.1038/nsmb.3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ratje AH, et al. Head swivel on the ribosome facilitates translocation by means of intra-subunit tRNA hybrid sites. Nature. 2010;468:713–716. doi: 10.1038/nature09547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao YG, et al. The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science. 2009;326:694–699. doi: 10.1126/science.1179709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramrath DJ, et al. Visualization of two transfer RNAs trapped in transit during elongation factor G-mediated translocation. Proc Natl Acad Sci USA. 2013;110:20964–20969. doi: 10.1073/pnas.1320387110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou J, Lancaster L, Donohue JP, Noller HF. How the ribosome hands the A-site tRNA to the P site during EF-G-catalyzed translocation. Science. 2014;345:1188–1191. doi: 10.1126/science.1255030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brilot AF, Korostelev AA, Ermolenko DN, Grigorieff N. Structure of the ribosome with elongation factor G trapped in the pretranslocation state. Proc Natl Acad Sci USA. 2013;110:20994–20999. doi: 10.1073/pnas.1311423110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peske F, Matassova NB, Savelsbergh A, Rodnina MV, Wintermeyer W. Conformationally restricted elongation factor G retains GTPase activity but is inactive in translocation on the ribosome. Mol Cell. 2000;6:501–505. doi: 10.1016/s1097-2765(00)00049-6. [DOI] [PubMed] [Google Scholar]
- 18.Horan LH, Noller HF. Intersubunit movement is required for ribosomal translocation. Proc Natl Acad Sci USA. 2007;104:4881–4885. doi: 10.1073/pnas.0700762104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frank J, Agrawal RK. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature. 2000;406:318–322. doi: 10.1038/35018597. [DOI] [PubMed] [Google Scholar]
- 20.Blanchard SC, Kim HD, Gonzalez RL, Jr, Puglisi JD, Chu S. tRNA dynamics on the ribosome during translation. Proc Natl Acad Sci USA. 2004;101:12893–12898. doi: 10.1073/pnas.0403884101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ermolenko DN, et al. Observation of intersubunit movement of the ribosome in solution using FRET. J Mol Biol. 2007;370:530–540. doi: 10.1016/j.jmb.2007.04.042. [DOI] [PubMed] [Google Scholar]
- 22.Cornish PV, Ermolenko DN, Noller HF, Ha T. Spontaneous intersubunit rotation in single ribosomes. Mol Cell. 2008;30:578–588. doi: 10.1016/j.molcel.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Borg A, et al. Fusidic acid targets elongation factor G in several stages of translocation on the bacterial ribosome. J Biol Chem. 2015;290:3440–3454. doi: 10.1074/jbc.M114.611608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou J, Lancaster L, Donohue JP, Noller HF. Crystal structures of EF-G-ribosome complexes trapped in intermediate states of translocation. Science. 2013;340:1236086. doi: 10.1126/science.1236086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang W, Dunkle JA, Cate JH. Structures of the ribosome in intermediate states of ratcheting. Science. 2009;325:1014–1017. doi: 10.1126/science.1175275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramrath DJ, et al. The complex of tmRNA-SmpB and EF-G on translocating ribosomes. Nature. 2012;485:526–529. doi: 10.1038/nature11006. [DOI] [PubMed] [Google Scholar]
- 27.Blanchard SC, Gonzalez RL, Kim HD, Chu S, Puglisi JD. tRNA selection and kinetic proofreading in translation. Nat Struct Mol Biol. 2004;11:1008–1014. doi: 10.1038/nsmb831. [DOI] [PubMed] [Google Scholar]
- 28.Wang L, et al. Allosteric control of the ribosome by small-molecule antibiotics. Nat Struct Mol Biol. 2012;19:957–963. doi: 10.1038/nsmb.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wasserman MR, et al. Chemically related 4,5-linked aminoglycoside antibiotics drive subunit rotation in opposite directions. Nat Commun. 2015;6:7896. doi: 10.1038/ncomms8896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blaha G, Stanley RE, Steitz TA. Formation of the first peptide bond: The structure of EF-P bound to the 70S ribosome. Science. 2009;325:966–970. doi: 10.1126/science.1175800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Geggier P, et al. Conformational sampling of aminoacyl-tRNA during selection on the bacterial ribosome. J Mol Biol. 2010;399:576–595. doi: 10.1016/j.jmb.2010.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Munro JB, Altman RB, O’Connor N, Blanchard SC. Identification of two distinct hybrid state intermediates on the ribosome. Mol Cell. 2007;25:505–517. doi: 10.1016/j.molcel.2007.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burnett BJ, et al. Elongation factor Ts directly facilitates the formation and disassembly of the Escherichia coli elongation factor Tu·GTP·aminoacyl-tRNA ternary complex. J Biol Chem. 2013;288:13917–13928. doi: 10.1074/jbc.M113.460014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johansson M, Zhang J, Ehrenberg M. Genetic code translation displays a linear trade-off between efficiency and accuracy of tRNA selection. Proc Natl Acad Sci USA. 2012;109:131–136. doi: 10.1073/pnas.1116480109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang J, Ieong KW, Johansson M, Ehrenberg M. Accuracy of initial codon selection by aminoacyl-tRNAs on the mRNA-programmed bacterial ribosome. Proc Natl Acad Sci USA. 2015;112:9602–9607. doi: 10.1073/pnas.1506823112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Munro JB, et al. Spontaneous formation of the unlocked state of the ribosome is a multistep process. Proc Natl Acad Sci USA. 2010;107:709–714. doi: 10.1073/pnas.0908597107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khade PK, Joseph S. Messenger RNA interactions in the decoding center control the rate of translocation. Nat Struct Mol Biol. 2011;18:1300–1302. doi: 10.1038/nsmb.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Katunin VI, Savelsbergh A, Rodnina MV, Wintermeyer W. Coupling of GTP hydrolysis by elongation factor G to translocation and factor recycling on the ribosome. Biochemistry. 2002;41:12806–12812. doi: 10.1021/bi0264871. [DOI] [PubMed] [Google Scholar]
- 39.Caliskan N, Katunin VI, Belardinelli R, Peske F, Rodnina MV. Programmed -1 frameshifting by kinetic partitioning during impeded translocation. Cell. 2014;157:1619–1631. doi: 10.1016/j.cell.2014.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davies J, Davis BD. Misreading of ribonucleic acid code words induced by aminoglycoside antibiotics. The effect of drug concentration. J Biol Chem. 1968;243:3312–3316. [PubMed] [Google Scholar]
- 41.Fast R, Eberhard TH, Ruusala T, Kurland CG. Does streptomycin cause an error catastrophe? Biochimie. 1987;69:131–136. doi: 10.1016/0300-9084(87)90245-8. [DOI] [PubMed] [Google Scholar]
- 42.Kurland CG. Translational accuracy and the fitness of bacteria. Annu Rev Genet. 1992;26:29–50. doi: 10.1146/annurev.ge.26.120192.000333. [DOI] [PubMed] [Google Scholar]
- 43.Feldman MB, Terry DS, Altman RB, Blanchard SC. Aminoglycoside activity observed on single pre-translocation ribosome complexes. Nat Chem Biol. 2010;6:54–62. doi: 10.1038/nchembio.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Misumi M, Nishimura T, Komai T, Tanaka N. Interaction of kanamycin and related antibiotics with the large subunit of ribosomes and the inhibition of translocation. Biochem Biophys Res Commun. 1978;84:358–365. doi: 10.1016/0006-291x(78)90178-x. [DOI] [PubMed] [Google Scholar]
- 45.Cabañas MJ, Vázquez D, Modolell J. Inhibition of ribosomal translocation by aminoglycoside antibiotics. Biochem Biophys Res Commun. 1978;83:991–997. doi: 10.1016/0006-291x(78)91493-6. [DOI] [PubMed] [Google Scholar]
- 46.Fredrick K, Noller HF. Catalysis of ribosomal translocation by sparsomycin. Science. 2003;300:1159–1162. doi: 10.1126/science.1084571. [DOI] [PubMed] [Google Scholar]
- 47.Studer SM, Feinberg JS, Joseph S. Rapid kinetic analysis of EF-G-dependent mRNA translocation in the ribosome. J Mol Biol. 2003;327:369–381. doi: 10.1016/s0022-2836(03)00146-3. [DOI] [PubMed] [Google Scholar]
- 48.Hirokawa G, et al. Post-termination complex disassembly by ribosome recycling factor, a functional tRNA mimic. EMBO J. 2002;21:2272–2281. doi: 10.1093/emboj/21.9.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Green R, Noller HF. Ribosomes and translation. Annu Rev Biochem. 1997;66:679–716. doi: 10.1146/annurev.biochem.66.1.679. [DOI] [PubMed] [Google Scholar]
- 50.Puglisi JD, et al. Aminoglycoside antibiotics and decoding. In: Garrett RA, et al., editors. The Ribosome: Structure, Function, Antibiotics and Cellular Interactions. ASM Press; Washington, DC: 2000. [Google Scholar]
- 51.Ogle JM, Carter AP, Ramakrishnan V. Insights into the decoding mechanism from recent ribosome structures. Trends Biochem Sci. 2003;28:259–266. doi: 10.1016/S0968-0004(03)00066-5. [DOI] [PubMed] [Google Scholar]
- 52.Peske F, Savelsbergh A, Katunin VI, Rodnina MV, Wintermeyer W. Conformational changes of the small ribosomal subunit during elongation factor G-dependent tRNA-mRNA translocation. J Mol Biol. 2004;343:1183–1194. doi: 10.1016/j.jmb.2004.08.097. [DOI] [PubMed] [Google Scholar]
- 53.Jerinic O, Joseph S. Conformational changes in the ribosome induced by translational miscoding agents. J Mol Biol. 2000;304:707–713. doi: 10.1006/jmbi.2000.4269. [DOI] [PubMed] [Google Scholar]
- 54.Peil L, et al. Distinct XPPX sequence motifs induce ribosome stalling, which is rescued by the translation elongation factor EF-P. Proc Natl Acad Sci USA. 2013;110:15265–15270. doi: 10.1073/pnas.1310642110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ude S, et al. Translation elongation factor EF-P alleviates ribosome stalling at polyproline stretches. Science. 2013;339:82–85. doi: 10.1126/science.1228985. [DOI] [PubMed] [Google Scholar]
- 56.Doerfel LK, et al. EF-P is essential for rapid synthesis of proteins containing consecutive proline residues. Science. 2013;339:85–88. doi: 10.1126/science.1229017. [DOI] [PubMed] [Google Scholar]
- 57.Doerfel LK, et al. Entropic contribution of elongation factor P to proline positioning at the catalytic center of the ribosome. J Am Chem Soc. 2015;137:12997–13006. doi: 10.1021/jacs.5b07427. [DOI] [PubMed] [Google Scholar]
- 58.Marman HE, Mey AR, Payne SM. Elongation factor P and modifying enzyme PoxA are necessary for virulence of Shigella flexneri. Infect Immun. 2014;82:3612–3621. doi: 10.1128/IAI.01532-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Navarre WW, et al. PoxA, yjeK, and elongation factor P coordinately modulate virulence and drug resistance in Salmonella enterica. Mol Cell. 2010;39:209–221. doi: 10.1016/j.molcel.2010.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dinman JD. Mechanisms and implications of programmed translational frameshifting. Wiley Interdiscip Rev RNA. 2012;3:661–673. doi: 10.1002/wrna.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Qu X, et al. The ribosome uses two active mechanisms to unwind messenger RNA during translation. Nature. 2011;475:118–121. doi: 10.1038/nature10126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fredrick K, Noller HF. Accurate translocation of mRNA by the ribosome requires a peptidyl group or its analog on the tRNA moving into the 30S P site. Mol Cell. 2002;9:1125–1131. doi: 10.1016/s1097-2765(02)00523-3. [DOI] [PubMed] [Google Scholar]
- 63.Phelps SS, Malkiewicz A, Agris PF, Joseph S. Modified nucleotides in tRNA(Lys) and tRNA(Val) are important for translocation. J Mol Biol. 2004;338:439–444. doi: 10.1016/j.jmb.2004.02.070. [DOI] [PubMed] [Google Scholar]
- 64.Curran JF, Yarus M. Reading frame selection and transfer RNA anticodon loop stacking. Science. 1987;238:1545–1550. doi: 10.1126/science.3685992. [DOI] [PubMed] [Google Scholar]
- 65.Ranjan N, Rodnina MV. Thio-modification of tRNA at the wobble position as regulator of the kinetics of decoding and translocation on the ribosome. J Am Chem Soc. 2017;139:5857–5864. doi: 10.1021/jacs.7b00727. [DOI] [PubMed] [Google Scholar]
- 66.Demeshkina N, Jenner L, Westhof E, Yusupov M, Yusupova G. A new understanding of the decoding principle on the ribosome. Nature. 2012;484:256–259. doi: 10.1038/nature10913. [DOI] [PubMed] [Google Scholar]
- 67.Yan S, Wen JD, Bustamante C, Tinoco I., Jr Ribosome excursions during mRNA translocation mediate broad branching of frameshift pathways. Cell. 2015;160:870–881. doi: 10.1016/j.cell.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen J, et al. Dynamic pathways of -1 translational frameshifting. Nature. 2014;512:328–332. doi: 10.1038/nature13428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hinnebusch AG, Lorsch JR. The mechanism of eukaryotic translation initiation: New insights and challenges. Cold Spring Harb Perspect Biol. 2012;4:a011544. doi: 10.1101/cshperspect.a011544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kaushik S, et al. The mode of inhibitor binding to peptidyl-tRNA hydrolase: Binding studies and structure determination of unbound and bound peptidyl-tRNA hydrolase from Acinetobacter baumannii. PLoS One. 2013;8:e67547. doi: 10.1371/journal.pone.0067547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Singh A, et al. Structural and binding studies of peptidyl-tRNA hydrolase from Pseudomonas aeruginosa provide a platform for the structure-based inhibitor design against peptidyl-tRNA hydrolase. Biochem J. 2014;463:329–337. doi: 10.1042/BJ20140631. [DOI] [PubMed] [Google Scholar]
- 72.Arenz S, et al. The stringent factor RelA adopts an open conformation on the ribosome to stimulate ppGpp synthesis. Nucleic Acids Res. 2016;44:6471–6481. doi: 10.1093/nar/gkw470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Himeno H, Kurita D, Muto A. tmRNA-mediated trans-translation as the major ribosome rescue system in a bacterial cell. Front Genet. 2014;5:66. doi: 10.3389/fgene.2014.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Griffin MA, Davis JH, Strobel SA. Bacterial toxin RelE: A highly efficient ribonuclease with exquisite substrate specificity using atypical catalytic residues. Biochemistry. 2013;52:8633–8642. doi: 10.1021/bi401325c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.An G, Glick BR, Friesen JD, Ganoza MC. Identification and quantitation of elongation factor EF-P in Escherichia coli cell-free extracts. Can J Biochem. 1980;58:1312–1314. doi: 10.1139/o80-177. [DOI] [PubMed] [Google Scholar]
- 76.Schmidt A, et al. The quantitative and condition-dependent Escherichia coli proteome. Nat Biotechnol. 2016;34:104–110. doi: 10.1038/nbt.3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Reynolds NM, Lazazzera BA, Ibba M. Cellular mechanisms that control mistranslation. Nat Rev Microbiol. 2010;8:849–856. doi: 10.1038/nrmicro2472. [DOI] [PubMed] [Google Scholar]
- 78.Ribas de Pouplana L, Santos MA, Zhu JH, Farabaugh PJ, Javid B. Protein mistranslation: Friend or foe? Trends Biochem Sci. 2014;39:355–362. doi: 10.1016/j.tibs.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 79.Rajkovic A, et al. Cyclic rhamnosylated elongation factor P establishes antibiotic resistance in Pseudomonas aeruginosa. MBio. 2015;6:e00823. doi: 10.1128/mBio.00823-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zou SB, et al. Loss of elongation factor P disrupts bacterial outer membrane integrity. J Bacteriol. 2012;194:413–425. doi: 10.1128/JB.05864-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Karimi R, Ehrenberg M. Dissociation rate of cognate peptidyl-tRNA from the A-site of hyper-accurate and error-prone ribosomes. Eur J Biochem. 1994;226:355–360. doi: 10.1111/j.1432-1033.1994.tb20059.x. [DOI] [PubMed] [Google Scholar]
- 82.Zaher HS, Green R. Quality control by the ribosome following peptide bond formation. Nature. 2009;457:161–166. doi: 10.1038/nature07582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Keiler KC. Mechanisms of ribosome rescue in bacteria. Nat Rev Microbiol. 2015;13:285–297. doi: 10.1038/nrmicro3438. [DOI] [PubMed] [Google Scholar]
- 84.Ortego BC, Whittenton JJ, Li H, Tu SC, Willson RC. In vivo translational inaccuracy in Escherichia coli: Missense reporting using extremely low activity mutants of Vibrio harveyi luciferase. Biochemistry. 2007;46:13864–13873. doi: 10.1021/bi602660s. [DOI] [PubMed] [Google Scholar]
- 85.Kramer EB, Farabaugh PJ. The frequency of translational misreading errors in E. coli is largely determined by tRNA competition. RNA. 2007;13:87–96. doi: 10.1261/rna.294907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Manickam N, Nag N, Abbasi A, Patel K, Farabaugh PJ. Studies of translational misreading in vivo show that the ribosome very efficiently discriminates against most potential errors. RNA. 2014;20:9–15. doi: 10.1261/rna.039792.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ciryam P, Morimoto RI, Vendruscolo M, Dobson CM, O’Brien EP. In vivo translation rates can substantially delay the cotranslational folding of the Escherichia coli cytosolic proteome. Proc Natl Acad Sci USA. 2013;110:E132–E140. doi: 10.1073/pnas.1213624110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kimchi-Sarfaty C, et al. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315:525–528. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- 89.Tsai CJ, et al. Synonymous mutations and ribosome stalling can lead to altered folding pathways and distinct minima. J Mol Biol. 2008;383:281–291. doi: 10.1016/j.jmb.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang G, Hubalewska M, Ignatova Z. Transient ribosomal attenuation coordinates protein synthesis and co-translational folding. Nat Struct Mol Biol. 2009;16:274–280. doi: 10.1038/nsmb.1554. [DOI] [PubMed] [Google Scholar]
- 91.Schlebach JP, Sanders CR. The safety dance: Biophysics of membrane protein folding and misfolding in a cellular context. Q Rev Biophys. 2015;48:1–34. doi: 10.1017/S0033583514000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Davis BD. Mechanism of bactericidal action of aminoglycosides. Microbiol Rev. 1987;51:341–350. doi: 10.1128/mr.51.3.341-350.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Borg A, Ehrenberg M. Determinants of the rate of mRNA translocation in bacterial protein synthesis. J Mol Biol. 2015;427:1835–1847. doi: 10.1016/j.jmb.2014.10.027. [DOI] [PubMed] [Google Scholar]
- 94.Schuller AP, Wu CC, Dever TE, Buskirk AR, Green R. eIF5A functions globally in translation elongation and termination. Mol Cell. 2017;66:194–205.e5. doi: 10.1016/j.molcel.2017.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Munro JB, Altman RB, Tung CS, Sanbonmatsu KY, Blanchard SC. A fast dynamic mode of the EF-G-bound ribosome. EMBO J. 2010;29:770–781. doi: 10.1038/emboj.2009.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dave R, Terry DS, Munro JB, Blanchard SC. Mitigating unwanted photophysical processes for improved single-molecule fluorescence imaging. Biophys J. 2009;96:2371–2381. doi: 10.1016/j.bpj.2008.11.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Aitken CE, Marshall RA, Puglisi JD. An oxygen scavenging system for improvement of dye stability in single-molecule fluorescence experiments. Biophys J. 2008;94:1826–1835. doi: 10.1529/biophysj.107.117689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang L, Altman RB, Blanchard SC. Insights into the molecular determinants of EF-G catalyzed translocation. RNA. 2011;17:2189–2200. doi: 10.1261/rna.029033.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Juette MF, et al. Single-molecule imaging of non-equilibrium molecular ensembles on the millisecond timescale. Nat Methods. 2016;13:341–344. doi: 10.1038/nmeth.3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Qin F, Auerbach A, Sachs F. Estimating single-channel kinetic parameters from idealized patch-clamp data containing missed events. Biophys J. 1996;70:264–280. doi: 10.1016/S0006-3495(96)79568-1. [DOI] [PMC free article] [PubMed] [Google Scholar]