Significance
To maintain cellular organization, many chaperones and targeting factors escort nascent proteins to membrane destinations. It was generally thought that substrate proteins preferably bind conformationally closed chaperones. Here, we used single-molecule spectroscopy to study the conformation and dynamics of the ATPase guided entry of tail-anchored protein 3 (Get3), which delivers an essential class of tail-anchored proteins (TAs) to the endoplasmic reticulum. Contrary to previous models, TA-bound Get3 rapidly fluctuates between open and closed conformations, forming a “protean clamp” that stably traps substrates. Biochemical data showed that this dynamic opening primes Get3 for targeting of TAs to membrane receptor sites. Analogous protean traps may operate in other chaperones, targeting factors, and transporters, providing a dual mechanism to both stably retain substrates and vectorially drive cellular processes.
Keywords: protein targeting, chaperones, protein dynamics, single-molecule spectroscopy, ATPases
Abstract
Proper localization of proteins to target membranes is a fundamental cellular process. How the nature and dynamics of the targeting complex help guide substrate proteins to the target membrane is not understood for most pathways. Here, we address this question for the conserved ATPase guided entry of tail-anchored protein 3 (Get3), which targets the essential class of tail-anchored proteins (TAs) to the endoplasmic reticulum (ER). Single-molecule fluorescence spectroscopy showed that, contrary to previous models of a static closed Get3•TA complex, Get3 samples open conformations on the submillisecond timescale upon TA binding, generating a fluctuating “protean clamp” that stably traps the substrate. Point mutations at the ATPase site bias Get3 toward closed conformations, uncouple TA binding from induced Get3•Get4/5 disassembly, and inhibit the ER targeting of the Get3•TA complex. These results demonstrate an essential role of substrate-induced Get3 dynamics in driving TA targeting to the membrane, and reveal a tightly coupled channel of communication between the TA-binding site, ATPase site, and effector interaction surfaces of Get3. Our results provide a precedent for large-scale dynamics in a substrate-bound chaperone, which provides an effective mechanism to retain substrate proteins with high affinity while also generating functional switches to drive vectorial cellular processes.
Over 35% of proteins need to be localized to the correct cellular destinations after their initial synthesis in the cytosol. These protein-targeting processes are essential for the establishment and maintenance of compartmentalization in all cells and pose complex mechanistic challenges to targeting machineries. To minimize improper exposure of substrate proteins in the cytosol, targeting factors must bind substrate proteins with high stability. This requirement is especially stringent during the targeting of integral membrane proteins, whose high aggregation propensity in the cytosol and other aqueous cellular environments demands that targeting factors also serve as effective chaperones to protect substrates from aggregation. Further, to minimize futile cycling of targeting factors, loading of substrates on the targeting factor must be tightly coupled to their delivery to membrane receptor sites. Finally, once at the target membrane, the targeting machinery must readily switch to a low-affinity state to release substrate proteins to receptor complexes, translocases, or the phospholipid bilayer. With a few exceptions (1, 2), the nature and dynamics of protein targeting complexes and how their biophysical properties help meet these complex functional demands are not well understood, especially for posttranslational protein targeting pathways.
The targeting of tail-anchored proteins (TAs) provides an excellent system to address these questions. TAs, defined by a single transmembrane domain (TMD) near the C terminus, comprise up to 5% of the eukaryotic membrane proteome and mediate diverse key cellular functions, including protein translocation across multiple organelle membranes, vesicular fusion, protein quality control, and apoptosis (3). In eukaryotic cells, TAs are targeted to the endoplasmic reticulum (ER) by the conserved guided entry of tail-anchored protein (GET) pathway, in which the Get3 ATPase captures TAs with help of the cytosolic Get4/5 complex and then delivers TAs to the Get1/2 receptor complex at the ER membrane (4–6). During the targeting cycle, Get3 undergoes extensive changes in conformation and activity in response to nucleotides, effector proteins, and the TA substrate (Fig. 1A). In the cytosol, ATP binding drives the Get3 homodimer from an open conformation, in which the helical domains of the two Get3 subunits are apart, to a closed conformation in which the helical domains are close together (7, 8) (Fig. 1B). Closed ATP•Get3 is preferentially bound by the Get4/5 complex (9, 10), which bridges between Get3 and the upstream cochaperone Sgt2 and stimulates TA transfer from Sgt2 onto Get3 (11–13). After dissociation from Get4/5, the Get3•TA complex hydrolyzes ATP and interacts with the Get1/2 membrane receptors (10, 14). Get1 drives Get3 into an open conformation, enabling TA release and insertion into the membrane (11, 15–17).
Fig. 1.
Direct observation of Get3 conformational changes using μs-ALEX. (A) Current model for Get3 conformations throughout the GET pathway. Question marks (“?”) highlight unresolved questions. (B) Approximate positions of donor and acceptor dyes (green and red stars, respectively) in the structures of open Get3 (Left; PDB ID code 3H84) and closed Get3 (Right; PDB ID code 2WOJ). The bound ADP•AlF4− in closed Get3 is in space-fill. (C–F) FRET histograms for Get3 in apo-, ADP-, AMPPNP-, and Get1CD-bound states, respectively. Lighter shaded molecules depict alternative conformations sampled by Get3. The letter “n” denotes the number of observed double-labeled Get3 used to generate each FRET histogram. (G–J) BVAs for apo- and ADP-, AMPPNP-, and Get1CD-bound Get3, respectively. The red curves indicate the expected SD for shot-noise-limited E* (static limit). Triangles denote the mean SD for individual FRET bins used to calculate the dynamic score (DS) and weighted dynamic score (WDS) (SI Materials and Methods).
Despite these advances, the conformation and dynamics of the Get3•TA complex during targeting remain an outstanding question (5, 6, 18). Crystallographic analyses showed that Get3 “closing” generates a contiguous hydrophobic groove in its helical domains (7, 8), which provides a binding site for the TMD of TA substrates. The cocrystal structure of Get3 bound with a TA-TMD (13) also shows a closed Get3 similar to that in the ATP•Get3•Get4/5 complex (9). It was proposed that closed Get3 stably binds TA substrates and protects the TMD from exposure to the cytosol (7, 13). However, an exclusively closed Get3•TA complex poses a fundamental dilemma for targeting (Fig. 1A, “?”). Based on thermodynamic coupling, if both Get4/5 and TA prefer closed Get3, TA loading would strengthen the Get3–Get4/5 interaction. In contrast, multiple downstream events in the pathway require Get3 to dissociate from Get4/5 after TA loading. These include the interaction of Get3 with Get1/2, whose binding sites on Get3 overlap with Get4/5 (9, 10, 15–17), and ATP hydrolysis by Get3, which is inhibited by Get4/5 (14). These considerations predict that substrate-loaded Get3 must adopt conformation(s) that are different from the highly closed structures observed previously. Indeed, biochemical studies showed that a TA substrate destabilizes the interaction of Get3 with Get4/5 and activates the Get3 ATPase activity (10, 14), implying that the substrate induces Get3 into distinct conformational state(s).
The GET pathway provides a salient example of the complex functional demands on the targeting machinery during a targeting cycle, as well as the conceptual and experimental challenges in understanding how these demands are met. The model of an exclusively closed Get3•TA complex also exemplifies the typical view of targeting complexes as static structures, wherein substrates fit into well-defined grooves or pockets in conformationally closed targeting factors or chaperones. Recent NMR studies began to challenge this view, demonstrating that client proteins populate a dynamic ensemble of conformational states and transit between multiple short-lived interaction sites when bound to chaperones such as Skp, SurA, and Spy during trafficking through the bacterial periplasm (19–21). Nevertheless, the nature and dynamics of the targeting factors/chaperones in these complexes, and how these properties help guide substrate proteins to the target membrane, have not been addressed.
To address these questions, we studied the conformation and dynamics of Get3 using single-molecule spectroscopy with microsecond time resolution. These analyses show that, contrary to previous models, the TA substrate destabilizes a closed Get3 and induces the ATPase to sample open conformations on the submillisecond timescale. Biochemical analyses demonstrate that these changes in the TA-binding domain are transmitted via the ATPase active site to drive the dissociation of Get3 from Get4/5, and are essential for the targeting of TA substrates to the Get1/2 receptors at the ER membrane. These results provide a unifying model to explain how the TA substrate drives the switch of Get3 from a substrate-loading mode to a membrane-targeting mode. Moreover, they demonstrate how rapid protein motions allow a targeting factor/chaperone to stably retain its substrate protein while undergoing changes in structure and function to vectorially drive a cellular pathway.
Results
Diffusion-Based Single-Molecule Spectroscopy Detects Global Structure and Dynamics of Get3.
To measure the global conformational changes of Get3, we used diffusion-based single-molecule Förster resonance energy transfer (FRET) between donor (Cy3B) and acceptor (ATTO 647N) dyes site-specifically incorporated in the two subunits of the Get3 dimer (Fig. 1B). Fluorophores were incorporated at a nonconserved loop in the Get3 helical domain, and labeling does not affect the activity of Get3 (12). We used confocal microscopy with alternating laser excitation with microsecond time resolution (μs-ALEX) to detect and quantify the fluorescence of single molecules transiting through a femtoliter-scale observation volume, and extracted relative FRET efficiencies (E*) for individual molecules (22) (Fig. S1 A–C). The distances between the dye pair are estimated to be ∼75 Å and ∼30 Å in open and closed Get3, respectively (Fig. 1B). Hence, a significant difference in FRET between the open and closed conformations of Get3 is expected for this dye pair, allowing us to monitor transitions along these states.
Fig. S1.
Monitoring Get3 conformational changes using μs-ALEX. (A) Schematic depiction of the μs-ALEX method. Donor and acceptor dyes labeled on Get3 were alternatively excited as they diffused through a confocal volume. Both the donor-to-acceptor S and relative E* were determined for individual Get3 molecules, allowing for optical purification of doubly labeled Get3 (S ∼ 0.5; cyan bracket) and differentiation of Get3 conformations displaying different E* values. FRET histograms were obtained by 1D projection of 2D E*-S histograms onto the E* axis after isolating double-labeled Get3. (B) Representative E*-S histogram of stochastically double-labeled apo-Get3 showing both single- and double-labeled Get3 populations. (C) Representative E*-S plot of apo-Get3 after a dual-channel burst search to isolate doubly labeled Get3. (D and E) Representative E*-S plots for a 1:1 mixture of donor-only and acceptor-only labeled Get3 before (D) and after (E) a dual-channel burst search. The absence of substantial colocalization of the donor and acceptor fluorophores indicates no significant exchange of Get3 subunits on our experimental timescale.
We first used μs-ALEX to visualize the conformations of Get3 under well-established conditions. Apo-Get3 displayed a broad FRET distribution, with E* maxima ranging from 0.5–0.7 (Fig. 1C and reproducibility of data in Fig. S2). The distribution for ADP-bound Get3 was also broad but peaked at higher FRET (Fig. 1D). When Get3 was bound to the nonhydrolyzable ATP analog adenosine 5′-(β,γ-imido)triphosphate (AMPPNP), the FRET distribution was narrower and peaked at an E* of ∼0.8 (Fig. 1E). In contrast, the cytosolic domain of Get1 (Get1CD) shifted the distribution to lower FRET (peak E* of ∼0.3; Fig. 1F). These data agree with previous work showing that ATP induces Get3 to closed conformations (7, 8), whereas Get1CD induces the open state of Get3 (15–17). To exclude photophysical artifacts, we repeated these measurements using another FRET pair, ATTO 550 and ATTO 647N, which yielded the same nucleotide- and Get1CD-induced changes in FRET distributions (Fig. S3 A–D). In addition, the presence of various ligands and interaction partners did not affect the dye photophysics in a way that would alter the FRET distributions (Fig. S3 E–G). We also confirmed that Get3 dimers do not exchange subunits during measurements (Fig. S1 D and E). Thus, our labeling strategy coupled with μs-ALEX can monitor the conformational transitions of Get3.
Fig. S2.
Replicates of the FRET histograms for apo-Get3. The orange outline shows the histogram generated by combining all of the data.
Fig. S3.
Controls for dye photophysics in μs-ALEX measurements of Get3. (A–D) FRET histograms of Get3 in the apo- and ADP-, ATP-, and Get1CD-bound states, respectively, for Get3 double-labeled with ATTO 550 and ATTO 647N. All of the FRET histograms are shifted to higher E* compared with Cy3B- and ATTO 647N-labeled Get3, as expected from the longer Förster radius of the ATTO 550-ATTO 647N pair. Nevertheless, the ligand-induced changes in the FRET histograms follow the same trends as those measured with the Cy3B-ATTO 647N pair. The letter “n” denotes the number of observed doubly labeled molecules used to generate each FRET histogram. (E and F) Effects of ligands on the steady-state fluorescence spectra of Cy3B-labeled Get3 and ATTO 647N-labeled Get3, respectively, measured using a Fluorolog 3-22 spectrofluorometer. Saturating amounts of each interaction partner (2 mM ATP, 4 mM ADP, 10 μM Get1CD, 4 μM Get4/5) were used. Most interaction partners did not significantly affect Cy3B or ATTO 647N fluorescence. ATP modestly reduced the fluorescence for both Cy3B and ATTO 647N, which should not significantly affect calculated FRET. arb., arbitrary. (G) TA substrate did not significantly affect the peak photon rate for both Cy3B and ATTO 647N, determined using the μs-ALEX setup for single-labeled Get3. This suggests that the TA substrate does not affect the photophysics of the FRET pair on Get3.
The FRET distributions of Get3 are broad, suggesting conformational heterogeneity. To distinguish whether this arises from the coexistence of multiple static structures or from dynamic conformational sampling, we performed burst variance analysis (BVA), which detects dynamics by comparing the SD of E* over time with the SD expected from shot noise (23). If the FRET distribution arises solely from static species, the SD is limited by shot noise and would lie on the static limit curve (Fig. 1 G–J, red lines). In contrast, if multiple conformations interconvert on the submillisecond or faster timescale, the observed SD would be higher than the static limit. Apo- and ADP-bound Get3 displayed substantially higher SDs than the static limit, especially for molecules that exhibit intermediate E* values (Fig. 1 G and H). In contrast, the SDs for AMPPNP- and Get1CD-bound Get3 are close to the static limit, especially for molecules at the peak E* values (Fig. 1 I and J). Thus, apo-Get3 and ADP-bound Get3 sample a range of conformations on the submillisecond timescale, as suggested by molecular dynamics simulations (24), and the peak E* values of 0.4–0.6 exhibited by apo-Get3 arise from conformational averaging between states with higher and lower FRET. Further, different interaction partners lock Get3 into distinct and more defined conformations.
The TA Substrate Induces Get3 to Dynamically Open.
To determine the conformation of Get3 when bound to the TA substrate, we assembled Get3•TA complexes by in vitro translation of Bos1, a model GET substrate (12), in Escherichia coli lysate in the presence of Get3 and affinity-purified Get3•TA complexes via the 3xStrep-tag on Bos1 (12) (Fig. 2A). Get3•TA complexes generated by this procedure were kinetically stable and highly efficient in TA targeting and insertion into the ER membrane (12) (Fig. 2B and Fig. S4 A and B). In μs-ALEX measurements, AMPPNP•Get3•TA displayed a broader FRET distribution shifted toward lower E* values compared with AMPPNP•Get3 (Fig. 2C). The distributions shifted further to lower FRET and peaked at E* values of ∼0.55–0.6 with ADP•Get3•TA and Get3•TA (Fig. 2 D and E). In addition, BVA showed that, in contrast to AMPPNP•Get3, Get3•TA complexes exhibiting intermediate E* values displayed higher SD than the static limit in all nucleotide states (Fig. 2 F–H). These observations indicate that Get3 also becomes more dynamic upon TA binding, and the observed E* values of 0.4–0.6 result from averaging of Get3•TA complexes that interconvert between lower (<0.4) and higher (>0.6) FRET states on the submillisecond or faster timescale. Thus, contrary to the highly closed Get3•TA structure observed crystallographically (13), the TA substrate induces Get3 to dynamically sample open conformations.
Fig. 2.
The TA substrate induces Get3 to dynamically sample open conformations. (A) Scheme for generation and purification of Get3•TA complexes for μs-ALEX experiments. Green and red asterisks denote donor and acceptor dyes labeled on Get3. A 100-fold excess of unlabeled Get3 was included to ensure that, statistically, intradimer FRET of Get3 was measured even in cases of potential Get3 tetramerization (8, 10, 14). Experimental details are provided in Materials and Methods. IVT, in vitro translation. (B) Get3•TA complexes prepared using the procedure in A are active in targeting and insertion into the ER. Representative SDS/PAGE autoradiograph (Top) and its quantification (Bottom) for the time courses of Get3•TA targeting and insertion are shown for wild-type Get3 (black solid line) and mutant Get3(Δα8) (cyan). The dotted curve depicts the insertion efficiency for Get3•TA generated with Sgt2 and Get4/5 included during translation as described previously (12), and shows that the inclusion of Sgt2 and Get4/5 is not necessary for generation of a functional Get3•TA complex. glyc, glycosylated; yRM, yeast rough microsome. (C–E) FRET histograms of Get3•TA complexes in AMPPNP-bound, ADP-bound, and nucleotide-free states, respectively. The outlines depict the FRET histograms of Get3 in the same nucleotide state without TA substrate and are shown for comparison; “n” denotes the number of observed double-labeled Get3 used to generate each FRET histogram. (F–H) BVAs of Get3•TA in AMPPNP-bound, ADP-bound, and nucleotide-free states, respectively. The red curves represent the SD expected for shot-noise-limited E*. Triangles denote the mean SD for individual FRET bins used to calculate the dynamic score (DS) and weighted dynamic score (WDS).
Fig. S4.
(A) Scheme of the TA targeting and insertion assay. Purified Get3•TA was presented to the ER from Δget3 yeast with or without nucleotide and/or Get4/5. Successful TA insertion results in glycosylation (glyc) of its C-terminal opsin tag, which can be visualized as a molecular weight shift after SDS/PAGE. (B) Purified Get3BDP•TACM (12) was presented to indicated concentrations of a TA trap, intein-cpSRP43 (28), and TA dissociation from Get3 was monitored through loss of FRET. The observed rate of TA dissociation is independent of trap concentration, indicating that intein-cpSRP43 acts as a passive trap to measure the intrinsic rate of Get3•TA spontaneous dissociation. The reported kdissociation value represents mean ± SD from the three measurements. (C) Structure of apo-Get3 (PDB ID code 3A36) highlighting helix α8 (cyan) that is resolved in one of the Get3 subunits (yellow). The residues preceding and following α8 are highlighted in cyan in the other Get3 subunit (salmon). (D) Visualization of purified Get3•TA complexes, assembled with wild-type Get3 or mutant Get3(Δα8), by in gel fluorescence (for Get3) and autoradiography (for TA). (E) Thermodynamic cycle showing the coupling of the TA-binding equilibrium (denoted by KTA and KTA′ for closed and open Get3, respectively) to the conformational equilibrium of Get3 opening (denoted by Kopen and Kopen′ for free and TA-loaded Get3, respectively).
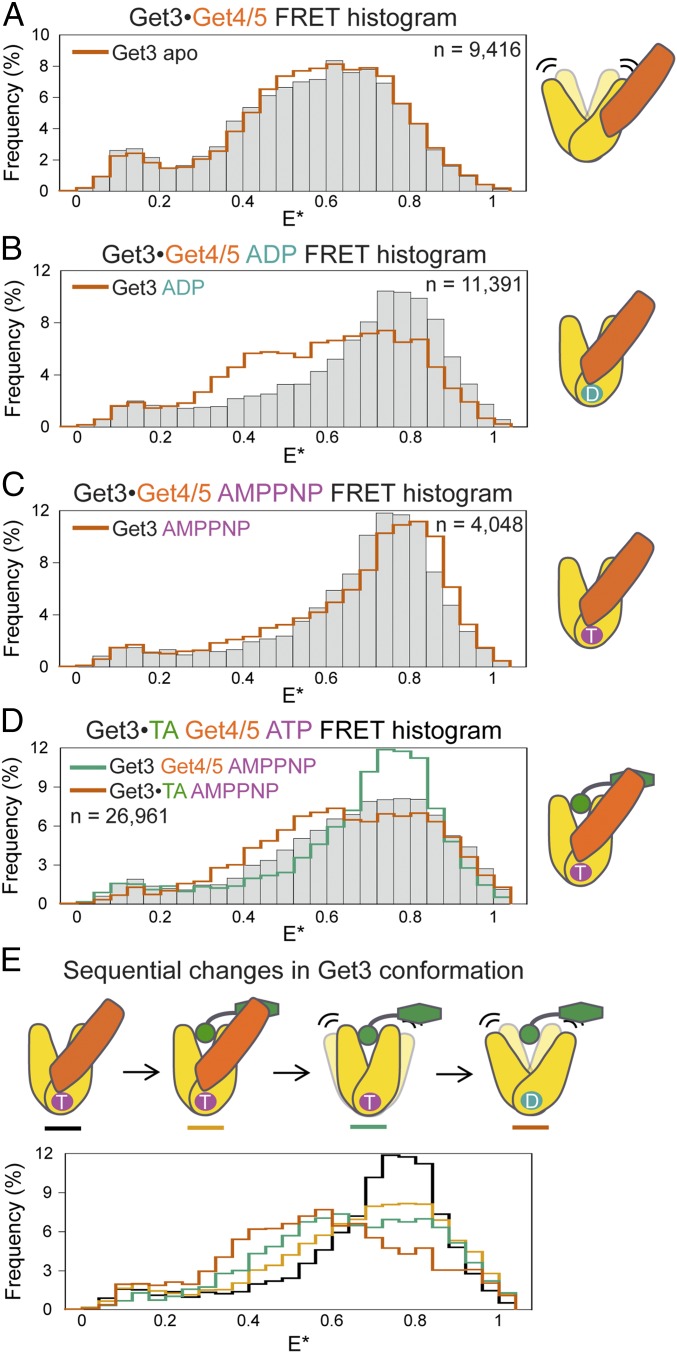
The Get3•TA complex was crystallized using a synthetic antibody (sAB), which binds at similar surfaces on Get3 as does Get4/5 (13); it also specifically recognizes ATP-bound Get3, as does Get4/5 (9, 10, 13). Thus, a potential explanation for the difference between the crystallographic and single-molecule FRET data is that Get4/5 stabilizes a more closed conformation of Get3•TA, and this effect was mimicked by the sAB. We therefore tested the effect of Get4/5 on Get3 conformation using μs-ALEX. With apo-Get3, to which Get4/5 binds at a different surface than ATP•Get3 (25), Get4/5 did not significantly change the FRET histogram (Fig. 3A and Fig. S5G). With ADP•Get3, which is distributed between low and high FRET states, Get4/5 shifted the distribution to predominantly high E* values (Fig. 3B and Fig. S5G). With AMPPNP•Get3, which is already closed, Get4/5 induced a modest but statistically significant shift of the FRET distribution to a distinct high-FRET state (Fig. 3C and Fig. S5G). These results agree well with previous work showing that Get4/5 preferentially binds closed Get3 (9, 10). Importantly, binding of Get4/5 also shifted the distribution of ATP•Get3•TA to higher E* (Fig. 3D, bars vs. orange outline). Compared with the FRET distribution of AMPPNP•Get3•Get4/5 before TA loading, the distribution after TA loading peaked at the same E* value but was substantially broader (Fig. 3D, bars vs. teal outline). Thus, Get4/5 also biases Get3•TA to more closed conformations. Moreover, these data illustrate sequential changes in the conformation and dynamics of Get3 during the targeting pathway (Fig. 3E): Starting with a static, closed AMPPNP•Get3•Get4/5 complex (black), TA loading induces conformational “breathing” of Get3 (light orange); upon dissociation from Get4/5, Get3 more frequently samples open conformations (teal) that become more dominant after ATP hydrolysis (dark orange).
Fig. 3.
Sequential opening of Get3 upon TA loading and Get4/5 release. (A–C) Effects of Get4/5 on the FRET histograms of apo-, ADP-, and AMPPNP-bound Get3, respectively. (D) FRET histogram of Get3•TA bound with Get4/5 and ATP. The complex was generated by supplementing Get4/5 and excess ATP throughout the preparation to capture the conformation of the physiological initial loading complex (Materials and Methods). Since Get4/5 inhibits ATP hydrolysis (14) and excess ATP is used, ATP turnover has no significant effect on the results. (E) Comparisons of FRET histograms of Get3 at different stages of the GET pathway. In A–D, the outlines depict the FRET histograms of indicated Get3 complexes and are shown for comparison, and “n” denotes the number of observed double-labeled Get3 used to generate each FRET histogram.
Fig. S5.
Differences between FRET histograms of indicated wild-type and mutant Get3 complexes (A–D) and between replicates of μs-ALEX measurements for Get3•TA and Get3(E251A)•TA samples (E and F) are shown. The differences in the FRET histograms shown in A, C, and D between wild-type and mutant Get3 are systematic. In comparison, the differences in the FRET histograms between replicates of data shown in E and F are randomly distributed across E*, and the difference in the FRET histograms shown in B is smaller and more random. (G and H) Adapted K-S tests to evaluate the significance of the differences between the FRET histograms of the indicated samples (SI Materials and Methods). The F (P < 0.05) values at each sample size m denote the fraction of 10 K-S tests from randomly selected subsets of data that yielded a statistically significant difference (P < 0.05), and were reported as mean ± SD, with n = 5. Adapted K-S tests for molecules from the same sample usually result in P > 0.05 regardless of m (pink and dark blue lines in H), and serve as negative controls for the absence of a significant difference. Histograms for which the F (P < 0.05) value rises slowly with m are considered similar (black line in G and light blue line in H). Histograms for which the F (P < 0.05) value rises quickly with m are interpreted as significantly different (navy and red lines in G and black, orange, and green lines in H).
TA Is Stably Bound by the Dynamically Fluctuating Get3.
The dynamic opening of the Get3•TA complex is unexpected, as crystallographic analyses suggested that in open Get3, the hydrophobic groove for TMD binding becomes discontiguous (7, 26). Intriguingly, our recent (12) and current measurements showed that Get3•TA complexes exhibit high kinetic stability: Using a membrane protein chaperone, cpSRP43 (27, 28), as an inert TA trap, the timescale for spontaneous dissociation of Bos1 from Get3 was measured to be ∼4 h (Fig. S4B). The lower limit for the lifetime of other Get3•TA complexes was 35–60 min (12). Thus, TAs are stably bound to Get3 despite the large-scale conformational fluctuations of Get3.
The simplest modification of the current model to explain these observations is that a 15-aa sequence termed helix α8, which was unresolved in most Get3 structures and proposed to form a lid over the TA-binding groove (7, 13), could prevent the escape of TA substrates during opening. Nevertheless, a structure of apo-Get3 [Protein Data Bank (PDB) ID code 3A36], in which α8 was resolved (29), suggests that the dimension of α8 is unlikely to be sufficient to completely shield the helical domains and block TA escape in open Get3 (Fig. S4C, α8 highlighted in cyan). To experimentally test the importance of α8, we replaced the conserved hydrophobic residues (199PMLNSFM) in this sequence with a GS linker (GGSGGGS) to generate mutant Get3(Δα8). Get3•TA complexes assembled with mutant Get3(Δα8) could be purified as a stable complex (Fig. S4D) and are fully functional in TA targeting and insertion into the ER membrane (Fig. 2B, cyan curve). These results show that shielding by α8 is insufficient to explain the high stability of the Get3•TA complex if the current model of Get3–TA interaction were the only possible interaction mode. Following the principle of thermodynamic coupling, the observation of TA-induced Get3 opening further predicts that the TA substrate can explore alternative modes for interacting with a more open Get3 that are energetically more favorable than the established interaction mode observed with closed Get3 (Fig. S4E). The implications of these observations are elaborated in Discussion.
TA-Induced Get3 Opening Drives Membrane Targeting.
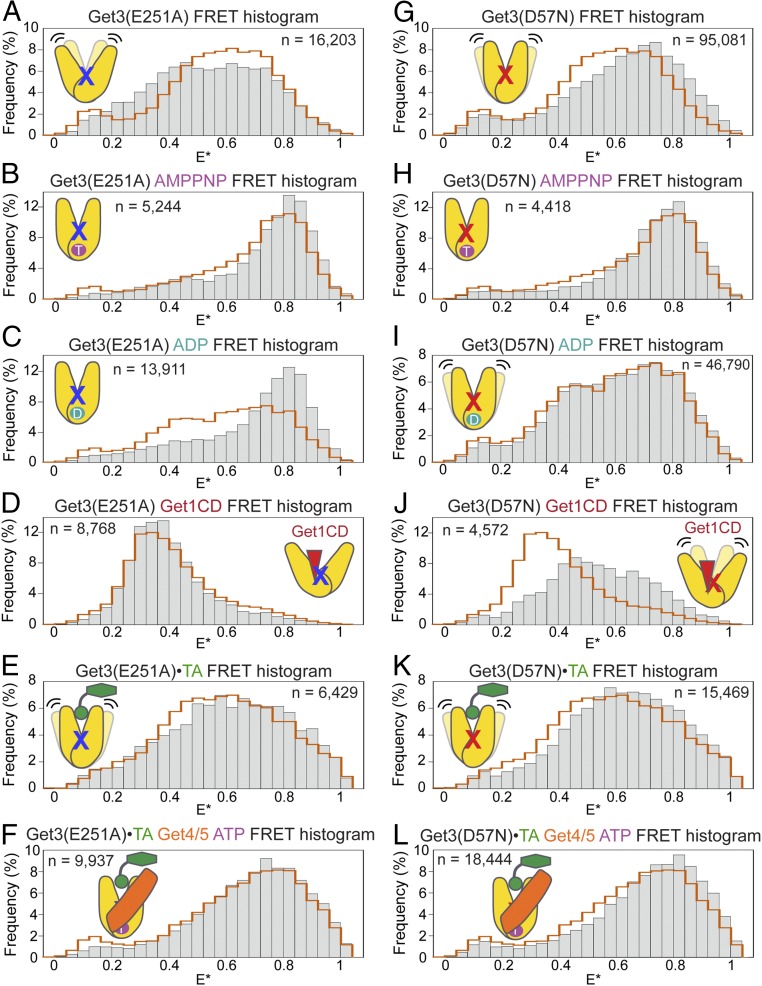
At the junction of the Get3 helical and ATPase domains is a network of residues that interact across the dimer interface and contribute catalytic interactions with ATP (7, 8, 26, 29) (Fig. S6A), raising the possibility that disruption of this network would interfere with the ability of Get3 to undergo regulated conformational changes. We tested two mutations in this network, D57N and E251A, which cause severe yeast growth defects under stress conditions (7, 26). Both mutants bind ATP tightly but displayed 102-fold slower ATPase activity than wild-type Get3 (7) (Fig. S6 C–F). Surprisingly, μs-ALEX measurements showed that these mutations bias Get3 toward closed conformations under different conditions. Get3(E251A) is more closed than Get3(WT) when bound with nucleotides (Fig. 4 A–D), whereas Get3(D57N) is more closed than Get3(WT) in apo- and Get1CD-bound states (Fig. 4 G–J). In the Get3•TA complex, the FRET distributions of mutant Get3(D57N) were shifted to higher E* with and without ATP and Get4/5 present (Fig. 4 K and L), whereas only the FRET distribution of free Get3(E251A)•TA was shifted (Fig. 4 E and F). Although the mutational effects on the FRET distributions of Get3•TA were modest, the changes are systematic compared with variations between replicates of data and are statistically significant (Figs. S2 and S5). The different effects of the two mutations on the FRET distributions of Get3•TA also agreed well with biochemical analyses of these mutants described later (Fig. 6). Thus, point mutations at the Get3 catalytic site alter the conformation and regulation of its helical domains.
Fig. S6.
Interactions at the Get3 ATPase site and the Get3–Get4 interface. (A) Network of interacting residues at the catalytic site across the Get3 dimer interface. Get3-D57 is positioned near the γ-phosphate and coordinates the nucleophilic water. Get3-N61 forms a salt bridge with Get3-E251 from the opposing Get3 monomer (PDB ID code 2WOJ). ADP•AlF4− is shown in space-fill. (B) Get3 interacts with Get4 via two interfaces: an anchoring interface (magenta spheres, which highlight residues whose mutations reduce the affinity between ATP-bound Get3 and Get4/5) and a regulatory interface (blue spheres, which highlight conserved residues that may contact Get4/5) (9). (C–E) ATP concentration dependences of observed ATP hydrolysis rate constants for wild-type Get3 and the mutants Get3(E251A) and Get3(D57N), respectively. (F) Summary of Km and kcat values for wild-type and mutant Get3s derived from the data in C–E. All values are reported as mean ± SD (n ≥ 2). Get3(E251A) and Get3(D57N) retain high-affinity ATP binding but are defective in ATP hydrolysis.
Fig. 4.
Point mutations at the ATPase active site bias Get3 to more closed conformations. (A–D) FRET histograms of Get3(E251A) in apo- and AMPPNP-, ADP-, and Get1CD-bound states, respectively. (E and F) FRET histograms of Get3(E251A)•TA without (E) or with (F) Get4/5 and ATP bound. (G–J) FRET histograms of Get3(D57N) in apo- and AMPPNP-, ADP-, and Get1CD-bound states, respectively. (K and L) FRET histograms of Get3(D57N)•TA without (K) and with (L) Get4/5 and ATP bound. In all histograms, the orange outlines depict the FRET histograms of wild-type Get3 under the same conditions, and “n” denotes the number of observed double-labeled molecules used to generate each FRET histogram.
Fig. 6.
The TA substrate regulates the Get3–Get4 interaction. (A and B) Representative equilibrium titrations to measure the binding of Get4/5 to wild-type and mutant Get3 in the ATP-bound (A) and TA-bound (B) states. arb., arbitrary. (C) Get3-K69 forms a putative salt bridge (dotted line) with Get4-D74 at the regulatory interface (PDB ID code 4PWX). Mutation of Get3-K69 or Get4-D74 did not substantially reduce Get3-Get4/5 binding but disrupted the ability of Get4/5 to regulate ATP hydrolysis by Get3 (9). (D) Summary of the effects of the Get4(D74K) mutation on the binding affinity of Get4/5 for wild-type and mutant Get3 (Kbinding = 1/Kd) in the indicated complexes. Data are from Table S1 and are reported as mean ± propagated error, with n ≥ 2. (E) Representative equilibrium titrations to measure the binding of Get4(D74K)/5 to wild-type and mutant Get3•TA complexes. Dashed lines are the binding curves of wild-type Get4/5 to the corresponding Get3 variant (from B) and are shown for comparison. (F) Time courses for targeting and insertion of Get3(D57N)•TA in the presence of ATP and indicated Get4/5 variants (n = 2).
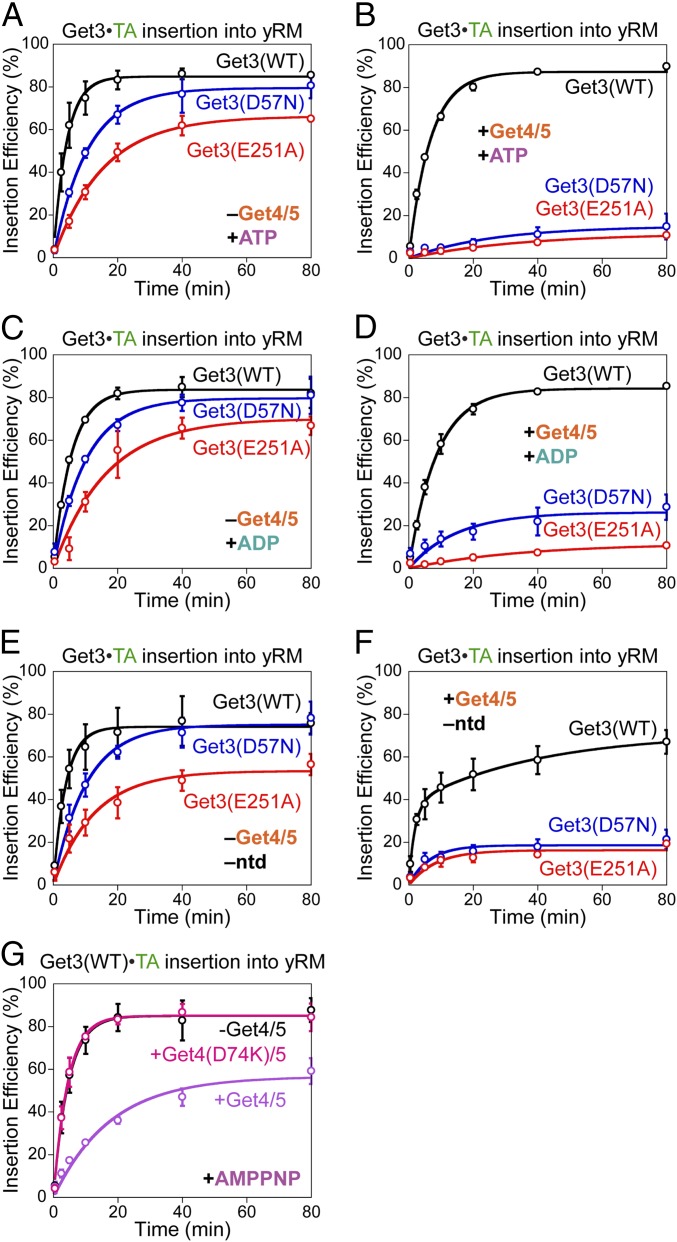
To assess if the conformational bias in Get3(D57N) and Get3(E251A) disrupts Get3 function, we tested the activities of these mutants in mediating TA targeting and insertion into the ER membrane. Without Get4/5 present, Get3(D57N)•TA and Get3(E251A)•TA were up to threefold and fivefold slower, respectively, than wild-type Get3•TA in targeting and insertion (Fig. 5 A, C, and E). However, physiological amounts of Get4/5 (30) nearly abolished TA insertion with both mutants, without substantially affecting TA insertion by wild-type Get3 (Fig. 5 B, D, and F). These mutational defects were observed regardless of the nucleotide state of Get3•TA complexes (Fig. 5 A–F); the slower phase of the insertion reactions with apo-Get3 was due to the nucleotide requirement for recycling Get3 from the ER membrane during multiple rounds of TA targeting (10, 11, 16). Together, these results show that TA-induced Get3 opening is essential for membrane targeting of the Get3•TA complex under conditions that mimic the physiological situation, where the Get1/2 receptor and Get4/5 complex must compete for binding to Get3.
Fig. 5.
Get3 active site mutants block the targeting of Get3•TA complexes. Time courses for targeting and insertion of wild-type and mutant Get3•TA complexes in the absence (A, C, and E) and presence (B, D, and F) of 0.5 μM Get4/5. The nucleotides used in the insertion reactions are as follows: 2 mM ATP in A and B, 2 mM ADP in C and D, and no nucleotides in E and F. (G) Time courses for targeting and insertion of wild-type AMPPNP•Get3•TA with and without Get4/5 or mutant Get4(D74K)/5 present. All experiments were repeated on different days (n ≥ 2) and are plotted as mean ± SD. yRM, yeast rough microsome.
The following data indicate that the effects of these Get3 mutants did not arise solely from the failure to hydrolyze ATP, but rather from defects in undergoing TA-induced conformational changes. Even with AMPPNP bound, targeting and insertion from wild-type Get3•TA was efficient (compare black lines in Fig. 5 A vs. G); thus, ATP hydrolysis per se is not required for membrane targeting (8). Wild-type AMPPNP•Get3•TA also tolerated the presence of Get4/5 and allowed TA insertion (Fig. 5G, purple), albeit three- to fivefold more slowly than the reaction of ATP•Get3•TA with Get4/5 present (Fig. 5B, black). Thus, ATP hydrolysis contributes three- to fivefold, but is not obligatory for the exchange of Get4/5 with Get1/2 on the Get3•TA complex. Further, the ATP occupancy of Get3(D57N)•TA and Get3(E251A)•TA complexes was measured to be 39% and 5.8%, respectively (Fig. S7D); even if only the ADP- or nucleotide-free targeting complexes were active, these ATP occupancies predict that the two mutants would retain 61% and 94%, respectively, of the targeting activity of wild-type Get3, which were insufficient to explain their targeting defects. Finally, the Get4/5-specific targeting defect of both Get3 mutants was observed regardless of the added nucleotide (Fig. 5 A–F). Assuming the simplest model in which the observed insertion occurs from the fraction of Get3•TA complexes that acquired a targeting-competent conformation, the biochemical data suggested that the defect of mutant Get3(E251A)•TA in attaining the active conformation was ∼70-fold after ATP hydrolysis [Fig. 5D, compare the time required for 11% insertion with ADP•Get3•TA vs. ADP•Get3(E251A)•TA]. These results underscore the essential role of TA-induced Get3 conformational change in driving membrane targeting.
Fig. S7.
Purification of wild-type and mutant Get3•TA complexes, and determination of ATP content in Get3 and Get3•TA. (A–C) Gel filtration chromatograms for recombinantly expressed and purified Get3•2xStrep-Sbh1 complexes made with Get3(WT), Get3(E251A), and Get3(D57N), respectively. Green and purple lines denote A280 and A260 readings, respectively, normalized to the peak A280 value. Fractions corresponding to the dominant peak were collected for each complex. The difference in elution volumes of recombinant wild-type and mutant Get3•TA has been noted previously (13). Get3(D57N)•TA displays a higher ratio of A280 to A260 compared with Get3(WT)•TA and Get3(E251A)•TA, consistent with the higher ATP retention in this mutant complex determined in D. (D) Luminescent ATP detection kit was used to determine the ATP concentrations for different Get3 and Get3•TA samples at 1 μM. Get3(D57N) shows higher nucleotide retention compared with Get3(E251A) and Get3(WT). All values are reported as mean ± SD, with n = 3.
The Get3 Active Site Couples the TA-Binding Site to the Get3–Effector Interaction Surface.
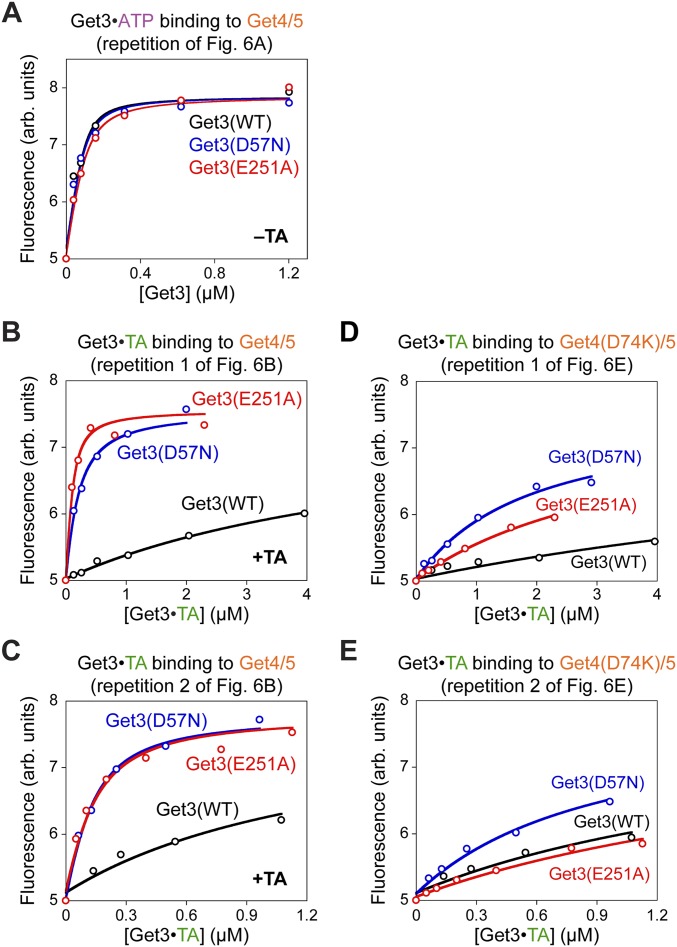
The Get4/5-specific targeting defects of Get3(D57N) and Get3(E251A) suggest that these mutants fail to dissociate from Get4/5 after TA loading, and thus block the subsequent interaction of Get3 with the Get1/2 receptor complex at the ER membrane. To test this hypothesis, we measured the Get3–Get4/5 interaction using an established assay based on the fluorescence enhancement of acrylodan labeled at Get4(S48C/C177T) upon Get3 binding (10). Equilibrium titrations showed that while wild-type Get3•TA bound Get4/5 much more weakly than ATP•Get3, as observed previously (10), mutant Get3(D57N)•TA and Get3(E251A)•TA retained high-affinity binding to Get4/5 (Fig. 6 A and B, Fig. S8 A–C, and Table S1). Thus, small defects in Get3 opening (Fig. 4 E, F, K, and L) could severely block the TA-induced disassembly of the Get3•Get4/5 complex.
Fig. S8.
Replicates of equilibrium titrations to measure the binding affinity of Get4/5 for wild-type and mutant Get3 in the ATP-bound state (A), for wild-type and mutant Get3 loaded with the TA substrate (B and C), and for the binding of Get4(D74K)/5 to wild-type and mutant Get3•TA complexes (D and E) are shown. The measurements in B and D were performed in parallel, as were the measurements in C and E. arb., arbitrary.
Table S1.
Summary of equilibrium binding affinities between Get4/5 variants and Get3 variants in different substrate and nucleotide states
| Get4/5 construct | Get3 complex | Nucleotide | Kd, μM |
| Wild type | Wild type | ATP | 0.016 ± 0.002 |
| D57N | 0.017 ± 0.003 | ||
| E251A | 0.032 ± 0.008 | ||
| Wild type•Sbh1 | None added | 1.5 ± 0.2 | |
| D57N•Sbh1 | 0.072 ± 0.006 | ||
| E251A•Sbh1 | 0.062 ± 0.03 | ||
| Wild type•Sbh1 | AMPPNP | 0.017 ± 0.004 | |
| D57N•Sbh1 | 0.060 ± 0.001 | ||
| E251A•Sbh1 | 0.035 ± 0.007 | ||
| D74K | Wild type | ATP | 0.075 ± 0.002 |
| D57N | 0.12 ± 0.04 | ||
| E251A | 0.14 ± 0.09 | ||
| Wild type•Sbh1 | None added | 2.2 ± 0.3 | |
| D57N•Sbh1 | 0.78 ± 0.08 | ||
| E251A•Sbh1 | 1.8 ± 0.8 | ||
| Wild type•Sbh1 | AMPPNP | 0.35 ± 0.01 | |
| D57N•Sbh1 | 0.42 ± 0.2 | ||
| E251A•Sbh1 | 0.70 ± 0.4 |
All values are reported as mean ± SD (n ≥ 2).
How do TAs induce the dissociation of Get3 from Get4/5? Structural work showed that Get4 contacts ATP-bound Get3 at two interfaces: an anchoring interface, which enables stable binding, and a regulatory interface, which regulates Get3’s ATPase activity (9) (Fig. S6B). We introduced the Get4(D74K) mutation that disrupts the salt bridge between Get4-D74 and Get3-K69 at the regulatory interface (9) (Fig. 6C). The affinity of Get3 for wild-type Get4/5 relative to mutant Get4(D74K)/5 provides a measure for the energetic contribution of this salt bridge to Get3-Get4/5 binding. While the Get4(D74K) mutation weakened the binding to ATP•Get3 fivefold (9), the mutational effect was >20-fold with AMPPNP•Get3•TA and then became negligible with Get3•TA (Fig. 6D, black bars and Table S1). These results illustrate sequential changes at the Get3–Get4 regulatory interface, where the Get3(K69)-Get4(D74) salt bridge is ancillary before TA binding, becomes stronger upon TA loading, and then dissolves after nucleotide hydrolysis and release from Get3•TA.
Importantly, while the TA substrate failed to weaken the binding of Get3(D57N) and Get3(E251A) with wild-type Get4/5 (Fig. 6B), it did with mutant Get4(D74K)/5 (Fig. 6E and Fig. S8 D and E), suggesting that the Get3 active site mutants exert their effects by blocking changes at the Get3–Get4 regulatory interface. Analysis of the mutational effects of Get4(D74K) on the Get3 active site mutants further showed that Get3(E251A) is primarily defective in removing the Get3-Get4 contact at the regulatory interface after nucleotide hydrolysis and release, while Get3(D57N) is defective in rearranging this interface both before and after ATP hydrolysis (Fig. 6D, red and blue bars and Table S1). These data agreed well with the different effects of these mutations on the FRET distributions of Get3•TA and ATP•Get3•TA (Fig. 4 E and F vs. K and L), and together they showed that the catalytic residues at the Get3 active site provide key functional links between the TA-binding site and the Get4/5 interaction surface of Get3. Finally, the Get4(D74K) mutation abolished the inhibitory effect of Get4/5 on the targeting of Get3(D57N)•TA (Fig. 6F) and wild-type AMPPNP•Get3•TA (Fig. 5G), indicating that the TA-induced rearrangements at the Get3–Get4 regulatory interface directly impact targeting efficiency.
Discussion
Across all organisms, targeting complexes guide nascent proteins to diverse cellular destinations. These complexes must bind substrate proteins with high overall stability, while also readily switching structure and function to drive these vectorial processes. In this work, single-molecule fluorescence spectroscopy coupled with biochemical analyses provide a unifying model to explain how these complex demands are met during TA targeting by the Get3 ATPase. In contrast to previous models, TA loading destablizes a static, closed Get3•TA complex and induces Get3 to rapidly sample open conformations, and TA substrates are stably trapped in the rapidly fluctuating Get3. Point mutations at the ATPase active site bias Get3 toward closed conformations, uncouple TA binding from substrate-induced Get3•Get4/5 disassembly, and inhibit the targeting of the Get3•TA complex to the ER membrane. These data demonstrate how the substrate-induced dynamic opening of Get3 provides a dual mechanism that allows the targeting complex to both retain substrates with high affinity and drive the exchange of Get3’s interaction partners required for the ER targeting of TAs.
The substrate-induced opening of Get3 is unexpected, given the large and contiguous hydrophobic groove in closed Get3 that appears highly conducive to TA binding (7, 8, 13) as well as the high kinetic stability of Get3•TA complexes (12) (Fig. S4B). The TA-induced opening of Get3 further suggests that the TA substrate explores alternative sites and conformations for interacting with Get3 besides the previously observed docking of the TA-TMD at the well-defined hydrophobic groove in closed Get3, and that, collectively, these alternative Get3–TA interaction modes are energetically more favorable than the established mode. In analogy to membrane protein substrates bound to the Skp and SurA chaperones (19, 20), it is plausible that TA substrates sample multiple transient interaction sites during Get3 opening, and TAs could be retained because interaction with alternative sites in Get3 is more favorable than with solvent. This model would explain, in part, the weak electron density for TA-TMDs in previous Get3•TA structures (13). Alternatively or in addition, the rapid conformational fluctuations of Get3 could enable its reclosing to kinetically outcompete potential TA dissociation from the targeting complex during Get3 opening. Potential tetramerization of Get3 upon TA binding (8, 14) could provide another mechanism to retain substrate in a more open Get3; as a recent study did not detect stable tetrameric Get3•TA complexes and showed that dimeric Get3•TA is active in mediating TA insertion (13), the precise roles of Get3 tetramerization remain to be determined. Finally, increased conformational entropy in these more dynamic models could contribute to the overall stability of the targeting complex. Regardless of the specific mechanism of substrate retention, our results collectively show that substrate proteins can be stably bound to a targeting factor/chaperone while the latter undergoes large-scale fluctuations between open and closed states.
The targeting pathway demands extensive changes in the activities of Get3 before and after substrate loading. Before TA binding, Get3 must be ATP-bound and tightly bound to Get4/5, whereas after TA loading, Get3 must hydrolyze ATP and detach from Get4/5 so that it can instead interact with the Get1/2 receptors at the ER membrane. The results from this and previous work (9, 10, 14) demonstrate that nucleotide, together with the Get4/5 complex, is responsible for inducing a highly closed conformation of Get3. Although these results provide an attractive mechanism to explain how Get3 efficiently captures TA substrates at early stages of the pathway, they fail to explain the functional switches of Get3 required for the targeting phase of the pathway. The substrate-induced opening of Get3 provides an attractive mechanism to drive this functional switch, and thus resolves this dilemma.
Experimentally, the important role of substrate-induced Get3 opening is demonstrated by the D57N and E251A mutations at the Get3 ATPase site, both of which bias Get3 toward closed conformations. These mutants uncouple TA binding from TA-induced changes in Get3’s biochemical activities, including dissociation from Get4/5 and efficient targeting of TA substrates to the ER membrane. Although the targeting defect of Get3(D57N) was previously attributed to failed ATP hydrolysis (7, 11, 16), complete analysis of the targeting reaction in all nucleotide states, including the nonhydrolyzable ATP analog AMPPNP, showed that the targeting defects of Get3 active site mutants are largely conformational in origin (Fig. 7, step 5). The majority of the defects of Get3(D57N) and Get3(E251A) can be attributed to their failures to undergo TA-induced dissociation from Get4/5, which competes with the Get1/2 receptors for binding Get3. In addition, the three- to sixfold defects of these mutants in ER targeting in the absence of Get4/5 suggest that TA-induced Get3 opening facilitates the binding and remodeling of the targeting complex by Get1/2. By combining these mutants with disruptions at the Get3–Get4 regulatory interface, we further demonstrate sequential TA-induced adjustments at the Get3–Get4 interaction surface that drive their disassembly, and how Get3 active site mutations disrupt this relay of signal. Together, these data reveal a tightly coupled channel of communication between the TA-binding site, ATPase catalytic site, and effector interaction surfaces on Get3.
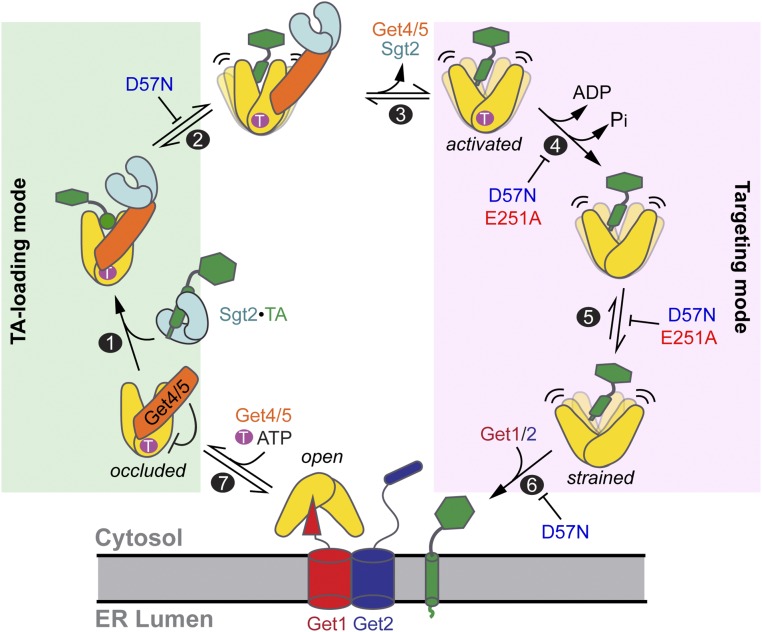
Fig. 7.
Model for how TA-induced Get3 opening drives the membrane targeting of the Get3•TA complex. Step 1: ATP•Get3 is bound to Get4/5, which induces Get3 to a closed, occluded conformation that inhibits ATP hydrolysis. TA is transferred from Sgt2 to Get3 in the Sgt2•Get4/5•Get3•TA complex, in which Get4/5 bridges Sgt2 and Get3. Step 2: TA loading induces Get3 to sample more open conformations, with concomitant adjustment at the Get3•Get4/5 interface that causes the regulatory interface to become the predominant stabilizing interaction. Step 3: Get4/5 spontaneously and reversibly dissociates from the conformationally adjusted Get3•TA complex. Step 4: TA induces Get3 to rapidly hydrolyze ATP after Get4/5 dissociation. Step 5: After ATP hydrolysis, Get3•TA becomes more open, attaining a strained conformation that prevents rebinding of Get4/5. Step 6: The strained Get3•TA complex is primed for targeting to and remodeling by the Get1/2 receptors at the ER membrane. Step 7: Get1/2 facilitates TA disassembly from Get3 and TA insertion into the membrane. Binding of ATP and Get4/5 releases Get3 from the membrane receptors, recycling it for additional rounds of targeting. The inhibition marks denote steps inhibited by the E251A and D57N mutations, with inhibition of step 5 being responsible for the majority of defects of these mutants on the overall targeting reaction under physiological conditions.
Collectively, our results illustrate how substrate-induced dynamic opening switches Get3 from a TA-loading mode to a targeting mode, thus initiating downstream steps in the GET pathway (Fig. 7). At early stages, ATP and Get4/5 lock Get3 into an occluded conformation, in which Get3 is closed and ATPase-inhibited (9, 14), and thus primed for TA capture (step 1). The closed Get3•TA-TMD structure, obtained with mutant Get3(D57N) and an sAB mimicking the interaction sites and biochemical activities of Get4/5 (13), likely represents the initial ATP•Get3•TA•Get4/5 complex. TA loading destabilizes this static structure and induces Get3 to undergo rapid conformational fluctuations to explore the more open state (step 2). These changes in the TA-binding domain are transmitted via the Get3 catalytic site to induce rearrangements at the Get3–Get4 interface, shifting stabilizing interactions from the anchoring interface to the regulatory interface (step 2). At this stage, spontaneous Get4/5 dissociation from Get3 (step 3), which is rapid despite the high stability of the complex (10), allows Get3•TA to further open and enables TA-induced ATPase activation (step 4). Opening of Get3 becomes more extensive after ATP hydrolysis (step 5), rendering the dissociation of Get4/5 irreversible and priming Get3 for interaction with the Get1/2 receptors instead (step 6). In addition, dynamic opening of the Get3•TA complex could provide a facile pathway for its remodeling by the Get1/2 receptor complex, facilitating TA release and insertion into the ER membrane (step 6); this may explain the modest but still significant defects of the Get3 active site mutants in TA insertion in the absence of Get4/5. Importantly, while Get1 was primarily responsible for opening Get3 in most previous models (11, 15–17), our findings show that the TA substrate itself initiates this opening to vectorially drive late stages of the targeting cycle.
Current models of substrate interactions with targeting factors and molecular chaperones often fall into the category of lock-and-key mechanisms, in which substrate proteins fit into preorganized, well-structured grooves or pockets in the substrate-binding domain (31–35). Although these mechanisms provide excellent explanations for how substrate proteins are captured, they also generate thermodynamic sinks that inhibit subsequent steps in the pathway, analogous to the situation described for Get3 (18). Our observations provide a precedent for a distinct class of models, in which rapid conformational fluctuations of a targeting factor/chaperone generate a “protean trap” that can retain substrates with high stability, and the dynamic nature of the complex enables functional switches to guide progression of the pathway. Analogous protean traps may provide an effective mechanism in other targeting factors, chaperones, and transporters that need to retain substrates with high affinity, while driving vectorial pathways that require distinct functions to be switched “on” and “off” in a sequential and coordinated manner.
Materials and Methods
Plasmids.
Plasmids for recombinant expression of Get3, Get4/5, Get1CD, and a superactive mutant of the cpSRP43 chaperone (intein-cpSRP43) and for in vitro translation of 3xStrep-SUMOnc-Bos1-opsin have been described (10, 12, 14, 28). DNA encoding 2xStrep-Sbh1 was in the pACYCDuet-1 vector. Plasmids encoding mutant proteins were generated using QuikChange site-directed mutagenesis (Stratagene) or FastCloning (36), and were confirmed with DNA sequencing (Laragen).
Biochemical Reagents.
Get3, Get4/5, Get1CD, and intein-cpSRP43 were expressed and purified as described (10, 12, 14, 28). E. coli S30 lysate and T7 RNA polymerase were prepared as described (12, 37). Microsomes were prepared from Δget3 yeast cells as described (14, 38, 39).
Get3•TA complexes for μs-ALEX and insertion assays were generated as described (12) with modifications. For μs-ALEX, the model TA 3xStrep-Bos1-opsin (12) was in vitro-translated in E. coli S30 lysate for 2 h at 30 °C in the presence of 2 μM Get3 (1:100 double-labeled to unlabeled), with or without 2 μM Sgt2 and 2 μM Get4/5 present. Complexes containing 3xStrep-tagged TA were purified using Strep-Tactin Sepharose (IBA Life Sciences) as described (12). Omission of Sgt2 and Get4/5 in this procedure does not change the targeting and insertion activity of Get3•TA (Fig. 2B). To characterize the effects of Get4/5 and ATP on Get3•TA conformation (Figs. 3D and 4 F and L), 0.5 μM Get4/5 and 2 mM ATP were present throughout the purification, and excess Get4/5 and ATP were supplemented to the purified complex. Get3BDP•TACM complexes used in Fig. S4B were generated as described (12).
Recombinant Get3•Sbh1 complexes were purified as described (13) with modifications. Untagged Get3 and 2xStrep-Sbh1 were coexpressed in One Shot BL21 Star (DE3) (Invitrogen) for 6 h at 26 °C after induction with 0.1 mM isopropyl β-d-1-thiogalactopyranoside at OD600 ∼ 0.8. Cells were disrupted by sonication in lysis buffer [50 mM Tris⋅HCl (pH 7.6), 300 mM NaCl, 5 mM β-mercaptoethanol, protease inhibitors]. Complexes containing 2xStrep-Sbh1 were purified using Strep-Tactin Sepharose followed by Superdex 200 10/300 GL (GE Healthcare) in GET buffer [50 mM KHEPES (pH 7.4), 150 mM KOAc, 5 mM Mg(OAc)2, 1 mM DTT, 10% glycerol] (Fig. S7 A–C).
Fluorescence Labeling.
We have described a strategy to site-specifically label Get3 at a ybbR tag inserted between residues 110 and 111 via Sfp-catalyzed incorporation of dye-CoA conjugates; ybbR insertion and fluorescence labeling do not perturb Get3 function (12). Using this strategy, we stochastically double-labeled Get3 dimers with a 1:1 ratio of CoA-conjugated Cy3B-maleimide (GE Healthcare) or ATTO 550-maleimide (ATTO-TEC) and CoA-conjugated ATTO 647N-maleimide (ATTO-TEC) at the α4-α5 loop. Donor- or acceptor-only Get3s were generated by labeling with one of the dyes. CoA-conjugated BODIPY FL maleimide (Thermo Fisher Scientific) was used to generate the Get3BDP used in Fig. S4B. Get4(C177T/S48C)/5 and Get4(C177T/S48C/D74K)/5 were labeled with acrylodan as described (10).
Biochemical Assays.
Fifty-microliter TA targeting and insertion reactions were initiated by adding 10 μL of Δget3 microsomes to purified Get3•TA complexes in which [35S]-methionine–labeled TA was normalized to 40,000 dpm. Two millimolar nucleotide and/or 0.5 μM Get4/5 was included where indicated. Reactions were incubated at 26 °C; at indicated time points, 6-μL samples were removed and quenched by addition of 2× SDS buffer and flash-freezing in liquid nitrogen. Samples were analyzed by SDS/PAGE and autoradiography.
Equilibrium-binding measurements between Get3 and acrylodan-labeled Get4/5 were performed as described (10). Get3BDP•TACM dissociation experiments were performed as described (12), except that indicated concentrations of intein-cpSRP43 were used as a chase instead of unlabeled Get3 (which we found to modestly accelerate TA dissociation). Multisite, multiturnover ATPase rate constants for Get3 were measured as described (14). The ATP concentrations in Get3 and Get3•TA samples were quantified using a luminescent ATP detection assay kit (ab113849; Abcam) per the manufacturer’s instructions.
μs-ALEX Measurements.
All proteins were ultracentrifuged in a TLA 100 rotor (Beckman Coulter) at 100,000 rpm for 1 h at 4 °C to remove aggregates before all measurements. Get3 samples were diluted to ∼100 pM in GET buffer containing 0.3 mg/mL BSA and indicated interaction partners. Based on previously determined Kd values (10, 14), saturating amounts of each interaction partner (2 mM AMPPNP, 4 mM ADP, 10 μM Get1CD, 4 μM Get4/5) were used to ensure that all observed Get3 molecules were ligand-bound. Samples were placed in a closed chamber made by sandwiching a perforated silicone sheet (Grace Bio-Labs) with two coverslips to prevent evaporation. Data were collected over 30–60 min using an ALEX–fluorescence-aided molecule sorting setup (40) with two single-photon Avalanche photodiodes (PerkinElmer) and 532-nm and 638-nm continuous wave lasers (Coherent) operating at 135 μW and 80 μW, respectively. Details of single-molecule data analysis are described in SI Materials and Methods.
SI Materials and Methods
All single molecule FRET (smFRET) data analyses, including burst search, burst selection, and BVA, were performed using FRETBursts, a Python-based open-source burst analysis toolkit for confocal smFRET (41). A dual-channel burst search (42) was performed to isolate the photon streams from species containing FRET pairs versus background noise and species containing donor or acceptor only. Each burst was identified as a minimum of 10 consecutive detected photons with a photon count rate at least 15-fold higher than the background photon count rate during both donor and acceptor excitation periods. Since the background rate can fluctuate within a measurement, the background rate was computed for every 50-s interval according to maximum likelihood fitting of the interphoton delay distribution. The identified bursts were further selected according to the following criteria: (i) 25 and (ii) 25, where is the number of photons detected from donor during donor excitation, is the number of photons detected from acceptor during donor excitation, and is the number of photons detected from acceptor during acceptor excitation.
The relative E* and stoichiometry (S) for each burst were calculated using the following equations:
| [S1] |
| [S2] |
E* corresponds to actual FRET efficiency if three conditions are met: (i) no donor fluorescence leaks into the acceptor detection channel (lk = 0), (ii) no acceptor is directly excited by the donor excitation laser (dir = 0), and (iii) the quantum yields and detection efficiencies for donor and acceptor are the same (γ = 1) (43). Although these conditions are not met in most cases, the contributions of lk, dir, and γ to the actual FRET efficiency are constant as long as the same optical setup and FRET pair are used throughout all measurements. Importantly, we did not observe significant changes in the quantum yields of donor and acceptor dyes depending on local environments (Fig. S3 E–G). Therefore, conformational changes in Get3 that change the actual FRET efficiency will also change the E* value, and the trend of the changes with different binding partners will be the same. FRET histograms were obtained by 1D projection of 2D E*-S plots onto the E* axis after dual-channel burst search.
To determine the statistical significance of the differences between FRET histograms, we adapted the nonparametric two-sample Kolmogorov–Smirnov (K-S) test (44, 45). As noted earlier, the K-S test tends to be overly sensitive to data with a large sample size, and we found the same to be true if this test was applied without modification to single-molecule data; moreover, it is important to distinguish between statistical versus biological significance of the difference between histograms for different sets of data, which contain intrinsic variability from instrument, sample preparation, and other factors (46, 47). To obtain more reliable comparisons, we regenerated FRET histograms from randomly selected subsets (with sample size m) of the complete data for each experimental condition and performed the K-S test. Ten repetitions of this process generated a statistical significance score, defined as the fraction of comparisons that gave P < 0.05, for each value of m. This process was repeated five more times for each value of m and for data subsets with different m values, and the dependence of the statistical significance on sample size was plotted. Molecules with E* < 0.3 were excluded from the histograms in this analysis, as the low FRET bursts could arise from photophysical artifacts and were not interpreted in this study. As shown in Fig. S5 G and H, the statistical significance of the difference remains low even at high m (>4,000) for histograms that are the same or similar, whereas the statistical significance of the difference increases rapidly with increasing m for histograms that are different.
BVA was performed to investigate submillisecond dynamics of Get3 as described (23). Eq. S3 was used to compute the static limit, defined as the expected SD of a FRET distribution due to photon statistics (shot noise) for a given E*:
| [S3] |
where . The observed mean FRET SD (SD of E*) for each molecule was computed using Eq. S4:
| [S4] |
in which e* and E* are relative FRET efficiencies (calculated from Eq. S1) of a subburst (a subset of each burst that contains a constant number of consecutive photons) and a burst, respectively, and M is the number of subbursts in the burst. A subburst size of five was used in this study.
To quantitatively represent the dynamics of molecules from the BVA, we calculated a dynamic score (DS) for each sample as follows. To reduce error that could arise from individual bursts, which contain a small number of subbursts, we first binned bursts along the E* axis into 20 bins with a bin width of 0.05. All of the subbursts within bursts in each bin were then used to calculate the mean FRET SD for each bin (SDE*) using Eq. S5 (23):
| [S5] |
where , L is the lower bound (E* − 0.025) of the bin, U is the upper bound (E* + 0.025) of the bin, Mi is the number of subbursts in the ith burst, and eij* is the relative FRET efficiency of the jth subburst in the ith burst. The DS was calculated using Eq. S6 (23):
| [S6] |
where SDE*,static is the SDE* of simulated static molecules using the Monte Carlo method. To ensure that the score reports on the dynamics of the majority of the molecules in a given sample, only bins with ≥8% of the total number of bursts in the entire FRET histogram were used to calculate DS, and the SDE* values for each of these bins are denoted with triangles in BVA plots. To take into account the different numbers of bursts in each bin, we also computed a weighted dynamic score (WDS) to weight bins according to their size using Eq. S7:
| [S7] |
where NE* is the number of bursts in each bin.
Acknowledgments
We thank Xavier Michalet and Antonino Ingargiola for help with analysis of μs-ALEX data; H. Cho, M. Rao, and S. Wang for reagents; the D. A. Dougherty laboratory for use of HPLC; and W. M. Clemons, A. Hoelz, R. Deshaies, M. Zimmer, T. F. Miller, M. Rome, and members of the S.-o.S. and S.W. laboratories for critical discussions and comments on the manuscript. This work was supported by NIH Grant GM107368, Gordon and Betty Moore Foundation Grant GBMF2939, and a fellowship from the Weston Havens Foundation (to S.-o.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1708731114/-/DCSupplemental.
References
- 1.Akopian D, Shen K, Zhang X, Shan SO. Signal recognition particle: An essential protein-targeting machine. Annu Rev Biochem. 2013;82:693–721. doi: 10.1146/annurev-biochem-072711-164732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tsirigotaki A, De Geyter J, Šoštaric N, Economou A, Karamanou S. Protein export through the bacterial Sec pathway. Nat Rev Microbiol. 2017;15:21–36. doi: 10.1038/nrmicro.2016.161. [DOI] [PubMed] [Google Scholar]
- 3.Borgese N, Fasana E. Targeting pathways of C-tail-anchored proteins. Biochim Biophys Acta. 2011;1808:937–946. doi: 10.1016/j.bbamem.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 4.Hegde RS, Keenan RJ. Tail-anchored membrane protein insertion into the endoplasmic reticulum. Nat Rev Mol Cell Biol. 2011;12:787–798. doi: 10.1038/nrm3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Denic V, Dötsch V, Sinning I. Endoplasmic reticulum targeting and insertion of tail-anchored membrane proteins by the GET pathway. Cold Spring Harb Perspect Biol. 2013;5:a013334. doi: 10.1101/cshperspect.a013334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chartron JW, Clemons WM, Jr, Suloway CJM. The complex process of GETting tail-anchored membrane proteins to the ER. Curr Opin Struct Biol. 2012;22:217–224. doi: 10.1016/j.sbi.2012.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mateja A, et al. The structural basis of tail-anchored membrane protein recognition by Get3. Nature. 2009;461:361–366. doi: 10.1038/nature08319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bozkurt G, et al. Structural insights into tail-anchored protein binding and membrane insertion by Get3. Proc Natl Acad Sci USA. 2009;106:21131–21136. doi: 10.1073/pnas.0910223106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gristick HB, et al. Crystal structure of ATP-bound Get3-Get4-Get5 complex reveals regulation of Get3 by Get4. Nat Struct Mol Biol. 2014;21:437–442. doi: 10.1038/nsmb.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rome ME, Chio US, Rao M, Gristick H, Shan SO. Differential gradients of interaction affinities drive efficient targeting and recycling in the GET pathway. Proc Natl Acad Sci USA. 2014;111:E4929–E4935. doi: 10.1073/pnas.1411284111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang F, Whynot A, Tung M, Denic V. The mechanism of tail-anchored protein insertion into the ER membrane. Mol Cell. 2011;43:738–750. doi: 10.1016/j.molcel.2011.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rao M, et al. Multiple selection filters ensure accurate tail-anchored membrane protein targeting. eLife. 2016;5:e21301. doi: 10.7554/eLife.21301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mateja A, et al. Protein targeting. Structure of the Get3 targeting factor in complex with its membrane protein cargo. Science. 2015;347:1152–1155. doi: 10.1126/science.1261671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rome ME, Rao M, Clemons WM, Shan SO. Precise timing of ATPase activation drives targeting of tail-anchored proteins. Proc Natl Acad Sci USA. 2013;110:7666–7671. doi: 10.1073/pnas.1222054110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stefer S, et al. Structural basis for tail-anchored membrane protein biogenesis by the Get3-receptor complex. Science. 2011;333:758–762. doi: 10.1126/science.1207125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mariappan M, et al. The mechanism of membrane-associated steps in tail-anchored protein insertion. Nature. 2011;477:61–66. doi: 10.1038/nature10362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kubota K, Yamagata A, Sato Y, Goto-Ito S, Fukai S. Get1 stabilizes an open dimer conformation of Get3 ATPase by binding two distinct interfaces. J Mol Biol. 2012;422:366–375. doi: 10.1016/j.jmb.2012.05.045. [DOI] [PubMed] [Google Scholar]
- 18.Shan SO. ATPase and GTPase tangos drive intracellular protein transport. Trends Biochem Sci. 2016;41:1050–1060. doi: 10.1016/j.tibs.2016.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burmann BM, Wang C, Hiller S. Conformation and dynamics of the periplasmic membrane-protein-chaperone complexes OmpX-Skp and tOmpA-Skp. Nat Struct Mol Biol. 2013;20:1265–1272. doi: 10.1038/nsmb.2677. [DOI] [PubMed] [Google Scholar]
- 20.Thoma J, Burmann BM, Hiller S, Müller DJ. Impact of holdase chaperones Skp and SurA on the folding of β-barrel outer-membrane proteins. Nat Struct Mol Biol. 2015;22:795–802. doi: 10.1038/nsmb.3087. [DOI] [PubMed] [Google Scholar]
- 21.He L, Sharpe T, Mazur A, Hiller S. A molecular mechanism of chaperone-client recognition. Sci Adv. 2016;2:e1601625. doi: 10.1126/sciadv.1601625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kapanidis AN, et al. Alternating-laser excitation of single molecules. Acc Chem Res. 2005;38:523–533. doi: 10.1021/ar0401348. [DOI] [PubMed] [Google Scholar]
- 23.Torella JP, Holden SJ, Santoso Y, Hohlbein J, Kapanidis AN. Identifying molecular dynamics in single-molecule FRET experiments with burst variance analysis. Biophys J. 2011;100:1568–1577. doi: 10.1016/j.bpj.2011.01.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wereszczynski J, McCammon JA. Nucleotide-dependent mechanism of Get3 as elucidated from free energy calculations. Proc Natl Acad Sci USA. 2012;109:7759–7764. doi: 10.1073/pnas.1117441109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gristick HB, et al. Mechanism of assembly of a substrate transfer complex during tail-anchored protein targeting. J Biol Chem. 2015;290:30006–30017. doi: 10.1074/jbc.M115.677328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suloway CJM, Chartron JW, Zaslaver M, Clemons WM., Jr Model for eukaryotic tail-anchored protein binding based on the structure of Get3. Proc Natl Acad Sci USA. 2009;106:14849–14854. doi: 10.1073/pnas.0907522106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jaru-Ampornpan P, et al. ATP-independent reversal of a membrane protein aggregate by a chloroplast SRP subunit. Nat Struct Mol Biol. 2010;17:696–702. doi: 10.1038/nsmb.1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liang FC, et al. Conformational dynamics of a membrane protein chaperone enables spatially regulated substrate capture and release. Proc Natl Acad Sci USA. 2016;113:E1615–E1624. doi: 10.1073/pnas.1524777113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamagata A, et al. Structural insight into the membrane insertion of tail-anchored proteins by Get3. Genes Cells. 2010;15:29–41. doi: 10.1111/j.1365-2443.2009.01362.x. [DOI] [PubMed] [Google Scholar]
- 30.Ghaemmaghami S, et al. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 31.Keenan RJ, Freymann DM, Walter P, Stroud RM. Crystal structure of the signal sequence binding subunit of the signal recognition particle. Cell. 1998;94:181–191. doi: 10.1016/s0092-8674(00)81418-x. [DOI] [PubMed] [Google Scholar]
- 32.Janda CY, et al. Recognition of a signal peptide by the signal recognition particle. Nature. 2010;465:507–510. doi: 10.1038/nature08870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ferbitz L, et al. Trigger factor in complex with the ribosome forms a molecular cradle for nascent proteins. Nature. 2004;431:590–596. doi: 10.1038/nature02899. [DOI] [PubMed] [Google Scholar]
- 34.Merz F, et al. Molecular mechanism and structure of trigger factor bound to the translating ribosome. EMBO J. 2008;27:1622–1632. doi: 10.1038/emboj.2008.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu Z, Horwich AL, Sigler PB. The crystal structure of the asymmetric GroEL-GroES-(ADP)7 chaperonin complex. Nature. 1997;388:741–750. doi: 10.1038/41944. [DOI] [PubMed] [Google Scholar]
- 36.Li C, et al. FastCloning: A highly simplified, purification-free, sequence- and ligation-independent PCR cloning method. BMC Biotechnol. 2011;11:92. doi: 10.1186/1472-6750-11-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saraogi I, Zhang D, Chandrasekaran S, Shan SO. Site-specific fluorescent labeling of nascent proteins on the translating ribosome. J Am Chem Soc. 2011;133:14936–14939. doi: 10.1021/ja206626g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schuldiner M, et al. The GET complex mediates insertion of tail-anchored proteins into the ER membrane. Cell. 2008;134:634–645. doi: 10.1016/j.cell.2008.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rothblatt JA, Meyer DI. Secretion in yeast: Reconstitution of the translocation and glycosylation of α-factor and invertase in a homologous cell-free system. Cell. 1986;44:619–628. doi: 10.1016/0092-8674(86)90271-0. [DOI] [PubMed] [Google Scholar]
- 40.Kapanidis AN, et al. Fluorescence-aided molecule sorting: Analysis of structure and interactions by alternating-laser excitation of single molecules. Proc Natl Acad Sci USA. 2004;101:8936–8941. doi: 10.1073/pnas.0401690101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ingargiola A, Lerner E, Chung S, Weiss S, Michalet X. FRETBursts: An open source toolkit for analysis of freely-diffusing single-molecule FRET. PLoS One. 2016;11:e0160716. doi: 10.1371/journal.pone.0160716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nir E, et al. Shot-noise limited single-molecule FRET histograms: Comparison between theory and experiments. J Phys Chem B. 2006;110:22103–22124. doi: 10.1021/jp063483n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee NK, et al. Accurate FRET measurements within single diffusing biomolecules using alternating-laser excitation. Biophys J. 2005;88:2939–2953. doi: 10.1529/biophysj.104.054114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Young IT. Proof without prejudice: Use of the Kolmogorov-Smirnov test for the analysis of histograms from flow systems and other sources. J Histochem Cytochem. 1977;25:935–941. doi: 10.1177/25.7.894009. [DOI] [PubMed] [Google Scholar]
- 45.Feller W. On the Kolmogorov-Smirnov limit theorems for empirical distributions. Ann Math Stat. 1948;19:177–189. [Google Scholar]
- 46.Cox C, Reeder JE, Robinson RD, Suppes SB, Wheeless LL. Comparison of frequency distributions in flow cytometry. Cytometry. 1988;9:291–298. doi: 10.1002/cyto.990090404. [DOI] [PubMed] [Google Scholar]
- 47.Lampariello F. On the use of the Kolmogorov-Smirnov statistical test for immunofluorescence histogram comparison. Cytometry. 2000;39:179–188. doi: 10.1002/(SICI)1097-0320(20000301)39:3<179::AID-CYTO2>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]