Significance
The incorporation of a small organic molecule into a protein core opens the door to create previously inaccessible three-dimensional structures. When combined with modern computational methods, we show that CovCore proteins can be designed with predictable folds. The small organic molecule is incorporated as an intrinsic part of the protein core, forming both covalent and noncovalent interactions, which help define the unique tertiary structures. The design methodology and experimental strategies are compatible with combinatorial library screening methods and hence hold promise for a variety of applications including inhibitors of protein–protein interactions.
Keywords: covalent core, protein design, mesosize protein, chemical protein synthesis, computational design
Abstract
The folding of natural proteins typically relies on hydrophobic packing, metal binding, or disulfide bond formation in the protein core. Alternatively, a 3D structure can be defined by incorporating a multivalent cross-linking agent, and this approach has been successfully developed for the selection of bicyclic peptides from large random-sequence libraries. By contrast, there is no general method for the de novo computational design of multicross-linked proteins with predictable and well-defined folds, including ones not found in nature. Here we use Rosetta and Tertiary Motifs (TERMs) to design small proteins that fold around multivalent cross-linkers. The hydrophobic cross-linkers stabilize the fold by macrocyclic restraints, and they also form an integral part of a small apolar core. The designed CovCore proteins were prepared by chemical synthesis, and their structures were determined by solution NMR or X-ray crystallography. These mesosized proteins, lying between conventional proteins and small peptides, are easily accessible either through biosynthetic precursors or chemical synthesis. The unique tertiary structures and ease of synthesis of CovCore proteins indicate that they should provide versatile templates for developing inhibitors of protein–protein interactions.
Advances in chemical synthesis technologies have opened the possibility of creating small proteins with new chemical compositions and folds inaccessible to natural proteins. However, designing such mesosize proteins to incorporate nonnatural structural motifs is an outstanding challenge. Previously, nonnatural cross-linking strategies have been used with combinatorial selection methods to discover cyclic peptides that block protein–protein interactions (1–6). The macrocyclic restraints combined with high-throughput library-screening and display techniques enabled developing such cross-linked peptides into effective protein inhibitors (1, 2, 7–9). However, these efforts focused on the use of cross-linkers to stabilize simple secondary structures such as α-helices (2, 3, 10, 11) or to discover bicyclic polypeptides without any predetermined 3D structures (1, 3). There have been no general computational strategies to design highly cross-linked proteins with predetermined tertiary structures that incorporate small molecule cross-links as an integral part of their cores. Such an endeavor would require new computational methods for placing cross-links during the process of computationally sampling backbone conformations and designing sequences and would require the expansion of existing protein energy functions to permit accurate modeling of the conformational flexibility of a small-molecule cross-link. We extended the use of Rosetta software suite (12) to permit such computational design and also developed complementary approaches using the Tertiary Motifs (TERMs) software package (13) to create well-structured mesoscaled proteins that incorporate the covalent cross-linker as an integral part of the folded core. Using these approaches, we designed multicyclic proteins that incorporate both side chain–side chain as well as backbone cyclization strategies, demonstrating the generality of the methods employed.
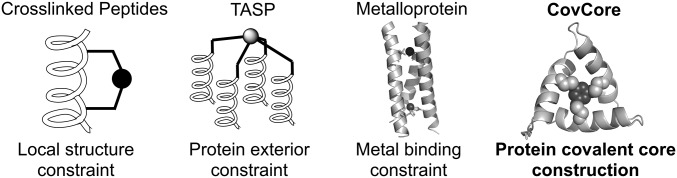
The folding of natural proteins typically depends on the formation of a well-packed core dictating the relative orientations of pieces of secondary and supersecondary structure (14–16). Small, disulfide-rich peptides, on the other hand, can derive much of their structural stability from covalent cross-links, which are sometimes augmented by a small hydrophobic core. However, disulfides are not always synthetically accessible, because correct cysteine pairing can be difficult to achieve. Furthermore, disulfides are reductively labile, presenting limitations for in vivo applications (17). Thus, considerable effort has been expended to design alternate strategies to stabilize the folded conformations of proteins and related biomimetic polymers such as foldamers (18). To stabilize simple secondary structures, covalent cross-links involving multifunctional small molecule linkers (19) have been used to considerable advantage (10, 11, 20–24) (shown in Fig. 1). Alternatively, to stabilize the cooperatively folded cores of proteins, Marsh and Tirrell have pioneered the use of fluorinated side chains (25, 26). The template assembled synthetic proteins (TASP) method has also been widely explored to facilitate the predictable assembly of helical bundles; hemes (27), porphyrins (28), cyclotribenzylene (29), metal chelating groups such as bipyridyl derivatives (30), and small cyclic peptides (31) have been used as templates for this class of proteins (31). However, in TASP proteins, the template serves as an appendage rather than being an integral part of the folding core. Metal-binding sites have also been inserted into the core of proteins (32–35), although metal ligand exchange reactions and endogenous metal chelators might limit their applicability in vivo.
Fig. 1.
Different strategies to covalently constrain peptides/proteins. CovCore focuses on protein covalently cross-linked core to achieve unique tertiary structures; shown is a CovCore molecule stabilized by a trivalent cross-linker.
The fully covalent cross-linking method described by Winter and coworkers (1, 3), which utilizes bifunctional and trifunctional benzylic cross-linkers, has already shown impressive applications for the selection of small peptide inhibitors of protein–protein interactions when combined with combinatorial phage display methods. However, the ability to design somewhat larger proteins with predictable tertiary structures incorporating these covalently linked cores (CovCore) has not been achieved. As initial targets, we designed roughly C2- and C3-symmetric protein folds, stabilized by bivalent or trivalent linkers (xylyl and mesityl, respectively). The cross-link is formed via thioether formation between the appropriate benzylic bromide and Cys thiolates of the protein, allowing use of either chemically synthesized or biosynthetically derived precursors, compatible with a variety of screening methods and either chemically or biosynthetically produced libraries. We also explored the incorporation of backbone cyclization, which could add additional conformational restriction and stability as needed.
The structures of the designed proteins were determined by NMR and X-ray crystallography and were, in most cases, in good agreement with design. The overall folds of the proteins were well defined in each case, and they conform well to the design. Some variability in the fine-tuned details was observed in one case. In fact, some flexibility and conformational variability might be desirable for future applications in which these CovCore proteins are used as templates for computational or experimental selection of variants that bind a given target.
Results and Discussion
Design and Characterization of Cyclic Antiparallel Helical Hairpins.
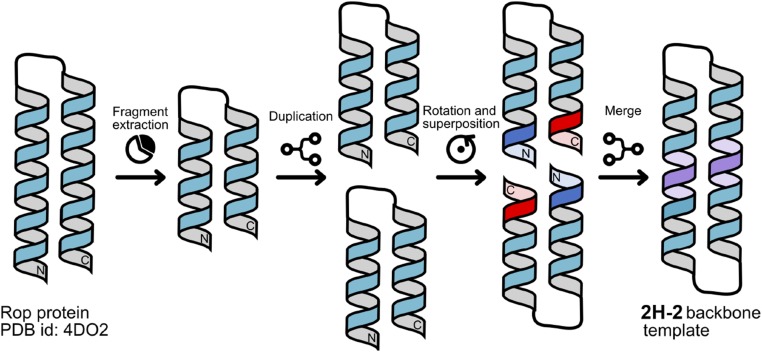
Given the frequent occurrence of helical hairpins in protein tertiary structures, we initially explored the design of a helix–turn–helix motif, stabilized by backbone cyclization as well as side chain cross-linking. Arora and coworkers (36, 37) reported the design of similar helical hairpin motifs using an unnatural cross-linker in combination with disulfides previously. The helix–loop–helix motif from the Rop protein (38) (PDB ID 4DO2) was used as the starting template and was symmetrized to define the main chain of a backbone-cyclized helical hairpin (2H-2) as described in Fig. 2 and SI Appendix. This protein was further constrained through small molecule-mediated side chain–side chain cross-linking via thioether bonds, yielding a bicyclic structure (2H) (Fig. 3).
Fig. 2.
Design procedure of a helical hairpin (2H-2) backbone. Crystal structure of the Rop protein (PDB ID 4DO2) was extracted and duplicated, and the two duplicates were merged at the terminal helices (i.e., the blue helix was merged with the red helix to produce the purple helix) to generate 2H2-2 backbone template.
Fig. 3.
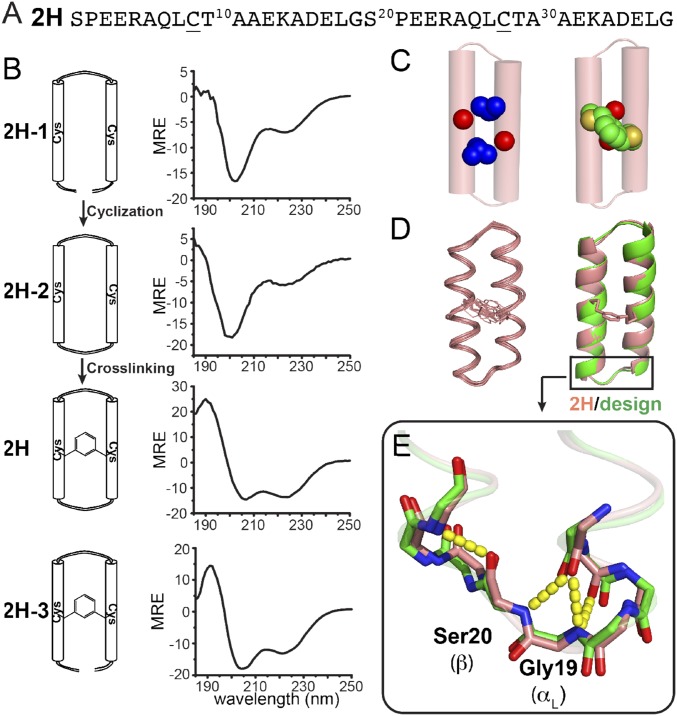
Structural characterization of the covalently constrained helical hairpins. (A) Primary sequence of 2H. Underlined residues are the cross-linking sites. (B) CD spectra of different helical dimer constructs. Unit for mean residue ellipticity (MRE) is 103 deg cm2 dmol−1. (C) (Left) Key Alacoil packing in the solution NMR structure of 2H (red spheres represent Ala11 and Ala30 side chains, and blue spheres represent Leu8 and Leu27 side chains). (Right) The semicovalent core in 2H. (D) (Left) Ensembles of 20 lowest-energy NMR structures (only the backbone is shown). Root-mean-square deviation (RMSD) is 0.3 ± 0.1 Å. (Right) Structure comparison of designed 2H model (green; only backbone is shown) and lowest-energy 2H NMR model (light pink) (Cα RMSD 0.6 Å). (E) A zoomed-in view of the αL-β motif in 2H and designed model. Yellow dash indicates hydrogen bonds in the αL-β motif; color code is the same as in D.
The sequence design of 2H-2 was guided by TERMs-based structural database mining (13), which guides selection of residues that stabilize both the main chain conformation as well as the interhelical packing (39, 40). The final design (2H-2) has an alanine coil conformation (41) with alanine residues packing tightly between the two helices (Fig. 3C, Left). Additionally, C-cap glycine and N-cap serine residues ensure helix stabilization by capping interactions. An interhelical xylyl cross-link was introduced between Cys residues at positions conductive to forming the cross-link in low-energy rotamers (2H).
The effects of backbone and side chain cyclization on the conformational stability of the peptide were evaluated by circular dichroism (CD) spectroscopy (Fig. 3B). The linear peptide (2H-1) displayed very little helical content before introduction of the xylyl thioether cross-linker. However, the introduction of the xylyl cross-linker in 2H-3 led to a significant increase in helical content as assessed from the double minimum at 222 and 208 nm in the CD spectrum and a cooperative unfolding transition (SI Appendix, Fig. S5). The bicyclic peptide (2H) had a CD spectrum very similar to the side chain cross-linked peptide (2H-3), indicating that the additional backbone cyclic restraint was structurally well accommodated, although it was not required to achieve a helical conformation. However, the converse was not true, because the backbone cyclized peptide (2H-2) showed low helicity in the absence of the xylyl cross-link. Thus, in future applications, variants of the side chain cross-linked peptide can be used to discover binders, and the resulting monocyclic peptides could be further stabilized toward proteolytic degradation through the introduction of an additional main chain cyclic restraint.
The solution NMR structure of 2H is in excellent agreement with the designed model (Fig. 3D). The backbone heavy atom root-mean-square deviation (RMSD) of 20 lowest-energy structures (Fig. 3D) is 0.3 ± 0.1 Å for 2H. The overlay of the designed model (green) and the lowest-energy NMR structure is almost within the precision of the experimentally determined structural ensemble, with a Cα RMSD of 0.6 Å (Fig. 3D). Finally, the hydrogen-bonded capping interactions of the serine and glycine residues of 2H (SI Appendix, Table S3) at the helical ends were precisely as specified in the design (Fig. 3E).
To explore further the role of the xylyl linker in 2H, we synthesized a variant in which the two Cys residues used in the cross-linking reaction were replaced with a pair of hydrophobic Leu residues that could form a favorable interfacial packing between the two helices. This side chain substitution was evaluated in both the linear as well as the main chain-cyclized form of the peptide. The linear peptide, 2H-4, showed little helical content, although helicity was restored in main chain cyclized peptide, 2H-5. The NMR structure of 2H-5, also determined by solution NMR, was found to conform well to the guiding structure (RMSD = 1.2 Å; SI Appendix, Fig. S7).
Together, these studies show that a side chain cross-link can serve as a powerful restraint to stabilize the fold of a relatively short helical hairpin, smaller than those seen in isolation in natural proteins. Thus, xylyl and related side chain cross-linking strategies might be used to stabilize minimal versions natural proteins. We next focused on the design of a more highly cross-linked protein with an entirely novel fold stabilized by a trivalent cross-linker, which forms an integral part of the hydrophobic core.
The Design of Tricyclic CovCore Protein 3H1.
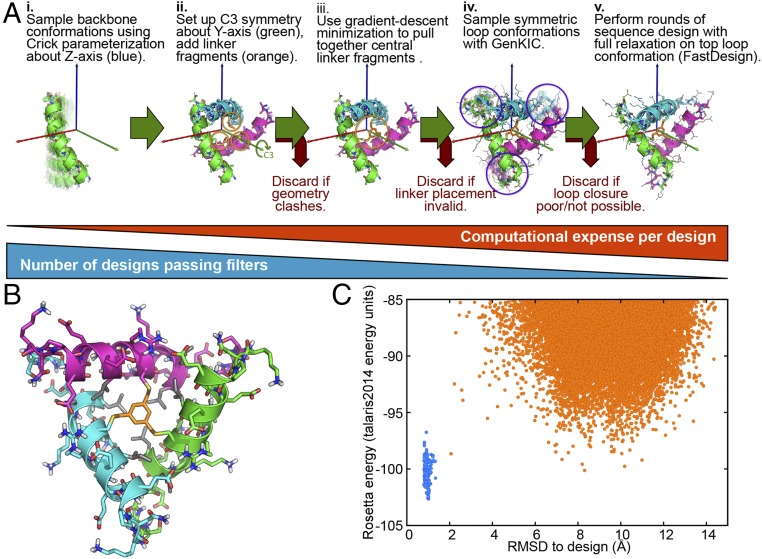
We used the Rosetta software suite (12) to design a three-helix structure not previously seen in nature. In the designed protein 3H1, a 1,3,5-trimethyl benzene group forms three thioether bonds, which nucleates a small hydrophobic core near the center of the structure (Fig. 4). We first made use of parametric backbone generation strategies to create various helical conformations of a monomer, which were symmetry-replicated to create C3-symmetrical trimers. The orientation of the helices within the trimeric structure was varied by sampling different orientations of the monomer relative to the threefold symmetry axis, using the Rosetta symmetry machinery to impose C3 symmetry on the system. Rosetta’s generalized kinematic closure module was next used to sample loop conformations connecting the helices (42). For each loop conformation sampled, we employed Rosetta symmetric sequence design tools to design a sequence for the helices and loops, making use of the LayerDesign tool to enforce a hydrophobic core and polar surface (43). We chose the most promising candidates by Rosetta energy and by visual inspection. Final sequences were validated by Rosetta ab initio structure prediction. Although we used symmetry to simplify the initial designs, fully asymmetric structures can easily be generated using a simple modification of the search procedure. Full details of the design protocol and design scripts are included in SI Appendix. 3H1 was prepared using three segments one-pot native chemical ligation methods as described in SI Appendix (44–47).
Fig. 4.
The design of 3H1, a tricyclic, C3-symmetric protein. (A) Steps in the design process. (B) The designed 3H1 model. (C) 3H1 energy landscape predicted by Rosetta ab initio structure prediction. Blue points represent relaxation of the designed model, and orange points are from independent conformation sampling trajectories using Rosetta’s ab initio structure prediction module.
Biophysical Characterization of Tricyclic CovCore Protein 3H1.
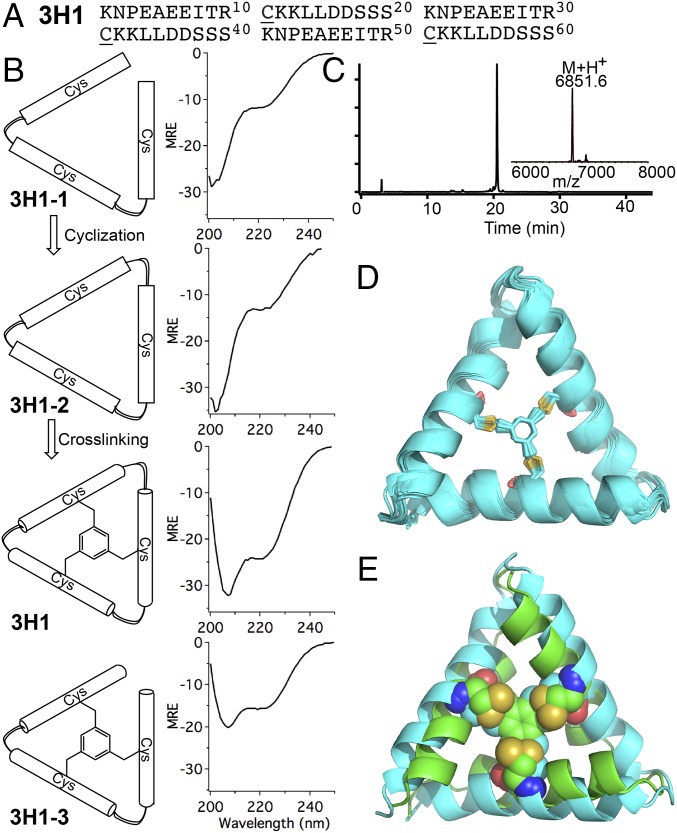
To differentiate between the importance of the backbone versus side chain cyclic restraints, we characterized the structures and stability of the linear precursor (3H1-1), the backbone cyclized precursor (3H1-2), the asymmetric side chain cross-linked bicyclic peptide (3H1-3), and the fully symmetrical backbone and side chain cross-linked tricyclic peptide (3H1). CD spectroscopy showed that the linear precursor (3H1-1) and backbone cyclized precursor (3H1-2) had very low helical content (Fig. 5B). By contrast, the side chain cross-linked bicyclic and tricyclic CovCore protein were highly helical, indicating that the backbone cyclization is not required for structure formation, an important consideration for methods such as phage display where head-to-tail main chain cyclization cannot be easily achieved. Thermal unfolding curves of 3H1 show only a small linear change in the folded baseline, indicating that the protein remains folded up to 95 °C (SI Appendix).
Fig. 5.
Characterization of tricyclic CovCore protein 3H1. (A) 3H1 sequence. Underlined residues are the cross-linking sites. (B) CD spectra of linear precursor 3H1-1, backbone cyclized precursors 3H1-2, tricyclic 3H1, and side chain cross-linked bicyclic 3H1-3. Unit for mean residue ellipticity (MRE) is 103 deg cm2 dmol−1. (C) Analytical HPLC and mass spectrometry analysis of 3H1, Obsd. 6,850.6 ± 0.5 Da, Calc. 6,850.6 Da (average isotope composition). (D) Twenty lowest-energy NMR models of 3H1. (E) Structure comparison between the designed 3H1 model (green) and lowest-energy 3H1 NMR model (cyan; RMSD 2.9 Å).
The solution NMR structure of 3H1 is in reasonable agreement with its designed structure. Only 20 unique assignable amino acid residues were observed, indicating 3H1 is indeed threefold symmetric as in the design. The most significant difference between the observed and designed structures (RMSD = 2.9 Å) is that the α-helices are straighter in the NMR solution structure. The straightening of the helices leads to a slight expansion of its triangular shape and somewhat looser packing of the apolar side chains around the covalent cross-linker. Overall, however, the experimentally determined topology matches the designed topology. Significantly, because 3H1 has a very small hydrophobic core responsible for stabilizing its structure (consisting of one Cys and two hydrophobic residues per helix, shown in gray in Fig. 4B) the great majority of the sequence can be varied for molecular recognition of targets.
The Design of Tricyclic CovCore Protein 3H2.
We next performed the redesign of 3H1 to increase its structure stability and also to reduce its hydrophilicity (to facilitate crystallization). We first used Rosetta and a loop-searching algorithm to search the best loops for bridging the adjacent helices. We searched the PDB for close structural matches (<1 Å) for the four terminal and four initial residues of each helix (e.g., 16DDSS19 and 23PEAE26 in 3H1) and found that the most common loop sequence, excluding those with proline, was –NGD– followed by –GGD–. We then used Rosetta’s fixed backbone side chain design application (48) to redesign the sequence for more compact hydrophobic core packing as well as to reduce the hydrophilicity of 3H1. At solvent-exposed positions, we selected amino acids to reduce the protein’s charge and surface-exposed side chain entropy. At hydrophobic core positions, we allowed Rosetta to sample apolar amino acids. The final redesigned sequence (3H2) is shown in Fig. 6. Due to the expected difficulties of -Asn-Gly- aspartimide formation during synthesis, -GGD– was chosen as the final loop sequence instead of –NGD– (detailed sequence redesign is described in SI Appendix).
Fig. 6.
3H2 sequence comparison with 3H1. Amino acids colored in red are in the loop regions based on 3H1 NMR solution structure (sequence redesign details shown in SI Appendix).
Biophysical Characterization of the Tricyclic CovCore Protein 3H2.
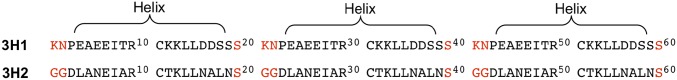
Both linear precursor (3H2-1) and backbone cyclized precursor (3H2-2) were predominantly α-helical based on CD spectra (Fig. 7B) with thermal melting temperature of 55.8 °C for linear precursor (3H2-1) and 64.9 °C for cyclized precursor (3H2-2) (SI Appendix, Fig. S5). After cross-linking with a mesityl group, the mean residue ellipticity at both 208 and 222 nm increased considerably irrespective of the main chain backbone cyclization (Fig. 7B). Furthermore, 3H2 remained essentially fully folded up to 95 °C, indicating that the covalent core indeed improved the stability of 3H2 dramatically. Such high thermal stability is very frequently observed in de novo designed helical proteins (49–51). 3H2 thermal melting also displays high thermal stability; the mean residue ellipticity of 3H2 is higher than that of 3H1 at all temperatures tested (SI Appendix, Fig. S5), demonstrating that the thermal stability of 3H2 is indeed enhanced from 3H1.
Fig. 7.
Characterization of tricyclic Covcore protein 3H2. (A) 3H2 sequence. Underlined residues are the cross-linking sites. (B) CD spectra of linear precursor 3H2-1, backbone cyclized precursors 3H2-2, tricyclic 3H2, and side chain cross-linked 3H2-3. Unit for mean residue ellipticity (MRE) is 103 deg cm2 dmol−1. (C) Analytical HPLC and mass spectrometry analysis of 3H2, Mass: Obsd. 6,280.4 ± 0.5 Da, Calc. 6,280.2 Da (average isotope composition). (D) Representative electron density maps of the amide bond between Gly1and Ser60. (E) The electron density maps of the covalent core. σA-weighted 2Fo − Fc electron density map at a σ level of 1.
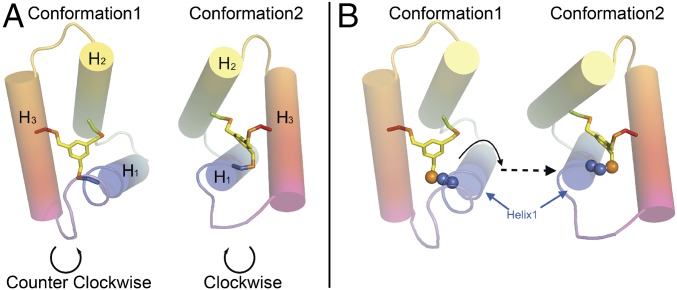
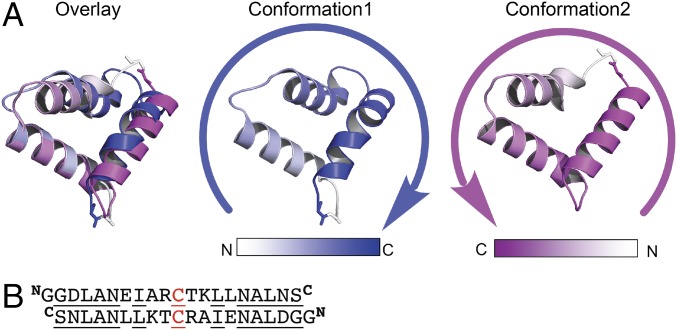
The crystallographic structure of 3H2 at 1.20 Å resolution was solved through direct methods using Arcimboldo (52) to a crystallographic R-factor of 0.144 (R-free 0.178) in CCP4 (53). The 3H2 X-ray structure showed interesting deviations from the symmetrical, designed model. There are six 3H2 protein molecules in the asymmetric unit, with four (chains A–D) molecules adopting one conformation and the other two molecules (chains E and F) adopting a different conformation. In each conformer, the three helices differ in length. The flexibility to adopt multiple asymmetric sequences appears to be in part due to the inclusion of a highly flexible diglycine linker. The topology of conformation 1 is opposite to that of conformation 2. When viewed from the bottom of the bundle, the three helices (H1–H3) are arranged in a counterclockwise direction in conformation 1, whereas they are arranged in a clockwise direction in conformation 2 as shown in Fig. 8A.
Fig. 8.
Overall structure comparisons of the two conformations of 3H2. (A) Opposite topologies are observed in these two different conformations. The three helices (H1–H3) from conformation 1 are arranged in a counterclockwise direction, and the same three helices (H1–H3) in conformation 2 are arranged in a clockwise direction. (B) Visualization of the translation and rotation of H3, and rotation of H1, to convert one conformation into the other (cysteine side chain of H1 is shown as spheres).
To adopt these two different topologies, the molecule must accommodate several critical structural changes. Although helix 2 essentially remains constant, with its cysteine side chain pointing in the same orientation, helix 1 rolls about its axis by 125° from one conformation to the other (Fig. 8B), so that its cysteine side chain is pointing in nearly the opposite direction. Helix 1 also shifts its crossing angle relative to helix 2 by 18.3°.
Another interesting aspect of this molecule is the fact that these two different conformations of 3H2 are retropeptides to one other. That is, conformation 1 and conformation 2 can nearly be superimposed if they are overlaid in reverse directions (shown in Fig. 9A). One possible explanation for this observation is the binary pattern of hydrophobic and polar residues in the sequence of 3H2 is largely invariant, irrespective of whether the sequence is read in the N-to-C or C-to-N direction (after aligning on the central cross-linking cysteine) (Fig. 9B).
Fig. 9.
3H2 is a retropeptide. (A) The two conformations of 3H2 can be overlaid when the peptide chain running in opposite directions. In conformation 1, white to blue color gradient indicates N–C direction as is shown with blue arrow; in conformation 2, white to magenta color gradient indicates N–C direction as is shown with magenta arrow. (B) Sequence alignment of 3H2 in opposite directions. The sequence is aligned on the cross-linking Cys residue. Only 20 residues are shown here. Residues underscored are either same or very similar amino acids.
To confirm that this asymmetric structure is not due to crystal packing alone, we performed NMR experiments to determine the solution behavior of 3H2. 3H2 has three isoleucine residues in its sequence, which would be in identical chemical environments in a C3-symmetric structure. From the natural abundance 13C-HSQC spectrum (SI Appendix, Fig. S8), we can clearly see three different isoleucine Cδ1–Hδ1 peaks, confirming that these three isoleucine residues are indeed in different chemical environments and that the 3H2 structure also lacks threefold symmetry in solution. The fact that we only observed three isoleucine peaks from NMR measurement suggests there is only one conformation present in solution structure. From the crystal structure, we found that four residues in conformation 2 (chain E) adopted left-handed helix φ and ψ angles: Arg10 (59.4° 36.6°), Asp23 (66.2°, 21.7°), Asn39 (47.1°, 36.0°), and Asn59 (39.4°, 53.7°). This observation suggests that conformation 2 is in a high-energy state, which might convert to conformation 1 in solution. However, due to the sequence degeneracy, we were unable to determine the solution NMR structure of 3H2.
The fact that 3H2 has this asymmetric structure presents a practical advantage because structures of this type are not easily accessible in conventional proteins. In fact, this asymmetric scaffold serendipitously opens new doors in terms of inhibiting protein–protein interactions by providing several unique surfaces that would not have been present in a symmetric structure. Thus, depending on the shapes of different protein targets, PPI inhibitor designers can choose different surfaces from the asymmetric scaffold to design to complement targeted proteins, leading to better chances of generating specific inhibitors.
Conclusions
The design of novel proteins often relies on hydrophobic core packing, metal binding, or disulfides to achieve tertiary structures. By comparison, computational design of proteins with covalently bonded molecules has been more challenging. In this work, we employed computational design approaches to create nonnatural CovCore protein scaffolds that have not been accessible previously. Chemical protein synthesis was used to prepare and validate the structures of the designed molecules efficiently. The inclusion of xylyl and mesityl cross-linkers, both to provide covalent constraints and to form a hydrophobic core, permits these molecules to adopt distinctive tertiary structures that are not easily accessible to conventional proteins. Also, these cross-links are fully compatible with display technologies for high-throughput screening. The predetermined overall folds of these CovCore proteins should allow integration of computational design capability for the redesign of these molecules to inhibit protein–protein interactions.
Materials and Methods
All materials used in the experiments were obtained through commercial sources that are described in the SI Appendix in detail. Detailed protein design procedures are also described in SI Appendix. All proteins designed were prepared using chemical synthesis.
Supplementary Material
Acknowledgments
We thank Lijun Liu for helping with X-ray structure determination. We thank Taia Wu for helping with differential scanning fluorimetry experiments. Beamline 8.3.1 at the Advanced Light Source is operated by the University of California Office of the President, Multicampus Research Programs and Initiatives Grant MR-15-328599, and Program for Breakthrough Biomedical Research, which is partially funded by the Sandler Foundation. Additional support comes from National Institutes of Health (NIH) (GM105404, GM073210, GM082250, and GM094625), National Science Foundation (1330685), Plexxikon Inc., and the M.D. Anderson Cancer Center. The Advanced Light Source is a national user facility operated by Lawrence Berkeley National Laboratory on behalf of the US Department of Energy (US DOE) under Contract DE-AC02-05CH11231, Office of Basic Energy Sciences, through the Integrated Diffraction Analysis Technologies program, supported by the US DOE of Biological and Environmental Research. This work was supported in part by Grants GM54616 and GM122603 from the NIH (to W.F.D.). An award of computer time was provided by the Innovative and Novel Computational Impact on Theory and Experiment program. This research used resources of the Argonne Leadership Computing Facility, which is a US DOE Office of Science User Facility supported under Contract DE-AC02-06CH11357.
Footnotes
The authors declare no conflict of interest.
Data deposition: NMR structures have been deposited in Protein Data Bank [PDB ID codes 5WOC (2H), 5WOD (2H-5), and 5V2G (3H1)] and in Biological Magnetic Resonance Bank [ID codes 30319 (2H), 30320 (2H-5), and 30267 (3H1)]. The 3H2 structure has been deposited in Protein Data Bank (PDB ID code 5V2O).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1710695114/-/DCSupplemental.
References
- 1.Heinis C, Rutherford T, Freund S, Winter G. Phage-encoded combinatorial chemical libraries based on bicyclic peptides. Nat Chem Biol. 2009;5:502–507. doi: 10.1038/nchembio.184. [DOI] [PubMed] [Google Scholar]
- 2.Diderich P, et al. Phage selection of chemically stabilized α-helical peptide ligands. ACS Chem Biol. 2016;11:1422–1427. doi: 10.1021/acschembio.5b00963. [DOI] [PubMed] [Google Scholar]
- 3.Heinis C, Winter G. Encoded libraries of chemically modified peptides. Curr Opin Chem Biol. 2015;26:89–98. doi: 10.1016/j.cbpa.2015.02.008. [DOI] [PubMed] [Google Scholar]
- 4.Jackson S, et al. Template-constrained cyclic-peptides—Design of high-affinity ligands for GpIIb/IIIa. J Am Chem Soc. 1994;116:3220–3230. [Google Scholar]
- 5.Hill TA, Shepherd NE, Diness F, Fairlie DP. Constraining cyclic peptides to mimic protein structure motifs. Angew Chem Int Ed Engl. 2014;53:13020–13041. doi: 10.1002/anie.201401058. [DOI] [PubMed] [Google Scholar]
- 6.Yu CX, Taylor JW. A new strategy applied to the synthesis of an alpha-helical bicyclic peptide constrained by two overlapping i, i+7 side-chain bridges of novel design. Tetrahedron Lett. 1996;37:1731–1734. [Google Scholar]
- 7.Angelini A, et al. Bicyclic peptide inhibitor reveals large contact interface with a protease target. ACS Chem Biol. 2012;7:817–821. doi: 10.1021/cb200478t. [DOI] [PubMed] [Google Scholar]
- 8.Baeriswyl V, et al. Development of a selective peptide macrocycle inhibitor of coagulation factor XII toward the generation of a safe antithrombotic therapy. J Med Chem. 2013;56:3742–3746. doi: 10.1021/jm400236j. [DOI] [PubMed] [Google Scholar]
- 9.Diderich P, Heinis C. Phage selection of bicyclic peptides binding Her2. Tetrahedron. 2014;70:7733–7739. [Google Scholar]
- 10.Phelan JC, Skelton NJ, Braisted AC, McDowell RS. A general method for constraining short peptides to an alpha-helical conformation. J Am Chem Soc. 1997;119:455–460. [Google Scholar]
- 11.Lau YH, de Andrade P, Wu Y, Spring DR. Peptide stapling techniques based on different macrocyclisation chemistries. Chem Soc Rev. 2015;44:91–102. doi: 10.1039/c4cs00246f. [DOI] [PubMed] [Google Scholar]
- 12.Das R, Baker D. Macromolecular modeling with Rosetta. Annu Rev Biochem. 2008;77:363–382. doi: 10.1146/annurev.biochem.77.062906.171838. [DOI] [PubMed] [Google Scholar]
- 13.Zheng F, Zhang J, Grigoryan G. Tertiary structural propensities reveal fundamental sequence/structure relationships. Structure. 2015;23:961–971. doi: 10.1016/j.str.2015.03.015. [DOI] [PubMed] [Google Scholar]
- 14.Dill KA, et al. Principles of protein folding—A perspective from simple exact models. Protein Sci. 1995;4:561–602. doi: 10.1002/pro.5560040401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kauzmann W. Some factors in the interpretation of protein denaturation. Adv Protein Chem. 1959;14:1–63. doi: 10.1016/s0065-3233(08)60608-7. [DOI] [PubMed] [Google Scholar]
- 16.Nick Pace C, Scholtz JM, Grimsley GR. Forces stabilizing proteins. FEBS Lett. 2014;588:2177–2184. doi: 10.1016/j.febslet.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trivedi MV, Laurence JS, Siahaan TJ. The role of thiols and disulfides on protein stability. Curr Protein Pept Sci. 2009;10:614–625. doi: 10.2174/138920309789630534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng RP, Gellman SH, DeGrado WF. Beta-peptides: From structure to function. Chem Rev. 2001;101:3219–3232. doi: 10.1021/cr000045i. [DOI] [PubMed] [Google Scholar]
- 19.Henchey LK, Jochim AL, Arora PS. Contemporary strategies for the stabilization of peptides in the alpha-helical conformation. Curr Opin Chem Biol. 2008;12:692–697. doi: 10.1016/j.cbpa.2008.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumita JR, Smart OS, Woolley GA. Photo-control of helix content in a short peptide. Proc Natl Acad Sci USA. 2000;97:3803–3808. doi: 10.1073/pnas.97.8.3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schafmeister CE, Po J, Verdine GL. An all-hydrocarbon cross-linking system for enhancing the helicity and metabolic stability of peptides. J Am Chem Soc. 2000;122:5891–5892. [Google Scholar]
- 22.Jo H, et al. Development of α-helical calpain probes by mimicking a natural protein-protein interaction. J Am Chem Soc. 2012;134:17704–17713. doi: 10.1021/ja307599z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yin H. Constrained peptides as miniature protein structures. ISRN Biochem. 2012;2012:692190. doi: 10.5402/2012/692190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Assem N, Ferreira DJ, Wolan DW, Dawson PE. Acetone-linked peptides: A convergent approach for peptide macrocyclization and labeling. Angew Chem Int Ed Engl. 2015;54:8665–8668. doi: 10.1002/anie.201502607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang Y, et al. Fluorinated coiled-coil proteins prepared in vivo display enhanced thermal and chemical stability. Angew Chem Int Ed Engl. 2001;40:1494–1496. doi: 10.1002/1521-3773(20010417)40:8<1494::AID-ANIE1494>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 26.Marsh EN. Fluorinated proteins: From design and synthesis to structure and stability. Acc Chem Res. 2014;47:2878–2886. doi: 10.1021/ar500125m. [DOI] [PubMed] [Google Scholar]
- 27.Sasaki T, Kaiser ET. Helichrome—Synthesis and enzymatic-activity of a designed hemeprotein. J Am Chem Soc. 1989;111:380–381. [Google Scholar]
- 28.Akerfeldt KS, et al. Tetraphilin—A four-helix proton channel built on a tetraphenylporphyrin framework. J Am Chem Soc. 1992;114:9656–9657. [Google Scholar]
- 29.Causton AS, Sherman JC. Design of proteins using rigid organic macrocycles as scaffolds. Bioorg Med Chem. 1999;7:23–27. doi: 10.1016/s0968-0896(98)00224-7. [DOI] [PubMed] [Google Scholar]
- 30.Ghadiri MR, Soares C, Choi C. A convergent approach to protein design—Metal ion-assisted spontaneous self-assembly of a polypeptide into a triple-helix bundle protein. J Am Chem Soc. 1992;114:825–831. [Google Scholar]
- 31.Ernest I, Vuilleumier S, Fritz H, Mutter M. Synthesis of a 4-helix bundle-like template-assembled synthetic protein (Tasp) by condensation of a protected peptide on a conformationally constrained cyclic carrier. Tetrahedron Lett. 1990;31:4015–4018. [Google Scholar]
- 32.Salgado EN, et al. Metal templated design of protein interfaces. Proc Natl Acad Sci USA. 2010;107:1827–1832. doi: 10.1073/pnas.0906852107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zastrow ML, Peacock AFA, Stuckey JA, Pecoraro VL. Hydrolytic catalysis and structural stabilization in a designed metalloprotein. Nat Chem. 2011;4:118–123. doi: 10.1038/nchem.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu F, et al. Protein design: Toward functional metalloenzymes. Chem Rev. 2014;114:3495–3578. doi: 10.1021/cr400458x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Churchfield LA, Medina-Morales A, Brodin JD, Perez A, Tezcan FA. De novo design of an allosteric metalloprotein assembly with strained disulfide bonds. J Am Chem Soc. 2016;138:13163–13166. doi: 10.1021/jacs.6b08458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Watkins AM, Wuo MG, Arora PS. Protein-protein interactions mediated by helical tertiary structure motifs. J Am Chem Soc. 2015;137:11622–11630. doi: 10.1021/jacs.5b05527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wuo MG, Mahon AB, Arora PS. An effective strategy for stabilizing minimal coiled coil mimetics. J Am Chem Soc. 2015;137:11618–11621. doi: 10.1021/jacs.5b05525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Amprazi M, et al. Structural plasticity of 4-α-helical bundles exemplified by the puzzle-like molecular assembly of the Rop protein. Proc Natl Acad Sci USA. 2014;111:11049–11054. doi: 10.1073/pnas.1322065111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Engel DE, DeGrado WF. Amino acid propensities are position-dependent throughout the length of alpha-helices. J Mol Biol. 2004;337:1195–1205. doi: 10.1016/j.jmb.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 40.Engel DE, DeGrado WF. Alpha-alpha linking motifs and interhelical orientations. Proteins. 2005;61:325–337. doi: 10.1002/prot.20522. [DOI] [PubMed] [Google Scholar]
- 41.Gernert KM, Surles MC, Labean TH, Richardson JS, Richardson DC. The Alacoil: A very tight, antiparallel coiled-coil of helices. Protein Sci. 1995;4:2252–2260. doi: 10.1002/pro.5560041102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bhardwaj G, et al. Accurate de novo design of hyperstable constrained peptides. Nature. 2016;538:329–335. doi: 10.1038/nature19791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fleishman SJ, et al. RosettaScripts: A scripting language interface to the Rosetta macromolecular modeling suite. PLoS One. 2011;6:e20161. doi: 10.1371/journal.pone.0020161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dawson PE, Muir TW, Clark-Lewis I, Kent SB. Synthesis of proteins by native chemical ligation. Science. 1994;266:776–779. doi: 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]
- 45.Bang D, Kent SB. A one-pot total synthesis of crambin. Angew Chem Int Ed Engl. 2004;43:2534–2538. doi: 10.1002/anie.200353540. [DOI] [PubMed] [Google Scholar]
- 46.Blanco-Canosa JB, Dawson PE. An efficient Fmoc-SPPS approach for the generation of thioester peptide precursors for use in native chemical ligation. Angew Chem Int Ed Engl. 2008;47:6851–6855. doi: 10.1002/anie.200705471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fang GM, et al. Protein chemical synthesis by ligation of peptide hydrazides. Angew Chem Int Ed Engl. 2011;50:7645–7649. doi: 10.1002/anie.201100996. [DOI] [PubMed] [Google Scholar]
- 48.Kuhlman B, et al. Design of a novel globular protein fold with atomic-level accuracy. Science. 2003;302:1364–1368. doi: 10.1126/science.1089427. [DOI] [PubMed] [Google Scholar]
- 49.Jacobs TM, et al. Design of structurally distinct proteins using strategies inspired by evolution. Science. 2016;352:687–690. doi: 10.1126/science.aad8036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang PS, et al. High thermodynamic stability of parametrically designed helical bundles. Science. 2014;346:481–485. doi: 10.1126/science.1257481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Polizzi NF, et al. De novo design of a hyperstable, non-natural protein-ligand complex with sub-Å accuracy. Nat Chem. 2017 doi: 10.1038/nchem.2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Millán C, Sammito M, Usón I. Macromolecular ab initio phasing enforcing secondary and tertiary structure. IUCrJ. 2015;2:95–105. doi: 10.1107/S2052252514024117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Murshudov GN, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.