Significance
Understanding the processes that generate or breakdown reproductive isolation between species is essential to understanding evolution. Assortative mating mediates reproductive isolation between species, but its dynamics in natural populations are poorly understood. Here we show that strong assortative mating maintains reproductive isolation in a natural hybrid population following an initial breakdown when the hybrid population formed, and strongly shaped the genetic structure of this population over ∼25 generations. Intriguingly, although in the wild these mate preferences result in nearly 100% of matings occurring between similar genotypes, this barrier breaks down in the laboratory. Our results highlight the importance of assortative mating in shaping hybrid population evolution and imply that short-term breakdown in assortative mating can have long-term evolutionary consequences.
Keywords: hybridization, population structure, assortative mating, reproductive isolation
Abstract
The emergence of new species is driven by the establishment of mechanisms that limit gene flow between populations. A major challenge is reconciling the theoretical and empirical importance of assortative mating in speciation with the ease with which it can fail. Swordtail fish have an evolutionary history of hybridization and fragile prezygotic isolating mechanisms. Hybridization between two swordtail species likely arose via pollution-mediated breakdown of assortative mating in the 1990s. Here we track unusual genetic patterns in one hybrid population over the past decade using whole-genome sequencing. Hybrids in this population formed separate genetic clusters by 2003, and maintained near-perfect isolation over 25 generations through strong ancestry-assortative mating. However, we also find that assortative mating was plastic, varying in strength over time and disappearing under manipulated conditions. In addition, a nearby population did not show evidence of assortative mating. Thus, our findings suggest that assortative mating may constitute an intermittent and unpredictable barrier to gene flow, but that variation in its strength can have a major effect on how hybrid populations evolve. Understanding how reproductive isolation varies across populations and through time is critical to understanding speciation and hybridization, as well as their dependence on disturbance.
Understanding what drives reproductive isolation between populations is one of the major puzzles in evolutionary biology. Premating isolation can play an important role in the early stages of speciation, allowing for the accumulation of other isolating mechanisms (1–3). Furthermore, premating isolation can evolve in response to selection against hybridization between diverging populations (4, 5), and thus is a key mechanism of reproductive isolation at multiple stages of speciation.
On the other hand, behavioral data suggest that these mechanisms can be plastic (6). Preferences for conspecifics can be abolished or even reversed depending on social and environmental context (7–9). Thus, while premating isolation plays a primary role in divergence between populations, its breakdown also may facilitate gene flow between species. Reconciling the possible context dependence of assortative mating with its importance in reproductive isolation is crucial to understanding the mechanisms of speciation. Few studies to date have investigated how these mechanisms operate in natural populations over time.
Genomic analyses of species not currently known to hybridize, including humans, reveal the footprints of past hybridization (10–15). These patterns suggest that the evolutionary history of many species is characterized by the frequent breakdown and reestablishment of reproductive isolation. Specifically, the episodic disruption of assortative mating is likely a frequent trigger of hybridization (9, 16). Conversely, assortative mating can be an important isolating mechanism in hybrid species (17). Thus, assortative mating should be of key importance in shaping the evolution of populations immediately after hybridization (18). However, further complexity is introduced by the fact that preferences can be plastic. Preferences that are learned (19, 20), environmentally dependent (8), or frequency dependent (21, 22) could generate distinct outcomes in hybridizing populations (6, 23)—even those formed between the same parental species. Since episodes of hybridization determine the extent of gene flow between lineages, understanding them and how they are influenced by assortative mating is central to understanding reproductive isolation between species.
Here we tracked hybrid populations through time to explore these questions. Species of swordtail fish (Xiphophorus) can hybridize, but reproductive isolation in sympatry is maintained by conspecific mate preferences (24). Nevertheless, these preferences are sensitive to particular ecological conditions (7), and their breakdown may play a major role in the rampant genetic exchange observed throughout the evolutionary history of the genus (13). Hybrid populations derived from the sister species Xiphophorus birchmanni and Xiphophorus malinche first formed 35–56 generations ago, or ∼15–30 y ago (25), likely due to human-mediated disruption of pheromonal communication (7).
We combine time series genomic data from hybrid populations, genetic analysis of mother-offspring pairs, and behavioral studies from the laboratory and the wild to understand the importance of assortative mating as a barrier to gene flow in these hybrid populations. We find that one population (“Aguazarca”) is composed of two types of hybrid individuals that have developed reproductive isolation through assortative mating. By tracking populations through time, we show that assortative mating can be a strong barrier to gene flow between these hybrid clusters, but also that assortative mating is variable over time, across populations, and environments. Such variation in the strength of assortative mating may implicate behavioral plasticity or an important role of environmental factors in assortative mating in this system. Regardless of its causes, the context dependence of assortative mating is likely an important factor shaping hybrid ancestry in modern genomes.
Results
Genome-Wide Sequencing of the Aguazarca Population.
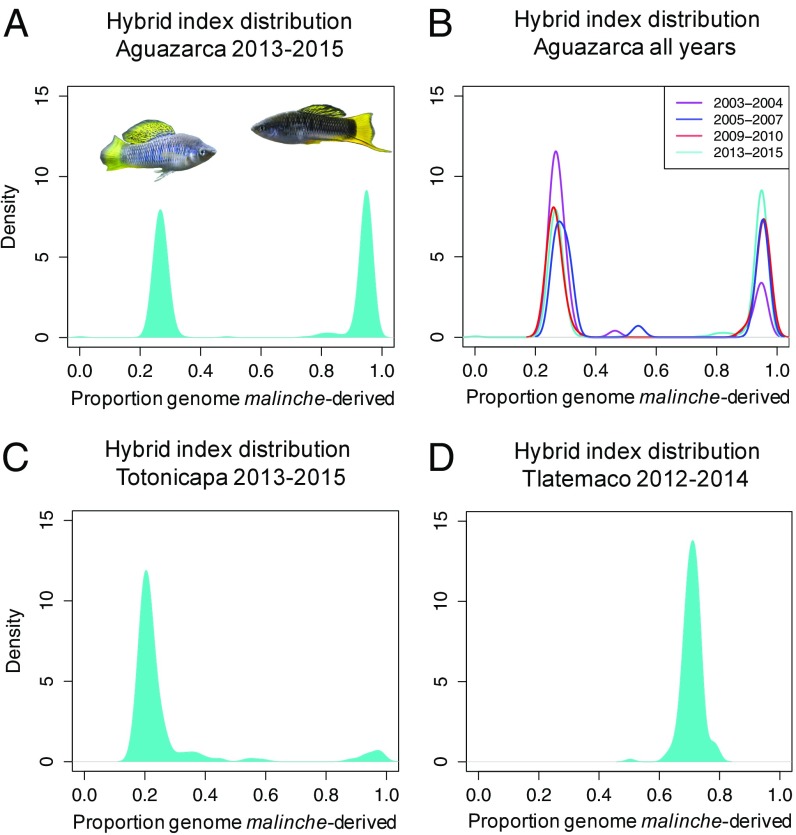
We applied a genome-wide genotyping approach known as multiplexed shotgun genotyping (MSG) (26) to characterize ancestry throughout the genomes of 642 adults that we sampled from a hybrid population, Aguazarca (SI Appendix, Fig. S1), over the course of more than a decade. We calculated the hybrid index for each individual from ∼1.3 million ancestry-informative sites. Based on these data, we show that the ancestry distribution at Aguazarca is clearly bimodal (Hartigan’s dip statistic for unimodality in 2013–2015: D = 0.21; P < 10−6) (Fig. 1A). Strikingly, adults fall into one of two hybrid clusters: a birchmanni-biased cluster with 72 ± 3% of the genome derived from X. birchmanni or a malinche-biased cluster with 95 ± 5% of the genome derived from X. malinche. Although the malinche-biased cluster has only 5% hybrid ancestry on average, comparisons to pure parentals indicate that both clusters have hybrid ancestry (SI Appendix, Fig. S2).
Fig. 1.
Ancestry distributions in three independently formed hybrid populations. Ancestry distributions in sampled hybrid populations differ dramatically. (A and B) The bimodal population structure observed at Aguazarca in 2013–2015 has been stable over the last 13 y or ∼25 generations. (C and D) This pattern contrasts markedly with the minimal population structure in two other hybrid populations. Although the Aguazarca population shows strong bimodality (A: Hartigan’s dip statistic, D = 0.21, P < 10−6), hybrid populations in other rivers show more subtle population structure (C: D = 0.024, P = 0.89; D: D = 0.02, P = 0.87).
Despite their genetic differentiation, the two clusters are fully sympatric. Individuals of each subpopulation were collected in every collection site and from the same traps over multiple years (SI Appendix, 1 and 2 and Fig. S3). Furthermore, underwater videos demonstrate that individuals from the two subpopulations come into frequent contact (SI Appendix, 2 and Movie S1), demonstrating that the subpopulations are not separated into distinct microhabitats.
Hybrid Subpopulations Have Persisted Over ∼25 Generations.
Furthermore, these distinct hybrid subpopulations have persisted at Aguazarca over at least ∼25 generations, or approximately two-thirds of the estimated hybrid population age, with no detectable change in hybrid ancestry distribution within genotype clusters over this period (Kolmogorov–Smirnov test, 2003 vs. 2015; birchmanni-biased: D = 0.17, P = 0.49; malinche-biased: D = 0.28, P = 0.39) (Fig. 1B). In fact, pairwise comparisons of ancestry distributions between years demonstrate that within-subpopulation ancestry has been remarkably stable over time (Fig. 1B). Only one pairwise comparison, between birchmanni-like subpopulations in 2005–2007 and 2009–2010, showed a significant difference in ancestry distribution (P = 0.03 after Bonferroni correction for multiple tests; average malinche ancestry 2% lower in 2005). These results suggest that there has not been substantial gene flow between subpopulations, despite continued sympatry.
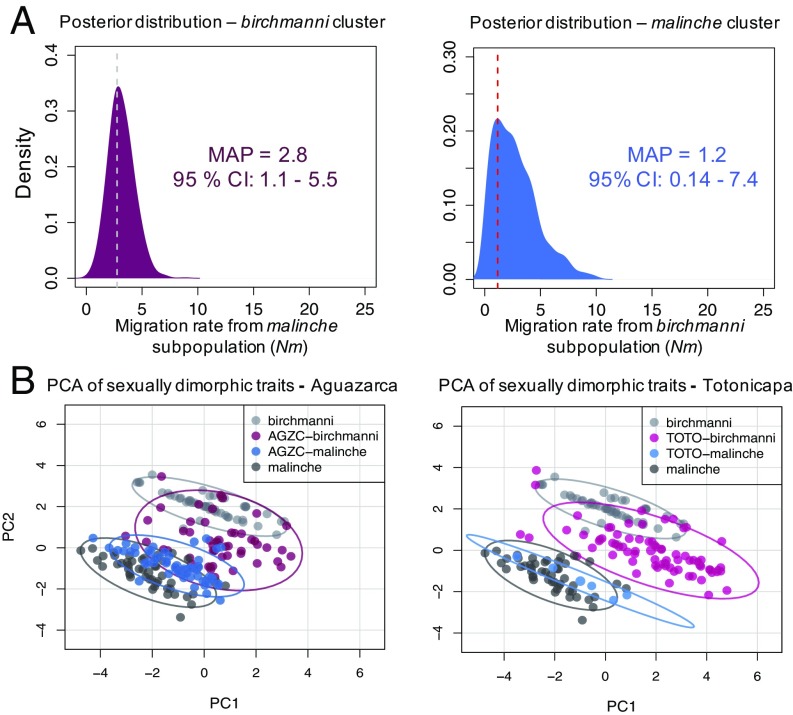
We took advantage of our extensive historical sampling to estimate the level of gene flow between subpopulations at Aguazarca. Using an approximate Bayesian approach to fit the distribution of ancestry proportions over time, we recovered well-resolved posterior estimates of the amount of gene flow between hybrid subpopulations (Fig. 2A and SI Appendix, 3). We found that gene flow between clusters has been remarkably low over the last 25 generations, with an estimated Nm of <3 in both directions. Higher rates would result in a less extreme population structure than what we observed. These results demonstrate that despite living in sympatry, the two Aguazarca subpopulations have successfully mated only rarely. Consistent with this observation, only two adult individuals out of 642 sampled from Aguazarca since 2003 were produced by recent mating events between subpopulations (intermediate ancestry individuals shown in Fig. 1B).
Fig. 2.
ABC simulations support near-complete reproductive isolation between the two hybrid clusters at Aguazarca, despite phenotypic and genetic similarity to another population lacking reproductive isolation. (A) Results of ABC simulations focusing on the Aguazarca population demonstrate that gene flow (Nm) between subpopulations in Aguazarca has been low over the past 25 generations (Nm from the malinche cluster, ∼2.8; Nm from the birchmanni cluster, ∼1.2). (B) Despite low levels of cross-cluster gene flow in Aguazarca, males of the birchmanni-like cluster are similar phenotypically in Aguazarca and Totonicapa, as are males in the malinche-like cluster. Shown are results of principal component analysis of male hybrids in these populations and individuals of the pure parental species. Discriminant function analysis identifies sword length and dorsal fin length as the phenotypic traits most differentiated between the two subpopulations in Aguazarca (SI Appendix, 1 and 2). However, these traits do not differ in their distribution between birchmanni cluster hybrids in Aguazarca and Totonicapa (SI Appendix, Fig. S14). This suggests that preference differences, environmental differences, or differences in traits not captured by our phenotyping underlie differences in assortative mating between populations.
We also used an ABC approach to ask whether gene flow between the two hybrid clusters in Aguazarca changed over time. Although such simulations are often difficult because of the need to explore a large parameter space, we used historical reports and our own estimates of the hybrid zone age in Aguazarca from previous work (25, 27, 28) to ground these simulations. Our results suggest that the data are consistent with early migration rates between clusters approximately 3- to 4-fold higher than the levels we inferred between 2003 and 2015, but likely 6- to 16-fold higher (SI Appendix, Figs. S4 and S5). Specifically, the 95% confidence intervals of the migration rate posterior distributions inferred for this earlier period do not overlap with the maximum a posteriori estimates for contemporary migration. These results imply that the strength of assortative mating in Aguazarca has changed over time, consistent with behavioral results suggesting that the mechanisms underlying assortative mating are context-dependent (7, 21).
Hybrid Population Structure at Aguazarca is Unusual.
The bimodal population structure that we observed at Aguazarca is unusual. Earlier data based on just 4 SNPs hinted at the population structure in Aguazarca and suggested that other birchmanni-malinche hybrid populations lack structure (29), but had limited power (30). For comparison, we examined genome-wide ancestry patterns at 0.47–0.79 million ancestry-informative sites in two other hybrid populations, Tlatemaco (25) (Fig. 1C) and Totonicapa (n = 245; Fig. 1D). Neither population differed significantly from a unimodal distribution of hybrid ancestry (Tlatemaco: D = 0.02, P = 0.87; Totonicapa: D = 0.024, P = 0.89).
However, phenotypes and hybrid index ranges in Totonicapa were qualitatively similar to those in Aguazarca, including a birchmanni cluster of similar ancestry and a small subpopulation of malinche-like individuals composing 6 ± 2% of the population (Figs. 1C and 2B and SI Appendix, 1). Thus, Totonicapa is a useful comparison with the extreme population structure observed in Aguazarca. In contrast to the patterns observed in Aguazarca, several samples from Totonicapa included first-generation (F1) offspring of matings between the two subpopulations. In addition, the distribution of hybrid indices in Totonicapa was significantly right-skewed (P < 0.001 based on bootstrapping), as expected if the birchmanni cluster in this population has continually mated with malinche-cluster individuals (SI Appendix, 4). Although a lack of sufficient historical samples from Totonicapa precluded direct comparisons with the inferred Nm in Aguazarca, the proportion of sampled individuals over all years that are cross-cluster hybrids was an order of magnitude higher in Totonicapa (5 ± 1% vs. 0.3 ± 0.2%), despite a greater opportunity for such mating events in Aguazarca (Fig. 1).
Strong Assortative Mating Explains Differences in Structure.
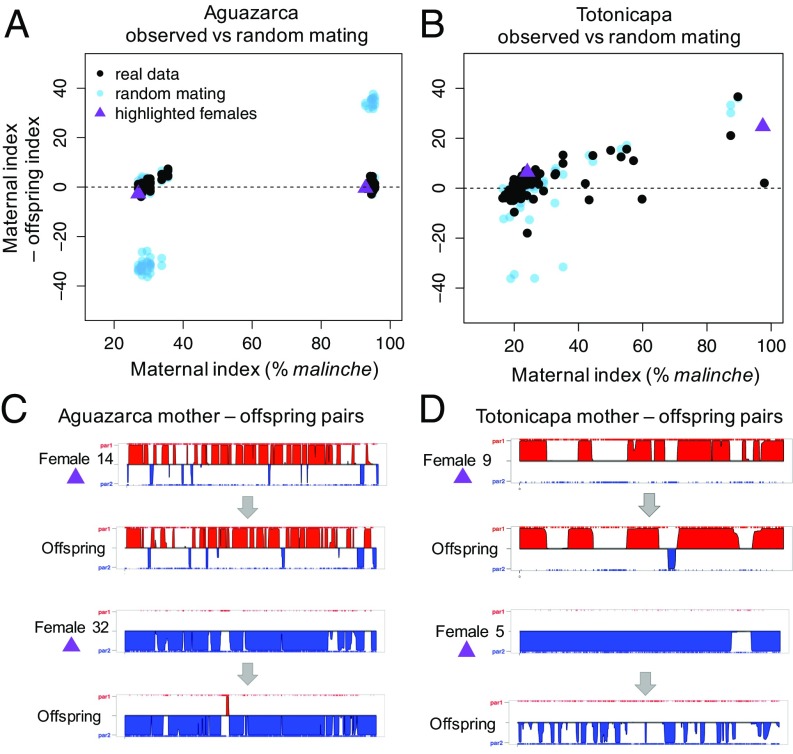
These results suggest that strong reproductive isolation exists between sympatric hybrid subpopulations at Aguazarca, but not at Totonicapa. To determine whether assortative mating is the cause of isolation in Aguazarca, we genotyped wild-caught mothers and their embryos and compared observed mating events with expectations under random mating in both populations. Under random mating, we would expect ∼50% of mating events at Aguazarca to occur between subpopulations (Fig. 3A and SI Appendix, Fig. S6); however, we did not detect a single mating event between subpopulations in 60 distinct mating events (Fig. 3A; P < 10−4 by simulation). At Totonicapa, in contrast, we used the same approach but found no evidence for strong assortative mating (Fig. 3B), despite sufficient power (SI Appendix, 5–7). Instead, we directly observed mating events between malinche-cluster and birchmanni-cluster hybrids (Fig. 3 B and D), at a frequency consistent with the proportion of individuals of each type found in the population (SI Appendix, 7). Furthermore, individuals produced by these types of mating events were detected in samples collected from Totonicapa ∼15 generations ago (SI Appendix, Table S1), demonstrating that such mating events have been persistent in this population. These differences are surprising given the similarity in ancestry, male trait distributions, and other features between the two hybrid populations (Fig. 2B and SI Appendix, 1).
Fig. 3.
Evidence for assortative mating and low rates of cross-cluster hybridization in Aguazarca, but not Totonicapa. (A) Observed differences between maternal and offspring indices in Aguazarca are tightly clustered around zero (black points), indicating that females mate with males of similar ancestry. Cross-cluster matings are predicted to occur at high frequency in simulations of random mating (blue points in Lower Left and Upper Right corners) but are absent from the population. (B) In contrast, in Totonicapa, observed mating events more closely match random mating. (C and D) Examples of the raw data used to generate ancestry estimates. The plots show ancestry across chromosome 1 for representative mother-offspring pairs from each population (purple triangles in the upper panels). The solid red regions are homozygous for X. birchmanni, whereas the solid blue regions are homozygous for X. malinche. Unshaded regions are heterozygous. The height of each region indicates the posterior probability of the ancestry call. Mothers and offspring sampled from Aguazarca have strongly similar ancestry across the chromosome (C), whereas in some cases mothers and offspring from Totonicapa show strikingly different ancestry patterns (D).
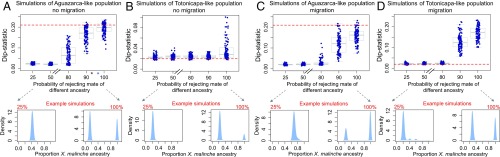
To show in principle that variation in the strength of ancestry-based assortative mating can be a key parameter in generating the variation in population structure that we observe, we perform a series of simulations. As expected, strong assortative mating was required to generate the bimodal population structure that we observed in Aguazarca (Fig. 4 and SI Appendix, 8 and Figs. S7 and S8), with such structure observed only in simulations with low estimated Nm between subpopulations (Nm ∼0–10; SI Appendix, 9). Conversely, simulations of weaker assortative mating generated results qualitatively similar to those seen for Totonicapa (Fig. 4), particularly at realistic parental migration rates (SI Appendix, 8). Although these simulations represent a simplified scenario and only a subset of the many possible mechanisms that may promote assortative mating between subpopulations, they suggest that differences in assortative mating between Aguazarca and Totonicapa play important roles in shaping the observed differences in population structure.
Fig. 4.
Simulations show that assortative mating can play a key role in shaping hybrid population structure. (A) Simulations varying the strength of assortative mating from 25% to 100%, with starting population conditions based on present-day Aguazarca. (B) Results with starting conditions based on present-day Totonicapa. (C and D) Results for the same simulation conditions for Aguazarca (C) and Totonicapa (D), but with ongoing migration from the X. malinche parent (m = 2%; SI Appendix, 8). The population structure in 100 replicate simulations of each scenario is summarized with Hartigan’s dip statistic. Note that the exact value of Hartigan’s dip statistic is sensitive both to the existence of multiple modes in a distribution and to the number of samples in each mode. The observed dip statistic in Aguazarca is shown by a red dotted line in A and C, and the dip statistic in Totonicapa is shown by a red dotted line in B and D. Asterisks on the x-axis in A indicate the level of assortative mating consistent with inference from ABC simulations (red, 93.5% assortative mating in birchmanni cluster; blue, 95.5% assortative mating in malinche cluster; Fig. 2 and SI Appendix, 9). Example hybrid index distributions from simulations drawn from the median of the dip statistic distribution for 25% and 100% assortative mating are shown below each set of main plots. The results are robust to a variety of approaches for simulating assortative mating (SI Appendix, Fig. S7).
Another factor likely influencing the degree of population structure is migration from upstream parental populations, which is evident in both Aguazarca and Totonicapa, but apparently absent in Tlatemaco (SI Appendix, 10). Although we see evidence of parental migration, at least from upstream populations of X. malinche, into both Aguazarca and Totonicapa (SI Appendix, 10), we are currently unable to directly compare parental migration rates between populations. However, we note that although continuous migration from parental populations will clearly influence population structure, our simulations demonstrate that in the absence of assortative mating, migration is not expected to generate the stable subpopulations found in Aguazarca (compare strengths of assortative mating in Fig. 4C).
Selection Does Not Explain Population Structure.
One alternative cause of the mother-offspring patterns that we observe in Aguazarca could be strong selection against cross-cluster offspring at the embryonic stage. Several lines of evidence point to assortative mating rather than selection as the primary cause of population structure, including viable and fertile F1 hybrids between species in the laboratory (SI Appendix, Fig. S9), cross-cluster hybrids at both the embryonic and adult stages in Totonicapa (Figs. 1 and 3), and viable and fertile hybrids between Aguazarca clusters generated in the laboratory (SI Appendix, Fig. S10). Although population-specific patterns of divergent ecological selection could explain these observations, and may indeed be present in wild populations, we consider it unlikely that ecological selection would completely remove cross-cluster offspring from the wild at the embryo stage when cross-cluster hybrids are readily produced in the laboratory. Thus, our results strongly suggest that it is prezygotic mechanisms, rather than selection, that have dramatically restricted gene flow between hybrid subpopulations at Aguazarca. These results are intriguing, because the time depth of our sampling suggests that prezygotic isolating mechanisms might have existed only a few generations after the Aguazarca hybrid population formed.
Assortative Mating Is Easily Disrupted.
Despite the persistence of reproductive isolation between clusters at Aguazarca over 25 generations, other evidence suggests that assortative mating is easily disrupted. We genotyped adults and first- and second-generation offspring from a laboratory mesocosm initially seeded with a sample of 16 individuals from Aguazarca (including six males, split evenly between the two clusters). Seventy percent of juveniles collected from this tank (n = 14) are either first- or second-generation hybrids between the two subpopulations (SI Appendix, Fig. S10), consistent with previous work showing that sympatric Xiphophorus will mate in the laboratory (24). These results highlight the context dependence of population structure driven by assortative mating, an inference also reflected by ABC simulations, suggesting that the strength of assortative mating has varied over time in Aguazarca (SI Appendix, Figs. S4 and S5).
Behavioral Trials Do Not Identify the Cause of Assortative Mating.
We performed extensive mate preference trials to try to understand the behavioral mechanisms driving assortative mating between clusters at Aguazarca. In parental species, behavioral preferences are mediated by urine-borne pheromones (31, 32), and individuals show strong preferences for conspecific olfactory signals (7) (SI Appendix, Fig. S11). We tested each subpopulation for their preferences for pheromones of each cluster (n = 23–57; SI Appendix, Figs. S11 and S12), as well as for preferences for a suite of visual cues (SI Appendix, 11 and 12). Despite predicted power to detect preferences if they are as strong as those parental individuals display (SI Appendix, 11 and 12), we found no evidence of a strong preference in Aguazarca females for males of their own cluster in any visual or olfactory test. This puzzling observation contrasts with patterns in the wild, where Aguazarca females mate exclusively with males of their own genotype cluster (Fig. 3A). These results highlight the complexity of understanding variation in assortative mating in these hybrid populations. Possible interpretations of the data include that hybrid preferences could be more context-dependent than parental preferences, an interpretation also potentially supported by cross-cluster mating in the laboratory mesocosm. Alternately, hybrids could use different cue combinations than parentals to mate assortatively, since the cues that we tested only included those shown to be important in parental preferences (SI Appendix, 11 and 12). Furthermore, power calculations based on parental preferences (SI Appendix, 11 and 12) might not be a good proxy for hybrid preferences. These behavioral trials and results are discussed in more detail in SI Appendix, 12.
Discussion
What mechanisms drive the divergence of populations and species? Positive assortative mating is predicted to accelerate reproductive isolation between lineages (33–35), and is a key component of many models of speciation. Conspecific mate preferences are ubiquitous (36–38), and may be one of the earliest reproductive isolating mechanisms to evolve between populations (39, 40); however, behavioral studies suggest that the underlying mechanisms are highly context-sensitive (6–9), predicting hybridization under a range of conditions, a finding at odds with the importance of assortative mating as a reproductive barrier in the empirical and theoretical literature on speciation. Thus, understanding the resilience of assortative mating as a barrier to gene flow, and how this factor varies across populations, is crucial for understanding its evolutionary consequences.
We used genome-wide sequencing to survey three independently formed hybrid populations of swordtail fish and discovered surprisingly strong population structure in one population. Individuals in this population fall into one of two hybrid subpopulations that differ by ∼70% in their genome-wide ancestry (Fig. 1). This bimodal hybrid population structure is rare and not observed in any other birchmanni-malinche hybrid populations sampled to date. This result highlights the surprising finding that diverse hybrid population structures can result from independent admixture events between the same parental species. Interestingly, a bimodal population structure has been reported in early studies of other hybrid systems (41) and this could suggest a role of assortative mating in generating a bimodal population structure in these hybrid populations.
We find that strong assortative mating has generated and maintained this unusual population structure in Aguazarca following hybridization. Our results suggest that assortative mating has been a persistent barrier over 25 generations, limiting gene flow between subpopulations to Nm ∼1–3. Because our sampling spans approximately two-thirds of the time since the population formed (25), this suggests that assortative mating between hybrid subpopulations closely coincided with the initial hybridization between species. Importantly, our mother-offspring genotyping directly shows that assortative mating is maintaining hybrid population structure in Aguazarca. In contrast, Totonicapa exhibits only weak departures from random mating, and mother-offspring genomic analyses demonstrated mating between clusters. These results implicate variation in the strength of assortative mating in shaping the evolutionary trajectories of hybrid populations (Fig. 4).
Although assortative mating has shaped the evolution of this hybrid population, we also find that it is context-dependent and not evident in other hybrid populations. The sensitivity of mating preferences provides a likely explanation for the diverse outcomes of hybridization between the same parental species. What factors explain this sensitivity? We speculate that environmental differences between Aguazarca and other populations could have influenced the reestablishment of assortative mating. Ecological factors have been shown to be important in plasticity in mate preferences in several systems (42), and can even promote hybridization (8, 9, 16). In particular, disruption of sensory channels used in mate recognition is a likely cause of hybridization between X. birchmanni and X. malinche. Thus, differences among sites in the sensory environment may be the simplest mechanistic explanation for the variable strength of assortative mating that we infer across populations and over time. Specific environmental conditions conducive to assortative mating in Aguazarca also could explain both the lack of preferences in laboratory trials in Aguazarca hybrids and the breakdown of assortative mating in laboratory populations. However, available data suggest that Aguazarca is likely the population most impacted by humans (SI Appendix, 1), which is puzzling given that assortative mating remains strong within this population.
Alternately, variation in the interaction between mate preferences and the social or phenotypic environment could drive differences in structure across populations. As in other species, because some mate preferences in swordtails are learned (43–45) and learning occurs differently in the two parental species (46, 47), learned preferences could have a large number of potential impacts on hybrid population structure, particularly given the complex social and phenotypic environment in hybrid populations (6, 20). Although hybrid clusters in Totonicapa and Aguazarca are phenotypically similar (Fig. 2), they differ in frequency, resulting in different distributions of social stimuli in the two populations. In this regard, frequency-dependent plasticity in preference could generate social conditions that result in different mating dynamics (19). Another possibility is that genetic differences, through drift or selection, allowed the maintenance of assortment in Aguazarca, but not in Totonicapa. However, the collapse of population structure in a mesocosm established from Aguazarca suggests that assortative mating can change over short time scales, making it likely that plasticity also impacts variation in the strength of assortative mating.
Finally, structure could be influenced by migration from other populations. Most directly, immigrants from X. malinche may have stronger mating preferences in some populations (e.g., Aguazarca) than in others. In addition, migration rates from the two parental species may differ from site to site. Strong asymmetric gene flow from X. birchmanni in Totonicapa could, for example, overwhelm the ecological forces that allow both clusters to coexist, although migration from X. birchmanni may be rare (SI Appendix, 10). Alternately, given the strong asymmetric migration from X. malinche detected in Totonicapa, gene flow could be preventing local adaptation. In contrast, Aguazarca might have lower immigration rates from the parental species.
Despite the various possible causes of assortative mating—including social, environmental, and genetic factors—the observed variation in assortative mating across hybrid populations, and plasticity in assortative mating in Aguazarca individuals, have important implications. These results suggest that even strong assortative mating observed over many generations may be an incomplete barrier to gene flow between populations. Furthermore, our findings highlight the importance of understanding the context dependence of assortative mating in understanding its potential impacts on reproductive isolation.
The genomes of many species, including Xiphophorus (13), are characterized by past gene flow. The ubiquity of hybrid ancestry in the genomes of contemporary species suggests that short-term, ecological-scale disturbances, such as those that disrupt mate preferences, can have long-term evolutionary consequences. Thus, the context dependence of assortative mating may play a fundamental role in hybridization and speciation.
Materials and Methods
The data analyzed for this study came from three independently formed X. birchmanni-X. malinche hybrid zones in different river systems in Hidalgo, Mexico. In the Río Calnali, individuals (n = 642) were collected from the Aguazarca stream reach (29). Other samples came from the Río Huazalingo (Totonicapa locality; n = 245) and the Río Claro (Tlatemaco locality; n = 170) (25). Because of extensive sampling by our group, time-transect data were available from Aguazarca populations spanning from 2003 to 2015, or ∼25 generations. We also sequenced 25 first- and second-generation offspring from a 280-L laboratory mesocosm tank stocked with individuals originally collected from the Aguazarca hybrid population. The tank was stocked in March 2014 with three adult males from each subpopulation and 10 females of unknown genotype. All individuals in this tank were genotyped in May 2016.
All wild-caught samples collected for this experiment were collected under a Mexican federal collector’s permit to Scott Monks (FAUT-217) and a scientific collecting permit to Guillermina Alcaraz (PPF/DGOPA-173/14). Fish were caught using baited minnow traps and lightly anesthetized in MS-222. We photographed each specimen and clipped ∼25% of the upper caudal fin for DNA extraction. Fin clips were stored in 95% ethanol until extraction. Based on the results of our population structure analysis (Results), we also collected gravid females and their offspring from both the Aguazarca (n = 27 females; 24 individuals for which offspring were successfully sequenced) and Totonicapa populations (n = 110 females; 97 individuals for which mother-offspring pairs were successfully sequenced). Females were killed with an overdose of MS-222, and embryos were dissected from females postmortem. All animal procedures followed Texas A&M Institutional Animal Care and Use Committee Protocols 2013–0271 and 2012–164. DNA was extracted using either an Agencourt bead-based purification kit (Beckman Coulter) following the manufacturer’s instructions or an in-house protein-kinase digestion protocol. For females and offspring from Aguazarca, microsatellite genotyping was performed to identify full and half siblings (SI Appendix, 13). This analysis was not performed for Totonicapa because the large number of samples made microsatellite genotyping impractical.
For genome-wide genotyping of hybrids, we used the MSG approach of Andolfatto et al. (26), which has been validated for X. birchmanni-X. malinche hybrids (25). Libraries were prepared as described previously (26) with custom barcodes (SI Appendix, Dataset S1). All raw data are available through the NCBI Sequence Read Archive (accession nos. SRX544941 and SRA607213) and parsed data are available at genomics.princeton.edu/AndolfattoLab/Links.html. For a subset of 85 individuals, a different library preparation protocol was used, as described in SI Appendix, 14. Raw reads were parsed by index and barcode and trimmed to remove low-quality base pairs (Phred quality score <20); reads with <30 bp of a high-quality contiguous sequence were discarded. The number of reads per individual ranged from 0.3 to 7.9 million (coverage and accuracy simulations in SI Appendix, 15–17), but reads in excess of 2 million were excluded to improve pipeline speed. The following MSG parameters were specified for the analysis: recRate = 420; rfac = 1; X. birchmanni error (deltapar1) = 0.05; X. malinche error (deltapar2) = 0.05. The recombination rate was set based an expectation of 0.0018 cM/Mb in Xiphophorus and ∼35 generations of recombination (25, 48), and the error rate was set based on observed error rates in parental individuals (25). Each population was analyzed separately. We performed analyses to evaluate the sensitivity of our results to other MSG parameters (SI Appendix, 15–17). MSG reports genotypes in the form of posterior probabilities. To calculate the hybrid index, we treated posterior probabilities ≥0.95 as support for a particular genotype, and for each individual divided the total number of malinche alleles by the total number of alleles with ≥0.95 posterior probability support. For calculations of hybrid index, we used only markers that were sampled in ≥70% of individuals (1,342,246 ancestry informative markers in Aguazarca and 790,577 in Totonicapa). To evaluate whether our data from Aguazarca and Totonicapa significantly departed from expectations under random mating, we collected gravid females from both hybrid zones and performed genome-wide genotyping of both mothers and embryos. We compared the observed similarity in mother and embryo hybrid index with data generated by simulating random mating. The simulations are described in detail in SI Appendix, 6.
To quantify how low levels of gene flow would need to be between Aguazarca subpopulations to generate the population structure that we observe, we conducted simulations and used a rejection sampling approach to approximate Bayesian computation (49). We based our starting population conditions for these simulations on individuals sampled from Aguazarca in 2003–2004, ∼25 generations ago. Hybrid population simulations were performed using admix’em (50). Summary statistics included Hartigan’s dip statistic (51), the coefficient of variation in hybrid index, and the average hybrid index for each subpopulation. Summary statistics were calculated from 500 neutral markers distributed uniformly on 24 chromosomes, because simulating 1.3 million markers would be computationally intractable. For comparison with the real data, we resampled 500 markers from the MSG data 10,000 times, recalculating summary statistics to obtain a distribution for comparison with the simulations (SI Appendix, Fig. S13). A detailed description of the simulations is provided in SI Appendix, 3. ABC simulations to infer historical migration rates were performed as described in SI Appendix, 18.
Supplementary Material
Acknowledgments
We thank the pertinent authorities in Mexico for permission to collect fish, and thank Gaston Jofre, Santiago Forero, Abigail Glueck, Mason Matheny, and Victoria Rodriguez for assistance. This manuscript was greatly improved by the comments of the editor, Daniel Matute, and two anonymous reviewers. This project was supported by the National Science Foundation (NSF) Graduate Research Fellowship Program (M. Schumer, D.L.P., and P.J.D.), an NSF Doctoral Dissertation Improvement Grant DEB-1405232 (to M. Schumer), and an NSF Long-Term Research in Environmental Biology Grant IOS-1354172 (to G.G.R.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Sequence data have been deposited in the National Center for Biotechnology Information’s Sequence Read Archive (accession nos. SRX544941 and SRA607213) and parsed data are available at genomics.princeton.edu/AndolfattoLab/Links.html.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1711238114/-/DCSupplemental.
References
- 1.Coyne JA, Orr HA. Speciation. Sinauer Associates; Sunderland, MA: 2004. [Google Scholar]
- 2.Mendelson TC. Sexual isolation evolves faster than hybrid inviability in a diverse and sexually dimorphic genus of fish (Percidae: Etheostoma) Evolution. 2003;57:317–327. doi: 10.1111/j.0014-3820.2003.tb00266.x. [DOI] [PubMed] [Google Scholar]
- 3.Puniamoorthy N. Behavioural barriers to reproduction may evolve faster than sexual morphology among populations of a dung fly. Anim Behav. 2014;98:139–148. [Google Scholar]
- 4.Hopkins R, Guerrero RF, Rausher MD, Kirkpatrick M. Strong reinforcing selection in a Texas wildflower. Curr Biol. 2014;24:1995–1999. doi: 10.1016/j.cub.2014.07.027. [DOI] [PubMed] [Google Scholar]
- 5.Servedio MR, Noor MAF. The role of reinforcement in speciation: Theory and data. Annu Rev Ecol Evol Syst. 2003;34:339–364. [Google Scholar]
- 6.Rosenthal GG. Individual mating decisions and hybridization. J Evol Biol. 2013;26:252–255. doi: 10.1111/jeb.12004. [DOI] [PubMed] [Google Scholar]
- 7.Fisher HS, Wong BBM, Rosenthal GG. Alteration of the chemical environment disrupts communication in a freshwater fish. Proc Biol Sci. 2006;273:1187–1193. doi: 10.1098/rspb.2005.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pfennig KS. Facultative mate choice drives adaptive hybridization. Science. 2007;318:965–967. doi: 10.1126/science.1146035. [DOI] [PubMed] [Google Scholar]
- 9.Seehausen O, vanAlphen JJM, Witte F. Cichlid fish diversity threatened by eutrophication that curbs sexual selection. Science. 1997;277:1808–1811. [Google Scholar]
- 10.Sankararaman S, et al. The genomic landscape of Neanderthal ancestry in present-day humans. Nature. 2014;507:354–357. doi: 10.1038/nature12961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schumer M, Cui R, Powell DL, Rosenthal GG, Andolfatto P. Ancient hybridization and genomic stabilization in a swordtail fish. Mol Ecol. 2016;25:2661–2679. doi: 10.1111/mec.13602. [DOI] [PubMed] [Google Scholar]
- 12.Stukenbrock EH, Christiansen FB, Hansen TT, Dutheil JY, Schierup MH. Fusion of two divergent fungal individuals led to the recent emergence of a unique widespread pathogen species. Proc Natl Acad Sci USA. 2012;109:10954–10959. doi: 10.1073/pnas.1201403109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cui R, et al. Phylogenomics reveals extensive reticulate evolution in Xiphophorus fishes. Evolution. 2013;67:2166–2179. doi: 10.1111/evo.12099. [DOI] [PubMed] [Google Scholar]
- 14.Heliconius Genome Consortium Butterfly genome reveals promiscuous exchange of mimicry adaptations among species. Nature. 2012;487:94–98. doi: 10.1038/nature11041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hermansen JS, et al. Hybrid speciation through sorting of parental incompatibilities in Italian sparrows. Mol Ecol. 2014;23:5831–5842. doi: 10.1111/mec.12910. [DOI] [PubMed] [Google Scholar]
- 16.Taylor EB, et al. Speciation in reverse: Morphological and genetic evidence of the collapse of a three-spined stickleback (Gasterosteus aculeatus) species pair. Mol Ecol. 2006;15:343–355. doi: 10.1111/j.1365-294X.2005.02794.x. [DOI] [PubMed] [Google Scholar]
- 17.Mavárez J, et al. Speciation by hybridization in Heliconius butterflies. Nature. 2006;441:868–871. doi: 10.1038/nature04738. [DOI] [PubMed] [Google Scholar]
- 18.Gilman RT, Behm JE. Hybridization, species collapse, and species reemergence after disturbance to premating mechanisms of reproductive isolation. Evolution. 2011;65:2592–2605. doi: 10.1111/j.1558-5646.2011.01320.x. [DOI] [PubMed] [Google Scholar]
- 19.Magurran AE, Ramnarine IW. Learned mate recognition and reproductive isolation in guppies. Anim Behav. 2004;67:1077–1082. [Google Scholar]
- 20.Grant PR, Grant BR. Hybridization, sexual imprinting, and mate choice. Am Nat. 1997;149:1–28. [Google Scholar]
- 21.Willis PM, Ryan MJ, Rosenthal GG. Encounter rates with conspecific males influence female mate choice in a naturally hybridizing fish. Behav Ecol. 2011;22:1234–1240. [Google Scholar]
- 22.Fernandez AA, Morris MR. Mate choice for more melanin as a mechanism to maintain a functional oncogene. Proc Natl Acad Sci USA. 2008;105:13503–13507. doi: 10.1073/pnas.0803851105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Verzijden MN, et al. The impact of learning on sexual selection and speciation. Trends Ecol Evol. 2012;27:511–519. doi: 10.1016/j.tree.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 24.Clark E, et al. Mating behavior patterns in two sympatric species of xiphophorin fishes: Their inheritance and significance in sexual isolation. Bull Am Mus Nat Hist. 1954;103:139–225. [Google Scholar]
- 25.Schumer M, et al. High-resolution mapping reveals hundreds of genetic incompatibilities in hybridizing fish species. Elife. 2014;3:e02535. doi: 10.7554/eLife.02535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andolfatto P, et al. Multiplexed shotgun genotyping for rapid and efficient genetic mapping. Genome Res. 2011;21:610–617. doi: 10.1101/gr.115402.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosenthal GG, et al. Dissolution of sexual signal complexes in a hybrid zone between the swordtails Xiphophorus birchmanni and Xiphophorus malinche (Poeciliidae) Copeia. 2003;2:299–307. [Google Scholar]
- 28.Rauchenberger M, Kallman KD, Morizot DC. Monophyly and geography of the Rio Panuco basin Mexico swordtails genus Xiphophorus with descriptions of four new species. Am Mus Novit. 1990;2975:1–41. [Google Scholar]
- 29.Culumber ZW, et al. Replicated hybrid zones of Xiphophorus swordtails along an elevational gradient. Mol Ecol. 2011;20:342–356. doi: 10.1111/j.1365-294X.2010.04949.x. [DOI] [PubMed] [Google Scholar]
- 30.Culumber Z, Ochoa O, Rosenthal GGR. Mating and the maintenance of population structure in a natural hybrid zone. Am Nat. 2014;184:225–232. doi: 10.1086/677033. [DOI] [PubMed] [Google Scholar]
- 31.McLennan DA, Ryan MJ. Responses to conspecific and heterospecific olfactory cues in the swordtail Xiphophorus cortezi. Anim Behav. 1997;54:1077–1088. doi: 10.1006/anbe.1997.0504. [DOI] [PubMed] [Google Scholar]
- 32.McLennan DA, Ryan MJ. Interspecific recognition and discrimination based upon olfactory cues in northern swordtails. Evolution. 1999;53:880–888. doi: 10.1111/j.1558-5646.1999.tb05382.x. [DOI] [PubMed] [Google Scholar]
- 33.Dobzhansky T. Genetics of the Evolutionary Process. Columbia Univ Press; New York: 1970. [Google Scholar]
- 34.Servedio MR. The evolution of premating isolation: Local adaptation and natural and sexual selection against hybrids. Evolution. 2004;58:913–924. doi: 10.1111/j.0014-3820.2004.tb00425.x. [DOI] [PubMed] [Google Scholar]
- 35.Funk DJ, Nosil P, Etges WJ. Ecological divergence exhibits consistently positive associations with reproductive isolation across disparate taxa. Proc Natl Acad Sci USA. 2006;103:3209–3213. doi: 10.1073/pnas.0508653103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Crapon de Caprona MD, Ryan MJ. Conspecific mate recognition in swordtails, Xiphophorus nigrensis and X. pygmaeus: Olfactory and visual cues. Anim Behav. 1990;39:290–296. [Google Scholar]
- 37.Saether SA, et al. Sex chromosome-linked species recognition and evolution of reproductive isolation in flycatchers. Science. 2007;318:95–97. doi: 10.1126/science.1141506. [DOI] [PubMed] [Google Scholar]
- 38.Turissini DA, Liu G, David JR, Matute DR. The evolution of reproductive isolation in the Drosophila yakuba complex of species. J Evol Biol. 2015;28:557–575. doi: 10.1111/jeb.12588. [DOI] [PubMed] [Google Scholar]
- 39.Coyne JA, Orr HA. Patterns of speciation in Drosophila. Evolution. 1989;43:362–381. doi: 10.1111/j.1558-5646.1989.tb04233.x. [DOI] [PubMed] [Google Scholar]
- 40.Jennings JH, Snook RR, Hoikkala A. Reproductive isolation among allopatric Drosophila montana populations. Evolution. 2014;68:3095–3108. doi: 10.1111/evo.12535. [DOI] [PubMed] [Google Scholar]
- 41.Jiggins CD, Mallet J. Bimodal hybrid zones and speciation. Trends Ecol Evol. 2000;15:250–255. doi: 10.1016/s0169-5347(00)01873-5. [DOI] [PubMed] [Google Scholar]
- 42.Heuschele J, Mannerla M, Gienapp P, Candolin U. Environment-dependent use of mate choice cues in sticklebacks. Behav Ecol. 2009;20:1223–1227. [Google Scholar]
- 43.Westerman EL, Chirathivat N, Schyling E, Monteiro A. Mate preference for a phenotypically plastic trait is learned, and may facilitate preference-phenotype matching. Evolution. 2014;68:1661–1670. doi: 10.1111/evo.12381. [DOI] [PubMed] [Google Scholar]
- 44.Kozak GM, Head ML, Boughman JW. Sexual imprinting on ecologically divergent traits leads to sexual isolation in sticklebacks. Proc Biol Sci. 2011;278:2604–2610. doi: 10.1098/rspb.2010.2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kozak GM, Boughman JW. Learned conspecific mate preference in a species pair of sticklebacks. Behav Ecol. 2009;20:1282–1288. [Google Scholar]
- 46.Cui R, Delclos P, Schumer M, Rosenthal G. Early social learning triggers neurogenomic expression changes in a swordtail fish. Proc R Soc B. 2017;284:20170701. doi: 10.1098/rspb.2017.0701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Verzijden MN, Culumber ZW, Rosenthal GG. Opposite effects of learning cause asymmetric mate preferences in hybridizing species. Behav Ecol. 2012;23:1133–1139. [Google Scholar]
- 48.Amores A, et al. A RAD-tag genetic map for the platyfish (Xiphophorus maculatus) reveals mechanisms of karyotype evolution among teleost fish. Genetics. 2014;197:625–641. doi: 10.1534/genetics.114.164293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Beaumont MA, Zhang W, Balding DJ. Approximate Bayesian computation in population genetics. Genetics. 2002;162:2025–2035. doi: 10.1093/genetics/162.4.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cui R, Schumer M, Rosenthal GG. Admix’em: A flexible framework for forward-time simulations of hybrid populations with selection and mate choice. Bioinformatics. 2016;32:1103–1105. doi: 10.1093/bioinformatics/btv700. [DOI] [PubMed] [Google Scholar]
- 51.Hartigan JA, Hartigan PM. The dip test of unimodality. Ann Stat. 1985;13:70–84. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.