Significance
Many important bacterial pathogens such as Vibrio cholerae and Helicobacter pylori use a sheathed flagellum as the main organelle for motility. Although bacterial flagella have been extensively studied in model systems such as Escherichia coli and Salmonella, relatively little is known about sheathed flagella. In this study, we use high-throughput cryoelectron tomography to visualize thousands of polar flagella in Vibrio alginolyticus. We not only reveal unprecedented details of sheathed flagella in V. alginolyticus but also uncover key differences between sheathed flagella and unsheathed flagella in the same Vibrio species. Our studies provide insight into the unique aspects of sheathed flagella, which are effectively used by many bacterial pathogens to establish infection and disseminate in humans and other mammalian hosts.
Keywords: nanomachine, electron tomography, vibrio, flagellum
Abstract
Vibrio species are Gram-negative rod-shaped bacteria that are ubiquitous and often highly motile in aqueous environments. Vibrio swimming motility is driven by a polar flagellum covered with a membranous sheath, but this sheathed flagellum is not well understood at the molecular level because of limited structural information. Here, we use Vibrio alginolyticus as a model system to study the sheathed flagellum in intact cells by combining cryoelectron tomography (cryo-ET) and subtomogram analysis with a genetic approach. We reveal striking differences between sheathed and unsheathed flagella in V. alginolyticus cells, including a novel ring-like structure at the bottom of the hook that is associated with major remodeling of the outer membrane and sheath formation. Using mutants defective in flagellar motor components, we defined a Vibrio-specific feature (also known as the T ring) as a distinctive periplasmic structure with 13-fold symmetry. The unique architecture of the T ring provides a static platform to recruit the PomA/B complexes, which are required to generate higher torques for rotation of the sheathed flagellum and fast motility of Vibrio cells. Furthermore, the Vibrio flagellar motor exhibits an intrinsic length variation between the inner and the outer membrane bound complexes, suggesting the outer membrane bound complex can shift slightly along the axial rod during flagellar rotation. Together, our detailed analyses of the polar flagella in intact cells provide insights into unique aspects of the sheathed flagellum and the distinct motility of Vibrio species.
Motility is often important for the virulence of bacterial pathogens, and the flagellum is the main organelle for motility in many bacteria. In most externally flagellated bacteria, such as Escherichia coli and Salmonella enterica, the flagella function as propellers: counterclockwise rotation of the flagella results in bundling together of the flagella and smooth swimming of the cell (running), whereas clockwise rotation of the flagella causes random turning of the cell with little translational motion (tumbling) (1–3). A sophisticated chemotactic signaling system allows the bacterium to sense chemical stimuli and effectively swim toward favorable environments by a biased random walk, a combination of “runs” and “tumbles” (1, 4).
The flagellum is a large macromolecular assembly composed of a long filament, a hook, and a motor. The flagellar motor is a remarkable nanomachine embedded in the bacterial cell envelope. More than 20 different proteins are required for the assembly of the motor, which can be divided into several morphological domains. The MS ring is embedded in the cytoplasmic membrane. The C ring, known as the switch complex, and the export apparatus are located in the cytoplasmic side of the MS ring. The rod connects the MS ring and the hook and is commonly divided into the distal rod and the proximal rod. The L and P rings on the rod function as bushings at the outer membrane and at the peptidoglycan layer, respectively. The stator is the torque generator embedded in the cytoplasmic membrane. In E. coli and Salmonella, a two-membrane protein complex consisting of MotA and MotB functions as the stator by being anchored to the peptidoglycan layer. Powered by the proton motive force, the stator generates the torque required to rotate the motor, the hook, and the filament. The stator shares several common features despite the difference in ions and proteins involved. Typically, multiple stator units work together (5), although a single stator is enough to generate rotational torque (6). Stepwise photo-bleaching of a functioning motor region revealed that the stator is highly dynamic and can associate into or dissociate from the rotor rapidly (5). Recent reports revealed that the stator–rotor association is dependent on conducting ions (7, 8) and torque load (9–11).
Vibrio species are highly motile and possess polar sheathed flagella (12), which are quite different from the unsheathed flagella found in E. coli and Salmonella. The membranous sheath appears as an extension of the outer membrane, but its function remains poorly understood. An earlier study indicated that Vibrio with a sheath flagellum could potentially prevent the host immune response from recognizing the filament (13). A more recent study suggested that rotation of sheathed flagella releases lipopolysaccharide that could promote immune recognition during Vibrio fischeri infection (14). Recently, a fast and high-resolution fluorescent imaging study was used to visualize in vivo assembly of sheathed flagella (15). The studies provide evidence in support of synchronized growth between the sheath and the filament. However, the mechanism underlying sheath formation remains unknown.
Vibrios use sodium ions as the energy source for flagellar rotation, and PomA and PomB proteins comprise the Na+-driven stator (16). Unlike the MotA/B stator complex, the PomA/B stator complex needs additional Vibrio components, MotX and MotY. They form a ring-like structure called the T ring, which appears to be specific to the sodium-driven flagellar motor (17–20). Another Vibrio specific ring-like structure (named as the H ring) is located on the top of the T ring. FlgT is the main protein component of the H ring. Evidence suggests that both the rings are required for stator assembly and function of the PomA/B stator complex (19, 21).
To better understand the structure and function of the flagella from different species, cryoelectron tomography (cryo-ET) has been extensively used to visualize intact flagella in their cellular context (22–24). Despite having common themes, these analyses have demonstrated striking structural diversity of these nanomachines in different bacteria (22). For example, a Vibrio motor has 13 stators assembled around the rotor (25), whereas a Borrelia burgdorferi motor has 16 stators and a distinctive periplasmic feature (26). To further our understanding of the sheathed flagellum as well as structural features specific to Vibrios, we chose Vibrio alginolyticus as a model system. Extensive genetic, functional, and structural studies have made the V. alginolyticus polar flagellum one of the mostly characterized sheathed flagella. Although V. alginolyticus possesses a single Na+-driven polar flagellum, mutations have resulted in multiple polar flagellar phenotypes (27–30), which are ideal for high-resolution in situ structural characterization. Here, we use high-throughput cryo-ET and subtomogram averaging to extensively analyze the motors in several V. alginolyticus mutants, providing insights into aspects of the sheathed flagella and Vibrio motility.
Results
Visualization of Vibrio Sheathed Flagella in Intact Cells.
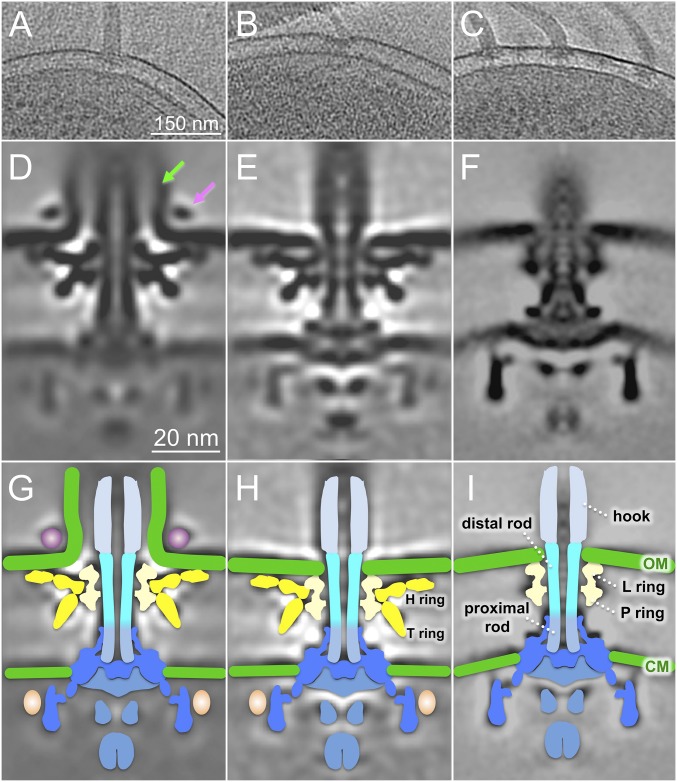
V. alginolyticus VIO5 typically has a single polar sheathed flagellum (Fig. 1A). To our surprise, we also observed an unsheathed flagellum after visualizing more than 30 wild-type cells (Fig. 1B), suggesting it is rare but possible to assemble unsheathed flagella in V. alginolyticus. As expected, the diameter of the unsheathed filament is considerably smaller than that of the sheathed filament, and the motors are also noticeably different in the original cell reconstructions (Fig. 1 A and B). To better reveal the difference between the two distinct flagellar types, we wanted to compare them side by side in the same cell. To do this, we used an flhG mutant, which is known to assemble multiple polar flagella (27, 28). Importantly, multiple polar flagella commonly form a large cluster at one cell pole; the small cell diameter near the pole is optimal for high-resolution cryo-ET analysis (Fig. 1C).
Fig. 1.
In situ V. alginolyticus flagellar structures: sheathed and unsheathed. (A) A representative slice of a 3D reconstruction of a wild-type V. alginolyticus VIO5 shows a single sheathed flagellum at the cell pole. (B) Another slice shows a single unsheathed flagellum. (C) A tomographic slice of the strain (KK148) shows multiple polar flagella. (D) A class average shows the sheathed flagellar motor structure. (E) Another class average shows the unsheathed flagellar motor structure. (F) An averaged structure of the E. coli motor is used as a reference for comparison. (G–I) Schematic models are overlaid on cryo-ET density maps of D–F, respectively. The key differences between Vibrio and E. coli motor structures are highlighted, with the O ring in purple, the H ring and T ring in yellow, and the extra density (colored in pink) located outside the C ring. CM, cytoplasmic membrane; OM, outer membrane. The stator density is not visible in the globally averaged structures of the Vibrio and E. coli motors likely because of the highly dynamic nature of the stators.
Novel Vibrio Flagellar Motor Structures.
From ∼600 tomograms of different bacterial cells, we generated ∼4,000 reconstructions from motors embedded in cell envelope. After extensive subtomogram alignment and averaging, we obtained two distinct Vibrio motor structures with unprecedented detail (Fig. 1 D and E) and compared these structures with the motor structure from E. coli (Fig. 1F). The motor structures shared several common themes: they are similarly embedded in the cytoplasmic and the outer membranes and possess a C ring, MS ring, export apparatus, and rod (including the proximal rod and the distal rod) and its surrounding L/P rings (Fig. 1 G–I).
Despite these similarities, the Vibrio motors exhibit several notable differences from the E. coli structures. For example, the Vibrio motors possess additional densities (colored in yellow) around the L/P rings (Fig. 1 G and H) that likely correspond to the Vibrio-specific structures such as the H and T rings. We also observed additional structures around the C ring (Fig. 1 D and G) in Vibrio that are absent in the E. coli motor. There is an external ring-like structure only associated with the sheathed flagellum (Fig. 1 D and G), but not with unsheathed flagellum (Fig. 1 E and F), that we named the O ring (outer membrane ring). Interestingly, the presence of this ring is correlated with a striking 90° outer membrane bend and the formation of a membrane sheath surrounding the filament, suggesting the O ring may play a critical role in sheath formation and function. In contrast, in the absence of the O ring, the outer membrane clearly intersects the L ring in the unsheathed Vibrio flagellum (Fig. 1 E and H), as well as the E. coli flagellum (Fig. 1 F and I).
Dynamic Nature of the Sheathed Flagellar Motor in Vibrio.
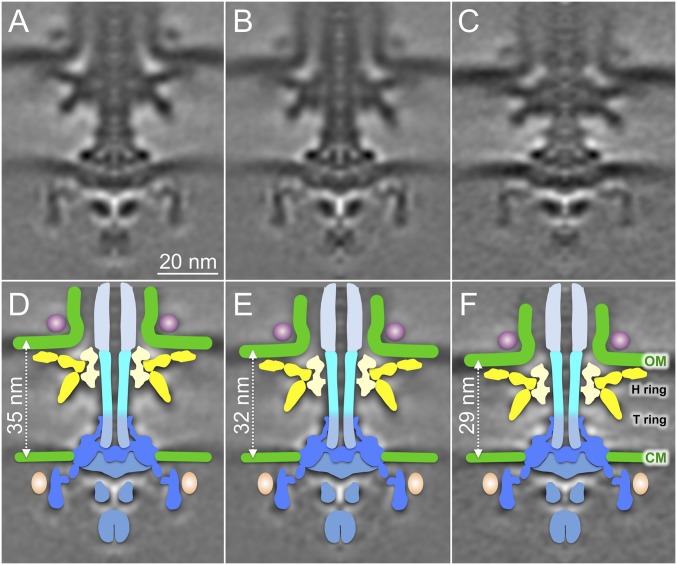
Comparison of sheathed and unsheathed flagellar motors also suggests intrinsic flexibility, as indicated by the relative fuzzy feature of the MS ring and the C ring in the sheathed flagellar motor (Fig. 1D). To further examine this potential flexibility, we generated three distinct class averages and then aligned them based on the MS ring and the C ring (Fig. 2 A–C). Notably, the distance between the cytoplasmic membrane and the outer membrane varied from 35 to 29 nm (Fig. 2 A, C, D, and F), although the most common class was 32 nm (Fig. 2 B and E). Interestingly, the outer membrane and the Vibrio-specific features (the T/H/O rings) all shifted their positions coordinately, suggesting this complex has the flexibility to slide along the distal portion of the flagellar rod. In contrast, the unsheathed flagellum in both Vibrio and E. coli appears to be rigid, likely because of the intimate interaction between the outer membrane and the L ring.
Fig. 2.
Conformational flexibility of sheathed flagellar motors in Vibrio. Sheathed flagellar motor structures from three distinct class averages show variation in spacing between CM and OM (A–C). Respective schematic models (D–F) show the outer membrane bound complex shifting slightly along the distal rod (cyan).
Characterization of T-Ring Components in Situ.
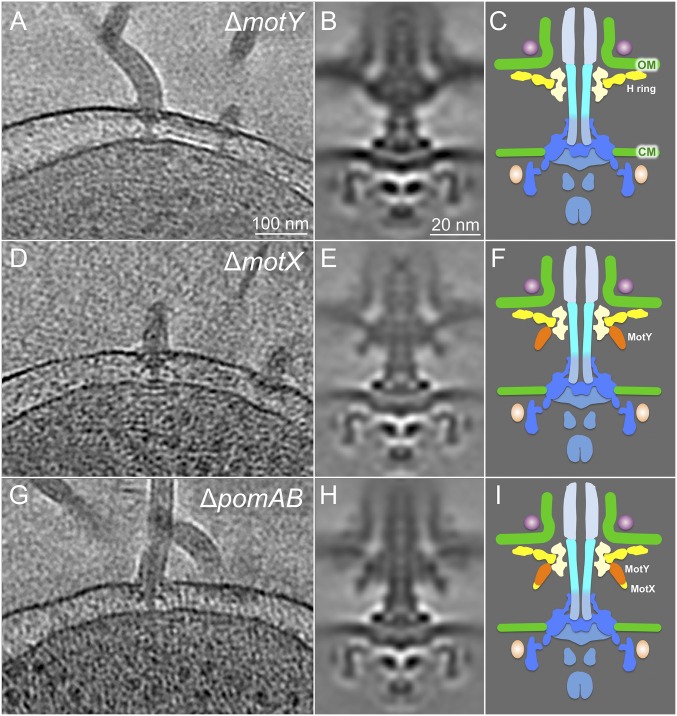
The major Vibrio specific features are similar in both sheathed and unsheathed flagella (Fig. 1). Although the T ring is the major Vibrio-specific feature required for stator assembly around the rotor (19), its structure and composition remain undefined. To characterize the T ring, we used cryo-ET to analyze ∆motX and ∆motY mutants in the flhG background. In a ∆motY mutant, we found that most of the T-ring density located beneath the P ring is not visible compared with the wild-type strain and the ∆pomAB mutant that lacks stators (Fig. 1 G and H for wild type; Fig. 3 G and H for ∆pomAB; Fig. 3 B and C for ∆motY). In MotX-defective cells, no significant difference in the T ring was observed (Fig. 3 D–F). A comparison between ∆motX and ∆pomAB flagella suggests MotX protein is likely located beneath the MotY in situ (Fig. 3 E, F, H, and I). Furthermore, the densities corresponding to the O ring and the H ring remain visible in these T-ring mutants, suggesting their formation is independent of T-ring assembly.
Fig. 3.
Characterization of the T-ring components in situ. (A) Representative slice of a tomogram from a ∆motY strain. (B) Averaged structure of the ∆motY motor. (C) A schematic model of the motor lacking MotY. (D) Representative slice of a tomogram from a ∆motX mutant. (E) Averaged structure of the ∆motX motor. (F) A schematic model of the motor lacking MotX. MotY is colored in orange. (G) A representative slice of a tomogram from a ∆pomAB mutant. (H) Averaged structure of the ∆pomAB motor. (I) A schematic model of the motor lacking PomA and PomB. A plausible location for MotX, which is largely based on previous analysis (19), is labeled in yellow.
Architecture of the Vibrio-Specific T Ring.
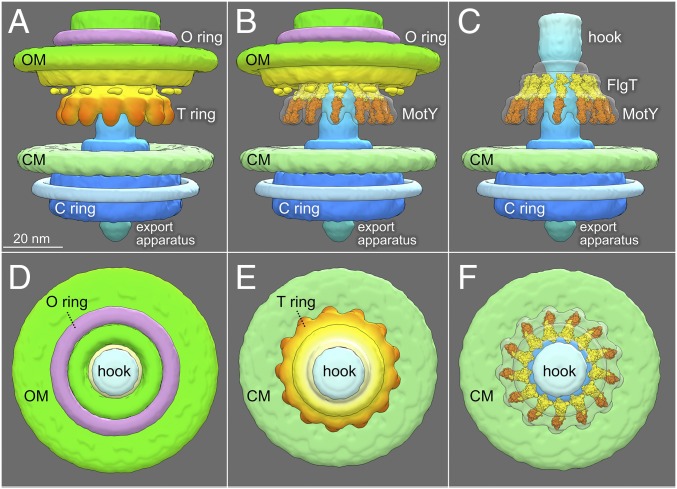
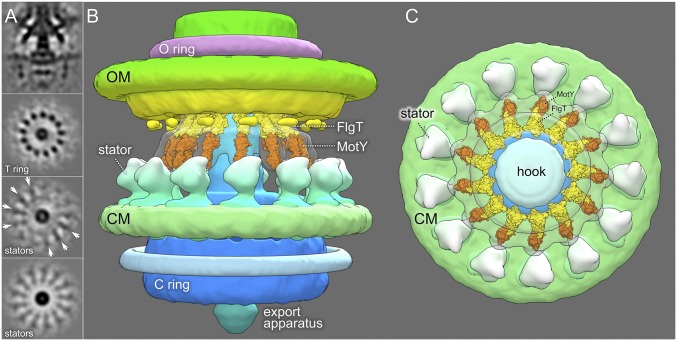
Our data suggest MotY is the major component of the T ring. In fact, 13 copies of MotY monomer fit well into the distal portion of the T ring (Fig. 4), based on the MotY crystal structure and biochemical characterization reported previously (31). Each MotY monomer consists of two distinct domains. The C-terminal domain of MotY contains a putative peptidoglycan-binding motif, whereas the N-terminal domain MotY is essential for stator assembly (31). One monomer can be fitted into each subunit of the T ring. Similarly, we fit 13 copies of the FlgT crystal structures into the densities on top of the MotY ring, based on structural and functional analysis (21). Together with MotY, they form a unique Vibrio-specific structure around the P ring.
Fig. 4.
Architecture of the Vibrio-specific T ring. (A) The Vibrio motor structure with different domains is highlighted in a 3D surface rendering. (B) Crystal structures of MotY and FlgT were fitted into the map of the T ring and the small portion of the H ring, respectively. (C) Outer membrane and its associated flagellar components were removed to have a better view of the T ring. (D) A top view of the Vibrio motor shows the O ring, the hook, and the OM. (E) Another top view after removing the OM-associated structures shows the CM, the hook, and the T ring. (F) Crystal structures of FlgT and MotY fit well into the T ring.
Stator Assembles Around the T Ring and C Ring.
Both MotY and MotX are involved in stator assembly and function. However, all the averaged structures from wild-type motors and the T-ring mutants did not show any obvious densities that might be contributed by the stators. In fact, the structure of the wild-type motor is similar to that of the ∆pomAB motor, in which the stator proteins PomA and PomB are absent. Therefore, there is no well-defined stator structure in V. alginolyticus or E. coli. Our observation is consistent with the notion that the stators are highly dynamic in these species.
To investigate the structure and location of stators in V. alginolyticus, we used 3D classification to analyze the densities around the T ring and the MS ring. In one of 20 class averages, there are obvious extra densities surrounding the T ring (Fig. 5A), whereas most class averages from wild type and all class averages from ∆pomAB do not show these structures, suggesting the extra densities are indeed formed by the stators (Fig. 5A). To visualize the extra densities with more clarity, we generated a symmetric structure of the stator complex (Fig. 5A), which was then mapped back onto the overall motor structure, as shown in Fig. 5 B and C.
Fig. 5.
Overall architecture of the Vibrio flagellar motor. (A) One class average of the wild-type motor. The first panel is a central section. The second panel is a cross-section from the T ring. The third panel is a cross-section at the top of the cytoplasmic membrane (CM). The fourth panel is the corresponding cross-section of the third panel after applying 13-fold symmetry. A side view (B) and a top view (C) of the overall structure of the Vibrio flagellar motor are shown.
Overall Architecture of Vibrio Flagellar Motor.
The T ring exhibits unique 13-fold symmetry in both sheathed and unsheathed flagella, and likely functions as a platform to recruit 13 highly dynamic stators assembled around the rotor (Fig. 5B). MotY, potentially anchored to the cell wall, might function as a scaffold for the T ring. The periplasmic domain of the stator appears to directly interact with MotY (Fig. 5 B and C). In comparison with the well-studied unsheathed flagellar motor in E. coli (Fig. 1 F and I), these Vibrio-specific components not only reflect the complexity of Vibrio sheathed flagellum (Fig. 5C) but also provide a solid foundation for establishing the characteristic high velocity and robustness of Vibrio motility.
Discussion
A Vibrio flhG Strain as a Model System for in Situ Structural Analysis of Sheathed Flagella.
The V. alginolyticus polar flagellum has been extensively studied by genetic, biochemical, functional, and structural approaches, and is a recognized model for a sheathed flagellum (32). However, despite the success of cryo-ET in characterizing many other bacterial flagellar structures (23–25, 33–38), this method was not previously used to visualize the intact wild-type V. alginolyticus flagellar motor, in part because the single polar flagellum in large cells of V. alginolyticus is suboptimal for high-throughput cryo-ET analysis. Fortunately, a defective flhG locus results in multiple polar flagella in V. cholerae and V. alginolyticus (27–29), as well as Pseudomonas and Shewanella (39, 40). Recently, a defect in hubP also conferred a multiple flagella phenotype in V. alginolyticus (30). Although V. alginolyticus cells are still larger than optimal, the cell pole region with multiple flagella is often thin enough for cryo-ET imaging. Together with genetic approaches available for making additional mutations in the flhG defective background, the multiple polar flagellated strain of V. alginolyticus is an ideal model system for high-resolution structural analysis of sheathed flagella in situ.
Comparison of Sheathed and Unsheathed Flagella Reveals Major Remodeling of the Outer Membrane.
As expected, most polar flagella observed in V. alginolyticus cells are sheathed. However, to our surprise, unsheathed flagella were detectable in wild-type cells of V. alginolyticus. The combination of high-throughput cryo-ET and cells with multiple polar flagella enabled us to generate sufficient data to determine two distinct structures from sheathed and unsheathed flagella in the same cell. The major components of the two distinct Vibrio flagella are comparable to those in E. coli flagella, including the C ring, the export apparatus, the MS ring, the rod, the P/L rings, and the hook. Furthermore, the outer membrane appears to interact directly with the L ring of the unsheathed flagellum in V. alginolyticus and E. coli, although the unsheathed Vibrio flagellum possesses the T ring and the H ring, both specific to Vibrios.
Importantly, the V. alginolyticus sheathed flagellum is strikingly different from the unsheathed flagellum at the point where the flagellum intersects the outer membrane. Notably, the sheathed flagellum possesses a novel O ring, which is associated with a 90° bend of the outer membrane that forms the sheath around the hook and the filament. We speculate that the O ring may form a scaffold for the dramatic remodeling of the outer membrane to form the sheath. Notably, the O ring has not yet been found in recently reported structures of sheathed flagella from V. fischeri (25) and H. pylori (36). Nevertheless, the sheathed flagella from different species share a common theme: the outer membrane does not intersect directly with the L ring, which functions as a bushing embedded in the outer membrane in model systems such as E. coli and Salmonella. FlgO is an outer membrane protein required for flagellar motility in Vibrio cholerae (41). FlgO is well conserved in Vibrio species. We therefore speculated that the O ring might be composed of FlgO. Mutagenesis and GFP-tag labeling together with Cryo-ET will be required to identify genes responsible for O-ring formation.
Structure and Function of the Unique Vibrio-Specific Feature.
The V. alginolyticus flagellum possesses two Vibrio-specific features in the periplasm: the T ring and the H ring. The T-ring structure is the essential component of the sodium ion-driven flagellar motor in addition to PomA/B (19). Our comparative analysis of motor structures from wild-type cells and several T ring mutants (∆motX∆motY, ∆motX, ∆motY) revealed that the T ring has a 13-fold symmetry. MotY is the major protein responsible for its assembly, as 13 copies of MotY fit well into the T ring. In contrast, the location of MotX is not well defined, because deletion of the motX gene did not change the overall shape of the T ring significantly. As MotX interacts with MotY, and both proteins are essential for stator assembly (19, 42), we postulate that MotX assembles at the outer rim of the T ring.
The H ring appears to be associated with the L ring and is important for motor assembly and function. FlgT plays a critical role in the formation of the H ring, which is likely composed of other flagellar proteins, such as FlgO and FlgP. However, the overall structure of the H ring is far more complex. We propose that FlgT forms a ring on top of the MotY ring. Our model is supported by previous biochemical data and the observation that ∆flgT mutant cells lose both the T and H rings (43).
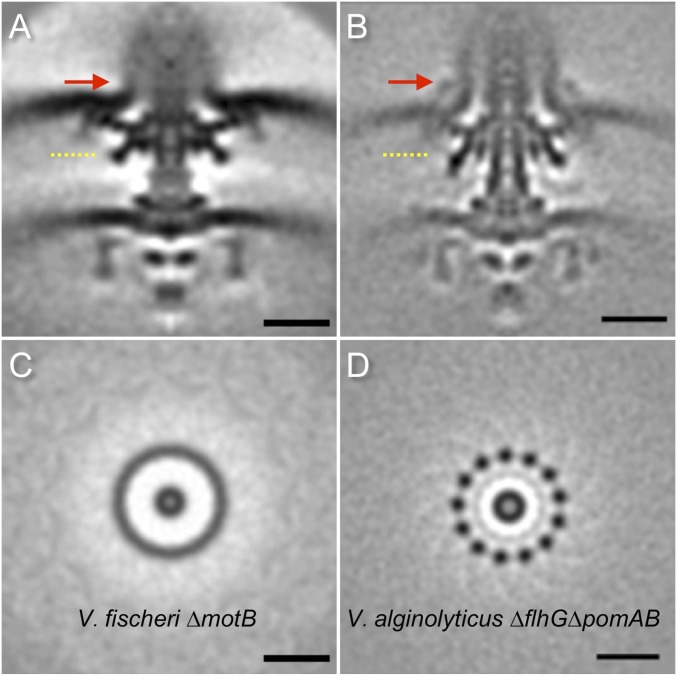
Both H and T rings are intimately associated with the L/P rings. They form a large Vibrio-specific complex underneath the outer membrane. Importantly, we demonstrated that the 13-fold symmetry of T rings is maintained in both wild-type and ∆pomAB strains, indicating that the symmetric features of the T ring are independent of the stators. In contrast, 13-fold symmetry of the T ring is not visible in the V. fischeri motor structure (25), although there are 13 stators associated with the V. fischeri motor (Fig. S1). One possibility is that the stators in V. fischeri are more statically localized around the motors, similar to those found in spirochetes (24, 37). It is reasonable to propose that in V. alginolyticus, the 13-fold symmetric T ring provides a platform for recruiting up to 13 stators in dynamic fashion, perhaps in response to environmental factors (44).
Fig. S1.
Flagellar motor comparison between V. fischeri and V. alginolyticus. (A) A central section of the averaged structure (EMD-3156) from 251 motor particles in a ∆pomB mutant of V. fischeri. (B) A central section of the averaged structure the averaged motor structure from 999 motor particles in V. alginolyticus ∆flhG∆pomAB. Red arrows highlight the difference in the O ring. (C) A cross-section of the T ring as indicated by the yellow dashed line in A. (D) A cross-section of the T ring shows the 13-fold symmetric feature in the T ring. No apparent symmetry is visible in the T ring of the motor structure from V. fischeri ∆pomB. (Scale bar, 20 nm.)
Indeed, whereas stator densities in V. alginolyticus can be localized outside of the T ring in some of the class averages, they are not observed in a global average, consistent with the notion that the stators are highly dynamic in the cytoplasmic membrane, as demonstrated previously (9–11). The dynamic nature of the Vibrio stators are dependent on sodium ions (7), FliL, and other proteins (45). Indeed, the stators appear considerably different in different species. The maximum stator number in E. coli is at least 11, as measured using total internal reflection fluorescence microscopy (5). A pioneering cryo-ET study of the Treponema primitia flagellar motor revealed a putative stator ring with 16-fold symmetry (24). Cryo-ET study of B. burgdorferi revealed that 16 stators are embedded in the highly curved spirochete cytoplasmic membrane (26, 37). A recent study of the V. fischeri flagellar motor revealed 13 stators (25), whereas H. pylori can possess 18 stators around the rotor (36).
Intrinsic Plasticity in Sheathed Flagella.
We found a surprising and significant variation in distance between the MS ring and the outer membrane-bound complex in our imaged V. alginolyticus flagellar motors. As flagellar rods in wild-type cells are constant in length, we propose that the length variation we observe is a result of the ability of the L/P/H/T/O rings to slide along the distal rod along with the associated outer membrane. Although it is generally thought that the rod rotates inside the P and L rings, which function as bushings in the PG layer and the outer membrane, respectively, it has not been reported previously that the P/L rings can slide along the rod. Multiple P rings were shown to assemble around a longer polyrod; however, the P rings were not evenly spaced (46), suggesting the P ring can interact differentially with the underlying protein component FlgG of the distal rod. In fact, a recent structural study showed that the distal rod has a rigid and smooth surface ∼13 nm in diameter and 18 nm in length (47). These overall properties of the distal rod are consistent with its main function as a drive shaft and may allow the outer membrane-bound complex to move flexibly with the membranous sheath during flagellar rotation. Alternatively, the distance variations may reflect structures that are fixed for each motor, but differ in length among different motors.
In conclusion, our results demonstrate that the V. alginolyticus system is ideal for studying sheathed flagellar architecture in situ. We reveal an outer membrane ring and a large conformational change of the outer membrane associated with sheath formation and flagellar assembly. We also uncovered a significant variation in motor positioning relative to the flagellar rod, suggesting a surprising level of conformational flexibility in sheathed flagella. Furthermore, we demonstrate a 13-fold symmetry in the motor, providing a basis to better understand specific rotor–stator interactions in Vibrio. Finally, our studies provide insights into the unique aspects of sheathed flagella, which are widely used by many bacterial pathogens to establish infection and disseminate in humans and other mammalian hosts.
Materials and Methods
Bacterial Growth and Sample Preparation.
Bacterial strains used in this study are listed in Table 1. V. alginolyticus strains were cultured overnight at 30 °C on VC medium [0.5% (wt/vol) polypeptone, 0.5% (wt/vol) yeast extract, 3% (wt/vol) NaCl, 0.4% (wt/vol) K2HPO4, 0.2% (wt/vol) glucose]. The overnight culture were diluted 100× with fresh VC medium and cultured at 30 °C at 250 rpm (Taitec, BioShaker BR-23FH). After 5 h, cells were collected and washed 2× and finally diluted with TMN500 medium (50 mM Tris·HCl at pH 7.5, 5 mM glucose, 5 mM MgCl, and 500 mM NaCl) (48). Colloidal gold solution (10 nm diameter) was added into the diluted Vibrio samples to yield a 10× dilution and then deposited on a freshly glow-discharged, holey carbon grid for 1 min. The grid was blotted with filter paper and rapidly plunge-frozen in liquid ethane in a homemade plunger apparatus, as described (49).
Table 1.
Bacterial strains
| Strain | Genotype or descriptions | Source |
| V. alginolyticus | ||
| VIO5 | VIK4 laf (Rifr Pof+ Laf−) | Ref. 18 |
| KK148 | Vibrio alginolyticus laf, flhG (Rifr Laf− multi-Pof+) | Ref. 27 |
| NMB300 | KK148 ∆pomAB (Mot−) | This study |
| TH1 | KK148 ∆motX (Mot−) | Ref. 19 |
| TH2 | KK148 ∆motY (Mot−) | Ref. 19 |
| TH3 | KK148 ∆motX∆motY (Mot−) | Ref. 19 |
| E. coli | ||
| WM4011 | WM3433 (pBAD30-flhDC) | Ref. 56 |
A: Rifr, rifampin resistant; Laf−, defective in lateral flagellar formation; Pof+, normal polar flagellar formation; multi-Pof+, multiple polar flagellar formation; Mot−, nonmotile.
Cryo-ET Data Collection and Image Processing.
The frozen-hydrated specimens were transferred to a Polara G2 electron microscope (FEI) equipped with a 300-kV field emission gun and a Direct Electron Detector (Gatan K2 Summit). Images were observed at 15,400× magnification, resulting in 0.25 nm/pixel. SerialEM (50) was used to collect tilt series at ∼8 µm defocus. A total dose of 50 e−/Å2 is distributed among 35 tilt images covering angles from −51° to +51° at tilt steps of 3°. For every single tilt series collection, dose-fractionated mode was used to generate 8–10 frames per projection image. Collected dose-fractionated data were first subjected to the motioncorr program to generate drift-corrected stack files (51). The stack files were aligned using gold fiducial markers and volumes reconstructed by the weighted back-projection method, using IMOD software (52) to generate tomograms. In total, 1,293 tomograms of wild-type and mutant cells were generated (Table 2).
Table 2.
Cryo-ET data
| Strain | No. of cellular tomograms | No. of motor subtomograms |
| V. alginolyticus | ||
| VIO5 | 95 | 90 |
| KK148 | 596 | 3,939 |
| NMB300 | 270 | 1,951 |
| TH1 | 132 | 1,184 |
| TH2 | 152 | 1,196 |
| TH3 | 143 | 491 |
| E. coli | ||
| WM4011 | 1,024 | 1,200 |
Subtomogram Analysis.
Bacterial flagellar motors were detected manually, using the i3 program (53, 54). We selected two points on each motor: one point at the C-ring region, and another near the flagellar hook. The orientation and geographic coordinates of selected particles were then estimated. In total, 8,761 subtomograms of Vibrio motors and 1,200 subtomograms of E. coli motors were used to subtomogram analysis. The i3 tomographic package was used on the basis of the “alignment by classification” method with missing wedge compensation (53, 54) for generating the averaged structure of the motor, as described previously (33, 49).
3D Visualization and Modeling.
Tomographic reconstructions were visualized using IMOD (52). UCSF Chimera software was used for 3D surface rendering of subtomogram averages and molecular modeling (55). The crystal structures of MotY (PDB ID: 2ZF8) and FlgT (PDB ID: 3W1E) from V. alginolyticus were docked into our density maps, using the function “fit in map” in UCSF Chimera.
Acknowledgments
We thank William Margolin for critically reading the manuscript before submission. This work was supported by GM107629 from the National Institute of General Medicine, and AU-1714 from the Welch Foundation (to J.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1712489114/-/DCSupplemental.
References
- 1.Berg HC. The rotary motor of bacterial flagella. Annu Rev Biochem. 2003;72:19–54. doi: 10.1146/annurev.biochem.72.121801.161737. [DOI] [PubMed] [Google Scholar]
- 2.Terashima H, Kojima S, Homma M. Flagellar motility in bacteria structure and function of flagellar motor. Int Rev Cell Mol Biol. 2008;270:39–85. doi: 10.1016/S1937-6448(08)01402-0. [DOI] [PubMed] [Google Scholar]
- 3.Chevance FF, Hughes KT. Coordinating assembly of a bacterial macromolecular machine. Nat Rev Microbiol. 2008;6:455–465. doi: 10.1038/nrmicro1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wadhams GH, Armitage JP. Making sense of it all: Bacterial chemotaxis. Nat Rev Mol Cell Biol. 2004;5:1024–1037. doi: 10.1038/nrm1524. [DOI] [PubMed] [Google Scholar]
- 5.Leake MC, et al. Stoichiometry and turnover in single, functioning membrane protein complexes. Nature. 2006;443:355–358. doi: 10.1038/nature05135. [DOI] [PubMed] [Google Scholar]
- 6.Lo C-J, Sowa Y, Pilizota T, Berry RM. Mechanism and kinetics of a sodium-driven bacterial flagellar motor. Proc Natl Acad Sci USA. 2013;110:E2544–E2551. doi: 10.1073/pnas.1301664110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fukuoka H, Wada T, Kojima S, Ishijima A, Homma M. Sodium-dependent dynamic assembly of membrane complexes in sodium-driven flagellar motors. Mol Microbiol. 2009;71:825–835. doi: 10.1111/j.1365-2958.2008.06569.x. [DOI] [PubMed] [Google Scholar]
- 8.Paulick A, et al. Two different stator systems drive a single polar flagellum in Shewanella oneidensis MR-1. Mol Microbiol. 2009;71:836–850. doi: 10.1111/j.1365-2958.2008.06570.x. [DOI] [PubMed] [Google Scholar]
- 9.Tipping MJ, Delalez NJ, Lim R, Berry RM, Armitage JP. Load-dependent assembly of the bacterial flagellar motor. MBio. 2013;4:e00551-13. doi: 10.1128/mBio.00551-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Che YS, et al. Load-sensitive coupling of proton translocation and torque generation in the bacterial flagellar motor. Mol Microbiol. 2014;91:175–184. doi: 10.1111/mmi.12453. [DOI] [PubMed] [Google Scholar]
- 11.Lele PP, Hosu BG, Berg HC. Dynamics of mechanosensing in the bacterial flagellar motor. Proc Natl Acad Sci USA. 2013;110:11839–11844. doi: 10.1073/pnas.1305885110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCarter LL. Polar flagellar motility of the Vibrionaceae. Microbiol Mol Biol Rev. 2001;65:445–462. doi: 10.1128/MMBR.65.3.445-462.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoon SS, Mekalanos JJ. Decreased potency of the Vibrio cholerae sheathed flagellum to trigger host innate immunity. Infect Immun. 2008;76:1282–1288. doi: 10.1128/IAI.00736-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brennan CA, et al. A model symbiosis reveals a role for sheathed-flagellum rotation in the release of immunogenic lipopolysaccharide. Elife. 2014;3:e01579. doi: 10.7554/eLife.01579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen M, et al. Length-dependent flagellar growth of Vibrio alginolyticus revealed by real time fluorescent imaging. Elife. January 18, 2017 doi: 10.7554/eLife.22140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Asai Y, et al. Putative channel components for the fast-rotating sodium-driven flagellar motor of a marine bacterium. J Bacteriol. 1997;179:5104–5110. doi: 10.1128/jb.179.16.5104-5110.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCarter LL. MotX, the channel component of the sodium-type flagellar motor. J Bacteriol. 1994;176:5988–5998. doi: 10.1128/jb.176.19.5988-5998.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okunishi I, Kawagishi I, Homma M. Cloning and characterization of motY, a gene coding for a component of the sodium-driven flagellar motor in Vibrio alginolyticus. J Bacteriol. 1996;178:2409–2415. doi: 10.1128/jb.178.8.2409-2415.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Terashima H, Fukuoka H, Yakushi T, Kojima S, Homma M. The Vibrio motor proteins, MotX and MotY, are associated with the basal body of Na-driven flagella and required for stator formation. Mol Microbiol. 2006;62:1170–1180. doi: 10.1111/j.1365-2958.2006.05435.x. [DOI] [PubMed] [Google Scholar]
- 20.Okabe M, Yakushi T, Kojima M, Homma M. MotX and MotY, specific components of the sodium-driven flagellar motor, colocalize to the outer membrane in Vibrio alginolyticus. Mol Microbiol. 2002;46:125–134. doi: 10.1046/j.1365-2958.2002.03142.x. [DOI] [PubMed] [Google Scholar]
- 21.Terashima H, et al. Insight into the assembly mechanism in the supramolecular rings of the sodium-driven Vibrio flagellar motor from the structure of FlgT. Proc Natl Acad Sci USA. 2013;110:6133–6138. doi: 10.1073/pnas.1222655110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen S, et al. Structural diversity of bacterial flagellar motors. EMBO J. 2011;30:2972–2981. doi: 10.1038/emboj.2011.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao X, Norris SJ, Liu J. Molecular architecture of the bacterial flagellar motor in cells. Biochemistry. 2014;53:4323–4333. doi: 10.1021/bi500059y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murphy GE, Leadbetter JR, Jensen GJ. In situ structure of the complete Treponema primitia flagellar motor. Nature. 2006;442:1062–1064. doi: 10.1038/nature05015. [DOI] [PubMed] [Google Scholar]
- 25.Beeby M, et al. Diverse high-torque bacterial flagellar motors assemble wider stator rings using a conserved protein scaffold. Proc Natl Acad Sci USA. 2016;113:E1917–E1926. doi: 10.1073/pnas.1518952113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moon KH, et al. Spirochetes flagellar collar protein FlbB has astounding effects in orientation of periplasmic flagella, bacterial shape, motility, and assembly of motors in Borrelia burgdorferi. Mol Microbiol. 2016;102:336–348. doi: 10.1111/mmi.13463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kusumoto A, et al. Regulation of polar flagellar number by the flhF and flhG genes in Vibrio alginolyticus. J Biochem. 2006;139:113–121. doi: 10.1093/jb/mvj010. [DOI] [PubMed] [Google Scholar]
- 28.Correa NE, Peng F, Klose KE. Roles of the regulatory proteins FlhF and FlhG in the Vibrio cholerae flagellar transcription hierarchy. J Bacteriol. 2005;187:6324–6332. doi: 10.1128/JB.187.18.6324-6332.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kusumoto A, et al. Collaboration of FlhF and FlhG to regulate polar-flagella number and localization in Vibrio alginolyticus. Microbiology. 2008;154:1390–1399. doi: 10.1099/mic.0.2007/012641-0. [DOI] [PubMed] [Google Scholar]
- 30.Takekawa N, Kwon S, Nishioka N, Kojima S, Homma M. HubP, a polar landmark protein, regulates flagellar number by assisting in the proper polar localization of FlhG in Vibrio alginolyticus. J Bacteriol. 2016;198:3091–3098. doi: 10.1128/JB.00462-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kojima S, et al. Insights into the stator assembly of the Vibrio flagellar motor from the crystal structure of MotY. Proc Natl Acad Sci USA. 2008;105:7696–7701. doi: 10.1073/pnas.0800308105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li N, Kojima S, Homma M. Sodium-driven motor of the polar flagellum in marine bacteria Vibrio. Genes Cells. 2011;16:985–999. doi: 10.1111/j.1365-2443.2011.01545.x. [DOI] [PubMed] [Google Scholar]
- 33.Zhao X, et al. Cryoelectron tomography reveals the sequential assembly of bacterial flagella in Borrelia burgdorferi. Proc Natl Acad Sci USA. 2013;110:14390–14395. doi: 10.1073/pnas.1308306110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kudryashev M, Cyrklaff M, Wallich R, Baumeister W, Frischknecht F. Distinct in situ structures of the Borrelia flagellar motor. J Struct Biol. 2010;169:54–61. doi: 10.1016/j.jsb.2009.08.008. [DOI] [PubMed] [Google Scholar]
- 35.Charon NW, et al. The flat-ribbon configuration of the periplasmic flagella of Borrelia burgdorferi and its relationship to motility and morphology. J Bacteriol. 2009;191:600–607. doi: 10.1128/JB.01288-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qin Z, Lin WT, Zhu S, Franco AT, Liu J. Imaging the motility and chemotaxis machineries in Helicobacter pylori by cryo-electron tomography. J Bacteriol. November 14, 2016 doi: 10.1128/JB.00695-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu J, et al. Intact flagellar motor of Borrelia burgdorferi revealed by cryo-electron tomography: Evidence for stator ring curvature and rotor/C-ring assembly flexion. J Bacteriol. 2009;191:5026–5036. doi: 10.1128/JB.00340-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raddi G, et al. Three-dimensional structures of pathogenic and saprophytic Leptospira species revealed by cryo-electron tomography. J Bacteriol. 2012;194:1299–1306. doi: 10.1128/JB.06474-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dasgupta N, Ramphal R. Interaction of the antiactivator FleN with the transcriptional activator FleQ regulates flagellar number in Pseudomonas aeruginosa. J Bacteriol. 2001;183:6636–6644. doi: 10.1128/JB.183.22.6636-6644.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schuhmacher JS, et al. MinD-like ATPase FlhG effects location and number of bacterial flagella during C-ring assembly. Proc Natl Acad Sci USA. 2015;112:3092–3097. doi: 10.1073/pnas.1419388112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martinez RM, Dharmasena MN, Kirn TJ, Taylor RK. Characterization of two outer membrane proteins, FlgO and FlgP, that influence vibrio cholerae motility. J Bacteriol. 2009;191:5669–5679. doi: 10.1128/JB.00632-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Okabe M, Yakushi T, Homma M. Interactions of MotX with MotY and with the PomA/PomB sodium ion channel complex of the Vibrio alginolyticus polar flagellum. J Biol Chem. 2005;280:25659–25664. doi: 10.1074/jbc.M500263200. [DOI] [PubMed] [Google Scholar]
- 43.Terashima H, Koike M, Kojima S, Homma M. The flagellar basal body-associated protein FlgT is essential for a novel ring structure in the sodium-driven Vibrio motor. J Bacteriol. 2010;192:5609–5615. doi: 10.1128/JB.00720-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baker AE, O’Toole GA. Bacteria, rev your engines: Stator dynamics regulate flagellar motility. J Bacteriol. 2017;199:e00088-17. doi: 10.1128/JB.00088-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhu S, Kumar A, Kojima S, Homma M. FliL associates with the stator to support torque generation of the sodium-driven polar flagellar motor of Vibrio. Mol Microbiol. 2015;98:101–110. doi: 10.1111/mmi.13103. [DOI] [PubMed] [Google Scholar]
- 46.Chevance FF, et al. The mechanism of outer membrane penetration by the eubacterial flagellum and implications for spirochete evolution. Genes Dev. 2007;21:2326–2335. doi: 10.1101/gad.1571607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fujii T, et al. Identical folds used for distinct mechanical functions of the bacterial flagellar rod and hook. Nat Commun. 2017;8:14276. doi: 10.1038/ncomms14276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu S, et al. Conformational change in the periplamic region of the flagellar stator coupled with the assembly around the rotor. Proc Natl Acad Sci USA. 2014;111:13523–13528. doi: 10.1073/pnas.1324201111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhu S, Qin Z, Wang J, Morado DR, Liu J. In situ structural analysis of the spirochetal flagellar motor by cryo-electron tomography. Methods Mol Biol. 2017;1593:229–242. doi: 10.1007/978-1-4939-6927-2_18. [DOI] [PubMed] [Google Scholar]
- 50.Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 51.Li X, et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods. 2013;10:584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 53.Winkler H. 3D reconstruction and processing of volumetric data in cryo-electron tomography. J Struct Biol. 2007;157:126–137. doi: 10.1016/j.jsb.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 54.Winkler H, et al. Tomographic subvolume alignment and subvolume classification applied to myosin V and SIV envelope spikes. J Struct Biol. 2009;165:64–77. doi: 10.1016/j.jsb.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pettersen EF, et al. UCSF chimera–A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 56.Liu J, et al. Molecular architecture of chemoreceptor arrays revealed by cryoelectron tomography of Escherichia coli minicells. Proc Natl Acad Sci USA. 2012;109:E1481–E1488. doi: 10.1073/pnas.1200781109. [DOI] [PMC free article] [PubMed] [Google Scholar]