Significance
In in vitro fertilization, it is difficult, if not impossible, with current methods to determine whether an embryo carries a chromosomal translocation. We have established a method for diagnosing chromosome abnormality named “Mapping Allele with Resolved Carrier Status” (MaReCs), which enables simultaneous screening of chromosomal ploidy and translocation in an embryo by next-generation sequencing. We demonstrate and validate that MaReCs allows accurate selection of translocation-free embryos, preventing the transmission of chromosomal translocations to future generations.
Keywords: chromosomal abnormality, PGD, IVF, translocation, MALBAC
Abstract
Reciprocal translocations (RecT) and Robertsonian translocations (RobT) are among the most common chromosomal abnormalities that cause infertility and birth defects. Preimplantation genetic testing for aneuploidy using comprehensive chromosome screening for in vitro fertilization enables embryo selection with balanced chromosomal ploidy; however, it is normally unable to determine whether an embryo is a translocation carrier. Here we report a method named “Mapping Allele with Resolved Carrier Status” (MaReCs), which enables chromosomal ploidy screening and resolution of the translocation carrier status of the same embryo. We performed MaReCs on 108 embryos, of which 96 were from 13 RecT carriers and 12 were from three RobT carriers. Thirteen of the sixteen patients had at least one diploid embryo. We have confirmed the accuracy of our carrier status determination in amniotic fluid karyotyping of seven cases as well as in the live birth we have thus far. Therefore, MaReCs accurately enables the selection of translocation-free embryos from patients carrying chromosomal translocations. We expect MaReCs will help reduce the propagation of RecT/RobT in the human population.
Reciprocal translocation (RecT) is a category of chromosomal abnormality in which reciprocal exchange occurs between partial arms of any two chromosomes. Robertsonian translocations (RobT) are a special form of RecT in which the breakpoints of the reciprocal exchange occur in the centromere region where the long arms of the two involved chromosomes are joined together. Unlike RecT, RobT occurs exclusively in chromosomes 13, 14, 15, and 21. The origin of translocations is presumably related to errors in recombination events ocurring during gamete formation, as demonstrated recently in both oocytes (1, 2) and sperms (3, 4).
RecT/RobT carriers often do not display any apparent abnormality in daily life, as in most cases no key genes are lost in these translocations. However, RecT and RobT are common anomalies that cause birth defects and infertility (5, 6) and account for ∼5% of recurrent pregnancy loss (7–11). Patients carrying an RecT or RobT karyotype are known to have a much higher rate of chromosomal abnormalities in their gametes or offspring (12, 13), such abnormalities are often the cause of recurrent pregnancy loss or birth defects such as Down or Patau syndrome, among many others (14, 15).
Normal embryos from chromosomal disease carriers can be selected by preimplantation genetic diagnosis (PGD). FISH was first used in PGD to target the two most common chromosomes involved in translocation. Although valuable, FISH is limited to the detection of a very few chromosomes, and since it relies on fluorescent markers, the results are sometimes inconclusive due to ambiguous optical signals and complex sample preparation procedures (16–19). In the past five years, comprehensive chromosome screening (CCS) has been performed on all 24 chromosomes by using comparative genomic hybridization array (CGH) or next-generation sequencing (NGS). Multiple clinical trials suggest improved per-transfer-cycle pregnancy and live-birth rates using CCS (20, 21). In particular, the combination of CCS and NGS has been shown to be more sensitive than the sole use of array-based methods in detecting chromosome abnormalities (22, 23). However, while current NGS-based CCS methods are valuable, they are still limited to preimplantation genetic testing for aneuploidy. For example, CCS is not able to resolve whether an embryo with balanced ploidy has normal karyotype or is a carrier of a chromosomal translocation (18, 19, 21, 24, 25). Transferring a translocation-carrying embryo of balanced ploidy causes propagation of the translocation karyotype into the next generation, in addition to all the associated pregnancy risks (26, 27). Therefore, developing an NGS-based CCS method that can resolve the translocation-carrier status of the embryo is highly desirable in treating translocation-carrying patients in PGD.
Efforts have been made toward using NGS to select translocation-free embryos with normal ploidy. Liang et al. (28) used a long-range mate-pair approach to identify translocation breakpoints in NGS. A breakpoint is a common phenomenon that occurs during chromosomal recombination in the process of meiosis and that can generate hereditary abnormalities. Translocation breakpoints are difficult to identify since they are often associated with highly repetitive sequences, causing ambiguity in their identification. Hu et al. (29) used laser microdissection to identify SNP markers that are associated with the translocation-affected allele. Although this method can identify a translocation-affected allele, it requires highly specialized equipment and a complex procedure for sample preparation, making it unlikely to become accessible for high-throughput daily clinical use. The SNP array-based linkage analysis method is also used to identify translocation-free embryos (30). However, to our knowledge, no live births have been reported with this method as yet, likely due to its limited accuracy in breakpoint identification and in linkage analysis.
Here we present an NGS-based method named “Mapping Allele with Resolved Carrier Status” (MaReCs) that enables the identification of the allele associated with translocation by using an already widely used linkage analysis method (31). In brief, a trophectoderm biopsy is performed for each embryo and is subjected to whole-genome amplification (WGA) and sequencing by multiple annealing and looping-based amplification cycle (MALBAC) and NGS (24, 32, 33). Translocation breakpoints in the chromosome-imbalanced embryos are first identified with high resolution (∼200 kb) by locating the copy number change. SNP markers located within 1 Mb of the detected breakpoints are then used to identify the translocation-carrying allele in a manner similar to our recently published linkage analysis method, MARSALA (34). After the translocation-carrying allele is identified in the chromosomal-imbalanced embryos, the same groups of SNP markers are used to identify whether a chromosomally balanced embryo carries the translocation. In this way, a translocation-free chromosomally balanced embryo is selected for implantation. We have validated the MaReCs method in 16 cases of translocation PGD. Using our MaReCs method, we identified a translocation-free embryo and transferred it into the uterus of a consenting patient, resulting in a healthy live birth on Dec. 28, 2016. The normal karyotypes of the other six ongoing pregnancies were confirmed by amniocentesis.
Results
Identification of a Reference Embryo.
We have developed the MaReCs technique to accurately resolve the translocation-carrier status in embryos with balanced chromosomal ploidy. The availability of at least one chromosomally imbalanced embryo is essential in the MaReCs pipeline; this embryo is defined as the reference embryo (Fig. 1). A reference embryo carries at least one abnormal allele inherited from the translocation-carrying parent. The haplotype of informative SNPs with linkage to the abnormal allele can then be mapped from the reference embryo. However, the haplotype mapping can be performed only when the specific karyotype is known in detail. In the MaReCs pipeline, the only way to determine the specific karyotype of a given embryo is by its chromosomal copy number variation (CNV) profile obtained from the NGS-based CCS. This requirement is easily met with MALBAC-NGS, in which CCS is performed with high resolution (∼200 kb) to identify translocation breakpoints, and the SNPs are selected within ∼1 Mb of the breakpoints to avoid the allele misidentification resulting from meiotic recombination during gamete formation.
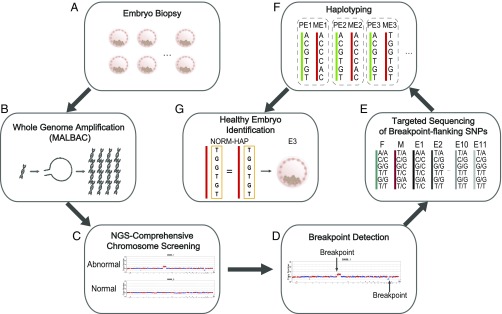
Fig. 1.
Workflow of MaReCs. (A and B) MALBAC-based WGA followed by NGS-based CCS is performed in the biopsy of each embryo. The WGA DNA products generate sufficient DNA that the second round of targeting sequencing can be carried out without a second trophectoderm biopsy of the embryo. Samples are saved for retrospective MaReCs analysis. (C) The embryos with abnormal CNV are identified and excluded from implantation at this stage. (D) Further analysis of the abnormal CNVs of the reference embryo allows identification of the breakpoint of the reciprocal translocation. (E) Multiple SNPs flanking the breakpoint are then targeted and sequenced by NGS for each embryo and the parents. (F) By linkage analysis of the informative SNPs, the haplotypes linked to the normal allele and to the translocation allele of the parental RecT carrier can be mapped separately. (G) Using the haplotype linkage information, the carrier status of the embryos with balanced ploidy can be resolved. Detailed haplotype linkage analysis to resolve the carrier status of embryos is illustrated in Fig. 4. E, embryo; F, father; M, mother; ME, maternal allele of the embryo; PE, paternal allele of the embryo.
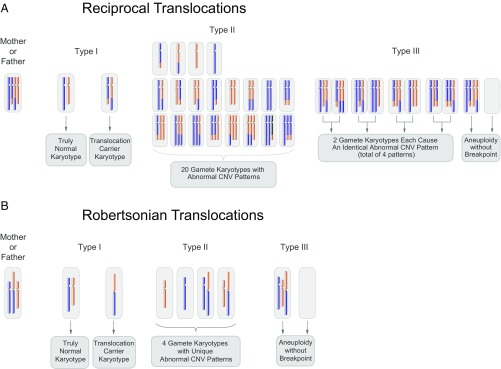
By a systematic review of the 32 possible gamete karyotypes generated by a parental RecT carrier (35), we found that the 32 karyotypes can be classified into three types. Type I contains the candidates for MaReCs selection, which include normal ploidy (a truly normal embryo: normal chromosomes) and balanced ploidy (an RecT carrier in which both chromosomes have the translocation). Type II, which can be used as reference in MaReCs, contains 20 abnormal karyotypes of imbalanced translocation. These karyotypes are biunique with their corresponding CNV patterns. Type III, however, has an abnormal karyotype and cannot be used as reference. Among the 10 type III karyotypes, eight correspond to four CNV patterns; the abnormal karyotypes are not biunique to their CNV patterns, and therefore the gamete karyotype cannot be specifically deduced by the detection of a CNV pattern. Even though the other two type III karyotypes are biunique to their corresponding CNV patterns, the chromosomes involved in translocation are aneuploid without breakpoints and thus are not suitable for SNP linkage analysis (Fig. 2A).
Fig. 2.
Possible karyotypes of gametes generated by an RecT or RobT carrier. (A) An RecT carrier may generate gametes with 32 different possible karyotypes. The karyotype of the mother or father in its diploid (somatic) state is shown at the far left. After fertilization with a normal gamete, the 32 resulting zygotic karyotypes may be categorized into three types. Type I gamete karyotypes show balanced ploidy in either an RecT carrier or a normal embryo. These embryos are candidates for MaReCs selection. Type II gamete karyotypes contain 20 abnormal karyotypes, and each of these karyotypes is biunique with its corresponding CNV pattern. These abnormal embryos can be functional and used as the reference for allelic haplotype mapping in MaReCs. Type III gamete karyotypes contain eight abnormal karyotypes corresponding to only four CNV patterns and two abnormal karyotypes with aneuploidy without a breakpoint; thus they cannot be used as reference embryos in MaReCs. (B) Gamete karyotypes from a RobT carrier are not as complex as those of an RecT carrier. The karyotype of mother or father in its diploid (somatic) state is shown at the far left. While the type I gamete also contains two karyotypes (normal and RobT), the type II gamete includes only four abnormal karyotypes, and the type III gamete includes two abnormal karyotypes.
In the RobT cases, the reference embryo is not as complex as that of RecT. There are eight different karyotypes that possibly exist in the gamete of a RobT carrier (35), two of which have balanced ploidy (type I). Four can be used as reference embryos in MaReCs (type II), and the other two are type III due to their aneuploid CNV patterns and therefore cannot be used as reference (Fig. 2B).
Live-Birth Case of an RecT Carrier Using MaReCs.
The MaReCs procedure was first successfully applied to a couple in which the female carried an RecT of chr 9 and chr 21 [46,XX,t (9, 21)(q24;q22.1)]. While seven embryos were obtained in the in vitro fertilization (IVF) procedure, only one showed normal ploidy by MALBAC-NGS–based CCS (embryo B, Fig. 3). The other six embryos were all type II abnormal embryos with or without aneuploidy in addition to translocated Chrs 9 and 21 (Fig. 3). Once the breakpoint was identified by CCS data, NGS-based targeted SNP examination flanking the breakpoint was followed by linkage analysis to the corresponding type II (reference) embryo to identify the SNP haplotypes linked to the translocation-carrying chromosome and the normal chromosome (9 or 21) (Fig. 4). When the karyotype of the reference embryo was deduced from its CNV pattern, the allele-specific haplotype with linkage to the abnormal (translocation) allele was mapped by logically subtracting the informative SNP haplotype inherited from the healthy parent from the heterozygous SNPs measured in the reference embryo. As the informative SNPs in the parental RecT carrier are heterozygous by definition, the haplotype linked to the normal allele can be further mapped by logically subtracting the abnormal allele-linked haplotype from the heterozygous SNPs measured in the parental RecT carrier. By doing this, the translocation-carrier status of the embryos with balanced ploidy can be resolved by matching the embryo’s allelic haplotype with that of the reference embryo. In this case, only one embryo was found to have balanced ploidy, which fortunately was free of translocation. This embryo was then transferred into the uterus of the consenting female patient, and the normal karyotype was confirmed by amniocentesis. This IVF case resulted in normal ploidy and a healthy live birth (Fig. 4).
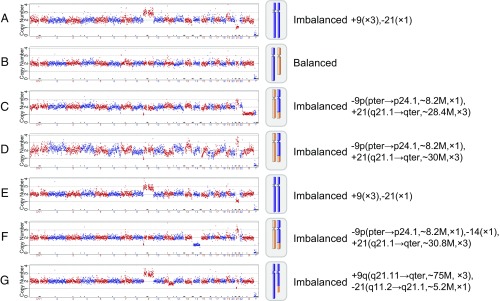
Fig. 3.
Chromosome ploidy results of the embryos from a patient (case no. 38744). In this clinical case, seven blastocyst embryos, A–G, were obtained and biopsied for chromosomal analysis. One embryo, B, was identified as having normal ploidy; the other six embryos were all type II embryos with abnormal Chrs 9 and 21. These abnormal embryos were used as reference embryos to identify the translocation breakpoint. The translocation breakpoint was identified at chr9:8,200 ± 200 kbp and chr21:19,600 ± 200 kbp with the consensus of the reference embryos. Subsequently, for each embryo, the SNPs flanking the breakpoint were examined, and then haplotype linkage analysis was performed. Detailed haplotype linkage analysis to resolve the translocation-carrier status of the embryos is illustrated in Fig. 4.
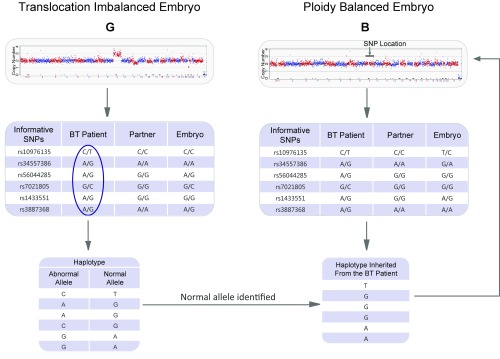
Fig. 4.
Allelic haplotype mapping to resolve the carrier status of the embryos. Shown is a representative example of a reference embryo (here embryo G of case 38744) in haplotype linkage analysis. By measuring the informative SNPs flanking the breakpoint from the parents and reference embryos, the haplotype with linkage to the translocation allele, CAACGG, can be deduced. Therefore, the other haplotype, TGGGAA, must be linked with the normal allele. In the same manner, the haplotype of the ploidy-balanced embryo B inherited from the parental RecT carrier can be identified, and the carrier status of embryo can be identified. The analysis indicates that embryo B inherited the haplotype TGGGAA and was identified as a normal embryo. The embryo was implanted into the uterus of the patient, resulting in a successful pregnancy and live birth.
The Efficacy of MaReCs.
We analyzed 108 embryos by MaReCs from 13 RecT and three RobT carriers with karyotype-normal partners. The background information of the patients and the analytical results of the embryos are summarized in Table 1. At least one reference embryo (Table 1, type II embryos) was available for each case, and we obtained a total of 49 ploidy-balanced embryos (Table 1, type I embryos). We identified at least one embryo as normal with the MaReCs method in 13 of the 16 cases, and subsequently one normal embryo was transferred into the consenting patient. No normal embryos, but only RecT/RobT-carrying embryos, were available for the other three cases (Table 1, cases 35518, 38606, and 41416); in these cases, at the request and with the informed consent of the patients, one carrier embryo without aneuploidy was implanted. Clinical pregnancies were confirmed in all three patients by a human chorionic gonadotropin test at 2 wk post implantation. At the time of this manuscript preparation, seven pregnancies had reached the 20th gestational week; thus the correctness of MaReCs examination (five noncarriers, two carriers) was confirmed by amniocentesis and karyotyping (Table 1, double daggers).
Table 1.
RecT/RobT carriers and embryos tested by MaReCs
| Translocation | Patient ID | Sex | Karyotype of the parental translocation carrier | Total no. embryos tested | Breakpoint location | Type I | Type II | Type III | C* | N† |
| RecT | 34825‡ | F | 46,XX,t (1, 8)(q25;q21.2) | 9 | chr1:161500001; chr8:68850001 | 3 | 5 | 1 | 1 (1) | 2 (2) |
| 38098 | F | 46,XX,t (12, 19)(q13;q13.3) | 9 | chr12:44200001; chr19:42500001 | 4 | 3 | 2 | 2 (1) | 2 (1) | |
| 38591‡ | M | 46,XY,t (6, 12)(q22;q21) | 6 | chr6:13040; chr12:81500001 | 3 | 3 | 0 | 1 (0) | 2 (2) | |
| 38606 | M | 46,XY,t (5, 10)(p15;p11.1) | 6 | chr5:11900001; chr10:28600000 | 2 | 4 | 0 | 1 (1) | 1 (0) | |
| 38744‡,§ | F | 46,XX,t (9, 21)(q24;q22.1) | 7 | chr9:8200000; chr21:19600000 | 1 | 6 | 0 | 0 (0) | 1 (1) | |
| 38888 | M | 46,XY,t (7, 20)(p21;q13.2) | 8 | chr7:45800001; chr20:43100000 | 4 | 4 | 0 | 1 (0) | 3 (2) | |
| 39960‡ | M | 46,XY,t (7, 15)(p22,q24) | 3 | chr7:3300001; chr15:79200001 | 1 | 2 | 0 | 0 (0) | 1 (1) | |
| 40116‡ | M | 46,XY,t (16, 18)(p13.1;q11.2) | 10 | chr16:13700001; chr18:25300001 | 3 | 7 | 0 | 0 (0) | 3 (3) | |
| 41416‡ | M | 46,XY,t (7, 15)(p15;q24) | 3 | chr7:34800001; chr15:74000001 | 2 | 1 | 0 | 2 (1) | 0 (0) | |
| 41476 | M | 46,XY,t (2, 18)(q37;q22) | 5 | chr2:209400001; chr18:42100001 | 3 | 1 | 1 | 2 (2) | 1 (1) | |
| 42309 | M | 46,XY,t (3, 7)(q13.2;q34) | 8 | chr3:127000001; chr7:150700001 | 3 | 5 | 0 | 2 (2) | 1 (1) | |
| 42606 | M | 46,XY,t (5, 17)(q35;p13) | 7 | chr5:qter; chr17:10600001 | 3 | 4 | 0 | 2 (2) | 1 (1) | |
| 42723 | F | 46,XX,t (5, 16)(p13;q13) | 15 | chr5:31260001; chr16:61640001 | 9 | 5 | 1 | 5 (4) | 4 (3) | |
| RobT | 35518‡ | F | 45,XX,rob (14, 21)(q10;q10) | 4 | chr14:q10; chr21:q10 | 2 | 2 | 0 | 2 (1) | 0 (0) |
| 40135 | M | 45,XY,rob (14, 21)(q10,q10) | 5 | chr14:q10; chr21:q10 | 4 | 1 | 0 | 3 (2) | 1 (1) | |
| 41375 | M | 45,XY,rob (13, 15)(q10,q10) | 3 | chr13:q10; chr15:q10 | 2 | 1 | 0 | 0 (0) | 2 (2) | |
| Total | M: 11 | — | 108 | — | 49 | 54 | 5 | 24 (17) | 25 (21) | |
| F: 5 | ||||||||||
F, female; M, male.
Embryos identified as carrier. Parenthetical number indicates embryos without any chromosomal abnormality.
Embryos identified as noncarrier. Parenthetical number indicates embryos without any chromosomal abnormality.
Cases confirmed by karyotyping with amniocentesis.
Live birth with normal ploidy.
The present study also allowed us to look at the translocation-carrying ratio of RecT/RobT patients from the view of a population of human embryos. In this study 108 embryos, 96 from RecT patients and 12 from RobT patients, were successfully examined by MaReCs. About half of the embryos (49/108) were ploidy balanced on chromosome translocations. From those 49 embryos, 38 were ploidy balanced over all chromosomes, having the potential to lead to pregnancy. More than half of the 38 ploidy-balanced embryos (21/38) were confirmed to be truly normal ploidy, which covers most of the patients in this study (11 of the 13 RecT patients and two of the three RobT patients). A significant chance of detecting a truly normal embryo in a large majority of RecT/RobT patients requires a highly efficient embryo-selection strategy, such as the proposed MaReCs. This method allows the avoidance of cross-generation transmission of translocations that are known to increase the chances of birth defects and pregnancy difficulties.
Discussion
Chromosome translocation can cause infertility and birth defects, including disorders such as Down and Patau syndromes. Increasing adoption of CCS without resolving the embryo’s translocation-carrier status may lead to the propagation and accumulation of translocation-carrying karyotypes in a population. Therefore resolving the carrier status of the preimplantation embryos simultaneously with the CCS procedure is highly desirable.
FISH was first used in PGD to target the two most common chromosomes involved in translocation. Although valuable, FISH is limited to the detection of a very few chromosomes, and since it relies on fluorescent markers, the results are sometimes inconclusive due to ambiguous optical signals and complex sample preparation procedures (16–19). Recently, Hu et al. (29) established a strategy to identify translocation breakpoints and flanking SNP haplotypes by using NGS of a microdissected junction region of a specific chromosome. This method of microdissection-NGS allows the identification of normal embryos in PGD. However, the method is complex and requires advanced operational skills and specialized equipment, making its adaptation as a routine PGD procedure difficult. Alternatively, Treff et al. (30) proposed a different strategy to distinguish normal embryos from reciprocal-translocation carriers. However, this method has a limited accuracy of breakpoint determination of 2.36 Mb. This wide range leads to poor information about the location of SNPs within a 5-Mbp region flanking the breakpoint (30). It is known that meiotic recombination occurs at a frequency of ∼1% per megabase on a chromosome. The 5-Mb region on each side of the breakpoint may contain recombination events at an incidence of ∼10%, possibly causing confusion or even error in identifying the translocation allele.
In the present study, we have shown that by using MALBAC-NGS we can improve the accuracy of breakpoint determination to ∼200 kbp, reducing the current range limitation of informative SNPs to ∼1 Mbp flanking the breakpoint. This improvement reduces the recombination incidence from ∼10 to ∼2% of the embryos analyzed. Indeed, among the 108 embryos analyzed in this work, one recombination event between allelic chromosomes was observed at ∼600 kbp from the breakpoint, which could not have been detected accurately with any of the previously existing methods. This advancement significantly improves the clinical applicability of the current method, which potentially could be applied successfully to all patients with translocations.
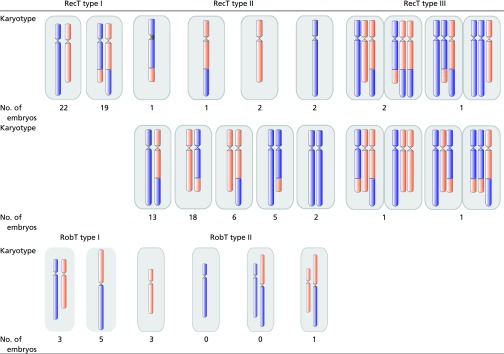
Our method also presents some limitations. Among the 30 possible varieties of imbalanced karyotypes generated by a parental RecT carrier, 10 were not suitable for use as reference embryos (type III embryos in Fig. 2). Even though the NGS technique has the potential to distinguish the two karyotypes from their identical CNV patterns by counting the read-depth ratio between the heterogeneous SNP alleles, allele-dropout events that could occur during the WGA process may affect the read-depth counts. While a systematic study to reveal the frequency of the 32 karyotype varieties is not yet available, our preliminary studies may shed light into it. We found that the 50 type II embryos from RecT patients analyzed in this work belonged to less than half of the possible karyotype varieties (Table 2). This is not surprising, as the gametes with severely abnormal karyotypes may not have survived to the point of fertilization. We also noticed that the type III karyotype embryos were presented as rare events (Table 1), indicating that MaReCs inapplicability in actual clinical situations is slim. It is noteworthy that in the cases mentioned in this work all five type III embryos were always accompanied by at least one sister type II embryo that was sufficient for MaReCs analysis. Furthermore, embryos arrested during development are common in clinical practice. Even though the potential for implantation is lost, these embryos may still be useful as references in MaReCs.
Table 2.
Karyotypes of embryos from RecT and RobT patients
 |
The limitation of the MaReCs strategy is that it requires the availability of a reference embryo. Fortunately, with recent progress in embryo cryopreservation technology, it may be possible to overcome this limitation. When a reference embryo is not available, the ploidy-normal embryos and their WGA product can be cryopreserved temporarily. Then, by an additional cycle of ovarian stimulation, there is a good chance of obtaining at least one functional reference embryo. Also, all the abnormal embryos during PGD can be collected for WGA to identify the breakpoint. Once the breakpoint location is known, and informative SNPs and allelic haplotype mapping are worked out from this reference embryo, MaReCs can be performed in a retrospective manner using the leftover WGA products of the cryopreserved embryos, without the need of thawing and potentially damaging the cryopreserved embryos. This method will allow the accurate identification of a normal embryo, which can be thawed and transferred.
In summary, we have established a practical and easy-to-adopt clinical strategy, MaReCs, to resolve the carrier status of ploidy-balanced embryos from a parental RecT carrier. The MaReCs method is an extension of the well-established MALBAC-NGS–based CCS (32, 34). Theoretically, any WGA method is applicable for MaReCs with the precondition that the amplification is sufficiently even to allow high-resolution (in this case ∼200 kb). MaReCs does not require specialized equipment or complex experimental procedures; therefore it can be fully adapted for routine use in a molecular diagnostic laboratory. We anticipate that, once MaReCs is widely used, it may contribute greatly to overcoming infertility due to RecT and in the long term will decrease translocation-carrying karyotypes in the population by blocking their propagation.
Materials and Methods
Patients and IVF.
Sixteen RecT carriers (including three RobT cases) from the Center for Reproductive Medicine of The First Affiliated Hospital of Zhengzhou University were recruited in this study. The study was approved by the Internal Review Board of The First Affiliated Hospital of Zhengzhou University (approval 2015KY-NO.06), and informed consent was obtained from all patients before MaReCs was applied to biopsy embryos. Intracytoplasmic single-sperm injection was performed for fertilization following a standard protocol (36, 37). Embryo culture, biopsy, and cryopreservation were all performed following the NGS-based CCS protocol as described previously (34).
Detection of CNVs.
Detection of CNVs for each embryo was performed as previously described (37). Briefly, reads from the sequencing results were trimmed to remove adapters and low-quality bases. Cleaned reads were aligned to the human reference genome hg19 (University of California, Santa Cruz Genome Browser; genome.ucsc.edu/). Unique mapped reads were counted in 1,000-kb bins and then were subjected to GC correction. GC correction was performed as follows: Reads of bin i with GC content i were assigned a weight wi = M/MGCi, where M is the average number of sequencing reads in each bin on autosomes and MGCi is the average number of sequencing reads in each bin, which is calculated for every 1% GC content. The GC-corrected reads number is RNGCi = RNri*wi, where RNri is the raw reads number of bin i. After GC correction, the unique mapped read counts were normalized to relative read numbers for the bin. A Perl script (available at https://www.perl.org/) was used to determine CNVs, and the CNVs were visualized by the R programming language (available at https://www.r-project.org/); detailed scripts have been previously submitted to GitHub.
MaReCs Analysis.
The workflow of the MaReCs method is shown in Fig. 1. First, MALBAC-based WGA (24) was applied to each embryo’s biopsy sample and was followed by an NGS-based CCS assay (23). The position of the translocation breakpoint was identified by the CCS data of the corresponding reference embryo. Subsequently, the SNPs flanking the breakpoint were examined for all the embryos and their parents by targeted NGS assay. By linkage analysis to the reference embryos (Fig. 3), the haplotypes linked to the abnormal and normal alleles in the parental RecT carrier were identified. By reviewing the haplotype of the embryos with balanced ploidy, the carrier status of the embryo was deduced.
Translocation Breakpoint Identification.
Identification of the translocation breakpoint location is the first crucial step of the MaReCs analysis. Specifically, MALBAC-NGS–based CCS was performed in all embryos at coverage depth of ∼5 million unique mapped reads. The embryos that showed a specific CNV pattern of imbalanced translocation corresponding to type II were chosen as reference embryos (type II embryos, Fig. 2). Translocation breakpoints of each parental RecT carrier were identified by CNV analysis of the reference embryos. The analysis was performed with a bin size of 100 kbp. The relative reads number (ratio) is calculated by R = r/M, where r is the number of uniquely mapped reads in each bin, and M is the average number of uniquely mapped reads in each bin on autosomes. The copy number value of each bin of a given sample was calculated after GC correction and was normalized to a reference sample with normal karyotype. The diploid copy number threshold for the bins was determined as (meanCN ± SDCN), where meanCN is the average copy number of autosome bins, and SDCN is the SD of the copy number of autosome bins. The identification of the breakpoint location in a specific chromosome translocation first required calculating the trimean of the copy number values among 10 consecutive bins. The calculation was performed in sliding window with a step length of one bin. The first bin in which the trimean value was offset from the diploid copy number threshold was recorded, and the calculation continued until it was encountered a bin in which the trimean value was the same as the diploid copy number. The region between the first and the last offset bin was defined as the level 1 region. Starting from the first bin of the level 1 region, the arithmetic mean of the copy number values of three consecutive bins was calculated, also in a sliding manner, with a step length of one bin. The calculation continued until two consecutive bins were both higher or lower than the threshold of being determined to be diploid. The first of these two bins was determined to be the location of the translocation breakpoint. In this manner, the breakpoint could be identified with an accuracy equivalent to the bin length, namely 200 kbp.
Resolving Carrier Status by Allelic Haplotype Mapping.
Once the location of the breakpoint in the chromosomes was determined, PCR primers were designed by targeting 60 randomly selected SNPs within the 1-Mbp region flanking the breakpoint in each of the two chromosomes involved in the translocation. For RobT, the target SNP was chosen within 1 Mbp flanking the centromere of the involved chromosomes. Targeted SNP amplifications were performed using the leftover WGA (MALBAC) products from previous CCS assays. The SNPs, which are heterozygous in the parental RecT carrier and homozygous in the healthy partner, were used as informative SNPs in subsequent MaReCs analysis. The allelic haplotype mapping of the informative SNPs was performed in the reference embryos to identify the haplotype linkage of the translocation allele. Then, the heterozygosity of the informative SNPs measured in the embryos with balanced ploidy was examined by matching with the information obtained from the reference embryos, thus determining whether the embryo carried the translocation (Fig. 3).
Validation of the MaReCs Results.
MaReCs results were confirmed by routine karyotyping examination of amniotic fluid at gestational weeks 20 (cases 34825, 35518, 38591, 38744, 39960, 40116, and 41416) and have been reconfirmed by amniocentesis in case 38744.
Acknowledgments
We thank Dr. Patricia Purcell of Harvard University for her critical reading and editing of the manuscript. This work was supported by the National Key R&D Program of China Grant 2016YFC0900300 (to Y. Sun), National Natural Science Foundation Grants 31501205 (to J.X.) and 31471404 (to Y. Sun), Clinical Medical Research Fund of Chinese Medical Association–Reproductive Medicine Research and Development Projects for Youth Grant 16020270643 (to J.X.), National Key Technology Research and Development Program Grant 2016YFC0900100 (to L. Huang), and the Beijing Advanced Innovation Center for Genomics at Peking University (X.S.X.).
Footnotes
Conflict of interest statement: S.L. and X.S.X. are cofounders and shareholders of Yikon Genomics Company, Ltd. Z.Z., Y. Gao, W.L., S.B., M.H., and J.R. receive research funding from Yikon Genomics Company, Ltd.
References
- 1.Ottolini CS, et al. Genome-wide maps of recombination and chromosome segregation in human oocytes and embryos show selection for maternal recombination rates. Nat Genet. 2015;47:727–735. doi: 10.1038/ng.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hou Y, et al. Genome analyses of single human oocytes. Cell. 2013;155:1492–1506. doi: 10.1016/j.cell.2013.11.040. [DOI] [PubMed] [Google Scholar]
- 3.Lu S, et al. Probing meiotic recombination and aneuploidy of single sperm cells by whole-genome sequencing. Science. 2012;338:1627–1630. doi: 10.1126/science.1229112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang J, Fan HC, Behr B, Quake SR. Genome-wide single-cell analysis of recombination activity and de novo mutation rates in human sperm. Cell. 2012;150:402–412. doi: 10.1016/j.cell.2012.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thornhill AR, et al. ESHRE PGD Consortium ESHRE PGD consortium ‘best practice guidelines for clinical preimplantation genetic diagnosis (PGD) and preimplantation genetic screening (PGS)’. Hum Reprod. 2005;20:35–48. doi: 10.1093/humrep/deh579. [DOI] [PubMed] [Google Scholar]
- 6.Ethics Committee of American Society for Reproductive Medicine Use of preimplantation genetic diagnosis for serious adult onset conditions: A committee opinion. Fertil Steril. 2013;100:54–57. doi: 10.1016/j.fertnstert.2013.02.043. [DOI] [PubMed] [Google Scholar]
- 7.Kupferminc MJ. Thrombophilia and pregnancy. Reprod Biol Endocrinol. 2003;1:111. doi: 10.1186/1477-7827-1-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arumugam B, Samuel CR, Thyagarajan SS. Balanced autosomal translocations in two women reporting recurrent miscarriage. J Clin Diagn Res. 2016;10:GD01–GD03. doi: 10.7860/JCDR/2016/23828.9075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hotaling J, Carrell DT. Clinical genetic testing for male factor infertility: Current applications and future directions. Andrology. 2014;2:339–350. doi: 10.1111/j.2047-2927.2014.00200.x. [DOI] [PubMed] [Google Scholar]
- 10.Fragouli E, Wells D, Delhanty JD. Chromosome abnormalities in the human oocyte. Cytogenet Genome Res. 2011;133:107–118. doi: 10.1159/000323801. [DOI] [PubMed] [Google Scholar]
- 11.Kutteh WH. Novel strategies for the management of recurrent pregnancy loss. Semin Reprod Med. 2015;33:161–168. doi: 10.1055/s-0035-1552586. [DOI] [PubMed] [Google Scholar]
- 12.Munné S, Howles CM, Wells D. The role of preimplantation genetic diagnosis in diagnosing embryo aneuploidy. Curr Opin Obstet Gynecol. 2009;21:442–449. doi: 10.1097/GCO.0b013e32832fad73. [DOI] [PubMed] [Google Scholar]
- 13.Lalioti MD. Can preimplantation genetic diagnosis overcome recurrent pregnancy failure? Curr Opin Obstet Gynecol. 2008;20:199–204. doi: 10.1097/GCO.0b013e3282f88e0c. [DOI] [PubMed] [Google Scholar]
- 14.Hernández-Gómez M, Meléndez-Hernández R, Rojas-Saiz WH, Maya-Goldsmit D, Mayén-Molina DG. [Recurrent aneuploidy in first trimester gestational loss: A case report and review of the literature] Ginecol Obstet Mex. 2013;81:733–737. [PubMed] [Google Scholar]
- 15.Engels H, et al. Genetic counseling in Robertsonian translocations der(13;14): Frequencies of reproductive outcomes and infertility in 101 pedigrees. Am J Med Genet A. 2008;146A:2611–2616. doi: 10.1002/ajmg.a.32500. [DOI] [PubMed] [Google Scholar]
- 16.Veselá K, et al. [Preimplantation genetic diagnosis (PGD) of chromosomal aberrations using the fluorescence in situ hybridization method (FISH)–Introduction to problems, sampling methods and examination techniques] Ceska Gynekol. 2003;68:89–94. [PubMed] [Google Scholar]
- 17.Pehlivan T, et al. Preimplantation genetic diagnosis by fluorescence in situ hybridization: Clinical possibilities and pitfalls. J Soc Gynecol Investig. 2003;10:315–322. doi: 10.1016/s1071-5576(03)00123-0. [DOI] [PubMed] [Google Scholar]
- 18.Cassel MJ, Munné S, Fung J, Weier HU. Carrier-specific breakpoint-spanning DNA probes: An approach to preimplantation genetic diagnosis in interphase cells. Hum Reprod. 1997;12:2019–2027. doi: 10.1093/humrep/12.9.2019. [DOI] [PubMed] [Google Scholar]
- 19.Willadsen S, et al. Rapid visualization of metaphase chromosomes in single human blastomeres after fusion with in-vitro matured bovine eggs. Hum Reprod. 1999;14:470–475. doi: 10.1093/humrep/14.2.470. [DOI] [PubMed] [Google Scholar]
- 20.Traeger-Synodinos J. Pre-implantation genetic diagnosis. Best Pract Res Clin Obstet Gynaecol. 2017;39:74–88. doi: 10.1016/j.bpobgyn.2016.10.010. [DOI] [PubMed] [Google Scholar]
- 21.Lu L, et al. Recent advances in preimplantation genetic diagnosis and screening. J Assist Reprod Genet. 2016;33:1129–1134. doi: 10.1007/s10815-016-0750-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lukackova R, Gerykova Bujalkova M, Majerova L, Mladosievicova B. Molecular genetic methods in the diagnosis of myelodysplastic syndromes. A review. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2014;158:339–345. doi: 10.5507/bp.2013.084. [DOI] [PubMed] [Google Scholar]
- 23.Kung A, Munné S, Bankowski B, Coates A, Wells D. Validation of next-generation sequencing for comprehensive chromosome screening of embryos. Reprod Biomed Online. 2015;31:760–769. doi: 10.1016/j.rbmo.2015.09.002. [DOI] [PubMed] [Google Scholar]
- 24.Zong C, Lu S, Chapman AR, Xie XS. Genome-wide detection of single-nucleotide and copy-number variations of a single human cell. Science. 2012;338:1622–1626. doi: 10.1126/science.1229164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gui B, et al. Chromosomal analysis of blastocysts from balanced chromosomal rearrangement carriers. Reproduction. 2016;151:455–464. doi: 10.1530/REP-16-0007. [DOI] [PubMed] [Google Scholar]
- 26.Vozdova M, Oracova E, Gaillyova R, Rubes J. Sperm meiotic segregation and aneuploidy in a 46,X,inv(Y),t(10;15) carrier: Case report. Fertil Steril. 2009;92:1748 e9–1748.e13. doi: 10.1016/j.fertnstert.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 27.Gutiérrez-Mateo C, et al. Aneuploidy 12 in a Robertsonian (13;14) carrier: Case report. Hum Reprod. 2005;20:1256–1260. doi: 10.1093/humrep/deh751. [DOI] [PubMed] [Google Scholar]
- 28.Liang D, et al. Copy number variation sequencing for comprehensive diagnosis of chromosome disease syndromes. J Mol Diagn. 2014;16:519–526. doi: 10.1016/j.jmoldx.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 29.Hu L, et al. Reciprocal translocation carrier diagnosis in preimplantation human embryos. EBioMedicine. 2016;14:139–147. doi: 10.1016/j.ebiom.2016.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Treff NR, et al. SNP array-based analyses of unbalanced embryos as a reference to distinguish between balanced translocation carrier and normal blastocysts. J Assist Reprod Genet. 2016;33:1115–1119. doi: 10.1007/s10815-016-0734-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simpson JL, Rechitsky S. Preimplantation diagnosis and other modern methods for prenatal diagnosis. J Steroid Biochem Mol Biol. 2017;165:124–130. doi: 10.1016/j.jsbmb.2016.03.022. [DOI] [PubMed] [Google Scholar]
- 32.Huang J, et al. Validation of a next-generation sequencing-based protocol for 24-chromosome aneuploidy screening of blastocysts. Fertil Steril. 2016;105:1532–1536. doi: 10.1016/j.fertnstert.2016.01.040. [DOI] [PubMed] [Google Scholar]
- 33.Li N, et al. The performance of whole genome amplification methods and next-generation sequencing for pre-implantation genetic diagnosis of chromosomal abnormalities. J Genet Genomics. 2015;42:151–159. doi: 10.1016/j.jgg.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 34.Yan L, et al. Live births after simultaneous avoidance of monogenic diseases and chromosome abnormality by next-generation sequencing with linkage analyses. Proc Natl Acad Sci USA. 2015;112:15964–15969. doi: 10.1073/pnas.1523297113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scriven PN, Handyside AH, Ogilvie CM. Chromosome translocations: Segregation modes and strategies for preimplantation genetic diagnosis. Prenat Diagn. 1998;18:1437–1449. [PubMed] [Google Scholar]
- 36.Neri QV, Lee B, Rosenwaks Z, Machaca K, Palermo GD. Understanding fertilization through intracytoplasmic sperm injection (ICSI) Cell Calcium. 2014;55:24–37. doi: 10.1016/j.ceca.2013.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu J, et al. Noninvasive chromosome screening of human embryos by genome sequencing of embryo culture medium for in vitro fertilization. Proc Natl Acad Sci USA. 2016;113:11907–11912. doi: 10.1073/pnas.1613294113. [DOI] [PMC free article] [PubMed] [Google Scholar]