Abstract
Importance
Low-density lipoprotein cholesterol (LDL-C) is causally related to coronary artery disease (CAD), but the relevance of HDL-C and triglycerides is uncertain. LDL-C lowering by statins modestly increases diabetes risk, but it is unknown if this effect is specific to statins.
Objective
To investigate the relationships of three routinely measured lipid fractions with CAD and diabetes through Mendelian randomization (MR), using conventional MR and making use of newer approaches such as multivariate MR and MR-Egger that addresses pleiotropy of genetic instruments.
Design
We used published data from genome wide association studies to construct genetic instruments and applied them to investigate associations between lipid fractions and risk of CAD and diabetes using MR approaches that took into account pleiotropy of genetic instruments.
Main outcomes and measures
coronary artery disease and diabetes
Results
We constructed genetic instruments composed of 130 SNPs for LDL-C (explaining 7.9% of its variance), 140 SNPs for HDL-C (6.6% of variance) and 140 SNPs for triglycerides (5.9% of variance). A 1-SD (genetically instrumented) elevation in LDL-C (equivalent to 38 mg/dL) and triglycerides (equivalent to 89 mg/dL) were associated with higher CAD risk: odds ratios (OR) and 95% confidence intervals (CI) were 1.68 (1.51-1.87) for LDL-C and 1.28 (1.13-1.45) for triglycerides. The corresponding OR for HDL-C (equivalent to a 16mg/dL increase) was 0.95 (0.85-1.06). All three lipid traits were associated with a lower risk of diabetes. The OR and 95% CI limits were 0.79 (0.71-0.88) for LDL-C and 0.83 (0.76-0.90) for HDL-C.
Conclusions and Relevance
Routinely measured lipid fractions exhibit contrasting relationships with risk of CAD and diabetes. Increased LDL-C, HDL-C and TG are associated with reduced risk of diabetes. This information will be relevant to design of clinical trials of lipid modifying agents, which should carefully monitor for dysglycemia and incidence of diabetes.
Keywords: lipids, diabetes, coronary artery disease, epidemiology, Mendelian randomization
Introduction
Understanding the interplay between circulating lipids and risk of type 2 diabetes (T2D) and coronary artery disease (CAD) is of emerging public health importance and has implications for drug development for cardiovascular disease prevention.1,2 For example, a causal influence of low-density lipoprotein cholesterol (LDL-C) on CAD is widely accepted 3–5 and the proposed causal role of triglycerides (TG) in CAD is gaining acceptance.6,7 In contrast, the role of high-density lipoprotein cholesterol (HDL-C) in CAD remains in doubt.7–9
However, evidence has emerged that LDL-C reduction with statin therapy results in a modest increase in risk of T2D10,11 (outweighed by the benefit of statins in protecting from CAD).12 Whether this diabetogenic effect is a general consequence of LDL-C lowering or if it is specific to inhibition of HMG-CoA reductase remains unclear.13 Moreover, the role of TG and HDL-C in the aetiology of T2D remains unclear.14
Residual confounding and reverse causality can limit causal inference from observational studies. Where a genetic instrument can be used as an instrument for an exposure, Mendelian randomization (MR) generates unbiased, unconfounded effect estimates that are sometimes interpreted as evidence of a causal role. This is because genotype is not modifiable by disease, and the random allocation of alleles at gametogenesis helps avoid bias from reverse causality and confounding, respectively.
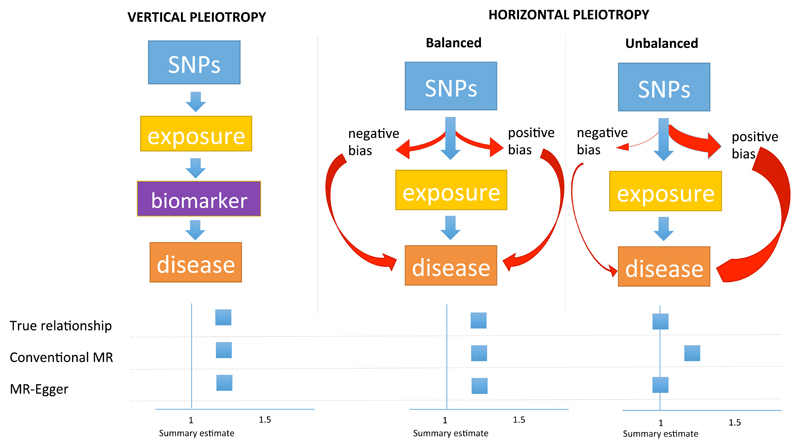
A critical assumption of the MR paradigm is that the genetic instrument influences disease risk exclusively through the exposure of interest. However, genetic variant(s) used to proxy the exposure of interest can also associate with other traits, a phenomenon termed ‘pleiotropy’. When pleiotropy arises as a downstream consequence of genetic perturbation of the biomarker of interest, it is referred to as vertical pleiotropy and the MR assumption is preserved.15 However, when pleiotropy arises because of the association of genetic variant(s) with additional phenotypes in alternative disease pathways (termed horizontal pleiotropy), the assumption is compromised. When MR analysis is based on multiple SNPs drawn from different regions of the genome selected systematically for their association with the biomarker of interest, additional non-systematic effects on any other biomarkers might be ‘balanced’ and the MR effect estimate could still be valid. However, if horizontal pleiotropy is unbalanced, as might occur when the set of biomarkers concerned come from closely connected pathways, MR estimates may become systematically biased (termed ‘unbalanced’ or ‘directional’ pleiotropy16), resulting in invalid effect estimates (see Figure 1 for more details).
Figure 1. Pleiotropy and the validity of estimates derived from Mendelian randomization.
SNPs are used in a genetic instrument for an exposure to assess the association with risk of disease. For each exposure there is a ‘true relationship’, which we try to approximate from Mendelian randomization. For the purposes of simplicity, conventional MR is compared to MR-Egger.
Vertical pleiotropy explains where the genetic instrument associates with biomarkers (other than the exposure) that are on the causal pathway from exposure through to disease. Horizontal pleiotropy is where the genetic instrument associates with additional traits not on the causal pathway of the exposure of interest. When horizontal pleiotropy is balanced, there should be no bias in the effect derived from MR. In this scenario, the estimate obtained from conventional MR is similar to that from MR-Egger.
When horizontal pleiotropy is unbalanced (also termed ‘directional pleiotropy’), the pleiotropy systematically biases the estimate (which can be exaggerated or diminished) in a naïve analysis using conventional MR. In the example in Figure 1, the unbalanced pleiotropy exaggerates the magnitude of the association. Conventional MR will derive a biased estimate, whereas MR-Egger, correcting for unbalanced pleiotropy, should yield a valid estimate. An example of unbalanced horizontal pleiotropy is the relationship of HDL-C and risk of CAD; the association derived from conventional MR is different to that of MR-Egger with the latter indicating that, once unbalanced pleiotropy is accounted for, there is no effect of HDL-C on risk of CAD (see Figure 3).
Recent methodological advances in MR analysis, including ‘multivariate’ MR17 and ‘MR-Egger’,16 provide new approaches for dealing with pleiotropic genetic instruments. In multivariate MR, adjustment is made for genetic associations with measured traits, but may not fully account for unbalanced pleiotropy18. In contrast, MR-Egger can detect and correct for unbalanced pleiotropy of the genetic instrument, even when unbalanced pleiotropy is mediated through unmeasured or unknown traits.
We used summary data from multiple major cardiometabolic genome wide association studies (GWAS) to investigate the underlying relationships between lipids, T2D and CAD using three MR approaches: (i) conventional MR that does not account for pleiotropy, (ii) multivariate MR, that adjusts for traits that may mediate unbalanced pleiotropy, and (iii) MR-Egger, that more fully accounts for unbalanced pleiotropy.
Methods
Data sources
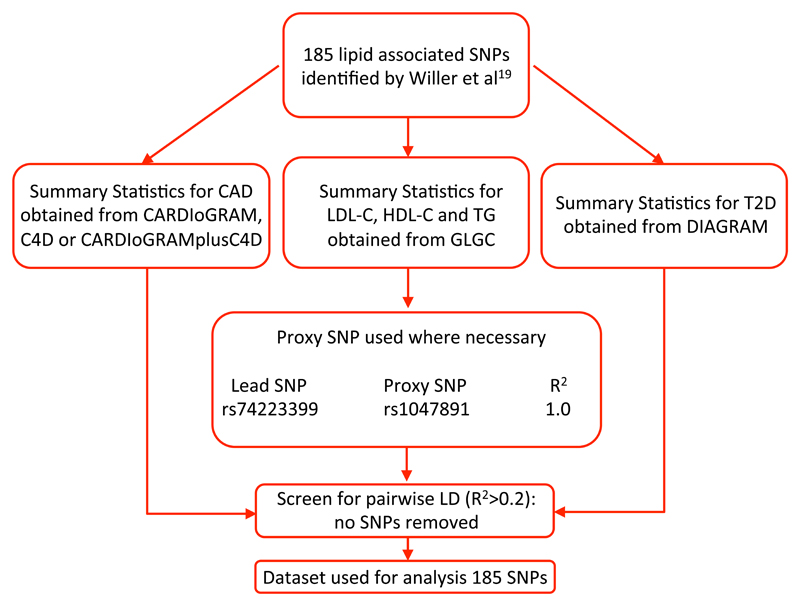
We used summary-level data for lipids from the Global Lipids Genetics Consortium (GLGC),19 T2D data from the DIAbetes Genetics Replication And Meta-analysis (DIAGRAM),20 and CAD data from the Coronary ARtery DIsease Genome-wide Replication And Meta Analysis (CARDIoGRAM) plus The Coronary Artery Disease (C4D) Genetics, collectively known as CARDIoGRAMplusC4D consortium.21 Details of the consortia and webpages for data download are provided in Table 1. All datasets were limited to individuals of European ancestry. Beta coefficients and standard errors were obtained for the per allele association of each SNP with all exposures and outcomes from these data sources. Where SNPs were not present in a dataset we used proxies (R2>0.9) as indicated in Figure 2.
Table 1. Details of the consortia.
| Consortium name | Trait/Disease | Numbers | Data Source; file |
|---|---|---|---|
| GLGC 19 | LDL-C, HDL-C, TG | 188,577 | http://www.sph.umich.edu/csg/abecasis/public/lipids2013 |
| CARDIoGRAMplusC4D 21 | CAD | 63,746 CAD cases, 130,681 controls | http://www.cardiogramplusc4d.org |
| DIAGRAM 20 | T2D | 34,840 T2D cases and 114,981 controls | http://diagram-consortium.org (v3 dataset) |
Abbreviations: CAD: coronary artery disease; CARIoGRAMplusC4D: Coronary ARtery DIsease Genome-wide Replication And Meta Analysis (CARDIoGRAM) plus The Coronary Artery Disease (C4D) Genetics; DIAGRAM: DIAbetes Genetics Replication And Meta-analysis; GLGC: Global Lipids Genetic Consortium; T2D: type 2 diabetes
Figure 2. Pipeline for derivation of the dataset used for Mendelian randomization analyses of lipid subtypes with risk of coronary artery disease and diabetes.
Selection of SNPs
We used 185 lipid-associated SNPs identified by Willer et al19 to generate a series of genetic instruments for each of the exposures: LDL-C, HDL-C and TG. This was conducted by first restricting to a set of SNPs in low linkage disequilibrium (pairwise R2<0.2). We then organized these SNPs by descending order of proportional variance (R2, estimated from the summary statistics using the gtx() package in R) between SNP with the corresponding lipid exposure to generate a range of instruments from 5 to 150 SNPs. The process used to determine the final tally of SNPs for inclusion in a genetic instrument for each lipid trait is described below.
Handling of SNPs
We matched SNPs across the data sources by aligning them to the same effect allele. Effect allele frequencies were checked for concordance.
Mendelian randomization analyses
We used three MR approaches.
First, we used conventional 2-sample instrumental variable (IV) analyses, which does not make any allowance for pleiotropy. Basing our approach on the method first proposed by Johnson,22 we incorporated the bootstrap suggested by Bowden,16 as a way to incorporate the error in the published estimates of SNP effect on both exposure and outcome.
Second, we conducted multivariate MR analyses, which statistically adjusts for pleiotropy with additional phenotypes measured in the dataset.23 Multivariate MR is an extension of the conventional weighted regression in which the betas for additional phenotypes are included as covariates. In this case we used all three lipid traits in the model (e.g. for the HDL-C instrument we included, thereby adjusting for, LDL-C and TG).
Third, we used MR-Egger,16 which accounts for unbalanced pleiotropy of a genetic instrument. MR-Egger is a linear regression of estimated SNP effects (for the exposure-raising allele) on exposure against the corresponding estimates of SNP on outcome, weighted by the inverse variance of the SNP on outcome effect estimates. This differs from conventional 2-sample MR in that the regression line is not forced through the origin. Bowden et al16 show that the MR-Egger estimate is unaffected by net pleiotropic effects of the instrument and, indeed, the presence of unbalanced pleiotropy can be inferred if the intercept term is not zero.
For all three approaches (conventional MR, multivariate MR and MR-Egger), we conducted 10,000 bootstraps, and our effect estimate is the mean of the bootstraps with the confidence interval (CI) determined empirically and set to Bonferroni adjusted (for 6 tests) 95% (i.e. 99.2%).
Quantifying the proportion of variance explained by the genetic instruments
i). R-Trend
The proportion of variance (R2) of the trait explained by the genetic instrument will rise with the addition of more SNPs. However, the improvement beyond the optimum number of SNPs in the instrument will come increasingly as a result of model over-specification. We examined the ratio of R2 for the current instrument to R2 for an instrument comprising 30 more SNPs (we term this the ‘R-Trend’). The trend in the ratio gives an indication of the transition from useful additional information to over-specification since it becomes asymptotic when each new SNP adds the same amount of information than the last. We judged the beginning of the asymptotic phase of the line to mark the largest useful instrument obtainable from the available data. The 30 SNP window was chosen empirically because it emphasises trend; a smaller window gave a more erratic line, obscuring the trend. Calculation and use of the R-trend effectively limited the analysis to instruments comprising 155 or fewer SNPs, further restricted to 150 for presentational purposes.
ii). Gain from adding a SNP to the instrument
We estimated the benefit to R2 from adding the current SNP to the previous instrument by bootstrapping the summary statistics and calculating R2 for the instruments with n and (n-1) SNPs. Over 10,000 bootstraps we noted the number of occasions when the current instrument gave higher R2 than the previous instrument. This value was summarised as a percentage; the point at which the current instrument was no better than the previous instrument being when 50% of the runs showed a benefit.
Selection of optimal number of SNPs in genetic instrument
The optimum number of SNPs was chosen by consideration of the R-ratio and the gain from adding the current SNP when presented graphically (eFigures 1-6 in the Supplement). The optimum instrument was identified when both estimates of R2 gain were asymptotic. As we discuss later, the exact point (± 20 SNPs) makes little difference to the conclusions. Two authors (JW and MVH) considered this independently and reached a consensus as to the number of SNPs to include for each genetic instrument for each lipid trait.
Selection of MR model to derive estimates of the underlying relationship
Once we determined the optimal number of SNPs to incorporate in each instrument, we used the following decision-tree to select the MR approach to derive the estimate:
-
(i)
if there was no evidence of unbalanced pleiotropy using the intercept derived from MR-Egger, we selected the conventional MR instrumental variable estimate as the most reliable indicator to the underlying relationship (as it retains maximal power and makes fewest assumptions)
-
(ii)
if there was evidence of unbalanced pleiotropy, we used the estimate from MR-Egger
-
(iii)
in cases where there was discordance between conventional MR and MR-Egger, we used multivariate MR to inform whether differences could arise from pathways shared between the three lipids traits.
The inSIDE assumption
Because the underlying models assume a linear dose-response, instrumental variable (IV) effect estimates must be independent of the exposure effect in MR analysis (the so-called ‘inSIDE rule’).16 We tested the null hypothesis that the instrumental variable effect (derived from the ratio of outcome to exposure) estimates for the SNPs in an instrument were independent of the exposure (lipid) effect estimates for the same SNPs for both CAD and T2D. In all scenarios, the ‘inSIDE’ assumption was satisfied (eTable 1 in the Supplement).
Power
We followed the method of Brion et al24 implemented at http://cnsgenomics.com/shiny/mRnd/. Using the average number of individuals and estimated R2 for the instrument together with the reported proportion of cases, we adjusted the estimate of the true effect of exposure on outcome to obtain the value for which we had 80% power at a Bonferroni adjusted alpha of 0.05/6.
Ethical Review of Study and Informed Consent of Study Participants
As this report used published GWAS data available in the public domain, specific ethical review and/or consent from study participants was not sought (and had been obtained in the original studies).
Results
The pooled dataset included up to 188,577 individuals with measures of blood lipids, 63,199 CAD cases and 34,840 T2D cases. The optimal number of SNPs for each lipid traits was 130 for LDL-C (explaining 7.9% of its variance), 140 for HDL-C (6.6% of HDL-C variance) and 140 for TG (5.9% of TG variance) (eFigures 1-6 in the Supplement).
LDL-C
The genetic instrument for LDL-C showed unbalanced pleiotropy for CAD and T2D. For CAD, the estimate derived from MR-Egger was OR 1.68 (95%CI: 1.51, 1.87) per 1-SD (equivalent to 38 mg/dL) genetically-instrumented higher LDL-C. This was of greater magnitude, but directionally consistent with conventional and multivariate MR estimates (Figure 3 and eFigure 1 in the Supplement).
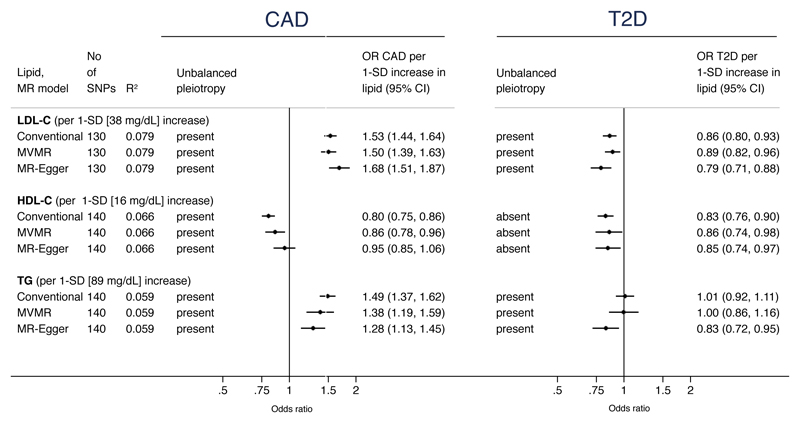
Figure 3. Associations of routinely measured lipids with risk of coronary artery disease (CAD) and type 2 diabetes (T2D) from Mendelian randomization analyses.
See Methods for description of the three Mendelian randomization (MR) models. Estimates for conventional MR are derived from two-sample MR that forces the slope through the origin, thereby not accounting for pleiotropy. Multivariate MR (MVMR) statistically adjusts for other lipid traits, and MR-Egger adjusts for unbalanced pleiotropy. R2 refers to proportion of variance of lipid trait explained by the genetic instrument. 95% confidence intervals (CI) are Bonferroni-adjusted. To convert HDL-C and LDL-C to mmol/L, multiply by 0.0259; to convert triglycerides to mmol/L, multiply by 0.0113
For T2D, the OR was 0.79 (95%CI: 0.71, 0.88) per 1-SD higher LDL-C from MR-Egger, which, was again of greater magnitude yet directionally consistent with conventional and multivariate MR estimates (Figure 3 and eFigure 2 in the Supplement).
HDL-C
A 1-SD genetically instrumented elevation in HDL-C (equivalent to 16 mg/dL) did not provide conclusive evidence of a relationship between HDL-C and risk of CAD. There was evidence of unbalanced pleiotropy of the HDL-C genetic instrument and the estimate for CAD from MR-Egger was OR 0.95 (95%CI: 0.85, 1.06). There was a step-wise weakening of the effect towards the null from conventional MR (OR 0.80; 95%CI: 0.75, 0.86), through adjusting for LDL-C and TG in multivariate MR (OR 0.86; 95%CI: 0.78, 0.96) to the MR-Egger estimate (Figure 3 and eFigure 3 in the Supplement).
For T2D, there was no evidence of unbalanced pleiotropy of the genetic instrument comprising 140 SNPs. The conventional MR provided an estimate of OR 0.83 (95%CI: 0.76, 0.90), consistent with estimates from both multivariate MR and MR-Egger (Figure 3 and eFigure 4 in the Supplement).
Triglycerides
The TG genetic instrument showed unbalanced pleiotropy for both CAD and T2D. A 1-SD genetically instrumented increase in TG (equivalent to 89 mg/dL) yielded an OR for CAD from MR-Egger of 1.28 (95%CI: 1.13, 1.45), weaker than the multivariate MR estimate and roughly half the magnitude of the conventional MR estimate (OR 1.49) (Figure 3 and eFigure 5 in the Supplement).
TG was associated with reduced risk of T2D (OR 0.83; 95%CI: 0.72, 0.95 from MR-Egger). This was dissimilar to both conventional and multivariate MR estimates (Figure 3). The scatter plot identified that the intercept of the MR-Egger slope was positive (eFigure 6 in the Supplement).
Power
There was adequate powered to detect the reported estimates (eTable 2 in the Supplement), making it unlikely that associations arose from the play of chance.
Putting the pieces together: framework of relationships
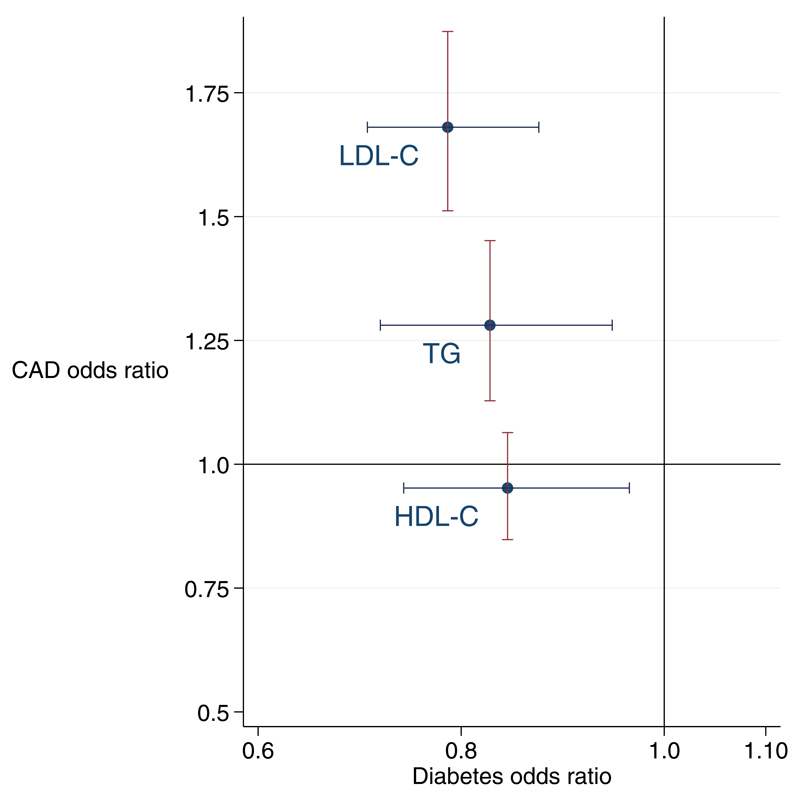
We demonstrate that elevations in LDL-C, TG and HDL-C are associated with reduced risk of T2D, with the magnitude (per 1-SD increase) being greatest for LDL-C, then TG followed by HDL-C (although the 95%CI for the effect on T2D for the three lipids overlap) (Figure 4). In contrast, only LDL-C and TG were associated with increased risk of CAD (with the magnitude again stronger for LDL-C than TG).
Figure 4. Cross-hair plot of a one standard deviation increase in lipids and risk of CAD and T2D.
All estimates derived from MR-Egger. Error bars represent 95% confidence intervals (CI) that are Bonferroni-adjusted.
Discussion
We exploited data from multiple GWAS to conduct MR analyses exploring the relationships between lipids and risk of T2D and CAD. Our findings reveal a series of relationships that will help inform on potential downstream consequences of pharmacological modification of lipid levels.
While all three lipids were associated with reduced risk of T2D, it does not necessarily follow that lowering of LDL-C or TG through inhibition of specific druggable proteins (such as PCSK9) will alter risk of T2D. Large-scale genetic and clinical investigations are needed to clarify the effects of pharmacological lowering of LDL-C and TG to gauge dysglycaemic associations.25,26
Our findings are complimentary to a study by Fall et al13, that, to address pleiotropy, excluded SNPs showing strong associations with T2D, glycaemia-related traits or potential confounders such as adiposity. This manual pruning weakened the associations, yielding inconsistent conclusions. In our study, we applied novel approaches for: (i) SNP selection (to optimize the SNPs in each genetic instrument); (ii) MR (using MR-Egger, obviating the need to manually prune SNPs); that collectively allows us to make more robust conclusions about the role of lipids in T2D.
The protective effect of TG and risk of T2D that we report is novel, yet potentially counter-intuitive. Observational studies report that increases in TG are associated with an increase in risk of T2D27, however insulin resistance results in perturbations in TG metabolism, 28 meaning that the direction of the casual relationship is not clear. While our data (suggesting TG may be protective of T2D) should be interpreted with caution, our findings are consistent with recent genetic studies in both Europeans and African-Americans. 29,30 Further investigations are needed to identify which TG pathways, if any, may lead to a reduction in risk of T2D.
LDL-C and TG showed robust effects on risk of CAD, however the evidence for HDL-C was far less convincing, with the estimate from MR-Egger failing to identify an effect. This is in keeping with prior MRs7,8, including the paper by Voight et al8 that manually pruned pleiotropic SNPs. However, selecting only non-pleiotropic SNPs could introduce selection bias in the genetic instrument by focusing on a subset of SNPs that is not representative of any meaningful proxy of HDL-C, with the removal of potentially informative HDL-C related pathways. Our data show that adjusting for TG and LDL-C in multivariate MR does not fully account for the unbalanced pleiotropy of the HDL-C genetic instrument. MR-Egger identifies that the likely underlying relationship is that a genetically-determined higher HDL-C does not result in a reduced risk of CHD. While these findings are consistent with recent trials of therapeutics targeting HDL-C,9,31 this does not preclude the possibility that a drug modifying HDL-C (or HDL particles) could reduce risk of CAD or other outcomes such as stroke.
The association of TG with CAD recapitulates findings from several prior MR and genetic studies.6,7 Of note, the MR-Egger estimate for TG was less than half the magnitude for an equivalent increase in LDL-C (ORs 1.28 and 1.68 for TG and LDL-C, respectively, per 1-SD increment). Specific triglyceride-lowering approaches have had, at best, modest efficacy whereas statin trials have had consistently and potently positive results.32,33 Our data suggest that pharmacological lowering of TG should translate into CAD benefit.
This study has several advantages. First, we use the most up-to-date data available for lipids to generate the most comprehensive genetic instruments available. Second, MR-Egger enabled inclusion of GWAS-identified lipid-related SNPs in the genetic instruments, irrespective of presence of unbalanced pleiotropy. Third, using summary-level data from different sources represents an efficient study design to facilitate original investigations such as these without the cost or need for de novo pheno-/genotyping.
Some limitations are also worthy of note. First, estimates could be sensitive to SNPs included in the genetic instruments. However, MR estimates were stable at the point at which we selected the genetic instrument. Second, our MR analyses pertain to biomarkers rather than specific drug targets. Third, patients targeted for lipid modification may be at risk for other diseases such as heart failure or atrial fibrillation – the relevance and direction of effects on these and other endpoints could be important but were not evaluated here. Fourth, we are not able to account for statin treatment in the analyses; given that we detect the protective effect of LDL-C on risk of T2D (the scenario that statins are most likely to confound), major bias is unlikely to arise in this setting. Finally, while our data casts yet further doubt on the relevance of HDL-C in the aetiology of CAD, it remains possible that HDL lipoproteins and/or lipid compositions could play a role in the aetiology of CAD. New methods, such as 1H-NMR metabolomics,34 that quantify lipoprotein subclasses and lipid compositions are likely to facilitate future MR studies of HDL subclasses.
In conclusion, our comprehensive MR investigations identify distinct relationships of major lipid subfractions and risk of CAD and T2D. LDL-C and TG increase risk of CAD. In contrast, LDL-C and HDL-C are very likely to be protective of T2D with new evidence suggesting that TG may also play a protective role. While further studies are needed to examine if specific pathways or lipid subtypes are implicated, our findings inform on potential expected downstream consequences of intervening on lipid traits and provide cautionary evidence that therapeutics that lower LDL-C and TG may have dysglycaemic effects.
Supplementary Material
Key points.
Question: Do routinely measured lipids affect risk of cardiometabolic disease?
Findings: In this Mendelian randomization analysis, a lifelong higher low density lipoprotein-cholesterol (LDL-C) or triglycerides was found to be associated with higher risk of coronary artery disease. In contrast, higher levels of all three lipid traits (LDL-C, high density lipoprotein-cholesterol [HDL-C] and triglycerides) associated with a reduced risk of diabetes.
Meaning: Lower LDL-C and TG levels may increase risk of diabetes; clinical trials of lipid-modifying agents should carefully monitor for the incidence of diabetes
Acknowledgements
Funding/Support
ZFH is supported by a Genomic Medicine and Statistics Wellcome Trust DPhil Studentship. FWA is supported by a Dekker scholarship-Junior Staff Member 2014T001 – Netherlands Heart Foundation and UCL Hospitals NIHR Biomedical Research Centre. SEH is a BHF Professor and is funded by PG08/008, and by the National Institute for Health Research University College London Hospitals Biomedical Research Centre. DIS is supported by a NIHR Academic Clinical Fellowship. ADH is supported by the UCL Hospitals NIHR Biomedical Research Centre and work in his laboratory is supported by BHF Programme and Special Project Grants.
Role of the funding source
The funding sources had no role in: design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Footnotes
Conflicts of Interest
CTSU (University of Oxford) is the central co-ordinating centre for the REVEAL trial of anacetrapib; REVEAL is funded through a grant to the University of Oxford by Merck Sharp & Dohme Corp but was designed and is being conducted independently of the funder. DIS is a consultant to Pfizer on work unrelated to the present analysis. All other co-authors report no conflicts of interest. NS reports having received honoraria for advisory boards or lectures for Amgen, Sanofi, Boehringer Ingelheim, Novo Nordisk, Merck, Janssen and Astrazeneca.
Access to data and Data Analysis
JW had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Author contributions
All authors satisfy ICMJE requirements for author contribution.
Originality of Content
All information and materials in the manuscript are original.
References
- 1.Stein EA, Mellis S, Yancopoulos GD, et al. Effect of a monoclonal antibody to PCSK9 on LDL cholesterol. The New England journal of medicine. 2012;366:1108–18. doi: 10.1056/NEJMoa1105803. [DOI] [PubMed] [Google Scholar]
- 2.Akdim F, Stroes ES, Sijbrands EJ, et al. Efficacy and safety of mipomersen, an antisense inhibitor of apolipoprotein B, in hypercholesterolemic subjects receiving stable statin therapy. Journal of the American College of Cardiology. 2010;55:1611–8. doi: 10.1016/j.jacc.2009.11.069. [DOI] [PubMed] [Google Scholar]
- 3.Cannon CP, Blazing MA, Giugliano RP, et al. Ezetimibe Added to Statin Therapy after Acute Coronary Syndromes. The New England journal of medicine. 2015 doi: 10.1056/NEJMoa1410489. [DOI] [PubMed] [Google Scholar]
- 4.Cholesterol Treatment Trialists C. Baigent C, Blackwell L, et al. Efficacy and safety of more intensive lowering of LDL cholesterol: a meta-analysis of data from 170,000 participants in 26 randomised trials. Lancet. 2010;376:1670–81. doi: 10.1016/S0140-6736(10)61350-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cholesterol Treatment Trialists C. Mihaylova B, Emberson J, et al. The effects of lowering LDL cholesterol with statin therapy in people at low risk of vascular disease: meta-analysis of individual data from 27 randomised trials. Lancet. 2012;380:581–90. doi: 10.1016/S0140-6736(12)60367-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Do R, Willer CJ, Schmidt EM, et al. Common variants associated with plasma triglycerides and risk for coronary artery disease. Nature genetics. 2013;45:1345–52. doi: 10.1038/ng.2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holmes MV, Asselbergs FW, Palmer TM, et al. Mendelian randomization of blood lipids for coronary heart disease. European heart journal. 2014 doi: 10.1093/eurheartj/eht571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Voight BF, Peloso GM, Orho-Melander M, et al. Plasma HDL cholesterol and risk of myocardial infarction: a mendelian randomisation study. Lancet. 2012;380:572–80. doi: 10.1016/S0140-6736(12)60312-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwartz GG, Olsson AG, Abt M, et al. Effects of dalcetrapib in patients with a recent acute coronary syndrome. The New England journal of medicine. 2012;367:2089–99. doi: 10.1056/NEJMoa1206797. [DOI] [PubMed] [Google Scholar]
- 10.Preiss D, Seshasai SR, Welsh P, et al. Risk of incident diabetes with intensive-dose compared with moderate-dose statin therapy: a meta-analysis. Jama. 2011;305:2556–64. doi: 10.1001/jama.2011.860. [DOI] [PubMed] [Google Scholar]
- 11.Swerdlow DI, Preiss D, Kuchenbaecker KB, et al. HMG-coenzyme A reductase inhibition, type 2 diabetes, and bodyweight: evidence from genetic analysis and randomised trials. Lancet. 2014 doi: 10.1016/S0140-6736(14)61183-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ridker PM, Pradhan A, MacFadyen JG, Libby P, Glynn RJ. Cardiovascular benefits and diabetes risks of statin therapy in primary prevention: an analysis from the JUPITER trial. Lancet. 2012;380:565–71. doi: 10.1016/S0140-6736(12)61190-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fall T, Xie W, Poon W, et al. Using Genetic Variants to Assess the Relationship Between Circulating Lipids and Type 2 Diabetes. Diabetes. 2015;64:2676–84. doi: 10.2337/db14-1710. [DOI] [PubMed] [Google Scholar]
- 14.Haase CL, Tybjaerg-Hansen A, Nordestgaard BG, Frikke-Schmidt R. High-density lipoprotein cholesterol and risk of type 2 diabetes: a Mendelian randomization study. Diabetes. 2015 doi: 10.2337/db14-1603. [DOI] [PubMed] [Google Scholar]
- 15.Lawlor DA, Harbord RM, Sterne JA, Timpson N, Davey Smith G. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat Med. 2008;27:1133–63. doi: 10.1002/sim.3034. [DOI] [PubMed] [Google Scholar]
- 16.Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44:512–25. doi: 10.1093/ije/dyv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burgess S, Dudbridge F, Thompson SG. Re: "Multivariable Mendelian randomization: the use of pleiotropic genetic variants to estimate causal effects". Am J Epidemiol. 2015;181:290–1. doi: 10.1093/aje/kwv017. [DOI] [PubMed] [Google Scholar]
- 18.Day FR, Loh PR, Scott RA, Ong KK, Perry JR. A Robust Example of Collider Bias in a Genetic Association Study. American journal of human genetics. 2016;98:392–3. doi: 10.1016/j.ajhg.2015.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Global Lipids Genetics C. Willer CJ, Schmidt EM, et al. Discovery and refinement of loci associated with lipid levels. Nature genetics. 2013;45:1274–83. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morris AP, Voight BF, Teslovich TM, et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nature genetics. 2012;44:981–90. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Consortium CAD. Deloukas P, Kanoni S, et al. Large-scale association analysis identifies new risk loci for coronary artery disease. Nature genetics. 2013;45:25–33. doi: 10.1038/ng.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson T. Efficient Calculation for Multi-SNP Genetic Risk Scores. presented at the American Society of Human Genetics Annual Meeting; San Francisco. November 6–10, 2012; 2012. http://cran.r-project.org/web/packages/gtx/vignettes/ashg2012.pdf. [Google Scholar]
- 23.Burgess S, Thompson SG. Multivariable Mendelian randomization: the use of pleiotropic genetic variants to estimate causal effects. Am J Epidemiol. 2015;181:251–60. doi: 10.1093/aje/kwu283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brion MJ, Shakhbazov K, Visscher PM. Calculating statistical power in Mendelian randomization studies. Int J Epidemiol. 2013;42:1497–501. doi: 10.1093/ije/dyt179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bonnefond A, Yengo L, Le May C, et al. The loss-of-function PCSK9 p.R46L genetic variant does not alter glucose homeostasis. Diabetologia. 2015 doi: 10.1007/s00125-015-3659-8. [DOI] [PubMed] [Google Scholar]
- 26.Tragante V, Asselbergs FW, Swerdlow DI, et al. Harnessing publicly available genetic data to prioritize lipid modifying therapeutic targets for prevention of coronary heart disease based on dysglycemic risk. Hum Genet. 2016 doi: 10.1007/s00439-016-1647-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tirosh A, Shai I, Bitzur R, et al. Changes in triglyceride levels over time and risk of type 2 diabetes in young men. Diabetes care. 2008;31:2032–7. doi: 10.2337/dc08-0825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ginsberg HN, Zhang YL, Hernandez-Ono A. Regulation of plasma triglycerides in insulin resistance and diabetes. Archives of medical research. 2005;36:232–40. doi: 10.1016/j.arcmed.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 29.Klimentidis YC, Wineinger NE, Vazquez AI, Campos G. Multiple Metabolic Genetic Risk Scores and Type 2 Diabetes Risk in Three Racial/Ethnic Groups. The Journal of Clinical Endocrinology & Metabolism. 2014;99:E1814–E8. doi: 10.1210/jc.2014-1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klimentidis YC, Chougule A, Arora A, Frazier-Wood AC, H C-H. Triglyceride-Increasing Alleles Associated with Protection against Type-2 Diabetes. PLoS genetics. 2015:e1005204. doi: 10.1371/journal.pgen.1005204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lilly to Discontinue Development of Evacetrapib for High-Risk Atherosclerotic Cardiovascular Disease. 2015 https://investor.lilly.com/releasedetail.cfm?ReleaseID=936130.
- 32.Preiss D, Sattar N. Lipids, lipid modifying agents and cardiovascular risk: a review of the evidence. Clin Endocrinol (Oxf) 2009;70:815–28. doi: 10.1111/j.1365-2265.2008.03490.x. [DOI] [PubMed] [Google Scholar]
- 33.Lee M, Saver JL, Towfighi A, Chow J, Ovbiagele B. Efficacy of fibrates for cardiovascular risk reduction in persons with atherogenic dyslipidemia: A meta-analysis. Atherosclerosis. 217:492–8. doi: 10.1016/j.atherosclerosis.2011.04.020. [DOI] [PubMed] [Google Scholar]
- 34.Soininen P, Kangas AJ, Wurtz P, Suna T, Ala-Korpela M. Quantitative serum nuclear magnetic resonance metabolomics in cardiovascular epidemiology and genetics. Circ Cardiovasc Genet. 2015;8:192–206. doi: 10.1161/CIRCGENETICS.114.000216. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.