Abstract
IMPORTANCE
Cerebrospinal fluid (CSF) and positron emission tomographic (PET) amyloid biomarkers have been proposed for the detection of Alzheimer disease (AD) pathology in living patients and for the tracking of longitudinal changes, but the relation between biomarkers needs further study.
OBJECTIVE
To determine the association between CSF and PET amyloid biomarkers (cross-sectional and longitudinal measures) and compare the cutoffs for these measures.
DESIGN, SETTING, AND PARTICIPANTS
Longitudinal clinical cohort study from 2005 to 2014 including 820 participants with at least 1 florbetapir F-18 (hereafter referred to as simply florbetapir)–PET scan and at least 1 CSF β-amyloid 1–42 (Aβ1–42) sample obtained within 30 days of each other (501 participants had a second PET scan after 2 years, including 150 participants with CSF Aβ1–42 measurements). Data were obtained from the Alzheimer’s Disease Neuroimaging Initiative database.
MAIN OUTCOMES AND MEASURES
Four different PET scans processing pipelines from 2 different laboratories were compared. The PET cutoff values were established using a mixture-modeling approach, and different mathematical models were applied to define the association between CSF and PET amyloid measures.
RESULTS
The values of the CSF Aβ1–42 samples and florbetapir-PET scans showed a nonlinear association (R2 = 0.48–0.66), with the strongest association for values in the middle range. The presence of a larger dynamic range of florbetapir-PET scan values in the higher range compared with the CSF Aβ1–42 plateau explained the differences in correlation with cognition (R2 = 0.36 and R2 = 0.25, respectively). The APOE genotype significantly modified the association between both biomarkers. The PET cutoff values derived from an unsupervised classifier converged with previous PET cutoff values and the established CSF Aβ1–42 cutoff levels. There was no association between longitudinal Aβ1–42 levels and standardized uptake value ratios during follow-up.
CONCLUSIONS AND RELEVANCE
The association between both biomarkers is limited to a middle range of values, is modified by the APOE genotype, and is absent for longitudinal changes; 4 different approaches in 2 different platforms converge on similar pathological Aβ cutoff levels; and different pipelines to process PET scans showed correlated but not identical results. Our findings suggest that both biomarkers measure different aspects of AD Aβ pathology.
Alzheimer disease (AD) pathology is defined by the deposition of extracellular β-amyloid (Aβ) plaques and intracellular tau neurofibrillary tangles in the brain.1 These deposits correlate with Aβ positron emission tomographic (PET) radiotracer retention2–4 and cerebrospinal fluid (CSF) Aβ levels.5–7 As expected, the CSF Aβ1–42 levels and the standardized uptake value ratios (SUVRs) of the different PET Aβ ligands are associated8–19 and show similar classification accuracy and diagnostic agreement. Conversely, plasma Aβ levels show a weak association with these biomarkers8,18 and cannot predict the clinical diagnosis.20 Whereas recent larger studies have noted a nonlinear association between CSF and PET measures of Aβ pathology, which was less obvious in smaller cohorts,15,16 most studies have centered on diagnostic utility or have assumed a linear association and applied parametric models without a value transformation. The goal of our study was to (1) assess the presence of nonlinear associations between CSF Aβ1–42 samples and florbetapir F-18 (hereafter referred to as simply florbetapir)–PET scans processed using different pipelines, (2) compare amyloid cutoffs across platforms, and (3) study the association between longitudinal measures of both amyloid biomarkers in a large longitudinal cohort study.
Methods
Participants
A total of 820 Alzheimer’s Disease Neuroimaging Initiative (ADNI) participants with CSF Aβ1–42 and florbetapir-PET Aβ imaging measurement values obtained with in 30 days were included in our study (Table 1). Florbetapir was not available at the baseline ADNI 1 visit, and therefore some of these participants had their first florbetapir-PET scan performed during subsequent visits. The number of visits in which PET scans were performed were 739 at baseline, 23 at 24 months, 3 at 36 months, 25 at 48 months, 25 at 60 months, and 5 at 72 months. For the CSF Aβ1–42 mixture model analysis, 1005 participants with a CSF Aβ1–42 measurement were included (ie, all participants with at least 1 CSF Aβ1–42 measurement to estimate the CSF Aβ1–42 cutoff level). Data were downloaded on September 12, 2014. A total of 501 participants had a second PET amyloid scan performed within 2 years, and a total of 150 participants also had CSF samples obtained within 30 days of the second PET scan. The CSF Aβ1–42 data used in the preparation of this article were obtained from UPENNBIOMK and UPENNBIOMK5–7 data generated by the ADNI Biomarker Core.
Table 1.
Characteristics of the ADNI Participants Included in the Study at the Time of the Scan
| Characteristic | Cognitively Normal Participants (n = 259) |
Participants With MCI (n = 415) |
Participants With AD (n = 146) |
P Value |
|---|---|---|---|---|
| Age at time of scan, mean (SD), y | 72.8 (5.9) | 71.3 (7.4) | 73.5 (8.5) | .002 |
| Male sex, % | 45.2 | 56.0 | 58.3 | .009 |
| ADAS-cog score, mean (SD) | 9.0 (4.4) | 15.0 (6.8) | 30.9 (8.9) | <.001 |
| SUVR, median (Q1-Q3) | ||||
| Average CB | 1.17 (1.12–1.32) | 1.27 (1.13–1.53) | 1.54 (1.37–1.68) | <.001 |
| Average WM | 0.66 (0.63–0.72) | 0.74 (0.66–0.84) | 0.87 (0.83–0.90) | <.001 |
| Summary CB | 1.06 (1.00–1.17) | 1.18 (1.02–1.39) | 1.42 (1.27–1.53) | <.001 |
| Summary composite | 0.73 (0.70–0.82) | 0.83 (0.72–0.99) | 1.03 (0.94–1.09) | <.001 |
| Aβ1–42 level, median (Q1-Q3), pg/mL | 209.3 (159.2–237.6) | 160.9 (131.9–214.4) | 131.8 (114.4–150.7) | <.001 |
Abbreviations: Aβ, β-amyloid; AD, Alzheimer disease; ADAS-cog, Alzheimer Disease Assessment Scale–cognitive subscale; ADNI, Alzheimer’s Disease Neuroimaging Initiative; CB, cerebellum; MCI, mild cognitive impairment; Q1, first quarter; Q3, third quarter; SUVR, standardized uptake value ratio; WM, white matter.
The ADNI (http://www.adni-info.org) was launched in 2004 by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, the US Food and Drug Administration, private pharmaceutical companies, and nonprofit organizations, and has been extensively reviewed elsewhere21 (eAppendix in the Supplement). A diagnosis of mild cognitive impairment or AD was established based on the criteria by Petersen et al22,23 for mild cognitive impairment and the National Institute of Neurological and Communicative Disorders and Stroke–Alzheimer’s Disease and Related Disorders Association criteria24 for probable AD. Protocols were submitted to institutional review boards for each participating location and their written unconditional approval obtained and submitted to Regulatory Affairs at the ADNI Coordinating Center (ADNI-CC) prior to commencement of the study. Written informed consent for the study was obtained from all participants and/or authorized representatives.
CSF Collection and Aβ1–42 Measurement
The CSF samples were obtained in the morning after an overnight fast and processed as previously described25,26 (eAppendix in the Supplement). The Aβ1–42 level was measured using the multiplex xMAP Luminex platform (Luminex Corp) with Innogenetics (INNO-BIA AlzBio3, for research use–only reagents) immunoassay kit–based reagents. The capture and detection antibodies for Aβ1–42 were 4D7A3 and 3D6, respectively.26 All longitudinal CSF samples belonging to the same participant were measured in the same plate to avoid assay-to-assay variation.
Florbetapir-PET Scan Processing
Florbetapir image data were acquired from a variety of PET scanners at ADNI sites nationwide. Image data were acquired in four 5-minute frames 50 to 70 minutes after injection of approximately 10 mCi, and the 4 frames were coregistered to each other, averaged, interpolated to a uniform image (160 × 106 × 96) and voxel size (1.5 mm3), and smoothed to a uniform resolution (8-mm full width at half-maximum) to account for differences between scanners.27
We included florbetapir SUVRs developed in 2 different laboratories (the University of Utah in Salt Lake City and the University of California, Berkeley), each including 2 different measures obtained using a different reference (eAppendix in the Supplement). Both laboratories used the same scans that were preprocessed as already detailed. From the University of Utah analysis, we included averaged regional values from medial and lateral frontal, temporal, and parietal cortices that were normalized either using the cerebellar region (the average cerebellum) or the white matter (the average white matter) as reference region. Two summary measures were obtained at the University of California, Berkeley using Aβ deposition in frontal, cingulate, lateral parietal, and temporal cortices and either the whole cerebellum as reference (the summary cerebellum) or the whole cerebellum, brainstem/pons, and eroded subcortical white matter (the summary composite) as reference region. There were 450 and 501 participants who had 2 PET scans obtained within 2 years at the University of Utah and University of California, Berkeley laboratories, respectively.
Statistical Analysis
For univariate group comparisons, analysis of variance and χ2 tests were applied for quantitative and qualitative variables. Power transformations were applied to normalize distributions in the analyses performed for the demographic variables included in Table 1. We used 5 different models to test which one better explained the association between CSF Aβ1–42 levels and PET SUVRs: lineal, polynomial, exponential, hyperbolic, and multivariate adaptive regression splines (MARSs). An MARS creates piecewise regression models (hinges) for each variable in the model, and these models are separated by knots to capture changes in the association according to different ranges of the measures, using a data-driven approach. To test the models, the sample was divided into a training set and a test set, which included two-thirds and one-third of the participants, respectively. Each participant was only included once in this analysis. The different statistical models were developed in the training set using a 10-fold cross-validation and afterward applied to the test set. The coefficient of determination (R2) is reported to summarize the goodness of fit of each model. Cutoffs for amyloid biomarkers were obtained using a previously reported strategy that uses finite mixture models (eTable 1 and eFigure 1 in the Supplement).28,29
Results
Cross-sectional Association Between Individual CSF and Florbetapir-PET Aβ Measures
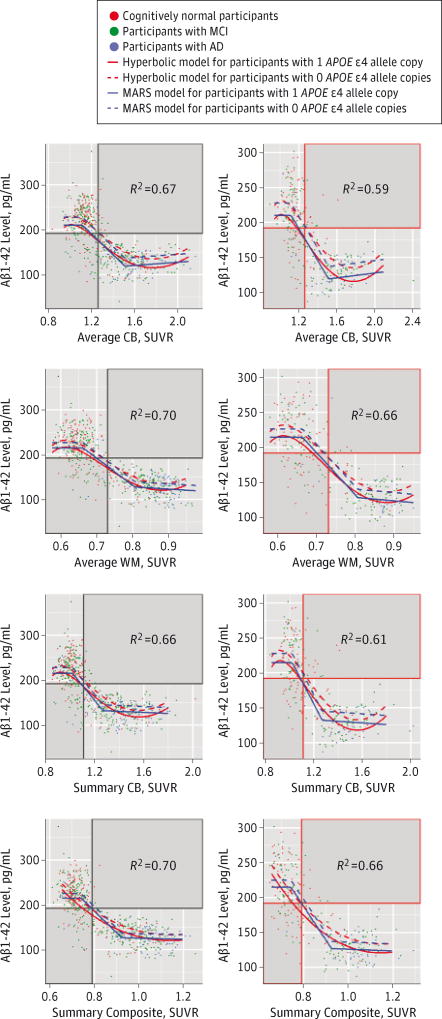
The first and second columns in Figure 1 show the CSF and PET Aβ levels for the participants included in the training and test sets, respectively, and the fitted models (the solid gray areas show disagreement in participant classification between both biomarkers). Coefficients of determination (R2) for the different models are summarized in eTable 2 in the Supplement. In all comparisons, the linear model showed the worst performance in the training and test sets, whereas the hyperbolic and MARS models showed overall the best performance.
Figure 1. Association Between CSF Aβ1–42 Levels and Florbetapir F-18 PET SUVRs.
A model for participants with 2 APOE ε4 copies is not included. The solid gray areas indicate disagreement in the classification based on the pair of Aβ measures. Aβ indicates β-amyloid; AD, Alzheimer disease; CB, cerebellum; CSF, cerebrospinal fluid; MARS, multivariate adaptive regression spline; MCI, mild cognitive impairment; PET, positron emission tomographic; SUVR, standardized uptake value ratio; and WM, white matter.
APOE genotype influenced the relationship; an increasing number of ε4 copies were associated with lower CSF Aβ1–42 levels for the same PET SUVR in all models. In all MARS models, the first hinge was located in a narrow range of Aβ1–42 levels (225–288 pg/mL for participants with 0 copies of the APOE ε4 allele and 208–214 pg/mL for participants with 1 copy of the APOE ε4 allele), and the second hinge showed a slightly higher variability (137–144 pg/mL for participants with 0 copies of the APOE ε4 allele and 119–132 pg/mL for participants with 1 copy of the APOE ε4 allele). The PET SUVRs could not accurately predict CSF Aβ1–42 levels before the first hinge (R2 = 0.01–0.10) and after the second hinge (R2 = 0.11–0.26). We tested whether clinical diagnosis was a significant predictor, but it was not selected in any of the MARS models. Similar results that included 2 hinges in the MARS were obtained when CSF Aβ1–42 level was selected as the predictor and the florbetapir measures were selected as outcomes (data not shown).
eTable 3 in the Supplement shows PET SUVRs that corresponded to the CSF Aβ1–42 cutoff level of 192 pg/mL for participants with 0 copies or 1 copy of the APOE ε4 allele. Table 2 summarizes the κ coefficients and overall percentage agreement for each pair of biomarkers. There was a substantial agreement between the CSF Aβ1–42–defined groups and the groups that were defined based on the different florbetapir-derived measures (κ = 0.69–0.76), but it was lower than the excellent agreement observed for the different florbetapir-PET measures (κ = 0.80–0.91). Most of the participants who were classified differently by CSF and PET Aβ measures presented with abnormal CSF Aβ1–42 levels and normal PET SUVRs (8.9%-12.5%) compared with participants with normal CSF Aβ1–42 levels and abnormal PET SUVRs (0.7%-4.5%). We compared clinical characteristics in the groups with mismatched biomarker results (eTables 4 and 5 in the Supplement). Although there were a larger number of participants who were cognitively impaired in the group that had only abnormal CSF Aβ1–42 levels compared with the group that had only abnormal summary cerebellum values, the differences were not significant (P = .50). Whereas there were no differences in the Alzheimer’s Disease Assessment Scale– cognitive subscale (ADAS-cog) scores between groups at the 12-month follow-up, the participants who had only abnormal CSF Aβ1–42 levels showed memory decline, and the participants who had only abnormal summary cerebellum values showed executive decline.
Table 2.
Matrix Showing Agreement Between the Different Aβ Measuresa
| CSF Aβ1–42 Level | Average CB | Average WM | Summary CB | Summary Composite |
|
|---|---|---|---|---|---|
| CSF Aβ1–42 level | 84.5 | 86.7 | 86.6 | 88.2 | |
|
| |||||
| Average CB | 0.69 | 90.2 | 92.4 | 90.8 | |
| (0.64–0.74) | |||||
|
| |||||
| Average WM | 0.74 | 0.80 | 89.9 | 95.3 | |
| (0.69–078) | (0.76–0.84) | ||||
|
| |||||
| Summary CB | 0.73 | 0.85 | 0.80 | 92.9 | |
| (0.68–0.77) | (0.81–0.89) | (0.74–0.84) | |||
|
| |||||
| Summary Composite | 0.76 | 0.82 | 0.91 | 0.86 | |
| (0.72–0.81) | (0.78–0.86) | (0.88–0.94) | (0.82–0.89) | ||
Abbreviations: Aβ, β-amyloid; CB, cerebellum; CSF, cerebrospinal fluid; WM, white matter.
Values below the diagonal space show the κ coefficient, and values above the diagonal space show percentage agreement.
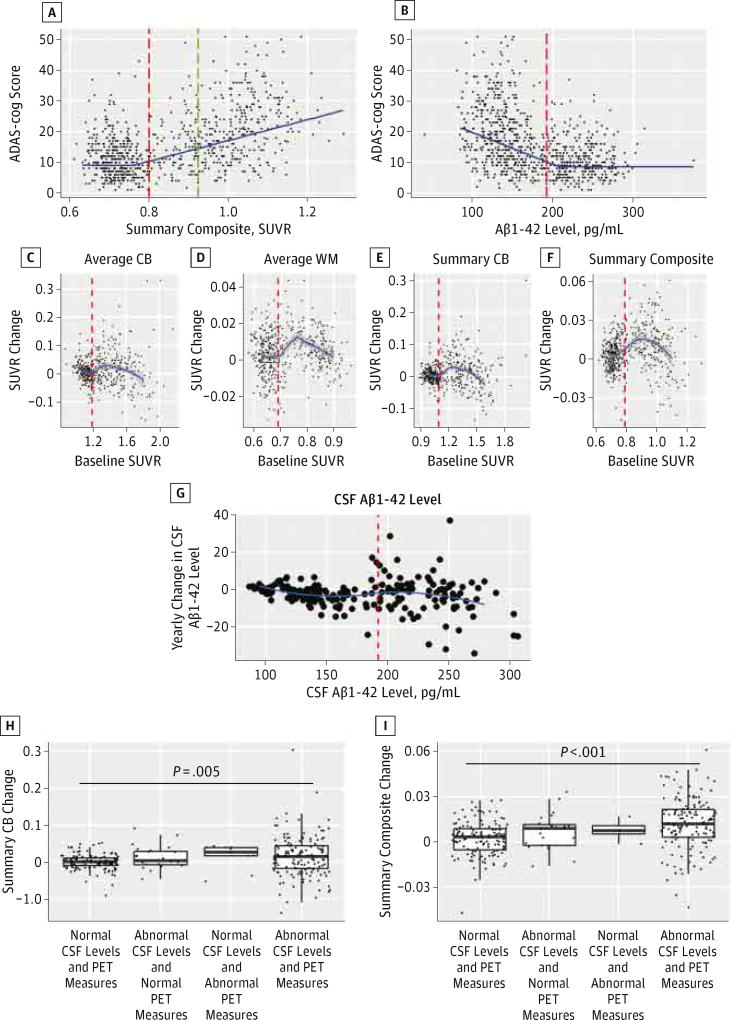
The different PET SUVRs obtained with the different references and pipelines were highly correlated (eFigure 2 in the Supplement), with correlation coefficients between 0.81 and 0.95, although the values were not comparable and needed a transformation between pipelines (eTable 6 in the Supplement). When we tested the ability of florbetapir-PET measures and CSF Aβ1–42 levels to predict the ADAS-cog score, the summary composite measure (R2 = 0.36) outperformed the Aβ1–42 level (R2 = 0.25) in a cross-validated MARS model that included age as a covariate (Figure 2A and B).The MARS model fits calculated for each of the clinical diagnostic groups are summarized in eTable 7 in the Supplement. Results were similar for other PET Aβ measures.
Figure 2. Clinical Associations and Longitudinal Changes of PET SUVRs.
Scatterplots showing the association between Alzheimer’s Disease Assessment Scale–cognitive subscale (ADAS-cog) scores on the y-axis and the summary composite standardized uptake value ratios (SUVRs) (A) and the cerebrospinal fluid (CSF) β-amyloid 1–42 (Aβ1–42) levels (B) on the x-axis. The blue continuous line represents the multivariate adaptive regression spline for a 65-year-old participant. The red dashed line represents the cutoff for the biomarker represented in the plot, and the green dashed line represents the value at which the CSF Aβ1–42 level plateaus. Longitudinal SUVR yearly changes (after a 2-year follow-up) for the average cerebellum (CB) (C), the average white matter (WM) (D), the summary CB (E), and the summary composite (F) are shown on the y-axes, with the x-axes representing baseline SURVs. G, Longitudinal changes in CSF Aβ1–42 level after a 2-year follow-up are shown. The red dashed line represents the value that corresponds to the CSF Aβ1–42 level of 192 pg/mL. Yearly changes in the summary CB (H) and the summary composite (I) values during follow-up (y-axis) are based on the presence of normal or abnormal baseline CSF Aβ1–42 levels and florbetapir F-18–positron emission tomographic (PET) measures. The horizontal line in each box indicates the median, while the top and bottom borders of the box mark the 75th and 25th percentiles, respectively. The whiskers above and below the box mark the 90th and 10th percentiles, respectively. The points beyond the whiskers are outliers beyond the 90th and 10th percentiles.
Longitudinal CSF and PET Aβ Measurements
Figure 2C–F shows baseline SUVRs for each of the PET measurements (x-axis) and the corresponding yearly change (y-axis) for participants with 2 PET measurements, and Figure 2G shows the changes in the CSF Aβ1–42 level for the same period. For 304 participants who had 2 PET scans and CSF samples obtained during the baseline visit, only the group with abnormal Aβ1–42 levels and abnormal PET SUVR summary measures showed a greater increase during follow-up (Figure 2H and I).
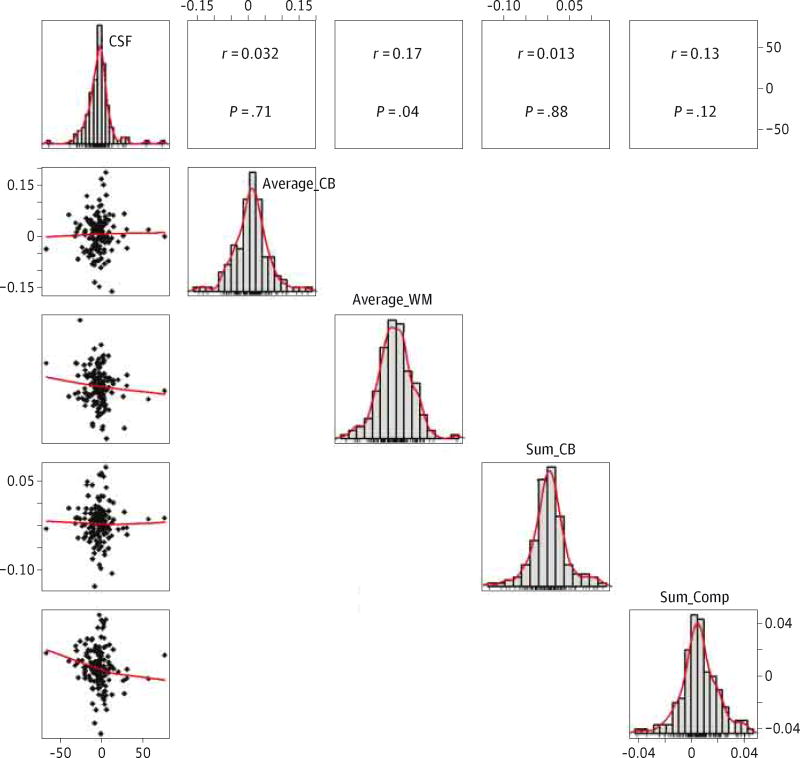
A total of 150 participants (53 cognitively normal participants, 90 participants with mild cognitive impairment, and 7 participants with AD) had 2 CSF and PET Aβ measurements obtained during the same visits, with the second set of measurements occurring within 2 years (ie, mean [SD], 729.7 [20.8] days) of the first. Figure 3 displays scatter plots with the yearly value changes during follow-up for the CSF Aβ1–42 and florbetapir-PET measurements below the diagonal and their correlation above the diagonal (eFigure 3 in the Supplement also shows associations between the PET SUVRs). There was no correlation between CSF and PET amyloid value changes, while the different PET Aβ amyloid measurements correlated with a higher degree. The correlation between CSF Aβ1–42 level and florbetapir-PET measure did not improve when only participants with Aβ1–42 levels between both MARS hinges (140–215 pg/mL) were included (data not shown).
Figure 3. Association Between CSF Aβ1–42 Level and Florbetapir F-18–PET Measure Longitudinal Changes.
Matrix showing the individual scatterplots depicting the association between cerebrospinal fluid (CSF) β-amyloid 1–42 (Aβ1–42) level (x-axis) and florbetapir F-18–positron emission tomographic (PET) standardized uptake value ratio (y-axis) changes during a 2-year follow-up (below the diagonal) and the corresponding Pearson correlation coefficient and P value (above the diagonal). The panels in the diagonal direction depict histograms showing the distribution of CSF Aβ1–42 levels and PET SUVRs. Average_CB indicates average cerebellum; Average_WM, average white matter; Sum_CB, summary cerebellum; and Sum_Comp, summary composite.
Discussion
Cross-sectional CSF Aβ1–42 levels and florbetapir-PET measures were associated for a limited middle range of values that included the cutoffs, and they were consistent with AD. The association was significantly modified by the number of APOE ε4 alleles. Nevertheless, there was a large agreement for the classification of participants as having an AD-like Aβ burden between the different measures. Different approaches converged on a similar cutoff for pathological Aβ deposition across platforms. However, there was no correlation between longitudinal changes observed after 2 years of follow-up.
Previous studies16,19 have mainly analyzed the agreement between CSF and PET Aβ measures in the same cohort using a single florbetapir-PET measure or using Pearson correlation or linear regressions assuming a linear association.18 Good agreement between CSF Aβ1–42 level and florbetapir-PET SUVR has been previously reported using a single pipeline for the latter.16,19 In the present study, we found an excellent correlation-classification agreement using 4 separate SUVRs obtained in 2 different laboratories using 2 distinct pipelines. Including different processing pipelines used in the 2 laboratories allowed us to analyze how the use of different pipelines and references can affect comparisons across studies. We showed that cross-sectional SUVRs were highly correlated and that different processing pipelines and choices of references led to a disagreement of 5% to 10%, and κ coefficients between 0.80 and 0.91 in a large sample of participants processed in 4 different ways, which could be a potential important source of variability between studies. Thus, each pipeline needs to establish its own cutoffs. Recently, a new method has been proposed to compare values across different PET ligands and processing pipelines.30 Nevertheless, CSF Aβ1–42 levels and florbetapir-PET measures showed much higher agreement and much higher κ coefficients than the ones observed when the different neuronal injury biomarkers were studied in the same cohort.31
The validity of the cutoffs has been previously demonstrated in a 3-fold manner: (1) the CSF Aβ1–42 level cutoff was initially demonstrated using autopsy-validated diagnoses25 to prevent biases due to clinical diagnostic uncertainties,5 (2) this cutoff was then validated using a “diagnosis-free”–driven mixture model analysis of CSF Aβ1–42 levels,28 and (3) for florbetapir-PET SUVRs, investigators used young controls32 and autopsy cases.2 In the mixture model analysis of summary CB values that we performed, 1.12 was designated as the cutoff that corresponds to an SUVR of 1.11 using the semiautomated quantification applied by Avid and, therefore, overlaps with their validated cutoff32 (eFigure 4 in the Supplement). Furthermore, the 1.12 summary CB cutoff value is close to the average of the transformation of the CSF Aβ1–42 autopsy-validated cutoff level for participants with 0 or 1 APOE ε4 allele. In addition, using a mixture-modeling approach in a sample of 1005 levels of CSF Aβ1–42, we reached the same level as the one previously described in our autopsy study.25
Therefore, we confirmed the previous florbetapir-PET cutoff established based on young controls using an unsupervised classification method, in a sample that included a large number of cognitively normal participants, participants with mild cognitive impairment, and participants with AD, and the CSF Aβ1–42 autopsy-validated cutoff level in a larger sample using the approach applied by De Meyer et al28 in a larger sample. Most importantly, we demonstrated that the conversion of the values across different platforms and methods converges robustly on the similar burden of Aβ pathology. However, we emphasize that recommended SUVR cutoffs vary according to the pipeline that was used, and therefore any modification in the pipeline must be followed by a validation of new cutoffs. Previous studies16,19 have described groups of participants that show disagreement in classification between CSF Aβ1–42 levels and florbetapir-PET measures. The size of these groups varies depending on the reference region, and the disagreement decreases when white matter regions are used as a reference. This might be explained by the fact that the cerebellum is affected in latter stages of AD,33 and therefore reference regions might be affected differently in later stages of disease.
Recently, a lower cutoff level for CSF Aβ1–42 (ie, 157 pg/mL) and an average cerebellum with a cutoff SUVR of 1.26 that is skewed toward more abnormal values of pathological Aβ biomarkers were suggested.19 These values were obtained using clinical diagnosis as the gold standard and contradict evidence from previous autopsy-based studies using unsupervised diagnosis-independent methods.25,28,32 This can be explained by our current understanding of the pathological Aβ biomarker model for AD,34 which describes biomarker and neuropathological changes that precede cognitive changes and that are being used for the staging of preclinical AD in cognitively normal participants,35 and by autopsy studies36–38 showing that 44.2% of cognitively normal participants have Consortium to Establish a Registry for Alzheimer’s Disease B and C scores and that 22.5% of cognitively normal participants have an intermediate probability of AD neuropathological changes. Hence, these and other studies2 emphasize that optimizing cutoffs based on clinical diagnosis to classify all participants with normal cognition as healthy controls will contradict the neuropathological findings for many participants and prevent an accurate preclinical diagnosis of the underlying Aβ pathology. This is of critical importance for the design and conduct of clinical trials of new therapies targeting pathological Aβ biomarkers in participants with underlying AD pathology who are cognitively normal.39–41
One previous study9 pursued efforts to transform CSF Aβ1–42 levels to Pittsburg Compound B–PET SUVRs, and vice versa, and used a log2 transformation for both values owing to the lack of a linear association. However, the goal of our study was to transform the values between the different methods and to understand how both are related (in order to interpret differences in the timing of the biomarker changes for both biomarkers across the whole clinical spectrum) and the implications there of. Based on the MARS models, it can be concluded that there is only a strong association between CSF and florbetapir-PET Aβ values for the midrange values of both measures, which include the currently applied CSF Aβ1–42 level measured using the multiplex xMAP Luminex platform25 and the florbetapir-PET Aβ amyloid measure normalized to cerebellum2,32 cutoff values. It is also in this range where most of the discrepant classification appears. This could be due, in part, to the variability inherent to any clinical measure that can have an important effect for cases with values close to a dichotomic cutoff.
Another explanation for this disagreement might be the lower affinity of PET amyloid ligands for diffuse plaques2,42 and the differential effect of the APOE genotype on biomarker values. Different amyloid PET ligands share their binding site and show a higher affinity for neuritic amyloid plaques compared with diffuse amyloid plaques, which can lead to false negatives.3,42–45 While it is thought that the decrease in CSF Aβ1–42 levels, but not in other Aβ levels,18 reflects brain Aβ deposition, more mature forms might not be in equilibrium with CSF and, therefore, might lead to the plateau observed in the CSF Aβ1–42 level, or later stages might represent Aβ levels that are not in equilibrium with the CSF. Therefore, the wider range of CSF Aβ1–42 levels in the lower range of florbetapir-PET SUVRs might imply a stronger association of the CSF Aβ1–42 level with diffuse amyloid plaques, which appear in earlier phases without the presence of neuritic plaques.46
Another explanation is that different sensitivities or ceiling effects of the assays could account for the strong association between CSF Aβ1–42 levels and florbetapir-PET Aβ measures only for the midrange values of these 2 most widely used measures of pathological Aβ deposition. However, the CSF Aβ1–42 level plateau is well above the lower detection limit of the Luminex assay. Furthermore, APOE ε4 is associated with a higher proportion of fibrillar amyloid and neurotic plaques,47,48 which show a higher affinity for amyloid PET ligands and, therefore, would explain the higher SUVRs, and this would explain why the presence of APOE ε4 had different effects on both the CSF Aβ1–42 levels and the florbetapir-PET measures across all pipelines. For the same CSF Aβ1–42 levels, participants with no ε4 alleles had lower florbetapir-PET SUVRs. We performed several analyses to assess the clinical correlations of participants who showed either abnormal CSF Aβ1–42 levels or abnormal florbetapir-PET measures and found differences in the cognitive changes, but a longer follow-up (including autopsies) will be needed to characterize these small groups of participants.
Surprisingly, there was no correlation between the changes in CSF Aβ1–42 levels and the changes in florbetapir-PET measures after a 2-year follow-up. There are several non– mutually exclusive explanations for this finding: (1) the changes are small and might not be detected owing to the inherent variability of the measurements, (2) a longer follow-up is needed to see larger changes, and (3) the different dynamic ranges of these 2 biomarkers could lead to different rates of changes in them across the biomarker spectrum. This might be due to the fact that these 2 biomarkers reflect different aspects of disease mechanisms, leading to Aβ fibrillation and deposition, as well as different floor and ceiling effects as already noted. However, another factor that might explain these differences is the sensitivity of the measures of CSF Aβ1–42 level and florbetapir-PET SUVR to track small changes during a 2-year follow-up. In any case, it is not surprising that the methods used to measure CSF Aβ1–42 level and brain Aβ amyloid deposits do, in fact, measure different aspects of pathological Aβ amyloid as previously discussed.
Florbetapir-PET SUVRs showed a stronger association with ADAS-cog scores, which can be explained by the absence of the floor effect observed for CSF Aβ1–42 levels, and thus can offer a larger dynamic range along disease progression. Nevertheless, the association with cognition is lower than the one observed for neuronal injury neuroimaging biomarkers.49
Conclusions
Thus, in conclusion, although CSF Aβ1–42 levels and florbetapir-PET Aβ measures show a high-classification agreement for dementia due to underlying AD pathology, these are clearly different measures of pathological Aβ amyloidosis that converge to similar diagnostic cutoffs across different cohorts, methods, and amyloid biomarkers, but they do not closely correlate in the cross-sectional low and high range of values. Notably, this extends to a lack of correlation for the longitudinal changes in these 2 biomarkers during a 2-year follow-up. Hence, our novel findings are significant for understanding how to interpret CSF Aβ1–42 levels and florbetapir-PET Aβ measures for diagnosis and for understanding the mechanisms of Aβ amyloidosis.
Supplementary Material
Acknowledgments
Dr Trojanowski may accrue revenue as coinventor in the future on patents submitted by the University of Pennsylvania, and he received revenue from the sale of Avid to Eli Lily as coinventor of patented imaging-related technology submitted by the University of Pennsylvania. Dr Weiner reports stock/stock options from Elan and Synarc; travel support from Novartis, Tohoku University, Fundacio Ace, eDreams, MCI Group, Neuroscience School of Advanced Studies, Danone Trading, ANT CONGRES, NeuroVigil, Centre Hospitalier Régional Universitaire–Hôpital Roger Salengro, Siemens, AstraZeneca, Geneva University Hospitals, Lilly, University of California (UC), San Diego–ADNI, Paris University, Institut Catala de Neurociencies Aplicades, University of New Mexico School of Medicine, Ipsen, Clinical Trials on Alzheimer’s Disease, Pfizer, International Conference on Alzheimer’s and Parkinson’s Diseases, and Paul Sabatier University; board membership to Lilly, Araclon, Institut Catala de Neurociencies Aplicades, Gulf War Veterans Illnesses Advisory Committee, Vaco, Biogen Idec, and Pfizer; consultancy fees from AstraZeneca, Araclon, Medivation/Pfizer, Ipsen, TauRx Therapeutics, Bayer Healthcare, Biogen Idec, ExonHit Therapeutics, Servier, Synarc, Pfizer, and Janssen; honoraria from NeuroVigil, Insitut Catala de Neurociencies Aplicades, Pharmaceuticals and Medical Devices/Japanese Ministry of Health, Labour, and Welfare, and Tohoku University; commercial research support from Merck and Avid; and government research support from the US Department of Defense and the Department of Veterans Affairs, all outside the submitted work. Dr Jack has provided consulting services for Janssen Research and Development, LLC, and Eli Lily. Dr Landau has consulted for Biogen Idec, Synarc, and Avid. Dr Shaw serves as a consultant for Janssen Alzheimer Immunotherapy Research and Development and Lilly, outside the submitted work. Dr Jagust has served as a consultant for Genentech, Synarc, Siemens, F. Hoffman–La Roche, TauRx Therapeutics, and Janssen Alzheimer Immunotherapy, all outside the submitted work. Dr Foster reports other support from Janssen Alzheimer Immunotherapy, Alzheimer’s Disease Cooperative Study, Baxter Bioscience, Bristol-Myers Squibb, and GE Healthcare; grants from the Department of Veterans Affairs Office of Rural Health, the National Institutes of Health Small Business Technology Transfer program, and the Northern California Institute for Research and Education; and personal fees from Sanofi, Lilly USA, GE Healthcare, and Piramal, all outside the submitted work.
Funding/Support: Dr Toledo is supported by grants P01 AG032953, P01 AG017586, P30 AG010124, and P50 NS053488. Dr Bjerke is supported by the Swedish Brain Foundation and the Sweden-America Foundation. Data collection and sharing for this project was funded by the ADNI (National Institutes of Health grant U01 AG024904) and US Department of Defense ADNI (Department of Defense award W81XWH-12-2-0012). Dr Jack receives research funding from the National Institutes of Health (grants R01-AG011378, U01-HL096917, U01-AG024904, R01 AG041851, R01 AG37551, R01AG043392, and U01-AG06786) and the Alexander Family Alzheimer’s Disease Research Professorship of the Mayo Foundation.
Role of the Funder/Sponsor: The funders had no role in the design and conduct of the study; collection, management, analysis, or interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Group Information
The ADNI investigators were Michael Weiner, MD (UC San Francisco, principal investigator [PI] of ADNI), Paul Aisen, MD (UC San Diego, Alzheimer’s Disease Cooperative Study PI and Director of Coordinating Center Clinical Core), Michael Weiner, MD (UC San Francisco, Executive Committee), Paul Aisen, MD (UC San Diego, Executive Committee), Ronald Petersen, MD, PhD (Mayo Clinic, Rochester, Executive Committee), Clifford R. Jack Jr, MD (Mayo Clinic, Rochester, Executive Committee), William Jagust, MD (UC Berkeley, Executive Committee), John Q. Trojanowki, MD, PhD (University of Pennsylvania, Executive Committee), Arthur W. Toga, PhD (UCLA [University of California, Los Angeles], Executive Committee), Laurel Beckett, PhD (UC Davis, Executive Committee), Robert C. Green, MD, MPH (Brigham and Women’s Hospital/Harvard Medical School, Executive Committee), Andrew J. Saykin, PsyD (Indiana University, Executive Committee), John Morris, MD (Washington University St Louis, Executive Committee), Enchi Liu, PhD (Janssen Alzheimer Immunotherapy, ADNI 2 Private Partner Scientific Board (Chair), Robert C. Green, MD, MPH (Brigham and Women’s Hospital/Harvard Medical School, Data and Publication Committee [Chair]), Tom Montine, MD, PhD (University of Washington, Resource Allocation Review Committee), Ronald Petersen, MD, PhD (Mayo Clinic, Rochester, Clinical Core Leaders [Core PI]), Paul Aisen, MD (UC San Diego, Clinical Core Leaders), Anthony Gamst, PhD (UC San Diego, Clinical Informatics and Operations), Ronald G. Thomas, PhD (UC San Diego, Clinical Informatics and Operations), Michael Donohue, PhD (UC San Diego, Clinical Informatics and Operations), Sarah Walter, MSc (UC San Diego, Clinical Informatics and Operations), Devon Gessert (UC San Diego, Clinical Informatics and Operations), Tamie Sather (UC San Diego, Clinical Informatics and Operations), Laurel Beckett, PhD (UC Davis, Biostatistics Core Leaders and Key Personnel [Core PI]), Danielle Harvey, PhD (UC Davis, Biostatistics Core Leaders and Key Personnel), Anthony Gamst, PhD (UC San Diego, Biostatistics Core Leaders and Key Personnel), Michael Donohue, PhD (UC San Diego, Biostatistics Core Leaders and Key Personnel), John Kornak, PhD (UC Davis, Biostatistics Core Leaders and Key Personnel), Clifford R. Jack Jr, MD (Mayo Clinic, Rochester, Magnetic Resonance Imaging [MRI] Core Leaders and Key Personnel [Core PI]), Anders Dale, PhD (UC San Diego, MRI Core Leaders and Key Personnel), Matthew Bernstein, PhD (Mayo Clinic, Rochester, MRI Core Leaders and Key Personnel), Joel Felmlee, PhD (Mayo Clinic, Rochester, MRI Core Leaders and Key Personnel), Nick Fox, MD (University of London, MRI Core Leaders and Key Personnel), Paul Thompson, PhD (UCLA School of Medicine, MRI Core Leaders and Key Personnel), Norbert Schuff, PhD (UC San Francisco MRI, MRI Core Leaders and Key Personnel), Gene Alexander, PhD (Banner Alzheimer’s Institute, MRI Core Leaders and Key Personnel), Charles DeCarli, MD (UC Davis, MRI Core Leaders and Key Personnel), William Jagust, MD (UC Berkeley, PET Core Leaders and Key Personnel [Core PI]), Dan Bandy, MS, CNMT (Banner Alzheimer’s Institute, PET Core Leaders and Key Personnel), Robert A. Koeppe, PhD (University of Michigan, PET Core Leaders and Key Personnel), Norm Foster, MD (University of Utah, PET Core Leaders and Key Personnel), Eric M. Reiman, MD (Banner Alzheimer’s Institute, PET Core Leaders and Key Personnel), Kewei Chen, PhD (Banner Alzheimer’s Institute, PET Core Leaders and Key Personnel), Chet Mathis, MD (University of Pittsburgh, PET Core Leaders and Key Personnel), John Morris, MD (Washington University St Louis, Neuropathology Core Leaders), Nigel J. Cairns, PhD, MRCPath (Washington University St Louis, Neuropathology Core Leaders), Lisa Taylor-Reinwald, BA, HTL (Washington University St Louis, Neuropathology Core Leaders), John Q. Trojanowki, MD, PhD (University of Pennsylvania School of Medicine, Biomarkers Core Leaders and Key Personnel [Core PI]), Les Shaw, PhD (University of Pennsylvania School of Medicine, Biomarkers Core Leaders and Key Personnel), Virginia M. Y. Lee, PhD, MBA (University of Pennsylvania School of Medicine, Biomarkers Core Leaders and Key Personnel), Magdalena Korecka, PhD (University of Pennsylvania School of Medicine, Biomarkers Core Leaders and Key Personnel), Arthur W. Toga, PhD (UCLA, Informatics Core Leaders and Key Personnel [Core PI]), Karen Crawford (UCLA, Informatics Core Leaders and Key Personnel), Scott Neu, PhD (UCLA, Informatics Core Leaders and Key Personnel), Andrew J. Saykin, PsyD (Indiana University, Genetics Core Leaders and Key Personnel), Tatiana M. Foroud, PhD (Indiana University, Genetics Core Leaders and Key Personnel), Steven Potkin, MD (UC Irvine, Genetics Core Leaders and Key Personnel), Li Shen, PhD (Indiana University, Genetics Core Leaders and Key Personnel), Zaven Kachaturian, PhD (Khachaturian, Radebaugh & Associates, Inc, Early Project Development), Richard Frank, MD, PhD (General Electric, Early Project Development), Peter J. Snyder, PhD (University of Connecticut, Early Project Development), Susan Molchan, PhD (National Institute on Aging/National Institutes of Health, Early Project Development), Jeffrey Kaye, MD (Oregon Health and Science University, Site Investigator), Joseph Quinn, MD (Oregon Health and Science University, Site Investigator), Betty Lind, BS (Oregon Health and Science University, Site Investigator), Sara Dolen, BS (Oregon Health and Science University, Past Site Investigator), Lon S. Schneider, MD (University of Southern California, Site Investigator), Sonia Pawluczyk, MD (University of Southern California, Site Investigator), Bryan M. Spann, DO, PhD (University of Southern California, Site Investigator), James Brewer, MD, PhD (UC San Diego, Site Investigator), Helen Vanderswag, RN (UC San Diego, Site Investigator), Judith L. Heidebrink, MD, MS (University of Michigan, Site Investigator), Joanne L. Lord, LPN, BA, CCRC (University of Michigan, Site Investigator), Ronald Petersen, MD, PhD (Mayo Clinic, Rochester, Site Investigator), Kris Johnson, RN (Mayo Clinic, Rochester, Site Investigator), Rachelle S. Doody, MD, PhD (Baylor College of Medicine, Site Investigator), Javier Villanueva-Meyer, MD (Baylor College of Medicine, Site Investigator), Munir Chowdhury, MBBS, MS (Baylor College of Medicine, Site Investigator), Yaakov Stern, PhD (Columbia University Medical Center, Site Investigator), Lawrence S. Honig, MD, PhD (Columbia University Medical Center, Site Investigator), Karen L. Bell, MD, (Columbia University Medical Center, Site Investigator), John C. Morris, MD (Washington University, St Louis, Site Investigator), Beau Ances, MD (Washington University, St Louis, Site Investigator), Maria Carroll, RN, MSN (Washington University, St Louis, Site Investigator), Sue Leon, RN, MSN (Washington University, St Louis, Site Investigator), Mark A. Mintun, MD (Washington University, St Louis, Past Site Investigator), Stacy Schneider, APRN, BC, GNP (Washington University, St Louis, Past Site Investigator), Daniel Marson, JD, PhD (University of Alabama–Birmingham, Site Investigator), Randall Griffith, PhD, ABPP (University of Alabama–Birmingham, Site Investigator), David Clark, MD (University of Alabama–Birmingham, Site Investigator), Hillel Grossman, MD (Mount Sinai School of Medicine, Site Investigator), Effie Mitsis, PhD (Mount Sinai School of Medicine, Site Investigator), Aliza Romirowsky, BA (Mount Sinai School of Medicine, Site Investigator), Leyla deToledo-Morrell, PhD (Rush University Medical Center, Site Investigator), Raj C. Shah, MD (Rush University Medical Center, Site Investigator) (Wein Center, Site Investigator), Ranjan Duara, MD (Wein Center, Site Investigator), Daniel Varon, MD (Wein Center, Site Investigator), Peggy Roberts, CNA (Wein Center, Site Investigator), Marilyn Albert, PhD (Johns Hopkins University, Site Investigator), Chiadi Onyike, MD, MHS (Johns Hopkins University, Site Investigator), Stephanie Kielb, MD (Johns Hopkins University, Site Investigator), Henry Rusinek, PhD (New York University, Site Investigator), Mony J de Leon, EdD (New York University, Site Investigator), Lidia Glodzik, MD, PhD (New York University, Site Investigator), Susan De Santi, PhD (New York University, Past Site Investigator), P. Murali Doraiswamy, MD (Duke University Medical Center, Site Investigator), Jeffrey R. Petrella, MD (Duke University Medical Center, Site Investigator), R. Edward Coleman, MD (Duke University Medical Center, Site Investigator), Steven E. Arnold, MD (University of Pennsylvania, Site Investigator), Jason H. Karlawish, MD (University of Pennsylvania, Site Investigator), David Wolk, MD (University of Pennsylvania, Site Investigator), Charles D. Smith, MD (University of Kentucky, Site Investigator), Greg Jicha, MD (University of Kentucky, Site Investigator) , Peter Hardy, PhD (University of Kentucky, Site Investigator), Oscar L. Lopez, MD (University of Pittsburgh, Site Investigator), MaryAnn Oakley, MA (University of Pittsburgh, Site Investigator), Donna M. Simpson, CRNP, MPH (University of Pittsburgh, Site Investigator), Anton P. Porsteinsson, MD (University of Rochester Medical Center, Site Investigator), Bonnie S. Goldstein, MS, NP (University of Rochester Medical Center, Site Investigator), Kim Martin, RN (University of Rochester Medical Center, Site Investigator), Kelly M. Makino, BS (University of Rochester Medical Center, Past Site Investigator), M. Saleem Ismail, MD (University of Rochester Medical Center, Past Site Investigator), Connie Brand, RN (University of Rochester Medical Center, Past Site Investigator), Ruth A. Mulnard, DNSc, RN, FAAN (UC Irvine, Site Investigator), Gaby Thai, MD (UC Irvine, Site Investigator), Catherine McAdams-Ortiz, MSN, RN, A/GNP (UC Irvine, Site Investigator), Kyle Womack, MD (University of Texas Southwestern Medical School, Site Investigator), Dana Mathews, MD, PhD (University of Texas Southwestern Medical School, Site Investigator), Mary Quiceno, MD (University of Texas Southwestern Medical School, Site Investigator), Ramon Diaz-Arrastia, MD, PhD (University of Texas Southwestern Medical School, Past Site Investigator), Richard King, MD (University of Texas Southwestern Medical School, Past Site Investigator), Myron Weiner, MD (University of Texas Southwestern Medical School, Past Site Investigator), Kristen Martin-Cook, MA (University of Texas Southwestern Medical School, Past Site Investigator), Michael DeVous, PhD (University of Texas Southwestern Medical School, Past Site Investigator), Allan I. Levey, MD, PhD (Emory University, Site Investigator), James J. Lah, MD, PhD (Emory University, Site Investigator), Janet S. Cellar, DNP, PMHCNS-BC (Emory University, Site Investigator), Jeffrey M. Burns, MD (University of Kansas, Medical Center, Site Investigator), Heather S. Anderson, MD (University of Kansas, Medical Center, Site Investigator), Russell H. Swerdlow, MD (University of Kansas, Medical Center, Site Investigator), Liana Apostolova, MD (UCLA, Site Investigator), Po H. Lu, PsyD (UCLA, Site Investigator), George Bartzokis, MD (UCLA, Past Site Investigator), Daniel H.S. Silverman, MD, PhD (UCLA, Past Site Investigator), Neill R. Graff-Radford, MBBCH, FRCP(London) (Mayo Clinic, Jacksonville, Site Investigator), Francine Parfitt, MSH, CCRC (Mayo Clinic, Jacksonville, Site Investigator), Heather Johnson, MLS, CCRP (Mayo Clinic, Jacksonville, Site Investigator), Martin R. Farlow, MD (Indiana University, Site Investigator), Ann Marie Hake, MD, Brandy R. Matthews, MD (Indiana University, Site Investigator), Scott Herring, RN (Indiana University, Past Site Investigator), Christopher H. van Dyck, MD (Yale University School of Medicine, Site Investigator), Richard E. Carson, PhD (Yale University School of Medicine, Site Investigator), Martha G. MacAvoy, PhD (Yale University School of Medicine, Site Investigator), Howard Chertkow, MD (McGill University, Montreal-Jewish General Hospital, Site Investigator), Howard Bergman, MD (McGill University, Montreal-Jewish General Hospital, Site Investigator), Chris Hosein, MEd (McGill University, Montreal-Jewish General Hospital, Site Investigator), Sandra Black, MD, FRCPC (Sunnybrook Health Sciences, Ontario, Site Investigator), Bojana Stefanovic, PhD (Sunnybrook Health Sciences, Ontario, Site Investigator), Curtis Caldwell, PhD (Sunnybrook Health Sciences, Ontario, Site Investigator), Ging-Yuek Robin Hsiung, MD, MHSc, FRCPC (UBC Clinic for AD & Related Disorders, Site Investigator), Howard Feldman, MD, FRCPC (UBC Clinic for AD & Related Disorders, Site Investigator), Benita Mudge, BS (UBC Clinic for AD & Related Disorders, Site Investigator), Michele Assaly, MA (UBC Clinic for AD & Related Disorders, Past Site Investigator), Andrew Kertesz, MD (Cognitive Neurology–St Joseph’s, Ontario, Site Investigator), John Rogers, MD (Cognitive Neurology–St Joseph’s, Ontario, Site Investigator), Dick Trost, PhD (Cognitive Neurology–St Joseph’s, Ontario, Site Investigator), Charles Bernick, MD (Cleveland Clinic Lou Ruvo Center for Brain Health, Site Investigator), Donna Munic, PhD (Cleveland Clinic Lou Ruvo Center for Brain Health, Site Investigator), Diana Kerwin, MD (Northwestern University, Site Investigator), Marek-Marsel Mesulam, MD (Northwestern University, Site Investigator), Kristina Lipowski, BA (Northwestern University, Site Investigator), Chuang-Kuo Wu, MD, PhD (Northwestern University, Past Site Investigator), Nancy Johnson, PhD (Northwestern University, Past Site Investigator), Carl Sadowsky, MD (Premiere Research Institute [Palm Beach Neurology], Site Investigator), Walter Martinez, MD (Premiere Research Institute [Palm Beach Neurology], Site Investigator), Teresa Villena, MD (Premiere Research Institute [Palm Beach Neurology], Site Investigator), Raymond Scott Turner, MD, PhD (Georgetown University Medical Center, Site Investigator), Kathleen Johnson, NP (Georgetown University Medical Center, Site Investigator), Brigid Reynolds, NP (Georgetown University Medical Center, Site Investigator), Reisa A. Sperling, MD (Brigham and Women’s Hospital, Site Investigator), Keith A. Johnson, MD (Brigham and Women’s Hospital, Site Investigator), Gad Marshall, MD (Brigham and Women’s Hospital, Past Site Investigator), Meghan Frey (Brigham and Women’s Hospital, Past Site Investigator), Jerome Yesavage, MD (Stanford University, Site Investigator), Joy L. Taylor, PhD (Stanford University, Site Investigator), Barton Lane, MD (Stanford University, Site Investigator), Allyson Rosen, PhD (Stanford University, Past Site Investigator), Jared Tinklenberg, MD (Stanford University, Past Site Investigator), Marwan Sabbagh, MD, FAAN, CCRI (Banner Sun Health Research Institute, Site Investigator), Christine Belden, PsyD (Banner Sun Health Research Institute, Site Investigator), Sandra Jacobson, MD (Banner Sun Health Research Institute, Site Investigator), Neil Kowall, MD (Boston University, Site Investigator), Ronald Killiany, PhD (Boston University, Site Investigator), Andrew E. Budson, MD (Boston University, Site Investigator), Alexander Norbash, MD (Boston University, Past Site Investigator), Patricia Lynn Johnson, BA (Boston University, Past Site Investigator), Thomas O. Obisesan, MD, MPH (Howard University, Site Investigator), Saba Wolday, MSc (Howard University, Site Investigator), Salome K. Bwayo, PharmD (Howard University, Past Site Investigator), Alan Lerner, MD (Case Western Reserve University, Site Investigator), Leon Hudson, MPH (Case Western Reserve University, Site Investigator), Paula Ogrocki, PhD (Case Western Reserve University, Site Investigator), Evan Fletcher, PhD (UC Davis–Sacramento, Site Investigator), Owen Carmichael, PhD (UC Davis–Sacramento, Site Investigator), John Olichney, MD (UC Davis– Sacramento, Site Investigator), Charles DeCarli, MD (UC Davis–Sacramento, Past Site Investigator), Smita Kittur, MD (Neurological Care of Central New York, Site Investigator), Michael Borrie, MB, ChB (Parkwood Hospital, Site Investigator), T.-Y. Lee, PhD (Parkwood Hospital, Site Investigator), Rob Bartha, PhD (Parkwood Hospital, Site Investigator), Sterling Johnson, PhD (University of Wisconsin, Site Investigator), Sanjay Asthana, MD (University of Wisconsin, Site Investigator), Cynthia M. Carlsson, MD (University of Wisconsin, Site Investigator), Steven G. Potkin, MD (UC Irvine–Brain Imaging Center, Site Investigator), Adrian Preda, MD (UC Irvine–Brain Imaging Center, Site Investigator), Dana Nguyen, PhD (UC Irvine–Brain Imaging Center, Site Investigator), Pierre Tariot, MD (Banner Alzheimer’s Institute, Site Investigator), Adam Fleisher, MD (Banner Alzheimer’s Institute, Site Investigator), Stephanie Reeder, BA (Banner Alzheimer’s Institute, Site Investigator), Vernice Bates, MD (Dent Neurologic Institute, Site Investigator), Horacio Capote, MD (Dent Neurologic Institute, Site Investigator), Michelle Rainka, PharmD, CCRP (Dent Neurologic Institute, Site Investigator), Douglas W. Scharre, MD (Ohio State University, Site Investigator), Maria Kataki, MD, PhD (Ohio State University, Site Investigator), Earl A. Zimmerman, MD (Albany Medical College, Site Investigator), Dzintra Celmins, MD (Albany Medical College, Site Investigator), Alice D. Brown, FNP (Albany Medical College, Past Site Investigator), Godfrey D. Pearlson, MD (Hartford Hospital, Olin Neuropsychiatry Research Center, Site Investigator), Karen Blank, MD (Hartford Hospital, Olin Neuropsychiatry Research Center, Site Investigator), Karen Anderson, RN (Hartford Hospital, Olin Neuropsychiatry Research Center, Site Investigator), Andrew J. Saykin, PsyD (Dartmouth-Hitchcock Medical Center, Site Investigator), Robert B. Santulli, MD (Dartmouth-Hitchcock Medical Center, Site Investigator), Eben S. Schwartz, PhD (Dartmouth-Hitchcock Medical Center, Site Investigator), Kaycee M. Sink, MD, MAS (Wake Forest University Health Sciences, Site Investigator), Jeff D. Williamson, MD, MHS (Wake Forest University Health Sciences, Site Investigator), Pradeep Garg, PhD (Wake Forest University Health Sciences, Site Investigator), Franklin Watkins, MD (Wake Forest University Health Sciences, Past Site Investigator), Brian R. Ott, MD (Rhode Island Hospital, Site Investigator), Henry Querfurth, MD (Rhode Island Hospital, Site Investigator), Geoffrey Tremont, PhD (Rhode Island Hospital, Site Investigator), Stephen Salloway, MD, MS (Butler Hospital, Site Investigator), Paul Malloy, PhD (Butler Hospital, Site Investigator), Stephen Correia, PhD (Butler Hospital, Site Investigator), Howard J. Rosen, MD (UC San Francisco, Site Investigator), Bruce L. Miller, MD (UC San Francisco, Site Investigator), Jacobo Mintzer, MD, MBA (Medical University South Carolina, Site Investigator), Crystal Flynn Longmire, PhD (Medical University South Carolina, Site Investigator), Kenneth Spicer, MD, PhD (Medical University South Carolina, Site Investigator), Elizabether Finger, MD (St Joseph’s Health Care, Site Investigator), Irina Rachinsky, MD (St Joseph’s Health Care, Site Investigator), John Rogers, MD (St Joseph’s Health Care, Site Investigator), Andrew Kertesz, MD (St Joseph’s Health Care, Past Site Investigator), Dick Drost, MD (St Joseph’s Health Care, Past Site Investigator), Nunzio Pomara, MD (Nathan Kline Institute, Site Investigator), Raymundo Hernando, MD (Nathan Kline Institute, Site Investigator), Antero Sarrael, MD (Nathan Kline Institute, Site Investigator), Susan K. Schultz, MD (University of Iowa, Site Investigator), Laura L. Boles Ponto, PhD (University of Iowa, Site Investigator), Hyungsub Shim, MD (University of Iowa, Site Investigator), Karen Elizabeth Smith, RN (University of Iowa, Site Investigator), Norman Relkin, MD, PhD (Cornell University), Gloria Chaing, MD (Cornell University), Lisa Raudin, PhD (Cornell University), Amanda Smith, MD (University of South Florida), Kristin Fargher, MD (University of South Florida), Balebail Ashok Raj, MD (University of South Florida).
Footnotes
Author Contributions: Dr Trojanowski had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: Toledo.
Acquisition, analysis, or interpretation of data: All authors.
Drafting of the manuscript: Toledo, Trojanowski.
Critical revision of the manuscript for important intellectual content: All authors.
Statistical analysis: Toledo.
Obtained funding: Foster, Jagust, Jack, Weiner, Trojanowski.
Administrative, technical, or material support: Weiner, Shaw.
Study supervision: Trojanowski.
Conflict of Interest Disclosures: No other disclosures are reported.
Additional Information: Data used in preparation of this article were obtained from the ADNI database (http://adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in the analysis or writing of this report. The ADNI is funded by the National Institute on Aging and the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica; Biogen Idec; Bristol-Myers Squibb; Eisai; Elan Pharmaceuticals; Eli Lilly; EuroImmun; F. Hoffmann–La Roche and its affiliated company Genentech; Fujirebio; GE Healthcare; IXICO; Janssen Alzheimer Immunotherapy Research and Development, LLC; Johnson & Johnson Pharmaceutical Research and Development, LLC; Medpace; Merck; Meso Scale Diagnostics, LLC; NeuroRx Research; Neurotrack Technologies; Novartis; Pfizer; Piramal Imaging; Servier; Synarc; and Takeda. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (http://www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Disease Cooperative Study at the University of California, San Diego. The ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California. Dr Trojanowski is the William Maul Measey–Truman G. Schnabel Jr Professor of Geriatric Medicine and Gerontology.
References
- 1.Montine TJ, Phelps CH, Beach TG, et al. National Institute on Aging; Alzheimer’s Association. National Institute on Aging-Alzheimer’s Association guidelines for the neuropathologic assessment of Alzheimer’s disease: a practical approach. Acta Neuropathol. 2012;123(1):1–11. doi: 10.1007/s00401-011-0910-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clark CM, Schneider JA, Bedell BJ, et al. AV45-A07 Study Group. Use of florbetapir-PET for imaging β-amyloid pathology [published correction appears JAMA. 2011;305(11):1096] JAMA. 2011;305(3):275–283. doi: 10.1001/jama.2010.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ikonomovic MD, Klunk WE, Abrahamson EE, et al. Post-mortem correlates of in vivo PiB-PET amyloid imaging in a typical case of Alzheimer’s disease. Brain. 2008;131(Pt 6):1630–1645. doi: 10.1093/brain/awn016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wolk DA, Grachev ID, Buckley C, et al. Association between in vivo fluorine 18-labeled flutemetamol amyloid positron emission tomography imaging and in vivo cerebral cortical histopathology. Arch Neurol. 2011;68(11):1398–1403. doi: 10.1001/archneurol.2011.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Toledo JB, Brettschneider J, Grossman M, et al. CSF biomarkers cutoffs: the importance of coincident neuropathological diseases. Acta Neuropathol. 2012;124(1):23–35. doi: 10.1007/s00401-012-0983-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tapiola T, Alafuzoff I, Herukka SK, et al. Cerebrospinal fluid β-amyloid 42 and tau proteins as biomarkers of Alzheimer-type pathologic changes in the brain. Arch Neurol. 2009;66(3):382–389. doi: 10.1001/archneurol.2008.596. [DOI] [PubMed] [Google Scholar]
- 7.Strozyk D, Blennow K, White LR, Launer LJ. CSF Aβ 42 levels correlate with amyloid-neuropathology in a population-based autopsy study. Neurology. 2003;60(4):652–656. doi: 10.1212/01.wnl.0000046581.81650.d0. [DOI] [PubMed] [Google Scholar]
- 8.Toledo JB, Vanderstichele H, Figurski M, et al. Alzheimer’s Disease Neuroimaging Initiative. Factors affecting Aβ plasma levels and their utility as biomarkers in ADNI. Acta Neuropathol. 2011;122(4):401–413. doi: 10.1007/s00401-011-0861-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weigand SD, Vemuri P, Wiste HJ, et al. Alzheimer’s Disease Neuroimaging Initiative. Transforming cerebrospinal fluid Aβ42 measures into calculated Pittsburgh Compound B units of brain Aβ amyloid. Alzheimers Dement. 2011;7(2):133–141. doi: 10.1016/j.jalz.2010.08.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Palmqvist S, Zetterberg H, Blennow K, et al. Accuracy of brain amyloid detection in clinical practice using cerebrospinal fluid β-amyloid 42: a cross-validation study against amyloid positron emission tomography. JAMA Neurol. 2014;71(10):1282–1289. doi: 10.1001/jamaneurol.2014.1358. [DOI] [PubMed] [Google Scholar]
- 11.Jagust WJ, Landau SM, Shaw LM, et al. Alzheimer’s Disease Neuroimaging Initiative. Relationships between biomarkers in aging and dementia. Neurology. 2009;73(15):1193–1199. doi: 10.1212/WNL.0b013e3181bc010c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grimmer T, Riemenschneider M, Förstl H, et al. Beta amyloid in Alzheimer’s disease: increased deposition in brain is reflected in reduced concentration in cerebrospinal fluid. Biol Psychiatry. 2009;65(11):927–934. doi: 10.1016/j.biopsych.2009.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tolboom N, van der Flier WM, Yaqub M, et al. Relationship of cerebrospinal fluid markers to 11C-PiB and 18F-FDDNP binding. J Nucl Med. 2009;50(9):1464–1470. doi: 10.2967/jnumed.109.064360. [DOI] [PubMed] [Google Scholar]
- 14.Forsberg A, Almkvist O, Engler H, Wall A, Långström B, Nordberg A. High PIB retention in Alzheimer’s disease is an early event with complex relationship with CSF biomarkers and functional parameters. Curr Alzheimer Res. 2010;7(1):56–66. doi: 10.2174/156720510790274446. [DOI] [PubMed] [Google Scholar]
- 15.Fagan AM, Xiong C, Jasielec MS, et al. Dominantly Inherited Alzheimer Network. Longitudinal change in CSF biomarkers in autosomal-dominant Alzheimer’s disease. Sci Transl Med. 2014;6(226):226ra30. doi: 10.1126/scitranslmed.3007901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Landau SM, Lu M, Joshi AD, et al. Disease Neuroimaging Initiative. Comparing positron emission tomography imaging and cerebrospinal fluid measurements of β-amyloid. Ann Neurol. 2013;74(6):826–836. doi: 10.1002/ana.23908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fagan AM, Shaw LM, Xiong C, et al. Comparison of analytical platforms for cerebrospinal fluid measures of β-amyloid 1–42, total tau, and p-tau181 for identifying Alzheimer disease amyloid plaque pathology. Arch Neurol. 2011;68(9):1137–1144. doi: 10.1001/archneurol.2011.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fagan AM, Mintun MA, Shah AR, et al. Cerebrospinal fluid tau and ptau181 increase with cortical amyloid deposition in cognitively normal individuals: implications for future clinical trials of Alzheimer’s disease. EMBO Mol Med. 2009;1(8–9):371–380. doi: 10.1002/emmm.200900048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mattsson N, Insel PS, Landau S, et al. Alzheimer’s Disease Neuroimaging Initiative. Diagnostic accuracy of CSF Ab42 and florbetapir PET for Alzheimer’s disease. Ann Clin Transl Neurol. 2014;1(8):534–543. doi: 10.1002/acn3.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Toledo JB, Shaw LM, Trojanowski JQ. Plasma amyloid beta measurements—a desired but elusive Alzheimer’s disease biomarker. Alzheimers Res Ther. 2013;5(2):8. doi: 10.1186/alzrt162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weiner MW, Veitch DP, Aisen PS, et al. Alzheimer’s Disease Neuroimaging Initiative. The Alzheimer’s Disease Neuroimaging Initiative: a review of papers published since its inception. Alzheimers Dement. 2013;9(5):e111–e194. doi: 10.1016/j.jalz.2013.05.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petersen RC, Smith GE, Waring SC, Ivnik RJ, Tangalos EG, Kokmen E. Mild cognitive impairment: clinical characterization and outcome. Arch Neurol. 1999;56(3):303–308. doi: 10.1001/archneur.56.3.303. [DOI] [PubMed] [Google Scholar]
- 23.Petersen RC, Aisen PS, Beckett LA, et al. Alzheimer’s Disease Neuroimaging Initiative (ADNI): clinical characterization. Neurology. 2010;74(3):201–209. doi: 10.1212/WNL.0b013e3181cb3e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM. Clinical diagnosis of Alzheimer’s disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer’s Disease. Neurology. 1984;34(7):939–944. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- 25.Shaw LM, Vanderstichele H, Knapik-Czajka M, et al. Alzheimer’s Disease Neuroimaging Initiative. Cerebrospinal fluid biomarker signature in Alzheimer’s disease neuroimaging initiative subjects. Ann Neurol. 2009;65(4):403–413. doi: 10.1002/ana.21610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shaw LM, Vanderstichele H, Knapik-Czajka M, et al. Alzheimer’s Disease Neuroimaging Initiative. Qualification of the analytical and clinical performance of CSF biomarker analyses in ADNI. Acta Neuropathol. 2011;121(5):597–609. doi: 10.1007/s00401-011-0808-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Joshi A, Koeppe RA, Fessler JA. Reducing between scanner differences in multi-center PET studies. Neuroimage. 2009;46(1):154–159. doi: 10.1016/j.neuroimage.2009.01.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Meyer G, Shapiro F, Vanderstichele H, et al. Alzheimer’s Disease Neuroimaging Initiative. Diagnosis-independent Alzheimer disease biomarker signature in cognitively normal elderly people. Arch Neurol. 2010;67(8):949–956. doi: 10.1001/archneurol.2010.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Toledo JB, Xie SX, Trojanowski JQ, Shaw LM. Longitudinal change in CSF Tau and Aβ biomarkers for up to 48 months in ADNI. Acta Neuropathol. 2013;126(5):659–670. doi: 10.1007/s00401-013-1151-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klunk WE, Koeppe RA, Price JC, et al. The Centiloid Project: standardizing quantitative amyloid plaque estimation by PET. Alzheimers Dement. 2015;11(1):1–15. doi: 10.1016/j.jalz.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Toledo JB, Weiner MW, Wolk DA, et al. Alzheimer’s Disease Neuroimaging Initiative. Neuronal injury biomarkers and prognosis in ADNI subjects with normal cognition. Acta Neuropathol Commun. 2014;2:26. doi: 10.1186/2051-5960-2-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Joshi AD, Pontecorvo MJ, Clark CM, et al. Florbetapir F 18 Study Investigators. Performance characteristics of amyloid PET with florbetapir F 18 in patients with Alzheimer’s disease and cognitively normal subjects. J Nucl Med. 2012;53(3):378–384. doi: 10.2967/jnumed.111.090340. [DOI] [PubMed] [Google Scholar]
- 33.Thal DR, Rüb U, Orantes M, Braak H. Phases of A beta-deposition in the human brain and its relevance for the development of AD. Neurology. 2002;58(12):1791–1800. doi: 10.1212/wnl.58.12.1791. [DOI] [PubMed] [Google Scholar]
- 34.Jack CR, Jr, Knopman DS, Jagust WJ, et al. Tracking pathophysiological processes in Alzheimer’s disease: an updated hypothetical model of dynamic biomarkers. Lancet Neurol. 2013;12(2):207–216. doi: 10.1016/S1474-4422(12)70291-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sperling RA, Aisen PS, Beckett LA, et al. Toward defining the preclinical stages of Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011;7(3):280–292. doi: 10.1016/j.jalz.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hyman BT, Phelps CH, Beach TG, et al. National Institute on Aging-Alzheimer’s Association guidelines for the neuropathologic assessment of Alzheimer’s disease. Alzheimers Dement. 2012;8(1):1–13. doi: 10.1016/j.jalz.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morris JC, Price JL. Pathologic correlates of nondemented aging, mild cognitive impairment, and early-stage Alzheimer’s disease. J Mol Neurosci. 2001;17(2):101–118. doi: 10.1385/jmn:17:2:101. [DOI] [PubMed] [Google Scholar]
- 38.Hof PR, Glannakopoulos P, Bouras C. The neuropathological changes associated with normal brain aging. Histol Histopathol. 1996;11(4):1075–1088. [PubMed] [Google Scholar]
- 39.Sperling RA, Rentz DM, Johnson KA, et al. The A4 study: stopping AD before symptoms begin? Sci Transl Med. 2014;6(228):228fs13. doi: 10.1126/scitranslmed.3007941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Da X, Toledo JB, Zee J, et al. Alzheimer’s Neuroimaging Initiative. Integration and relative value of biomarkers for prediction of MCI to AD progression: spatial patterns of brain atrophy, cognitive scores, APOE genotype and CSF biomarkers. Neuroimage Clin. 2014;4:164–173. doi: 10.1016/j.nicl.2013.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Worley S. After disappointments, Alzheimer’s researchers seek out new paths: biomarkers and combination therapies may lead to disease-modifying treatments, experts say. P T. 2014;39(5):365–374. [PMC free article] [PubMed] [Google Scholar]
- 42.Cairns NJ, Ikonomovic MD, Benzinger T, et al. Absence of Pittsburgh compound B detection of cerebral amyloid beta in a patient with clinical, cognitive, and cerebrospinal fluid markers of Alzheimer disease: a case report. Arch Neurol. 2009;66(12):1557–1562. doi: 10.1001/archneurol.2009.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ikonomovic MD, Abrahamson EE, Price JC, et al. Early AD pathology in a [C-11]PiB-negative case: a PiB-amyloid imaging, biochemical, and immunohistochemical study. Acta Neuropathol. 2012;123(3):433–447. doi: 10.1007/s00401-012-0943-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Clark VH, Resnick SM, Doshi J, et al. Longitudinal imaging pattern analysis (SPARE-CD index) detects early structural and functional changes before cognitive decline in healthy older adults. Neurobiol Aging. 2012;33(12):2733–2745. doi: 10.1016/j.neurobiolaging.2012.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ikonomovic M, Price J, Abrahamson E, et al. Direct correlations of [H-3]flutemetamol binding with [H-3]PiB binding and amyloid-beta concentration and plaque load in [C-11]PiB imaged brains. Neurology. 2012;78:S34.002. doi: 10.1212/WNL.78.1_MeetingAbstracts.S34.002. [DOI] [Google Scholar]
- 46.Boluda S, Toledo JB, Irwin DJ, et al. A comparison of Aβ amyloid pathology staging systems and correlation with clinical diagnosis. Acta Neuropathol. 2014;128(4):543–550. doi: 10.1007/s00401-014-1308-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rebeck GW, Reiter JS, Strickland DK, Hyman BT. Apolipoprotein E in sporadic Alzheimer’s disease: allelic variation and receptor interactions. Neuron. 1993;11(4):575–580. doi: 10.1016/0896-6273(93)90070-8. [DOI] [PubMed] [Google Scholar]
- 48.Holtzman DM, Bales KR, Tenkova T, et al. Apolipoprotein E isoform-dependent amyloid deposition and neuritic degeneration in a mouse model of Alzheimer’s disease. Proc Natl Acad Sci U S A. 2000;97(6):2892–2897. doi: 10.1073/pnas.050004797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Toledo JB, Da X, Bhatt P, et al. Alzheimer’s Disease Neuroimaging Initiative. Relationship between plasma analytes and SPARE-AD defined brain atrophy patterns in ADNI. PLoS One. 2;8:e55531. doi: 10.1371/journal.pone.0055531. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.