Abstract
Blockade of the pathway including Programmed death-ligand 1 (PD-L1) and its receptor Programmed cell death protein 1 (PD-1) has produced clinical benefits in patients with a variety of cancers. Elevated levels of soluble PD-L1 (sPD-L1) have been associated with worse prognosis in renal cell carcinoma and multiple myeloma. However, the regulatory roles and function of sPD-L1 particularly in connection with immune checkpoint blockade treatment are not fully understood. We identified four splice variants of PD-L1 in melanoma cells, and all of them are secreted. Secretion of sPD-L1 resulted from alternate splicing activities, cytokine induction, cell stress, cell injury and cell death in melanoma cells. Pretreatment levels of sPD-L1 were elevated in stage IV melanoma patient sera compared to healthy donors. High pre-treatment levels of sPD-L1 were associated with increased likelihood of progressive disease in patients treated by CTLA-4 or PD-1 blockade. Although changes in circulating sPD-L1 early after treatment could not distinguish responders from those with progressive disease, after five months of treatment by CTLA-4 or PD-1 blockade patients who had increased circulating sPD-L1 had greater likelihood of developing a partial response. Induction of sPD-L1 was associated with increased circulating cytokines after CTLA-4 blockade but not following PD-1 blockade. Circulating sPD-L1 is a prognostic biomarker that may predict outcomes for subgroups of patients receiving checkpoint inhibitors.
Keywords: Melanoma, soluble PD-L1, CTLA-4 blockade, PD-1 blockade
INTRODUCTION
Programmed death-ligand 1 (PD-L1) is a membrane-bound protein primarily expressed on dendritic cells (DC) and monocytes (1). Its receptor, Programmed cell death protein 1 (PD-1), is expressed on activated T cells and B cells, DC, and monocytes (1). As T cells engage with the antigen/MHC complex, binding of PD-L1 to PD-1 inhibits T cell activation, leading to immune suppression (1, 2). A wide range of tumors express PD-L1 (3–6). Although conflicting reports exist for melanoma (7), PD-L1 expression is associated with worse clinical outcomes in some tumor types (8, 9). Blocking the PD-1/PD-L1 interaction increases antigen-specific T-cell activity while decreasing Treg suppressive function (10, 11). Antibodies that block either PD-1 or PD-L1 improve clinical responses as well as patients’ overall survival in many tumor types (12–16).
Soluble PD-L1 (sPD-L1) in sera is associated with aggressive renal cell carcinoma and shorter survival in multiple myeloma and diffuse large B-cell lymphoma (17–19). sPD-L1 can be produced by cytokine-activated mature dendritic cells in vitro (20). However, the mechanisms by which sPD-L1 in patients are generated remain poorly understood. The clinical significance of circulating sPD-L1 in melanoma is unknown. A splice variant of PD-L1, which lacks the IgV domain by splicing out exon 2 (21), is likely not secreted nor functional, because it retains the transmembrane domain but lacks the PD-1 binding site within the IgV domain. An additional variant with splicing regions in exons 3 and 4 is documented in Genbank (Accession: AY714881).
Here, we identify four splice variants of PD-L1 in melanoma, and investigated production of sPD-L1 in patients receiving immune checkpoint blockade.
MATERIAL AND METHODS
Cell lines
A375, K008, K028, K029, K033, M34, and UACC257 melanoma cell lines were cultured in DMEM medium with 10% fetal bovine serum. 293T cells were cultured in complete DMEM. Human melanoma cells were isolated from tumor biopsies of melanoma patients, and the human melanoma cell lines were developed approximately 25 years ago in accordance with Dana-Farber/Harvard Cancer Center Institutional Review Board approved protocols. UACC257 cells were kindly provided by Dr. David E. Fisher from Massachusetts General Hospital, Boston, 11 years ago. A375 cells were obtained from ATCC 10 years ago. The cell lines have been used in current project for 5 years. All cell lines were confirmed to express MITF and melanocytic markers. Cell line authentication was performed using short tandem repeat profiling and profiling data were compared with known cell line DNA profiles in the end of current project in 2016.
Plasma and sera of healthy donors and melanoma patients
Peripheral blood samples were obtained from melanoma patients and healthy donors on Dana-Farber Cancer Center Institutional Review Board approved protocols. Peripheral blood was collected in heparinized and anticoagulant-free tubes. Plasma and serum supernatant were collected by centrifugation. Specimens were further analyzed from 42 patients receiving combination ipilimumab plus bevacizumab in a clinical trial [NCT00790010] (22), from 23 patients receiving ipilimumab, and from 35 patients receiving pembrolizumab (anti–PD-1) at DFCI. Peripheral blood samples were obtained from melanoma patients in the NCI-sponsored Eastern Cooperative Group Trial E1608 comparing ipilimumab plus sargramostim versus ipilimumab [NCT01134614] (23). Peripheral blood was collected in red top, anticoagulant-free tubes, shipped overnight from clinical sites to the ECOG-ACRIN immunology reference Lab at the University of Pittsburgh Cancer Institute, where it was processed upon receipt for serum. Serum supernatant was collected by centrifugation and stored at a −80°C in monitored freezer. Specimens were further analyzed from 151 patients. Among them, seventy-eight patients were treated with ipilimumab plus sargramostim (arm A), and seventy-three patients received ipilimumab (control arm B).
RT-PCR and human PD-L1 variant cloning
Total RNA of melanoma cells lines was generated with RNeasy Mini kit (Qiagen, Valencia, CA). RNA (1 µg) of each melanoma cell line was reverse-transcripted to cDNA with SuperScript reverse transcriptase (Life Technologies, Grand Island, NY). PD-L1 transcripts from A375 and M34 melanoma cell lines were cloned by PCR with a XbaI restriction site tagged forward primer: GCGTCGTCTAGAGCCACCATGAGGATATTTGCTGTCT encompassing the translational start site and a SalI tagged reverse primer: Sal1 GCGCCAGTCGACTTACGTCTCCTCCAAATGTGT encompassing the translational stop site of full-length PD-L1. The PCR products were cloned into a TA TOPO vector (Life Technologies) for sequencing analysis. The variants of PD-L1 were further inserted into a lentiviral transfer vector pELNS, which was kindly provided by Dr. Michael P. Riley from University of Pennsylvania.
To detect mRNA splicing variants of PD-L1 in melanoma cell lines, primers were designed to contain both ends of splice donor and acceptor, and were specific for PD-L1-1, 3/12, and 9 variants (Supplemental Fig. S1). The specific primers of PD-L1-1, PD-L1-3/12, and PD-L1-9 were CCAAATGAAAGGACTCACTTG/CGTCTCCTCCAAATGTGTATCTT, AAGTCCTGAGTGGAGATTAGATC/CATTCTCCCAAGTGAGTCC, and ACCAGCACACTGAGAATCAAC/CACATCCATCATTCTCCCAAG, respectively. The sizes of PCR products were 103, 104, and 161 bps, respectively. The identities of the PCR products were confirmed by sequencing (Eton Bioscience Inc. Boston, MA).
Transfection, lentiviral production, and lentiviral transduction
The pELNS expressing PD-L1 variants were cotransfected into 293T cells with three packaging plasmids expressing gag/pol, VSV-g, and REV using TransfectIT-293 (Mirus, Madison, WI). Lentiviral supernatants were collected and filtered. PD-L1-1, PD-L1-3, and PD-L1-9 were transduced into 1 × 105 A375 cells with the supernatant in the presence of 8 µg/ml Polybrene (EMD Millipore, Billerica, MA).
Immnuoprecipitation, SDS-PAGE, and immunoblotting
For BRAF resistant cell line studies, approximately 8 × 107 PLX4032 resistant A375 and 1 × 107 PLX4032 resistant M34 melanoma cell lines were cultured in complete DMEM medium in the presence of 1 µg/ml PLX4032 for 2 days. After washing with PBS three times, the cells were further cultured in Opti-MEM reduced serum medium (Life Technologies, Grand Island, NY) in the presence of 1 µg/ml PLX4032 for 2 days. For cytokine induction studies, approximately 5 × 106 to 30 × 106 cells (A375, K008, K028, and UACC257) were cultured in complete DMEM medium for 2 days. After washing with PBS three times, the melanoma cell lines were cultured in Opti-MEM reduced serum medium in the presence of either 200 U/ml IFNγ (Biolegend, San Diego, CA), or 2000 U/ml IFNα (EMD Millipore, Billerica, MA), or 10 ng/ml TNFα (R&D systems, Minneapolis, MN) for an additional two days. The culture media were collected, and concentrated with a 3K cutoff Centriprep spin column (EMD Millipore, Billerica, MA). Samples were normalized by cell numbers. Concentrated supernatant was incubated with 0.5 µg anti-human PD-L1 mAb (clone 29E.2A3, Biolegend) and 10 µl protein G plus agarose (Santa Cruz Biotechnology, Santa Cruz, CA) at 4 °C overnight. After washing with PBS, the agarose beads were resuspended in Laemmli’s reducing buffer (Boston Bioproducts, Worcester, MA), and further heated. Immunoprecipitated proteins were subjected to 12% SDS-polyacrylamide gel electrophoresis (PAGE), and transferred onto PVDF membranes. The membranes were immunoblotted overnight at 4 °C with a biotinylated goat anti-human PD-L1 at 0.1 µg/ml (R&D systems), and further incubated with HRP conjugated streptavidin at 2.5 µg/ml (Jackson ImmunoResearch, West Grove, PA) at room temperature for 2 hours. The protein bands were detected by chemiluminescence (PerkinElmer, Waltham, MA).
PD-L1-3/Fc fusion protein
CH2 and CH3 domains of human IgG1 was fused to PD-L1-3, PD-L1-9, and PD-L1-1 in pELNS vector. PD-L1-3/Fc, PD-L1-9/Fc, and PD-L1-1/Fc were transduced into CHO-S cells by lentiviral supernatant in the presence of 8 µg/ml Polybrene (EMD Millipore), respectively. PD-L1-3/Fc, PD-L1-9/Fc, and PD-L1-1/Fc were purified with protein A agarose (Life Technologies). It was further confirmed by SDS-PAGE and Coomassie staining, and by immunoblotting with anti-human PD-L1 mAb (29E.1D5) (Supplemental Fig. S2A and S2B).
Proliferation assay
To generate activated T cells, human naive CD4+ or CD8+ T cells were treated with 10 µg/ml PHA for 3 days and further rested overnight. 1×105 cells/well of PHA-activated CD4+ or CD8+ T cells were stimulated with 5 µg/ml coated anti-CD3 (BD Biosciences, San Jose, CA) in the absence or presence of 10 µg/ml coated either recombinant fusion proteins of PD-L1 variants or human IgG for 3 days, and further pulsed with [3H]thymidine (0.25 µCi H3/well) for 6 hours. The incorporated radioactivity was measured in a liquid scintillation counter (Wallac 1450 Microbeta Trilux, Perkin Elmer, Waltham, MA).
sPD-L1 ELISA
To assay sPD-L1 variants, 0.1 µg/well of mouse anti-human PD-L1 mab (130021, R&D systems) or 0.2 µg/well anti-human PD-L1 mAb (29E.12B1) were coated on Costar ELISA plates overnight at 4°C. Plates were then washed with PBS and blocked with protein-free blocking buffer (Pierce, Rockford, IL) for 4 hours. Patient sera or plasma were diluted with PBS in 1:1 volume ratio. 100 µl per well of diluted patient sera or plasma were added and incubated overnight at 4°C. Plates were washed with PBS containing Tween-20, and incubated with 100 µl per well of 0.1µg/well biotinylated anti-PD-L1 mAb (29E.2A3, Biolegend) in protein-free blocking buffer at room temperature for 2 hours. Plates were then washed and incubated with 1mg/ml streptavidin-HRP (Jackson ImmunoResearch) diluted 1:40,000 in protein-free blocking buffer for 2 hours. Plates were washed and treated with biotinyl tyramide (Perkin Elmer) for 30 min, and then washed and incubated with 1 mg/ml streptavidin-HRP (Jackson ImmunoResearch) diluted 1:400,000 in protein-free blocking buffer for 2 hours with further development with TMB (Pierce). Plates were read at an optical density (O.D.) of 450 nm. All samples were assayed in duplicate. A standard curve using recombinant human PD-L1-HIS (Novoprotein, Summit, NJ) was also performed with each assay.
Cytokine Luminex assay
IFNα, IFNγ, and TNFα in plasma of melanoma patients were quantified by a Luminex beads kit (EMD Millipore). 25 µl/well of antibody coupled beads and 50 µl/well of diluted patient plasma were added into a 96 well plate and incubated overnight at 4°C. Plates were processed following manufacturer’s instruction and read by Luminex 200 (Luminex Corporation, Austin, TX). All samples were performed in duplicate. The standard curves were also performed with each assay.
Statistical analyses
ELISAs were conducted in duplicate to examine sPD-L1 in patient serum samples. A value of 0.1 ng/ml was determined to be the lower limit of detection based on sensitivity of the ELISA assay. If assay values were < 0.1ng/ml, 0.01 ng/ml was substituted. Clinical comparisons addressed two hypotheses: (1) high levels of sPD-L1 were associated with progressive disease; and, (2) long-term or delayed increases in sPD-L1 were associated with favorable clinical response. Data from the ipilimumab plus bevacizumab trial were used as a test set; data from E1608 and from patients treated with ipilimumab and PD-1 were validation sets. Division points in pre-treatment sPD-L1 in sPD-L1 used in the analysis were based on the test set and were selected to determined using the algorithm of Contal-O’Quigley (24). Division points for pre-treatment sPD-L1L and sPD-L1all were 0.5 ng/ml and 1.4 ng/ml, respectively. An increase of at least 1.5-fold relative to pre-treatment was considered meaningful.
Pre-treatment comparisons were based on data from all patients. Five-month landmark samples were used to assess the effects of change in sPD-L1 upon response or survival. Patients who were alive and had post-treatment samples collected between 5 and 7 months (ipilimumab plus bevacizumab trial) or 5 and 11 months (E1608 Control Arm B or PD-1) were followed forward in time. If multiple samples were collected for a patient during the landmark window, data from the first sample were used in the analysis. Two sample Student’s t-tests with unequal variances were conducted to assess sPD-L1 pretreatment level differences in sera between healthy donors and melanoma patients. The comparisons of clinical response according to categories of pretreatment levels or on-treatment increases in sPD-L1 were carried out with Fisher’s exact tests. Comparisons of sPD-L1 levels according to response were based on Kruskal-Wallis tests. The distribution of overall survival was summarized using the method of Kaplan-Meier; comparisons of survival according to pre-treatment or on-treatment sPD-L1 classes were based on log-rank tests. All tests were two-sided, and P < 0.05 was considered statistically significant.
RESULTS
Splice variants of PD-L1 in melanoma
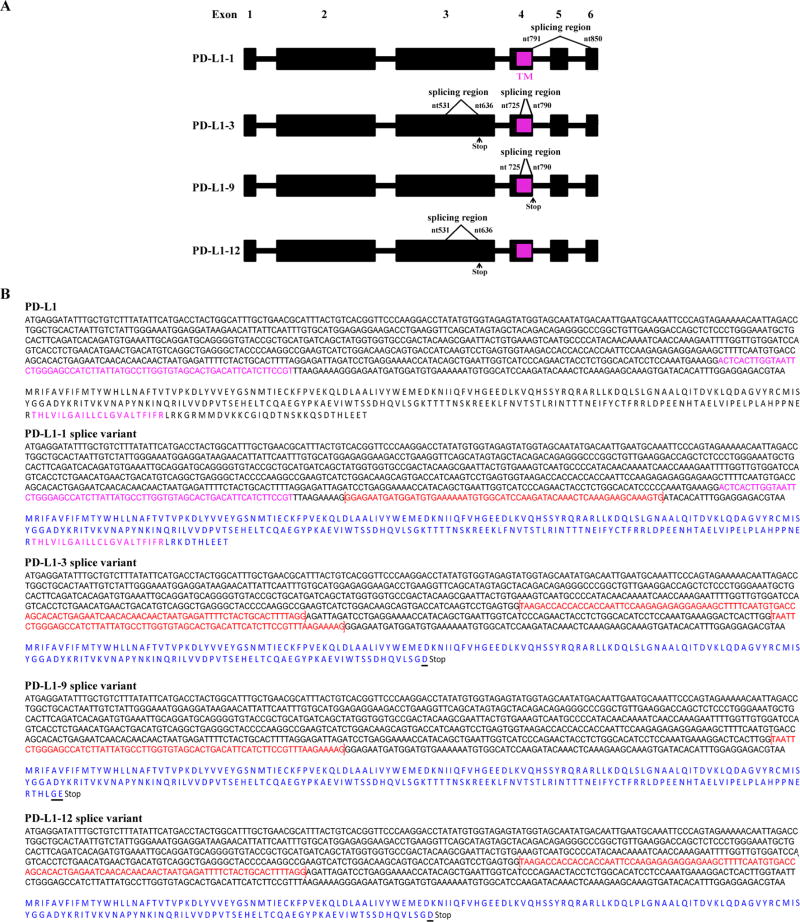
We identified four splice variants in addition to full-length PD-L1 in both A375 and M34 melanoma cell lines (Fig. 1). The PD-L1-3 variant has been previously reported in Genebank (Accession: AY714881). The PD-L1-1 variant has a 60 bp deletion from nucleotide (nt)-791 to 850 of PD-L1 (Fig. 1). This deletion removes 20 amino acids of the intracellular domain. The splice occurs from the end of exon 4 to the middle of exon 6, deleting exon 5 and half of exon 6. The PD-L1-12 variant has a splice in the extracellular domain resulting in a 106 bp deletion from nt-531 to 636 within the exon 3 region, resulting in a frame shift leading to a stop codon 4 nt after nt-530 in exon 3. The resulting protein is truncated before the transmembrane domain and terminates with a different amino acid. PD-L1-9 has lost a 66 bp region from nt-725 to 790 in exon 4. This results in a frame shift leading to a stop codon 4 nt after nt-724 before the transmembrane domain, and adding two additional amino acids at the end. Variant PD-L1-3 has both the splices of PD-L1-9 and PD-L1-12, but encodes the same protein as PD-L1-12, as the second splice occurs after the stop codon of PD-L1-3.
Figure 1.
A. Schematic diagram of identified splice variants of PD-L1. The full-length of PD-L1 consists of six exons. The transmembrane domain (TM) is located in exon 4 and marked in pink. Spliced-out regions of PD-L1-1, 3, 9, 12 are indicated with bracket symbols. Stop represents a stop codon. B. Identification of splice variants of human PD-L1. PD-L1 transcripts from A375 and M34 melanoma cell lines were generated by RT-PCR and cloned into a TA TOPO vector. Four unique PD-L1 splice variants were identified by sequencing. Three splice variants PD-L1-1, PD-L1-9, and PD-L1-12 have not been previously reported. The full-length nucleotide and amino acid sequence for membrane-bound PD-L1 is represented (top). The trans-membrane domain is shown in pink. Spliced-out regions are indicated with bracket symbols and shown in red. In each of the identified splice variants, the amino acid sequences are shown in blue. Underlines indicate additional different amino acids.
We used primers specific to the splice variants to examine their expression in six melanoma cell lines. Five of the melanoma cell lines expressed all of the variants. The sixth line, K029, lacked variant PD-L1-9 (Supplemental Fig. S2C). Thus PD-L1 splice variants are generally expressed in melanoma.
Secretion of PD-L1 variants
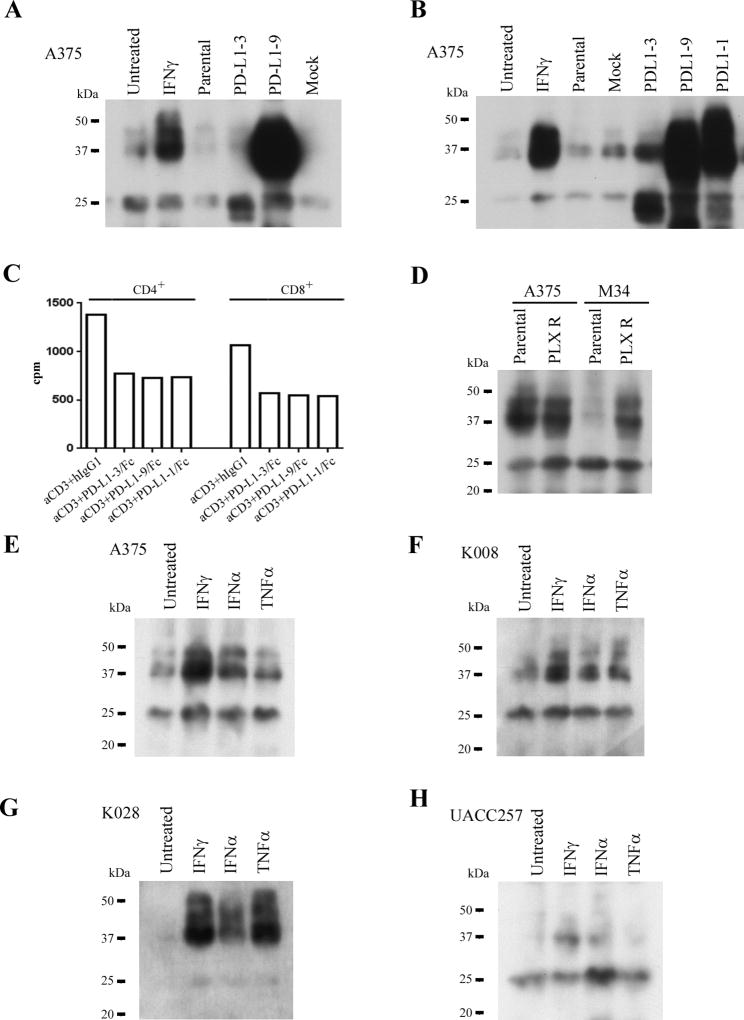
Because splice variants lacking the transmembrane domain could be secreted, we assayed culture supernatants of cell lines A375, K008, K028, M34, and UACC257 for sPD-L1. Three PD-L1 bands of 24, 38, and 45kDa were detected in the culture medium of all cell lines. PD-L1-3 and PD-L1-9 corresponded to the 24 and 38 kDa bands, respectively. A375 cells, in which PD-L1-3 and PD-L1-9 were overexpressed, secreted more sPD-L1 than did parental or mock infected cells (Fig. 2A, B). Thus, these melanoma cell lines secrete sPD-L1 variants.
Figure 2.
Secretion of spliced variants of PD-L1 from melanoma cell lines, suppression of T cell activation by sPD-L1, effects of BRAF inhibitor on sPD-L1 secretion in resistant melanoma cell lines, and differetial secretion of sPD-L1 variants in response to cytokine stimulations. A and B. A375 cell line was transduced with lentiviral vectors of PD-L1-1, 3, and 9 variants. sPD-L1 variants from culture medium were examined by immunoprecipitation, SDS-PAGE and immunoblotting assay. Loading samples were normalized by cell numbers. C. Effects of sPD-L1 on the proliferation of human CD4+ and CD8+ T cells. PHA-activated human CD4+ and CD8+ T cells were stimulated with anti CD3 (5µg/ml) in the absence or presence of either PD-L1-3/Fc, or PD-L1-9/Fc, or PD-L1-1/Fc (all at 10µg/ml). Proliferation of the T cells were examined by [3H]thymidine incorperation assay. Human IgG1 was used as a control. The results represent one out of two independent experiments. D. Secretion of sPD-L1 by BRAF inhibitor resistant melanoma cell lines. sPD-L1 from culture medium of either parental or PLX resistant A375 and M34 cell lines were analyzed by immunoprecipitation, SDS-PAGE and immunoblotting assay. Loading samples were normalized by cell numbers. The culture medium were from aproximately 8 × 107 cells of A375 and 1 × 107 cells of M34, respectively. PLX R respresents PLX4032 resistant. E to H. Melanoma cell lines were cultured in the absence and presence of IFNγ (200 U/ml) or IFNα (2000 U/ml), or TNFα (10 ng/ml). sPD-L1 in culture medium was analyzed by immunoprecipitation, SDS-PAGE and immunoblotting assay. Loading samples were normalized by cell numbers.
Over-expression of PD-L1-1 in A375 cells not only increased the membrane-bound form (Supplemental Fig. S2D), but also increased the soluble 38 and 45 kDa bands in the culture medium (Fig. 2B). Secretion of sPD-L1 may result from alternative spliced variants of the PD-L1 transcript.
Biologic activity of sPD-L1
PD-L1 inhibits T cell activation. To assess the function of the sPD-L1 variants, we stimulated PHA-activated human CD4+ and CD8+ T cells with anti-CD3 in the absence or presence of either fusion proteins of PD-L1 variants (Supplemental Fig. S2A and S2B) or human IgG1 for 3 days. Activated CD4+ and CD8+ T cells showed less activation when treated with PD-L1 variants than with human IgG1 (Fig. 2C). Membrane-bound full-length PD-L1 abrogated ML-IAP antigen-specific CD8+ T cell (25) activation and proliferation in A375 and K028 cells (Supplemental Fig. S3A–C). Although not as potent as full-length membrane-bound PD-L1, sPD-L1 variants exhibit inhibitory functions on T cell activation and proliferation.
Alternative spliced variants of V600E BRAF have been reported in BRAF inhibitor resistant melanoma tumors (26). To examine whether BRAF inhibitor resistant melanoma cell lines induced sPD-L1 expression, culture medium from PLX4032-resistant A375 and M34 cells were analyzed. The M34 parental cells produced modest amounts of the 38 and 45kDa sPD-L1, which was increased about 4.6 fold in the PLX4032 resistant M34 cells (Fig. 2D). Although concentrations of the 38 and 45 kDa sPD-L1 variants were relatively high in parental supernatants, expression of the 24 kDa sPD-L1 variant increased about 2-fold in supernatants from PLX4032-resistant A375 cells (Fig. 2D).
To assess the effects of cytokines such as IFNγ (which induces PD-L1) on the secretion of sPD-L1 variants, cell lines A375, K008, K028, and UACC257 were cultured in the absence or presence of IFNγ (200 U/ml ), IFNα (2000 U/ml), or TNFα (10 ng/ml) for 2 days. The 38 and 45 kDa bands of sPD-L1 were increased 4 or 2.3-fold by IFNγ treatment, 2.7 or 1.3-fold by IFNα, and 1.6 or 1.2-fold by TNFα, in supernatants from cell lines A375 and K008, respectively (Fig. 2E and 2F). The 38- and 45-kDa bands of PD-L1 in supernatant from K028 were increased by about 14-fold with IFNγ or TNFα treatments and by 6.6-fold with IFNα treatment (Fig. 2G). The 38- and 45-kDa bands increased in cell line UACC257 supernatants 6.3- or 4.7-fold with IFNγ or IFNα treatments, respectively, and the 24-kDa band increased 2.2-fold with IFNα treatment (Fig. 2H). Different melanoma cell lines appeared to produce different sPD-L1 variants in response to different cytokine treatment.
Cytokine treatment of A375, K028, and UACC257 melanoma cell lines resulted in increased secretion of sPD-L1, as well as increased expression of cell surface PD-L1 and decreased cell proliferation (Fig. 2E, 2G, and 2H, Supplemental Fig. S4A to S4C). K008 cells constitutively express PD-L1, but express sPD-L1 in response to cytokine treatment (Fig. 2F, Supplemental Fig. S4D). Thus, cytokines induced expression of both sPD-L1 and PD-L1 in a dose-dependent manner in melanoma cell lines (Supplemental Fig. S5A to S5F).
The expression of cell surface PD-L1 parallels expression of sPD-L1 in melanoma cell lines without cytokine treatment (Supplemental Fig. S4 Supplemental Fig. S5G), and is inducible by sodium azide (Supplemental Fig. S6). We suggest that secretion or release of sPD-L1 can result from BRAF-resistance mechanisms, cytokine induction, cell stress or cell injury, or cell death.
Development of sPD-L1 ELISA
We developed ELISA assays with two capture antibodies that detect PD-L1 variants. Specificity was assayed with concentrations of recombinant PD-L1 (Novoprotein), PDL2 (Novoprotein), ML-IAP (R&D systems), and human IgG1 (SouthernBiotech, Birmingham, AL) ranging from 0.001 ng/ml to 100 ng/ml (Fig. S7A and S7B). Mouse anti PD-L1 monoclonal antibodies, clone 29E.12B1 and 130021 were both specific and sensitive capture mAbs for PD-L1. The lower limit of detection sensitivity was 0.1 ng/ml of recombinant PD-L1.
The specificity of the two capture ELISA assays for the sPD-L1 variants was also investigated. The pretreatment secretion of sPD-L1 in A375 was below the level of detection. Thus, A375 cells were transduced with either PD-L1-3, or PD-L1-9, or PD-L1-1 and the supernatants were assayed by ELISA (Supplemental Fig. S7C and S7D). Amino acid regions of sPD-L1 variants, amino acid regions of recombinant PD-L1-3/Fc and PD-L1-HIS, and antibody recognition regions are shown in Supplemental Table S1A and S1B, and Supplemental Fig. S8A, respectively. The detection mAb 29E.2A3 recognizes an epitope in the IgV domain that is non-overlapping with the 29E.12B1 epitope (G. Freeman, unpublished). Clone 130021 mAb was able to detect sPD-L1-9 and sPD-L1-1 variants, which are the two longer forms (PD-L1L) with an intact IgC domain, whereas clone 29E.12B1 mAb recognized all three sPD-L1 variants (PD-L1all). To confirm the different recognitions of these two antibodies, recombinant PD-L1-3/Fc and PD-L1-HIS were detected with the antibodies by SDS-PAGE and immunoblotting. Clone 130021 only recognized PD-L1-HIS, which contains the complete extracellular domain found in the long soluble forms (PD-L1L) (Supplemental Fig. S8A to S8D). On the other hand, clone 29E.12B1 detected PD-L1-3/Fc, the shortest form, recognizing an epitope in the IgV domain. Thus clone 29E.12B1 and clone 130021 can distinguish sPD-L1 variants.
Quantitative detection of sPD-L1 concentrations between sera and plasma from the same patient showed no differences in these assays (Supplemental Fig. S9A and S9B).
sPD-L1 levels in melanoma patients
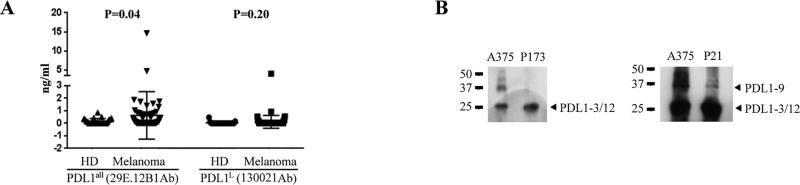
sPD-L1 in samples from twenty-five healthy donors and sixty-five untreated stage IV melanoma patients were analyzed. Concentrations of sPD-L1all were elevated in the plasma of melanoma patients in comparison with healthy donors (P = 0.04, Fig. 3A). Circulating sPD-L1L was undetectable (< 0.1 ng/ml) in most, but not all, melanoma patients, similar to healthy donors.
Figure 3.
sPD-L1 in plasma of melanoma patients. A. sPD-L1 detected in both healthy donors and melanoma patients by ELISA. Data were represented as mean ± SE. B. Immunoprecipitation, SDS-PAGE and immunoblotting analyses on sPD-L1 in plasma of melanoma patients. sPD-L1 from the culture medium of A375 melanoma cell line were used as a positive control.
To confirm the presence of sPD-L1 variants in melanoma patient plasma, immunoblot analysis of the immunoprecipitant from plasma was performed. For example, ELISA assay on patient P173 detected a high concentration (1.87 ng/ml) of sPD-L1all but no (< 0.1 ng/ml) sPD-L1L in pre-treatment plasma. This is consistent with the banding pattern of sPD-L1-3 or sPD-L1-12 (Fig. 3B, left panel). In pretreatment plasma from patient P21, both isoforms of sPD-L1all (14.63 ng/ml) and sPD-L1L (0.94 ng/ml) were detected, which is consistent with the banding pattern when sPD-L1-3, sPD-L1-12, and sPD-L1-9 were assayed by immunoblot (Fig. 3B, right panel).
sPD-L1 levels in melanoma patients on immune checkpoint blockade
To explore the clinical significance of sPD-L1 in patients with melanoma on ipilimumab-based therapy, we first analyzed plasma specimens collected from forty-two patients on the ipilimumab (3mg/kg or 10mg/kg) combined with bevacizumab (7.5mg/kg or 15mg/kg) clinical trial [NCT00790010] (22). Pretreatment levels of sPD-L1 were determined before starting checkpoint immunotherapy. Based on sensitivities and the lower limit of detection of the ELISA assays, pretreatment levels of sPD-L1, and clinical outcomes, the patients were divided into three groups: undetectable/low, moderate, and high sPD-L1. For sPD-L1all, groups were divided into < 0.5 ng/ml (low), ≥ 0.5ng/ml and < 1.4 ng/ml (moderate), and ≥ 1.4 ng/ml (high). For sPD-L1L, groups were divided into < 0.1 ng/ml (low), ≥ 0.1 ng/ml and < 0.5 ng/ml (moderate), and ≥ 0.5 ng/ml (high). All patients with high pretreatment levels of either sPD-L1all or sPD-L1L experienced progressive disease (PD) (Fisher’s exact test P = 0.0015 and 0.025, respectively; Table 1A and 1B, Supplemental Fig. S10).
Table 1.
Associations between pretreatment levels of sPD-L1 and clinical responses. A and B. ipilimumab plus bevacizumab treatment. C and D. Either ipilimumab or ipilimumab plus sargramostim treatment in ECOG 1608 trial. “unevaluable” indicated that the patients deceased in short time and didn’t received evaluation. Statistic results between analyses with and without “unevaluable” were similar. E and F. Anti–PD-1 treatment.
| Association between pretreatment levels of sPD-L1all and clinical responses | ||||
|---|---|---|---|---|
| A Ipilimumab plus bevacizumab | ||||
| sPD-L1all of pre-treatment (baseline, ng/ml) |
PR | SD | PD | Total |
| < 1.4 | 8(21%) | 21(57%) | 8(22%) | 37 |
| ≥ 1.4 | 0(0%) | 0(0%) | 5(100%) | 5 |
| Fisher exact test P=0.0015 | ||||
| C Ipilimumab or ipilimumab plus sargramostim | ||||
|---|---|---|---|---|
| sPD-L1all of pre-treatment (baseline, ng/ml) |
PR | SD | PD | Total |
| < 1.4 | 32(23%) | 32(23%) | 77(54%) | 141 |
| ≥ 1.4 | 0(0%) | 1(10%) | 9(90%) | 10 |
| Fisher exact test P=0.04 | ||||
| E Anti PD-1 antibody | ||||
|---|---|---|---|---|
| sPD-L1all of pre-treatment (baseline, ng/ml) |
PR | SD | PD | Total |
| < 1.4 | 11(34%) | 8(25%) | 13(41%) | 32 |
| ≥ 1.4 | 1(33%) | 0(0%) | 2(67%) | 3 |
| Association between pretreatment levels of sPD-L1L and clinical responses | ||||
|---|---|---|---|---|
| B Ipilimumab plus bevacizumab | ||||
| sPD-L1L of pre-treatment (baseline, ng/ml) |
PR | SD | PD | Total |
| < 0.5 | 8(20%) | 21(54%) | 10(26%) | 39 |
| ≥ 0.5 | 0(0%) | 0(0%) | 3(100%) | 3 |
| Fisher exact test P=0.025 | ||||
| D Ipilimumab or ipilimumab plus sargramostim | ||||
|---|---|---|---|---|
| sPD-L1L of pre-treatment (baseline, ng/ml) |
PR | SD | PD | Total |
| < 0.5 | 30(22%) | 31(22%) | 78(56%) | 139 |
| ≥ 0.5 | 2(17%) | 2(17%) | 8(66%) | 12 |
| Fisher exact test P=0.55 | ||||
| F Anti PD-1 antibody | ||||
|---|---|---|---|---|
| sPD-L1L of pre-treatment (baseline, ng/ml) |
PR | SD | PD | Total |
| < 0.5 | 11(33%) | 8(24%) | 14(43%) | 33 |
| ≥ 0.5 | 1(50%) | 0(0%) | 1(50%) | 2 |
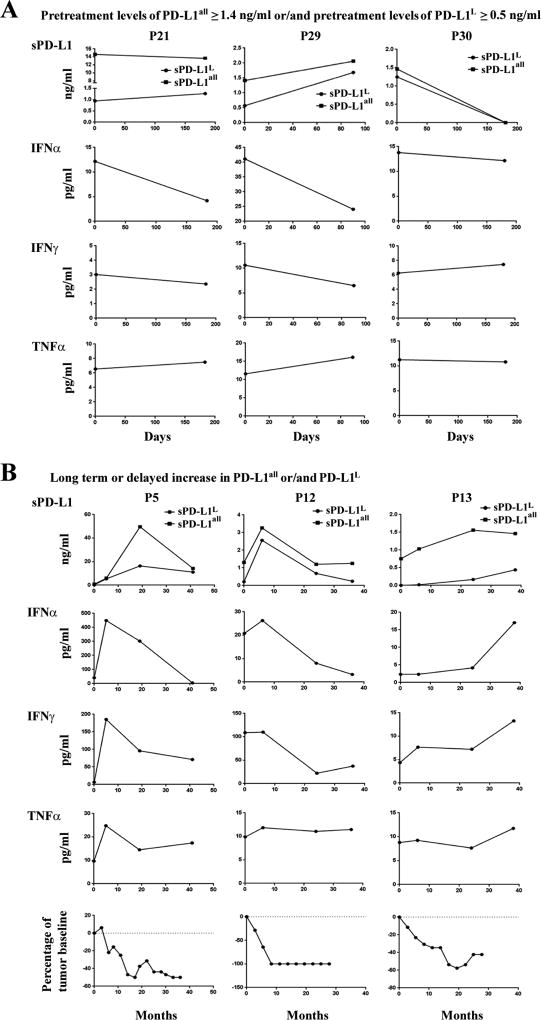
We next sought to investigate kinetic changes of sPD-L1 in patients as a function of treatment. We began with a cohort of patients treated with ipilimumab plus bevacizumab [NCT00790010] (22). Among the five patients with high pretreatment concentrations of sPD-L1all and the three patients with high pretreatment sPD-L1L, patients after treatment showed either unchanged, or increased, or decreased sPD-L1all and sPD-L1L (Fig. 4A, and Supplemental Fig. S10A and S10B). To investigate the associations between sPD-L1 and immunologic responses, we examined the kinetic changes of cytokines in plasma samples in relation to changes in sPD-L1. Patients with high pretreatment concentrations of sPD-L1 showed either decreased or no change in circulating cytokine production after treatment (Fig. 4A, Supplemental Fig. S10A to S10H, and Supplemental Fig. S11), suggesting that high pretreatment concentrations of sPD-L1 may suppress effective antitumor immunity directly or may be a proxy for a state of dominant immune inhibition.
Figure 4.
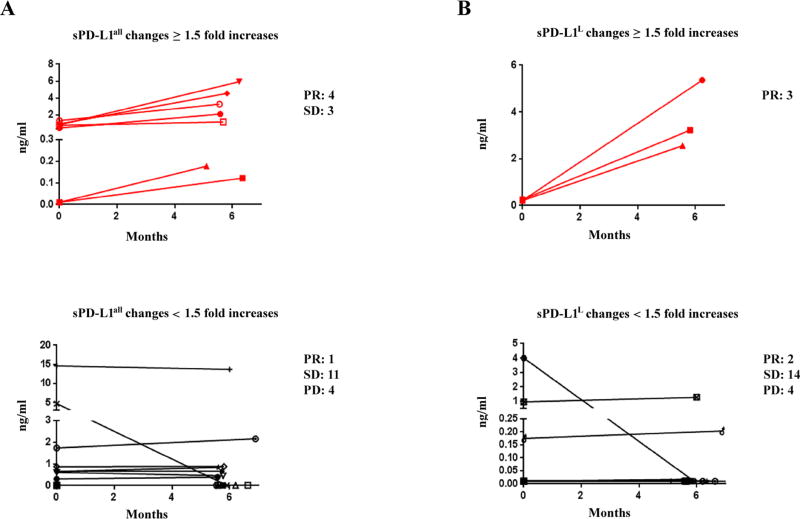
Kinetic changes of sPD-L1 and cytokines, and kinetic radiographic reduction in tumor size after ipilimumab plus bevacizumab treatment. A. Patients with pretreatment levels ≥ 1.4 ng/ml sPD-L1all and/or ≥ 0.5 ng/ml sPD-L1L. B. Patients with ≥ 1.5-fold increases in sPD-L1all and/or sPD-L1L after 5 months of treatment.
Whether the increases in sPD-L1 may be a pharmacodynamic marker and related to clinical outcomes regardless of the pretreatment concentration of sPD-L1 was next explored. Fourteen patients experienced ≥ 1.5-fold increases in either sPD-L1all or sPD-L1L following treatment with ipilimumab-bevacizumab (Fig. 4B, Fig. 5, Supplemental Fig. S10A to S10F). Four of five patients who had ≥ 1.5-fold increases in either sPD-L1all or sPD-L1L within 4.5 months showed progressive disease (Supplemental Fig. S10A to S10F). Eight patients who had increases in either sPD-L1all or PD-L1L after 5 months of treatment experienced favorable clinical responses and corresponding increases in cytokines, and patients with <1.5-fold increases in sPD-L1 experienced less favorable outcomes (Fig. 4B, Fig. 5, Supplemental Fig. S10A to S10F, and Supplemental Fig. S11). Landmark analyses indicated significant associations between the increases of sPD-L1 and partial responses (Fisher’s exact test P = 0.02 and 0.006 for sPD-L1all and sPD-L1L analyses, respectively, Fig. 5A and 5B, and Supplemental Fig. S10G and S10H). The peak post-treatment sPD-L1 corresponded with clinical disease regressions (Fig. 4B, bottom). In patients without increasing sPD-L1, increases in cytokines after 5 months of treatment were associated with stable disease (Supplemental Fig. S10 and Supplemental Fig. S11). We suggest that functional immune activation as assessed by induction of sPD-L1, can be associated with cytokine production from the immune activation that resulted from treatment.
Figure 5.
sPD-L1 in plasma of melanoma patients receiving ipilimumab plus bevacizumab treatment. A and B. Comparision between long term or delayed increases and non increase in sPD-L1 in melanoma patients receiving ipilimumab plus bevacizumab treatment. Red line indicates a greater than 1.5-fold increases in sPD-L1 after 5 months of treatment. Black line indicates < 1.5-fold increases in sPD-L1 after 5 months of treatment. Fisher exact tests were performed based on window from 5 to 7 months post-treatment (Supplemental Fig. S10G and S10H). P < 0.05 are considered statistically significant.
To further investigate the role of sPD-L1 related to ipilimumab alone, twenty-three patients who received ipilimumab (dose 3 mg/kg) were studied. Two patients with high pretreatment concentrations of sPD-L1all experienced progressive disease. The single patient with high pretreatment sPD-L1L experienced stable disease, but only had 12 months survival (Supplemental Fig. S12A to D). Only two patients had partial responses: patients P187 and P169 (Supplemental Fig. S12C to 12H). Patient P187 had a >1.5-fold increase of sPD-L1all at 4 months. Although this patient survived 29 months, no further samples were available. Patient P169’s sPD-L1all/L both decreased at 8 months after therapy. Upon closer exploration of the levels of sPD-L1all/L after the initial analyses, the treatment resulted in a 1.5-fold increase in the calculated shortest variant of sPD-L1 (red line, Supplemental Fig. S12I). Despite limits of sample availability, these data support our findings in the ipilimumab-bevacizumab treated cohort.
The associations between sPD-L1 and clinical responses were further validated in 151 patients treated on the ECOG 1608 trial (randomized phase II of ipilimumab 10mg/kg plus sargramostim versus ipilimumab 10mg/kg, NCT01134614) (23). Nine of ten patients with high pretreatment levels of sPD-L1all experienced progressive disease. Eight of twelve patients with high pretreatment levels of sPD-L1L experienced progressive disease (Fisher’s exact test P = 0.04 and P = 0.55, respectively, Table 1C and 1D).
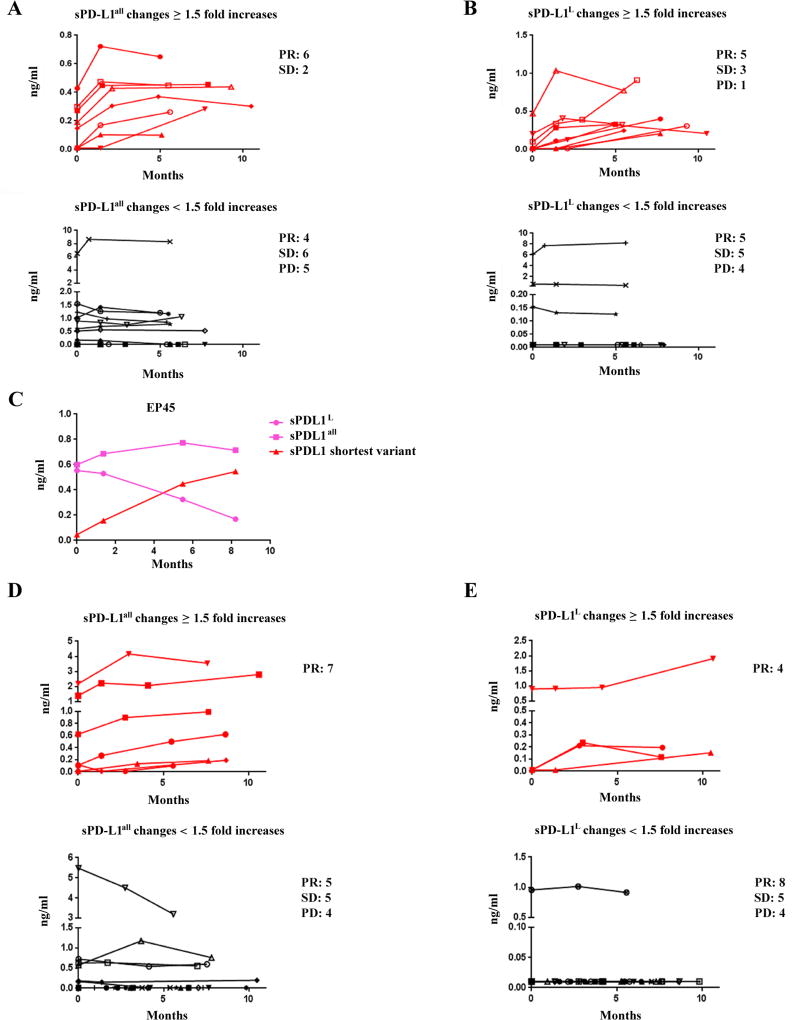
Next, we focused on kinetic changes of sPD-L1 in within the ipilimumab-only arm (73 patients). Many patients with ≥ 1.5-fold increase in sPD-L1 within 4.5 months suffered progressive disease (Supplemental Fig. S13A to S13F). Patients with ≥ 1.5-fold increases in sPD-L1all after 5 month of treatment were significantly associated with partial responses, compared to patients with < 1.5-fold increases (75% versus 27%, Fisher’s exact test P = 0.039, Fig. 6A and Supplemental Fig. S13G). Patients with ≥ 1.5-fold increases in sPD-L1L after 5 months of treatment did not show significant associations with partial responses (56% versus 36%, Fisher’s exact test P = 0.42, Fig. 6B and Supplemental Fig. S13H). These data are consistent with our findings in the ipilimumab-bevacizumab cohort. Patient EP45, who had a subtle increase of sPD-L1all (< 1.5-fold increase), developed a partial response. Secondary review noted a relative decrease in sPD-L1L, which resulted in ≥ 1.5-fold increase in the calculated shortest variant of sPD-L1 (shown in Fig. 6C).
Figure 6.
sPD-L1 in sera of melanoma patients receiving either ipilimumab in ECOG 1608 trial or anti–PD-1 antibody. A and B. Comparision between long term or delayed increases and non increase in sPD-L1 in melanoma patients receiving ipilimumab. C. Different charectors in secretion of sPD-L1 in patient EP45 after ipilimumab treatment. D and E. Comparision between long term or delayed increases and non increase in sPD-L1 in melanoma patients receiving anti–PD-1 antibody. Red line indicates a greater than 1.5-fold increases in sPD-L1 after 5 months of treatment, and black line indicates < 1.5-fold increases in sPD-L1 after 5 months of treatment (shown in A, B, D, and E). Pink lines represent changes of sPD-L1all and sPD-L1L, and a red line stands for a greater than 1.5-fold increases in the shortest sPD-L1 variant (shown in C). Fisher exact tests were performed based on window from 5 to 11 months post-treatment (Supplemental Fig. S13G and S13H, and Supplemental Fig. S14G and S14H). P < 0.05 are considered statistically significant.
To further investigate whether the sPD-L1 relationship between clinical responses was restricted to ipilimumab-based treatment, samples were assayed from 35 patients treated with PD-1 blocking antibody. Four patients had high pretreatment concentrations of either sPD-L1all or sPD-L1L, and two of these patients experienced progressive disease (Table 1E and 1F; Supplemental Fig. S14A and S14B). Kinetic changes of sPD-L1 as a function of treatment were also examined. All eight patients with ≥ 1.5-fold increases in sPD-L1all after 5 months of treatment experienced partial responses, and all four patients with ≥ 1.5-fold increases in sPD-L1L after 5 months of treatment experienced partial responses (Fisher’s exact test P = 0.007 and P = 0.103, respectively; Fig. 6D and 6E; Supplemental Fig. S14A to S14H). These data are consistent with our findings from the ipilimumab treatment cohorts. However, in contrast to that seen following ipilimumab, there were no increases in circulating cytokines in these patients whether or not they had increases in sPD-L1 (Supplemental Fig. S15).
DISCUSSION
Great advances in the treatment of patients with metastatic melanoma have been realized over the last decade, in large part due to the development of immune checkpoint inhibitors. Patients that survive three years after ipilimumab therapy are likely to experience continued benefit with up to ten years of follow up (27). PD-1 blockade produces higher response rates than ipilimumab (28). Prognostic and predictive biomarkers for which patients will fail therapy early and which patients will develop durable clinical benefit are needed. The development of future treatments will need to account for the dynamic changes in immune responses as a function of treatment.
Elevated sPD-L1 has been associated with worse clinical outcomes in both renal cell carcinoma and multiple myeloma. Studying metastatic melanoma, here we identified multiple splice variants of PD-L1 and report elevated sPD-L1 in sera from metastatic melanoma patients. Patients with the highest pretreatment sPD-L1 concentrations had a tendency toward rapidly progressive disease after ipilimumab-based therapy. Early changes in sPD-L1 concentrations after checkpoint inhibition therapy did not correspond with benefit. However, rise in sPD-L1 after 5 months of treatment correlated with partial responses in cohorts of patients treated with ipilimumab-based therapy. Rise in sPD-L1 after anti–PD-1 treatment was also associated with partial responses. Changes in circulating cytokines, corresponding with changes in sPD-L1 after anti-CTLA-4-based therapy, were not seen in patients receiving PD-1 blockade, suggesting different mechanisms for increasing circulating sPD-L1 are associated with different immune checkpoint inhibitors.
The sources of sPD-L1 in patients with cancer remain unclear. We found that alternative splicing of PD-L1 occurs in all melanoma cell lines. Alternative splicing of membrane proteins can result in new in-frame stop codons before the transmembrane domain and lead to the secretion of splice variants (29–32). Indeed, three splice variants lack the transmembrane domain and result in the secretion of sPD-L1. In one variant, loss of the intracellular domain due to splicing also led to secretion, suggesting that the intracellular domain may stabilizePD-L1 on the cell surface.
Aberrant alternative splicing activities occur in a variety of cancers, and these events impact the expression of transcription factors, cell signaling factors, and membrane proteins (29). Functional changes of these spliced proteins are involved in the development, proliferation, and metastasis of cancers (29, 33, 34). We show here that such splicing activities for PD-L1 affect metastatic melanoma. High pretreatment concentrations of sPD-L1 that were associated with worse outcomes to checkpoint blockade may reflect the extent of increased splicing activities in melanoma cells as well as tumor burden. This idea is supported by the increased splice variants of sPD-L1 observed in BRAF inhibitor resistant melanoma cell lines. Selective pressures from BRAF inhibitors are also associated with induced splicing variants of BRAF V600E (26).
The balance between a pro-tumor inflammation and anti-tumor immune response is complex. Inflammation is a hallmark of cancer (35) and can cause tumor progression (36, 37). Cytokines also can produce an anti-tumor immune response and are essential for anti-tumor immune effector function (36, 38). As part of the regulatory homeostatic response, cytokines such as IFNγ and TNFα induce expression of PD-L1 in a variety of cancer cells (39–42). CD8+ T cells at tumor sites correlate with PD-L1 expression and clinical responses (43, 44). Our data demonstrated that cytokines such as IFNα, IFNγ, and TNFα increase splicing activities of PD-L1 leading to secretion of sPD-L1 directly by tumor cells. PD-L1 cell surface expression was parallel to sPD-L1 secretion in response to cytokines in melanoma cancer cells, and patients with moderate pretreatment levels of sPD-L1 experienced favorable clinical outcomes. In addition, most patients with favorable clinical responses showed detectable levels of cytokines in their pre-treatment sera. Moderate pretreatment levels of sPD-L1 may indicate existing anti-tumor immune responses in some patients. Pretreatment sPD-L1 concentrations may be related to pre-existing protumor inflammatory responses.
Circulating sPD-L1 in the sera of patients may have multiple sources produced by distinct mechanisms. These include intrinsic splicing activities in tumor cells, protumor inflammatory responses, and antitumor immune responses. Patients with high pretreatment sPD-L1 had poor outcomes, perhaps due to large tumor burden, increased aberrant splicing activities in tumor cells, or an exhausted antitumor immune response, which may be difficult to overcome with single checkpoint blockade. As a result, qualitative differences in pretreatment sPD-L1 concentrations could represent either favorable or unfavorable factors for clinical outcomes depending on the source of sPD-L1. Favorable clinical outcomes with immune checkpoint blockade occurred predominately in patients with moderate to low pretreatment sPD-L1. Moderate sPD-L1 may reflect pre-existing antitumor immune responses. Although clinical responses in patients with moderate pretreatment sPD-L1 concentrations were most impressive in the ipilimumab plus bevacizumab trial, this was not seen in the ipilimumab arm of the ECOG 1608 trial. However, cross trial comparisons in the cohorts may not be appropriate, given that it would be comparing different combinations and different treatment doses in nonrandomized cohorts. Multiplex analysis of pretreatment sPD-L1 and cytokine concentrations in patients may help to distinguish PD-L1 splicing activities from tumor cells versus inflammatory responses.
We show that circulating sPD-L1 levels frequently increase as the result of treatment with checkpoint blockade. Patients with long term or delayed increase in sPD-L1 correspond with clinically beneficial outcomes. This scenario occurred in around 70% of total patients with partial responses. Furthermore, sPD-L1 concentrations are associated with concentrations of circulating cytokines after ipilimumab-based therapy. Since secretion of sPD-L1 can result from cytokine induction, cell stress, cell injury, and cell death, increases in sPD-L1 by checkpoint blockade may indirectly reflect antitumor immune responses. Long-term or delayed increases in sPD-L1 following treatment suggests that the anti-tumor immune responses can overcome the negative barriers of PD-L1 and other potential immune suppressive factors in the tumor microenvironment. Short term increases in sPD-L1 from treatment were associated with progressive disease and shorter survival in subsets of patients. This suggests a challenge in the early evaluation of sPD-L1 rise and discerning whether rising sPD-L1 is because of tumor progression or activated anti-tumor immune responses, and determining whether anti-tumor immune responses could overcome immune suppressive barriers in long term. Based on our initial analyses of four cohorts of patients, the minimum time needed to distinguish whether an increase in sPD-L1 increases the likelihood of clinical benefit is approximately 5 months. Further detailed collection of long term samples from both responding and non-responding patients is needed to investigate association of sPD-L1 changes and clinical outcomes. Preliminary studies on this were investigated (Supplemental Fig. S16 and S17). Increases in sPD-L1 concentration occurred with both CTLA-4 and PD-1 blockade, indicating that sPD-L1 concentration may be an indicator of beneficial clinical outcomes. Patients receiving CTLA-4 blockade showed associations between increases of sPD-L1 and cytokine production, whereas this association was not observed in patients receiving PD-1 blockade alone. This may be related to the mechanism of action for these drugs, with anti-CTLA-4 acting globally on immune function and anti–PD-1 affecting primarily the tumor microenvironment.
Although tumors that express PD-L1 may produce secreted splice variants detectable in the circulation, the tumor may not be the only source of sPD-L1. Our and others’ (20) data (Supplemental Fig. S18) also indicated that sPD-L1 can be secreted from human myeloid DC in the presence of cytokines and LPS during DC maturation. sPD-L1 from myeloid DC in patients without treatment can’t be ruled out. However, secretion of sPD-L1 results from cytokine presence, cell stress, cell injury and cell death, and long term or delayed increases in sPD-L1 were associated with favorable clinical outcomes during treatment with checkpoint inhibitors, and the peak post-treatment sPD-L1 corresponded with clinical tumor regression. In contrast, human myeloid DC expressed high levels of PD-L1 and suppressed T cell activation, and T cells conditioned with PD-L1 blocked myeloid DC inhibited human ovarian carcinoma growth (45). We suggest that sPD-L1 mainly originated from the tumor in patients who had favorable clinical responses after checkpoint blockade.
sPD-L1 secretion in melanoma is associated with three major splice variants that may be differentially secreted. In addition, secretion of sPD-L1 may more precisely reflect cytokine stimulations, since there are dissociations between constitutive expression of membrane-bound PD-L1 and increases in sPD-L1. PD-L1 variants should be taken into consideration in assay development as there may be difference in biological significance across variants, although sPD-L1all is more significantly associated with clinical outcomes. We found no associations of sPD-L1 with age, gender, M-stage and LDH in the current evaluated patient cohorts (Supplemental Table S2). These data suggest that high sPDL1 and LDH concentrations may be not related in terms of prognostic or predictive values. It remains unclear whether sPD-L1 is a clinically significant systemic immunosuppressant.
Immune therapy is not as effective in patients with symptomatic, rapidly progressive disease. Although the number of patients with high sPD-L1 prior to anti–PD-1 therapy is small, we found it interesting that some of these benefited from PD-1 blockade. In patients for whom we had plasma samples after 5 months on therapy, we found that those who had 1.5-fold increases in sPD-L1all plasma concentrations were more likely to achieve partial responses not only to ipilimumab or ipilimumab-bevacizumab treatment, but also anti–PD-1 therapy. Prospective validation of the prognostic and kinetics of sPD-L1 alone or in the context of a multiple assay incorporating cytokines such as IFNs and TNF may help establish whether kinetic changes in sPD-L1 after immune checkpoint are effective in predicting not only RECIST criteria responses, but also durable clinical benefit.
In summary, sPD-L1 exists as several variants that can originate from both tumor and immune cells. sPD-L1 may serve as a meaningful and practical dynamic biomarker for the prediction of durable efficacy to immunotherapy agents alone or in the context of additional factors. Soluble circulating factors offer a practical means for monitoring patient outcomes while improving our mechanistic understanding.
Supplementary Material
Acknowledgments
F.S.H has served as a consultant for Merck, Genentech, Novartis, and EMD Serono, and has received research support to institution from Bristol-Myers Squibb. Patent application for soluble PD-L1 as a biomarker is pending.
A portion of this study was coordinated by the ECOG-ACRIN Cancer Research Group (Robert L. Comis, MD and Mitchell D. Schnall, MD, PhD, Group Co-Chairs) and supported in part by Public Health Service Grants CA180794, CA180820, CA21115, CA66636, CA23318, CA180867, CA39229, CA180844, and P50CA101942 (GF) from the National Cancer Institute, National Institutes of Health and the Department of Health and Human Services. Its content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute. KM was supported by Claudia Adams Barr Program for Innovative Cancer Research, the 2014 AACR Basic Cancer Research Fellowship, Grant Number 14-40-01-MAHO, and the ASCO Young Investigator Award supported by Kidney Cancer Association. Funding. FSH was supported by NIH CA143832, the Melanoma Research Alliance, the Sharon Crowley Martin Memorial Fund for Melanoma Research and the Malcolm and Emily Mac Naught Fund for Melanoma Research at Dana-Farber Cancer Institute, Genentech/Roche, and Bristol-Myers Squibb.
Footnotes
No other authors report any conflicts of interests.
References
- 1.Keir ME, Butte MJ, Freeman GJ, Sharpe AH. PD-1 and its ligands in tolerance and immunity. Annual review of immunology. 2008;26:677–704. doi: 10.1146/annurev.immunol.26.021607.090331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sharpe AH, Freeman GJ. The B7-CD28 superfamily. Nature reviews Immunology. 2002;2:116–126. doi: 10.1038/nri727. [DOI] [PubMed] [Google Scholar]
- 3.Thompson RH, Gillett MD, Cheville JC, Lohse CM, Dong H, Webster WS, Krejci KG, Lobo JR, Sengupta S, Chen L, Zincke H, Blute ML, Strome SE, Leibovich BC, Kwon ED. Costimulatory B7-H1 in renal cell carcinoma patients: Indicator of tumor aggressiveness and potential therapeutic target. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:17174–17179. doi: 10.1073/pnas.0406351101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Inman BA, Sebo TJ, Frigola X, Dong H, Bergstralh EJ, Frank I, Fradet Y, Lacombe L, Kwon ED. PD-L1 (B7-H1) expression by urothelial carcinoma of the bladder and BCG-induced granulomata: associations with localized stage progression. Cancer. 2007;109:1499–1505. doi: 10.1002/cncr.22588. [DOI] [PubMed] [Google Scholar]
- 5.Hamanishi J, Mandai M, Iwasaki M, Okazaki T, Tanaka Y, Yamaguchi K, Higuchi T, Yagi H, Takakura K, Minato N, Honjo T, Fujii S. Programmed cell death 1 ligand 1 and tumor-infiltrating CD8+ T lymphocytes are prognostic factors of human ovarian cancer. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:3360–3365. doi: 10.1073/pnas.0611533104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ghebeh H, Tulbah A, Mohammed S, Elkum N, Bin Amer SM, Al-Tweigeri T, Dermime S. Expression of B7-H1 in breast cancer patients is strongly associated with high proliferative Ki-67-expressing tumor cells. International journal of cancer Journal international du cancer. 2007;121:751–758. doi: 10.1002/ijc.22703. [DOI] [PubMed] [Google Scholar]
- 7.Gadiot J, Hooijkaas AI, Kaiser AD, van Tinteren H, van Boven H, Blank C. Overall survival and PD-L1 expression in metastasized malignant melanoma. Cancer. 2011;117:2192–2201. doi: 10.1002/cncr.25747. [DOI] [PubMed] [Google Scholar]
- 8.Hino R, Kabashima K, Kato Y, Yagi H, Nakamura M, Honjo T, Okazaki T, Tokura Y. Tumor cell expression of programmed cell death-1 ligand 1 is a prognostic factor for malignant melanoma. Cancer. 2010;116:1757–1766. doi: 10.1002/cncr.24899. [DOI] [PubMed] [Google Scholar]
- 9.Chen BJ, Chapuy B, Ouyang J, Sun HH, Roemer MG, Xu ML, Yu H, Fletcher CD, Freeman GJ, Shipp MA, Rodig SJ. PD-L1 expression is characteristic of a subset of aggressive B-cell lymphomas and virus-associated malignancies. Clinical cancer research. 2013;19:3462–3473. doi: 10.1158/1078-0432.CCR-13-0855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wong RM, Scotland RR, Lau RL, Wang C, Korman AJ, Kast WM, Weber JS. Programmed death-1 blockade enhances expansion and functional capacity of human melanoma antigen-specific CTLs. International immunology. 2007;19:1223–1234. doi: 10.1093/intimm/dxm091. [DOI] [PubMed] [Google Scholar]
- 11.Wang W, Lau R, Yu D, Zhu W, Korman A, Weber J. PD1 blockade reverses the suppression of melanoma antigen-specific CTL by CD4+ CD25(Hi) regulatory T cells. International immunology. 2009;21:1065–1077. doi: 10.1093/intimm/dxp072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brahmer JR, Drake CG, Wollner I, Powderly JD, Picus J, Sharfman WH, Stankevich E, Pons A, Salay TM, McMiller TL, Gilson MM, Wang C, Selby M, Taube JM, Anders R, Chen L, Korman AJ, Pardoll DM, Lowy I, Topalian SL. Phase I study of single-agent anti-programmed death-1 (MDX-1106) in refractory solid tumors: safety, clinical activity, pharmacodynamics, and immunologic correlates. Journal of clinical oncology. 2010;28:3167–3175. doi: 10.1200/JCO.2009.26.7609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, Powderly JD, Carvajal RD, Sosman JA, Atkins MB, Leming PD, Spigel DR, Antonia SJ, Horn L, Drake CG, Pardoll DM, Chen L, Sharfman WH, Anders RA, Taube JM, McMiller TL, Xu H, Korman AJ, Jure-Kunkel M, Agrawal S, McDonald D, Kollia GD, Gupta A, Wigginton JM, Sznol M. Safety, activity, and immune correlates of anti–PD-1 antibody in cancer. The New England journal of medicine. 2012;366:2443–2454. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ansell SM, Lesokhin AM, Borrello I, Halwani A, Scott EC, Gutierrez M, Schuster SJ, Millenson MM, Cattry D, Freeman GJ, Rodig SJ, Chapuy B, Ligon AH, Zhu L, Grosso JF, Kim SY, Timmerman JM, Shipp MA, Armand P. PD-1 blockade with nivolumab in relapsed or refractory Hodgkin's lymphoma. The New England journal of medicine. 2015;372:311–319. doi: 10.1056/NEJMoa1411087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Powles T, Eder JP, Fine GD, Braiteh FS, Loriot Y, Cruz C, Bellmunt J, Burris HA, Petrylak DP, Teng SL, Shen X, Boyd Z, Hegde PS, Chen DS, Vogelzang NJ. MPDL3280A (anti–PD-L1) treatment leads to clinical activity in metastatic bladder cancer. Nature. 2014;515:558–562. doi: 10.1038/nature13904. [DOI] [PubMed] [Google Scholar]
- 16.Herbst RS, Soria JC, Kowanetz M, Fine GD, Hamid O, Gordon MS, Sosman JA, McDermott DF, Powderly JD, Gettinger SN, Kohrt HE, Horn L, Lawrence DP, Rost S, Leabman M, Xiao Y, Mokatrin A, Koeppen H, Hegde PS, Mellman I, Chen DS, Hodi FS. Predictive correlates of response to the anti–PD-L1 antibody MPDL3280A in cancer patients. Nature. 2014;515:563–567. doi: 10.1038/nature14011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang L, Wang H, Chen H, Wang WD, Chen XQ, Geng QR, Xia ZJ, Lu Y. Serum levels of soluble programmed death ligand 1 predict treatment response and progression free survival in multiple myeloma. Oncotarget. 2015;6:41228–41236. doi: 10.18632/oncotarget.5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frigola X, Inman BA, Lohse CM, Krco CJ, Cheville JC, Thompson RH, Leibovich B, Blute ML, Dong H, Kwon ED. Identification of a soluble form of B7-H1 that retains immunosuppressive activity and is associated with aggressive renal cell carcinoma. Clinical cancer research. 2011;17:1915–1923. doi: 10.1158/1078-0432.CCR-10-0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rossille D, Gressier M, Damotte D, Maucort-Boulch D, Pangault C, Semana G, Le Gouill S, Haioun C, Tarte K, Lamy T, Milpied N, Fest T Groupe Ouest-Est des Leucemies et Autres Maladies du S, Groupe Ouest-Est des Leucemies et Autres Maladies du S. High level of soluble programmed cell death ligand 1 in blood impacts overall survival in aggressive diffuse large B-Cell lymphoma: results from a French multicenter clinical trial. Leukemia. 2014;28:2367–2375. doi: 10.1038/leu.2014.137. [DOI] [PubMed] [Google Scholar]
- 20.Frigola X, Inman BA, Krco CJ, Liu X, Harrington SM, Bulur PA, Dietz AB, Dong H, Kwon ED. Soluble B7-H1: differences in production between dendritic cells and T cells. Immunology letters. 2012;142:78–82. doi: 10.1016/j.imlet.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He XH, Xu LH, Liu Y. Identification of a novel splice variant of human PD-L1 mRNA encoding an isoform-lacking Igv-like domain. Acta pharmacologica Sinica. 2005;26:462–468. doi: 10.1111/j.1745-7254.2005.00086.x. [DOI] [PubMed] [Google Scholar]
- 22.Hodi FS, Lawrence D, Lezcano C, Wu X, Zhou J, Sasada T, Zeng W, Giobbie-Hurder A, Atkins MB, Ibrahim N, Friedlander P, Flaherty KT, Murphy GF, Rodig S, Velazquez EF, Mihm MC, Jr, Russell S, DiPiro PJ, Yap JT, Ramaiya N, Van den Abbeele AD, Gargano M, McDermott D. Bevacizumab plus Ipilimumab in Patients with Metastatic Melanoma. Cancer immunology research. 2014;2:632–642. doi: 10.1158/2326-6066.CIR-14-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hodi FS, Lee S, McDermott DF, Rao UN, Butterfield LH, Tarhini AA, Leming P, Puzanov I, Shin D, Kirkwood JM. Ipilimumab plus sargramostim vs ipilimumab alone for treatment of metastatic melanoma: a randomized clinical trial. Jama. 2014;312:1744–1753. doi: 10.1001/jama.2014.13943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Contal C, O'Quigley J. An application of changepoint methods in studying the effect of age on survival in breast cancer. Computational Statistics & Data Analysis. 1999;30:253–270. [Google Scholar]
- 25.Schmollinger JC, Vonderheide RH, Hoar KM, Maecker B, Schultze JL, Hodi FS, Soiffer RJ, Jung K, Kuroda MJ, Letvin NL, Greenfield EA, Mihm M, Kutok JL, Dranoff G. Melanoma inhibitor of apoptosis protein (ML-IAP) is a target for immune-mediated tumor destruction. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:3398–3403. doi: 10.1073/pnas.0530311100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poulikakos PI, Persaud Y, Janakiraman M, Kong X, Ng C, Moriceau G, Shi H, Atefi M, Titz B, Gabay MT, Salton M, Dahlman KB, Tadi M, Wargo JA, Flaherty KT, Kelley MC, Misteli T, Chapman PB, Sosman JA, Graeber TG, Ribas A, Lo RS, Rosen N, Solit DB. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E) Nature. 2011;480:387–390. doi: 10.1038/nature10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schadendorf D, Hodi FS, Robert C, Weber JS, Margolin K, Hamid O, Patt D, Chen TT, Berman DM, Wolchok JD. Pooled Analysis of Long-Term Survival Data From Phase II and Phase III Trials of Ipilimumab in Unresectable or Metastatic Melanoma. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2015;33:1889–1894. doi: 10.1200/JCO.2014.56.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Larkin J, Hodi FS, Wolchok JD. Combined Nivolumab and Ipilimumab or Monotherapy in Untreated Melanoma. The New England journal of medicine. 2015;373:1270–1271. doi: 10.1056/NEJMc1509660. [DOI] [PubMed] [Google Scholar]
- 29.Venables JP. Aberrant and alternative splicing in cancer. Cancer research. 2004;64:7647–7654. doi: 10.1158/0008-5472.CAN-04-1910. [DOI] [PubMed] [Google Scholar]
- 30.Tang W, Gunn TM, McLaughlin DF, Barsh GS, Schlossman SF, Duke-Cohan JS. Secreted and membrane attractin result from alternative splicing of the human ATRN gene. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:6025–6030. doi: 10.1073/pnas.110139897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gower HJ, Barton CH, Elsom VL, Thompson J, Moore SE, Dickson G, Walsh FS. Alternative splicing generates a secreted form of N-CAM in muscle and brain. Cell. 1988;55:955–964. doi: 10.1016/0092-8674(88)90241-3. [DOI] [PubMed] [Google Scholar]
- 32.van der Voort R, Verweij V, de Witte TM, Lasonder E, Adema GJ, Dolstra H. An alternatively spliced CXCL16 isoform expressed by dendritic cells is a secreted chemoattractant for CXCR6+ cells. Journal of leukocyte biology. 2010;87:1029–1039. doi: 10.1189/jlb.0709482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frasca F, Pandini G, Scalia P, Sciacca L, Mineo R, Costantino A, Goldfine ID, Belfiore A, Vigneri R. Insulin receptor isoform A, a newly recognized, high-affinity insulin-like growth factor II receptor in fetal and cancer cells. Molecular and cellular biology. 1999;19:3278–3288. doi: 10.1128/mcb.19.5.3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vella V, Pandini G, Sciacca L, Mineo R, Vigneri R, Pezzino V, Belfiore A. A novel autocrine loop involving IGF-II and the insulin receptor isoform-A stimulates growth of thyroid cancer. The Journal of clinical endocrinology and metabolism. 2002;87:245–254. doi: 10.1210/jcem.87.1.8142. [DOI] [PubMed] [Google Scholar]
- 35.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 36.Dranoff G. Cytokines in cancer pathogenesis and cancer therapy. Nature reviews Cancer. 2004;4:11–22. doi: 10.1038/nrc1252. [DOI] [PubMed] [Google Scholar]
- 37.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chada S, Ramesh R, Mhashilkar AM. Cytokine- and chemokine-based gene therapy for cancer. Current opinion in molecular therapeutics. 2003;5:463–474. [PubMed] [Google Scholar]
- 39.Dong H, Strome SE, Salomao DR, Tamura H, Hirano F, Flies DB, Roche PC, Lu J, Zhu G, Tamada K, Lennon VA, Celis E, Chen L. Tumor-associated B7-H1 promotes T-cell apoptosis: a potential mechanism of immune evasion. Nature medicine. 2002;8:793–800. doi: 10.1038/nm730. [DOI] [PubMed] [Google Scholar]
- 40.Abiko K, Mandai M, Hamanishi J, Yoshioka Y, Matsumura N, Baba T, Yamaguchi K, Murakami R, Yamamoto A, Kharma B, Kosaka K, Konishi I. PD-L1 on tumor cells is induced in ascites and promotes peritoneal dissemination of ovarian cancer through CTL dysfunction. Clinical cancer research : an official journal of the American Association for Cancer Research. 2013;19:1363–1374. doi: 10.1158/1078-0432.CCR-12-2199. [DOI] [PubMed] [Google Scholar]
- 41.Lee SJ, Jang BC, Lee SW, Yang YI, Suh SI, Park YM, Oh S, Shin JG, Yao S, Chen L, Choi IH. Interferon regulatory factor-1 is prerequisite to the constitutive expression and IFN-gamma-induced upregulation of B7-H1 (CD274) FEBS letters. 2006;580:755–762. doi: 10.1016/j.febslet.2005.12.093. [DOI] [PubMed] [Google Scholar]
- 42.Liu J, Hamrouni A, Wolowiec D, Coiteux V, Kuliczkowski K, Hetuin D, Saudemont A, Quesnel B. Plasma cells from multiple myeloma patients express B7-H1 (PD-L1) and increase expression after stimulation with IFN-{gamma} and TLR ligands via a MyD88-, TRAF6-, and MEK-dependent pathway. Blood. 2007;110:296–304. doi: 10.1182/blood-2006-10-051482. [DOI] [PubMed] [Google Scholar]
- 43.Spranger S, Spaapen RM, Zha Y, Williams J, Meng Y, Ha TT, Gajewski TF. Up-regulation of PD-L1, IDO, and T(regs) in the melanoma tumor microenvironment is driven by CD8(+) T cells. Science translational medicine. 2013;5 doi: 10.1126/scitranslmed.3006504. 200ra116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tumeh PC, Harview CL, Yearley JH, Shintaku IP, Taylor EJ, Robert L, Chmielowski B, Spasic M, Henry G, Ciobanu V, West AN, Carmona M, Kivork C, Seja E, Cherry G, Gutierrez AJ, Grogan TR, Mateus C, Tomasic G, Glaspy JA, Emerson RO, Robins H, Pierce RH, Elashoff DA, Robert C, Ribas A. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515:568–571. doi: 10.1038/nature13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Curiel TJ, Wei S, Dong H, Alvarez X, Cheng P, Mottram P, Krzysiek R, Knutson KL, Daniel B, Zimmermann MC, David O, Burow M, Gordon A, Dhurandhar N, Myers L, Berggren R, Hemminki A, Alvarez RD, Emilie D, Curiel DT, Chen L, Zou W. Blockade of B7-H1 improves myeloid dendritic cell-mediated antitumor immunity. Nature medicine. 2003;9:562–567. doi: 10.1038/nm863. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.