Abstract
Genetic association studies have a long history of delivering insightful results for cardiovascular disease (CVD) research. Beginning with early candidate gene studies, to genome-wide association studies, and now on to newer whole-genome sequencing studies, research in human genetics has enriched our understanding of the pathobiology of CVD. As these studies continue to expand, the issue of statistical power plays an important role in study design as well as the interpretation of results. We provide an overview of the component parts that determine statistical power and preview the future of CVD genetic association studies through this lens.
Introduction
Genetic studies have contributed immensely to our knowledge of the etiology and mechanisms that underlie cardiovascular disease (CVD). Family-based studies of hypercholesterolemic patients led to the discovery of the LDLR gene and its role in lipid metabolism and CVD, ultimately leading to mechanistic insights that resulted in the development of statins.1 Other studies discovered rare-mutations in the PCSK9 gene associated with low-density lipoprotein (LDL) cholesterol levels2 and heart disease3,4 hastening the development of several PCKS9 inhibitors that are currently undergoing clinical trials.5 More recently, genome-wide association studies for a wide variety of cardiovascular diseases and traits including coronary artery disease6, atrial fibrillation7, plasma lipids8, and blood pressure9 among others have identified hundreds of associated loci, highlighting important regions of the genome implicated in CVD.
From a clinical perspective, findings from genetic studies can play an important role in genomic medicine by helping to stratify participants in clinical trials based on genotypes at specific markers.10 Genetic discoveries have also enabled disease sub-phenotyping based on an individual’s genetic profile.11 Ultimately, to fulfil the promise of disease prevention in the era of precision medicine, the goal is to build personalized risk-profiles based on genetics, environmental and life-style variables, and clinical measurements.
Implicit in these goals of target identification, drug development, genomic medicine, and precision prevention, is the continuing discovery and refinement of genetic variants that underlie inherited risk for CVD. In this article, we focus on a critical component of genetic association studies of CVD, namely statistical power. Specifically, we provide an historical perspective on the power of gene-mapping studies, an overview of the components of statistical power, and implications for current and future studies.
Fundamental Concepts in Human Genetics
The human genome is comprised of approximately 3.2 billion bases of double-stranded DNA. Humans carry two copies of the genome, one inherited from each parent. Although 99% of the human genome is identical between individuals, many millions of bases of DNA are variable (or “polymorphic”) within human populations.12 Though large, structural changes in DNA play an important role in human phenotypic variation (e.g., deletions and re-arrangements of large sections of a chromosome), the primary focus of many studies of genetic variation takes place at the single-nucleotide level. There are many million known single-nucleotide polymorphisms (SNPs) across the human genome.13 Though most SNPs exert neutral phenotypic consequences, many are thought to be “deleterious” and underlie an individual’s risk for diseases and disorders. Because humans carry two copies of the genome, a SNP is defined by its location on the genome and its two “versions” or alleles. For instance, the rs1260326 SNP is located at the 27,508,073th base on chromosome 2, and both the C and T alleles have been observed in human populations. An individual may carry two copies of the more common allele (i.e., homozygous for the common allele), one copy of the common allele and one copy of the rare allele (i.e., heterozygous), or two copies of the rare allele (i.e., homozygous for the rare allele).
Monogenic and Complex Diseases
In human genetics, a distinction is often drawn between “complex” disease and “Mendelian”, or “monogenic”, disease. “Monogenic” diseases are those in which the disease state in a family is determined by a single mutation (for a variety of reasons, mutations typically differ from family to family even for the same disease). Typically, these are highly penetrant mutations, meaning that individuals who inherit the rare allele have a very high likelihood of developing the disease. When this is the case, the disease “segregates” within a family pedigree in a manner consistent with Mendel’s Law of Segregation, hence the term “Mendelian” disease. In contrast, “complex” diseases are often highly “polygenic” with many genetic variants (that potentially interact) influencing susceptibility. In addition, the mutations that underlie monogenic disorders often occur within the portion of the genome that encodes for proteins. These mutations often “knock-out” the function of a gene, with sometimes drastic phenotypic consequences. Recently, we have found that the variants underlying complex diseases are typically found in non-protein-coding parts of the genome and are thought to affect disease risk through regulatory mechanisms determining how and when genes are expressed.14
The Components of Statistical Power
Genetic Variation
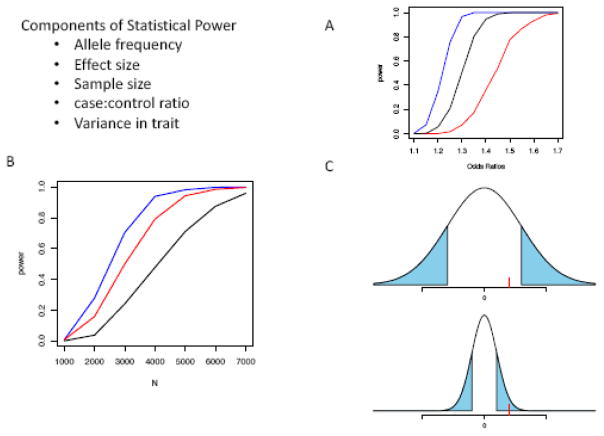
Most genetic variation in the human genome is very rare. For instance, in a recent large-scale DNA sequencing project, the minor allele was observed in only 3 or fewer individuals at approximately 72% of all variable sites.15 This result holds significant implications for gene-mapping studies, because – assuming all other parameters are held constant – the power to detect a true association between the variant and a phenotype of interest decreases as alleles become rarer. Here we define “power” to be the probability of detecting a true association between a SNP and a phenotype of interest. Figure 1 shows the relationship between allele frequency and other components of statistical power.
Figure 1.
Panel A shows the power to detect association (y-axis) by the effect sizes measured in odds-ratios (x-axis) at 3 different minor allele frequencies (0.2 in blue, 0.1, in black, 0.05 in red). Panel B shows powers on the y-axis by sample size (N) on the x-axis at 3 different case:control ratios (1:1 in blue, 1:2 in red, 1:4 in black). Panel C shows the variability in 2 different quantitative traits. The regions colored in blue are the effect sizes that could be detected at a given sample size, allele frequency. The red line represents an effect size of 1, which is easily detected in the trait with smaller variance (bottom) and undetected in the trait with larger variance (top).
Effect Size
In addition to the allele frequency, genetic effect sizes play an important role in statistical power. Effect sizes are typically represented as odds-ratios (for binary phenotypes such as cases and controls) or continuous variables (for quantitative phenotypes). For instance, an odds ratio of 2 implies that the odds of disease increase 2-fold with the addition of each risk allele. As effect sizes increase, power increases as well. In other words, it is easier to detect an association with genetic variants that have larger effects.
When considering statistical power with quantitative phenotypes (for instance, circulating lipids), it is helpful to interpret effect sizes in terms of phenotypic standard deviations. For example, in population-based samples of “healthy” individuals, the standard deviation in plasma LDL-cholesterol and HDL-cholesterol concentration is approximately 30mg/dl and 15mg/dl, respectively.16 Thus an effect size of approximately 30mg/dl for an LDL associated SNP corresponds to approximately one phenotypic standard deviation, whereas an effect size of 30mg/dl for an HDL associated SNP corresponds to approximately 2 phenotypic standard deviations. The HDL associated SNP would be considered as having a much larger effect size (scaled by the standard deviation) than the LDL associated SNP and would thus be much easier to detect in an association study.
Sample Size
Sample sizes also play a crucial role in determining statistical power. All other things being equal (e.g., effect sizes and allele frequencies), larger sample sizes result in higher power. This is because the effect sizes are estimated with higher precision as sample sizes grow, thus resulting in higher power. However, not all increases in sample size are the same (see Figure 1). For instance, given a fixed overall sample size of 1,000, a 1:1 case-control ratio (500 cases and 500 controls) is more powerful than a 1:2 case-control ratio (333 cases and 667 controls). Often, investigators will attempt to enhance the sample size by adding additional controls to a case-control design. However, there are diminishing returns to this strategy. A general rule of thumb is that beyond a 1:10 case-control ratio, it is typically “not worth the trouble” to keep adding controls to the study as any power gains will be minimal.17
Multiple Testing Correction
A final consideration regarding statistical power is the penalty for performing multiple statistical tests. The simplest and most widely used multiple testing penalty is called the “Bonferroni” correction. If a study includes n number of tests (e.g. n number of genetic variants being tested for association with disease) and one is willing to tolerate a 5% chance of observing a false positive (i.e., the typical 5 percent cutoff for declaring “statistical significance”), the cutoff for declaring statistical significance should be p=0.05/n. Because many SNPs are correlated (i.e., they are in linkage disequilibrium) this Bonferroni correction can be overly conservative. To address this issue, the HapMap Consortium used a technique called “permutation testing” to estimate an effective number of independent tests. They found that a significance threshold of 5.5×10−8 is an appropriate cutoff for a typical genome-wide association study (GWAS).18 For newer types of studies that go beyond standard GWAS to investigate many millions of variants (both common and rare in a population), a threshold of 6×10−9 has been suggested.19 These cutoffs are related to power in the following way: the lower the cutoff, the lower the power. Thus, a study that looks only at a single SNP (or a “candidate gene approach”) could declare significance at a cutoff of p=0.05, resulting in much higher power compared to a GWAS that was considering this SNP along with thousands of other SNPs.
Tools for Determining Statistical Power
The Genetic Power Calculator (http://pngu.mgh.harvard.edu/~purcell/gpc/) and the Genetic Association Study Power Calculator (http://csg.sph.umich.edu/abecasis/cats/gas_power_calculator/index.html) are two popular, easy-to-use tools that provide power calculations for a variety of different study designs and significance levels.
Power in Association Studies
Early Candidate Gene Studies
The theoretical and motivational underpinnings of complex genetic association studies have existed for nearly a century.20 However it was not until the development of gene cloning and sequencing that DNA variation maps (initially using restriction-length fragment polymorphisms and later with single nucleotide polymorphisms) could be used in association studies. The high cost and low throughput of genotyping, however, made true genome-wide studies impractical; as a result, the initial rounds of association studies focused on studying candidate genes. This type of study design relies on assumptions about the causal biological pathways underlying a disease and focuses only on testing association with disease for variants in these nominated “candidate genes.”
Limitations of the Candidate Gene Approach
With time, the candidate gene approach revealed substantial limitations. First, because only a few markers are genotyped in a candidate gene study there is essentially no ability to correct for a type of confounding known as population stratification. It is now widely appreciated that genetic variation unrelated to disease can be seemingly correlated to phenotype due to subtle population differences.21 Although genotypes across the genome can successfully be used to correct for this,22 population stratification can lead to false positive results in candidate gene studies because these genome-wide genotypes are not available. Second, given the fact that an individual locus for a complex trait accounts for a small fraction of the heritability of the trait, the statistical power for discovering modest effects is low. When coupled with liberal p-value thresholds (p<0.05 was often used in candidate gene studies), this increases the likelihood of making a false positive discovery. In fact, only a small fraction of candidate gene study results have been subsequently validated in replication studies.23 Finally, and likely most importantly, by design there was no ability to discover novel biology because candidate gene studies only focused on genes thought to be important in the disease. One of the powerful advantages of genome-wide association studies is the unbiased ascertainment and assessment of genetic variation, often revealing associations in unexpected genes thereby revealing novel insights into the pathobiology of disease.
Genome-wide association studies
Technological advances in genotyping technology enabled the first generation of GWAS. Compared to contemporary studies, the initial GWAS had far fewer samples and thus had limited power to detect associations at the 5×10−8 significance level.24,25 In contrast to candidate gene studies, GWAS assume an unbiased or “agnostic” approach across the genome, where all SNPs are weighted equally during association testing. In other words, prior information on the likelihood of a gene or region being related to the trait is ignored. The result of these unbiased scans has implicated many new regions of the genome for which little was previously known.16 In particular as mentioned above, GWAS have generally implicated non protein-coding portions of the genome, suggesting that genetic variation influences phenotypic variation through gene regulatory mechanisms.26
Advantages of GWAS
Along with enforcing standards for unbiased-ness and statistical significance, replication has become an important component of GWAS. The goals of replication can be thought of as 3-fold: to provide convincing statistical evidence for association, to rule out associations due to biases such as technical artifacts, and to refine estimates of effect sizes.27 As a result, GWAS do not suffer from the same “lack of replication” that plagued the candidate-gene era.
As the search for genetic loci underlying complex diseases has progressed, later iterations of GWAS have greatly expanded sample sizes. Because summary statistics can often easily be shared across institutions, meta-analyses have become quite common in this context. With hundreds of thousands of samples, these studies were well powered to discover common variants with large-to-modest effect sizes.8, 28
More recently, imputation-based studies have become a popular design for exploring genetic variants with allele frequencies as low as 0.5%. Genotype imputation (also called “in silico genotyping”) is a statistical technique for predicting genotypes at variants that were not directly measured.29 This technique utilizes a set of reference samples to identify segments of the genome (i.e., haplotypes) that are shared with the study or “target” samples. These shared haplotypes are then “imputed” from the reference samples into the target samples. In this manner, samples with genome-wide array data (representing hundreds of thousands of SNPs) can be extended to represent millions of imputed SNPs. Genotype imputation has been successfully applied to discover many new genetic loci that underlie cardiovascular disease and associated risk factors.30, 31 Imputation-based GWAS provide statistical power similar to traditional GWAS for common variants. The advantage of this approach is the ability to interrogate variants of less common allele frequency.32
Limitations of GWAS
Although meta-analysis and imputation have substantially contributed to the success of GWAS by permitting large-scale analyses of common markers, several limitations remain. First, GWAS have rarely identified the “causal variant” or functional allele that drives the observed association. By design, GWAS implicate genomic regions or loci associated with a trait or disease. They are not designed to identify the precise nucleotide change that is responsible for the effect on phenotype. Additionally, GWAS are not well-powered to detect associations of variants with rare allele frequencies, particularly if these rare alleles have only a modest effect on the phenotype of interest. Due to these limitations, recent attention has shifted beyond standard GWAS to utilize new technologies for understanding the role of rare-variation on human diseases.
Sequencing studies and rare-variant associations
Though GWAS were successful in finding common variant associations, many recent studies have become focused on rare-variant associations. Standard GWAS arrays were designed to capture only common genetic variation, and thus do a poor job of representing rare-variants. Although imputation represents a powerful approach for studying less common variants, the accuracy of this approach is dramatically reduced for variants below 0.1%–0.5% frequency. As a result, the gold-standard approach for testing rare-variant associations is through direct DNA sequencing.Comprehensive sequencing of an entire human genome is relatively expensive compared to alternative approaches such as GWAS arrays or whole-exome sequencing (WES). WES is a technique for capturing the protein-coding portion of the genome (i.e., the “exome”) and submitting the captured DNA to high-throughput sequencing. Because the exome represents less than 3% of the entire human genome, WES is considerably less expensive than whole-genome sequencing (WGS). Importantly, WES is able to capture very rare variants that reside in the exome, making them amenable to association analyses. Because population genetics predicts that rare variants should exert larger phenotypic effects compared with common variants, the sample sizes initially used in WES were significantly smaller than a standard GWAS. (Recall that because statistical power is related to effect size, the rationale for using smaller sample sizes in WES studies was sound.) WES data are also much more expensive to generate than GWAS-array data. On the other hand, because WES studies consider very rare variants, the power to detect associations is significantly compromised compared to studies of common variation (Figure 1).
Statistical Approaches to Detecting Rare-Variant Associations
Due to the lack of power in detecting individual rare-variant associations, a number of statistical approaches have been developed to enhance power in the analysis of WES data. Specifically, because WES data are restricted to gene-regions, the analysis can consider a gene as a specific unit of analysis and aggregate rare-variants within a gene. The simplest approach is to sum the number of rare alleles observed in a particular gene, and test for association between that summation and the phenotype of interest.33 A somewhat orthogonal approach is to test whether the rare-alleles in a particular gene display “heterogeneity” of effect.34 There are many different variations on these two types of approaches,35 and they gain power in two different ways. First, the number of genes (~20,000) is much smaller than the number of rare-variants. Therefore, the multiple testing penalty for these gene-level approaches is much more generous than for testing each variant individually. Second, by testing for association across a gene-region the cumulative effect of rare-variants within a gene are aggregated, rather than considered in isolation.
Array-Based Assays for Studying Rare Variation
Though WES is an effective approach for assaying rare, protein-coding variation, it is still expensive compared to array-based technologies. Recently, custom arrays have been designed that capture rare variation at much lower cost. For example, the Exome-Chip was designed based on information from WES studies, captures most nonsynonymous protein-coding variation at > 0.1% in European American populations, and is available at ~10% of the cost of WES, making it feasible to collect data from very large sample sizes. With the wide availability of commercial Exome-Chips, multiple studies have collected data on >100,000 individuals. Such studies have led to important discoveries on rare, protein-coding genetic variants that influence triglyceride levels and CVD36, circulating lipid levels,37 blood-pressure,9 and coronary disease.38
Discoveries such as these have been met with considerable excitement, as many of these variants “knock-out” the function of a gene, and exist in healthy individuals.36,39 Every disease associated variant contains two alleles, one that “protects” against the disease and one that is associated with risk. Protective alleles that “knock-out“ (i.e., are loss-of-function (LOF) alleles) the function of a gene are akin to the effects of a therapeutic agent and discovery of protective alleles has stimulated the investigation of strategies for implementing therapeutic gene-knockouts by multiple pharmaceutical companies.40 In this way, the discovery of LOF variants has the potential to transform drug development by accelerating drug target identification.
Interpreting Evidence from Genetic Association Studies
Finally, we offer a brief checklist of items that should be considered when interpreting the evidence from genetic association studies: (a) was the study adequately powered to detect the observed associations? Using any number of web-based tools, it is straightforward to calculate power given a study design, an allele frequency, and an effect size. If the study was under-powered, the results should be interpreted with caution as under-powered studies are more likely to report false positive associations; (b) did the study utilize a correction for multiple testing? Without a proper correction, the chance of a false positive association greatly increases; (c) what is the p-value of reported association? P-values less than 5×10−8 represent solid evidence for association while associations with p-values that do not meet this threshold should be interpreted with caution; (d) do the reported loci/genes match our understanding of the biological processes involved in the disease? If a study identifies a gene in a pathway seemingly un-related to the disease process, then the signal for the association should be very strong (e.g., p ≪ 5×10−8); and (e) have the results been replicated in an independent study? If the results from a study pass do not pass all of these criteria, then they should be regarded with caution. As we have learned from the candidate gene era, false positive associations are often reported from under-powered studies that do not rely on independent replication.
Implications for Future Studies and Clinical Research
As technological development in DNA sequencing continues to advance, WGS has started replacing WES for studies of complex human diseases. Indeed, the NIH has funded several very large-scale projects to sequence the whole genomes of many tens of thousands of individuals. These types of studies will permit comprehensive investigations of all variation across the genome, including rare and common, and single-nucleotide as well as structural variants. Though comprehensive in scope, these studies also face enormous challenges with statistical power. Sample sizes will remain somewhat limited due to cost (i.e., WGS on hundreds of thousands of individuals is still prohibitively expensive), and rare-variants will be assayed across the entire genome rather than just protein-coding regions (in comparison to WES). This will have two major implications for statistical power: (1) outside of gene-regions, there are no clear units of analysis in which to aggregate signal and implement the “gene-level” statistical approaches that were developed for WES data; and (2) WGS will supply data on tens of millions of variants, dramatically increasing the multiple testing penalty and reducing power for individual genetic variants. Future research on overcoming these obstacles to statistical power will be important for the future of WGS studies.
In addition to WGS, there is an expanding interest in performing genetic association studies in multiple ancestry groups. The majority of human genetic variation is currently unknown and represents an untapped resource for discovering genetic associations.12 To date, most GWAS and WES studies have been specific to populations of European ancestry,8, 41, 42 though recently this has started to change.43 Because many genetic variants are population specific (i.e., they are invariant in certain populations, but polymorphic in others), studies representing diverse ancestry groups are well situated to detect associations with these population specific alleles. As genetic studies expand beyond samples of European ancestry, the power to detect associations with population specific alleles will increase.
Although not always routinely considered, statistical power will continue to be a relevant factor for not only research but clinical practice as well. For example, to optimally realize the goal precision medicine, the full complement of genetic and non-genetic determinants of disease will need to be defined. The component parts determining statistical power will play a substantial role in sculpting our ability to fully define the genetic architecture underlying these traits and diseases of biomedical relevance. Through realizing cost savings and higher efficiencies of sequencing technologies in the future, investigators will be able to increase sample sizes of future studies, thereby increasing statistical power. In addition, creative study design choices focused on sub-populations of interest may increase relative genetic effects, also serving to increase the power of discovery. Finally, newer statistical approaches are likely to be developed allowing investigators to augment power in the analysis of existing data. Regardless of the specific approach, recognizing that statistical power is a relevant issue to be considered will allow investigators to implement optimal strategies focused on discovering genetic associations and translating those findings to improve clinical care.
Table 1.
Overview of population-based gene-mapping study designs and sentinel publications
| Type of Study | Noteworthy papers | Comments |
|---|---|---|
| Candidate gene studies | Brown et al. (1986)1
(established LDLR as a driver of cholesterol homeostasis) Cohen et al. (2005)3 (identified mutations in PCSK9 associated with LDL levels) Ioannidis et al. (2001)44 (critique of candidate gene studies and lack of replication) |
Generally under-powered; hypothesis driven; often susceptible to false positives |
| Genome-wide association studies | Deloukas et al. (2013)28
(recent large-scale GWAS on coronary artery diseases) Willer et al. (2013)45 (recent large-scale GWAS on lipids) Stitziel et al. (2016)38 (large-scale exome based study of coronary disease) |
Moderately-powered for common variants; hypothesis generating; low false positive rate; genome wide coverage of common variants |
| Whole-exome sequencing | Do et al. (2015)46
(exome-sequencing study on myocardial infarction) Lange et al. (2015)47 (exome-sequencing study on lipids) |
Under-powered; hypothesis generating; low false positive rate; coverage of common and rare protein coding variants |
| Whole-genome sequencing | The UK10K project48
(whole-genome sequencing of 3,781 individuals to find rare-variants associated complex disease) The GoNL project49 (whole-genome sequencing of 250 Dutch parent-offspring families) The SardiNIA project50 (whole-genome sequencing of 2,500 Sardinians to find variants associated with longevity and related traits). |
Currently under-powered; hypothesis generating; low false positive rate; genome wide coverage of common and rare variants |
Acknowledgments
NOS was supported, in part, by grants from the National Heart, Lung, and Blood Institute (R01HL131961 and K08HL114642), the National Human Genome Research Institute (UM1HG008853), and by The Foundation for Barnes-Jewish Hospital.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to disclose.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Brown MS, Goldstein JL. A receptor-mediated pathway for cholesterol homeostasis. Science. 1986;232:34–47. doi: 10.1126/science.3513311. [DOI] [PubMed] [Google Scholar]
- 2.Abifadel M, Varret M, Rabes JP, et al. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat Genet. 2003;34:154–6. doi: 10.1038/ng1161. [DOI] [PubMed] [Google Scholar]
- 3.Cohen J, Pertsemlidis A, Kotowski IK, et al. Low LDL cholesterol in individuals of African descent resulting from frequent nonsense mutations in PCSK9. Nat Genet. 2005;37:161–5. doi: 10.1038/ng1509. [DOI] [PubMed] [Google Scholar]
- 4.Cohen JC, Boerwinkle E, Mosley TH, Jr, et al. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N Engl J Med. 2006;354:1264–72. doi: 10.1056/NEJMoa054013. [DOI] [PubMed] [Google Scholar]
- 5.Dadu RT, Ballantyne CM. Lipid lowering with PCSK9 inhibitors. Nat Rev Cardiol. 2014;11:563–75. doi: 10.1038/nrcardio.2014.84. [DOI] [PubMed] [Google Scholar]
- 6.Nikpay M, Goel A, Won HH, et al. A comprehensive 1,000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet. 2015;47:1121–30. doi: 10.1038/ng.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ellinor PT, Lunetta KL, Glazer NL, et al. Common variants in KCNN3 are associated with lone atrial fibrillation. Nat Genet. 2010;42:240–4. doi: 10.1038/ng.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Global Lipids Genetics C. Willer CJ, Schmidt EM, et al. Discovery and refinement of loci associated with lipid levels. Nat Genet. 2013;45:1274–83. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu C, Kraja AT, Smith JA, et al. Meta-analysis identifies common and rare variants influencing blood pressure and overlapping with metabolic trait loci. Nat Genet. 2016;48:1162–70. doi: 10.1038/ng.3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pereira NL, Sargent DJ, Farkouh ME, et al. Genotype-based clinical trials in cardiovascular disease. Nat Rev Cardiol. 2015;12:475–87. doi: 10.1038/nrcardio.2015.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foulkes WD, Brunet JS, Stefansson IM, et al. The prognostic implication of the basal-like (cyclin E high/p27 low/p53+/glomeruloid-microvascular-proliferation+) phenotype of BRCA1-related breast cancer. Cancer Res. 2004;64:830–5. doi: 10.1158/0008-5472.can-03-2970. [DOI] [PubMed] [Google Scholar]
- 12.Genomes Project C. Auton A, Brooks LD, et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sherry ST, Ward MH, Kholodov M, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–11. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Edwards SL, Beesley J, French JD, et al. Beyond GWASs: illuminating the dark road from association to function. Am J Hum Genet. 2013;93:779–97. doi: 10.1016/j.ajhg.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tennessen JA, Bigham AW, O’Connor TD, et al. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337:64–9. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Teslovich TM, Musunuru K, Smith AV, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–13. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hodge SE, Subaran RL, Weissman MM, et al. Designing case-control studies: decisions about the controls. Am J Psychiatry. 2012;169:785–9. doi: 10.1176/appi.ajp.2012.11111686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.International HapMap C. A haplotype map of the human genome. Nature. 2005;437:1299–320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu C, Tachmazidou I, Walter K, et al. Estimating genome-wide significance for whole-genome sequencing studies. Genet Epidemiol. 2014;38:281–90. doi: 10.1002/gepi.21797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Plomin R, Haworth CM, Davis OS. Common disorders are quantitative traits. Nat Rev Genet. 2009;10:872–8. doi: 10.1038/nrg2670. [DOI] [PubMed] [Google Scholar]
- 21.Campbell CD, Ogburn EL, Lunetta KL, et al. Demonstrating stratification in a European American population. Nat Genet. 2005;37:868–72. doi: 10.1038/ng1607. [DOI] [PubMed] [Google Scholar]
- 22.Price AL, Zaitlen NA, Reich D, et al. New approaches to population stratification in genome-wide association studies. Nat Rev Genet. 2010;11:459–63. doi: 10.1038/nrg2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hirschhorn JN, Lohmueller K, Byrne E, et al. A comprehensive review of genetic association studies. Genet Med. 2002;4:45–61. doi: 10.1097/00125817-200203000-00002. [DOI] [PubMed] [Google Scholar]
- 24.Sladek R, Rocheleau G, Rung J, et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445:881–5. doi: 10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- 25.Wellcome Trust Case Control C. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maurano MT, Humbert R, Rynes E, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–5. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kraft P, Zeggini E, Ioannidis JP. Replication in genome-wide association studies. Stat Sci. 2009;24:561–573. doi: 10.1214/09-STS290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Consortium CAD. Deloukas P, Kanoni S, et al. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat Genet. 2013;45:25–33. doi: 10.1038/ng.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nat Rev Genet. 2010;11:499–511. doi: 10.1038/nrg2796. [DOI] [PubMed] [Google Scholar]
- 30.Iotchkova V, Huang J, Morris JA, et al. Discovery and refinement of genetic loci associated with cardiometabolic risk using dense imputation maps. Nat Genet. 2016 doi: 10.1038/ng.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Surakka I, Horikoshi M, Magi R, et al. The impact of low-frequency and rare variants on lipid levels. Nat Genet. 2015;47:589–97. doi: 10.1038/ng.3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCarthy S, Das S, Kretzschmar W, et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet. 2016 doi: 10.1038/ng.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morris AP, Zeggini E. An evaluation of statistical approaches to rare variant analysis in genetic association studies. Genet Epidemiol. 2010;34:188–93. doi: 10.1002/gepi.20450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu MC, Lee S, Cai T, et al. Rare-variant association testing for sequencing data with the sequence kernel association test. Am J Hum Genet. 2011;89:82–93. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee S, Abecasis GR, Boehnke M, et al. Rare-variant association analysis: study designs and statistical tests. Am J Hum Genet. 2014;95:5–23. doi: 10.1016/j.ajhg.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tg Hdl Working Group of the Exome Sequencing Project NHL Blood I et al. Loss-of-function mutations in APOC3, triglycerides, and coronary disease. N Engl J Med. 2014;371:22–31. doi: 10.1056/NEJMoa1307095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peloso GM, Auer PL, Bis JC, et al. Association of low-frequency and rare coding-sequence variants with blood lipids and coronary heart disease in 56,000 whites and blacks. Am J Hum Genet. 2014;94:223–32. doi: 10.1016/j.ajhg.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Myocardial Infarction G Investigators CAEC. Stitziel NO, et al. Coding Variation in ANGPTL4, LPL, and SVEP1 and the Risk of Coronary Disease. N Engl J Med. 2016;374:1134–44. doi: 10.1056/NEJMoa1507652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Myocardial Infarction Genetics Consortium I. Stitziel NO, Won HH, et al. Inactivating mutations in NPC1L1 and protection from coronary heart disease. N Engl J Med. 2014;371:2072–82. doi: 10.1056/NEJMoa1405386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harper AR, Nayee S, Topol EJ. Protective alleles and modifier variants in human health and disease. Nat Rev Genet. 2015;16:689–701. doi: 10.1038/nrg4017. [DOI] [PubMed] [Google Scholar]
- 41.Locke AE, Kahali B, Berndt SI, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Polfus LM, Khajuria RK, Schick UM, et al. Whole-Exome Sequencing Identifies Loci Associated with Blood Cell Traits and Reveals a Role for Alternative GFI1B Splice Variants in Human Hematopoiesis. Am J Hum Genet. 2016;99:785. doi: 10.1016/j.ajhg.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Auer PL, Reiner AP, Wang G, et al. Guidelines for Large-Scale Sequence-Based Complex Trait Association Studies: Lessons Learned from the NHLBI Exome Sequencing Project. Am J Hum Genet. 2016 doi: 10.1016/j.ajhg.2016.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ioannidis JP, Ntzani EE, Trikalinos TA, et al. Replication validity of genetic association studies. Nat Genet. 2001;29:306–9. doi: 10.1038/ng749. [DOI] [PubMed] [Google Scholar]
- 45.Global Lipids Genetics C. Willer CJ, Schmidt EM, et al. Discovery and refinement of loci associated with lipid levels. Nat Genet. 2013 doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Do R, Stitziel NO, Won HH, et al. Exome sequencing identifies rare LDLR and APOA5 alleles conferring risk for myocardial infarction. Nature. 2014 doi: 10.1038/nature13917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lange LA, Hu Y, Zhang H, et al. Whole-exome sequencing identifies rare and low-frequency coding variants associated with LDL cholesterol. Am J Hum Genet. 2014;94:233–45. doi: 10.1016/j.ajhg.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Consortium UK. Walter K, Min JL, et al. The UK10K project identifies rare variants in health and disease. Nature. 2015;526:82–90. doi: 10.1038/nature14962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Genome of the Netherlands C. Whole-genome sequence variation, population structure and demographic history of the Dutch population. Nat Genet. 2014;46:818–25. doi: 10.1038/ng.3021. [DOI] [PubMed] [Google Scholar]
- 50.Sidore C, Busonero F, Maschio A, et al. Genome sequencing elucidates Sardinian genetic architecture and augments association analyses for lipid and blood inflammatory markers. Nat Genet. 2015;47:1272–81. doi: 10.1038/ng.3368. [DOI] [PMC free article] [PubMed] [Google Scholar]