Abstract
This study was conducted to investigate the roles of different Toll-like receptor (TLR) signaling in Porphyromonas gingivalis (P. gingivalis)-induced and ligature-induced experimental periodontal bone resorption in mice. Wild-type (WT), TLR2 knockout (KO), TLR4KO, and TLR2&4 KO mice with C57/BL6 background were divided into three groups: control, P. gingivalis infection, and ligation. Live P. gingivalis or silk ligatures were placed in the sulcus around maxillary second molars over a 2-week period. Images were captured by digital stereomicroscopy, and the bone resorption area was measured with ImageJ software. The protein expression level of gingival RANKL was measured by ELISA. The gingival mRNA levels of RANKL, IL-1β, TNF-α, and IL-10 were detected by RT-qPCR. The results showed that P. gingivalis induced significant periodontal bone resorption in WT mice and TLR2 KO mice but not in TLR4 KO mice or TLR2&4 KO mice. For all four types of mice, ligation induced signifcant bone loss compared with that in control groups, and this bone loss was signifcantly higher than that in the P. gingivalis infection group. RANKL protein expression was signifcantly increased in the ligation group compared with that in the control group for all four types of mice, and in the P. gingivalis infection group of WT, TLR2 KO, and TLR4 KO mice. Expression patterns of RANKL, IL-1β, TNF-α, and IL-10 mRNA were different in the P. gingivalis infection group and the ligation group in different types of mice. In summary, P. gingivalis-induced periodontal bone resorption is TLR4-dependent, whereas ligation-induced periodontal bone resorption is neither TLR2- nor TLR4-dependent.
Descriptors: Toll-Like Receptors, Bone Resorption, Periodontitis
Introduction
Periodontal disease is one of the most common chronic diseases in humans and is induced by microbial pathogens that reside in the oral cavity.1,2 However, compelling evidence now indicates that periodontal disease is not only a conventional bacterial infectious disease but also an inflammatory disease triggered by the host immune response to microorganisms.3 In host immune response, innate immunity is considered to act as a sentinel for the immune system and is promptly activated after recognition of the diverse repertoire of microbial pathogens.4,5
Toll-like receptors (TLRs) were the first pattern-recognition receptors (PRRs) to be identified and the best-characterized class of PRRs in mammalian species.6,7 Bacteria consist of various pathogen-associated molecular patterns (PAMP) that are detected by TLRs.7,8 Two members of the TLR family, TLR2 and TLR4, have been identified as the principal signaling receptors for bacterial cell wall components.9 TLR4 recognizes lipopolysaccharide (LPS) from Gram-negative bacteria.10,11 TLR2recognizes a wide variety of PAMPs,12 such as lipoproteins13 and peptidoglycans (PGN),14 from both Gram-positive and -negative bacteria, as well as lipoteichoic acid from Gram-positive bacteria.15 Recently, the presence of TLR2 and TLR4 has been shown to be essential for the progression of periodontitis in animal models and human patients.16,17 However, the contributions of TLR2 and TLR4 to periodontitis induced by various antigens are still uncertain and complicated.
Traditional methods of inducing experimental periodontitis include bacterial [Porphyromonas gingivalis (P. gingivalis) or Aggregatibacter actinomycetemcomitans (A.a.)] application, silk ligature, or a combination thereof.18,19 Animal models with both P. gingivalis- induced and ligature-induced experimental periodontitis are commonly used, but they have different mechanisms to induce periodontitis.20 P. gingivalis is a human-periodontitis-associated microbiome21,22 and ligatures will induce dysbiosis of the resident mouse microbiome to mimic dysbiosis of the human biofilm in periodontitis.18,23 Thus, the purpose of the present study was to compare the different roles of TLR2 and TLR4 in P. gingivalis-induced vs. ligature-induced experimental periodontitis mouse models.
Methodology
Animals
Wild-type (WT) C57BL/6J mice were purchased from the Jackson Laboratory (Bar Harbor, USA). TLR2 Knock-out (KO) mice, TLR4 KO mice, and TLR2&4 KO mice with a C57BL/6 background were a gift from Dr. Kawai (The Forsyth Institute). All the mice used in the study were 8–10 weeks old and maintained in specific pathogen-free (SPF) units. The mice were kept on a 12-hour light/dark cycle. The experimental protocols were approved by the Institutional Animal Care and Use Committee of The Forsyth Institute.
Bacterial culture
P. gingivalis (strain ATCC 33277) was grown on anaerobic blood agar plates (NHK agar, Northeast Laboratory Services, Winslow, USA) in an anaerobic chamber with 85% N2, 5% H2, and 10% CO2. Bacterial numbers in culture medium were determined from optical density values read by spectrophotometry and compared with a curve derived from a standard plate count. After incubation at 37°C for 5 days, bacteria were collected, washed three times with sterile phosphate-buffered saline (PBS), and re-suspended in PBS at the concentration of 1×1010/mL.
Experimental periodontitis
C57BL/6J wild-type (WT), TLR2KO, TLR4KO, and TLR2&4KO mice were used for P. gingivalis-induced experimental periodontitis or ligature-induced experimental periodontitis. Kanamycin and ampicillin were added to the drinking water for 4 days, and the oral cavity was swabbed with a 0.12% chlorhexidine gluconate mouthrinse to suppress the native oral microbiota on the 4th day. After 24 h, experimental periodontitis was induced by the application of 10 μL of live P. gingivalis (1 × 1010/mL in PBS) in the sulcus of the maxillary second molars or by the wrapping of silk ligatures (7-0, Fisher Scientific, Waltham, USA) around maxillary second molars on both sides over a 2-week period. To enhance reproducibility for the P. gingivalis infection group, P. gingivalis was applied around the tooth for 4 consecutive days after initial infection.
Sample preparation
Animals were euthanized by CO2 inhalation 2 weeks after P. gingivalis infection or ligation. The maxillary teeth were removed from each group, and gingival tissues were isolated under a surgical microscope for homogenate. The maxillary teeth were then de-fleshed by a Dermestid beetle colony. After being bleached with 3% hydrogen peroxide, the bone was stained with 1% toluidine blue and mounted on microscope slides for bone resorption measurement. For each gingival tissue, half of the collected gingival tissues were subjected to RNA isolation to determine cytokine expression by RT-qPCR. The other half were used to measure RANKL expression by ELISA.
Measurements of bone resorption
Images were captured by digital stereomicroscopy on a custom-made stage-holder with maxillary teeth at a certain angle (15 degree) to enhance visualization of the cementoenamel junction (CEJ) and alveolar bone level. The bone resorption area was enclosed by the second molar CEJ, the lateral margins of the first molars distal to the exposed root and the third molars mesial to the exposed root, and the alveolar crest and ridge. The polygonal area was measured with ImageJ software (NIH, Bethesda, USA) on buccal and palatal surfaces for each segment, and a standard calibrator was used for calibration at the same magnification. All bone resorption measurements were performed without the operator having prior knowledge of the group designation of the mice, and the recordings were verified by a second examiner. The results were presented in mm2.
Real-time PCR
Gingival RNA was extracted with TRIzole reagent (Sigma, St. Louis, USA) reverse-transcribed with SuperScript II Reverse Transcriptase (Invitrogen, Carlsbad, USA). The primer (Sigma) sequences were as follows: RANKL, forward 5′-GGGTGTGTACAAGACCC-3′ and reverse 5′-CATGTGCCACTGAGAACCTTGAA-3′; IL-1β, forward 5′-CCAGCTTCAAATCTCACAGCAG-3′ and reverse 5′-CTTCTTTGGGTATTGCTTGGGATC-3′; TNFα, for wa rd 5′-CACAGAAAGCATGATCCGCGACGT-3′ and reverse 5′- CGGCAGAGAGGAGGTTGACTTTCT-3 ′; I L-10, forward 5′- GACCAGCTGGACAACATACTGCTAA-3′ and reverse 5′-GATAAGGCTTGGCAACCCAAGTAA-3′; GAPDH, forward 5′-CCCCAGCAAGGACACTGAGCAA-3′ and reverse 5′- GTGGGTGCAGCGAACTTTATTGATG-. Real-time PCR was conducted with the LightCycler® SYBR Green master solution and LightCycler® 480 system (Roche, Indianapolis, USA). mRNA expression levels were presented as fold changes relative to the GAPDH reference.
ELISA assay
The gingival tissues collected were homogenized with a Dounce glass homogenizer in RIPA buffer and proteinase inhibitor cocktail (Sigma). The secreted RANKL level in the gingival homogenate was detected by means of a murine RANKL ELISA development kit (PeproTech, Rocky Hill, USA) following the manufacturer's instructions. The absorbance values were read by means of a Synergy HT Microplate Reader (BioTek, Winooski, USA) at 450 nm, and RANKL concentrations were calculated according to the standard curve.
Statistical analysis
All the quantitative data were expressed as means ± standard error. Statistical analysis was performed by Student's t-test for comparisons of two groups and by nonparametric ANOVA (Kruskal-Wallis test) with post hoc adjustments for multiple comparisons. Statistical significance was set at p < 0.05.
Results
Bone resorption differences of P. gingivalis-induced and ligation-induced experimental periodontitis in WT, TLR2 KO, TLR4 KO, and TLR2&4 KO mice
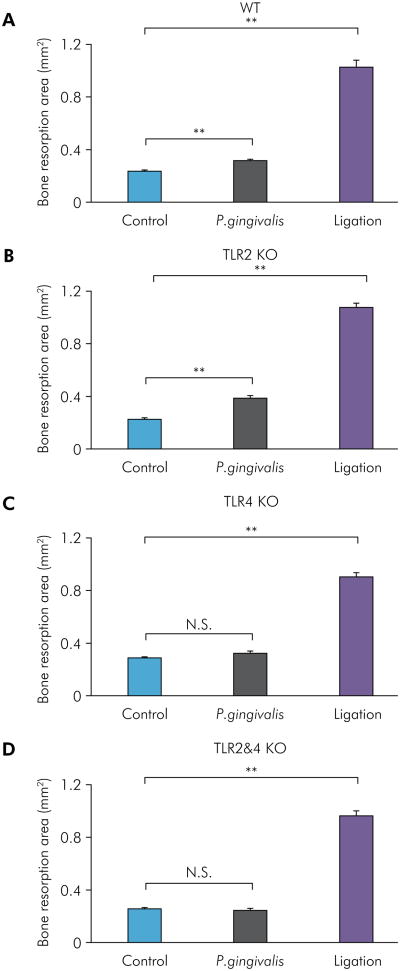
Experimental periodontitis was induced by the application of P. gingivalis in the sulcus of the maxillary second molars or by silk ligatures wrapped around maxillary second molars on both sides for 14 days in WT, TLR2 KO, TLR4 KO, and TLR2&4 KO mice. Images of the palatal surfaces of maxillae were captured (Figure 1), and bone loss in the polygonal area around maxillary second molars was measured with ImageJ software. The P. gingivalis infection group showed significantly higher bone loss compared with the control group for WT mice (Figure 2A) and TLR2 KO mice (Figure 2B), but there were no significant differences in TLR4 KO mice (Figure 2C) and TLR2&4 KO mice (Figure 2D). However, the ligation group revealed significantly higher bone loss in all four types of mice compared with the control group (Figure 2A-D). Taken together, bone loss induced by P. gingivalis showed a TLR4-dependence, and bone loss induced by ligation was TLR2- and TLR4-independent and significantly higher than in WT, TLR2 KO, TLR4 KO, and TLR2&4 KO control mice.
Figure 1.
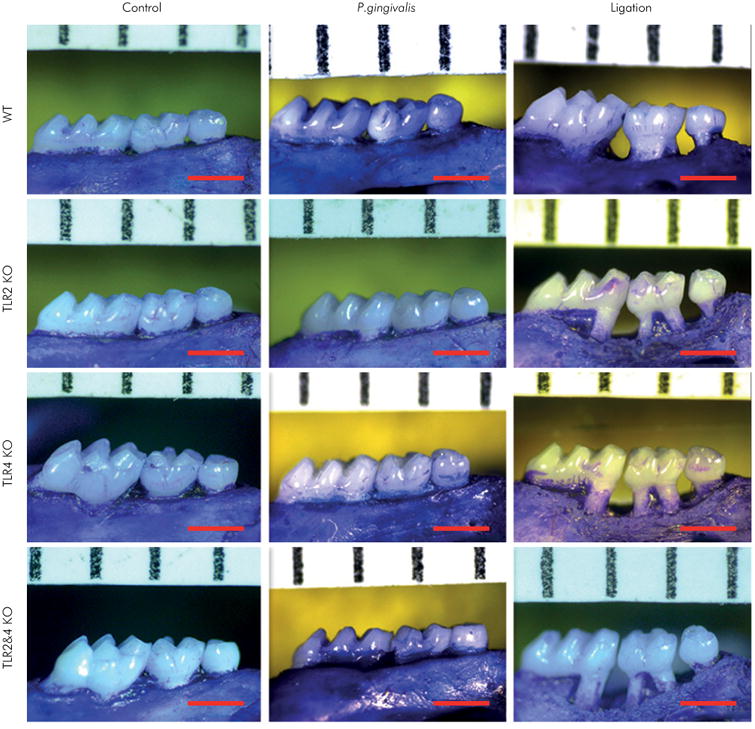
Maxillary images of P. gingivalis-induced and ligation-induced experimental periodontitis in WT, TLR2 KO, TLR4 KO, and TLR2&4 KO mice. Experimental periodontitis was induced by the application of live P. gingivalis in the sulcus of maxillary second molars or by silk ligatures wrapped around maxillary second molars on both sides over a 14-day period in WT, TLR2 KO, TLR4 KO, and TLR2&4 KO mice. Maxillae were collected on day 14 and de-fleshed by a Dermestid beetle colony. After being bleached with 3% hydrogen peroxide, the bone was stained with 1% toluidine blue. The alveolar bone resorption areas around maxillary secondary molars were viewed by microscopy (30×) for the control group, the P. gingivalis infection group, and the ligation group of WT, TLR2 KO, TLR4 KO, and TLR2&4 KO mice, respectively (scale bar, 1 mm).
Figure 2.
Bone resorption analysis of P. gingivalis-induced and ligation-induced experimental periodontitis in WT, TLR2 KO, TLR4 KO, and TLR2&4 KO mice. The bone resorption area of P. gingivalis-induced and ligation-induced experimental periodontitis was measured and analyzed with ImageJ software on buccal and palatal surfaces in WT mice (A), TLR2 KO mice (B), TLR4 KO mice (C), and TLR2&4 KO mice (D) (means ± SE, n = 5 mice per group, **p < 0.01, N.S. = no significant difference). For each segment, a standard calibrator was used for calibration at the same magnification for all images.
RANKL mRNA and protein expression differences of P. gingivalis-induced and ligation-induced experimental periodontitis in WT, TLR2 KO, TLR4 KO, and TLR2&4 KO mice
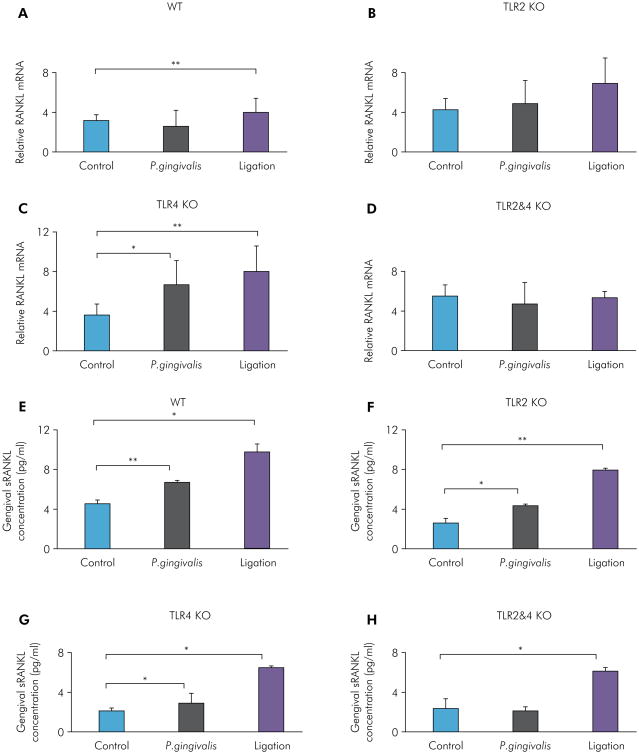
For investigation of the molecular mechanism of bone resorption in P. gingivalis-induced and ligation-induced experimental periodontitis in WT, TLR2 KO, TLR4 KO, and TLR2&4 KO mice, gingival RANKL mRNA levels and protein expression levels were measured by RT-qPCR and ELISA, respectively. For RANKL mRNA, the P. gingivalis infection group showed significantly higher levels compared with the control group for TLR4 KO mice (Figure 3C), with no significant differences in WT mice (Figure 3A), TLR2 KO mice (Figure 3B), and TLR2&4 KO mice (Figure 3D). The ligation group showed significantly higher levels compared with the control group for WT mice (Figure 3A) and TLR4 KO mice (Figure 3C), with no significant differences in TLR2 KO mice (Figure 3B) and TLR2&4 KO mice (Figure 3D). For gingivally secreted RANKL, the P. gingivalis infection group showed significantly higher expression compared with the control group for WT mice (Figure 3E), TLR2 KO mice (Figure 3F), and TLR4 KO mice (Figure 3G), with no significant differences in TLR2&4 KO mice (Figure 3H). However, the ligation group showed significantly higher expression compared with the control group in all four types of mice (Figure 3E-H). These results indicated that increased RANKL mRNA and protein expression induced by P. gingivalis was diminished when both TLR2 and TLR4 were lacking, suggesting that TLR2 was also involved in the regulation of RANKL-mediated bone loss in the P. gingivalis-induced experimental periodontitis model. Moreover, the ligation-induced increase of RANKL protein expression was TLR2- and TLR4-independent.
Figure 3.
Gingival RANKL mRNA and protein expression levels of P. gingivalis-induced and ligation-induced experimental periodontitis in WT, TLR2 KO, TLR4 KO, and TLR2&4 KO mice. Gingival tissues on the palatal side were collected under a surgical microscope from maxillae and then homogenized for RNA extraction or protein measurement. Gingival RANKL mRNA levels were determined by real-time PCR in the control group, the P. gingivalis infection group, and the ligation group of WT mice (A), TLR2 KO mice (B), TLR4 KO mice (C), and TLR2&4 KO mice (D) (means ± SE, n = 5 mice per group, *p < 0.05, **p < 0.01). Gingival RANKL protein expression levels were measured by ELISA in the control group, the P. gingivalis infection group, and the ligation group of WT mice (E), TLR2 KO mice (F), TLR4 KO mice (G), and TLR2&4 KO mice (H) (means ± SE, n = 5 mice per group, *p < 0.05, **p < 0.01).
I L-1β, TNF-α, and IL-10 mRNA differences in P. gingivalis-induced and ligation-induced experimental periodontitis in WT, TLR2 KO, TLR4 KO, and TLR2&4 KO mice
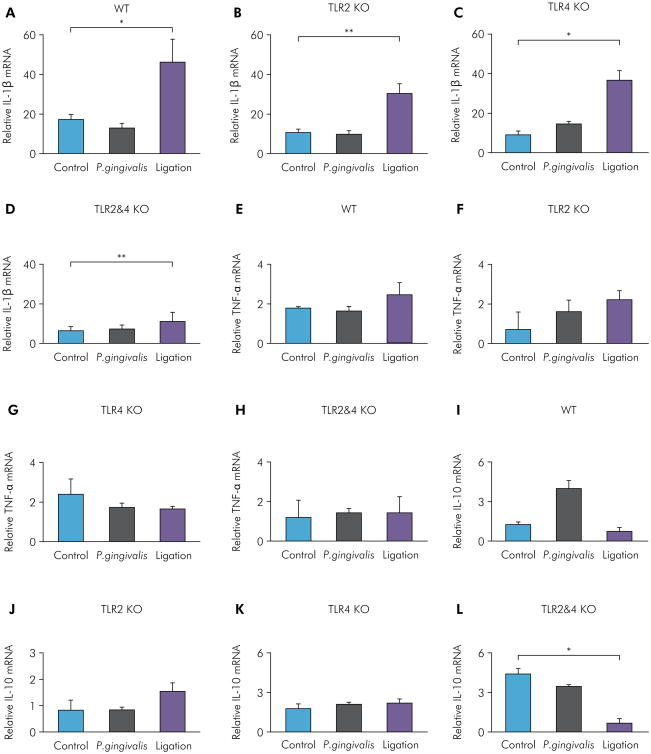
For comparison of the effects of TLR2 and TLR4 on inflammation in P. gingivalis-induced and ligation-induced experimental periodontitis in WT, TLR2 KO, TLR4 KO, and TLR2&4 KO mice, mRNA levels of the pro-infammatory cytokines IL-1β and TNF-α and the anti-inflammatory cytokine IL-10 were measured by RT-qPCR. For IL-1β mRNA, the P. gingivalis infection group showed no significant differences compared with the control group for WT mice (Figure 4A), TLR2 KO mice (Figure 4B), TLR4 KO mice (Figure 4C), and TLR2&4 KO mice (Figure 4D). The ligation group showed significantly higher levels of IL-1β expression compared with the control group in all four types of mice (Figure 4A-D). For TNF-α mRNA, both the P. gingivalis infection group and the ligation group showed no significant differences compared with the control group for WT mice (Figure 4E), TLR2 KO mice (Figure 4F), TLR4 KO mice (Figure 4G), and TLR2&4 KO mice (Figure 4H). For IL-10 mRNA, the P. gingivalis infection group showed significantly higher levels compared with the control group for WT mice (Figure 4I), and no differences for TLR2 KO mice (Figure 4J), TLR4 KO mice (Figure 4K), and TLR2&4 KO mice (Figure 4L). The ligation group showed no significant differences compared with the control group for WT, TLR2 KO, and T LR4 KO m ice (Figure 4I-K) but showed significantly lower levels in TLR2&4 KO mice (Figure 4L). These results indicated that P. gingivalis infection-induced and ligation-induced experimental periodontitis showed differential regulation of gingival IL-1β and IL-10 expression through TLR2/4 signaling.
Figure 4.
Gingival IL-1β, TNF-α, and IL-10 mRNA levels of P. gingivalis-induced and ligation-induced experimental periodontitis in WT, TLR2 KO, TLR4 KO, and TLR2&4 KO mice. Gingival IL-1β mRNA levels were determined by real-time PCR in the control group, the P. gingivalis infection group, and the ligation group of WT mice (A), TLR2 KO mice (B), TLR4 KO mice (C), and TLR2&4 KO mice (D) (means ± SE, n = 5 mice per group, *p < 0.05, **p < 0.01). Gingival TNF-α mRNA levels were determined by real-time PCR in the control group, the P. gingivalis infection group, and the ligation group of WT mice (E), TLR2 KO mice (F), TLR4 KO mice (G), and TLR2&4 KO mice (H) (means ± SE, n = 5 mice per group). Gingival IL-10 mRNA levels were determined by real-time PCR in the control group, the P. gingivalis infection group, and the ligation group of WT mice (I), TLR2 KO mice (J), TLR4 KO mice (K), and TLR2&4 KO mice (L) (means ± SE, n = 5 mice per group, *p < 0.05, **p < 0.01).
Discussion
The Toll-like receptor (TLR) family of innate immune recognition receptors plays a fundamental role in the induction of innate immunity, inflammation, cell survival, and proliferation.24,25 The presence of TLR2 and TLR4 has been shown to be essential for the progression of inflammation and related bone metabolism in periodontitis.16,26 However, little is known about the specific contributions of TLR2 and TLR4 signaling in different models of experimental periodontitis in mice. In the present study, we investigated the changes in bone resorption, RANKL (bone resorption marker) mRNA and protein level s, IL-1β, TNF-α (pro-inflammatory marker), and IL-10 (anti-infammatory marker) mRNA levels in P. gingivalis-induced and ligation-induced experimental periodontitis in WT, TLR2 KO, TLR4 KO, and TLR2&4 KO mice. The results showed that P. gingivalis-induced periodontal bone resorption is TLR4-dependent, whereas ligation-induced periodontal bone resorption is neither TLR2- nor TLR4-dependent. Also, TLR2 and TLR4 signaling plays different roles in gingival IL-1β, TNF-α, and IL-10 expression in P. gingivalis-induced and ligation-induced experimental periodontitis.
P. gingivalis-induced periodontitis is mediated by the disruption of the host tissue homeostasis and adaptive immune response.27,28 In the present study, P. gingivalis infection showed significant increases in periodontal bone resorption in WT mice and TLR2 KO mice but not in TLR4 KO mice and TLR2&4 KO mice (Figure 2A-D), suggesting that TLR4 is essential for bone loss associated with P. gingivalis infection. However, RANKL protein levels were significantly higher in the P. gingivalis infection group in TLR4 KO mice (Figure 3G) and showed no significant change in TLR2&4 KO mice (Figure 3H), indicating that not only TLR4 signaling but also TLR2 signaling is involved in the RANKL-mediated bone loss. Moreover, there could be cross-talk between TLR4 and TLR2 signaling to fully regulate periodontal bone resorption in P. gingivalis-induced periodontitis. These possible mechanisms need to be investigated in the future.
Ligature-induced periodontitis disrupts the resident mouse microbiome and mimics dysbiosis of the human biofilm in periodontitis.18,23,29,30 Analysis of our data showed that ligation-induced bone loss was observed in all types of mice tested, including WT, TLR2 KO, TLR4 KO, and TLR2&4 KO mice (Figure 2A-D). Analysis further suggested that ligation-induced periodontal bone loss is neither TLR2- nor TLR4-dependent. Other TLR-or non-TLR-related pathways are also involved in the pathogenesis of ligation-induced bone loss. Moreover, IL-1β mRNA levels were consistently elevated in the ligation groups compared with the control groups in all types of mice (Figure 4A-D), suggesting that the TLR-independent pathways activated in ligation-induced bone loss are mediated through the up-regulation of IL-1β. Also, analysis of our data showed that, in the presence of both TLR2 and TLR4 deficiency, the ligation group still demonstrated significantly increased bone loss and inflammatory cytokine expression compared with the control group (Figures 2D, 3H, 4D), further suggesting that TLR-independent pathways may be involved in the ligation-induced experimental periodontitis that regulates bone loss and inflammation. Taken together, for investigations focused on periodontal bone loss and inflammation, the ligation-induced experimental periodontitis mouse model may represent a broader spectrum of pathogenesis than the P. gingivalis infection mouse model, and results derived from these two models should be compared and interpreted with caution.
Conclusion
In summary, TLR2 and TLR4 signaling contributes differently to mouse models of P. gingivalis-induced and ligature-induced experimental periodontitis. P. gingivalis-induced periodontal bone resorption is TLR4-dependent, whereas ligation-induced periodontal bone resorption is neither TLR2- nor TLR4-dependent.
Acknowledgments
This study was supported by NIH NIDCR grants DE023807 and DE025255.
Footnotes
Declaration of Interest: The authors certify that they have no commercial or associative interest that represents a conflict of interest in connection with the manuscript.
References
- 1.Joshi D, Garg T, Goyal AK, Rath G. Advanced drug delivery approaches against periodontitis. Drug Deliv. 2016;23(2):363–77. doi: 10.3109/10717544.2014.935531. https://doi.org/10.3109/10717544.2014.935531. [DOI] [PubMed] [Google Scholar]
- 2.Kaur G, Mohindra K, Singla S. Autoimmunity: basics and link with periodontal disease. Autoimmun Rev. 2017;16(1):64–71. doi: 10.1016/j.autrev.2016.09.013. https://doi.org/10.1016/j.autrev.2016.09.013. [DOI] [PubMed] [Google Scholar]
- 3.Hajishengallis G. Periodontitis: from microbial immune subversion to systemic inflammation. Nat Rev Immunol. 2015;15(1):30–44. doi: 10.1038/nri3785. https://doi.org/10.1038/nri3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boller T, He SY. Innate immunity in plants: an arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science. 2009;324(5928):742–4. doi: 10.1126/science.1171647. https://doi.org/10.1126/science.1171647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Plüddemann A, Mukhopadhyay S, Gordon S. Innate immunity to intracellular pathogens: macrophage receptors and responses to microbial entry. Immunol Rev. 2011;240(1):11–24. doi: 10.1111/j.1600-065X.2010.00989.x. https://doi.org/10.1111/j.1600-065X.2010.00989.x. [DOI] [PubMed] [Google Scholar]
- 6.Bauer S, Müller T, Hamm S. Pattern recognition by Tolllike receptors. Adv Exp Med Biol. 2009;653:15–34. doi: 10.1007/978-1-4419-0901-5_2. https://doi.org/10.1007/978-1-4419-0901-5_2. [DOI] [PubMed] [Google Scholar]
- 7.Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. 2010;11(5):373–84. doi: 10.1038/ni.1863. https://doi.org/10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 8.Ozinsky A, Underhill DM, Fontenot JD, Hajjar AM, Smith KD, Wilson CB, et al. The repertoire for pattern recognition of pathogens by the innate immune system is defined by cooperation between toll-like receptors. Proc Natl Acad Sci USA. 2000;97(25):13766–771. doi: 10.1073/pnas.250476497. https://doi.org/10.1073/pnas.250476497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takeuchi O, Hoshino K, Kawai T, Sanjo H, Takada H, Ogawa T, et al. Differential roles of TLR2 and TLR4 in recognition of gram-negative and gram-positive bacterial cell wall components. Immunity. 1999;11(4):443–51. doi: 10.1016/s1074-7613(00)80119-3. https://doi.org/10.1016/S1074-7613(00)80119-3. [DOI] [PubMed] [Google Scholar]
- 10.Beutler B. Tlr4: central component of the sole mammalian LPS sensor. Curr Opin Immunol. 2000;12(1):20–6. doi: 10.1016/s0952-7915(99)00046-1. https://doi.org/10.1016/S0952-7915(99)00046-1. [DOI] [PubMed] [Google Scholar]
- 11.Lu YC, Yeh WC, Ohashi PS. LPS/TLR4 signal transduction pathway. Cytokine. 2008;42(2):145–51. doi: 10.1016/j.cyto.2008.01.006. https://doi.org/10.1016/j.cyto.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 12.Willcocks S, Offord V, Seyfert HM, Coffey TJ, Werling D. Species-specific PAMP recognition by TLR2 and evidence for species-restricted interaction with Dectin-1. J Leukoc Biol. 2013;94(3):449–58. doi: 10.1189/jlb.0812390. https://doi.org/10.1189/jlb.0812390. [DOI] [PubMed] [Google Scholar]
- 13.Schenk M, Belisle JT, Modlin RL. TLR2 looks at lipoproteins. Immunity. 2009;31(6):847–9. doi: 10.1016/j.immuni.2009.11.008. https://doi.org/10.1016/j.immuni.2009.11.008. [DOI] [PubMed] [Google Scholar]
- 14.Michelsen KS, Aicher A, Mohaupt M, Hartung T, Dimmeler S, Kirschning CJ, et al. The role of toll-like receptors (TLRs) in bacteria-induced maturation of murine dendritic cells (DCS). Peptidoglycan and lipoteichoic acid are inducers of DC maturation and require TLR2. J Biol Chem. 2001;276(28):25680–6. doi: 10.1074/jbc.M011615200. https://doi.org/10.1074/jbc.M011615200. [DOI] [PubMed] [Google Scholar]
- 15.Rashidi N, Mirahmadian M, Jeddi-Tehrani M, Rezania S, Ghasemi J, Kazemnejad S, et al. Lipopolysaccharide- and lipoteichoic acid-mediated pro-inflammatory cytokine production and modulation of TLR2, TLR4 and MyD88 expression in human endometrial cells. J Reprod Infertil. 2015;16(2):72–81. [PMC free article] [PubMed] [Google Scholar]
- 16.Berdeli A, Emingil G, Han Saygan B, Gürkan A, Atilla G, Köse T, et al. TLR2 Arg753Gly, TLR4 Asp299Gly and Thr399Ile gene polymorphisms are not associated with chronic periodontitis in a Turkish population. J Clin Periodontol. 2007;34(7):551–7. doi: 10.1111/j.1600-051X.2007.01092.x. https://doi.org/10.1111/j.1600-051X.2007.01092.x. [DOI] [PubMed] [Google Scholar]
- 17.Lin J, Bi L, Yu X, Kawai T, Taubman MA, Shen B, et al. Porphyromonas gingivalis exacerbates ligature-induced, RANKL-dependent alveolar bone resorption via differential regulation of Toll-like receptor 2 (TLR2) and TLR4. Infect Immun. 2014;82(10):4127–34. doi: 10.1128/IAI.02084-14. https://doi.org/10.1128/IAI.02084-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee CT, Teles R, Kantarci A, Chen T, McCafferty J, Starr JR, et al. Resolvin E1 Reverses Experimental Periodontitis and Dysbiosis. J Immunol. 2016;197(7):2796–806. doi: 10.4049/jimmunol.1600859. https://doi.org/10.4049/jimmunol.1600859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu X, Lin J, Yu Q, Kawai T, Taubman MA, Han X. Activation of Toll-like receptor 9 inhibits lipopolysaccharide-induced receptor activator of nuclear factor kappa- B ligand expression in rat B lymphocytes. Microbiol Immunol. 2014;58(1):51–60. doi: 10.1111/1348-0421.12129. https://doi.org/10.1111/1348-0421.12115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cai Y, Chen Z, Liu H, Xuan Y, Wang X, Luan Q. Green tea epigallocatechin-3-gallate alleviates Porphyromonas gingivalis-induced periodontitis in mice. Int Immunopharmacol. 2015;29(2):839–45. doi: 10.1016/j.intimp.2015.08.033. https://doi.org/10.1016/j.intimp.2015.08.033. [DOI] [PubMed] [Google Scholar]
- 21.Cugini C, Klepac-Ceraj V, Rackaityte E, Riggs JE, Davey ME. Porphyromonas gingivalis: keeping the pathos out of the biont. J Oral Microbiol. 2013;5(1):5. doi: 10.3402/jom.v5i0.19804. https://doi.org/10.3402/jom.v5i0.19804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mazumdar V, Snitkin ES, Amar S, Segrè D. Metabolic network model of a human oral pathogen. J Bacteriol. 2009;191(1):74–90. doi: 10.1128/JB.01123-08. https://doi.org/10.1128/JB.01123-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maekawa T, Abe T, Hajishengallis E, Hosur KB, DeAngelis RA, Ricklin D, et al. Genetic and intervention studies implicating complement C3 as a major target for the treatment of periodontitis. J Immunol. 2014;192(12):6020–7. doi: 10.4049/jimmunol.1400569. https://doi.org/10.4049/jimmunol.1400569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tu S, Zhong D, Xie W, Huang W, Jiang Y, Li Y. Role of Toll-Like Receptor Signaling in the Pathogenesis of Graft-versus-Host Diseases. Int J Mol Sci. 2016;17(8):17. doi: 10.3390/ijms17081288. https://doi.org/10.3390/ijms17081288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abdul-Cader MS, Amarasinghe A, Abdul-Careem MF. Activation of toll-like receptor signaling pathways leading to nitric oxide-mediated antiviral responses. Arch Virol. 2016;161(8):2075–86. doi: 10.1007/s00705-016-2904-x. https://doi.org/10.1007/s00705-016-2904-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oliveira NF, Andia DC, Planello AC, Pasetto S, Marques MR, Nociti FH, Jr, et al. TLR2 and TLR4 gene promoter methylation status during chronic periodontitis. J Clin Periodontol. 2011;38(11):975–83. doi: 10.1111/j.1600-051X.2011.01765.x. https://doi.org/10.1111/j.1600-051X.2011.01765.x. [DOI] [PubMed] [Google Scholar]
- 27.Franca M, Moura-Costa L, Meyer RJ, Trindade SC, Tunes UR, Freire SM. Humoral immune response to antigens of Porphyromonas gingivalis ATCC 33277 in chronic periodontitis. J Appl Oral Sci. 2007;15(3):213–9. doi: 10.1590/S1678-77572007000300011. https://doi.org/10.1590/S1678-77572007000300011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zadeh HH, Nichols FC, Miyasaki KT. The role of the cell-mediated immune response to Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis in periodontitis. Periodontol. 1999;200020(1):239–88. doi: 10.1111/j.1600-0757.1999.tb00163.x. https://doi.org/10.1111/j.1600-0757.1999.tb00163.x. [DOI] [PubMed] [Google Scholar]
- 29.Darzi Y, Jiao Y, Hasegawa M, Moon H, Núñez G, Inohara N, et al. The genomic sequence of the oral pathobiont strain ni1060 reveals unique strategies for bacterial competition and pathogenicity. PLoS One. 2016;11(7):e0158866. doi: 10.1371/journal.pone.0158866. https://doi.org/10.1371/journal.pone.0158866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zenobia C, Hasturk H, Nguyen D, Van Dyke TE, Kantarci A, Darveau RP. Porphyromonas gingivalis lipid A phosphatase activity is critical for colonization and increasing the commensal load in the rabbit ligature model. Infect Immun. 2014;82(2):650–9. doi: 10.1128/IAI.01136-13. https://doi.org/10.1128/IAI.01136-13. [DOI] [PMC free article] [PubMed] [Google Scholar]