Abstract
Deregulating the subcellular localization, functions and expression of Forkhead box (FOX) transcription factors that are critically involved in embryonic development and multiple biological processes is known to result in the development and progression of diseases, in particular cancer. Human FOXF transcription factors, including FOXF1 and FOXF2, are a subfamily of the FOX gene family. The recent findings from ours and others have linked FOXF2 to breast cancer development and progression. Our studies have shown that FOXF2 acts as a tumor-suppressive inhibitor of DNA replication in luminal and HER2-positive breast cancers and as an oncogenic activator of the epithelial-mesenchymal transition (EMT) in triple-negative/basal-like breast cancers (TN/BLBC), suggesting that FOXF2 plays a dual role in breast cancer. However, studies from Feng's research group have pointed out an opposite role of FOXF2 in TN/BLBC, which acts as an inhibitor of the EMT and as a promoter of cell proliferation in TN/BLBC. These discrepancies between our and Feng's studies have caused controversy in the role of FOXF2 in breast cancer. This article reviews both studies and discusses what causes might have led to these inconsistencies as well as what future experiments are needed to solve this debate.
Keywords: FOXF2, breast cancer, epithelial-mesenchymal transition, DNA replication
The overview of physiological and pathological roles of FOXF2
The forkhead box (FOX) genes are an evolutionally conserved gene family and encode transcription factors involved in regulating embryogenesis and pattern formation in multicellular organisms. FOX proteins bind DNA as a monomer through their forkhead domain. Forkhead box F2 (FOXF2), a gene member of the FOXF subfamily in the FOX gene family, is a mesenchymal regulator specifically expressed in the mesenchyme directly adjacent to the ectoderm-derived epithelium that develops into tongue and in the mesenchyme adjacent to the endoderm-derived epithelium that develop into the gastrointestinal (GI) tract, lungs, and genitalia (1). Genetic inactivation of Foxf2 caused defects in mouse development, including cleft palate and the abnormal development of tongue and gut (2–5). From studies of these Foxf2-deficient mice, Foxf2 mediates hedgehog signaling to activate Tgfβ signaling and inhibit Wnt as well as Fgf18 signaling during organogenesis (3–5). These regulatory events mediated by Foxf2 are critical for maintaining extracellular matrix (ECM) content and the functions of epithelium as well as mesenchyme for organogenesis (3–5).
An accumulating number of recent studies have linked the dysregulation of FOXF2 to various cancers, including breast, esophageal, lung, liver, prostate and colon cancers (6–19) (Table 1). Most of these studies indicate that FOXF2 is aberrantly downregulated in cancers through epigenetic mechanisms such as FOXF2 promoter hypermethylation and targeting by oncogenic microRNAs (e.g. miR-301, miR-182 and miR-519a). These lines of evidence suggest that FOXF2 functions as a tumor suppressor to inhibit tumorigenesis of various tissue types. However, FOXF2 has been reported to be an oncogenic factor in alveolar rhabdomyosarcoma and lung cancer (10, 20). Therefore, FOXF2 may play a dual role functioning as either a tumor suppressor or an oncogene in a tissue-context- and stage-specific manner. This dual FOXF2 function has led to its controversial role in breast cancer, which is raised by the inconsistency between our and Feng's studies (12, 16, 21). This review summarizes both studies and discusses possible causes leading to this controversy as well as future solutions to this debate.
Table 1.
Dysregulation and the functional role of the FOXF2 gene in various types of human cancer.
| Cancer Type | FOXF2 Dysregulation | FOXF2 Function | Reference |
|---|---|---|---|
| Prostate | Downregulation | FOXF2 expression is downregulated by miR-182-5p in prostate cancer. | Hirata et al. (7) |
| Breast | Downregulation | FOXF2 expression correlates with the early-onset metastasis and poor prognosis for patients with histological grade II and triple-negative breast cancer. | Kong et al. (8) |
| Colon | Downregulation | FOXF2 expression is inversely correlated with miR-182 expression in colorectal cancer (CRC). miR-182 downregulates FOXF2 expression to upregulate β-catenin expression, and promote cell growth and invasion. | Zhang et al. (9) |
| Lung | Upregulation | FOXF2 expression is aberrantly overexpressed in mesenchymal-like metastatic lung cancer cells and tightly correlates with the EMT transcription factor ZEB1. FOXF2 upregulation induces the EMT, migration, invasion and metastasis in lung cancer cells through inhibiting expression of E-cadherin and miR-200. | Kundu et al. (10) |
| Liver | Downregulation | Downregulation of FOXF2 expression is observed in hepatocellular carcinoma (HCC) and correlates with overall survival and recurrence-free survival of HCC patients. FOXF2 knockdown in a HCC cell line promotes proliferation and anti-apoptosis. | Shi et al. (11) |
| Breast | Downregulation | FOXF2 is specifically expressed in most basal-like breast cancer (BLBC) cell lines and tissues. FOXF2 deficiency promotes the metastatic ability of BLBC cells in vitro and in vivo through EMT inhibition. | Wang et al. (12) |
| Breast | Downregulation | DNA methylation is involved in silencing SP1-mediated FOXF2 expression in non-TNBC. SP1-induced FOXF2 expression promotes proliferation of BLBC cells. | Tian et al. (13) |
| Esophagus | Downregulation | FOXF2 downregulation is detected in esophageal squamous cell carcinoma (ESCC) and correlates with a higher frequency of lymph node metastasis and a decreased survival rate of ESCC patients. | Zheng et al. (14) |
| Liver | Downregulation | FOXF2 expression inversely correlates miR-519a in HCC. miR-519a promotes proliferation and inhibits apoptosis of HCC by repressing FOXF2 expression. | Shao et al. (15) |
| Breast | Downregulation/upregulation | FOXF2 is epigenetically silenced in luminal and HER2+ breast cancer, but is frequently overexpressed in BLBC. FOXF2 functions as an EMT promoter and DNA replication inhibitor. | Lo et al. (16) |
| Breast | Downregulation | miR-182 targets FOXF2 to promote cell proliferation and migration of TNBC. | Yu et al. (17) |
| Breast | Downregulation | FOXF2 mediates the function of Myc-associated zinc finger protein (MAZ) to promote proliferation and inhibit aggressiveness of BLBC. | Yu et al. (18) |
| Liver | Downregulation | FOXF2 deficiency facilitates the metastasis of HCC through inducing mesenchymal-epithelial transition (MET). | Dou et al. (19) |
The dual role of FOXF2 in breast cancer (Lo's studies)
Breast cancer has been classified into several molecular subtypes, including luminal, HER2-positive and basal-like, due to its heterogeneity. Approximately 70–80% of basal-like breast cancers (BLBC) lack expression of estrogen receptor (ER), progesterone receptor (PR) and HER2, which are generally called triple-negative breast cancers (TNBC). Our DNA methylation studies showed that FOXF2 expression was frequently silenced in luminal and HER2-positive breast cancers through epigenetic mechanisms, which predominantly resulted from the hypermethylation of the FOXF2 promoter (16). FOXF2 promoter methylation positively correlated with the tumor size and stage, but was inversely associated with patients' age and triple-negative status. Luminal-type breast cancers with low FOXF2 expression were associated with poor prognosis (16). Exogenous expression of FOXF2 in FOXF2-deficient luminal and HER2-positive breast cancer cell lines dramatically suppressed their tumorigenicity in vitro and in vivo (16). Ectopic expression of FOXF2 led to a blockage of cell cycle progression at the G1-S transition with or without induction of apoptosis in luminal and HER2-positive breast cancer cells. Mechanistically, FOXF2 blocks the G1-S transition and DNA replication through its inhibitory effect on the CDK2-RB-E2F cascade (16). Our FOXF2 knockdown studies demonstrate that the in vivo function of FOXF2 is to maintain the stringency of DNA replication, and its loss results in dysregulation of DNA replication, which in turn activates the p53 checkpoint pathway to arrest cell cycle progression. Therefore, knockdown of FOXF2 in human mammary epithelial cells induced cell cycle arrest at the G1 phase, which was due to the activation of the p53 checkpoint pathway, not to the intrinsic function of FOXF2 (16). These observed events in FOXF2 knockdown studies are analogous to the outcome from the knockdown of its paralog FOXF1 in HCT116 colon cancer cells (22). Besides its role in cell cycle regulation, FOXF2 is functionally required for the mobility and epithelial-to-mesenchymal transition (EMT) of normal breast epithelial cells (16).
In contrast, triple-negative/basal-like breast cancers (TN/BLBC), in particular the more malignant basal-B subgroup, frequently exhibit FOXF2 overexpression when compared to non-TN/BLBC and normal breast tissue (16). We also found that the tumor-suppressive role of FOXF2 in DNA replication is lost in TN/BLBC (16). This finding explains why TN/BLBC can tolerate the overexpressed levels of FOXF2. Moreover, FOXF2 is required for malignant characteristics of TN/BLBC, including cell migration, invasion and anchorage-independent growth (16). Our in silico analysis of public cancer gene expression databases indicates that FOXF2 expression in breast cancer is associated with basal-signature and stem-cell-related genes (16). Our microarray-based expression profiling, qRT-PCR and Western blot studies of FOXF2-knockdown MCF10A (an immortalized human mammary epithelial cell line) and MDA-MB-231 (a metastatic TN/BLBC line) further indicate that FOXF2 can regulate expression of genes functionally implicated in cell-cycle progression, EMT and TGFβ signaling (16).
Our studies for the first time have revealed that FOXF2 can function as either a tumor suppressor that negatively regulates DNA replication or an oncogenic factor that drives the EMT and metastatic progression in a tissue-context-dependent manner. Our proposed paradigm provides a rational explanation for the contradictory phenotypes of downregulated FOXF2 expression in luminal-type and HER2-positive breast cancers but upregulated FOXF2 expression in TN/BLBC. Our studies also suggest a hypothesis that during the development of TN/BLBC, the transcriptional function of FOXF2 is altered from a tumor suppressor to an oncogenic factor (16). However, our studies lack in vivo animal evidence to demonstrate the oncogenic role of FOXF2 in tumorigenicity and metastasis of TN/BLBC.
FOXF2 functions as an EMT inhibitor and a proliferation promoter in TN/BLBC (Feng's studies)
Feng's research group conducted RT-qPCR analysis of FOXF2 mRNA levels in their 305 primary breast cancer tissues and found that FOXF2 mRNA levels in primary breast cancer negatively correlated with breast tumor progression, including tumor size, the number of metastatic lymph nodes, and clinical stage (8). Their analyses also indicated that low FOXF2 mRNA expression predisposed patients with histological grade II and TN breast cancer to relapse and metastasis (8). Based on these findings, they concluded that FOXF2 underexpression can serve as a promising biomarker to indicate the early-onset metastasis and poor prognosis for patients with histological grade II and TN breast cancers.
Their following studies of breast cancer cell lines showed that FOXF2 expression was specifically detected in most TN/BLBC cell lines but silenced in non-TN/BLBC cell lines through DNA methylation of the FOXF2 promoter (12, 13). Their functional studies indicated that FOXF2 knockdown promoted the metastatic ability of TN/BLBC cells in vitro and in vivo (12). They further found that FOXF2 deficiency activated the EMT and concurrently inhibited cell proliferation of TN/BLBC cells (12). Therefore, their studies suggest that FOXF2 functions as an inhibitor of the EMT and a promoter of proliferation in TN/BLBC cells. Their mechanistic studies revealed that FOXF2 transcriptionally repressed the expression of two EMT transcription factors TWIST1 and FOXC2 (12, 21). Recently they further showed that FOXF2 is a transcriptional target of Myc-associated zinc finger protein (MAZ) and mediates the dual function of MAZ to inhibit aggressiveness and promote proliferation of TN/BLBC cells (18). The low expression of both MAZ and FOXF2 mRNA levels are significantly associated with poor prognosis of TN/BLBC patients (18), suggesting the combination of both markers is a potential prognostic predictor for TN/BLBC.
Based on these findings, they hypothesized that FOXF2 is an EMT-suppressing transcription factor in TN/BLBC and its deficiency promotes the metastatic ability of TN/BLBC cells through activating the transcription of TWIST1 and FOXC2, which in turn enhance the EMT program. However, in their studies it is unclear how FOXF2 promotes the proliferation of TN/BLBC cells. Although their studies also showed that FOXF2 is epigenetically silenced in non-TN/BLBC cell lines, they only focused on the role of FOXF2 in TN/BLBC and did not pursue the functional studies of FOXF2 in non-TN/BLBC. Therefore, the functional role of FOXF2 in non-TN/BLBC remains unclear in their studies.
The controversial role of FOXF2 in breast cancer
Findings from Feng's studies are the essential opposite of our findings. These discrepancies between our and Feng's findings might have been due to differences in experimental approaches, such as the strategies for FOXF2 knockdown and analysis of FOXF2 in different patient expression/survival datasets, and variations in the usage of experimental materials, such as the FOXF2 expression plasmid, shRNA/siRNA reagents and cell lines. For example, to create the HA-tagged FOXF2 expression construct for exogenous FOXF2 studies, as described in "Materials and Methods" of their paper (12), Feng's group obtained the pEV-FOXF2 plasmid from Dr. Peter Carlsson (University of Gothenburg, Department of Cell and Molecular Biology, Sweden) who first cloned the human FOXF2 cDNA and named this gene FREAC-2 (Forkhead RElated ACtivator-2) (23), and then they subcloned the FOXF2 cDNA into the pcDNA3.1-HA vector. Based on Dr. Carlsson's paper, the cloned FOXF2 cDNA was not full-length and contained the truncated open reading frame encoding 408 amino acids (23). According to the NCBI reference sequence NM_00145 (https://www.ncbi.nlm.nih.gov/nuccore/NM_001452.1), the full-length FOXF2 protein contains 444 amino acids. Therefore, the FOXF2 cDNA Feng's group used in their studies lacks the nucleotide sequence encoding the N-terminal 36 amino acids of FOXF2 protein. Therefore, it is uncertain whether the use of this truncated form of FOXF2 in exogenous expression or rescue studies can truly represent the function of endogenous wild-type FOXF2 in cells. Since the truncated FOXF2 still possesses the forkhead DNA-binding domain, it may cause dominant mutant phenotypes if this truncated form lacks the wild-type FOXF2 function and can compete with endogenous FOXF2 or other transcription factors to bind target genes. In contrast, we cloned the full-length FOXF2 cDNA from normal breast tissue to construct the HA-tagged FOXF2 expression plasmid for our exogenous expression studies of FOXF2 function in luminal-type and HER2-positive breast cancer cells (16). As aforementioned, studies of FOXF2 restoration in non-TN/BLBC showed that FOXF2 functions as a tumor suppressor to inhibit the proliferation of non-TN/BLBC cells through blocking DNA replication. This difference in the use of the FOXF2 expression plasmid might have led to the significant discrepancy between our and Feng's findings.
This tumor-suppressive role of FOXF2 we found (16) has raised a question about how TN/BLBC can tolerate overexpressed levels of FOXF2. FOXF2 knockdown in two TN/BLBC cell lines, MDA-MB-231 and MDA-MB-468, had no impact on their DNA replication and cell cycle progression (16). Given that both cell lines were p53-deficient, activation of the p53 checkpoint was lost in both cell lines when FOXF2 knockdown occurred. Moreover, genome-wide expression profiling of FOXF2-knockdown MCF10A and MDA-MB-231 cells indicated that FOXF2-mediated regulation of cell cycle genes found in MCF10A cells was lost in MDA-MB-231 cells (16). These findings together suggest that the cell-cycle-regulatory function of FOXF2 is likely lost in MDA-MB-231 cells. Our proposed model rationally explains why the significant portion of TN/BLBC cell lines exhibited the FOXF2-overexpressed phenotype (16). In contrast, Feng's studies suggest that FOXF2 is an EMT inhibitor and a cell proliferation promoter (12). This model seems not to be able to explain why TN/BLBC cell lines express high levels of FOXF2 and at the same time exhibit the basal/EMT phenotype. Moreover, since non-TN/BLBC, especially luminal breast cancers, generally possess epithelial features and lack basal/EMT phenotypes, their studies cannot explain why FOXF2 is epigenetically silenced in non-TN/BLBC if FOXF2 can promote cell proliferation and inhibit the EMT, both of which are supposed to preferentially support the development of non-TN/BLBC. Our model that FOXF2 functions as an EMT promoter and a cell proliferation inhibitor more matches the FOXF2 silencing phenotype found in non-TN/BLBC.
A recent study has shown that FOXF2 is the target of miR-182, which is expressed significantly higher in TNBC tissues compared to matched normal tissues (17). Even though the functional role of FOXF2 was not investigated in the paper, this study suggests that the oncogenic role of miR-182 in promoting migration and proliferation of TNBC cells is through inhibiting FOXF2 expression (17). These findings seem to support Feng's model. However, FOXF2 promotes proliferation of TNBC cells in Feng's model (12), inconsistent with the proposed mechanism that miR-182-promoted cell proliferation is mediated by downregulating FOXF2 expression (17). Moreover, expression of miR-182 mimics significantly enhanced proliferation and migration of luminal MCF7 breast cancer cells (17). It is doubtful that these observed oncogenic effects of miR-182 in MCF7 cells are related to FOXF2 as MCF7 cells are known to lack FOXF2 expression due to its epigenetic silencing (13, 16). In colorectal cancer (CRC), FOXF2 is also targeted and downregulated by miR-182, leading to increased β-catenin expression, proliferation and invasion of CRC (9). These two lines of studies of the miR-182/FOXF2 axis in breast and colorectal cancers suggest that FOXF2 functions as an inhibitor of proliferation and invasiveness. Since most of researchers perform migration and invasion studies using transwells that generally need 24-hr incubation, it is difficult to discriminate whether the observed suppression of migration/invasion results from inhibition of invasiveness, or from suppression of cell proliferation/survival when manipulation of gene function affects cell growth. Therefore, whether FOXF2 functions as an inhibitor of invasiveness in cancer cells still needs further study.
It has been reported elsewhere that FOXF transcription factors are involved in inducing and maintaining the mesenchymal/EMT phenotypes of stromal and basal-type cancer cells (3, 10, 24–26). These findings are consistent with our findings (16), but contradictory to Feng's findings (12). Our findings from studies of FOXF1 (22, 27) and FOXF2 (16) suggest a paradigm that the tumor-suppressive roles of FOXF factors result from their functions as the inhibitors of DNA replication, but not from their EMT functions. Consistently with our findings, Dou and colleagues also recently found that FOXF2 deficiency promoted the mesenchymal-epithelial transition (MET, a reverse process of the EMT) of hepatocellular carcinoma cells through inhibiting the EMT and enhancing cell proliferation (19). Moreover, Kundu and colleagues found that activation of TGFβ signaling led to the induction of FOXF2 expression, suggesting FOXF2 mediates the TGFβ signaling effect (10). Their studies further showed that the miR-200 family, functioning as EMT suppressors, target and repress FOXF2 expression (10). In our studies we also found that FOXF2 positively regulates the expression of TGFβ pathway components, suggesting that FOXF2 functions as the activator of TGFβ signaling (16). Our finding is consistent with a previously reported animal study showing that Foxf2 is required for Tgfβ signaling (4). Therefore, studies from ours and others suggest that FOXF2 forms a positive feedback regulation loop with TGFβ signaling. The functional connection of FOXF2 to TGFβ signaling suggests a paradigm wherein FOXF2 activates and mediates the dual functional role of TGFβ signaling in suppressing cell cycle progression (the tumor-suppressive function) and in promoting EMT and metastatic processes (the oncogenic function). In contrast, Feng's findings seem not to be compatible with this regulatory relationship between FOXF2 and TGFβ signaling.
Perspectives
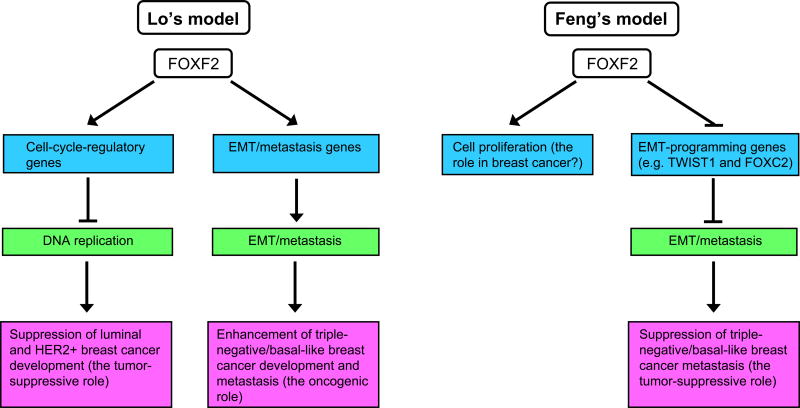
For the functional role of FOXF2 in breast cancer, our and Feng's studies have suggested two different models that are the opposite of each other (Figure 1). For understanding of pathological mechanisms involved in the development and progression of breast cancer, it is important to unravel which model really takes place in breast cancers in vivo. Evidence from xenograft studies is not sufficient to address this issue. Mouse models of breast cancer can fulfill this need. For example, mammary tumors formed in BLG-Cre;Brca1F22– 24/F22– 24; p53+/− mice or in Brca1fl/fl, MMTV-Cre; p53+/− mice resemble human TN/BLBC (28, 29). These TN/BLBC mouse models can cross with Foxf2 conditional knockout mice for studying the in vivo role of FOXF2 in the development and metastatic progression of TN/BLBC. Evidence from these animal studies would tell which model represents the in vivo function of FOXF2.
Figure 1.
Two different models for the functional role of FOXF2 in breast cancer have been proposed according to Lo's and Feng's studies.
Acknowledgments
The FOXF2 studies were supported by the National Institutes of Health (grant number: SPORE-1 P50 CA088843-08) and the Department of Defense (grant number: Center of Excellence-W81XWH-04-1-0595) to Dr. Saraswati Sukumar at the Johns Hopkins University.
Footnotes
Conflict of Interest
The author declares no conflict of interest.
References
- 1.Aitola M, Carlsson P, Mahlapuu M, Enerbäck S, Pelto-Huikko M. Forkhead transcription factor FoxF2 is expressed in mesodermal tissues involved in epithelio-mesenchymal interactions. Dev Dyn. 2000;218:136–149. doi: 10.1002/(SICI)1097-0177(200005)218:1<136::AID-DVDY12>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 2.Wang T, Tamakoshi T, Uezato T, Shu F, Kanzaki-Kato N, Fu Y, Koseki H, Yoshida N, Sugiyama T, Miura N. Forkhead transcription factor Foxf2 (LUN)-deficient mice exhibit abnormal development of secondary palate. Dev Biol. 2003;259:83–94. doi: 10.1016/s0012-1606(03)00176-3. [DOI] [PubMed] [Google Scholar]
- 3.Ormestad M, Astorga J, Landgren H, Wang T, Johansson BR, Miura N, Carlsson P. Foxf1 and Foxf2 control murine gut development by limiting mesenchymal Wnt signaling and promoting extracellular matrix production. Development. 2006;133:833–843. doi: 10.1242/dev.02252. [DOI] [PubMed] [Google Scholar]
- 4.Nik AM, Johansson JA, Ghiami M, Reyahi A, Carlsson P. Foxf2 is required for secondary palate development and Tgfβ signaling in palatal shelf mesenchyme. Dev Biol. 2016;415:14–23. doi: 10.1016/j.ydbio.2016.05.013. [DOI] [PubMed] [Google Scholar]
- 5.Xu J, Liu H, Lan Y, Aronow BJ, Kalinichenko VV, Jiang R. A Shh-Foxf-Fgf18-Shh Molecular Circuit Regulating Palate Development. PLoS Genet. 2016;12:e1005769. doi: 10.1371/journal.pgen.1005769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nik AM, Reyahi A, Pontén F, Carlsson P. Foxf2 in intestinal fibroblasts reduces numbers of Lgr5(+) stem cells and adenoma formation by inhibiting Wnt signaling. Gastroenterology. 2013;144:1001–1011. doi: 10.1053/j.gastro.2013.01.045. [DOI] [PubMed] [Google Scholar]
- 7.Hirata H, Ueno K, Shahryari V, Deng G, Tanaka Y, Tabatabai ZL, Hinoda Y, Dahiya R. MicroRNA-182-5p promotes cell invasion and proliferation by down regulating FOXF2, RECK and MTSS1 genes in human prostate cancer. PLoS One. 2013;8:e55502. doi: 10.1371/journal.pone.0055502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kong PZ, Yang F, Li L, Li XQ, Feng YM. Decreased FOXF2 mRNA expression indicates early-onset metastasis and poor prognosis for breast cancer patients with histological grade II tumor. PLoS One. 2013;8:e61591. doi: 10.1371/journal.pone.0061591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Y, Wang X, Wang Z, Tang H, Fan H, Guo Q. miR-182 promotes cell growth and invasion by targeting forkhead box F2 transcription factor in colorectal cancer. Oncol Rep. 2015;33:2592–2598. doi: 10.3892/or.2015.3833. [DOI] [PubMed] [Google Scholar]
- 10.Kundu ST, Byers LA, Peng DH, Roybal JD, Diao L, Wang J, Tong P, Creighton CJ, Gibbons DL. The miR-200 family and the miR-183~96~182 cluster target Foxf2 to inhibit invasion and metastasis in lung cancers. Oncogene. 2016;35:173–186. doi: 10.1038/onc.2015.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi Z, Liu J, Yu X, Huang J, Shen S, Zhang Y, Han R, Ge N, Yang Y. Loss of FOXF2 Expression Predicts Poor Prognosis in Hepatocellular Carcinoma Patients. Ann Surg Oncol. 2016;23:211–217. doi: 10.1245/s10434-015-4515-2. [DOI] [PubMed] [Google Scholar]
- 12.Wang QS, Kong PZ, Li XQ, Yang F, Feng YM. FOXF2 deficiency promotes epithelial-mesenchymal transition and metastasis of basal-like breast cancer. Breast Cancer Res. 2015;17:30. doi: 10.1186/s13058-015-0531-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tian HP, Lun SM, Huang HJ, He R, Kong PZ, Wang QS, Li XQ, Feng YM. DNA Methylation Affects the SP1-regulated Transcription of FOXF2 in Breast Cancer Cells. J Biol Chem. 2015;290:19173–19183. doi: 10.1074/jbc.M114.636126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng YZ, Wen J, Cao X, Yang H, Luo KJ, Liu QW, Huang QY, Chen JY, Fu JH. Decreased mRNA expression of transcription factor forkhead box F2 is an indicator of poor prognosis in patients with resected esophageal squamous cell carcinoma. Mol Clin Oncol. 2015;3:713–719. doi: 10.3892/mco.2015.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shao J, Cao J, Liu Y, Mei H, Zhang Y, Xu W. MicroRNA-519a promotes proliferation and inhibits apoptosis of hepatocellular carcinoma cells by targeting FOXF2. FEBS Open Bio. 2015;5:893–899. doi: 10.1016/j.fob.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Lo PK, Lee JS, Liang X, Sukumar S. The dual role of FOXF2 in regulation of DNA replication and the epithelial-mesenchymal transition in breast cancer progression. Cell Signal. 2016;28:1502–1519. doi: 10.1016/j.cellsig.2016.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu J, Shen W, Gao B, Zhao H, Xu J, Gong B. MicroRNA-182 targets FOXF2 to promote the development of triple-negative breast cancer. Neoplasma. 2017;64:209–215. doi: 10.4149/neo_2017_206. [DOI] [PubMed] [Google Scholar]
- 18.Yu ZH, Lun SM, He R, Tian HP, Huang HJ, Wang QS, Li XQ, Feng YM. Dual function of MAZ mediated by FOXF2 in basal-like breast cancer: Promotion of proliferation and suppression of progression. Cancer Lett. 2017;402:142–152. doi: 10.1016/j.canlet.2017.05.020. [DOI] [PubMed] [Google Scholar]
- 19.Dou C, Jin X, Sun L, Zhang B, Han M, Li T. FOXF2 deficiency promotes hepatocellular carcinoma metastasis by inducing mesenchymal-epithelial transition. Cancer Biomark. 2017 doi: 10.3233/CBM-170139. (in press) [DOI] [PubMed] [Google Scholar]
- 20.Milewski D, Pradhan A, Wang X, Cai Y, Le T, Turpin B, Kalinichenko VV, Kalin TV. FoxF1 and FoxF2 transcription factors synergistically promote rhabdomyosarcoma carcinogenesis by repressing transcription of p21(Cip1) CDK inhibitor. Oncogene. 2017;36:850–862. doi: 10.1038/onc.2016.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cai J, Tian AX, Wang QS, Kong PZ, Du X, Li XQ, Feng YM. FOXF2 suppresses the FOXC2-mediated epithelial-mesenchymal transition and multidrug resistance of basal-like breast cancer. Cancer Lett. 2015;367:129–137. doi: 10.1016/j.canlet.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 22.Lo PK, Lee JS, Sukumar S. The p53-p21WAF1 checkpoint pathway plays a protective role in preventing DNA rereplication induced by abrogation of FOXF1 function. Cell Signal. 2012;24:316–324. doi: 10.1016/j.cellsig.2011.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hellqvist M, Mahlapuu M, Samuelsson L, Enerback S, Carlsson P. Differential activation of lung-specific genes by two forkhead proteins, FREAC-1 and FREAC-2. J Biol Chem. 1996;271:4482–4490. doi: 10.1074/jbc.271.8.4482. [DOI] [PubMed] [Google Scholar]
- 24.Nilsson J, Helou K, Kovacs A, Bendahl PO, Bjursell G, Ferno M, Carlsson P, Kannius-Janson M. Nuclear Janus-activated kinase 2/nuclear factor 1-C2 suppresses tumorigenesis and epithelial-to-mesenchymal transition by repressing Forkhead box F1. Cancer Res. 2010;70:2020–2029. doi: 10.1158/0008-5472.CAN-09-1677. [DOI] [PubMed] [Google Scholar]
- 25.Saito RA, Micke P, Paulsson J, Augsten M, Pena C, Jonsson P, Botling J, Edlund K, Johansson L, Carlsson P, Jirstrom K, Miyazono K, Ostman A. Forkhead box F1 regulates tumor-promoting properties of cancer-associated fibroblasts in lung cancer. Cancer Res. 2010;70:2644–2654. doi: 10.1158/0008-5472.CAN-09-3644. [DOI] [PubMed] [Google Scholar]
- 26.Malin D, Kim IM, Boetticher E, Kalin TV, Ramakrishna S, Meliton L, Ustiyan V, Zhu X, Kalinichenko VV. Forkhead box F1 is essential for migration of mesenchymal cells and directly induces integrin-beta3 expression. Mol Cell Biol. 2007;27:2486–2498. doi: 10.1128/MCB.01736-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lo PK, Lee JS, Liang X, Han L, Mori T, Fackler MJ, Sadik H, Argani P, Pandita TK, Sukumar S. Epigenetic inactivation of the potential tumor suppressor gene FOXF1 in breast cancer. Cancer Res. 2010;70:6047–6058. doi: 10.1158/0008-5472.CAN-10-1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCarthy A, Savage K, Gabriel A, Naceur C, Reis-Filho JS, Ashworth A. A mouse model of basal-like breast carcinoma with metaplastic elements. J Pathol. 2007;211:389–398. doi: 10.1002/path.2124. [DOI] [PubMed] [Google Scholar]
- 29.Xu X, Wagner KU, Larson D, Weaver Z, Li C, Ried T, Hennighausen L, Wynshaw-Boris A, Deng CX. Conditional mutation of Brca1 in mammary epithelial cells results in blunted ductal morphogenesis and tumour formation. Nat Genet. 1999;22:37–43. doi: 10.1038/8743. [DOI] [PubMed] [Google Scholar]