Abstract
Streptomyces platensis MA7327 is a bacterium producing interesting antibiotics, which act by the novel mechanism of inhibiting fatty acid biosynthesis. The antibiotics produced by this actinomycete are platensimycin and platencin plus some minor related antibiotics. Platensimycin and platencin have activity against antibiotic-resistant bacteria such as methicillin-resistant Staphylococcus aureus and vancomycin-resistant Enterococcus; they also lack toxicity in animal models. Platensimycin also has activity against diabetes in a mouse model. We have been interested in studying the effects of primary metabolites on production of these antibiotics in our chemically defined production medium. In the present work, we tested 32 primary metabolites for their effect. They included 20 amino acids, 7 vitamins and 5 nucleic acid derivatives. Of these, only L-aspartic acid showed stimulation of antibiotic production. We conclude that the stimulatory effect of aspartic acid is due to its role as a precursor involved in the biosynthesis of aspartate-4-semialdehyde, which is the starting point for the biosynthesis of the 3-amino-2,4-dihydroxy benzoic acid portion of the platensimycin molecule.
INTRODUCTION
The world is in desperate need for new antibiotic compounds. Resistance to currently available drugs is growing and rapidly spreading.1 Every class of antibiotic drugs has at least one, and often more than one, pathogen that shows resistance against it; moreover, every major pathogen is resistant to at least one type of antibiotic.2 This situation is problematic because a myriad of diseases develop that are difficult to treat and are often easily spread. Two examples of problematic resistant pathogens are methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant Enterococcus (VRE), each of which kills tens of thousands of people per year.3,4 Because of resistant pathogens like these, infectious diseases remain a leading cause of death worldwide, even in developed countries.
Two compounds that have shown promising activity against resistant pathogens are platensimycin and platencin. These compounds work by inhibiting FabF and FabH, two key elongation enzymes in the fatty acid synthesis II pathway. This is an attractive antibiotic target due to a lack of similarity between the prokaryotic and eukaryotic fatty acid synthesis enzymes.5 Accordingly, platensimycin and platencin potentially have low cytotoxicity in humans, adding to their promise. Both compounds are natural products made by the actinomycete Streptomyces platensis MA7327. Platensimycin was discovered by Merck & Co. in 2006 in a soil sample from South Africa6 using a target-based whole-cell antisense differential sensitivity assay for inhibitors of fatty acid biosynthesis. It is characterized by a 3-amino-2,4-dihydroxybenzoic acid and C-17 pentacyclic enone with an ether ring joined by an amide bond and a hydrophobic region at the latter end of the molecule (Figure 1).6 Platencin was discovered using the same approach on soil samples from Spain in 2009. Its structure is very similar to platensimycin; the main structural difference is the presence of a ketolide portion lacking the cyclic ether ring (Figure 1).5
Figure 1.
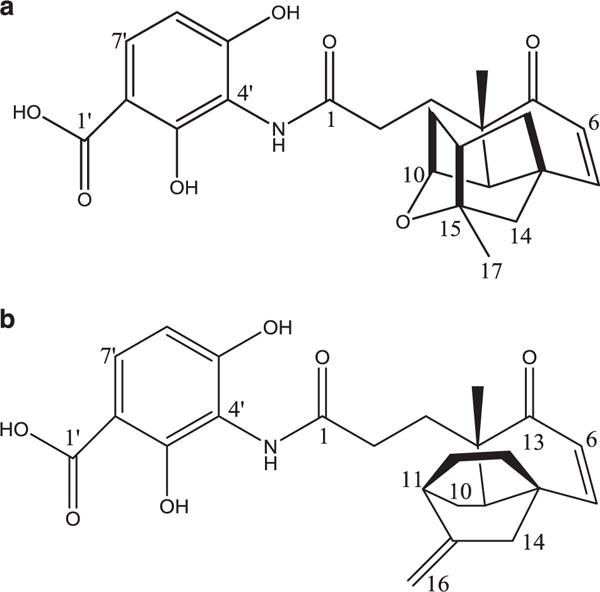
Structures of Platensimycin (a) and Platencin (b).
In addition to their activity against resistant pathogens (MRSA and VRE), both antibiotics exhibit potent antibacterial activity (MIC = 0.06–2 μg ml−1) against Gram-positive pathogens,7 including penicillin-resistant Streptococcus pneumoniae.8 They have also shown activity against mycobacteria such as Mycobacterium tuberculosis, Mycobacterium bovis, and Mycobacterium smegmatis.9 Platensimycin has the potential to be an effective treatment for tuberculosis because it inhibits biosynthesis of fatty acids and mycolic acid in M. tuberculosis.9,10 Of further interest, platensimycin has shown potential as an agent for treatment of diabetes in mouse models and type II diabetes mellitus.11 Despite problems with their pharmokinetic properties, platensimycin and platencin are promising because of their effective and non-toxic mode of action. These antibiotics have been further reviewed by Singh,12 and Martens and Demain.13
While the biosynthetic pathways of platensimycin and platencin and the enzymes involved have been elucidated,14–18 very little is known about the nutritional control and regulation of production. Following the discovery of platensimycin and platencin, Merck scientists produced these antibiotics in the laboratory by culturing S. platensis in a crude, insoluble production medium. Thereafter, we developed a semi-defined19 and then a completely chemically defined production medium (PM7).20 The chemically defined production medium has allowed us to begin to study the nutritional regulation of antibiotic production; to do this, we have added primary metabolites to chemically defined PM7 and assayed for changes in antibiotic production relative to PM7 without additives. In this way, we are able to observe if the addition of certain compounds to the production medium potentially regulates antibiotic production. The chemically defined production medium is crucial to unambiguously observe the effects of additives and to begin to understand how these effects are related to the biosynthesis. In an undefined medium, such as the original Merck medium, one cannot be certain that any observed effects on production are related to control of synthesis by the specifically added compounds. Understanding the nutritional control will allow for a better understanding of the antibiotic biosynthesis and its regulation. We tested a wide range of primary metabolites, including vitamins and amino acids and out of all compounds tested, only aspartic acid had a stimulatory effect on antibiotic production.
MATERIALS AND METHODS
Media and fermentation
All media were made using distilled water and the pH was adjusted to 7 using concentrated potassium hydroxide. The stock culture medium contained 30 g l−1 Difco Trypticase Soy Broth (Difco Laboratories, Detroit, MI, USA). The seed medium developed by Merck (Rahway, NJ, USA) and employed in this study contained (g l−1): 20 soluble starch, 10 dextrose, 5 NZ Amine type A, 3 beef extract, 5 Bacto-Peptone, 5 yeast extract and 1 CaCO3. All flasks were autoclaved at 121 °C for 30 min prior to use. The stock culture medium was inoculated with a sample of S. platensis MA7327 and incubated on the gyratory shaker at 220 r.p.m. at 28 °C for 2 days followed by storage in the refrigerator until use. The seed medium was inoculated with 0.5 ml of S. platensis stock culture and shaken at 220 r.p.m. and 28 °C for 5 days. The 5-day seed culture was spun down in a sterile 50-ml conical tube using a clinical one-speed centrifuge for 10 min. The supernatant fluid was discarded, replaced with an equal volume of sterile water and the pellet was resuspended. This washing process was repeated two more times. The culture was stored as a liquid in the refrigerator after the third addition of water. Fermentations were carried out in 125-ml Erlenmeyer flask containing 25 ml of fermentation broths on a rotary shaker at 28 °C and 250 r.p.m.
Compound testing
Flasks with chemically defined medium PM7 (g l−1: 30 glucose, 25 lactose, 0.5 ammonium sulfate and 5 MOPS buffer), with and without added compounds, were inoculated with 0.5 ml of washed seed culture per 25 ml of media. Each experimental condition was evaluated using duplicate flasks. Vitamins tested were ascorbic acid, folic acid, thiamine, inositol, D-biotin, riboflavin and vitamin B12. They were tested at 10 μg ml−1, except for vitamin B12 which was tested at 1 μg ml−1. Nucleic acid components, such as purines (adenine and guanine) and pyrimidines (cytosine, thymine and uracil), were examined at 0.5 mg ml−1. L-Amino acids were alanine, aminoadipic acid, arginine, asparagine, aspartic acid, citrulline, cysteine, glutamine, glycine, histidine, isoleucine, leucine, methionine, ornithine, phenylalanine, threonine, serine, tryptophan, tyrosine and valine. They were tested at 1 mg ml−1. After adding the additives, the pH was adjusted to 7.0. To evaluate effects of the additives, their zones of inhibition were compared directly to the PM7 control.
Agar diffusion assay
To test for antibiotic activity, zone of inhibition assays against S. aureus, Bacillus subtilis, and/or Bacillus megaterium were carried out in Petri plates.19 Stock cultures of these test organisms were made by inoculating 0.5 ml of a liquid culture into 25 ml of stock culture medium (see above) and incubating on the shaker at 35 °C for two days. 37 g l−1 of LB agar Miller was suspended in deionized water and autoclaved at 121 °C for 30 min. After autoclaving, the mixture was allowed to cool for a period of time and was then inoculated with 2.5 ml of stock culture for every 100 ml of agar. The inoculated agar was distributed into sterile Petri plates at 10 ml per plate. To assay, a small amount of the fermentation broths were aseptically removed from each experimental flask and placed into individual tubes. A paper disc, 6 mm in diameter, was dipped into each tube, dried briefly, and placed onto the agar plates. Plates were inverted and placed in the incubator at 35 °C for 2 days. Clear zones of inhibition around the discs were measured in mm across the diameter of two different sides and averaged. Each plate was read by two to three individuals, and the readings were averaged to generate a value for each flask. Values for identical flasks were also averaged, generating one value for each experimental condition. Conditions were compared by making a general comparison of the zone of inhibition size throughout the fermentation period, as well as by comparing the highest zone of inhibition observed, regardless of which day it occurred. Assays were conducted every 1 – 3 days, and zone sizes were compared throughout the length of the experiment. When evaluating for production stimulation (Table 1) compounds that consistently increased the zone of inhibition diameter by 2 mm or more, over several experiments were classified as positive (+) and compounds that yielded no change in zone size were classified as negative (−).
Table 1.
Stimulatory activity of the tested compounds against S. aureus, Bacillus subtilis, and/or Bacillus megaterium
| Compound | Concentration tested | Stimulatory effect on productiona |
|---|---|---|
| Adenine | 0.5 mg ml−1 | − |
| Alanine | 1 mg ml−1 | − |
| Aminoadipic acid | 1 mg ml−1 | − |
| Arginine | 1mg ml−1 | − |
| Ascorbic acid | 10 μg ml−1 | − |
| Aspartic acid | 1 mg ml−1 | + |
| Asparagine | 1 mg ml−1 | − |
| Biotin | 10 μg ml−1 | − |
| Citrulline | 1 mg ml−1 | − |
| Cysteine | 1 mg ml−1 | − |
| Cytosine | 0.5 mg ml−1 | − |
| Folic acid | 10 μg ml−1 | − |
| Guanine | 0.5 mg ml−1 | − |
| Glutamine | 1 mg ml−1 | − |
| Glycine | 1 mg ml−1 | − |
| Histidine | 1mg ml−1 | − |
| Isoleucine | 1mg ml−1 | − |
| Inositol | 10 μg ml−1 | − |
| Leucine | 1 mg ml−1 | − |
| Methionine | 1 mg ml−1 | − |
| Ornithine | 1 mg ml−1 | − |
| Phenylalanine | 1 mg ml−1 | − |
| Riboflavin | 10 μg ml−1 | − |
| Serine | 1 mg ml−1 | − |
| Thiamine | 10 μg ml−1 | − |
| Threonine | 1 mg ml−1 | − |
| Thymine | 0.5 mg ml−1 | − |
| Tryptophan | 1 mg ml−1 | − |
| Tyrosine | 1 mg ml−1 | − |
| Uracil | 0.5 mg ml−1 | − |
| Valine | 1 mg ml−1 | − |
| Vitamin B12 | 1 mg ml−1 | − |
Effect on production was determined by comparing zones of inhibition from fermentations with added compounds to PM7 with no compounds. To observe zones of inhibition, a paper disc, 6 mm in diameter, was dipped into a sample from each fermentation, dried briefly, and placed onto agar plates inoculated with S. aureus. Plates were inverted and placed in the incubator at 35 °C for 2 days. Clear zones of inhibition around discs were measured in mm across the diameter of two different sides and averaged. Each plate was read by two to three individuals, and the readings were averaged to generate a value for each flask. Values for identical flasks were also averaged, generating one value for each experimental condition. Compounds that consistently increased the zone of inhibition diameter by 2 mm or more over several experiments were classified as positive (+) and compounds that yielded no change in zone size were classified as negative (−).
RESULTS
Table 1 shows the results obtained with the 32 compounds tested (20 amino acids, 7 vitamins and 5 nucleic acid derivatives). The only active compound was L-aspartic acid. Addition of L-aspartic acid at 1 mg ml−1 consistently increased the zone of inhibition size compared to the PM7 control, an example of which is shown in Figure 2. After consistently observing the stimulation by aspartic acid at 1 mg ml−1, we tested it at other concentrations, both higher and lower, to find its optimal stimulatory concentration. Higher concentrations of aspartic acid, that is, 3 and 5 mg ml−1 did not increase the zone of inhibition size relative to PM7 plus 1 mg ml−1 aspartic acid. Lower concentrations, that is, 0.25 and 0.5 mg ml−1 were also tested. The optimal result was obtained with 0.5 mg ml−1 aspartic acid as compared with all concentrations tested.
Figure 2.
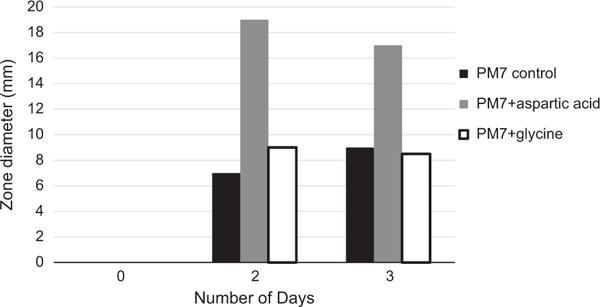
Stimulation of antibiotic formation by the addition of L-aspartic acid, assay against S. aureus, Bacillus subtilis, and/or Bacillus megaterium. Also shown is the lack of stimulation by glycine.
DISCUSSION
Using agar diffusion assays, we have shown that addition of aspartic acid to chemically defined production medium PM7 increases antibiotic production by S. platensis, which suggests a potential role for aspartic acid in the regulation of production. The optimal concentration for stimulation was 0.5 mg ml−1. The aspartic acid stimulation is interesting, given what we know about the biosynthesis of platensimycin and platencin. Aspartate-4-semialdehyde, which is synthesized from aspartic acid, is the starting material for the biosynthesis of the 3-amino-2,4-dihydroxybenzoic acid (AHDBA) portion of both molecules, as shown in Figure 3. Isotopic labeling studies have also connected this synthesis to the citric acid cycle, which is connected to aspartic acid synthesis.14,15 During the citric acid cycle, two molecules of acetyl CoA are incorporated into two molecules of oxaloacetate, which is involved in the synthesis of aspartic acid.16 Aspartic acid is then converted to aspartate-4-semialdehyde to serve as starting material for AHDBA synthesis.14,15 The link of the citric acid cycle to this synthesis highlights the importance of aspartic acid, which is reflected in our experimental studies, and indicates its potential regulatory role. Interestingly, none of the other compounds tested exhibited an effect on production, indicating potential minimal roles in regulation. These results are useful in better understanding nutritional regulation of platensimycin and platencin biosynthesis
Figure 3.
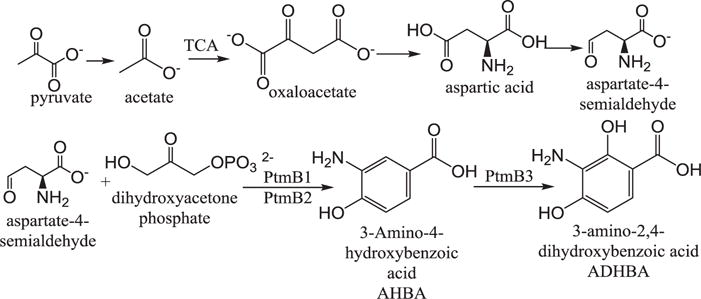
Biosynthetic pathway of the ADHBA moiety of platensimycin and platencin. Shows aspartate-4-semialdehyde formation from the citric acid cycle and its conversion to ADHBA. Ptm refers to genes found in the platensimycin gene cluster (adapted from references 14–18).
Acknowledgments
We acknowledge the encouragement by RISE Director Dr Jon Kettenring, and the assistance of RISE Administrative Assistant Miriam Donohue and Nilya Schaber, RISE/ResMed Program Coordinator.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
References
- 1.Laxminarayan R, et al. Antibiotic resistance-the need for global solutions. Lancet Inf Dis. 2013;13:1057–1098. doi: 10.1016/S1473-3099(13)70318-9. [DOI] [PubMed] [Google Scholar]
- 2.Allahverdiyev AM, et al. The use of platensimycin and platencin to fight antibiotic resistance. Infect Drug Res. 2013;6:99–114. doi: 10.2147/IDR.S25076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klevens RM, et al. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA. 2007;298:1763–1771. doi: 10.1001/jama.298.15.1763. [DOI] [PubMed] [Google Scholar]
- 4.Courvalin P. Vancomycin resistance in gram-positive cocci. Clin Infect Dis. 2006;42:S25–S34. doi: 10.1086/491711. [DOI] [PubMed] [Google Scholar]
- 5.Wang J, et al. Discovery of platencin, a dual FabF and FabH inhibitor with in vivo antibiotic properties. Proc Natl Acad Sci USA. 2007;104:7612–7616. doi: 10.1073/pnas.0700746104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang J, et al. Platensimycin is a selective FabF inhibitor with potent antibiotic properties. Nature. 2006;441:358–361. doi: 10.1038/nature04784. [DOI] [PubMed] [Google Scholar]
- 7.Singh SB, Young K. New antibiotic structures from fermentations. Expert Opin Ther Pat. 2010;20:1359–1371. doi: 10.1517/13543776.2010.509347. [DOI] [PubMed] [Google Scholar]
- 8.Tiefenbacher K, Mulzer J. A nine-step total synthesis of (−)-platencin. J Org Chem. 2009;74:2937–2941. doi: 10.1021/jo9001855. [DOI] [PubMed] [Google Scholar]
- 9.Brown AK, et al. Platensimycin activity against mycobacterial P-ketoacyl-ACP synthases. PLoS ONE. 2009;4:1–10. doi: 10.1371/journal.pone.0006306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ashforth EJ, et al. Bioprospecting for antituberculosis leads from microbial metabolites. Nat Prod Rep. 2010;27:1709–1719. doi: 10.1039/c0np00008f. [DOI] [PubMed] [Google Scholar]
- 11.Wu M, et al. Antidiabetic and antisteatotic effects of the selective fatty acid synthase (FAS) inhibitor platensimycin in mouse models of diabetes. Proc Natl Acad Sci USA. 2011;108:5378–5384. doi: 10.1073/pnas.1002588108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singh SB. In: Drug Discovery from Natural Products. Genilloud O, Vicente F, editors. Royal Society of Chemistry; Cambridge, UK: 2012. pp. 249–277. Ch. 12. [Google Scholar]
- 13.Martens E, Demain AL. Platensimycin and platencin: promising antibiotics for future application in human medicine. J Antibiot. 2011;64:705–710. doi: 10.1038/ja.2011.80. [DOI] [PubMed] [Google Scholar]
- 14.Herath K, et al. Biosynthetic studies of platencin. Tetrahedron Lett. 2008;49:6265–6268. [Google Scholar]
- 15.Herath K, et al. Biosynthetic studies of platensimycin. J Am Chem Soc. 2007;129:15422–15423. doi: 10.1021/ja0758943. [DOI] [PubMed] [Google Scholar]
- 16.Berg JM, et al. Biochemistry. 7th. W. H. Freeman and Company; New York, NY: 2012. pp. 499–511.pp. 710–714. [Google Scholar]
- 17.Smanski M, et al. Dedicated ent-kaurene and ent-atiserene synthases for platensimycin and platencin biosynthesis. Proc Natl Acad Sci USA. 2011;108:13498–13503. doi: 10.1073/pnas.1106919108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dairi T. Studies on the biosynthetic genes and enzymes of isoprenoids produced by actinomycetes. J Antibiot. 2005;58:227–243. doi: 10.1038/ja.2005.27. [DOI] [PubMed] [Google Scholar]
- 19.Aluotto S, et al. Development of a semi-defined medium supporting production of platensimycin and platencin by Streptomyces platensis. J Antibiot. 2013;66:51–54. doi: 10.1038/ja.2012.97. [DOI] [PubMed] [Google Scholar]
- 20.Falzone M, et al. Development of a chemically defined medium for the production of the antibiotic platensimycin by Streptomyces platensis. Appl Microbiol Biotechnol. 2013;97:9535–9539. doi: 10.1007/s00253-013-5201-6. [DOI] [PubMed] [Google Scholar]


