Abstract
Enterotoxigenic Escherichia coli (ETEC) is a major cause of diarrhoea in children below 5 years of age in endemic areas, and is a primary cause of diarrhoea in travellers visiting developing countries. Epidemiological analysis of E. coli pathovars is traditionally carried out based on the results of serotyping. However, genomic analysis of a global ETEC collection of 362 isolates taken from patients revealed nine novel O-antigen biosynthesis gene clusters that were previously unrecognized, and have collectively been called unclassified. When put in the context of all isolates sequenced, one of the novel O-genotypes, OgN5, was found to be the second most common ETEC O-genotype causing disease, after O6, in a globally representative ETEC collection. It’s also clear that ETEC OgN5 isolates have spread globally. These novel O-genotypes have now been included in our comprehensive O-genotyping scheme, and can be detected using a PCR-based and an in silico typing method. This will assist in epidemiological studies, as well as in ETEC vaccine development.
Keywords: enterotoxigenic Escherichia coli, O-antigen biosynthesis gene cluster, O genotype, PCR
Abbreviations
CF, colonization factor; ETEC, enterotoxigenic Escherichia coli; LT, heat-labile toxin; O-AGC, O-antigen biosynthesis gene cluster; OgUT, O-genotype untypeable; ST, heat-stable toxin; STEC, Shiga toxin-producing Escherichia coli.
Data Summary
1. Assembled draft genomes for 55 enterotoxigenic Escherichia coli isolates were deposited in GenBank/EMBL/DDBJ under the accession numbers shown in Table S1 (available with the online Supplementary Material).
2. Sequences of nine annotated O-antigen biosynthesis gene clusters and three wzy genes from positive control strains (OT-37 for OgN3, EHOUT43 for OgN5 and EH-OSB16 for OSB16) were deposited in GenBank/EMBL/DDBJ under the accession numbers LC177546–LC177554 and LC223608–LC223610, respectively.
Impact Statement
We identified nine novel O-genotypes (OgN) in a global collection of enterotoxigenic Escherichia coli (ETEC) isolates. The novel O-genotype OgN5 was found to be the second most common ETEC O-genotype globally, with no prior information regarding the contribution to the burden of disease. To gain more information about trends in ETEC OgN epidemiology, further studies of global OgN isolates are needed. The PCR method described in this study and an in silico typing method may help the surveillance and monitoring of the OgN groups.
Introduction
The first diarrhoeal illness that infants often experience in endemic areas is caused by enterotoxigenic Escherichia coli (ETEC) [1]. In 2010, annual mortality from illness due to ETEC was estimated at 157 000 deaths (9 % of all deaths attributed to diarrhoea) and approximately 1 % of all deaths in children 28 days to 5 years of age [2]. Additionally, ETEC is a primary cause of diarrhoea in travellers visiting developing countries. In our previous study, we sequenced 362 globally representative ETEC isolates collected between 1980 and 2011 from 20 countries, including isolates from children and adults in endemic areas, as well as from travellers visiting such areas [3]. The majority of the isolates were collected from patients with diarrhoea. Genome-wide analysis showed that, contrary to previous understanding, there are long-term stable associations of ETEC lineages with specific virulence factors, such as plasmid-encoded heat-labile toxin (LT) and/or heat-stable toxin (ST; including two subtypes, STh and STp), and colonization factors (CFs), and that these lineages are globally distributed.
O-serogrouping remains the gold standard for the subtyping of E. coli isolates, especially pathogenic E. coli, for taxonomical and epidemiological studies. Most of what we know about E. coli prevalence, outbreaks and surveillance is described in terms of the O-serogroup. Recently, sequence analyses show that phenotypic O-serogroup diversification can be correlated with differences in the gene content and genetic diversity of the O-antigen biosynthesis gene cluster (O-AGC) located on the chromosome [4]. In particular, sequences from O-antigen processing genes, such as wzx (encoding the O-antigen flippase), wzy (encoding the O-antigen polymerase), and the wzm and wzt genes (encoding components of the ABC transporter pathway) located on the O-AGCs are highly variable in sequence, and can be used as gene markers for the identification of O-serogroups via molecular approaches [4]. By applying an in silico blast analysis using a wzx/wzy and wzm/wzt sequence set extracted from O1 to O187 O-AGCs, we subtyped our 362 global ETEC isolates [3] into 48 O-genotypes, of which the top 6 were O6 (n=38), O25 (n=24), O27 (n=18), O114 (n=17), O115 and O159 (n=16, each). In addition to the ETEC isolates classified into 48 known O-genotypes, 55 isolates carried wzx/wzy genes that showed <50 % or no sequence identity to the previously defined sequences. These ETEC isolates were categorized as O-genotype untypeable (OgUT), but carried novel O-AGCs.
In this study, we focused on characterizing the OgUT ETEC isolates detected in our previous study [3]. Such ETEC isolates may not be recognized by public-health surveillance, because they cannot be assigned to any known O-genotype and/or O-serogroup. To determine the relative contribution of OgUT ETEC isolates, we defined the novel O-genotypes of these ETEC isolates and estimated their overall contribution to disease by screening our global collection of ETEC.
Methods
Source sequences
ETEC O-AGC sequences were obtained from draft genomes used in a previous study [3]. The wzx and wzy sequences were extracted from each O-AGC. The known wzx/wzy sequences from typical E. coli O-serogroups (171 groups, O1–O187 except for wzm/wzt-type groups) [4], Shigella O-serogroups (total 34 types; 13 from Shigella dysenteriae, 18 from Shigella boydii, 2 from Shigella flexnerii and a single type of Shigella sonnei) [5], E. coli OX serogroups (11 groups: OX6, OX9, OX10, OX13, OX18, OX19, OX21, OX25, OX28, OX38, OX43) [6] and 6 recently defined novel O genotypes from Shiga toxin-producing E. coli (OgN1, OgN8, OgN9, OgN10, OgN12, OgN31) [7] were used (except for wzy genes of OX25 and S. dysenteriae type 6, which are not found in the O-AGCs). A complete set of fliC sequences [8] was also used for molecular-based H-typing.
Sequence comparisons
Phylogenetic trees of wzx and wzy were constructed by using the neighbour-joining algorithm using mega5 software [9], following multiple alignments of nucleotide sequences by the clustal W program [10].
PCR
OgN-specific PCR primers targeting OgN3, OgN5 and OSB16 were designed based on each alignment of wzy sequences. PCR was performed as follows: the 30 µl reaction mixture contained 2 µl genomic DNA, 6 µl 5× Kapa Taq buffer, dNTP mix (final concentration 0.3 mM each), MgCl2 (final concentration 2.5 mM), primers (final concentration 0.5 µM each) and 0.8 U Kapa Taq DNA polymerase (Kapa Biosystems). The thermocycling conditions were: 25 cycles of 94 °C for 30 s, 58 °C for 30 s and 72 °C for 1 min. The PCR products were visualized following agarose gel (1.5 %) electrophoresis in 0.5x TBE (25 mM Tris borate, 0.5 mM EDTA) and staining with ethidium bromide (1 mg/ml). Three strains (OT-37 for OgN3, EHOUT43 for OgN5 and EH-OSB16 for OSB16) were used as positive-control strains for PCR.
Results
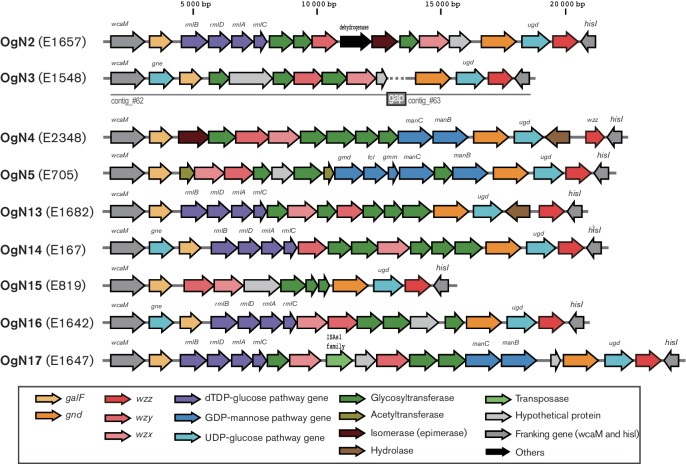
Comparative analyses revealed that chromosomal O-AGC sequences flanked by gene clusters of the colanic acid biosynthesis (wca) (upstream) and the histidine biosynthesis (his) (downstream) extracted from OgUT ETEC genomes contained nine novel O-AGCs, OgN2, OgN3, OgN4, OgN5, OgN13, OgN14, OgN15, OgN16 and OgN17 (Fig. 1), which all carried unique wzx/wzy genes, and showed <70 % nucleotide sequence identities among those from known E. coli O-AGCs (Fig. 2). One of the O-AGCs was similar (the same gene construction and ≥97 % sequence identity) to that of S. boydii type 16 (OSB16) (Fig. 2). Among the novel O-AGC sequences identified in our ETEC collection, it was apparent that the novel OgN5 was carried by more than half of ETEC OgUT isolates (29/55), followed in frequency by OgN3 and OSB16, 8 isolates each (Fig. 3). A summary of the epidemiological and genotypic data for the novel and SB16 O-genotype isolates is shown in Table S1. Sequences of wzx and wzy were highly conserved within each O-genotype (99.8–100 % sequence identity). ETEC OgN5 strains were isolated in five countries, Guatemala (n=12), Argentina (n=5), Egypt (n=4), Mexico (n=4) and Indonesia (n=4), between 1989 and 2003. All ETEC OgN5 isolates carried the ST-encoding gene, except for a single isolate (E1542) that carried the LT-encoding gene, and all OgN5 isolates were confirmed negative for all known CFs by dot-blot and PCR-based analyses in our previous study [3]. The fliC-based H-typing showed that the OgN5 isolates were classified into eight H-types: H5, H9, H10, H16, H18, H19, H32 and H39 (see Table S1). Phylogenetic analysis showed OgN5 isolates were spread across phylogenetically distinct lineages where most/some isolates grouped based on their H-types (see Fig. S1, Table S1). The major groups were OgN5 : H10 and OgN5 : H16 (n=6, each) belonging to a phylogenetic lineage L14 [3] of E. coli phylogroup A and a lineage L22 (designed in this study) of phylogroup B, respectively (see Fig. S1). Additionally, OgN5 : H18 (L11, n=1), OgN5 : H9 (L14, n=1), OgN5 : H32 (L15, n=4), OgN5 : H19 (L15, n=1), OgN5 : H34 (L16, n=4), OgN5 : H16 (L23, n=3) and OgN5 : H5 (L24, n=3) were observed. No significant association between lineages and geographical origins was observed (see Table S1).
Fig. 1.
Nine novel O-antigen biosynthesis gene clysters from the ETEC collection.
Fig. 2.
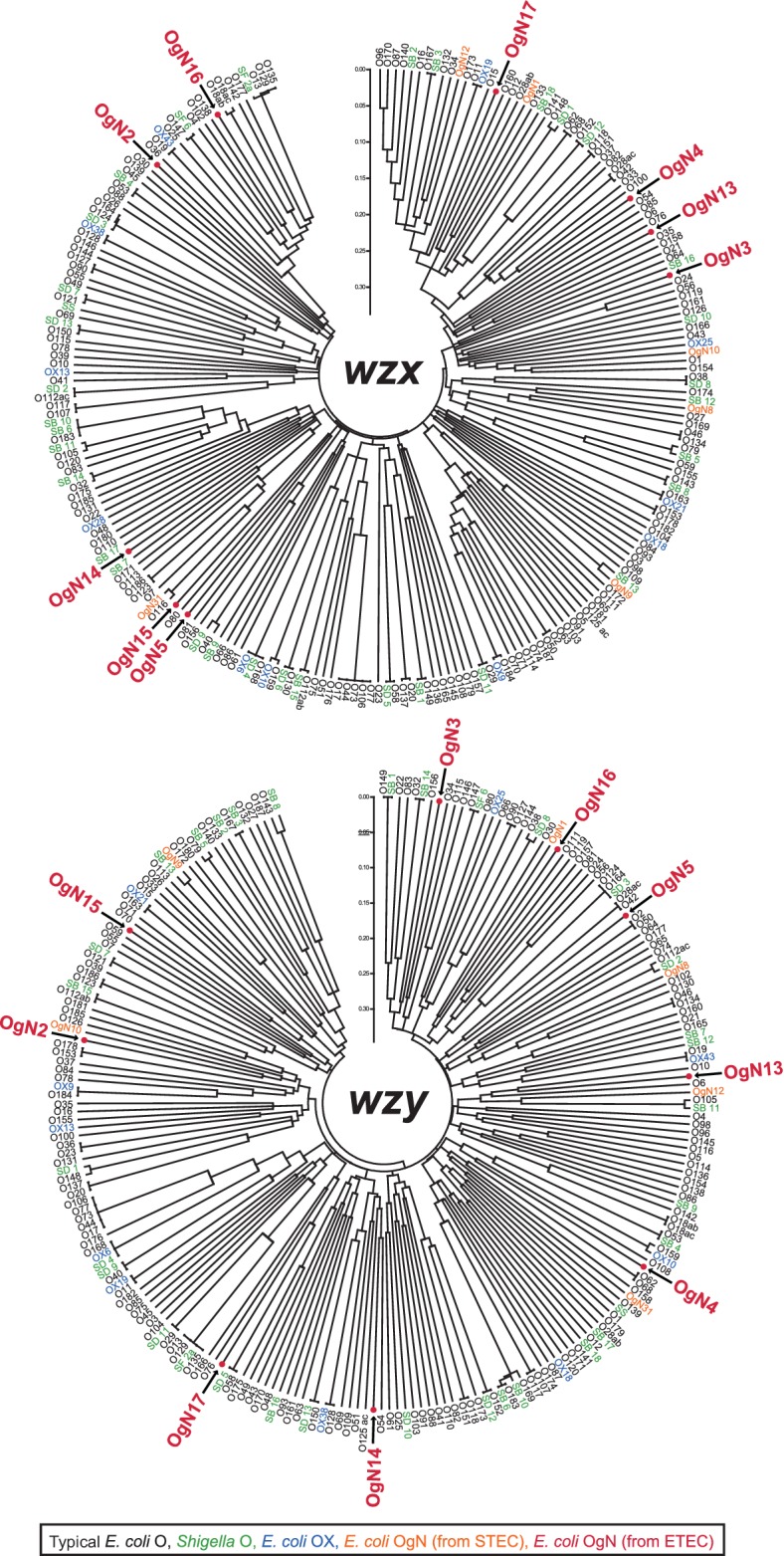
Phylogenetic analysis of wzx and wzy homologues from nine novel O-AGCs of ETEC and known E. coli and Shigella O-serogroups. STEC, Shiga toxin-producing E. coli.
Fig. 3.
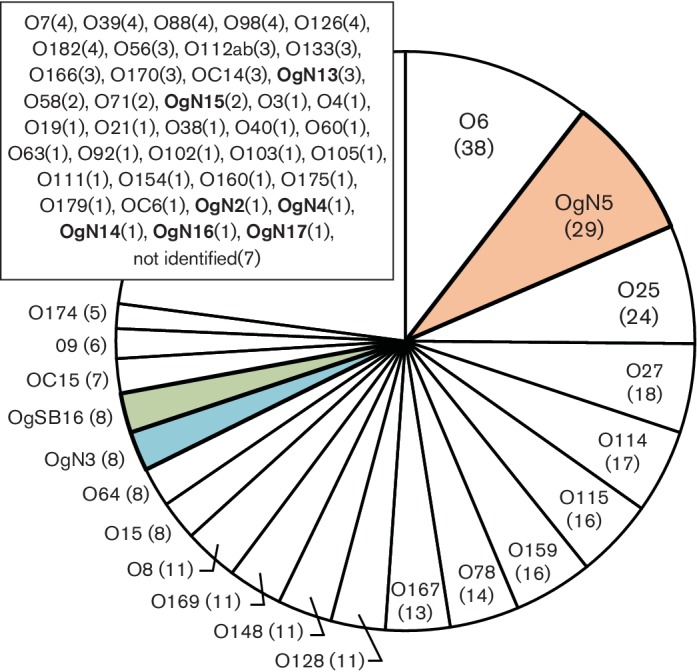
Molecular-based O-serogroup distribution of a global collection of 362 ETEC isolates. The numbers in parentheses indicate the number of isolates.
Six of eight OgN3 strains carried the fliC of H45, and were confirmed positive for STh and two CFs, CFA/I (a fimbria) and CS21 (a type IV pilus) (see Table S1). All OgN3 : H45 isolates were phylogenetically grouped into L6 of phylogroup A, and originated from Central and South America, including Mexico, Guatemala and Argentina (see Fig. S1, Table S1). Six of eight OSB16 strains carried the fliC of H32, and were confirmed positive for LT, LT+STh or LT+STp, and negative for all known CFs (see Table S1). All OSB16 : H32 and OSB16 : H2 isolates were phylogenetically grouped into L13 of phylogroup A, and originated from Guatemala and Argentina (see Fig. S1, Table S1). Additionally, three OgN13, two OgN15 and single OgN2, OgN4, OgN14, OgN16 and OgN17 strains were observed in our ETEC collection (see Table S1).
To aid in the identification of the predominant novel ETEC O-AGCs, we designed specific PCR primers for identifying OgN5, OgN3 and OSB16 targeting unique sequences on wzy genes (Table 1, Fig. 4), and their specificities were confirmed by using positive-control strains and all O-serogroup reference strains (O1–O188).
Table 1. PCR primers for identifying OgN3, OgN5 and OSB16.
| O genotype | Primer name | Sequence (5′→3′) | Target gene | Size (bp) |
|---|---|---|---|---|
| OgN3 | OgN3_PCR_F | GCTTGGCATCGTTGGGGATA | wzy | 189 |
| OgN3_PCR_R | TGCTACCAATCAGGCCGCTA | |||
| OgN5 | OgN5_PCR_F | GGTTTAAGCGACCCGTATCG | wzy | 650 |
| OgN5_PCR_R | CCAATTCCAGCCAGTGATGAG | |||
| OSB16 | OgSB16_PCR_F | AACCGCAGTGGAAACTGCA | wzy | 717 |
| OgSB16_PCR_R | AATTCCACATCAATCCACGGA |
Fig. 4.
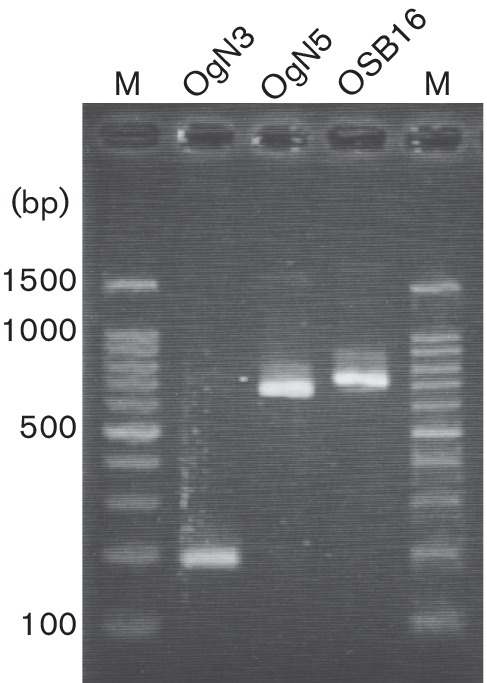
Gel image of PCR products obtained using the three primer pairs and positive control strains [OgN3, OT-37 (189 bp); OgN5, EHOUT43 (650 bp); OSB16, EH-OSB16 (717 bp)]. M, Marker.
Discussion
The results of the current study encapsulate problems with using a limited number of phenotypic markers for typing and tracking pathogenic bacteria of importance to human or animal health. It also highlights how, with whole-genome sequencing, there are still major discoveries to be made in identifying the isolates/lineages or types that are responsible for a significant burden of reported disease and yet untypeable by traditional methods. Since the O-serogroup is still used as a general marker for the subtyping of E. coli isolates, epidemiologically there can be an unawareness of the presence, emergence or spread of O-serogroup untypeable pathogenic E. coli groups. In this study, we used extracted O-AGC sequences from 362 ETEC genomes and by comparative genomics revealed nine novel O-genotypes that were previously unrecognized by the standard methods used for serotype detection and surveillance.
In recent surveillance studies, O6 has been shown to be the most common ETEC O-serogroup (mostly O6 : H16 serotype) associated with diarrhoeal patients in Egypt [11], Bolivia [12], Bangladesh [13], China [14] and Japan [15]. The O6-serogroup is mainly found in isolates with the CF profile CS1+CS2 and CS1+CS3, which are three of the most prevalent CFs identified in clinical isolates [16]. In our ETEC collection, the most prevalent O-serogroup was O6. O25 is also a well-known ETEC O-serogroup frequently isolated from ETEC patients, such as in Bosnia and Herzegovina [17], Bangladesh [13], China [14] and Japan [15]. Interestingly, in our representative ETEC collection, the novel O-genotype OgN5 was the second most common ETEC after O6 and before O25. Moreover, ETEC OgN5 isolates included those collected from children and adults in endemic regions such as Argentina, Egypt, Guatemala and Indonesia, as well as from travellers to Guatemala and Mexico, between 1989 and 2003. These data indicate that OgN5-positive ETEC isolates have spread both globally and over time.
Novel O-genotype ETEC isolates were found in several distinct phylogenetic lineages, and sequences of wzx/wzy from O-AGCs were highly conserved within each O-genotype, indicating that these O-AGCs have been spread across this species by horizontal gene transfer. Phylogenetic analysis in our previous study [3] showed that most ETEC isolates clustered based on their O-serogroups, such as O6 isolates belonging to L1+L2 (37/38), O25 belonging to L4 (22/24), O27 belonging to L9+L17 (18/18), O114 belonging to L4+L10 (14/17) and O115 belonging to L5 (16/16). In this study, ETEC OgN3 and OSB16 were mostly clustered in L16 and L13, respectively. However, OgN5 isolates were found in several distinct lineages, suggesting frequent exchanges of the O-AGC occur between lineages. Horizontal gene transfer-promoting sequences, such as transposase genes and repeat sequences, were not found in and around the OgN5 O-AGC.
Recently, we have been able to access whole-genome sequencing data due to the appearance of next-generation sequence technologies. A publicly available in silico-based serotyping web tool, SerotypeFinder, of the Center for Genomic Epidemiology (https://cge.cbs.dtu.dk/services/SerotypeFinder) [8], which has a database consisting of E. coli O-antigen associated genes (wzx, wzy, wzm and wzt) from O1 to O187 and H-antigen associated genes (fliC and its homologues) from H1 to H56 can be used to estimate E. coli O : H serotypes of sequenced isolates. If it is possible to organize and update the database with new wzx and wzy sequences, it becomes possible to meet the requirements for broader O-genotypes, including OgN5, OgN3 and OSB16 in the in silico typing.
In conclusion, by using detailed whole genome sequence-based analyses, it is clear that there is a hidden diversity of ETEC O-genotypes for which there is no prior information regarding their contribution to the burden of disease. It should be noted that even from our global collection, ETEC isolates from Africa are underrepresented. Hence, it is possible and perhaps likely that additional novel ETEC O-types and their associated lineages will continue to be discovered. These novel O-genotypes identified in this study have now been included in our comprehensive O-genotyping scheme and can be detected using the PCR-based method described in this study and by the in silico typing method.
Data bibliography
Bastin DA, Reeves PR. Sequence and analysis of the O antigen gene (rfb) cluster of Escherichia coli O111. Gene 1995;16:17–23. GenBank/EMBL/DDBJ accession no.: AF078736.
Beutin L, Kong Q, Feng L, Wang Q, Krause G et al.Development of PCR assays targeting the genes involved in synthesis and assembly of the new Escherichia coli O 174 and O 177 O antigens. J Clin Microbiol 2005;43:5143–5149. GenBank/EMBL/DDBJ accession no.: DQ008593.
Beutin L, Tao J, Feng L, Krause G, Zimmermann S et al. Sequence analysis of the Escherichia coli O15 antigen gene cluster and development of a PCR assay for rapid detection of intestinal and extraintestinal pathogenic E. coli O15 strains. J Clin Microbiol 2005;43:703–710. GenBank/EMBL/DDBJ accession no.: AY647261.
Cheng J, Wang Q, Wang W, Wang Y, Wang L et al. Characterization of E. coli O24 and O56 O antigen gene clusters reveals a complex evolutionary history of the O24 gene cluster. Curr Microbiol 2006;53:470–476. GenBank/EMBL/DDBJ accession nos: DQ220292, DQ220293.
Cheng J, Liu B, Bastin DA, Han W, Wang L et al. Genetic characterization of the Escherichia coli O66 antigen and functional identification of its wzy gene. J Microbiol 2007;45:69–74. GenBank/EMBL/DDBJ accession no.: DQ069297.
Cunneen MM, Reeves PR. The Yersinia kristensenii O11 O-antigen gene cluster was acquired by lateral gene transfer and incorporated at a novel chromosomal locus. Mol Biol Evol 2007;24:1355–1365. GenBank/EMBL/DDBJ accession no.: DQ180602.
D'Souza JM, Samuel GN, Reeves PR. Evolutionary origins and sequence of the Escherichia coli O4 O-antigen gene cluster. FEMS Microbiol Lett 2005;244:27–32. GenBank/EMBL/DDBJ accession no.: AY568960.
D'Souza JM, Wang L, Reeves P. Sequence of the Escherichia coli O26 O antigen gene cluster and identification of O26 specific genes. Gene 2002;297:123–127. GenBank/EMBL/DDBJ accession no.: AF529080.
DebRoy C, Fratamico PM, Roberts E, Davis MA, Liu Y. Development of PCR assays targeting genes in O-antigen gene clusters for detection and identification of Escherichia coli O45 and O55 serogroups. Appl Environ Microbiol 2005;71:4919–4924. GenBank/EMBL/DDBJ accession no.: AY771223.
DebRoy C, Fratamico PM, Yan X, Baranzoni G, Liu Y, et al. Comparison of O-antigen gene clusters of all O-serogroups of Escherichia coli and proposal for adopting a new nomenclature for O-typing. PLoS One 2016;11:e0147434. GenBank/EMBL/DDBJ accession nos: KJ778794, KJ778795, KJ778792, KP710591, KJ710507, KP868751, KJ739596, KP710594, KT207929, KJ739599, KP835691.
Feng L, Senchenkova SN, Yang J, Shashkov AS, Tao J, et al. Structural and genetic characterization of the Shigella boydii type 13 O antigen. J Bacteriol 2004;186:383–392. GenBank/EMBL/DDBJ accession no.: AY369140.
Feng L, Tao J, Guo H, Xu J, Li Y et al. Structure of the Shigella dysenteriae 7 O antigen gene cluster and identification of its antigen specific genes. Microb Pathog 2004;36:109–115. GenBank/EMBL/DDBJ accession no.: AY380835.
Feng L, Senchenkova SN, Yang J, Shashkov AS, Tao J et al. Synthesis of the heteropolysaccharide O antigen of Escherichia coli O52 requires an ABC transporter: structural and genetic evidence. J Bacteriol 2004;186:4510–4519. GenBank/EMBL/DDBJ accession no.: AY528413.
Feng L, Wang W, Tao J, Guo H, Krause G et al. Identification of Escherichia coli O114 O-antigen gene cluster and development of an O114 serogroup-specific PCR assay. J Clin Microbiol 2004;42:3799–3804. GenBank/EMBL/DDBJ accession no.: AY573377.
Feng L, Senchenkova SN, Wang W, Shashkov AS, Liu B et al. Structural and genetic characterization of the Shigella boydii type 18 O antigen. Gene 2005;355:79–86. GenBank/EMBL/DDBJ accession no.: AY948196.
Feng L, Senchenkova SN, Tao J, Shashkov AS, Liu B et al. Structural and genetic characterization of enterohemorrhagic Escherichia coli O145 O antigen and development of an O145 serogroup-specific PCR assay. J Bacteriol 2005;187:758-64. GenBank/EMBL/DDBJ accession no.: AY647260.
Feng L, Perepelov AV, Zhao G, Shevelev SD, Wang Q et al. Structural and genetic evidence that the Escherichia coli O148 O antigen is the precursor of the Shigella dysenteriae type 1 O antigen and identification of a glucosyltransferase gene. Microbiology 2007;153:139–147. GenBank/EMBL/DDBJ accession nos: DQ000313, DQ167407.
Fratamico PM, Briggs CE, Needle D, Chen CY, DebRoy, C. Sequence of the Escherichia coli O121 O-antigen gene cluster and detection of enterohemorrhagic E. coli O121 by PCR amplification of the wzx and wzy genes. J Clin Microbiol 2003;41:3379–3383. GenBank/EMBL/DDBJ accession no.: AY208937.
Fratamico PM, DebRoy C, Strobaugh TP Jr, Chen CY. DNA sequence of the Escherichia coli O103 O antigen gene cluster and detection of enterohemorrhagic E. coli O103 by PCR amplification of the wzx and wzy genes. Can J Microbiol 2005;51:515–522. GenBank/EMBL/DDBJ accession no.: AY532664.
Guo H, Feng L, Tao J, Zhang C, Wang L. Identification of Escherichia coli O172 O-antigen gene cluster and development of a serogroup-specific PCR assay. J Appl Microbiol 2004;97:181–190. GenBank/EMBL/DDBJ accession no.: AY545992.
Guo H, Yi W, Shao J, Lu Y, Zhang W, et al. Molecular analysis of the O-antigen gene cluster of Escherichia coli O86:B7 and characterization of the chain length determinant gene (wzz). Appl Environ Microbiol 2005;71:7995–8001. GenBank/EMBL/DDBJ accession no.: AY220982.
Guo H, Kong Q, Cheng J, Wang L, Feng L. Characterization of the Escherichia coli O59 and O155 O-antigen gene clusters: the atypical wzx genes are evolutionary related. FEMS Microbiol Lett 2005;15:153–161. GenBank/EMBL/DDBJ accession nos: AY654590, AY657020.
Han W, Liu B, Cao B, Beutin L, Krüger U et al. DNA microarray-based identification of serogroups and virulence gene patterns of Escherichia coli isolates associated with porcine postweaning diarrhea and edema disease. Appl Environ Microbiol 2007;73:4082–4088. GenBank/EMBL/DDBJ accession nos: DQ868764–DQ868766.
Hu B, Perepelov AV, Liu B, Shevelev SD, Guo D et al. Structural and genetic evidence for the close relationship between Escherichia coli O71 and Salmonella enterica O28 O-antigens. FEMS Immunol Med Microbiol 2010;59:161–169. GenBank/EMBL/DDBJ accession no.: GU445927.
Iguchi A, Iyoda S, Seto K, Ohnishi M, EHEC Study Group. Emergence of a novel Shiga toxin-producing Escherichia coli O serogroup cross-reacting with Shigella boydii type 10. J Clin Microbiol 2011;49:3678–3680. GenBank/EMBL/DDBJ accession no.: AB627352.
Iguchi A, Iyoda S, Kikuchi T, Ogura Y, Katsura K et al. A complete view of the genetic diversity of the Escherichia coli O-antigen biosynthesis gene cluster. DNA Res 2015;22:101–107. GenBank/EMBL/DDBJ accession nos: AB811596–AB811624, AB812020–AB812085, AB972413–AB972424.
Iguchi A, Iyoda S, Seto K, Nishii H, Ohnishi M et al. Six novel O genotypes from Shiga toxin-producing Escherichia coli. Front Microbiol 2016;7:765. GenBank/EMBL/DDBJ accession nos: LC125927–LC125932.
Kido N, Torgov VI, Sugiyama T, Uchiya K, Sugihara H et al. Expression of the O9 polysaccharide of Escherichia coli: sequencing of the E. coli O9 rfb gene cluster, characterization of mannosyl transferases, and evidence for an ATP-binding cassette transport system. J Bacteriol 1995;177:2178-2187. GenBank/EMBL/DDBJ accession no.: D43637.
Li D, Liu B, Chen M, Guo D, Guo X et al. A multiplex PCR method to detect 14 Escherichia coli serogroups associated with urinary tract infections. J Microbiol Methods 2010;82:71-77. GenBank/EMBL/DDBJ accession nos: GU299791, GU299792, GU299795.
Li X, Perepelov AV, Wang Q, Senchenkova SN, Liu B et al. Structural and genetic characterization of the O-antigen of Escherichia coli O161 containing a derivative of a higher acidic diamino sugar, legionaminic acid. Carbohydr Res 2010;345:1581–1587. GenBank/EMBL/DDBJ accession nos: GU220361, GU220362.
Li Y, Perepelov AV, Guo D, Shevelev SD, Senchenkova SN et al. Structural and genetic relationships of two pairs of closely related O-antigens of Escherichia coli and Salmonella enterica: E. coli O11/S. enterica O16 and E. coli O21/S. enterica O38. FEMS Immunol Med Microbiol 2011;61:258–268. GenBank/EMBL/DDBJ accession no.: HQ388393.
Liu B, Senchenkova SN, Feng L, Perepelov AV, Xu T et al. Structural and molecular characterization of Shigella boydii type 16 O antigen. Gene 2006;380:46–53. GenBank/EMBL/DDBJ accession no.: DQ371800.
Liu B, Knirel YA, Feng L, Perepelov AV, Senchenkova SN et al. Structure and genetics of Shigella O antigens. FEMS Microbiol Rev 2008;32:627–653. GenBank/EMBL/DDBJ accession nos: EU294162–EU294178, EU296402–EU296404, EU296406–EU296418, EU296421.
Liu B, Wu F, Li D, Beutin L, Chen M et al. Development of a serogroup-specific DNA microarray for identification of Escherichia coli strains associated with bovine septicemia and diarrhea. Vet Microbiol 2009;142:373-378. GenBank/EMBL/DDBJ accession nos: FJ940774, FJ940775, GQ499368.
Liu B, Perepelov AV, Li D, Senchenkova SN, Han Y et al. Structure of the O-antigen of Salmonella O66 and the genetic basis for similarity and differences between the closely related O-antigens of Escherichia coli O166 and Salmonella O66. Microbiology 2010;156:1642–1649. GenBank/EMBL/DDBJ accession no.: GU299794.
Liu Y, DebRoy C, Fratamico P. Sequencing and analysis of the Escherichia coli serogroup O117, O126, and O146 O-antigen gene clusters and development of PCR assays targeting serogroup O117-, O126-, and O146-specific DNA sequences. Mol Cell Probes 2007;21:295–302. GenBank/EMBL/DDBJ accession no.: DQ465247-DQ465249.
Marolda CL, Feldman MF, Valvano MA. Genetic organization of the O7-specific lipopolysaccharide biosynthesis cluster of Escherichia coli VW187 (O7:K1). Microbiology 1999;145:2485–2495. GenBank/EMBL/DDBJ accession no.: AF125322.
Morona R, Mavris M, Fallarino A, Manning PA. Characterization of the rfc region of Shigella flexneri. J Bacteriol 1994;176:733–747. GenBank/EMBL/DDBJ accession no.: X71970.
Paton AW, Paton JC. Molecular characterization of the locus encoding biosynthesis of the lipopolysaccharide O antigen of Escherichia coli serotype O113. Infect Immun 1999;67:5930–5937. GenBank/EMBL/DDBJ accession no.: AF172324.
Perelle S, Dilasser F, Grout J, Fach P. Identification of the O-antigen biosynthesis genes of Escherichia coli O91 and development of a O91 PCR serotyping test. J Appl Microbiol 2002;93:758–764. GenBank/EMBL/DDBJ accession no.: AY035396.
Perepelov AV, Li D, Liu B, Senchenkova SN, Guo D et al. Structural and genetic characterization of the closely related O-antigens of Escherichia coli O85 and Salmonella enterica O17. Innate Immun 2008;17:164–73. GenBank/EMBL/DDBJ accession no.: GU299798.
Perepelov AV, Li D, Liu B, Senchenkova SN, Guo D et al. Structural and genetic characterization of Escherichia coli O99 antigen. FEMS Immunol Med Microbiol 2009;57:80–87. GenBank/EMBL/DDBJ accession no.: FJ940773.
Perepelov AV, Ni Z, Wang Q, Shevelev SD, Senchenkova SN et al. Structure and gene cluster of the O-antigen of Escherichia coli O109; chemical and genetic evidences of the presence of L-RhaN3N derivatives in the O-antigens of E. coli O109 and O119. FEMS Immunol Med Microbiol 2010;61:47–53. GenBank/EMBL/DDBJ accession no.: HM485572.
Ren Y, Liu B, Cheng J, Liu F, Feng L et al. Characterization of Escherichia coli O3 and O21 O antigen gene clusters and development of serogroup-specific PCR assays. J Microbiol Methods 2008;75:329–334. GenBank/EMBL/DDBJ accession no.: EU694097, EU694097.
Senchenkova SN, Feng L, Yang J, Shashkov AS, Cheng J et al. Structural and genetic characterization of the Shigella boydii type 10 and type 6 O antigens. J Bacteriol 2005;187:2551-2554. GenBank/EMBL/DDBJ accession no.: AY693427.
Senchenkova SN, Feng L, Wang Q, Perepelov AV, Qin D et al. Structural and genetic characterization of Shigella boydii type 17 O antigen and confirmation of two new genes involved in the synthesis of glucolactilic acid. Biochem Biophys Res Commun 2006;349:289–295. GenBank/EMBL/DDBJ accession no.: DQ875941.
Shao J, Li M, Jia Q, Lu Y, Wang PG. Sequence of Escherichia coli O128 antigen biosynthesis cluster and functional identification of an alpha-1,2-fucosyltransferase. FEBS Lett 2003;553:99–103. GenBank/EMBL/DDBJ accession no.: AY217096.
Tao J, Feng L, Guo H, Li Y, Wang L. The O-antigen gene cluster of Shigella boydii O11 and functional identification of its wzy gene. FEMS Microbiol Lett 2004;234:125–132. GenBank/EMBL/DDBJ accession no.: AY529126.
Tao J, Wang L, Liu D, Li Y, Bastin DA et al. Molecular analysis of Shigella boydii O1 O-antigen gene cluster and its PCR typing. Can J Microbiol 2005;51:387–392. GenBank/EMBL/DDBJ accession no.: AY630255.
Wang L, Reeves PR. Organization of Escherichia coli O157 O antigen gene cluster and identification of its specific genes. Infect Immun 1998;66:3545–3551. GenBank/EMBL/DDBJ accession no.: AF061251.
Wang L, Qu W, Reeves PR. Sequence analysis of four Shigella boydii O-antigen loci: implication for Escherichia coli and Shigella relationships. Infect Immun 2001;69:6923–6930. GenBank/EMBL/DDBJ accession no.: AF402312–AF402315.
Wang L, Briggs CE, Rothemund D, Fratamico P, Luchansky JB et al. Sequence of the E. coli O104 antigen gene cluster and identification of O104 specific genes. Gene 2001;30:231–236. GenBank/EMBL/DDBJ accession no.: AF361371.
Wang L, Huskic S, Cisterne A, Rothemund D, Reeves PR. The O-antigen gene cluster of Escherichia coli O55:H7 and identification of a new UDP-GlcNAc C4 epimerase gene. J Bacteriol 2002;184:2620–2625. GenBank/EMBL/DDBJ accession no.: AF461121.
Wang L, Liu B, Kong Q, Steinrück H, Krause G et al. Molecular markers for detection of pathogenic Escherichia coli strains belonging to serogroups O 138 and O 139. Vet Microbiol 2005;111:181–90. GenBank/EMBL/DDBJ accession nos: DQ109551, DQ109552.
Wang Q, Perepelov AV, Feng L, Knirel YA, Li Y et al. Genetic and structural analyses of Escherichia coli O107 and O117 O-antigens. FEMS Immunol Med Microbiol 2009;55:47–54. GenBank/EMBL/DDBJ accession no.: EU694095.
Wang Q, Wang S, Beutin L, Cao B, Feng L et al. Development of a DNA microarray for detection and serotyping of enterotoxigenic Escherichia coli. J Clin Microbiol 2010;48:2066–2074. GenBank/EMBL/DDBJ accession nos: GU014554, GU014555.
Wang Q, Ruan X, Wei D, Hu Z, Wu L et al. Development of a serogroup-specific multiplex PCR assay to detect a set of Escherichia coli serogroups based on the identification of their O-antigen gene clusters. Mol Cell Probes 2010;24:286–290. GenBank/EMBL/DDBJ accession nos: GU068041, GU068044–GU068046.
Xu DQ, Cisar JO, Ambulos Jr N, Burr DH et al. Molecular cloning and characterization of genes for Shigella sonnei form I O polysaccharide: proposed biosynthetic pathway and stable expression in a live salmonella vaccine vector. Infect Immun 2002;70:4414–4423. GenBank/EMBL/DDBJ accession no.: AF294823.
Xu DQ, Cisar JO, Osorio M, Wai TT, Kopecko DJ. Core-linked LPS expression of Shigella dysenteriae serotype 1 O-antigen in live Salmonella Typhi vaccine vector Ty21a: preclinical evidence of immunogenicity and protection. Vaccine 2007;25:6167–6175. GenBank/EMBL/DDBJ accession no.: AY585348.
Funding information
This research was partially supported by Health Labor Sciences Research Grants from the Ministry of Health, Labor, and Welfare, Japan (H27-Syokuhin-Wakate-018), and by the Research Program on Emerging and Re-emerging Infectious Diseases from the Japan Agency for Medical Research and Development (AMED) (15fk0108008h0001).
Conflicts of interest
The authors declare that there are no conflicts of interest.
Supplementary Data
Supplementary Data
References
- 1.Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet. 2013;382:209–222. doi: 10.1016/S0140-6736(13)60844-2. [DOI] [PubMed] [Google Scholar]
- 2.WHO . Diarrhoeal diseases: enterotoxigenic Escherichia coli (ETEC) Geneva: World Health Organization; 2009. The Initiative for Vaccine Research. (Updated February 2009)www.who.int/vaccine_research/diseases/diarrhoeal/en/index4.html [Google Scholar]
- 3.von Mentzer A, Connor TR, Wieler LH, Semmler T, Iguchi A, et al. Identification of enterotoxigenic Escherichia coli (ETEC) clades with long-term global distribution. Nat Genet. 2014;46:1321–1326. doi: 10.1038/ng.3145. [DOI] [PubMed] [Google Scholar]
- 4.Iguchi A, Iyoda S, Kikuchi T, Ogura Y, Katsura K, et al. A complete view of the genetic diversity of the Escherichia coli O-antigen biosynthesis gene cluster. DNA Res. 2015;22:101–107. doi: 10.1093/dnares/dsu043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu B, Knirel YA, Feng L, Perepelov AV, Senchenkova SN, et al. Structure and genetics of Shigella O antigens. FEMS Microbiol Rev. 2008;32:627–653. doi: 10.1111/j.1574-6976.2008.00114.x. [DOI] [PubMed] [Google Scholar]
- 6.DebRoy C, Fratamico PM, Yan X, Baranzoni G, Liu Y, et al. Comparison of O-antigen gene clusters of all O-serogroups of Escherichia coli and proposal for adopting a new nomenclature for O-typing. PLoS One. 2016;11:e0147434. doi: 10.1371/journal.pone.0147434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iguchi A, Iyoda S, Seto K, Nishii H, Ohnishi M, et al. Six novel O genotypes from Shiga toxin-producing Escherichia coli. Front Microbiol. 2016;7:765. doi: 10.3389/fmicb.2016.00765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Joensen KG, Tetzschner AM, Iguchi A, Aarestrup FM, Scheutz F. Rapid and easy in silico serotyping of Escherichia coli isolates by use of whole-genome sequencing data. J Clin Microbiol. 2015;53:2410–2426. doi: 10.1128/JCM.00008-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary Genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 10.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shaheen HI, Khalil SB, Rao MR, Abu Elyazeed R, Wierzba TF, et al. Phenotypic profiles of enterotoxigenic Escherichia coli associated with early childhood diarrhea in rural Egypt. J Clin Microbiol. 2004;42:5588–5595. doi: 10.1128/JCM.42.12.5588-5595.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rodas C, Mamani R, Blanco J, Blanco JE, Wiklund G, et al. Enterotoxins, colonization factors, serotypes and antimicrobial resistance of enterotoxigenic Escherichia coli (ETEC) strains isolated from hospitalized children with diarrhea in Bolivia. Braz J Infect Dis. 2011;15:132–137. doi: 10.1590/S1413-86702011000200007. [DOI] [PubMed] [Google Scholar]
- 13.Ansaruzzaman M, Bhuiyan NA, Begum YA, Kühn I, Nair GB, et al. Characterization of enterotoxigenic Escherichia coli from diarrhoeal patients in Bangladesh using phenotyping and genetic profiling. J Med Microbiol. 2007;56:217–222. doi: 10.1099/jmm.0.46473-0. [DOI] [PubMed] [Google Scholar]
- 14.Chen Y, Chen X, Zheng S, Yu F, Kong H, et al. Serotypes, genotypes and antimicrobial resistance patterns of human diarrhoeagenic Escherichia coli isolates circulating in southeastern China. Clin Microbiol Infect. 2014;20:52–58. doi: 10.1111/1469-0691.12188. [DOI] [PubMed] [Google Scholar]
- 15.Konishi N, Obata H, Monma C, Nakama A, Kai A, et al. Bacteriological and epidemiological characteristics of enterotoxigenic Escherichia coli isolated in Tokyo, Japan, between 1966 and 2009. J Clin Microbiol. 2011;49:3348–3351. doi: 10.1128/JCM.02576-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Svennerholm AM, Lundgren A. Recent progress toward an enterotoxigenic Escherichia coli vaccine. Expert Rev Vaccines. 2012;11:495–507. doi: 10.1586/erv.12.12. [DOI] [PubMed] [Google Scholar]
- 17.Dedeić-Ljubović A, Hukić M, Bekić D, Zvizdić A. Frequency and distribution of diarrhoeagenic Escherichia coli strains isolated from pediatric patients with diarrhoea in Bosnia and Herzegovina. Bosn J Basic Med Sci. 2009;9:148–155. doi: 10.17305/bjbms.2009.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.