Abstract
lncRNA-X-inactive specific transcript (lncRNA XIST) has been demonstrated to be a tumor suppressor involved in the pathogenesis and development of various cancers. However, the function of XIST and its working mechanism in osteosarcoma (OS) remain enigmatic. Firstly, we determined the expression of XIST in OS tissues and cell lines by quantitative reverse transcription-PCR (qRT-PCR) and explored whether aberrant XIST expression was associated with recurrence and short overall survival. Furthermore, the effects of XIST on osteosarcoma cells were studied by lentivirus mediated overexpression approach in vitro and in vivo. Detection of a set of epithelial-mesenchymal transition (EMT) markers was performed to explore whether XIST is involved in EMT. Finally, we investigated the regulatory mechanism of XIST acting as a competitive endogenous RNA (ceRNA) of miR-21-5p in OS progression and metastasis. lncRNA XIST was significantly downregulated in osteosarcoma tissues and osteosarcoma cells, and associated with recurrence and short overall survival in OS patients. XIST overexpression remarkably inhibited the proliferation of OS cells as well as the xenograft tumor formation in vivo. Both cell invasion and migration were inhibited by XIST overexpression via suppressing the EMT process. These results indicated that XIST functioned as a tumor suppressor in OS. Moreover, we found that miR-21-5p interacted with XIST by directly targeting the miRNA-binding site in the XIST sequence, and qRT-PCR results showed XIST and miR-21-5p could affect each other's expression, respectively. The following assays verified that the tumor suppressor, PDCD4 was a functional target of miR-21-5p in OS cells. Finally, we affirmed that XIST regulated PDCD4 expression by competitively binding to miR-21-5p. XIST inhibited cell proliferation and cell mobility by competitively binding to miR-21-5p and upregulating PDCD4 in OS. Our study demonstrated that lncRNA-XIST, which acts as a miRNA sponge, impedes miR-21-5p to maintain the expression of PDCD4, which contributes to the progression of OS. Our findings suggest that the newly identified XIST/miR-21-5p/PDCD4 axis could be a potential biomarker or therapeutic target for OS.
Keywords: osteosarcoma, lncRNA-XIST, miR-21-5p, ceRNA, PDCD4
Introduction
Osteosarcoma (OS) is the most common primary tumor of bone with high mortality and poor prognosis (1). In recent years, the adjuvant chemotherapy and radiotherapy are common treatments for OS. However, owing to the recurrence and metastasis, the overall 5-year survival rate for osteosarcoma patients remains unsatisfactory (2-4). Therefore, the development of new diagnosis and treatment strategies is required to reduce recurrence and improve the survival rate.
Long non-coding RNAs (lncRNAs) are a class of transcripts longer than 200 nucleotides in length with little functional protein-coding ability. Emerging data have reported that lncRNAs are involved in a wide range of biological processes, such as proliferation, apoptosis, and differentiation (5,6). Several lncRNAs, such as MEG3 (maternally expressed gene 3) (7) and TUG1 (taurine upregulated gene 1) (8) have been reported to play crucial roles during the development and progression of OS. lncRNA XIST (X-inactive specific transcript) is a product of the XIST gene which has been found to be dysregulated in a variety of human cancers, such as hepatocellular carcinoma (HCC), gastric cancer (GC) and human nasopharyngeal carcinoma (NPC) (9-11). Furthermore, increasing evidence demonstrated that XIST can act as tumor suppressors, and play important roles in carcinogenesis and cancer development (12-15). Silence of XIST reduced cell proliferation, migration and invasion as well as inducing apoptosis in human glioblastoma stem cells (15). XIST was reported to inhibit HCC cell proliferation and metastasis (14). Recently, a study from Huang et al showed that XIST expression was decreased in breast tumor samples and breast cancer cell lines and functioned as a tumor suppressor through inhibition of AKT activation (13). However, it is unknown whether XIST plays a tumor suppressive role in OS.
Abundant evidence indicates that lncRNAs suppress the expression and biological functions of miRNAs by acting as a competitive endogenous RNA (16,17). For example, HOTAIR is specifically upregulated in gastric cancer and functions as a ceRNA to regulate human epithelial growth factor receptor 2 (HER2) expression by competitively binding to miR-331-3p (18). lncRNA H19 functions as a miRNA sponge to restrain the activity of many miRNAs, such as miR-138 and miR-200a, leading to the suppression of its target gene, vimentin, ZEB1, and ZEB2, thereby promoting EMT progression in colorectal cancer (CRC) (19). A recent study from Chen et al showed that knockdown of lncRNA XIST exerted its tumor-suppressive effect though downregulating the expression of EZH2 via miR-101 in gastric pathogenesis (20). However, whether lncRNA-XIST affects the biological behavior of OS cells by regulating miRNAs has not yet been reported.
In the present study, we found that XIST was significantly downregulated in both OS tissues and cell lines, and over-expression of XIST inhibited cell proliferation and metastasis in vitro as well as tumorigenesis in vivo. Then, we investigated the molecular mechanism of XIST in the progression of OS and the reciprocal regulation between XIST and miR-21-5p, a potent oncogenic microRNA. Our findings provide new insights into the molecular function of XIST/miR-21-5p/PDCD4 signaling pathway in OS, and will give a novel strategy for the treatment of OS.
Materials and methods
Cell culture and tissue samples
Osteosarcoma cell lines MG63, U2OS, Saos2, HOS, SOSP-9697 and SV40 immortalized human fetal osteoblastic cell line hFOB 1.19 were cultured in were grown in RPMI-1640 medium supplemented with 10% fetal bovine serum (Gibco, Beijing, China). Cultures were maintained at 37°C in a humidified atmosphere with 5% CO2. A total of 50 pairs of tumor tissues and adjacent normal tissues were collected from routine therapeutic surgery at our department. All samples were obtained with informed consent and approved by the hospital institutional review board.
Quantitative reverse transcription-PCR (qRT-PCR)
Total RNAs from frozen OS and paired non-cancerous tissues or cell lines were extracted using TRIzol (Invitrogen, Carlsbad, CA, USA) according to the instructions provided by the manufacturer. cDNA was synthesized by reverse transcribing the total RNA using PrimeScript™ RT Master Mix (Takara Biotechnology, Dalian, China). The expression level of XIST and miR-21-5p was detected by qRT-PCR using the Ultra SYBR Mixture with ROX (CWBio Co., Ltd.) and ABI7100 system (Applied Biosystems, Darmstadt, Germany). GAPDH and U6 were used as internal controls for XIST, PDCD4 and miR-21-5p. All qRT-PCR reactions were performed in triplicate. Relative quantification of gene expression was calculated by the 2−ΔΔCt method. The primers used in this study were: XIST forward 5′-CTAGCTAGCTTTTGTAGTGAGCTT GCTCCT-3′ and reverse 5′-GCTCTAGAATGTCTCCATCTCCATTTTGC-3′; PDCD4 forward 5′-CGACAGTGGGAGTGACGCCCTTA-3′; reverse 5′-CAGACACCTTTGCCTCC TGCACC-3′; GAPDH forward 5′-GCACCGTCAAGGCTG AGAAC-3′ and reverse 5′-TGGTGAAGACGCCAGTGGA-3′; miR-21-5p forward 5′-GTGCAGGGTCCGAGGT-3′, reverse 5′-GCCGCTAGCTTATCAGACTGATGT-3′; U6 forward 5′-TGCGGGTGCTCGCTTCGGCAGC-3′ and reverse 5′-CCA GTGCAGGGTCCGAGGT-3′.
Cell transfection
XIST overexpression lentivirus (Lv-XIST) vector as well as the NC lentivirus vector was obtained from GenePharma (Shanghai, China). The HEK293 cells were cotransfected with Lenti-Pac HIV Expression Packaging Mix and the lentiviral vectors (or the control lentivirus vectors) using Lipofectamine 2000 (Life Technologies Corp., Carlsbad, CA, USA). After 48 h, lentiviral particles in the supernatant were harvested and filtered by centrifugation at 600 × g for 10 min. The packaged lentiviruses were named Lv-XIST and Lv-control. The MG63 and Saos2 cells were then infected with Lv-XIST or Lv-control at a MOI of 20.
miR-21-5p mimics, miR-21-5p inhibitor and miR-21-5p negative control (NC), small interfering RNA (siRNAs) against PDCD4 (si-PDCD4) and si-NC were also employed and synthesized by GenePharma, and transfected into MG63 and Saos2 cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. All the transfections were repeated more than three times independently.
Immunohistochemical staining
The tumor tissues were paraffin-embedded and cut into 5-µm-thick slides for immuno-histochemical analysis. After incubating in 0.15% Triton X-100 at room temperature and blocking with 1% goat serum albumin in modified D-PBS Tween-20 for 1 h, the sections were incubated overnight with PDCD4 antibody (1:1500, Cell Signaling Technology, Danvers, MA, USA). After washing with PBS three times, the sections were incubated with secondary antibodies (anti-rabbit IgG antibodies) at 37°C for 30 min, and visualized with diaminobenzidine (DAB). Finally, the slides were counterstained with 10% hematoxylin, and analyzed under a light microscope with a digital camera. Image analysis was performed by Image-Pro Plus software.
CCK-8 assay
MG63 and Saos2 cells were seeded into a 96-well plate (5×103 cells/well) and cultured at 37°C and cell proliferation was measured by the Cell Counting Kit-8 (CCK-8; Dojindo, Kumamoto, Japan) according to the manufacturer's protocol. At different time points, 10 µl CCK-8 solution was added into each well and incubated for an additional 2 h at 37°C. The absorbance at 450 nm was measured using a microplate reader (ELx800; BioTek Instuments, Inc., Winooski, VT, USA).
Apoptosis assay
Cell apoptosis was measured by flow cytometry following the instructions of the Annexin V-FITC/PI apoptosis detection kit (KeyGEN, Nanjing, China). Cultured cells were harvested and washed twice in PBS, re-suspended in binding buffer, and then incubated with Annexin V-FITC and propidium iodide (PI) for 15 min at room temperature. Afterwards, flow cytometry was performed to determine rate of apoptosis on a FACSAria flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA).
In vitro invasion and migration assay
At 24 h after transfection, cell invasion of MG63 and Saos2 cells was assessed using Transwell chambers (8 µm, 24-well format; Corning Inc., Corning, NY, USA) as previously described in detail (14). The number of invaded cells was calculated by counting five random views under the microscope. The experiment was performed in triplicate and repeated three times.
At 24 h after transfection, MG63 and Saos2 cells were seeded into 96-well plates (3×103 cells/well), and wound healing assays was monitored to measure cell migration as previously described (14). The wound closure was observed and imaged under a microscope. Then, the wound area was measured and the percentage of the wound healing was calculated by ImageJ software (NIH, Bethesda, MD, USA).
Western blotting
Total cellular proteins were extracted using RIPA lysis buffer containing proteinase inhibitor (Sigma, St. Louis, MO, USA). Concentrations of total cellular protein were determined using a BCA assay kit (Pierce, Rockford, IL, USA). Total protein samples (25 µg) were analyzed by 8% SDS-PAGE gel. The protein was transferred to polyvinylidene difluoride (PVDF) membranes by a wet blotting procedure (100 V, 120 min, 4°C). After blocked with 5% blocking buffer, the membranes were incubated with primary antibodies at 4°C overnight using the following concentration: cleaved caspase-3, cleaved caspase-9, Bax, Bcl-2 (1:1000, Cell Signaling Technology), PDCD4, ZEB1, vimentin, N-cadherin, E-cadherin (1:2000; Cell Signaling Technology) and anti-β-actin (1:500; Santa Cruz Biotechnology, Santa Cruz, CA, USA), followed by horseradish peroxidase-conjugated secondary antibody (anti-rabbit, 1:2000, Cell Signaling Technology). Anti-β-actin antibody was used as an internal control. The protein bands were visualized by enhanced chemiluminescence detection reagents (Applygen Technologies, Inc., Beijing, China) according to the manufacturer's instructions. Relative band intensities were determined by densitometry using Scion image software (version 4.0).
Luciferase reporter assay
The fragment from XIST containing the predicted miR-21-5p binding site was amplified by PCR and cloned into a pmirGLO Dual-luciferase Target Expression Vector (Promega, Madison, WI, USA) to form the reporter vector XIST-wild-type (XIST-Wt). To test the binding specificity, the corresponding mutant was created by mutating the miR-21-5p seed region binding site (seed sequence binding fragment 5′-TATTCGA-3′ changed to 5′-CGTGAGC-3′), which were named as pmirGLO-XIST-Mt (XIST-MUT). pmirGLO-XIST-Mt or pmirGLO-XIST-WT was co-transfected with miR-21-5p mimics, miR-21-5p inhibitor or miR-21-5p NC into HEK 293T cells using Lipofectamine 2000. Luciferase reporter assay was performed using the Dual-Luciferase Reporter Assay System (Promega) 48 h later, the firefly luciferase activity was measured and normalized by Renilla luciferase activity.
OS xenograft mouse model
After infection with Lv-XIST (1.2×107 TU) or Lv-control, MG63 and Saos2 cells (5×106) were suspended in 100 µl PBS and injected subcutaneously to the right flank of the BALB/c nude mice purchased from the Shanghai Institute of Materia Medica (Shanghai, China). The tumor volume was measured every week and calculated as follows: tumor volume (mm3) = (length x width2)/2. On day 35 after cell implantation, the mice were sacrificed and the tumor specimens were removed and weighed. All animal studies were conducted in the Animal Institute of Shanghai Jiaotong University according to the protocols approved by the Medical Experimental Animal Care Commission of Shanghai Jiaotong University.
Statistical analysis
Statistical analyses were performed with SPSS 13.0 software. The results were evaluated by χ2 test and the other data were evaluated by Student's t-test and expressed as the mean ± SD from three independent experiments. A P-value of <0.05 was considered to indicate a statistically significant difference.
Results
Downregulation of XIST is correlated with poor outcome of OS patients
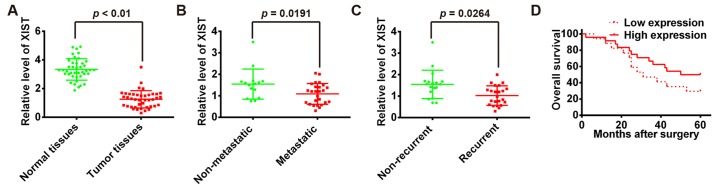
To elucidate the expression of XIST in OS, qRT-PCR was performed to detect the expression of XIST in 41 pairs of human osteosarcoma and adjacent normal tissues (among which 26 metastases and 15 non-metastases osteosarcoma tissues, 23 recurrent and 18 non-recurrent osteosarcoma tissues). The transcript level of XIST was lower in OS tissues when compared with normal tissues (Fig. 1A). In addition, the expression level of XIST was also significantly lower in metastatic group compared with non-metastatic group (Fig. 1B). It was noteworthy that XIST low-expression was significantly associated with tumor recurrence (Fig. 1C). Furthermore, the Kaplan-Meier method and log-rank test revealed that patients with high XIST expression in OS had significantly longer overall survival than those with low XIST expression (Fig. 1D). Taken together, these data suggest decreased XIST expression might be critically involved in OS progression.
Figure 1.
lncRNA XIST is expressed at low level in human OS. (A) qRT-PCR analysis of XIST expression levels in 41 pairs of human osteosarcoma and adjacent normal tissues. P<0.01 vs. normal tissue group. (B) Comparison of the expression of XIST between metastatic (n=26) and non-metastatic (n=15) osteosarcoma tissues. P=0.0191. (C) Comparison of the expression of XIST between tumor tissues from the recurrent (n=23) and non-recurrent (n=18) group. P=0.0264. (D) The 41 patients were classified into high (n=24) and low (n=17) XIST expression groups, according to the median value of XIST expression. Kaplan-Meier analyses of the associations between XIST expression level and overall survival of patients with OS (The log-rank test was used to calculate P-values). **P<0.01 vs. low XIST expression group.
Overexpression of XIST inhibits OS cell growth in vitro as well as tumorigenesis in vivo
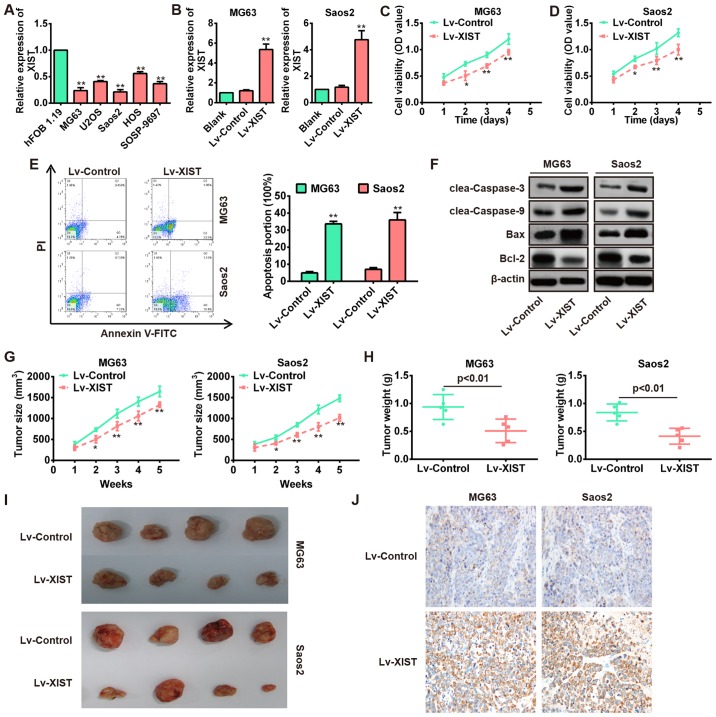
It has been reported that XIST exerted tumor-suppressive effects in many cancers (12,20). However, it is not clear whether XIST functions as a tumor suppressor in OS. To explore the biological functions of XIST in OS, we first measured the expression levels of XIST in five OS cell lines including mG63, U2OS, Saos2, HOS and SOSP-9697. Consistent with expression levels in OS tissues, the expression levels of XIST markedly decreased in OS cell lines, especially in MG63 and Saos2 cells compared with the human osteoblast cell line hFOB 1.19 (Fig. 2A). Then, the lentivirus containing XIST (MOI=20) was infected into both mG63 and Saos2 cells and its overexpression efficiency was remarkable compared with Lv-control (Fig. 2B). CCK-8 assays were performed to detect the impact of XIST over expression on cell viability of OS cell lines. As shown in Fig. 2C and D, overexpression of XIST significantly inhibited the cell proliferation compared with Lv-control group. Subsequently, flow cytometry showed that overexpression of XIST significantly increased the percentage of apoptotic cells in both MG63 and Saos2 cells (Fig. 2E). In addition, we detected the protein levels of classic apoptotic markers in MG63 and Saos2 cells. As expected, western blot analysis showed that XIST overexpression markedly increased the expression levels of cleaved-caspase-3, cleaved-caspase-9 and Bax, while that of anti-apoptotic protein Bcl-2 was decreased (Fig. 2F).
Figure 2.
Overexpression of XIST inhibits the proliferation, invasion and migration as well as promotes apoptosis of OS cells. (A) qRT-PCR analysis of XIST expression levels in five OS cell lines (mG63, U2OS, Saos2, HOS and SOSP-9697). **P<0.01 vs. hFOB 1.19. (B) qRT-PCR analysis of XIST in MG63 and Saos2 cells infected with lentivirus overexpressing XIST (Lv-XIST). **P<0.01 vs. Lv-control group. (C and D) CCK8 assay was performed to evaluate the OS cell growth, and the relative cell viability was determined at different times. Lv-XIST inhibited cell growth in MG63 and Saos2 cells. *P<0.05, **P<0.01 vs. Lv-control. (E) Flow cytometer was performed to evaluate the OS cell apoptosis, and the cell apoptosis was determined after transfection for 48 h. Lv-XIST promoted cell apoptosis in MG63 and Saos2 cells. **P<0.01 vs. Lv-control. (F) Apoptosis related proteins (cleaved caspase-3, cleaved caspase-9, Bax and Bcl-2) were detected by western blotting in MG63 and Saos2 cells, and Lv-XIST promoted the expression levels of cleaved caspase-3, cleaved caspase-9 and Bax, while inhibited the expression of Bcl-2. (G and H) Lv-control or Lv-XIST (MOI=20) was infected into MG63 and Saos2 cells, which were injected into nude mice, respectively. Tumor sizes were calculated every 7 days after injection (G). *P<0.05, **P<0.01 vs. Lv-control. Tumor weights were measured 35 days after injection (H). P<0.01 vs. Lv-control. Data are presented as mean ± SD from three independent experiments. (I) Representative images of tumors from xenografts with MG63 and Saos2 cells infected with Lv-XIST or Lv-control. (J) The expression of PDCD4 was detected by immunohistochemistry staining in cell-derived xenograft tumor model.
To explore whether overexpression of XIST affects tumor growth in vivo, MG63 and Saos2 cells infected with Lv-XIST or Lv-control were subcutaneously injected into the flank of nude mice. Tumor sizes were obviously smaller in the Lv-XIST group compared with the Lv-control group (Fig. 2G and I). In addition, tumor weights in the Lv-XIST group were significantly lower than in the Lv-control group (Fig. 2H). These results indicate that XIST plays a tumor suppressor role in the progression of OS.
Overexpression of XIST inhibits OS cell migration and invasion in vitro
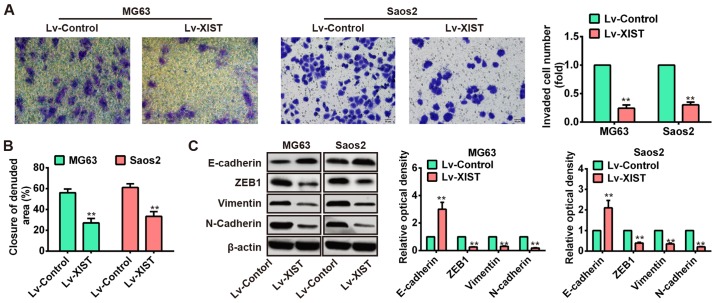
Next, we investigated the association of XIST and OS metastasis. Two OS cell lines, MG63 and Saos2, were injected with Lv-NC or Lv-XIST (MOI=20), and then Transwell and wound healing assays were carried out to explore the effects of XIST on OS cell metastasis. The results of Transwell assay showed that numbers of invaded cells were obviously attenuated in the Lv-XIST groups compared with Lv-control groups (Fig. 3A). In addition, wound healing assay showed that wound closure of both MG63 and Saos2 cells with ectopic expression of XIST was slower than that in Lv-control groups (Fig. 3B). It is well known that EMT is a crucial event in the invasion and migration of tumor cells (21). Therefore, we used western blotting to explore whether XIST can affect EMT in OS cells. Of interest, overexpression of XIST increased the level of epithelial marker E-cadherin while reduced the level of mesenchymal markers such as ZEB1, vimentin and N-cadherin (Fig. 3C). These data suggested that over expression of XIST could inhibit OS cell metastasis in vitro.
Figure 3.
Overexpression of XIST inhibited the migration and invasion of OS cells in vitro. (A) Invasion assay (use matrigel Transwell chambers) in MG63 and Saos2 cells was performed to determined cell invasiveness after infection with Lv-XIST (MOI=20). (B) Wound healing assay to evaluate the effect of XIST on cell migration in MG63 and Saos2 cells. (C) EMT-relatived proteins were detected by western blotting in MG63 and Saos2 cells, and Lv-XIST increased the expression levels of epithelial marker E-cadherin while reduced the level of mesenchymal markers such as ZEB1, vimentin and N-cadherin. **P<0.01 vs. Lv-control group. Data are presented as mean ± SD from three independent experiments.
XIST binds to miR-21-5p and represses its expression
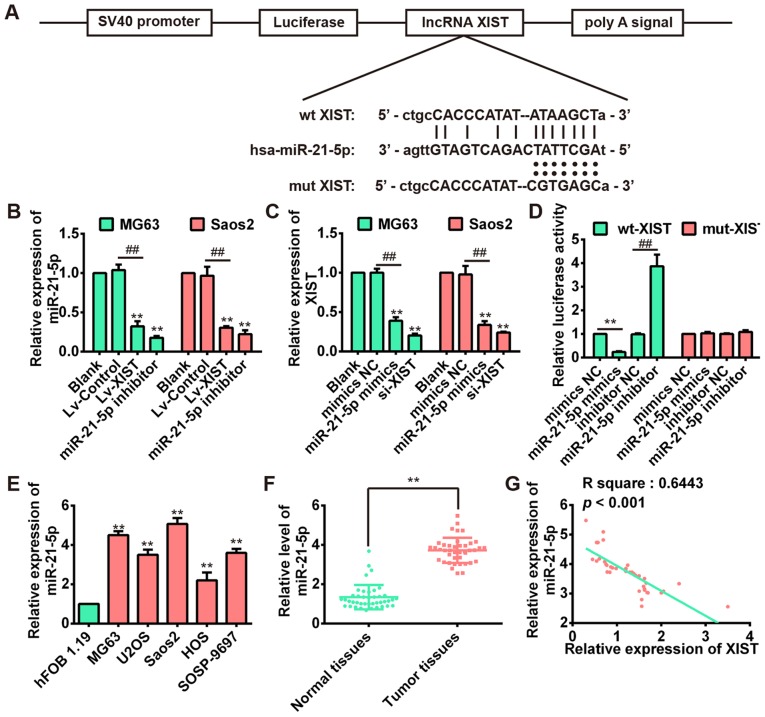
Recent studies have suggested lncRNA could communicate with miRNAs via shared common miRNA binding sites and regulate its expression and activity (22). To determine whether XIST could serve as a ceRNA, we used two bioinformatic databases (starBase and TargetScanS) to search for the potential miRNAs that can be regulated by XIST. Interestingly, miR-21-5p, a well-known oncogene in osteosarcoma (23), could bind to XIST. Fig. 4A revealed the presence of XIST binding sites in the 3′-UTR of miR-21-5p. We subsequently investigated the correlation between XIST and miR-21-5p in MG63 and Saos2 cell lines. The results of qRT-PCR indicated that miR-21-5p expression was downregulated after over-expression of XIST in both MG63 and Saos2 cells (Fig. 4B), whereas the XIST level could also be repressed by miR-21-5p overexpression (Fig. 4C). These results suggested that there was reciprocal repression between XIST and miR-21-5p.
Figure 4.
XIST negatively regulates miR-21-5p level in OS cells. (A) The predicted miR-21-5p binding sites on XIST. (B) After overexpression of XIST, a dramatically decreased expression of miR-21-5p was observed in MG63 and Saos2 cells. **P<0.01 vs. Blank group; ##P<0.01 vs. Lv-control. (C) After over-expression of miR-21-5p, a dramatically decreased XIST expression was observed in MG63 and Saos2 cells. **P<0.01 vs. Blank group; ##P<0.01 vs. mimics NC group. (D) Luciferase activity in HEK 293T cells co-transfected with miR-21-5p mimics, miR-21-5p inhibitor and luciferase reporters containing XIST wild-type or mutant type (MUT) 3′-UTR. Histogram indicates the values of luciferase measured 48 h after transfection. **P<0.01 vs. mimic NC group; ##P<0.01 vs. inhibitor NC group. (e) The miR-21-5p expression level was significantly higher in OS cell lines when compared with that in the osteoblastic hFOB1.19 cells. **P<0.01 vs. hFOB 1.19. (F) The expression level of miR-21-5p in 41 pairs of human osteosarcoma and adjacent normal tissues was detected by using qRT-PCR. **P<0.01 vs. Normal tissues group. (G) Pearson's correlation analysis of the relationship between miR-21-5p expression and XIST expression (R2=0.6443, P<0.001).
To confirm the direct binding relationship between XIST and miR-21-5p, a luciferase activity assay was conducted. The predicted miR-21-5p binding site (XIST-wt) and its mutant type (XIST-mt) were amplified and directly fused to the downstream of the luciferase reporter gene in the pmirGLO-basic vector. Co-transfection of miR-21-5p mimic and pmirGLO-XIST-wt significantly decreased the luciferase activity, whereas co-transfection of miR-21-5p inhibitor and pmirGLO-XIST-wt increased the luciferace activity (Fig. 4D). Likewise, cells cotransfected with miR-21-5p and pmirGLO-XIST-mut showed no obvious change in luciferase activity (Fig. 4D). These data indicated that the miR-21-5p binding site within XIST was functional.
Next, we investigated the expression of miR-21-5p in OS cell lines and 41 pairs of OS and correspondingly adjacent tissues. As shown in Fig. 4E and F, the expression of miR-21-5p was significantly increased compared with the human osteoblast cell line hFOB 1.19 and adjacent normal tissues. This result is consistent with a previous study (24). Moreover, we examined the potential correlation among the expression levels of XIST and miR-21-5p, and an inverse correlation between XIST and miR-21-5p expression levels was observed (Fig. 4G). These results demonstrated that there exist a negative regulation between XIST and miR-21-5p.
Knockdown of miR-21-5p inhibits cell proliferation and metastasis by targeting PDCD4
Previous studies demonstrated that miR-21-5p could play oncogenic roles in several types of cancer by regulating cell growth, EMT and metastasis (25,26). Moreover, a recent study showed that miR-21 promoted cell proliferation through the downregulation of PDCD4 expression in head and neck squamous carcinoma (27). However, it is unknown whether or not PDCD4 is a function target of miR 21-5p in OS. Analysis using targeting algorithms (TargetScan and microRNA.org) indicated that PDCD4 could be a potential target gene of miR-21-5p. Fig. 5A illustrates the predicted miR-21-5p binding site in the 3′-UTR of PDCD4. Western blot analysis showed that overexpression of miR-21-5p markedly reduced the protein levels of PDCD4 in the OS cell lines, whereas it was enhanced by knockdown of miR-21-5p (Fig. 5B). Consistent with previous research, we also demonstrated that knockdown of miR-21-5p inhibited cell proliferation and invasion as well as increased cell apoptosis (Fig. 5C–H). To determine whether PDCD4 acts as a functional target of miR-21-5p, miR 21-5p inhibitor together with si-PDCD4 was transfected into MG63 and Saos2 cells. Knockdown of PDCD4 by siRNA significantly restored reduced cell proliferation, migration, and invasion as well as increased apoptosis caused by inhibition of miR-21-5p in OS cells (Fig. 5C–H). These results indicate that inhibition of miR-21-5p suppresses the cell proliferation, invasion and promoted apoptosis of OS cells, at least in part by targeting PDCD4.
Figure 5.
Knockdown of miR-21-5p inhibits the proliferation, invasion and migration as well as promotes apoptosis by targeting PDCD4. (A) The predicted miR-21-5p binding sites in the 3′-UTR region of PDCD4 (PDCD4-3′-UTR-WT) and the corresponding mutant sequence (PDCD4-3′-UTR-MUT) are shown. (B) Relative expression of PDCD4 protein levels in MG63 and Saos2 cells after transfected with miR-21-5p mimics, miR-21-5p inhibitor, and si-PDCD4. (C and D) miR-21-5p inhibitor suppressed the cell proliferation, and this suppression was significantly reversed by PDCD4 knockdown. (e and F) miR-21-5p inhibitor promoted cell apoptosis, and this enhancement was significantly reversed by si-PDCD4. (G and H) miR-21-5p inhibitor suppressed the cell invasion, and this suppression was significantly reversed by si-PDCD4. *P<0.05, **P<0.01 vs. Blank group; ##P<0.01 vs. miR-21-5p inhibitor group. Data are presented as mean ± SD from three independent experiments.
miR-21-5p mediates the expression of PDCD4 involved in XIST-regulated antitumor effects of OS cells
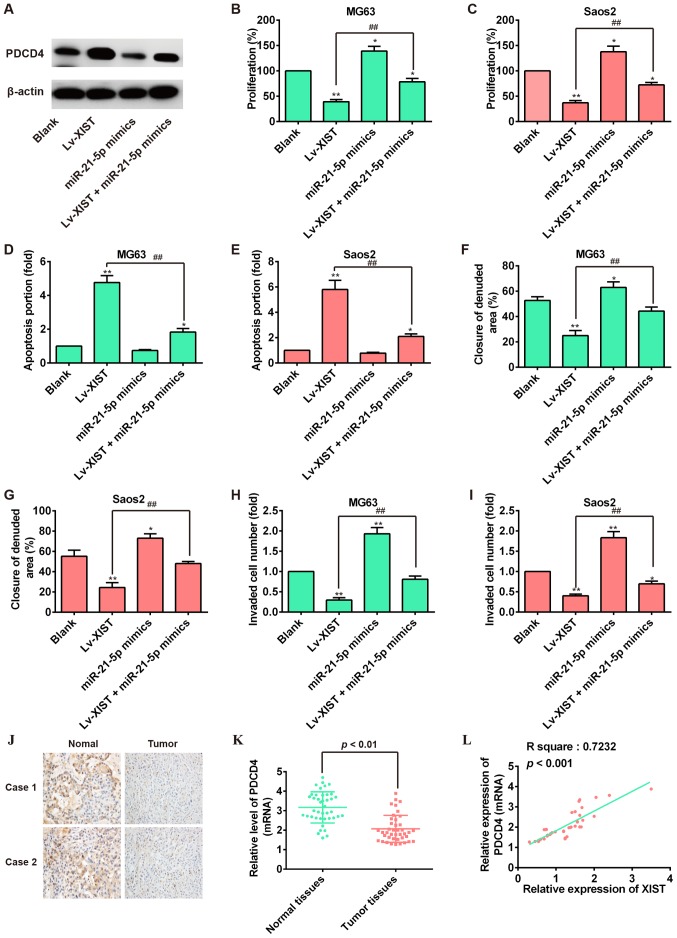
Increasing evidence suggests that PDCD4 functions as a tumor suppressor in several cancers including OS (28,29). Our finding that miR-21-5p exerted an oncogenic role through negatively regulating PDCD4 in OS cells led us to examine whether XIST inhibited OS cell growth and metastasis via suppression of miR-21-5p activity and promotion of PDCD4 expression. First, we explore whether miR-21-5p was involved in the effect of XIST in regulating the PDCD4 expression in OS cells. As shown in Fig. 6A, western blot analysis showed that overexpression of XIST could increase the expression of PDCD4, but miR-21-5p mimics reversed the promoting effect of XIST overexpression on PDCD4 expression. Subsequently, the cell function assay showed that miR-21-5p overexpression could restore the reduction of cell proliferation and reverse the enhancement of cell apoptosis induced by XIST overexpression (Fig. 6B–E). Importantly, the Transwell invasion assay and would healing assay revealed that the suppression of lv-XIST-regulated cell migration and invasion was attenuated by miR-21-5p over expression (Fig. 6F–I). We also examine the expression of PDCD4 in OS tissue samples by immunohistochemical staining and qRT-PCR. As expected, PDCD4 was down regulated in OS tissue samples (Fig. 6J and K). Additionally, the correlation of XIST and PDCD4 in OS tissue samples was investigated and a positive relationship was observed between XIST and PDCD4 (Fig. 6L). In cell-derived xenograft tumors model, we also found that overexpression of XIST positively regulated the expression of PDCD4 by immunohistochemical staining (Fig. 2J). Collectively, we concluded that XIST functions as a ceRNA to enhance the expression of PDCD4 by competitively binding miR-21-5p, leading to the inhibition of OS cell growth and metastasis.
Figure 6.
XIST suppresses cell proliferation and metastasis through downregulating miR-21-5p and promoting PDCD4 expression. MG63 and Saos2 cells were co-transfected with miR-21-5p mimics and Lv-XIST. (A) Relative expression of PDCD4 protein levels in MG63 cells. (B and C) Cell viability was measured by CCK-8 assay. (D and e) Cell apoptosis was analyzed by flow cytometer. (F and G) migration of OS cells was measured by wound healing assay (magnification, ×200; scale bar, 100 µm). (H and I) Invasion of OS cells was measured by invasion assay. *P<0.05, **P<0.01 vs. Blank group, ##P<0.01 vs. Lv-XIST group. Data are presented as mean ± SD from three independent experiments. (J) The expression of PDCD4 was detected by immunohistochemistry staining in 2 pairs of tumor tissues and normal tissues from patients with OS. (K) qRT-PCR analysis of PDCD4 expression levels in 41 pairs of human osteosarcoma and normal tissues. P<0.01 vs. Normal tissues group. (L) Pearson's correlation analysis of the relationship between PDCD4 and XIST expression (R2=0.7232, P<0.001).
Discussion
In the present study, we demonstrated that XIST expression was downregulated and associated with a poor clinical outcome in OS patients. Overexpression of XIST inhibited the cell proliferation, migration and invasion, and promoted apoptosis in vitro and reduced tumor growth in vivo. Furthermore, this study provided evidence that XIST exerted tumor-suppressive functions by downregulating miR-21-5p, thereby enhancing the expression of PDCD4, a well-known tumor suppressor. Our study not only revealed the important role of XIST/miR-21-5p/PDCD4 axis in OS pathogenesis but also implicated the potential role of XIST in the clinical diagnosis and treatment of OS.
Recently, more studies have reported that lncRNAs play considerable functional roles in the initiation and progression of multiple cancers, including OS (30,31). lncRNA XIST was a product of the XIST gene which was located in the X inactivation center (32). Previous studies have found that XIST was upregulated in glioma tissues and human glioblastoma stem cells (GSCs), and knockdown of XIST exerted tumor-suppressive roles by reducing cell proliferation, migration and invasion as well as inducing apoptosis (15). Song et al discovered that XIST was upregulated in NPC tissues and XIST overexpression enhanced, while XIST silencing hampered the cell growth in NPC (11). However, other researchers reported that XIST expression was lost in several cancers, including ovarian, cervical cancer and breast cell lines (13,33,34). These results demonstrated that XIST have different roles either as an oncogene or a tumor suppressor role in different tumors. In the present study, our results confirmed that XIST was markedly downregulated in OS, and low XIST expression was associated with a poor clinical outcome in OS patients. Moreover, we found that overexpression of XIST led to repressed cell proliferation, migration and invasion as well as increased apoptosis, which was consistent with the suppressive roles of XIST revealed by previous studies. In addition, the in vivo studies also confirmed that overexpression of XIST suppressed tumor growth in nude mice. These findings suggest that XIST may function as a tumor suppressor in OS progression.
It is worth noting that XIST, an lncRNA from the inactive X-chromosome, is required for the silencing of one X chromosome in female mammalian cells to achieve equivalent X-linked gene dosage between females and males (35-37). Previous studies showed that XIST was expressed mainly in female cells, and loss of XIST contributes to cancer progression in breast cancer (13), hematologic cancer (12) and hepatocellular carcinoma (38). A tumor suppressor role of XIST in breast cancer has been widely suggested (13,39-41), but still remains controversial (42,43). Similarly, we observed that a recent paper has claimed that the role of XIST as tumor progressor contributed to osteosarcoma cell proliferation and invasion and XIST exerted its function through the miR-320b/RAP2B axis (44). However, there are no animal models such as nude mice used in that study.
In our study, we confirmed that XIST remarkably inhibited the growth of OS cells in vitro as well as the xenograft tumor formation in vivo. Moreover, both cell invasion and migration were inhibited by XIST overexpression via suppressing the EMT process. Most importantly, our data show that loss of XIST associated with recurrence and short overall survival in OS patients. It was reported that multiple demographics including gender were related to the incidence rate and outcome of osteosarcoma (45,46). Since XIST was proved expressed mainly in female cells in other tumors, we suspect this may be because of gender differences of samples lead to different results. In the future, our work will focus the correlation between gender disparity of OS incidence and different expression patterns of XIST in a large number of samples.
It has been reported that EMT is associated with the acquisition of metastasis in cancer (47). During EMT, the epithelial protein level, such as E-cadherin, is downregu-lated, while mesenchymal protein such as N-cadherin and vimentin are upregulated (48). Zhuang et al found that high level of XIST seems to be associated with distant metastasis and poor prognosis in patients with hepatocellular carcinoma (HCC) (38). A recent study from Chen et al showed that XIST could affect the metastasis of human gastric cancer cell via inducing EMT (20). In this study, we found that overexpression of XIST promoted E-cadherin protein expression but inhibited the expression of ZEB1, vimentin and N-cadherin in OS cells in vitro, suggesting that XIST overexpression inhibited cell invasion and migration via suppressing the EMT process.
As a newly described regulatory mechanism, lncRNAs can antagonize miRNA function by sponging miRNAs via a competing endogenous mechanism (22). For example, Ma et al have shown that lncRNA-ATB acts as a ceRNA to regulate the expression of TGF-β by competing for miR-200a in glioma (49). Several recent studies have demonstrated that XIST functions as a ceRNA in many types of cancer, such as human nasopharyngeal carcinoma (NPC) (11), hepato cellular carcinoma (HCC) (9), and gastric cancer (GC) (10). XIST upregulated the expression of miR-34a-5p targeted gene E2F3 through acting as a ceRNA of miR-34a-5p in NPC (11). XIST upregulated EZH2 by competitively binding the miR-101 and then induced cell proliferation and invasion in GC (10). Based on these studies, we hypothesized that XIST may act as a ceRNA in OS and so we searched for potential interactions with miRNAs. In support of this notion, bioinformatics analysis and luciferase assays were performed to verify the direct binding ability of the predicted miRNA on the XIST transcript. As expected, we discovered that XIST is directly bound to miR-21-5p and there was reciprocal repression between XIST and miR-21-5p in OS cells. In addition, qRT-PCR analysis showed that expression of miR-21-5p was inversely correlated with XIST in OS tissues. Moreover, knockdown of miR-21-5p expression could arrest cell proliferation and invasion, which was consistent with results of overexpression of XIST expression in OS cells. Taken together, these data are consistent with our hypothesis and indicate that XIST affects the biological characteristic of OS cells by modulating the miR-21-5p function.
PDCD4 is a novel tumor suppressor that frequently exhibits downregulated expression in a number of cancers (50-52). Downregulation of PDCD4 was reported to be significantly associated with short overall survival of patients with head and neck, digestive system, and urinary system cancers (53-55). Lin et al previously reported that PDCD4 was identified as a target of miR-202 and involved in the inhibitory effect of miR-202 on cell apoptosis and drug resistance of osteosarcoma cells (28). Other research demonstrates that PDCD4 inhibition may not be affected by gene amplification alone, but is also likely to be influenced by transcriptional activation and/or post-transcriptional mechanisms in carcinomas (56). In previous studies, PDCD4 mRNA and protein over expression have been directly affected by miRNA-mediated post-transcriptional mechanisms in cancers (57-59). Our study also confirms PDCD4 as a direct target of miR-21-5p. Considering the interaction of XIST/miR-21-5p, we therefore hypothesize that XIST may also regulate PDCD4 expression in OS which signifies the role of XIST in the tumorigenesis-regulating network. As expected, we demonstrated that PDCD4 expression was positively associated with XIST in OS cells, and miR-21-5p reversed the enhancement of PDCD4 mediated by XIST overexpression. More importantly, we demonstrated overexpression of miR-21-5p could attenuate the inhibitory effects on cell proliferation, invasion and migration, as well as inhibit cell apoptosis induced by overexpression of XIST. These results support our hypothesis that XIST can antagonize miRNA-21-5p, thereby protecting PDCD4 from repression, finally inhibiting OS tumorigenesis.
In conclusion, our data demonstrated that XIST was downregulated in OS and its expression was associated with overall survival of OS patients. Furthermore, we determined that XIST suppressed OS cell proliferation and metastasis via downregulation of miR-21-5p and activation of PDCD4 function. Thus, XIST/miR-21-5p/PDCD4 axis may represent a novel prognostic biomarker and therapeutic target in OS.
References
- 1.Ottaviani G, Jaffe N. The epidemiology of osteosarcoma. Cancer Treat Res. 2009;152:3–13. doi: 10.1007/978-1-4419-0284-9_1. [DOI] [PubMed] [Google Scholar]
- 2.Meazza C, Luksch R, Daolio P, Podda M, Luzzati A, Gronchi A, Parafioriti A, Gandola L, Collini P, Ferrari A, et al. Axial skeletal osteosarcoma: A 25-year monoinstitutional experience in children and adolescents. Med Oncol. 2014;31:875. doi: 10.1007/s12032-014-0875-x. [DOI] [PubMed] [Google Scholar]
- 3.Ando K, Heymann MF, Stresing V, Mori K, Rédini F, Heymann D. Current therapeutic strategies and novel approaches in osteosarcoma. Cancers (Basel) 2013;5:591–616. doi: 10.3390/cancers5020591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wittig JC, Bickels J, Priebat D, Jelinek J, Kellar-Graney K, Shmookler B, Malawer MM. Osteosarcoma: A multidisciplinary approach to diagnosis and treatment. Am Fam Physician. 2002;65:1123–1132. [PubMed] [Google Scholar]
- 5.Shi X, Sun M, Liu H, Yao Y, Song Y. Long non-coding RNAs: A new frontier in the study of human diseases. Cancer Lett. 2013;339:159–166. doi: 10.1016/j.canlet.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 6.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 7.Tian ZZ, Guo XJ, Zhao YM, Fang Y. Decreased expression of long non-coding RNA MEG3 acts as a potential predictor biomarker in progression and poor prognosis of osteosarcoma. Int J Clin Exp Pathol. 2015;8:15138–15142. [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Q, Geng PL, Yin P, Wang XL, Jia JP, Yao J. Down-regulation of long non-coding RNA TUG1 inhibits osteosarcoma cell proliferation and promotes apoptosis. Asian Pac J Cancer Prev. 2013;14:2311–2315. doi: 10.7314/APJCP.2013.14.4.2311. [DOI] [PubMed] [Google Scholar]
- 9.Mo Y, Lu Y, Wang P, Huang S, He L, Li D, Li F, Huang J, Lin X, Li X, et al. Long non-coding RNA XIST promotes cell growth by regulating miR-139-5p/PDK1/AKT axis in hepatocellular carcinoma. Tumour Biol. 2017;39:1010428317690999. doi: 10.1177/1010428317690999. [DOI] [PubMed] [Google Scholar]
- 10.Ma L, Zhou Y, Luo X, Gao H, Deng X, Jiang Y. Long non-coding RNA XIST promotes cell growth and invasion through regulating miR-497/mACC1 axis in gastric cancer. Oncotarget. 2017;8:4125–4135. doi: 10.18632/oncotarget.13670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Song P, Ye LF, Zhang C, Peng T, Zhou XH. Long non-coding RNA XIST exerts oncogenic functions in human nasopharyngeal carcinoma by targeting miR-34a-5p. Gene. 2016;592:8–14. doi: 10.1016/j.gene.2016.07.055. [DOI] [PubMed] [Google Scholar]
- 12.Yildirim E, Kirby JE, Brown DE, Mercier FE, Sadreyev RI, Scadden DT, Lee JT. Xist RNA is a potent suppressor of hematologic cancer in mice. Cell. 2013;152:727–742. doi: 10.1016/j.cell.2013.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang YS, Chang CC, Lee SS, Jou YS, Shih HM. Xist reduction in breast cancer upregulates AKT phosphorylation via HDAC3-mediated repression of PHLPP1 expression. Oncotarget. 2016;7:43256–43266. doi: 10.18632/oncotarget.9673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang S, Chen B, Wang X, Wu K, Sun Y. Long non-coding RNA XIST regulates PTEN expression by sponging miR-181a and promotes hepatocellular carcinoma progression. BMC Cancer. 2017;17:248. doi: 10.1186/s12885-017-3216-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yao Y, Ma J, Xue Y, Wang P, Li Z, Liu J, Chen L, Xi Z, Teng H, Wang Z, et al. Knockdown of long non-coding RNA XIST exerts tumor-suppressive functions in human glioblastoma stem cells by up-regulating miR-152. Cancer Lett. 2015;359:75–86. doi: 10.1016/j.canlet.2014.12.051. [DOI] [PubMed] [Google Scholar]
- 16.John-Aryankalayil M, Palayoor ST, Makinde AY, Cerna D, Simone CB, II, Falduto MT, Magnuson SR, Coleman CN. Fractionated radiation alters oncomir and tumor suppressor miRNAs in human prostate cancer cells. Radiat Res. 2012;178:105–117. doi: 10.1667/RR2703.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu XH, Sun M, Nie FQ, Ge YB, Zhang EB, Yin DD, Kong R, Xia R, Lu KH, Li JH, et al. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol Cancer. 2014;13:92. doi: 10.1186/1476-4598-13-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liang WC, Fu WM, Wong CW, Wang Y, Wang WM, Hu GX, Zhang L, Xiao LJ, Wan DC, Zhang JF, et al. The lncRNA H19 promotes epithelial to mesenchymal transition by functioning as miRNA sponges in colorectal cancer. Oncotarget. 2015;6:22513–22525. doi: 10.18632/oncotarget.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen DL, Ju HQ, Lu YX, Chen LZ, Zeng ZL, Zhang DS, Luo HY, Wang F, Qiu MZ, Wang DS, et al. Long non-coding RNA XIST regulates gastric cancer progression by acting as a molecular sponge of miR-101 to modulate EZH2 expression. J Exp Clin Cancer Res. 2016;35:142. doi: 10.1186/s13046-016-0420-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bonnomet A, Brysse A, Tachsidis A, Waltham M, Thompson EW, Polette M, Gilles C. Epithelial-to-mesenchymal transitions and circulating tumor cells. J Mammary Gland Biol Neoplasia. 2010;15:261–273. doi: 10.1007/s10911-010-9174-0. [DOI] [PubMed] [Google Scholar]
- 22.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lv C, Hao Y, Tu G. MicroRNA-21 promotes proliferation, invasion and suppresses apoptosis in human osteosarcoma line mG63 through PTeN/Akt pathway. Tumour Biol. 2016;37:9333–9342. doi: 10.1007/s13277-016-4807-6. [DOI] [PubMed] [Google Scholar]
- 24.Xu B, Xia H, Cao J, Wang Z, Yang Y, Lin Y. MicroRNA-21 inhibits the apoptosis of osteosarcoma cell line SAOS-2 via targeting caspase 8. Oncol Res. 2017;25:1161–1168. doi: 10.3727/096504017X14841698396829. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Selcuklu SD, Donoghue MT, Spillane C. miR-21 as a key regulator of oncogenic processes. Biochem Soc Trans. 2009;37:918–925. doi: 10.1042/BST0370918. [DOI] [PubMed] [Google Scholar]
- 26.Dillhoff M, Liu J, Frankel W, Croce C, Bloomston M. MicroRNA-21 is overexpressed in pancreatic cancer and a potential predictor of survival. J Gastrointest Surg. 2008;12:2171–2176. doi: 10.1007/s11605-008-0584-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun Z, Li S, Kaufmann AM, Albers AE. miR-21 increases the programmed cell death 4 gene-regulated cell proliferation in head and neck squamous carcinoma cell lines. Oncol Rep. 2014;32:2283–2289. doi: 10.3892/or.2014.3456. [DOI] [PubMed] [Google Scholar]
- 28.Lin Z, Song D, Wei H, Yang X, Liu T, Yan W, Xiao J. TGF-β1-induced miR-202 mediates drug resistance by inhibiting apoptosis in human osteosarcoma. J Cancer Res Clin Oncol. 2016;142:239–246. doi: 10.1007/s00432-015-2028-9. [DOI] [PubMed] [Google Scholar]
- 29.Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S, Allgayer H. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128–2136. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 30.Dong Y, Liang G, Yuan B, Yang C, Gao R, Zhou X. MALAT1 promotes the proliferation and metastasis of osteosarcoma cells by activating the PI3K/Akt pathway. Tumour Biol. 2015;36:1477–1486. doi: 10.1007/s13277-014-2631-4. [DOI] [PubMed] [Google Scholar]
- 31.Wang B, Su Y, Yang Q, Lv D, Zhang W, Tang K, Wang H, Zhang R, Liu Y. Overexpression of long non-coding RNA HOTAIR promotes tumor growth and metastasis in human osteosarcoma. Mol Cells. 2015;38:432–440. doi: 10.14348/molcells.2015.2327. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32.Escamilla-Del-Arenal M, da Rocha ST, Heard E. Evolutionary diversity and developmental regulation of X-chromosome inactivation. Hum Genet. 2011;130:307–327. doi: 10.1007/s00439-011-1029-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Benoît MH, Hudson TJ, Maire G, Squire JA, Arcand SL, Provencher D, Mes-Masson AM, Tonin PN. Global analysis of chromosome X gene expression in primary cultures of normal ovarian surface epithelial cells and epithelial ovarian cancer cell lines. Int J Oncol. 2007;30:5–17. [PubMed] [Google Scholar]
- 34.Kawakami T, Zhang C, Taniguchi T, Kim CJ, Okada Y, Sugihara H, Hattori T, Reeve AE, Ogawa O, Okamoto K. Characterization of loss-of-inactive X in Klinefelter syndrome and female-derived cancer cells. Oncogene. 2004;23:6163–6169. doi: 10.1038/sj.onc.1207808. [DOI] [PubMed] [Google Scholar]
- 35.Xiao C, Sharp JA, Kawahara M, Davalos AR, Difilippantonio MJ, Hu Y, Li W, Cao L, Buetow K, Ried T, et al. The XIST noncoding RNA functions independently of BRCA1 in X inactivation. Cell. 2007;128:977–989. doi: 10.1016/j.cell.2007.01.034. [DOI] [PubMed] [Google Scholar]
- 36.Marahrens Y, Panning B, Dausman J, Strauss W, Jaenisch R. Xist-deficient mice are defective in dosage compensation but not spermatogenesis. Genes Dev. 1997;11:156–166. doi: 10.1101/gad.11.2.156. [DOI] [PubMed] [Google Scholar]
- 37.Penny GD, Kay GF, Sheardown SA, Rastan S, Brockdorff N. Requirement for Xist in X chromosome inactivation. Nature. 1996;379:131–137. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- 38.Zhuang LK, Yang YT, Ma X, Han B, Wang ZS, Zhao QY, Wu LQ, Qu ZQ. MicroRNA-92b promotes hepatocellular carcinoma progression by targeting Smad7 and is mediated by long non-coding RNA XIST. Cell Death Dis. 2016;7:e2203. doi: 10.1038/cddis.2016.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soudyab M, Iranpour M, Ghafouri-Fard S. The role of long non-coding RNAs in breast cancer. Arch Iran Med. 2016;19:508–517. [PubMed] [Google Scholar]
- 40.Salvador MA, Wicinski J, Cabaud O, Toiron Y, Finetti P, Josselin E, Lelièvre H, Kraus-Berthier L, Depil S, Bertucci F, et al. The histone deacetylase inhibitor abexinostat induces cancer stem cells differentiation in breast cancer with low Xist expression. Clin Cancer Res. 2013;19:6520–6531. doi: 10.1158/1078-0432.CCR-13-0877. [DOI] [PubMed] [Google Scholar]
- 41.Vincent-Salomon A, Ganem-Elbaz C, Manié E, Raynal V, Sastre-Garau X, Stoppa-Lyonnet D, Stern MH, Heard E. X inactive-specific transcript RNA coating and genetic instability of the X chromosome in BRCA1 breast tumors. Cancer Res. 2007;67:5134–5140. doi: 10.1158/0008-5472.CAN-07-0465. [DOI] [PubMed] [Google Scholar]
- 42.Schouten PC, Vollebergh MA, Opdam M, Jonkers M, Loden M, Wesseling J, Hauptmann M, Linn SC. High XIST and low 53BP1 expression predict poor outcome after high-dose alkylating chemotherapy in patients with a BRCA1-like breast cancer. Mol Cancer Ther. 2016;15:190–198. doi: 10.1158/1535-7163.MCT-15-0470. [DOI] [PubMed] [Google Scholar]
- 43.Sirchia SM, Tabano S, Monti L, Recalcati MP, Gariboldi M, Grati FR, Porta G, Finelli P, Radice P, Miozzo M. Misbehaviour of XIST RNA in breast cancer cells. PLoS One. 2009;4:e5559. doi: 10.1371/journal.pone.0005559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lv GY, Miao J, Zhang XL. Long non-coding RNA: Long non-coding RNA XIST promotes osteosarcoma progression by targeting Ras-related protein RAP2B via miR-320b. Oncol Res. 2017 Apr 12; doi: 10.3727/096504017X14920318811721. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lindsey BA, Markel JE, Kleinerman ES. Osteosarcoma overview. Rheumatol Ther. 2017;4:25–43. doi: 10.1007/s40744-016-0050-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smeland S, Müller C, Alvegard TA, Wiklund T, Wiebe T, Björk O, Stenwig AE, Willén H, Holmström T, Follerås G, et al. Scandinavian Sarcoma Group Osteosarcoma Study SSG VIII: Prognostic factors for outcome and the role of replacement salvage chemotherapy for poor histological responders. Eur J Cancer. 2003;39:488–494. doi: 10.1016/S0959-8049(02)00747-5. [DOI] [PubMed] [Google Scholar]
- 47.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A, Weinberg RA. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 49.Ma CC, Xiong Z, Zhu GN, Wang C, Zong G, Wang HL, Bian EB, Zhao B. Long non-coding RNA ATB promotes glioma malignancy by negatively regulating miR-200a. J Exp Clin Cancer Res. 2016;35:90. doi: 10.1186/s13046-016-0367-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang HS, Jansen AP, Nair R, Shibahara K, Verma AK, Cmarik JL, Colburn NH. A novel transformation suppressor, Pdcd4, inhibits AP-1 transactivation but not NF-kappaB or ODC transactivation. Oncogene. 2001;20:669–676. doi: 10.1038/sj.onc.1204137. [DOI] [PubMed] [Google Scholar]
- 51.Mudduluru G, Medved F, Grobholz R, Jost C, Gruber A, Leupold JH, Post S, Jansen A, Colburn NH, Allgayer H. Loss of programmed cell death 4 expression marks adenoma-carcinoma transition, correlates inversely with phosphorylated protein kinase B, and is an independent prognostic factor in resected colorectal cancer. Cancer. 2007;110:1697–1707. doi: 10.1002/cncr.22983. [DOI] [PubMed] [Google Scholar]
- 52.Yang HS, Knies JL, Stark C, Colburn NH. Pdcd4 suppresses tumor phenotype in JB6 cells by inhibiting AP-1 transactivation. Oncogene. 2003;22:3712–3720. doi: 10.1038/sj.onc.1206433. [DOI] [PubMed] [Google Scholar]
- 53.Feng G, Li P, You H, Liu W, Zhang X, Xu X, Sun G, Li F. The expression and clinical pathological significance of PDCD in laryngocarcinoma. Lin Chung Er Bi Yan Hou Tou Jing Wai Ke Za Zhi. 2011;25:16–19. In Chinese. [PubMed] [Google Scholar]
- 54.Zhen Y, Liu Z, Yang H, Yu X, Wu Q, Hua S, Long X, Jiang Q, Song Y, Cheng C, et al. Tumor suppressor PDCD4 modulates miR-184-mediated direct suppression of C-MYC and BCL2 blocking cell growth and survival in nasopharyngeal carcinoma. Cell Death Dis. 2013;4:e872. doi: 10.1038/cddis.2013.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li X, Xin S, Yang D, Li X, He Z, Che X, Wang J, Chen F, Wang X, Song X. Down-regulation of PDCD4 expression is an independent predictor of poor prognosis in human renal cell carcinoma patients. J Cancer Res Clin Oncol. 2012;138:529–535. doi: 10.1007/s00432-011-1121-y. [DOI] [PubMed] [Google Scholar]
- 56.Leupold JH, Asangani IA, Mudduluru G, Allgayer H. Promoter cloning and characterization of the human programmed cell death protein 4 (pdcd4) gene: Evidence for ZBP-89 and Sp-binding motifs as essential Pdcd4 regulators. Biosci Rep. 2012;32:281–297. doi: 10.1042/BSR20110045. [DOI] [PubMed] [Google Scholar]
- 57.Li JZ, Gao W, Lei WB, Zhao J, Chan JY, Wei WI, Ho WK, Wong TS. MicroRNA 744-3p promotes MMP-9-mediated metastasis by simultaneously suppressing PDCD4 and PTEN in laryngeal squamous cell carcinoma. Oncotarget. 2016;7:58218–58233. doi: 10.18632/oncotarget.11280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li C, Deng L, Zhi Q, Meng Q, Qian A, Sang H, Li X, Xia J. MicroRNA-183 functions as an oncogene by regulating PDCD4 in gastric cancer. Anticancer Agents Med Chem. 2016;16:447–455. doi: 10.2174/1871520615666150914114237. [DOI] [PubMed] [Google Scholar]
- 59.Ma QQ, Huang JT, Xiong YG, Yang XY, Han R, Zhu WW. MicroRNA-96 regulates apoptosis by targeting PDCD4 in human glioma cells. Technol Cancer Res Treat. 2017;16:92–98. doi: 10.1177/1533034616629260. [DOI] [PMC free article] [PubMed] [Google Scholar]