Abstract
Hepatitis B virus (HBV) is the dominant risk factor for hepatocellular carcinoma (HCC). HBV X protein (HBx) plays crucial roles in HCC carcinogenesis. HBx interferes with several signaling pathways including the Notch1 pathway in HCC. In this study, we found that Notch1 was highly expressed in HCC, especially in large HCCs. Notch1 and HBx co-localized in HCC and their levels were positively correlated with each other. Notch1 expression was more elevated in HepG2.2.15 cells than that in HepG2 cells. HBx activated the Notch1 pathway in HepG2.2.15 cells. Suppression of HBx and the Notch1 pathway attenuated the growth of HepG2.2.15 cells. Notch1, ERK, and AKT pathways were inhibited after γ-secretase inhibitor treatment. Dual-specificity phosphatase 1 (DUSP1) and phosphatase and tensin homolog (PTEN) were upregulated after γ-secretase inhibitor treatment and Hes1 inhibition. Luciferase reporter assays showed that Hes1 suppressed the promoters of DUSP1 and PTEN genes, which was reversed by γ-secretase inhibitor treatment. Western blotting demonstrated that DUSP1 dephosphorylated pERK and PTEN dephosphorylated pAKT. Collectively, we found a link among HBx, the Notch1 pathway, DUSP1/PTEN, and ERK/AKT pathways, which influenced HCC cell survival and could be a therapeutic target for HCC treatment.
Keywords: hepatitis B virus X protein, Notch1 pathway, Hes1, Dual-specificity phosphatase 1, PTEN, ERK, AKT, hepatocellular carcinoma
Introduction
Hepatocellular carcinoma (HCC) remains a major health concern worldwide (1). Hepatitis B virus (HBV) is the dominant risk factor for HCC (2), accounting for more than half of all cases (3). HBV X protein (HBx) is the only expressed HBV protein and plays critical roles in hepatocarcinogenesis (4). Previous reports have shown that HBx interferes with many signal transduction pathways including Hippo, nuclear factor-κB, WNT/β-catenin, and p53 pathways (2,4-7). However, the molecular mechanisms underlying the hepatocellular carcinogenesis induced by HBx remain unclear.
The Notch pathway plays crucial roles in organogenesis and morphogenesis, and influences various biological processes including apoptosis, proliferation, and differentiation (8). Emerging evidence demonstrates that dysregulation of the Notch pathway is associated with various types of malignancies (9,10). Persistent activation of the Notch pathway leads to liver malignancies (11). A link between HBx and the Notch pathway has been reported previously (12,13). Studies have demonstrated that HBx activates the Notch1 pathway that further upregulates ERK and AKT pathways to promote cell proliferation (14,15). However, the detailed molecular mechanisms whereby Notch1 activates ERK and AKT pathways in HCC are unresolved.
In this study, we further explored the link between HBx and Notch1 in HCC and elucidated the molecular mechanisms underlying Notch1/ERK and Notch1/AKT activations by HBx.
Materials and methods
Patients and clinical specimens
A total of 121 human liver tissue samples were collected from patients who underwent surgical resections at the Hepatic Surgery Centre, Tongji Hospital of Huazhong University of Science and Technology (Wuhan, China). Detailed clinicopathological parameters are listed in Table I. The procedure of human specimen collection was approved by the Ethics Committee of Tongji Hospital, Huazhong University of Science and Technology, and the study was conducted according to the Declaration of Helsinki.
Table I.
Correlation between the factors and clinicopathological parameters in HCC patients (n=121).
| Clinicopathological variables | Relative Notch1 expression
|
P-value | |
|---|---|---|---|
| Low | High | ||
| Sex | 0.322 | ||
| Male (n=91) | 16 | 75 | |
| Female (n=30) | 3 | 27 | |
| Age (years) | 0.506 | ||
| ≤50 (n=49) | 9 | 40 | |
| >50 (n=72) | 10 | 62 | |
| HBV | <0.001 | ||
| Negative (n=13) | 7 | 6 | |
| Positive (n=108) | 12 | 96 | |
| AFP (µg/l) | 0.410 | ||
| ≤20 (n=41) | 8 | 33 | |
| >20 (n=80) | 11 | 69 | |
| Cirrhosis | 0.044 | ||
| No (n=29) | 8 | 21 | |
| Yes (n=92) | 11 | 81 | |
| Tumor size (cm) | 0.023 | ||
| ≤5 (n=54) | 13 | 41 | |
| >5 (n=67) | 6 | 61 | |
Cell lines, cell culture, and reagents
The human hepatoma cell line HepG2 and HBV genome-transfected HepG2 (HepG2.2.15) cells were obtained from the China Center for Type Culture Collection (Wuhan, China). Cells were cultured in high glucose Dulbecco's modified Eagle's medium (Gibco, Carlsbad, CA, USA) containing 10% fetal bovine serum (Gibco). Dimethyl sulfoxide (DMSO) and γ-secretase inhibitor N-[N-(3,5-difluorophenacetyl)-l-alanyl]-S-phenylglycine t-butyl ester (DAPT) were purchased from Sigma-Aldrich (St. Louis, MO, USA). DAPT was dissolved in DMSO.
Immunohistochemical analysis
Immunohistochemistry (IHC) was performed as described previously (16). Primary antibodies against HBx and Notch1 were purchased from Merck-Millipore (Billerica, MA, USA) and Santa Cruz Biotechnology (Santa Cruz, CA, USA), respectively.
Western blotting
Western blotting was performed as described previously (17). The primary antibodies and their sources were as follows: anti-Notch1, anti-Hes1, anti-NICD, anti-pERK, anti-ERK, and anti-pAKT (Cell Signaling Technology, Beverly, MA, USA); anti-Jagged1, anti-β-actin, anti-AKT, and anti-DUSP1 (Santa Cruz Biotechnology); anti-HBx (Merck-Millipore); anti-PTEN (Proteintech Group, Chicago, IL, USA). Horseradish peroxidase-conjugated secondary antibodies were purchased from Jackson Immuno Research Laboratories (West Grove, PA, USA).
Double immunofluorescence analysis
Double immunofluorescence immunostaining was performed as described previously (18,19). Double-labeling immunofluorescence was used to detect HBx and Notch1 simultaneously. All sections were analyzed by confocal laser-scanning microscopy using a Nikon Digital Eclipse C1 system (Nikon Corp., Japan).
Reverse transcription and real-time quantitative PCR (qPCR) analysis
PCR was performed as described previously (20). The primer sequences for HBx were as follows: sense, 5′-GGCTGCTAGGCTGTGCTGCC-3′; antisense, 5′-GTTCCTGTGGGCGTTCACGG-3′. Images of electrophoresed PCR products were acquired using the Alpha Innotech Fluorochem Imaging system. Real-time PCR primers are listed in Table II. Cycle threshold values were reported relative to β-actin mRNA. Expression values were obtained in triplicate and normalized to β-actin expression. Results were calculated as fold induction relative to controls.
Table II.
Primer sequences for real-time PCR.
| Gene | Primer sequences |
|---|---|
| HBx | F:5′-CAC CTC TCT TTA CGC GGA CT-3′ |
| R: 5′-GGT CGT TGA CAT TGC AGA GA-3′ | |
| Hes1 | F: 5′-AAG AAA GAT AGC TCG CGG CAT-3′ |
| R: 5′-CCA GCA CAC TTG GGT CTG T-3′ | |
| DUSP1 | F: 5′-CCA GTA CAA GAG CAT CCC TGT-3′ |
| R: 5′-AGT GGA CAA ACA CCC TTC CTC-3′ | |
| PTEN | F:5′-AGC GTG CAG ATA ATG ACA AGG-3′ |
| R: 5′-TGG ATC AGA GTC AGT GGT GTC-3′ | |
| β-actin | F: 5′-CAA GGC CAA CCG CGA GAA GAT-3′ |
| R: 5′-CCA GAG GCG TAC AGG GAT AGC AC-3′ |
Transient RNA interference
Small interfering RNA (siRNA) duplexes targeting human HBx, Hes1, DUSP1, and PTEN sequences and a scrambled siRNA were designed as described previously (5,21-23). All siRNAs were synthesized by Ribobio (Guangzhou, China). Transfection of the siRNA duplexes was performed by jetPRIME (Polyplus-transfection SA, Illkirch, France) according to the manufacturer's instructions.
Cell viability assay
The cell viability assay was performed as described previously (24). HepG2.2.15 cells (2×103 per well) were seeded and cultured in 96-well plates for the indicated times. Cell Counting Kit-8 (CCK-8, Dojindo, Japan) was added to the cells, and the optical density value at 450 nm was measured after 2 h.
Transcriptional response assay
Luciferase assays were performed as described previously (25). Cell lysates was subjected to luciferase assays using the Dual-luciferase Reporter assay system (Promega) according to the manufacturer's instructions. DUSP1-luc reporter, PTEN-luc reporter, and Hes1 expression vectors were transfected as reported previously (26-28). Relative luciferase activities were determined by a Glomar 20/20 Illuminometer (Promega) and normalized against Renilla luciferase as an internal control.
Chromatin immunoprecipitation (ChIP)
Quantitative ChIP analysis was performed as described previously (29). Briefly, cells were crosslinked, sonicated, and immunoprecipitated with anti-Hes1 antibodies or IgG (negative control). A mixture of two anti-Hes1 antibodies (H140, Santa Cruz Biotechnology; 4H1, Novus Biologicals) was used for immunoprecipitation. Then, DNA was eluted and crosslinking was reversed, followed by purification and amplification for PCR analysis. The primers used in qPCR are listed in Table III.
Table III.
Primers used for ChIP.
| ChIP primers | Primer sequences |
|---|---|
| DUSP1 | F: 5′-AAC CGC AGA ATG TTC CTG AC-3′ |
| promoter A | R: 5′-CGT TAT AGG CCG AAA GCA AA-3′ |
| DUSP1 | F: 5′-GCT CGA GTC GGT CTT GGT AG-3′ |
| promoter B | R: 5′-CCC CTT TTC CTC ATT TCC TC-3′ |
| PTEN | F: 5′-GGG AGT GGG AAT TTG GAA AG-3′ |
| promoter A | R: 5′-TCA AAA GGA GGT GGA AGG AT-3′ |
| PTEN | F: 5′-TCC CTG CAT TTC CCT CTA CA-3′ |
| promoter B | R: 5′-GTG CGT TGA GCA GTG TCA CT-3″ |
Statistical analysis
Data were analyzed using GraphPad Prism 5.0 (La Jolla, CA, USA) or SPSS 13.0 (Chicago, IL, USA). All experiments were independently performed at least three times. The results are presented as the mean ± standard error of the mean. Comparisons between different groups were evaluated by one-way analysis of variance. P<0.05 was considered as statistically significant.
Results
Expression of Notch1 in human liver and HCC tissues
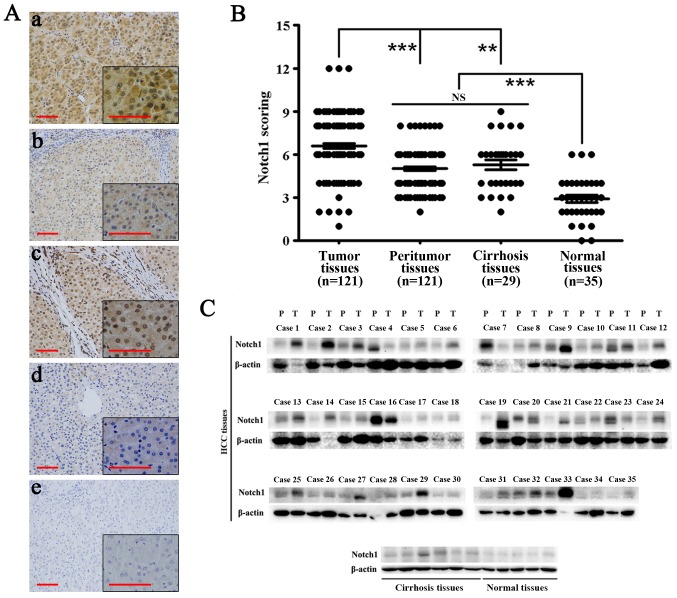
Immunohistochemical analysis showed that Notch1 was highly expressed in HCC tissues. Notch1 also exhibited expression in adjacent non-tumor and cirrhosis tissues. However, its expression was significantly lower than that in HCC tissues (P<0.01). Moreover, the expression of Notch1 in normal liver tissues was significantly lower than that in the above-mentioned tissues (P<0.001) (Fig. 1A and B). Western blotting showed similar results (Fig. 1C). In most HCC tissues, the expression of Notch1 was highly elevated compared with that in the other tissues. Notch1 was weakly expressed in all normal liver tissues.
Figure 1.
Notch1 is highly expressed in HCC tissues. (A) Immunohistochemical staining of Notch1 in HCC (a), adjacent non-tumor (b), cirrhosis (c) and normal liver tissues (d). (e) Negative control. Scale bar, 100 µm. Magnification, ×200 and ×400. (B) The immunohistochemistry score of Notch1 in each immunostained liver section. (C) Western blotting of Notch1 in HCC, adjacent non-tumor, cirrhosis and normal liver tissues. **P<0.01, ***P<0.001; NS, not significant.
Specifically, among 121 HCC tissues, 19 of them had weak expression and 102 of them had high expression. In the 121 corresponding peritumoral tissues, 59 of them had weak expression and 62 of them had high expression (P<0.001) (Table IV).
Table IV.
Expression of Notch1 in HCC and peritumor liver tissues.
| Cases tested | Relative Notch1 expression
|
P-value | ||
|---|---|---|---|---|
| Low | High | |||
| Tumor tissues | 121 | 19 | 102 | <0.001 |
| Peritumor tissues | 121 | 59 | 62 | |
Table I shows the correlation between the factors and clinicopathological parameters in the 121 HCC patients. As shown in the table, sex, age, and the AFP level were unrelated to Notch1 expression. The level of Notch1 was significantly more elevated in HBV-positive patients (P<0.001), cirrhosis patients, (P<0.05), and patients with large HCCs (P<0.05).
Co-localization and relationship of Notch1 with HBx in HCC tissues and the HepG2.2.15 cell line
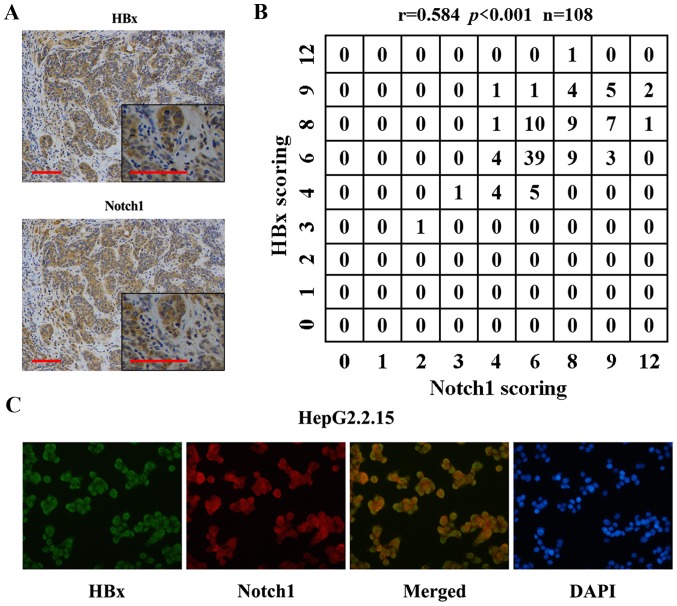
Previous reports have shown the relationship of Notch1 with HBx. Here, we found co-localization of the two proteins. IHC results showed that Notch1 and HBx were co-expressed in HCC tissues (Fig. 2A). Fig. 2B shows the correlation of Notch1 and HBx expression levels. Spearman's rho was 0.584, indicating that Notch1 levels were positively correlated with HBx expression in the 108 HBV-positive HCC tissues. Fig. 2C shows HBx (green) and Notch1 (red) staining in HepG2.2.15 cells. The yellow staining in dual labeling experiments indicated overlapping areas of HBx and Notch1 staining, suggesting co-expression of Notch1 with HBx in HepG2.2.15 cells.
Figure 2.
Notch1 and HBx co-expressed in HCC tissues and HepG2.2.15 cells. (A) Co-localization of Notch1 and HBx detected by immunohistochemical staining. Scale bar, 100 µm. Magnification, ×200 and ×400. (B) Correlation of expression levels of Notch1 and HBx in 108 HBV-positive HCC tissues. The Spearman's rho = 0.584, P<0.001. (C) Co-expression of Notch1 with HBx in HepG2.2.15 cells detected by double-fluorescence immunostaining.
Regulation of Notch1 by HBx in HepG2.2.15 cells
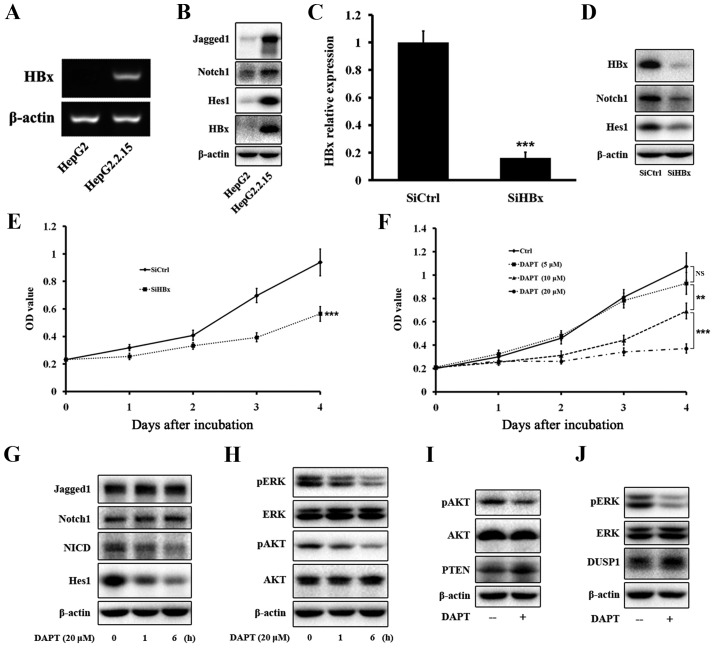
Because a relationship of HBx with Notch1 was found in HCC tissues and cells, we determined whether HBx could regulate the expression of Notch1 in HepG2.2.15 cells in vitro. To test this hypothesis, we compared the expression of HBx and some members of the Notch1 pathway between HepG2 and HepG2.2.15 cells, and between HepG2.2.15-SiCtrl and HepG2.2.15-SiHBx cells. Western blotting and PCR confirmed that HepG2 cells did not express HBx, whereas the HBx gene and protein were expressed in HepG2.2.15 cells (Fig. 3A and B). Next, we tested the expression of some members of the Notch1 pathway, such as Jagged1, Notch1, and Hes1. Western blotting showed that the protein levels of the three proteins were much more elevated in HepG2.2.15 cells than in HepG2 cells (Fig. 3B). Next, we knocked down HBx expression in HepG2.2.15 cells by HBx-specific siRNA. Western blotting and PCR confirmed that HBx expression was significantly decreased by the siRNA (Fig. 3C and D). Western blotting showed that the decrease of HBx expression led to a decrease of Notch1 and Hes1 expression in HepG2.2.15 cells (Fig. 3D), verifying that HBx regulated the expression of Notch1 in vitro.
Figure 3.
HBx activates the Notch1 pathway and DAPT attenuates cell growth via inhibition of ERK and AKT pathways. HBx gene (A) and protein (B) were expressed in HepG2.2.15 cells. (B) The Notch1 pathway was activated in HepG2.2.15. (C) HBx was knocked down by SiRNA and its mRNA level was tested by qRT-PCR. The changes were presented as fold change in comparison to the control. (D) The Notch1 pathway was inhibited after SiHBx. (E) Cell proliferation of HepG2.2.15 cells was inhibited after SiHBx. (F) Cell proliferation of HepG2.2.15 cells was inhibited after DAPT treatment. (G) Alteration of expression of the Notch1 pathway after DAPT treatment. (H) Expressions of pERK and pAKT were decreased after DAPT treatment. (I and J) Expression of DUSP1 and PTEN was increased after DAPT treatment. Data were collected in at least three independent experiments. **P<0.01, ***P<0.001; NS, not significant.
Suppression of HBx and the Notch1 pathway inhibits the growth of HepG2.2.15 cells
Our results indicated that Notch1 was highly expressed in HCC tissues, especially in large HCC tissues, and the Notch1 pathway was regulated by HBx, we therefore determined whether inhibition of the Notch1 pathway attenuated cell growth. To inhibit the Notch1 pathway, we used the γ-secretase inhibitor DAPT. HepG2.2.15 cells were treated with increasing concentrations of DAPT for 4 days, and then cell viability was evaluated by CCK-8 assays. As shown in Fig. 3E, inhibition of HBx attenuated the growth of HepG2.2.15 cells. Increasing concentrations and treatment times of DAPT resulted in progressive inhibition of HepG2.2.15 cell viability (Fig. 3F). At day 1, DAPT treatment at various concentrations did not reduce cell viability. However, DAPT treatment for more than 2 days triggered a significant time- and dose-dependent decrease in the viability of HepG2.2.15 cells. We ultimately selected a DAPT treatment concentration of 20 µM for further experiments.
Inhibition of Notch1, ERK, and AKT pathways after DAPT treatment in HepG2.2.15 cells
HepG2.2.15 cells treated with 20 µM DAPT for 1 and 6 h were assessed for inhibition levels of the Notch1 pathway by western blot analyses of Jagged1, Notch1, NICD, and Hes1 expression. DAPT treatment significantly decreased the amount of NICD and Hes1 in a time-dependent manner, but did not have any effect on Jagged1 or Notch1 (Fig. 3G). Our data showed that DAPT treatment greatly inhibited the growth of HepG2.2.15 cells. Therefore, we tested expression of ERK and AKT pathways that are closely related to cell proliferation. Western blotting indicated that DAPT greatly reduced the amount of pERK and pAKT in a time-dependent manner, but did not have any effect on total ERK or AKT (Fig. 3H).
Upregulation of DUSP1 and PTEN after DAPT treatment in HepG2.2.15 cells
Previous reports have revealed the regulatory circuit linking the pathway with DUSP1 expression and ERK activity (21). A link has also been found between the Notch1 pathway and PTEN and AKT activities (28). Our data showed that DUSP1 and PTEN were upregulated after DAPT treatment in HepG2.2.15 cells by western blotting (Fig. 3I and J).
Upregulation of DUSP1 and PTEN after SiHes1 treatment of HepG2.2.15 cells
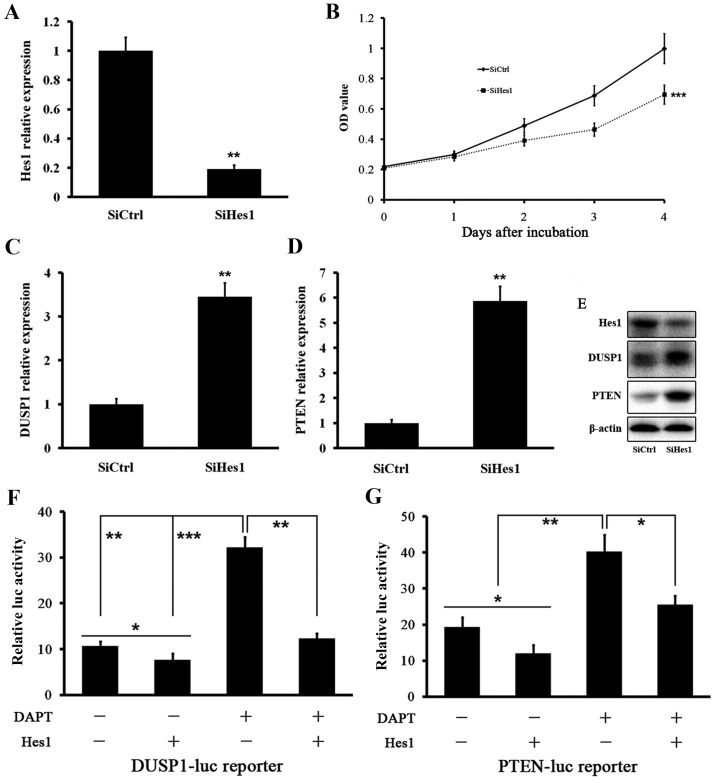
We tested the effect of Hes1 inhibition on DUSP1 and PTEN levels. Treatment of HepG2.2.15 cells with siRNA targeting Hes1 mRNA (SiHes1) effectively reduced the Hes1 mRNA level (Fig. 4A). Growth of HepG2.2.15 cells was inhibited after SiHes1 treatment (Fig. 4B). Next, we found that the mRNA levels of DUSP1 and PTEN were greatly increased after SiHes1 treatment (Fig. 4C and D). Western blot analysis showed the same results. The Hes1 protein level was signifi-cantly reduced and the protein levels of DUSP1 and PTEN were elevated significantly (Fig. 4E).
Figure 4.
Hes1 represses the expressions of DUSP1 and PTEN. (A) Hes1 mRNA level was effectively reduced after SiHes1. The changes were presented as fold change in comparison to the control. (B) Cell growth of HepG2.2.15 was inhibited after SiHes1. (C) DUSP1 mRNA level was increased after SiHes1. The changes were presented as fold change in comparison to the control. (D) PTEN mRNA level was increased after SiHes1. The changes were presented as fold change in comparison to the control. (E) Protein levels of DUSP1 and PTEN were increased after SiHes1. (F and G) Luciferase activity of HepG2.2.15 cells transfected with a DUSP1-luc reporter or PTEN-luc reporter together with a plasmid expressing HES1 or its corresponding empty control. Cells were treated with DAPT or its vehicle. Data were collected in at least three independent experiments. *P<0.05, **P<0.01, ***P<0.001.
Hes1 suppresses and directly binds to the promoters of DUSP1 and PTEN genes in HepG2.2.15 cells
Based on the above data, we investigated whether Hes1 suppressed DUSP1 and PTEN. We performed DUSP1 and PTEN promoter assays using a luciferase reporter. The basal activities of DUSP1 and PTEN gene promoters were reduced by Hes1. Treatment of HepG2.2.15 cells with DAPT induced DUSP1 and PTEN gene promoters, which was reversed by cotransfection of Hes1 (Fig. 4F and G). We next performed ChIP assays to determine whether Hes1 directly bound to the promoters of DUSP1 and PTEN genes using two mixed antibodies against Hes1. We designed several primers in the promoter regions of DUSP1 and PTEN genes (Fig. 5). ChIP assays showed that Hes1 bound to the regulatory sequences in the promoters of DUSP1 and PTEN genes in HepG2.2.15 cells (Fig. 6A and B). Quantitative ChIP assays revealed significant enrichment of two regions of the DUSP1 and PTEN gene promoters compared with control IgG immunoprecipitation, which was reversed by treatment with DAPT (Fig. 6C and D). These results further reinforce our previous findings that DAPT upregulated DUSP1 and PTEN expression by downregulating Hes1.
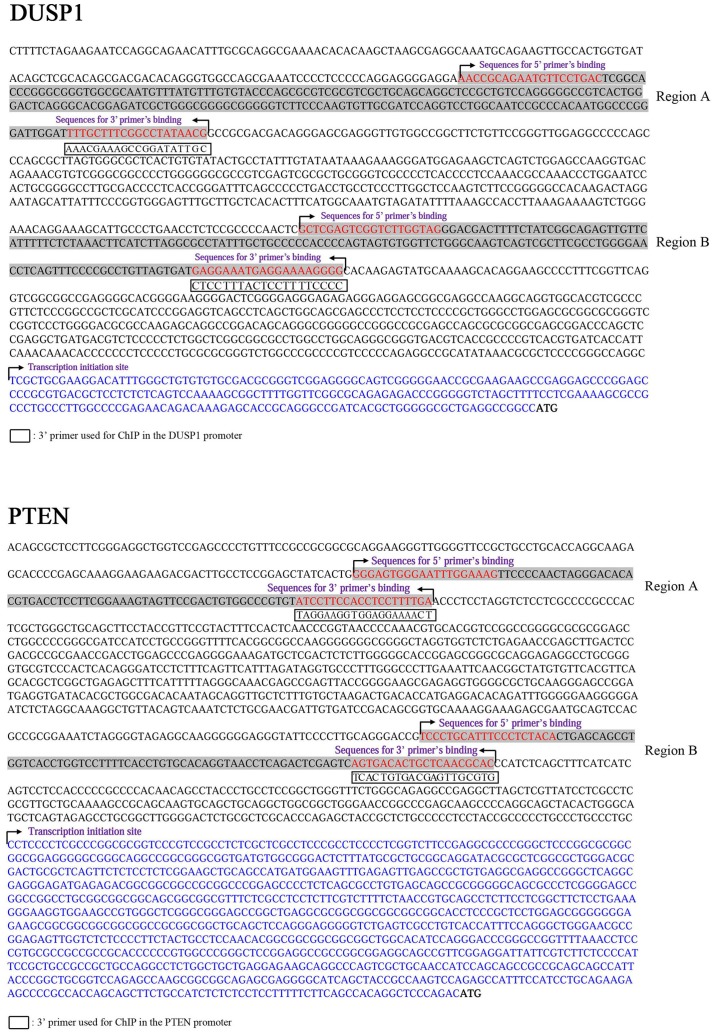
Figure 5.
Sequences of the upstream regions from the ATG (bold) of human DUSP1 and PTEN promoters. The 5′-UTRs of DUSP1 and PTEN are in blue letters. The primers used to amplify the PCR products of the human DUSP1 and PTEN promoters are in red letters and correspond to regions A and B.
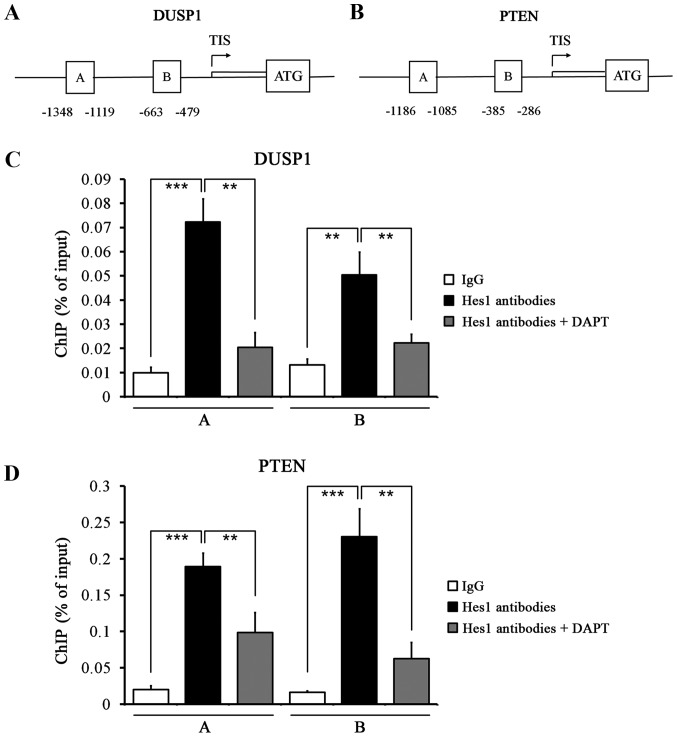
Figure 6.
Quantitative ChIP analysis of Hes1 binding to the promoter sequences of DUSP1 and PTEN. (A) Schematic diagram of the DUSP1 promoter region cantaining potential binding sites of Hes1. Immunoprecipitated DNA was amplified by PCR using primers specific for regions A and B. The arrow indicates the transcription initiation site (TIS). ATG, the translation start codon. (B) Schematic diagram of the PTEN promoter region cantaining potential binding sites of Hes1. (C) HepG2.2.15 cells were subjected to ChIP assay with mixed Hes1 antibodies or IgG antibody to identify Hes1 binding sites on the DUSP1 promoter. Two different DNA regions from the DUSP1 promoter were analyzed by qPCR. Cells were treated with DAPT or vehicle. (D) ChIP assay was performed using Hes1 antibodies or IgG to identify Hes1 binding sites on the PTEN promoter in HepG2.2.15 cells. **P<0.01, ***P<0.001.
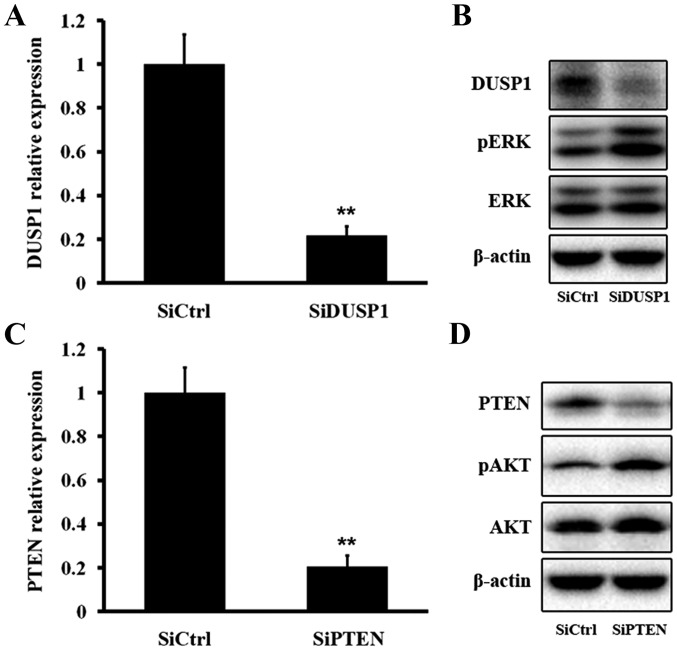
SiDUSP1/SiPTEN treatment of HepG2.2.15 cells elevates pERK/pAKT
We next tested the effect of DUSP1 inhibition on ERK phosphorylation. Treatment of HepG2.2.15 cells with siRNA targeting DUSP1 mRNA (SiDUSP1) effectively reduced DUSP1 mRNA and protein levels, resulting in an elevated pERK level (Fig. 7A and B). We then evaluated the effect of PTEN inhibition on AKT phosphorylation. Treatment with SiPTEN effectively decreased PTEN mRNA and protein levels, which induced an increase of the pAKT level (Fig. 7C and D). The above findings demonstrated that DUSP1 dephosphorylated pERK and PTEN dephosphorylated pAKT.
Figure 7.
DUSP1 regulates the ERK pathway and PTEN regulates the AKT pathway. (A and B) DUSP1 mRNA and protein levels were significantly reduced after SiDUSP1. (B) Expression of pERK was increased after SiDUSP1. (C and D) PTEN mRNA and protein levels were significantly reduced after SiPTEN. (D) Expression of pAKT was increased after SiPTEN. The changes of DUSP1 and PTEN mRNA levels were presented as fold change in comparison to the control. *P<0.05, **P<0.01, ***P<0.001.
Discussion
Notch1 is overexpressed in HCC (14). Consistent with the results of previous studies, we found that Notch1 expression in tumor tissues was much more elevated than that in peri-tumoral, cirrhosis, and normal tissues by IHC and western blot analyses. Previous reports have shown co-localization of Notch1 with HBx (14). In accordance with these findings, IHC and confocal analyses demonstrated that Notch1 morphologically co-localized with HBx in HCC tissues and HepG2.2.15 cells. Furthermore, there was a positive correlation between Notch1 and HBx expression. It has been demonstrated that HBx activates the Notch1 pathway. We also found that the Notch1 pathway was upregulated by HBx. The growth of HepG2.2.15 cells was inhibited by treatment with SiHBx or DAPT, indicating that HBx stimulated cell proliferation via the Notch1 pathway. A previous report has shown that HBx inhibition attenuates the phosphorylation of ERK and AKT in HepG2.2.15 cells (15). In our study, we found that inhibition of the Notch1 pathway downregulated the phosphorylated levels of ERK and AKT.
Previous studies have shown that Hes1, the key member of the Notch1 pathway, represses expression of DUSP1 that is active against pERK in non-small cell lung carcinoma (21). Another study has shown that Hes1 decreases expression of PTEN, a negative regulator of the AKT pathway in T-cell leukemia (28). Thus, we hypothesized that the pathway influenced expression of DUSP1 and PTEN, and further affected ERK and AKT pathways in HepG2.2.15 cells. To test this hypothesis, we performed PCR, western blotting, luciferase assays, and ChIP. We found that mRNA and protein levels of DUSP1 and PTEN were increased after Hes1 inhibition. To investigate the mechanisms that link inhibition of the Notch1 pathway with dephosphorylation of ERK and AKT, we analyzed the transcriptional changes induced by DAPT. Promoter induction of DUSP1 and PTEN genes after DAPT treatment were confirmed in HepG2.2.15 cells. Previous data have shown that ERK and AKT pathways are important to regulate the Notch1 pathway downstream of HBx. However, a direct link between Notch1 and ERK/AKT pathways has not been revealed (15).
Hes1 is a well-known transcriptional regulator of multiple genes (30,31). Based on our finding that the Hes1 level was reduced upon DAPT treatment of HepG2.2.15 cells, we hypothesized that dephosphorylation of ERK or AKT induced by DAPT could be regulated by Hes1-mediated decreases of DUSP1 or PTEN. Our luciferase reporter assays showed that Hes1 was a negative regulator of DUSP1 and PTEN. It directly repressed DUSP1 and PTEN gene promoters, which was reversed by DAPT treatment. Next, we found that increased levels of pERK and pAKT were induced by inhibition of DUSP1 and PTEN, respectively. This result could explain the decrease in ERK and AKT phosphorylations upon DAPT treatment.
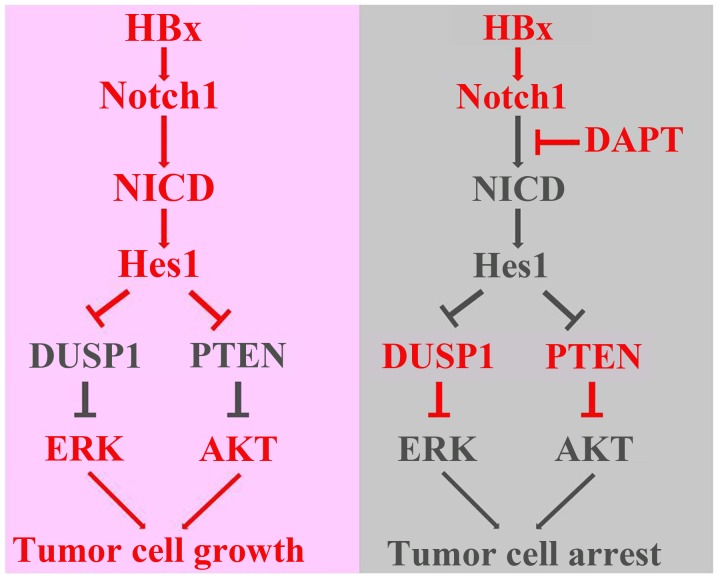
In conclusion, this study explored a direct link among HBx, the Notch1 pathway, DUSP1/PTEN, and ERK/AKT pathways (Fig. 8). We found that HBx activated the Notch1 pathway to promote cell growth, which was correlated with the capacity of Hes1 to increase ERK/AKT activities through decrease of DUSP1/PTEN expression. Nevertheless, we cannot exclude that other molecular mechanisms might take part in mediating the effects of DAPT. Therefore, the underlying mechanisms need to be elucidated further.
Figure 8.
Schematic illustrations of the role of HBx and the Notch1 pathway in HCC proliferation explored in this study. Detailed explanation can be seen in Discussion.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (nos. 81272421 and 81202300) and the Project of Hubei Natural Science Foundation of China (no. 2015CFB462).
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet. 2012;379:1245–1255. doi: 10.1016/S0140-6736(11)61347-0. [DOI] [PubMed] [Google Scholar]
- 3.Sherman M. Hepatocellular carcinoma: Epidemiology, surveillance, and diagnosis. Semin Liver Dis. 2010;30:3–16. doi: 10.1055/s-0030-1247128. [DOI] [PubMed] [Google Scholar]
- 4.Geng M, Xin X, Bi LQ, Zhou LT, Liu XH. Molecular mechanism of hepatitis B virus X protein function in hepatocar-cinogenesis. World J Gastroenterol. 2015;21:10732–10738. doi: 10.3748/wjg.v21.i38.10732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang T, Zhang J, You X, Liu Q, Du Y, Gao Y, Shan C, Kong G, Wang Y, Yang X, et al. Hepatitis B virus X protein modulates oncogene Yes-associated protein by CREB to promote growth of hepatoma cells. Hepatology. 2012;56:2051–2059. doi: 10.1002/hep.25899. [DOI] [PubMed] [Google Scholar]
- 6.Xu C, Zhou W, Wang Y, Qiao L. Hepatitis B virus-induced hepatocellular carcinoma. Cancer Lett. 2014;345:216–222. doi: 10.1016/j.canlet.2013.08.035. [DOI] [PubMed] [Google Scholar]
- 7.Levrero M, Zucman-Rossi J. Mechanisms of HBV-induced hepatocellular carcinoma. J Hepatol. 2016;64(Suppl):S84–S101. doi: 10.1016/j.jhep.2016.02.021. [DOI] [PubMed] [Google Scholar]
- 8.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: Cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 9.Miele L, Golde T, Osborne B. Notch signaling in cancer. Curr Mol Med. 2006;6:905–918. doi: 10.2174/156652406779010830. [DOI] [PubMed] [Google Scholar]
- 10.Rizzo P, Osipo C, Foreman K, Golde T, Osborne B, Miele L. Rational targeting of Notch signaling in cancer. Oncogene. 2008;27:5124–5131. doi: 10.1038/onc.2008.226. [DOI] [PubMed] [Google Scholar]
- 11.Geisler F, Strazzabosco M. Emerging roles of Notch signaling in liver disease. Hepatology. 2015;61:382–392. doi: 10.1002/hep.27268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang F, Zhou H, Xia X, Sun Q, Wang Y, Cheng B. Activated Notch signaling is required for hepatitis B virus X protein to promote proliferation and survival of human hepatic cells. Cancer Lett. 2010;298:64–73. doi: 10.1016/j.canlet.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 13.Wang F, Zhou H, Yang Y, Xia X, Sun Q, Luo J, Cheng B. Hepatitis B virus X protein promotes the growth of hepatocel-lular carcinoma by modulation of the Notch signaling pathway. Oncol Rep. 2012;27:1170–1176. doi: 10.3892/or.2012.1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao J, Xiong Y, Wang Y, Wang Y, Zheng G, Xu H. Hepatitis B virus X protein activates Notch signaling by its effects on Notch1 and Notch4 in human hepatocellular carcinoma. Int J Oncol. 2016;48:329–337. doi: 10.3892/ijo.2015.3221. [DOI] [PubMed] [Google Scholar]
- 15.Kongkavitoon P, Tangkijvanich P, Hirankarn N, Palaga T. Hepatitis B virus HBx activates Notch signaling via delta-like 4/Notch1 in hepatocellular carcinoma. PLoS One. 2016;11:e0146696. doi: 10.1371/journal.pone.0146696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ding ZY, Jin GN, Wang W, Chen WX, Wu YH, Ai X, Chen L, Zhang WG, Liang HF, Laurence A, et al. Reduced expression of transcriptional intermediary factor 1 gamma promotes metastasis and indicates poor prognosis of hepatocellular carcinoma. Hepatology. 2014;60:1620–1636. doi: 10.1002/hep.27273. [DOI] [PubMed] [Google Scholar]
- 17.Chen L, Zhang W, Zhou QD, Yang HQ, Liang HF, Zhang BX, Long X, Chen XP. HSCs play a distinct role in different phases of oval cell-mediated liver regeneration. Cell Biochem Funct. 2012;30:588–596. doi: 10.1002/cbf.2838. [DOI] [PubMed] [Google Scholar]
- 18.Xiang S, Dong HH, Liang HF, He SQ, Zhang W, Li CH, Zhang BX, Zhang BH, Jing K, Tomlinson S, et al. Oval cell response is attenuated by depletion of liver resident macrophages in the 2-AAF/partial hepatectomy rat. PLoS One. 2012;7:e35180. doi: 10.1371/journal.pone.0035180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dong HH, Xiang S, Chen XP, Liang HF, Zhang W, Jing K, Zhang W, Zhang WG, Chen L. The epithelial-mesenchymal transition promotes transdifferentiation of subcutaneously implanted hepatic oval cells into mesenchymal tumor tissue. Stem Cells Dev. 2009;18:1293–1298. doi: 10.1089/scd.2008.0321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen L, Zhang W, Liang HF, Zhou QF, Ding ZY, Yang HQ, Liu WB, Wu YH, Man Q, Zhang BX, et al. Activin A induces growth arrest through a SMAD-dependent pathway in hepatic progenitor cells. Cell Commun Signal. 2014;12:18. doi: 10.1186/1478-811X-12-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maraver A, Fernandez-Marcos PJ, Herranz D, Cañamero M, Muñoz-Martin M, Gómez-López G, Mulero F, Megías D, Sanchez-Carbayo M, Shen J, et al. Therapeutic effect of γ-secretase inhibition in KrasG12V-driven non-small cell lung carcinoma by derepression of DUSP1 and inhibition of ERK. Cancer Cell. 2012;22:222–234. doi: 10.1016/j.ccr.2012.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Calvisi DF, Pinna F, Meloni F, Ladu S, Pellegrino R, Sini M, Daino L, Simile MM, De Miglio MR, Virdis P, et al. Dual-specificity phosphatase 1 ubiquitination in extracellular signal-regulated kinase-mediated control of growth in human hepatocellular carcinoma. Cancer Res. 2008;68:4192–4200. doi: 10.1158/0008-5472.CAN-07-6157. [DOI] [PubMed] [Google Scholar]
- 23.Chen X, Song M, Chen W, Dimitrova-Shumkovska J, Zhao Y, Cao Y, Song Y, Yang W, Wang F, Xiang Y, et al. MicroRNA-21 Contributes to Liver Regeneration by Targeting PTEN. Med Sci Monit. 2016;22:83–91. doi: 10.12659/MSM.896157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liao B, Liang H, Chen J, Liu Q, Zhang B, Chen X. Suberoylanilide hydroxamic acid enhances chemosensitivity to 5-fluorouracil in hepatocellular carcinoma via inhibition of thymidylate synthase. Tumour Biol. 2015;36:9347–9356. doi: 10.1007/s13277-015-3497-9. [DOI] [PubMed] [Google Scholar]
- 25.Ding ZY, Jin GN, Liang HF, Wang W, Chen WX, Datta PK, Zhang MZ, Zhang B, Chen XP. Transforming growth factor β induces expression of connective tissue growth factor in hepatic progenitor cells through Smad independent signaling. Cell Signal. 2013;25:1981–1992. doi: 10.1016/j.cellsig.2013.05.027. [DOI] [PubMed] [Google Scholar]
- 26.Tchen CR, Martins JR, Paktiawal N, Perelli R, Saklatvala J, Clark AR. Glucocorticoid regulation of mouse and human dual specificity phosphatase 1 (DUSP1) genes: Unusual cis-acting elements and unexpected evolutionary divergence. J Biol Chem. 2010;285:2642–2652. doi: 10.1074/jbc.M109.037309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Real PJ, Tosello V, Palomero T, Castillo M, Hernando E, de Stanchina E, Sulis ML, Barnes K, Sawai C, Homminga I, et al. Gamma-secretase inhibitors reverse glucocorticoid resistance in T cell acute lymphoblastic leukemia. Nat Med. 2009;15:50–58. doi: 10.1038/nm.1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palomero T, Sulis ML, Cortina M, Real PJ, Barnes K, Ciofani M, Caparros E, Buteau J, Brown K, Perkins SL, et al. Mutational loss of PTEN induces resistance to NOTCH1 inhibition in T-cell leukemia. Nat Med. 2007;13:1203–1210. doi: 10.1038/nm1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ding ZY, Liang HF, Jin GN, Chen WX, Wang W, Datta PK, Zhang MZ, Zhang B, Chen XP. Smad6 suppresses the growth and self-renewal of hepatic progenitor cells. J Cell Physiol. 2014;229:651–660. doi: 10.1002/jcp.24488. [DOI] [PubMed] [Google Scholar]
- 30.Iso T, Kedes L, Hamamori Y. HES and HERP families: Multiple effectors of the Notch signaling pathway. J Cell Physiol. 2003;194:237–255. doi: 10.1002/jcp.10208. [DOI] [PubMed] [Google Scholar]
- 31.Sang L, Roberts JM, Coller HA. Hijacking HES1: How tumors co-opt the anti-differentiation strategies of quiescent cells. Trends Mol Med. 2010;16:17–26. doi: 10.1016/j.molmed.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]