Abstract
Human and bovine respiratory syncytial viruses (HRSV/BRSV) are major causes of severe lower respiratory tract infections in children and calves, respectively. Shared epidemiological, clinical, pathological and genetic characteristics of these viruses make comparative research highly relevant. To characterise the host response against BRSV infection, bronchoalveolar lavage supernatant (BAL) from i) non-vaccinated, BRSV-infected ii) vaccinated, BRSV-infected and iii) non-infected calves was analysed by tandem mass spectrometry. Proteins were semi-quantified and protein expression was validated by immunoblotting. Correlations between selected proteins and pathology, clinical signs and virus shedding were investigated. Calves with BRSV-induced disease had increased total protein concentrations and a decreased number of proteins identified in BAL. The protein profile was characterised by neutrophil activation and a reduction in identified antioxidant enzymes. The presence of neutrophils in alveolar septa, the expression level of neutrophil-related or antioxidant proteins and LZTFL1 correlated significantly with disease. Citrullinated histone 3, an indicator of extracellular traps (ETs), was only detected in non-vaccinated, BRSV-infected animals. By bringing disequilibrium in the release and detoxification of reactive oxygen species, generating ETs and causing elastine degradation, exaggerated neutrophil responses might exacerbate RSV-induced disease. Neutrophil-mitigating or antioxidant treatments should be further explored.
Introduction
Human and bovine respiratory syncytial viruses (HRSV and BRSV) are host-specific but genetically highly similar and induce comparable pathology in humans and cattle [1]. It is suspected that the clinical signs induced by RSV partly derive from inflammatory responses. In humans, severe RSV-induced inflammation was shown to additionally have long-term consequences including airway hyper-reactivity during childhood (reviewed by [2]). As recently reviewed by Dapat and Oshitani (2016), proteomic data based on mass spectrometry are sparse from in vivo RSV infections [3], but does exist on nasal aspirates from HRSV-infected children, lung tissue from HRSV-infected rats, and from mice with vaccine-induced enhanced HRSV disease [4–6]. Overall, proteins were separated by gel-electrophoresis. To complement these approaches, by using an experimental model of RSV infection in a natural host that reproduces severe clinical signs of disease [7] and by analysing the proteomic profile of samples collected from the lower respiratory tract at precise times post infection, we assumed that we could obtain new information about RSV pathogenesis. Thereby, targets for new treatments could potentially be identified. We used label free relative quantification by liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS), the shotgun method [8], to investigate the proteomes of bronchoalveolar lavage supernatants (BAL) obtained from calves at the peak of clinical signs and analysed data in relation to pathology, clinical score, and virus shedding.
Material and methods
Samples, animals and experimental design
Bronchoalveolar lavage (BAL) was collected post mortem, as previously described [9], from the left lung of 15 conventionally-reared, 2–4 months old calves of the Swedish red and white and Swedish Holstein breeds that had been infected with BRSV by aerosol in two independent studies (Table 1).
Table 1. Animals and experimental design.
| Study | No. calves | Treatment | BRSV strain | Virology | % lung lesions | Designation | Mean BAL protein conc. (μg/μl) | Mean no. proteins identified |
|---|---|---|---|---|---|---|---|---|
| I | 5 | Non-vaccinated | 9402022 | +++ |
15.7 ± 14.5 |
Susceptible_I |
0.49 |
394 |
| 5 |
BRSV-ISCOM-vaccinated | 9402022 |
± |
3.8 ± 2.9 |
Vaccinated |
0.19 | 547 | |
| II | 5 |
Non-vaccinated | Snook |
+++ |
48 ± 12.1 |
Susceptible_II |
0.79 |
159 |
| 5 | None | None | - | 0 | Non-infected | 0.37 | 296 |
The allocation to groups, the extent of macroscopic lung lesions, the clinical scores (prior and post infection) and the quantity of virus shed in nasal secretions in these calves has been documented in detail elsewhere [9, 10]. The clinical scoring system is available as supplemental data (S1 Table). From these animals tissue samples from the right cranial lobe, if present from an area of consolidation, were processed for routine diagnostic evaluation with hematoxylin/eosin-staining. The degree and characteristics of the inflammatory response was assessed in a blinded manner by a diagnostic pathologist. In case of histopathological lesions, including infiltration by inflammatory cells, they were subjectively graded mild, moderate to severe, e.g. the degree of neutrophils in alveolar septa was scored (in text represented as score 0–3, score 0 negative and 3 severe). BAL was also collected from 5 uninfected, clinically healthy calves of the same breeds and approximate age at slaughter. Calves with no or minimal macroscopic lung lesions were selected for this purpose.
Fresh BALs were kept on ice, filtered over sterile gauze, centrifuged at 200 x g for 10 minutes and the supernatants were stored at -80°C, before analyses in two separate experiments:
In experiment I, the BAL originated from calves that had been infected with BRSV (strain no. 9402022, kindly provided by Pr. L.E. Larsen, DTU, Denmark), 6 days previously, and were either a) non-vaccinated and showed clinical signs of disease and shed high quantities of virus (n = 5, called susceptible_I), or b) BRSV-ISCOM-vaccinated and showed little or no clinical signs of disease and shed little or no virus (n = 5, called vaccinated) (Table 1) [9]. As described in detail previously [11], this vaccine consisted of pleomorphic nanoparticles that contained purified, solubilized proteins from BRSV-infected cell culture, as well as lipids and Quillaja saponin.
In experiment II, the BAL originated from calves that had either been infected with BRSV (Snook strain), 7 days previously, and shed high quantities of virus, showed severe clinical signs of disease and had extensive macroscopic lung lesions (n = 5, called susceptible_II) [10], or were non-infected and healthy, at slaughter (n = 5, called non-infected) (Table 1). The clinical signs and pathology in the susceptible animals were more severe in experiment II than in experiment I. In experiment II, five calves showed marked to severe signs of illness, versus two calves in experiment I. BRSV had been isolated in cell culture by using BAL cells (i.e. not the supernatant), from all susceptible calves and one vaccinated calf, in experiment I and II [9, 10].
Animal care
Both experiments were carried out in compliance with the E.U. Directive 86/609, and approved by the Ethical Committee of the district court of Uppsala, Sweden (Refs. no. C68/10, C330/11). The health and behaviour of all animals were monitored at least twice daily throughout the experiments (6 and 7 weeks) and at least three times daily from the day of challenge to the day of euthanasia (day 6 or 7 post infection). The humane endpoint, which was used as criterion for euthanasia, was defined as follows: i) abdominal or forced breathing, or respiratory rate exceeding 100 min-1, in combination with severely depressed general condition, or ii) anorexia for more than 24 hours, or iii) rectal temperature exceeding 41°C for more than 36 hours. One calf in experiment II reached the humane endpoint on PID7 and was euthanized immediately after the clinical examination. None of the calves died before reaching the endpoint.
To minimize suffering and distress of the animals, the bedding was kept clean and thick and the air and food quality was optimal. The calves were fed hay and water ad libitum, and pellets and milk replacer twice daily. They were frequently handled, to reduce stress at sample collection. Clinical examinations and sample collections were performed by trained veterinarians, certified by individual licenses to perform animal experiments. The calves were euthanised by an overdose of general anesthesia (5 mg/kg ketamine and 15 mg/kg pentobarbital sodium) followed by exsanguination. The calves with the most severe clinical signs were euthanized first.
Liquid chromatography tandem mass spectrometry (LC-MS/MS) and data analyses
BAL supernatants (hereafter called BAL) containing 20 μg total protein, determined using the Bradford assay (BioRad), were diluted to contain 50 mM ammonium bicarbonate. The samples were reduced with dithiothreitol (DTT) and alkylated with iodoacetamide. Trypsin was added in a trypsin:protein ratio of 1:20 and digestion was performed overnight. Thereafter the samples were purified by Pierce C18 Spin Columns (Thermo Scientific), dried and resolved in 60μL 0.1% formic acid, and analysed by tandem mass spectrometry. Analyses were performed in a blind manner using peptide separation by reversed phase liquid chromatography and an QExactive Plus Orbitrap mass spectrometer (Thermo Fisher Scientific, Bremen, Germany) equipped with a nano electrospray ion source, as detailed in [12], with the following modifications: Protein identification was performed against a FASTA database containing proteins from Bos Taurus extracted from the SwissProt database (release Oct 2016, 44366 entries). Search parameters included a maximum of 4.5 ppm and 20 ppm error tolerances for the survey scan and MS/MS analysis, respectively. For protein identification, only peptides with a minimum of 7 amino acids were considered. A total label free intensity analysis was performed for each individual sample. Pathway analyses were carried out in Reactome v59 [13], (www.reactome.org). Proteins were classified as detected at calf treatment group level if identified in at least 4 out of 5 calves and venn diagrams were designed using the Interactivenn software [14], (www.interactivenn.net). Protein functions were analysed in Panther v11.1 [15], (www.pantherdb.org/).
Immunoblotting
Dotblots and Western blots were carried out as described previously [11], using polyclonal rabbit antibodies against myeloperoxidase or histone H3 citrulline 2+8+17 peptide (ab14323 and ab5103, Abcam,) and polyclonal HRP-conjugated sheep antibodies against rabbit IgG (Star 54, Bio-Rad). Controls using BAL antigen and the secondary antibodies alone were included. Protein bands were quantified using Image Lab 5.2.1 (BioRad).
Bacterial analyses
All BRSV-infected animals had been treated with penicillin prior to BRSV-infection. The presence of bacteria in the lungs was determined by aerobic culture of fresh BAL from BRSV-infected calves on bovine blood agar. Since all animals were conventionally reared, analyses of DNA specific for the bacterial opportunists Pasteurella multocida, Mannheimia haemolytica, Histophilus somni and Mycoplasma bovis were additionally performed by TaqMan® qPCR (LSI VetMAX™ Screening Pack, Life Technologies) on BAL from all animals but two (vaccinated calves b and d), due to lack of material from these individuals. The assay was performed according to the manufacurer’s instructions.
Statistical analyses
Statistical analyses were carried out in Minitab® software, version 17. For comparisons of label free quantification (LFQ) of intensities for relative protein expression level in BAL, a two-tailed student’s T-test was used, with assumed unequal variance. For correlations, the Spearman's rank and Pearson product moment correlation coefficients were used for data with non-normal and normal distribution, respectively (determined using the Anderson-Darling test). Samples from all animals were included in all analyses, if not otherwise specified.
Results
BRSV-infection increased extracellular protein levels and decreased the number of proteins identified in the lumen of the lower respiratory tract
In experiment I, 6 days post BRSV-infection (DPI), the total protein concentration in BAL was significantly higher in susceptible_I calves than in vaccinated animals which were protected against BRSV infection (mean 0.49 μg/μL (0.20–0.71) and 0.19 μg/μL (0.12–0.46), respectively; p = 0.04) (Table 1). Similarly, in experiment II, the total protein concentration was higher in BAL of susceptible_II calves 7 DPI than in that of non-infected animals, although this was not statistically significant (mean 0.79 μg/μL (0.07–1.34) and 0.37 μg/μL (0.10–0.68), respectively; p = 0.15) (Table 1).
The number of proteins identified by LC-MS/MS per BAL was significantly lower in the susceptible_II calves compared to non-infected animals (mean 159 (64–240) and 296 (124–349), respectively; p = 0.03) and tended to be lower in the susceptible_I calves than in the vaccinated animals (mean 394 (322–504) and 547 (249–702), respectively; p = 0.11) (Table 1).
BRSV-infection increased the expression of 96 proteins involved in several biological processes
In addition to differences in the total protein concentration and number of proteins identified, the pattern of proteins identified in BAL differed between susceptible calves with high virus shedding and vaccinated calves with very little or no virus shedding, or non-infected calves. Qualitatively, 39 and 41 proteins were uniquely detected in susceptible calves, in experiment I and II, respectively (Fig 1A and 1B, right side of right circles; Table 2, Qualitative).
Fig 1. Bronchoalveolar proteomes of calves.
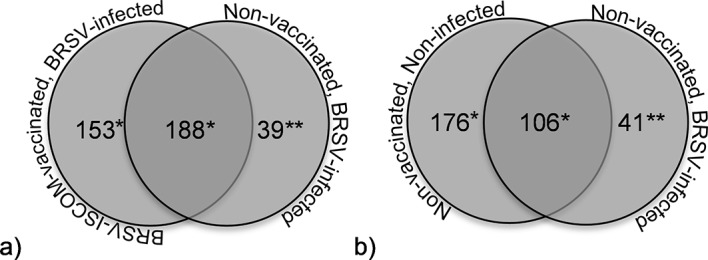
Proteins were identified by LC-MS/MS in bronchoalveolar lavage of calves and were classified as detected at group level if identified in at least 4 out of 5 calves per group. The groups consisted of non-vaccinated, BRSV-infected calves with clinical signs of disease and high levels of virus shedding (a and b, right circles), or ISCOM-vaccinated, BRSV-infected calves with no or little clinical signs of disease and no or low levels of virus shedding (a, experiment I, left circle), or non-vaccinated, non-infected calves (b, experiment II, left circle). Protein names are provided as supplemental data (*) or in Table 1 (**).
Table 2. Proteins associated with BRSV-induced disease.
| Qualitative: Proteins classified as uniquely detected in non-vaccinated, BRSV-infected, susceptible calves and not in controlsa,b |
Quantitative: Proteins identified at significantly higher expression levels in non-vaccinated, BRSV-infected, susceptible calves compared to controlsa,c |
||
|---|---|---|---|
| Calf experiment I | Calf experiment II | Calf experiment I | Calf experiment II |
| Actin, alpha cardiac muscle 1 | Actin, alpha cardiac muscle 1* | ||
| Actin-related protein 2/3 complex subunit 5 | Actin-related protein 2/3 complex subunit 5* | ||
| Adenylyl cyclase-associated protein** | |||
| Alpha-1B-glycoprotein | Alpha-1B-glycoprotein* | ||
| Annexin A1 * | |||
| Azurocidin | Azurocidin | Azurocidin* | |
| Beta-2-microglobulin* | |||
| Beta-defensin (2/8/10) | Beta-defensin (2/8/10/13) | Beta-defensin (10*) | Beta-defensin (2**/8*/10*/12**) |
| BRSV (M/M2-1/N /P) | BRSV(M*/M2-1*/N* /P**) | ||
| Calponin-2 | |||
| Cathelicidin (4/5/7) | Cathelicidin (5) | Cathelicidin (1***/3*/4*/5**) | Cathelicidin (1**/2*/3*/4**/5**/6*/7**) |
| Cathepsin (G/L2) | Cathepsin (G like) | Cathepsin (G**/L2*) | Cathepsin (G**) |
| CD14 | CD14** | ||
| CD177 | CD177** | ||
| CD44 | |||
| Complement factor properdin | Complement factor properdin** | ||
| Coronin-1A** | |||
| Cystatin-C** | |||
| ECM1 protein | |||
| EF-hand domain-containing protein D2 | EF-hand domain-containing protein D2** | ||
| Elastase | Elastase | Elastase* | Elastase* |
| Folate receptor alpha | Folate receptor alpha | Folate receptor alpha* | |
| Fructose-bisphosphate aldolase* | |||
| Glucose-6-phosphate isomerase* | |||
| Glyceraldehyde-3-phosphate dehydrogenase* | |||
| Granulin | Granulin | Granulin** | Granulin** |
| Haptoglobin* | Haptoglobin* | ||
| Hematopoietic cell-specific Lyn substrate 1 | Hematopoietic cell-specific Lyn substrate 1 | Hematopoietic cell-specific Lyn substrate 1* | |
| High mobility group protein B2* | |||
| Histone (H2A/H2AB/H2B) | Histone (H1e/H1.1/H1.2/H1.3/H2A.J/H2B 1/H2B 1-K/ H3.2) | Histone (H1.3*/H2A*/H2A 2-C*/H2B*/H3.1*/H4*) | Histone (H1e*/H1.1*/H1.2*/ H2AC*/H2AJ*) |
| Ig lambda chain V-I region BL2-like | Ig lambda chain V-I region BL2-like** | ||
| Ig like V-set | Ig like V-set* | ||
| Inter-alpha-trypsin inhibitor heavy chain H4 | |||
| Lactotransferrin** | Lactotransferrin* | ||
| Lingual antimicrobial peptide | Lingual antimicrobial peptide* | ||
| L-serine dehydratase/L-threonine deaminase | L-serine dehydratase/L-threonine deaminase | L-serine dehydratase/L-threonine deaminase** | |
| Lymphocyte cytosolic protein 1 | Lymphocyte cytosolic protein 1* | ||
| Lymphocyte-specific protein 1 | Lymphocyte-specific protein 1 | ||
| Macrophage migration inhibitory factor | |||
| Matrix metalloproteinase-9*** | |||
| MSLN protein | |||
| Myeloperoxidase | Myeloperoxidase** | Myeloperoxidase* | |
| Neutrophilic granule protein-like | Neutrophilic granule protein-like* | ||
| Nucleobindin 2 | Nucleobindin 2** | ||
| Olfactomedin 4** | |||
| Pentraxin-related protein PTX3 | Pentraxin-related protein PTX3** | ||
| Peptidoglycan recognition protein 1** | Peptidoglycan recognition protein 1* | ||
| Primary amine oxidase, liver isozyme | |||
| Protein FAM49B | |||
| Protein S100-A9 | Protein S100-A9 | Protein S100 (A8**/A9*) | |
| Proteinase 3 | Proteinase 3** | ||
| Ras-related C3 botulinum toxin substrate 2* | |||
| Regakine-1 | |||
| Resistin | Resistin* | Resistin** | |
| Rho GDP-dissociation inhibitor 2 | Rho GDP-dissociation inhibitor 2* | ||
| Secretoglobin family 1D member | |||
| SERPINB4 protein | SERPINB4 protein** | ||
| Serum amyloid A protein (3.2) | Serum amyloid A protein (1/3.2) | Serum amyloid A protein (1*/3.2**) | |
| Spleen trypsin inhibitor I | Spleen trypsin inhibitor I*** | ||
| Sulfhydryl oxidase* | |||
| Transaldolase | Transaldolase*** | ||
| Uteroglobin | |||
| Transcobalamin 1*** | |||
| Transketolase** | |||
| Uncharacterised (G5E604d)** | |||
| Uncharacterised (ENSEMBL:ENSBTAP00000014147d)* | |||
aProteins were detected in bronchoalveolar lavage from non-infected and/or BRSV-infected calves by LC-MS/MS and semi-quantified by label free analysis. Grey highlight; proteins associated with BRSV-induced disease, in both animal experiments I and II
bUniquely detected at group level in non-vaccinated, BRSV-infected, susceptible calves (i.e. detected in at least 4 out of 5 non-vaccinated, BRSV-infected, susceptible calves and in less than 4 ISCOM-vaccinated, BRSV-infected, protected calves (Experiment I) or non-infected calves (Experiment II)).
cDetected at significantly higher expression levels in non-vaccinated, BRSV-infected, susceptible calves compared to in ISCOM-vaccinated, BRSV-infected, protected calves (Experiment I) or non-infected calves (Experiment II)).
dProtein identification number, uncharacterized protein, no gene identified
Statistically significant differences are indicated by asterisks; p≤0.05(*); p≤0.01 (**); p≤0.001 (***).
Quantitatively, 57 and 35 proteins were expressed at significantly higher levels in BRSV-infected, susceptible calves compared to vaccinated or non-infected calves in experiments I and II, respectively (Table 2, Quantitative).
In total, 96 different proteins were associated with BRSV-disease, either qualitatively or quantitatively (Table 2), 44 of which were common between experiment I and II, or belonged to protein families in which one or several proteins were common between the experiments (Table 2, grey highlight). The common proteins associated with BRSV infection were related to the following cells and processes:
Neutrophil activation and chemotaxis, including neutrophil extracellular trap (NET) formation [16–21]: azurocidin, beta defensins, cathelicidins, cathepsins, elastase, granulin, hematopoietic cell-specific Lyn-substrate 1, histones, lactotransferrin, lymphocyte-specific protein 1, myeloperoxidase, peptidoglycan recognition protein, proteins S100-A9 and resistin.
Epithelial cells and epithelial-derived responses [16, 22, 23]: beta defensins, cathelicidins, folate receptor alpha.
Lymphocyte and natural killer cell activation and chemotaxis [24, 25]: hematopoietic cell-specific Lyn-substrate, lymphocyte-specific protein 1.
Macrophage activation [26]: beta defensins, cathelicidins, resistin.
Systemic acute-phase responses [27]: serum amyloid A, haptoglobin.
Cell necrosis and/or apoptosis [28]: histones.
Gluconeogeneis [29]: L-serine dehydratase/L-threonine deaminase.
BRSV proteins were identified in 5/5 susceptible_I calves and 2/5 susceptible_II calves but not in any of the vaccinated or non-infected calves. The immunoglobulin J chain was detected in all calves in both experiments and the heavy constant μ chain was detected in all calves in experiment II, but the expression levels of these proteins did not in differ significantly between groups.
Less antioxidant proteins were detected following BRSV-infection
Qualitatively, 153 and 176 proteins were uniquely detected in vaccinated and non-infected calves, respectively (Fig 1A and 1B, left side of left circles, supplemental data S2 Table), and quantitatively, 95 and 101 proteins were expressed at significantly lower levels in BRSV-infected, susceptible calves compared to vaccinated and non-infected calves, respectively (supplemental data S3 Table). Thirty-eight of the proteins identified at lower expression levels in the animals with BRSV disease were common between the two experiments and were submitted to Panther for classification [15]. In total, 36 proteins were classified and the major protein class was oxidoreductase (n = 9), followed by cytoskeletal proteins (n = 5), hydrolases (n = 4) and chaperone proteins (n = 4). The remaining nine protein classes contained 1–3 proteins.
Pathway analysis confirmed increased neutrophil degranulation and reduced detoxification of reactive oxygen species associated with BRSV infection
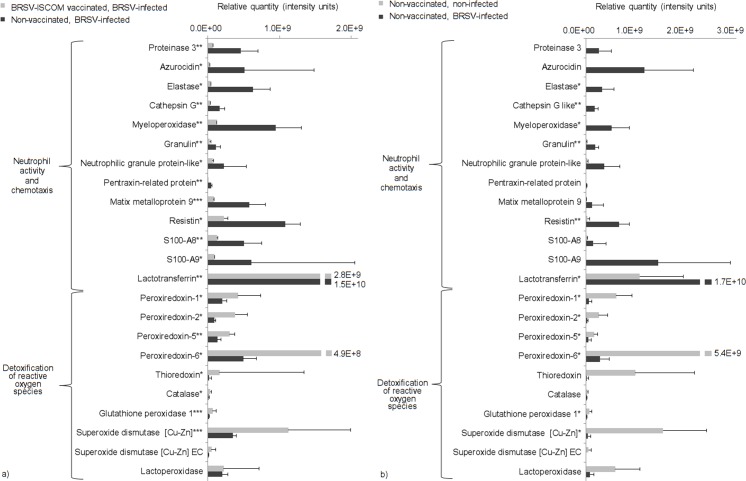
The four quantitative datasets, containing the 57 and 35 proteins identified at statistically significantly higher expression levels in BRSV-infected, susceptible calves (Table 2, Quantitative), as well as the 95 and 101 proteins identified at statistically significantly lower expression levels in these animals compared to those in their vaccinated or non-infected controls, in experiment I and II, respectively (S3 Table), were submitted to Reactome [13]. The numbers of proteins that were recognized and mapped were as follows: 29/57 (higher, experiment I), 10/35 (higher, experiment II), 56/95 (lower, experiment I) and 52/101 (lower, experiment II). The bovine cathelicidins 1–7, which were increased in animals with BRSV disease in experiments I and II (Table 2) were among the proteins not recognized by Reactome. Nevertheless, in both experiments, the pathway identified with the highest probability to be activated during BRSV infection was neutrophil degranulation. The pathway identified with the highest probability to be downregulated, inactive or not identified during BRSV infection was detoxification of reactive oxygen species. The relative expression levels of proteins included in these pathways followed the same pattern, which is illustrated in Fig 2 (see black bars for BRSV-infected, susceptible calves).
Fig 2. Semi-quantification of selected proteins involved in processes that were affected by BRSV-infection.
Proteins were detected in bronchoalveolar lavage from non-vaccinated, BRSV-infected calves with clinical signs of disease and high levels of virus shedding (black bars) and/or in BRSV-ISCOM-vaccinated, BRSV-infected calves with no or little clinical signs of disease and no or low levels of virus shedding (a, experiment I, grey bars), and/or non-vaccinated, non-infected calves (b, experiment II, grey bars). Proteins were identified by LC-MS/MS and semi-quantified by label-free analysis. Statistically significant differences are indicated by asterisks; p≤0.05(*); p≤0.01 (**); p≤0.001 (***). Proteins were selected based on being related to neutrophil activation and chemotaxis or detoxification of reactive oxygen species: biological processes identified by protein pathway analysis.
The presence of proteins related to neutrophil activity was confirmed by immunoblotting on serially diluted BAL. Citrullinated histone 3 and myeloperoxidase were detected in BAL from all or several BRSV-infceted, susceptible_I and susceptible_II calves, but not in their vaccinated or non-infected controls (Figs 3 and 4). Citrullinated histone 3, detected by western blot at 17 kDa, was semi-quantified within each of the two experiments. When compared to k (experiment I), or D1 (experiment II) the relative quantity was estimated to 100%, 5%, 8%, 82%, 20% (calves k-o) and 100%, 87%, 68%, 94%, 20% (calves D1-D5).
Fig 3. Association of citrullinated histone 3 with BRSV disease.
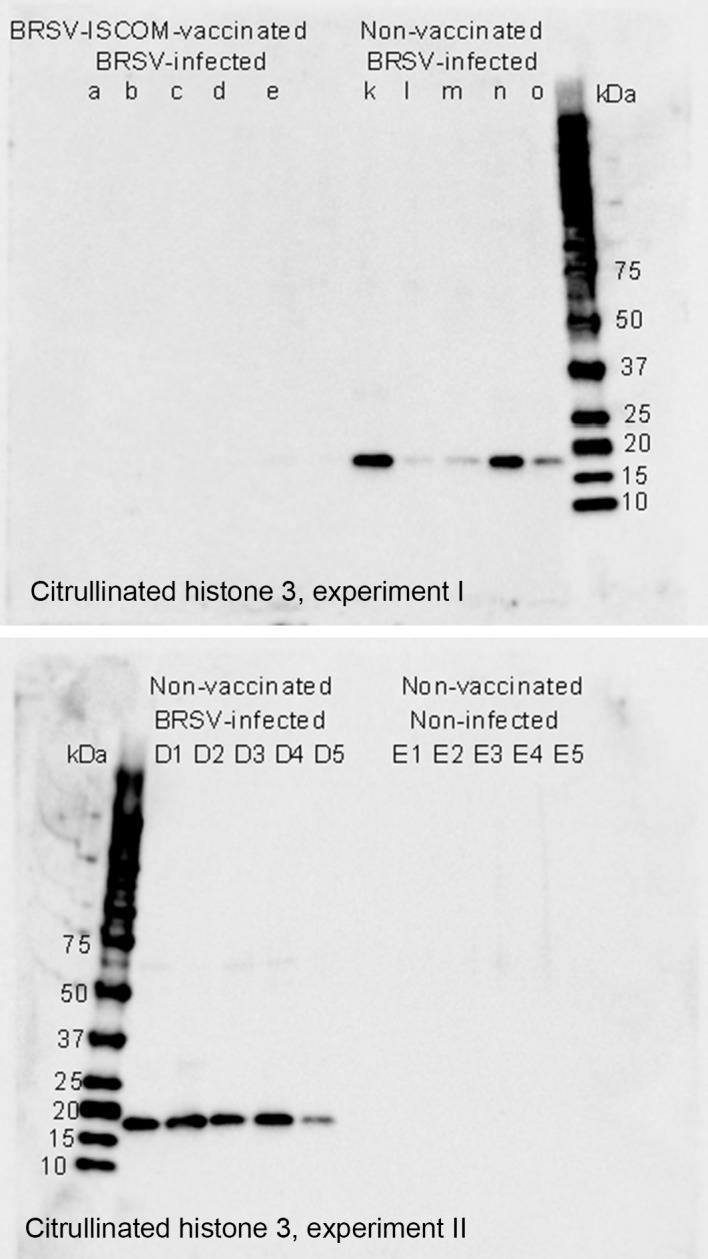
Detection of citrullinated histone 3 by Western blot in bronchoalveolar lavage from BRSV-infected calves with clinical signs of disease and high levels of virus shedding (k-o and D1-D5) compared to that in BRSV-ISCOM-vaccinated, BRSV-infected calves with no or little clinical signs of disease and no or low levels of virus shedding (a-e, Experiment I), or non-vaccinated, non-infected calves (E1-E5, Experiment II).
Fig 4. Detection of citrullinated histone 3 and myeloperoxidase in serially diluted BAL.
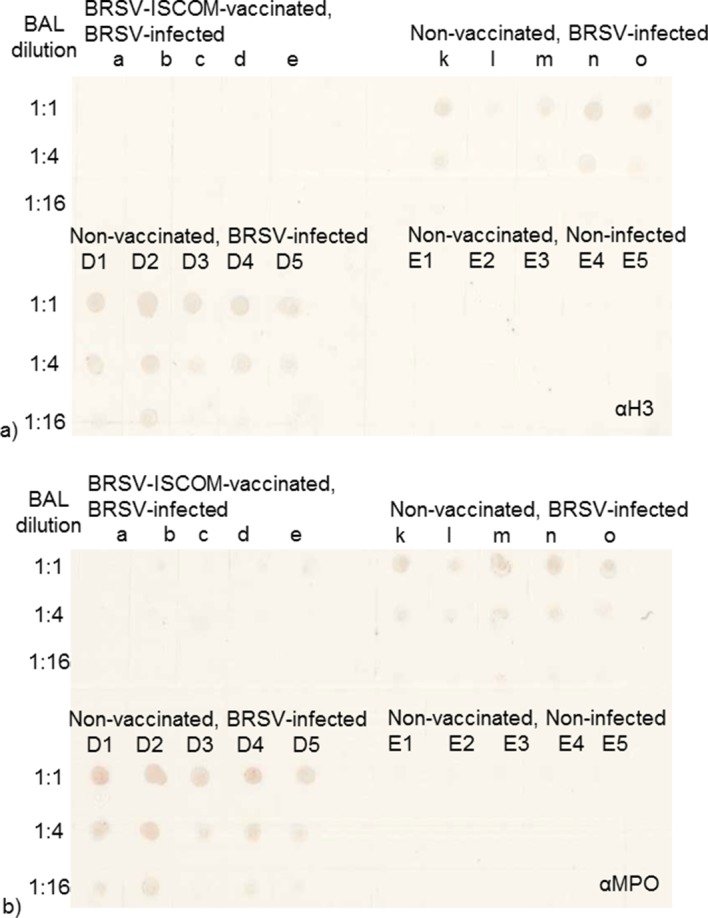
Detection of citrullinated histone 3 (a) and myeloperoxidase (b) by dotblot in bronchoalveolar lavage (BAL) from BRSV-infected calves with clinical signs of disease and high levels of virus shedding (k-o and D1-D5) compared to that in BRSV-ISCOM-vaccinated, BRSV-infected calves with no or little clinical signs of disease and no or low levels of virus shedding (a-e, Experiment I), or non-vaccinated, non-infected calves (E1-E5, Experiment II). The proteins were selected based on being present in neutrophil extracellular traps, incriminated to be important in the pathogenesis of RSV [30].
The level of neutrophil activation correlated with the degree of pathology, clinical signs and virus shedding
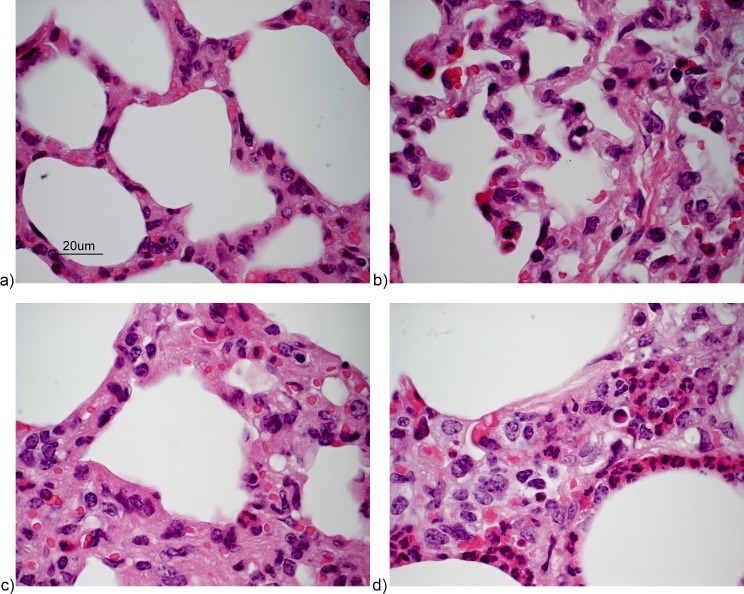
To determine if high neutrophil activity constituted a protective immune response in infected animals or, may have contributed to clinical signs of disease, the degree of pulmonary neutrophil-infiltration was evaluated in relation to BRSV-induced disease in experiment I, because the calves in this experiment had varying levels of disease and neutrophil scores [9]. The susceptible calves in experiment II were all very severely affected and all had the highest neutrophil score (for examples of neutrophil scoring, see Fig 5).
Fig 5. Scores of neutrophilic infiltration of alveolar septa in BRSV-infected calves.
HE stained sections. a) score 0 (vaccinated calf a, experiment I), b) score 1 (vaccinated calf b, experiment I), c) score 2 (non-vaccinated, BRSV-infected calf l, experiment I) and d) score 3 (non-vaccinated, BRSV infected calf D5, experiment II). Lesions are examples of neutrophil scores and not representative for the overall inflammation regarding exudate and consolidation.
In experiment I, the neutrophil score in alveolar septa ranged from 1–3 (median 2) in BRSV-infected, susceptible_I calves and between 0–1 (median 1) in vaccinated calves. Overall, there was a significant correlation between severity of clinical signs, pathology and viral load and the presence of neutrophils in alveolar septa (Spearmans rho 0.71, p = 0.02, for extent of lung lesions; 0.63, p = 0.05 for clinical score; and 0.87, p = 0.001 for virus shedding). Two of these correlations were also statistically significant within just the group of five BRSV-infected, susceptible_I animals, i.e. those for lung lesions (Pearson’s r: 0.89, p = 0.04) and virus shedding (Pearson’s r: 0.93, p = 0.02), but not for clinical score (Pearson’s r: 0.72, p = 0.17).
To confirm these data and to validate the mass spectrometric results for neutrophil-related proteins, the relative expression level of selected proteins detected by LC-MS/MS in BAL was analysed in relation to the degree of BRSV-induced gross pathology, clinical score, virus shedding and degree of neutrophils in alveolar septa. A significant positive correlation between these parameters and the relative expression level of elastase, myeloperoxidase, resisitin and S100-A9 was observed (Table 3).
Table 3. Correlations (Spearman’s rho) between relative expression level of selected proteins in bronchoalveolar lavage from calves six days post BRSV infection, BRSV-induced disease and presence of neutrophils in alveolar septa.
| Protein | Lung lesion exenta,b | Clinical scorea,c | Virus sheddinga,d | Neutrophils in septae |
|---|---|---|---|---|
| Elastase | 0.78, p = 0.007 | 0.77, p = 0.009 | 0.80, p = 0.006 | 0.70, p = 0.025 |
| Myeloperoxidase | 0.69, p = 0.028 | 0.67, p = 0.036 | 0.79, p = 0.006 | 0.80, p = 0.005 |
| Resistin | 0.78, p = 0.008 | 0.85, p = 0.002 | 0.88, p = 0.001 | 0.89, p = 0.001 |
| S100-A9 | 0.75, p = 0.013 | 0.85, p = 0.002 | 0.88, p = 0.001 | 0.83, p = 0.003 |
| Superoxide dismutase | -0.70, p = 0.024 | -0.79, p = 0.007 | -0.82, p = 0.004 | -0.78, p = 0.009 |
| Gluthathione peroxidase | n.s. | -0.64, p = 0.048 | -0.74, p = 0.014 | -0.73, p = 0.018 |
| Peroxiredoxin-6 | n.s. | -0.69, p = 0.027 | -0.74, p = 0.015 | n.s. |
| Leucine Zipper TFL1 | n.s. | -0.76, p = 0.010 | -0.71, p = 0.021 | n.s. |
aProteins were identified and semi-quantified by LC-MS/MS including label free analysis, in bronchoalveolar lavage (BAL) from non-vaccinated, BRSV-infected calves with clinical signs of disease and high levels of virus shedding (n = 5) and in BRSV-ISCOM-vaccinated, BRSV-infected animals with no or little clinical signs of disease and no or low levels of virus shedding (n = 5).
bCorrelation of protein with extent of pathologic lung lesions at the day of BAL collection, post infection day (PID) 6
cCorrelation of protein with sum of clinical scores PID 6
dCorrelation of protein with quantity of BRSV-RNA detected in nasal swabs PID 6
eCorrelation of protein with extent of neutrophils in alveolar septa on one histological section from the right cranial lobe (from areas with macroscopic consolidation, if present) on PID 6. The presence of neutrophils was graded between 0 and 3.
n.s; not satistically significant (p>0.05)
Resistin and S100-A9 correlated best with BRSV-induced clinical signs, virus shedding and neutrophils, whereas elastase and resistin correlated best with lung lesions (Table 3). In contrast, a significant negative correlation was observed between the disease parameters and several antioxidant proteins or the leucine zipper transcription factor-like protein 1 (Table 3). A significant negative correlation was also observed between the relative expression level of each of the antioxidant proteins and each of the neutrophil-related proteins listed in Table 3 (p<0.05 for all comparisons, Spearman’s rho, supplemental data S4 Table). In summary, high levels of resistin, S100-A9, and elastase and low levels of antioxidant proteins and leucine zipper transcription factor-like protein 1 correlated with the severity of BRSV disease.
The inflammation was characterised by mild to severe broncho-interstitial or interstitial pneumonia, typical of BRSV, in all BRSV-infected calves. Despite negative bacterial cultures, DNA from Pasteurella multocida was detected in BAL from 5 out of 8 tested calves in experiment I (calf a (ct-value 33), k (ct-value 26), l (ct-value 35), n (ct-value 40) and o (ct-value 29)). DNA from Pasteurella multocida was not detected in BAL from any of the calves in experiment II (D1-D5, E1-E5). Likewise, DNA from Histophilus somni, Mannheimia haemolytica or Mycoplasma bovis could not be detected by PCR in BAL from any animal.
Discussion
This is the first study that describes bronchoalveolar proteome profiles induced by BRSV in calves, which is considered as a very pertinent model of HRSV in humans. The absence of detectable DNA from bacterial opportunistic pathogens in the animals with the most severe clinical signs of disease and extensive lung lesions highlights the direct pathogenicity of BRSV. The data suggested that neutrophil responses are dominant during primary BRSV infections at the peak of clinical signs, at least at the protein level (detectable by LC-MS/MS) in the lumen of the respiratory tract. Although we cannot exclude that these responses were enhanced by the presence of low numbers of Pasteurella multocida in some of the calves in the mildest challenge experiment (calf experiment I), the protein pattern was very similar in the more severely ill calves included in experiment II, in the absence of this bacterium. Many of the proteins associated with BRSV-disease were thus related to neutrophils, such as those in azurophilic granules (e.g. azurocidin, cathepsin G, elastase, myeloperoxidase, and proteinase-3), the neutrophil-specific surface receptor CD177, major internal neutrophil proteins, S100-A8/A9, antimicrobial proteins (e.g. lactotransferrin, defensins and cathelicidins), histones and neutrophil chemoattractants (e.g. S100-A9, hematopoietic cell-specific Lyn-substrate 1 and resisitin) [16, 18–21, 31]. Nevertheless, some of these proteins might also have been derived from epithelial cells or macrophages (e.g. beta defensins, cathelicidins and histones [16, 22]), or monocytes (e.g. S100-A9 [31]). S100-A8 and S100-A9 were similarly two of the major proteins detected by proteomics in nasal secretions of HRSV-infected children, admitted for routine HRSV diagnosis [6].
In this study, other BRSV-disease associated proteins probably originated from epithelial cells (folate receptor alpha [23]), possibly due to sloughing of such cells into the respiratory lumen, or from systemic acute phase responses (serum amyloid A, haptoglobin) similarly detected by proteomic analyses of lung tissue from HRSV-infected rats and by ELISA in BRSV-infected calves [5, 32]. We additionally detected proteins related to gluconeogenesis possibly enhanced by a catabolic state generated by cell destruction (L-serine dehydratase/L-threonine deaminase), and to lymphocytes and natural killer cell activation and chemotaxis (hematopoietic cell-specific Lyn-substrate, lymphocyte-specific protein 1 [24, 25]). However, overall, compared to the neutrophil-related proteins, these proteins were less numerous, at least at days 6 to 7 after infection.
Neutrophils probably contribute to the clearance of BRSV infections, by removing virus-infected cells [33], but when present in excessive numbers, they appear to have negative effects [2]. In this study, we demonstrated a positive correlation between the number of neutrophils in alveolar septa or the relative expression level of neutrophil-related proteins and gross pathology, clinical signs of respiratory disease and virus shedding. This is in agreement with recent studies that showed a link between disease severity in HRSV-infected children and BAL neutrophil counts as well as interleukin (IL)-8 and IL-17, which, directly or indirectly, are neutrophil chemoattractants [34, 35]. Moreover, it has been suggested that the causal link between severe HRSV infection at an early age and recurrent wheeze [36] and the increased risk of developing asthma and atopy later in life in these individuals [37, 38] can be explained by a genetic predisposition for neutrophil recruitment and activity, through increased IL-8 and decreased IL-10 responses to infection [2, 39, 40]. Furthermore, the clinical signs of BRSV-infection in cattle, which start on PID 3–5, and peak on PID 6–9 [7, 10, 33, 41], are associated with a pulmonary neutrophil response, which peaks on PID 5–6 in the exudate of alveolar lumina [42] and constitutes the predominant cell type in BAL of non-vaccinated, BRSV-infected calves, on PID 6–8 [7, 33, 43].
The detrimental effect of neutrophils is partly due to their contribution to the physical obstruction of small airways that is central in RSV pathogenesis [33, 42, 44]. In susceptible animals with high levels of BRSV replication, we confirmed the presence of proteins associated with NETs, which agrees with previous findings [30]. It is known that the RSV fusion protein induces NETs through activation of toll like receptor 4, or immunoglobulin-complex activation of Fcγ receptors [45] and that these filamentous DNA containing structures can trap and kill microbes, but additionally adversely increase the viscosity of mucus [46]. Herein, NET-associated proteins were not detected by immunoblotting in non-infected animals, nor in vaccinated calves despite high levels of BRSV-specific IgA in BAL, which potentially induces NET formation through FcalphaRI triggering[9, 47]. However, such triggering requires antigen-antibody complex formation and vaccinated animals had no or low levels of detectable virus in BAL. Interleukins were not identified in any calves, probably because of the low concentration of these proteins in the post mortem BALs. We have previously demonstrated significantly higher concentrations of interferon gamma by ELISA in BAL, concentrated 20 x, from BRSV-infected calves compared to that in controls [7], but did not have enough material to continue these investigations in the present study.
As well as contributing to airway plugging, neutrophils contribute to lung remodeling. The neutrophil-derived serine protease elastase, which in this study most significantly correlated with pathology, causes elastine fiber degradation, induces epithelial to mesenchymal transition (EMT), goblet cell metaplasia and mucus production [48–50]. Elastine fibers are major components of pulmonary extracellular matrix, are essential for lung elasticity and are very difficult to repair [48]. During EMT, pneumocytes are transformed into migratory fibroblast-like cells, which following permanent stimuli can result in lung fibrosis [51]. Herein, BRSV disease correlated negatively with LZTFL1, which inhibits the EMT process [52]. Lung remodeling might thus contribute to the long term clinical consequences sometimes observed after RSV-infections, including clinical signs several weeks after viral clearance (personal observations) and reduced growth [53]. Mucins produced by goblet cells were detected in both experiments, in all groups, but were not significantly associated with disease. However, BAL of several susceptible calves contained large aggregates of mucus that were trapped in the gauze when the BAL was filtered.
On the other hand, numerous histones were associated with disease. These proteins probably derived from NETs, or were released to the extracellular space during RSV-induced cellular apoptosis, or cellular senescence induced by oxidative stress [28, 33, 54]. The oxidative stress in the lungs of animals with BRSV disease was probably high, since antioxidant enzymes appeared either consumed or downregulated, although we cannot exclude the possibility that these proteins were competed out in the LC-MS/MS analysis. Such competition could explain that fewer proteins were detected in animals with BRSV disease. Nevertheless, neutrophils are the most potent producers of reactive oxygen species in the lung and there was a negative correlation between antioxidant enzymes and neutrophil-related proteins. A consumption of antioxidants was previously observed in severe chronic obstructive pulmonary disease (COPD), in which neutrophils similarly play a key role and, against which, antioxidant/redox-modulating therapies are beneficial [55, 56]. Superoxide dismutase (SOD)-1, which correlated negatively with all disease parameters, is produced by a multitude of cells, including hepatocytes and T lymphocytes, and is involved in T cell receptor activation [57].
In conclusion, the results obtained in this paper have identified new pathways to target in order to reduce the excessive pulmonary inflammation in RSV-infected calves and man. Our findings suggest that treatments that mitigate neutrophilic responses or have antioxidant properties could improve RSV-induced clinical signs and pathology, in both the short and long terms. High similarities in RSV pathogenesis make the calf model an important complement in HRSV research.
Supporting information
Scoring system for respiratory signs of disease in calves.
(DOCX)
Names of proteins identified by LC-MS/MS in bronchoalveolar lavage as described in Fig 1.
(XLSX)
(XLSX)
(TXT)
Acknowledgments
We thank the technical staff at Swedish University of Agricultural Science (SLU) and the National Veterinary Institute, Sweden, for maintaining the animals and preparation of histological samples. A. Rikberg, A. Svensson and K. Selin-Wretling at SLU are acknowledged for logistic support and K. Hörneus, A. Falk and G. Shevchenko at the Science for Life Laboratory, Mass Spectrometry Based Proteomics Facility, Uppsala, Sweden, for technical assistance. We also wish to thank M. Andersson Franko, SLU, for his feedback on statistical analyses, and Pr. L. E. Larsen, DTU, Denmark, for generously sharing the BRSV isolate no. 9402022.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This project received funding from the European Union’s Horizon 2020 program for research, technological development and demonstration under the Grant Agreement no633184 (SAPHIR, https://ec.europa.eu/ This publication reflects the views only of the author, and not the European Commission (EC). The EC is not liable for any use that may be made of the information contained herein); the Swedish Research Council (Formas, Sweden), L’Agence Nationale de la Recherche (France) and the Biotechnology and Biological Sciences Research Council Institute Strategic Programme on Livestock Viral Diseases at The Pirbright Institute (United Kingdom), where GT is a Jenner Investigator, through the Emerging and Major Infectious Diseases of Livestock (EMIDA) project in the European Research Area Network (ERA-NET), grant FP#87, https://www.anihwa.eu/; Formas grant 2016-01770_3 http://www.formas.se/; the Swedish Farmers’ Foundation for Agricultural Research, grant H0750358 http://www.lantbruksforskning.se/ and Carl Trygger grant CST 15:57 http://www.carltryggersstiftelse.se/. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Valarcher JF and Taylor G. Bovine respiratory syncytial virus infection. Vet Res 2007; 38: 153–80. doi: 10.1051/vetres:2006053 [DOI] [PubMed] [Google Scholar]
- 2.Geerdink RJ, Pillay J, Meyaard L, Bont L. Neutrophils in respiratory syncytial virus infection: A target for asthma prevention. J Allergy Clin Immunol 2015; 136: 838–47. doi: 10.1016/j.jaci.2015.06.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dapat C and Oshitani H. Novel insights into human respiratory syncytial virus-host factor interactions through integrated proteomics and transcriptomics analysis. Expert Rev Anti Infect Ther 2016; 14: 285–97. doi: 10.1586/14787210.2016.1141676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Diepen A, Brand HK, de Waal L, Bijl M, Jong VL, Kuiken T, et al. Host proteome correlates of vaccine-mediated enhanced disease in a mouse model of respiratory syncytial virus infection. J Virol 2015; 89: 5022–31. doi: 10.1128/JVI.03630-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang XF, Zhang XY, Gao X, Liu XX, Wang YH. Proteomic Profiling of a Respiratory Syncytial Virus-Infected Rat Pneumonia Model. Jpn J Infect Dis 2016; 69: 285–92. doi: 10.7883/yoken.JJID.2015.244 [DOI] [PubMed] [Google Scholar]
- 6.Fornander L, Ghafouri B, Kihlstrom E, Akerlind B, Schon T, Tagesson C, et al. Innate immunity proteins and a new truncated form of SPLUNC1 in nasopharyngeal aspirates from infants with respiratory syncytial virus infection. Proteomics Clin Appl 2011; 5: 513–22. doi: 10.1002/prca.201100016 [DOI] [PubMed] [Google Scholar]
- 7.Blodörn K, Hägglund S, Gavier-Widen D, Eleouet JF, Riffault S, Pringle J, et al. A bovine respiratory syncytial virus model with high clinical expression in calves with specific passive immunity. BMC Vet Res 2015; 11: 76 doi: 10.1186/s12917-015-0389-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yates JR, Ruse CI, Nakorchevsky A. Proteomics by mass spectrometry: approaches, advances, and applications. Annu Rev Biomed Eng 2009; 11: 49–79. doi: 10.1146/annurev-bioeng-061008-124934 [DOI] [PubMed] [Google Scholar]
- 9.Hägglund S, Hu K, Vargmar K, Pore L, Olofson AS, Blodorn K, et al. Bovine respiratory syncytial virus ISCOMs-Immunity, protection and safety in young conventional calves. Vaccine 2011; 29: 8719–30. doi: 10.1016/j.vaccine.2011.07.146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blodörn K, Hägglund S, Fix J, Dubuquoy C, Makabi-Panzu B, Thom M, et al. Vaccine safety and efficacy evaluation of a recombinant bovine respiratory syncytial virus (BRSV) with deletion of the SH gene and subunit vaccines based on recombinant human RSV proteins: N-nanorings, P and M2-1, in calves with maternal antibodies. PLoS One 2014; 9: e100392 doi: 10.1371/journal.pone.0100392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hägglund S, Hu K, Blodörn K, Makabi-Panzu B, Gaillard AL, Ellencrona K, et al. Characterization of an experimental vaccine for bovine respiratory syncytial virus. Clin Vaccine Immunol 2014; 21: 997–1004. doi: 10.1128/CVI.00162-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hörnaeus K, Guillemant J, Mi J, Hernroth B, Bergquist J, Lind SB. Mass spectrometry data from a quantitative analysis of protein expression in gills of immuno-challenged blue mussels (Mytilus edulis). Data Brief 2016; 8: 470–3. doi: 10.1016/j.dib.2016.05.073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fabregat A, Sidiropoulos K, Garapati P, Gillespie M, Hausmann K, Haw R, et al. The Reactome pathway Knowledgebase. Nucleic Acids Res 2016; 44: D481–7. doi: 10.1093/nar/gkv1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heberle H, Meirelles GV, da Silva FR, Telles GP, Minghim R. InteractiVenn: a web-based tool for the analysis of sets through Venn diagrams. BMC Bioinformatics 2015; 16: 169 doi: 10.1186/s12859-015-0611-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mi H, Muruganujan A, Thomas PD. PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res 2013; 41: D377–86. doi: 10.1093/nar/gks1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meade KG, Cormican P, Narciandi F, Lloyd A, O'Farrelly C. Bovine beta-defensin gene family: opportunities to improve animal health? Physiol Genomics 2014; 46: 17–28. doi: 10.1152/physiolgenomics.00085.2013 [DOI] [PubMed] [Google Scholar]
- 17.Zawrotniak M and Rapala-Kozik M. Neutrophil extracellular traps (NETs)—formation and implications. Acta Biochim Pol 2013; 60: 277–84. [PubMed] [Google Scholar]
- 18.Kessel C, Holzinger D, Foell D. Phagocyte-derived S100 proteins in autoinflammation: putative role in pathogenesis and usefulness as biomarkers. Clin Immunol 2013; 147: 229–41. doi: 10.1016/j.clim.2012.11.008 [DOI] [PubMed] [Google Scholar]
- 19.Jiang S, Park DW, Tadie JM, Gregoire M, Deshane J, Pittet JF, et al. Human resistin promotes neutrophil proinflammatory activation and neutrophil extracellular trap formation and increases severity of acute lung injury. J Immunol 2014; 192: 4795–803. doi: 10.4049/jimmunol.1302764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wright HL, Moots RJ, Bucknall RC, Edwards SW. Neutrophil function in inflammation and inflammatory diseases. Rheumatology (Oxford) 2010; 49: 1618–31. [DOI] [PubMed] [Google Scholar]
- 21.Cavnar PJ, Mogen K, Berthier E, Beebe DJ, Huttenlocher A. The actin regulatory protein HS1 interacts with Arp2/3 and mediates efficient neutrophil chemotaxis. J Biol Chem 2012; 287: 25466–77. doi: 10.1074/jbc.M112.364562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Telcian AG, Zdrenghea MT, Edwards MR, Laza-Stanca V, Mallia P, Johnston SL, et al. Vitamin D increases the antiviral activity of bronchial epithelial cells in vitro. Antiviral Res 2017; 137: 93–101. doi: 10.1016/j.antiviral.2016.11.004 [DOI] [PubMed] [Google Scholar]
- 23.Yi YS. Folate Receptor-Targeted Diagnostics and Therapeutics for Inflammatory Diseases. Immune Netw 2016; 16: 337–343. doi: 10.4110/in.2016.16.6.337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pulford K, Jones M, Banham AH, Haralambieva E, Mason DY. Lymphocyte-specific protein 1: a specific marker of human leucocytes. Immunology 1999; 96: 262–71. doi: 10.1046/j.1365-2567.1999.00677.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mukherjee S, Kim J, Mooren OL, Shahan ST, Cohan M, Cooper JA. Role of cortactin homolog HS1 in transendothelial migration of natural killer cells. PLoS One 2015; 10: e0118153 doi: 10.1371/journal.pone.0118153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Merriman KE, Kweh MF, Powell JL, Lippolis JD, Nelson CD. Multiple beta-defensin genes are upregulated by the vitamin D pathway in cattle. J Steroid Biochem Mol Biol 2015; 154: 120–9. doi: 10.1016/j.jsbmb.2015.08.002 [DOI] [PubMed] [Google Scholar]
- 27.Angen O, Thomsen J, Larsen LE, Larsen J, Kokotovic B, Heegaard PM, et al. Respiratory disease in calves: microbiological investigations on trans-tracheally aspirated bronchoalveolar fluid and acute phase protein response. Vet Microbiol 2009; 137: 165–71. doi: 10.1016/j.vetmic.2008.12.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen R, Kang R, Fan XG, Tang D. Release and activity of histone in diseases. Cell Death Dis 2014; 5: e1370 doi: 10.1038/cddis.2014.337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ledgard AM, Smolenski GA, Henderson H, Lee RS. Influence of pathogenic bacteria species present in the postpartum bovine uterus on proteome profiles. Reprod Fertil Dev 2015; 27: 395–406. doi: 10.1071/RD13144 [DOI] [PubMed] [Google Scholar]
- 30.Cortjens B, de Boer OJ, de Jong R, Antonis AF, Sabogal Pineros YS, Lutter R, et al. Neutrophil extracellular traps cause airway obstruction during respiratory syncytial virus disease. J Pathol 2016; 238: 401–11. doi: 10.1002/path.4660 [DOI] [PubMed] [Google Scholar]
- 31.Foell D and Roth J. Proinflammatory S100 proteins in arthritis and autoimmune disease. Arthritis Rheum 2004; 50: 3762–71. doi: 10.1002/art.20631 [DOI] [PubMed] [Google Scholar]
- 32.Heegaard PM, Godson DL, Toussaint MJ, Tjornehoj K, Larsen LE, Viuff B, et al. The acute phase response of haptoglobin and serum amyloid A (SAA) in cattle undergoing experimental infection with bovine respiratory syncytial virus. Vet Immunol Immunopathol 2000; 77: 151–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Viuff B, Tjornehoj K, Larsen LE, Rontved CM, Uttenthal A, Ronsholt L, et al. Replication and clearance of respiratory syncytial virus: apoptosis is an important pathway of virus clearance after experimental infection with bovine respiratory syncytial virus. Am J Pathology 2002; 161: 2195–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stoppelenburg AJ, de Roock S, Hennus MP, Bont L, Boes M. Elevated Th17 response in infants undergoing respiratory viral infection. Am J Pathol 2014; 184: 1274–9. doi: 10.1016/j.ajpath.2014.01.033 [DOI] [PubMed] [Google Scholar]
- 35.Stoppelenburg AJ, Salimi V, Hennus M, Plantinga M, Huis in 't Veld R, Walk J, et al. Local IL-17A potentiates early neutrophil recruitment to the respiratory tract during severe RSV infection. PLoS One 2013; 8: e78461 doi: 10.1371/journal.pone.0078461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoshihara S, Kusuda S, Mochizuki H, Okada K, Nishima S, Simoes EA, et al. Effect of palivizumab prophylaxis on subsequent recurrent wheezing in preterm infants. Pediatrics 2013; 132: 811–8. doi: 10.1542/peds.2013-0982 [DOI] [PubMed] [Google Scholar]
- 37.Wenzel SE, Gibbs RL, Lehr MV, Simoes EA. Respiratory outcomes in high-risk children 7 to 10 years after prophylaxis with respiratory syncytial virus immune globulin. Am J Med 2002; 112: 627–33. [DOI] [PubMed] [Google Scholar]
- 38.Garcia-Garcia ML, Calvo Rey C, Del Rosal Rabes T. Pediatric Asthma and Viral Infection. Arch Bronconeumol 2016; 52: 269–73. doi: 10.1016/j.arbres.2015.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hull J, Thomson A, Kwiatkowski D. Association of respiratory syncytial virus bronchiolitis with the interleukin 8 gene region in UK families. Thorax 2000; 55: 1023–7. doi: 10.1136/thorax.55.12.1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Korppi M, Nuolivirta K, Lauhkonen E, Holster A, Terasjarvi J, Vuononvirta J, et al. IL-10 gene polymorphism is associated with preschool atopy and early-life recurrent wheezing after bronchiolitis in infancy. Pediatr Pulmonol 2016. [DOI] [PubMed] [Google Scholar]
- 41.Hägglund S, Hu KF, Larsen LE, Hakhverdyan M, Valarcher JF, Morein B, et al. Bovine respiratory syncytial virus ISCOMs—protection in the presence of maternal antibodies. Vaccine 2004; 23: 646–655. doi: 10.1016/j.vaccine.2004.07.006 [DOI] [PubMed] [Google Scholar]
- 42.Bryson DG, McConnell S, McAliskey M, McNulty MS. Ultrastructural features of alveolar lesions in induced respiratory syncytial virus pneumonia of calves. Vet Pathol 1991; 28: 286–92. doi: 10.1177/030098589102800404 [DOI] [PubMed] [Google Scholar]
- 43.Taylor G, Thomas LH, Stott EJ. Effect of vaccination on cell populations in lung washes from calves after infection with respiratory syncytial virus. Res Vet Sci 1989; 47: 231–5. [PubMed] [Google Scholar]
- 44.Johnson JE, Gonzales RA, Olson SJ, Wright PF, Graham BS. The histopathology of fatal untreated human respiratory syncytial virus infection. Mod Pathol 2007; 20: 108–19. doi: 10.1038/modpathol.3800725 [DOI] [PubMed] [Google Scholar]
- 45.Funchal GA, Jaeger N, Czepielewski RS, Machado MS, Muraro SP, Stein RT, et al. Respiratory syncytial virus fusion protein promotes TLR-4-dependent neutrophil extracellular trap formation by human neutrophils. PLoS One 2015; 10: e0124082 doi: 10.1371/journal.pone.0124082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheng OZ and Palaniyar N. NET balancing: a problem in inflammatory lung diseases. Front Immunol 2013; 4: 1 doi: 10.3389/fimmu.2013.00001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aleyd E, van Hout MW, Ganzevles SH, Hoeben KA, Everts V, Bakema JE, et al. IgA enhances NETosis and release of neutrophil extracellular traps by polymorphonuclear cells via Fcalpha receptor I. J Immunol 2014; 192: 2374–83. doi: 10.4049/jimmunol.1300261 [DOI] [PubMed] [Google Scholar]
- 48.Shifren A and Mecham RP. The stumbling block in lung repair of emphysema: elastic fiber assembly. Proc Am Thorac Soc 2006; 3: 428–33. doi: 10.1513/pats.200601-009AW [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mayer C, Darb-Esfahani S, Meyer AS, Hubner K, Rom J, Sohn C, et al. Neutrophil Granulocytes in Ovarian Cancer—Induction of Epithelial-To-Mesenchymal-Transition and Tumor Cell Migration. J Cancer 2016; 7: 546–54. doi: 10.7150/jca.14169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wagner CJ, Schultz C, Mall MA. Neutrophil elastase and matrix metalloproteinase 12 in cystic fibrosis lung disease. Mol Cell Pediatr 2016; 3: 25 doi: 10.1186/s40348-016-0053-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rogel MR, Soni PN, Troken JR, Sitikov A, Trejo HE, Ridge KM. Vimentin is sufficient and required for wound repair and remodeling in alveolar epithelial cells. FASEB J 2011; 25: 3873–83. doi: 10.1096/fj.10-170795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wei Q, Chen ZH, Wang L, Zhang T, Duan L, Behrens C, et al. LZTFL1 suppresses lung tumorigenesis by maintaining differentiation of lung epithelial cells. Oncogene 2016; 35: 2655–63. doi: 10.1038/onc.2015.328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Griffin D. The monster we don't see: subclinical BRD in beef cattle. Anim Health Res Rev 2014; 15: 138–41. doi: 10.1017/S1466252314000255 [DOI] [PubMed] [Google Scholar]
- 54.Campisi J. Cellular Senescence and Lung Function during Aging. Yin and Yang. Ann Am Thorac Soc 2016; 13: S402–S406. doi: 10.1513/AnnalsATS.201609-703AW [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rahman I and Kinnula VL. Strategies to decrease ongoing oxidant burden in chronic obstructive pulmonary disease. Expert Rev Clin Pharmacol 2012; 5: 293–309. doi: 10.1586/ecp.12.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Angelis N, Porpodis K, Zarogoulidis P, Spyratos D, Kioumis I, Papaiwannou A, et al. Airway inflammation in chronic obstructive pulmonary disease. J Thorac Dis 2014; 6 Suppl 1: S167–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Terrazzano G, Rubino V, Damiano S, Sasso A, Petrozziello T, Ucci V, et al. T cell activation induces CuZn superoxide dismutase (SOD)-1 intracellular re-localization, production and secretion. Biochim Biophys Acta 2014; 1843: 265–74. doi: 10.1016/j.bbamcr.2013.10.020 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Scoring system for respiratory signs of disease in calves.
(DOCX)
Names of proteins identified by LC-MS/MS in bronchoalveolar lavage as described in Fig 1.
(XLSX)
(XLSX)
(TXT)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.