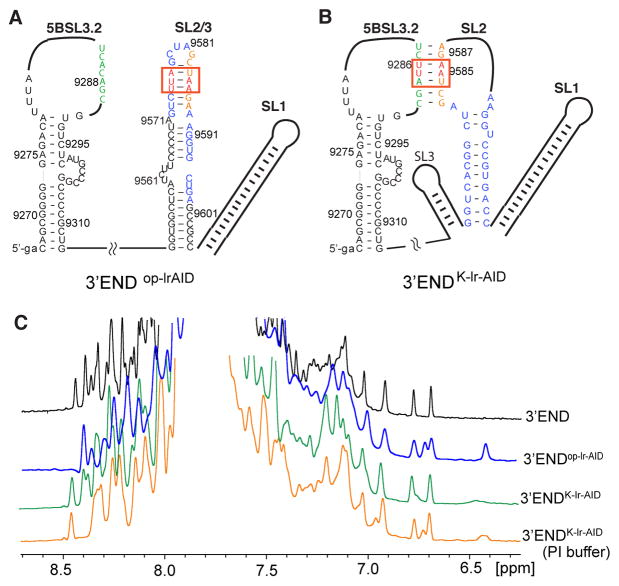
Figure 5.
Structural probing of the 3′-end RNA by the lr-AID NMR approach. (A) Proposed secondary structure of the 3′-end open conformation in which 3′X folds into the 2SL conformation. The lr-AID cassette was engineered into the top SL2/3 stem in 3′-endop-lrAID. (B) Proposed secondary structure of the kissing-loop conformation of the 3′-end. The lr-AID cassette was introduced into the kissing-loop base pairs of 3′-endK-lrAID to probe the long-range interaction. Lowercase letters denote non-native nucleotides added to the RNA constructs to promote in vitro RNA synthesis by T7 polymerase. (C) One-dimensional NOESY spectra of 3′-end (black), 3′-endop-lrAID (blue), 3′-endK-lrAID in low-ionic strength buffer (green), and 3′-endK-lrAID in PI buffer (orange).