Abstract
Background
A recent analysis of 25 historical candidate gene polymorphisms for schizophrenia in the largest genome-wide association study (GWAS) conducted to date suggested that these commonly studied variants were no more associated with the disorder than would be expected by chance. However, the same study identified other variants within those candidate genes that demonstrated genome-wide significant associations with schizophrenia. As such, it is possible that variants within historic schizophrenia candidate genes are associated with schizophrenia at levels above those expected by chance, even if the most studied specific polymorphisms are not.
Methods
The present study used association statistics from the largest schizophrenia GWAS conducted to date as input to a gene set analysis to investigate whether variants within schizophrenia candidate genes are enriched for association with schizophrenia.
Results
As a group, variants in the most studied candidate genes were no more associated with schizophrenia than variants in control sets of non-candidate genes. While a small subset of candidate genes did appear to be significantly associated with schizophrenia, these genes were not particularly noteworthy given the large number of more strongly associated non-candidate genes.
Conclusions
The history of schizophrenia research should serve as a cautionary tale to candidate gene investigators examining other phenotypes: our findings indicate that the most investigated candidate gene hypotheses of schizophrenia are not well supported by GWAS, and it is likely that this will be the case for other complex traits as well.
Keywords: schizophrenia, candidate genes, single nucleotide polymorphism (SNP), genome-wide association study (GWAS), gene set analysis, complex traits
INTRODUCTION
Schizophrenia is highly heritable(1), and since the 1960s, candidate gene studies have played a major role in research dedicated to understanding the genetic etiology of schizophrenia.(2, www.szgene.org) Most historical candidate genes were selected based on known drug treatment targets and corresponding neurobiological pathways.(3) As family-based genetics studies began to reveal regions of the genome that appeared to be associated with psychiatric disorders, researchers began to consider additional candidate genes located in chromosomal regions suggested by linkage analyses (e.g. NOTCH4 (4)). Within genes chosen in this manner, candidate gene analyses typically focused on specific variants in regions of the genome thought likely to be functional.
The SZGene database(2) (www.szgene.org), a curated catalog of findings from genetic association studies for schizophrenia, comprised of all studies published in a peer-reviewed English language journal from 1965 through 2012, lists over 1,500 published studies for schizophrenia, the majority of which were candidate gene studies. However, few clear results have emerged from these studies, with many studies reporting contradictory results for the same candidate gene polymorphisms. Factors that may underlie this inconsistency include lack of statistical power, different genetic or environmental backgrounds across studies, incomplete coverage of relevant genetic variation within candidate genes, and false positives arising from, e.g., publication bias (5,6). With the advent of genome-wide association studies (GWAS), investigators can now assess the vast majority of common genetic variation across the entire genome, enabling hypothesis-free exploration of the associations between common genetic variants and schizophrenia or other complex disorders. Due to sample sizes that are two to three orders of magnitude larger than most candidate gene studies, adherence to analytic procedures shared in common across the field, and conservative significance thresholds, associations discovered by GWAS have proven to be more robust, replicable, and reflective of the true effect sizes of common genetic variants than those based on candidate gene reports (7). In addition, the agnostic approach of GWAS mitigates incentives (i.e., findings are reported for all loci regardless of statistical significance) to selectively report results from just certain genes or polymorphisms of interest. Moreover, modern large-scale GWAS have ample statistical power to detect effect sizes typically reported in candidate gene studies. For example, the recent PGC GWAS (12) had > 99% power to detect genome-wide significant (α = 5e-08) associations that explain a mere four-one hundredths of one percent of the variation in schizophrenia liability, an effect size much smaller than any discovery reported in candidate gene studies of schizophrenia. For these reasons, GWAS results can be used to determine the plausibility of previously reported findings on common candidate gene polymorphisms.
Two reports in the past five years have compared GWAS and candidate gene study results of schizophrenia. In 2012, Collins et al.(8) employed a pathway analysis approach to test for enrichment of lower p-values for all (732) schizophrenia candidate genes identified by the SZGene database in the International Schizophrenia Consortium(9) (ISC) GWAS data (N = 6,909). They found no evidence for p-value enrichment in this set of 732 genes after correction for multiple testing. They also calculated a polygenic risk score (PRS) based on the SNPs located in the 732 candidate genes they examined, but did not see differences between cases and controls in an independent target sample (SCZ GAIN(10), N = 2,366).
Using the existing published results in the SZGene database, Farrell et al. (11) meta-analyzed the 25 most-studied schizophrenia candidate gene polymorphisms and found that none approached genome-wide significance (p < 5e-8) in the PGC (12) schizophrenia GWAS study (34,241 cases and 45,604 controls). Moreover, the odds ratios of the significantly associated loci in the PGC study (~1.10) imply that almost all previous candidate gene studies examining genetic associations with schizophrenia diagnosis (the largest of which had a sample size some 16 times smaller than the PGC dataset) have been severely underpowered to detect any true association, much less potential associations at specific candidate polymorphisms. Though four of the most studied candidate genes (DRD2, GRM3, NOTCH4, TNF) had genome-wide significant polymorphisms within 25 kb of their boundaries in the PGC study, only one of these associations (rs1800629 in TNF) was in linkage disequilibrium (LD) with the previously studied candidate polymorphism.
Still, the fact that four of the top 25 schizophrenia candidate genes contained significant GWAS signals raises the question of whether schizophrenia candidate genes themselves are supported by GWAS results, even if the specific candidate polymorphisms within them have not been. In other words, are SNPs within the most studied schizophrenia candidate genes more strongly associated with schizophrenia than expected by chance? Previous studies have not addressed this question. Farrell et al.(11) focused solely on candidate polymorphisms rather than candidate genes and did not perform a gene set test for enrichment of lower GWAS p-values for all variants within the candidate genes. Collins et al.(8) performed a gene set analysis for all 732 schizophrenia candidate genes identified in the SZGene database, but more than 75% of the genes currently listed in the SZGene database have been studied only once or twice, and most would not be considered “candidate genes” by researchers in the field. The current study used a gene set analysis approach and the latest PGC summary statistics (12) to determine whether polymorphisms within classic schizophrenia candidate genes are more related to schizophrenia risk than polymorphisms within other control sets of genes.
METHODS AND MATERIALS
Schizophrenia Candidate Genes
Our primary analysis focused on the same 25 top candidate genes examined by Farrell et al. (11) in their review (see Table 1). These 25 genes were either featured in previous reviews of schizophrenia research (13–16) or studied more than 20 times according to the SZGene database (2), and include what can be considered the “classic” candidate genes for schizophrenia (COMT, DISC1, DRD3, etc.). To ensure that no effects were missed, in a supplementary analysis we expanded this set to include all genes from SZGene that were (a) studied more than five times and (b) not originally motivated by GWAS. Eighty-six genes met both criteria (see Table S1), approximately 23% of which were motivated by prior linkage results with the remaining motivated by involvement in promising biological pathways or pharmacological hypotheses. The distribution of the number of studies per gene is shown in Supplementary Figure 1.
Table 1. Descriptive statistics of 25 historical candidate genes.
The average numbers of cases and controls were calculated from the SZGene database, excluding GWAS and family-based studies. Gene rankings are based on the genes’ z scores from MAGMA, which quantify each gene’s association with schizophrenia, with rankings calculated across all genes in the genome excluding the MHC region.
| Gene | NCBI ID |
Location | Size (Kb) |
Number of Studies |
Average Number of Cases |
Average Number of Controls |
Most-studied Polymorphism |
Type (SNP, repeat, etc.) |
Association Statistic (Z) from MAGMA |
Rank (excluding MHC) |
|---|---|---|---|---|---|---|---|---|---|---|
| NOTCH4a | 4855 | 6p21.3 | 29.2 | 24 | 347 | 792 | rs367398 | SNP | 8.78 | na |
| DRD2 | 1813 | 11q23 | 65.7 | 67 | 197 | 266 | rs1801028 | SNP | 5.92 | 129 |
| KCNN3b | 3782 | 1q21.3 | 172.8 | 23 | 159 | 154 | 1333T/C | SNP | 5.03 | 301 |
| GRM3 | 2913 | 7q21.1 | 221.0 | 15 | 590 | 681 | rs2228595 | SNP | 4.6 | 435 |
| TNFa | 7124 | 6p21.3 | 2.8 | 21 | 164 | 214 | rs1800629 | SNP | 4.28 | na |
| ZDHHC8 | 29801 | 22q11.21 | 16.2 | 9 | 308 | 401 | rs175174 | SNP | 4.11 | 670 |
| PPP3CC | 5533 | 8p21.3 | 100.0 | 9 | 683 | 763 | rs7837713 | SNP | 3.47 | 1,214 |
| BDNFb | 627 | 11p13 | 67.2 | 40 | 236 | 292 | 270C/T | SNP | 3.01 | 1,774 |
| DAO | 1610 | 12q24 | 20.9 | 10 | 440 | 542 | rs3918346 | SNP | 1.87 | 4,291 |
| SLC6A4 | 6532 | 17q11.2 | 39.6 | 32 | 173 | 207 | 5-HTTVNTR | VNTR | 1.64 | 5,101 |
| MTHFR | 4524 | 1p36.3 | 20.4 | 20 | 221 | 290 | rs1801133 | SNP | 1.2 | 6,926 |
| COMT | 1312 | 22q11.21 | 28.2 | 81 | 241 | 383 | rs4680 | SNP | 0.85 | 8,645 |
| RGS4 | 5999 | 1q23.3 | 8.2 | 22 | 401 | 493 | rs2661319 | SNP | 0.42 | 10,909 |
| DRD3 | 1814 | 3q13.3 | 50.3 | 71 | 168 | 198 | rs6280 | SNP | 0.11 | 12,652 |
| AKT1 | 207 | 14q32.32 | 26.4 | 13 | 478 | 539 | rs3730358 | SNP | 0.07 | 12,850 |
| DRD4 | 1815 | 11p15.5 | 3.4 | 45 | 202 | 230 | rs4646983 | Indel | 0.06 | 12,906 |
| NRG1 | 3084 | 8p12 | 1125.7 | 41 | 384 | 487 | rs62510682 | SNP | 0.03 | 13,040 |
| PRODH | 5625 | 22q11.21 | 23.8 | 10 | 235 | 320 | rs383964 | SNP | −0.16 | 14,059 |
| DTNBP1 | 84062 | 6p22.3 | 140.3 | 32 | 400 | 444 | rs3213207 | SNP | −0.3 | 14,745 |
| HTR2A | 3356 | 13q14 | 63.7 | 57 | 215 | 224 | rs6311 | SNP | −0.32 | 14,860 |
| CHRNA7 | 1139 | 15q14 | 139.7 | 12 | 315 | 316 | rs28531779 | SNP | −0.59 | 16,114 |
| DISC1 | 27185 | 1q42.1 | 414.5 | 22 | 348 | 410 | rs999710 | SNP | −0.66 | 16,384 |
| DAOA | 267012 | 13q33.2 | 25.2 | 27 | 406 | 526 | rs3916965 | SNP | −0.83 | 17,012 |
| SLC6A3 | 6531 | 5p15.3 | 52.6 | 22 | 176 | 234 | rs28363170 | VNTR | −0.91 | 17,295 |
| APOE | 348 | 19q13.2 | 3.6 | 32 | 143 | 211 | ε2/3/4 | triallelic | −1.3 | 18,167 |
Denotes genes within the MHC region
The polymorphisms within these genes were mistakenly designated “non-SNPs” in Farrell et al.(11), but they have been correctly labeled as SNP markers here
Choosing Comparison Gene Sets
To compare the overall association of schizophrenia candidate genes to other sets of control genes, we selected genes containing polymorphisms significantly associated with one of two non-psychiatric phenotypes genetically uncorrelated with schizophrenia according to LDHub (17): type 2 diabetes and height. There were a total of 258 height-associated genes (keyword “height” in the database of reported associations from the GWAS Catalog (18)) and 70 type 2 diabetes-related genes (keyword “type 2 diabetes”) that did not overlap with the list of candidate genes. A list of 1028 unique genes related to pre- and post-synapse processes, chosen as a positive control, were downloaded from http://ctg.cncr.nl/software/genesets (originally curated by Ruano et al.(19) and Lips et al.(20))
Psychiatric Genomics Consortium GWAS Data
We downloaded the summary statistics (association p-values for ~9.5 million imputed variants) from the PGC schizophrenia samples. Because the composition of the PGC schizophrenia sample is largely of European ancestry, we chose the 1000 Genomes(21) phase 1 European samples as a reference population to estimate LD between SNPs.
Statistical Analysis
We used the MAGMA software (22) to test whether the top 25 or top 86 schizophrenia candidate genes demonstrated enrichment of lower p-values in the PGC schizophrenia GWAS data. We also used VEGAS2 (23) software to assess consistency of results across methods. Results were highly consistent (see Supplemental Methods and Tables S5 and S6); for clarity, presented results are from MAGMA.
We calculated the overall strength of association for each gene, zi, by summing the −log(p) for all SNP p-values within each gene boundary. The distribution of this sum is unknown but is approximated by a scaled chi-square distribution with scaling and degrees of freedom a function of the squared SNP-SNP correlation matrix, which accounts for LD between SNPs within the gene. A gene-level p-value was derived from this scaled chi-square distribution, which was then converted to a z-value zi = Φ−1(1 − pi) where Φ−1 is the probit function. For an alternative set of analyses (primarily presented in the Supplement, Table S2), we derived zi using the minimum SNP p-value per gene instead of the sum of −log(p).
After calculating the strength of association for each gene in the genome, we grouped the 25 or 86 schizophrenia candidate genes into gene sets and ran one of two gene set tests in MAGMA. In our primary analysis, we used MAGMA’s “competitive” test to assess whether the candidate gene set was more associated with schizophrenia than all other genes not in the gene set, controlling for potentially confounding gene characteristics. To understand whether our set of candidate genes showed stronger or weaker association with schizophrenia than control sets of genes (genes involved in type 2 diabetes, height, or synaptic processes) we used MAGMA’s “relative” test (see Supplemental Methods for additional details of these tests). As recommended by the MAGMA authors (22), we report one-tailed p-values for competitive tests but two-tailed p-values for relative tests.
RESULTS
The importance of candidate genes as a group
We found no evidence that the 25 candidate genes of interest showed enrichment for lower p-values in the PGC GWAS compared to all other genes in the genome. This competitive test was non-significant, regardless of whether we controlled for gene size, SNP density, and minor allele count (β = 0.28, SE = 0.26, p = 0.14) or not (β = 0.34, SE = 0.26, p = 0.09). Because of the strong associations with schizophrenia previously shown to exist within the major histocompatibility complex (MHC) and the long-range LD which makes it difficult to know which genes drive the multiple associations in this region, we also repeated these analyses after removing all MHC genes, including NOTCH4 and TNF; none of our conclusions changed. All results are presented in Table 2. Further, using MAGMA’s relative test, the set of 25 schizophrenia candidate genes did not show a stronger association with schizophrenia than did genes associated with type 2 diabetes (β = 0.39, SE = 0.30, p = 0.19), genes associated with height (β = 0.23, SE = 0.27, p = 0.39), or genes involved in synaptic processes (β = 0.15, SE = 0.26, p = 0.57). No conclusions changed when we repeated the above analyses using strict gene boundaries (Supplementary Table S3) or the minimum SNP p-value in a gene as the gene-level statistic (Supplementary Table S2). Using a resampling approach, we also confirmed that the discrepancy in gene set sizes in these three relative tests (25 schizophrenia candidate genes vs. 258 height, 70 type 2 diabetes, and 1028 synaptic genes respectively) had no influence on our conclusions (see Supplement, pg. 6).
Table 2. MAGMA gene set analyses.
These analyses used the sum of the negative log of the p-values as the gene-level test statistic, defined the genes with extended gene boundaries (including the +/− 25kb regions upstream or downstream of gene start and end points), and controlled for gene size, SNP density, and minor allele count, as well as the log of each.
| Model | Target Gene Set | Comparison Gene Set |
Beta (SE) | P-value |
|---|---|---|---|---|
| 1 | Historical 25 candidate genes | All other genes | 0.28 (0.26) | 0.14 |
| 2 | Historical 25 candidate genes | Height associated genes | 0.23 (0.27) | 0.39 |
| 3 | Historical 25 candidate genes | Type 2 Diabetes associated genes | 0.39 (0.30) | 0.19 |
| 4 | Historical 25 candidate genes | Genes involved in synaptic processes | 0.15 (0.26) | 0.57 |
| 5 | 86 Most-studied candidate genes | All other genes | 0.27 (0.13) | 0.01* |
| 6 | 86 Most-studied candidate genes | Height associated genes | 0.22 (0.15) | 0.14 |
| 7 | 86 Most-studied candidate genes | Type 2 Diabetes associated genes | 0.48 (0.20) | 0.02* |
| 8 | 86 Most-studied candidate genes | Genes involved in synaptic processes | 0.13 (0.13) | 0.32 |
| 9 | Historical 25 candidate genes minus MHC genes | All other genes | 0.18 (0.27) | 0.24 |
| 10 | Historical 25 candidate genes minus MHC genes | Height associated genes | 0.12 (0.28) | 0.66 |
| 11 | Historical 25 candidate genes minus MHC genes | Type 2 Diabetes associated genes | 0.32 (0.31) | 0.30 |
| 12 | Historical 25 candidate genes minus MHC genes | Genes involved in synaptic processes | 0.05 (0.19) | 0.88 |
| 13 | 86 Most-studied candidate genes minus MHC genes | All other genes | 0.25 (0.13) | 0.02* |
| 14 | 86 Most-studied candidate genes minus MHC genes | Height associated genes | 0.19 (0.15) | 0.20 |
| 15 | 86 Most-studied candidate genes minus MHC genes | Type 2 Diabetes associated genes | 0.50 (0.21) | 0.02* |
| 16 | 86 Most-studied candidate genes minus MHC genes | Genes involved in synaptic processes | 0.11 (0.13) | 0.41 |
P-values in bold and starred are significant at α < 0.05. None of the four significant tests would survive multiple testing corrections.
When we expanded our gene set to include the 86 candidate genes that had been studied more than five times according to the SZGene database (not including GWAS results), the gene set was more associated with schizophrenia compared to all other genes (β = 0.27, SE = 0.13, p = 0.01). This larger gene set was more strongly associated with schizophrenia than the set of genes associated with type 2 diabetes (β = 0.48, SE = 0.20, p = 0.02), but no more so than genes associated with height (β = 0.22, SE = 0.15, p = 0.14) or genes involved in synaptic processes (β = 0.13, SE = 0.13, p = 0.32) (Table 2). Results were similar when using strict gene boundaries, except that the relative test comparing to type 2 diabetes-related genes was no longer significant (Table S3). When the same analyses were performed using the minimum p-value as the gene-level statistic, none of the gene set associations were significant except for the test relative to height-related genes when using strict gene boundaries (Table S2). In addition, we repeated these analyses with genes within the MHC removed; none of our conclusions changed (Table 2). Finally, conclusions did not change after excluding the twenty candidate genes motivated by prior linkage studies (Supplementary Table S4).
Contrary to initial expectations, the expanded set of 86 candidate genes yielded more significant associations (13 significant results out of 40 tests conducted) than the set consisting of the 25 most-studied candidate genes (0 of 40). This pattern of results might arise if less-studied candidate genes are more related to schizophrenia, or if the ability to correctly identify relevant candidate genes has increased over time (given that the most-studied candidate genes were typically first investigated longer ago). However, neither hypothesis was supported: there were no significant relationships between candidate genes’ strengths of association with schizophrenia (gene-wise z-values) and either the number of times each was studied (r = −0.06, p = 0.61; Supplementary Figure S2) or the first year each was studied (r = 0.11, p = 0.34; Supplementary Figure S3).
A more likely reason we observed more significant associations with the expanded gene set is differences in statistical power between analyses. Whereas adding genes unassociated with a trait to a gene set is known to decrease power, when each potential member of a gene set contributes a small amount of heritability, on average, to a trait, adding additional genes to the set increases power due in competitive tests due to increased variance explained by the set (24) (note that this is not the case with relative tests because gene set size is explicitly controlled in these tests). We confirmed this increased power with increased gene set size in our own data by permuting different set sizes of genes involved in synaptic processes (which, as a set, were significantly associated with schizophrenia; β = 0.152, SE = 0.04, p = 1.94e−05), and finding a strong, negative relationship between gene set size and average p-value in competitive tests (Supplementary Figure S4). Thus, the results from the 80 gene set analyses we performed are consistent with the hypothesis that schizophrenia candidate genes are weakly related to schizophrenia on average, and that tests involving larger sets were typically more significant only because the larger gene set contained more weakly related genes. This conclusion is supported by the similar βs (which estimate the predicted increase in average z-score per gene for being in the set versus not) for the competitive gene set tests for the top 25 (β25 = 0.28) and the top 86 (β86 = 0.27) candidate genes, with the difference in significance between the two tests being due to their different standard errors (SE25 = 0.26 vs. SE86 = 0.13).
The importance of specific candidate genes
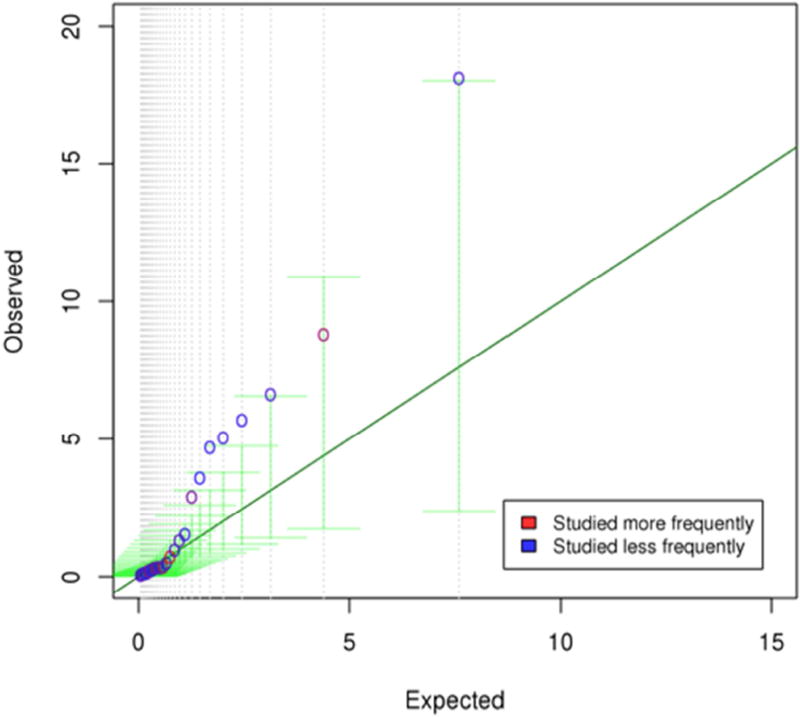
While we found little evidence to support the idea that the candidate genes as a group were more relevant to schizophrenia than control sets of genes, especially when compared to genes involved in synaptic processes, several of the most-studied candidate genes were significantly related to schizophrenia, some of them highly so (Figure 1). In particular, 9 genes in the set of 25 candidates were nominally (p < .05) associated with schizophrenia. To understand how surprising this result is for a highly polygenic trait such as schizophrenia, we permuted sets of 25 genes from the entire genome and observed 9 or more nominally significant genes in 25.2% of permutations (Supplementary Figure S6), suggesting that 9 significant genes out of 25 is not unexpected for a highly polygenic trait like schizophrenia. However, when we performed a relative test in MAGMA of the 9 significant candidate genes versus all other genes significantly (p < .05) related to schizophrenia in the genome, we found evidence that the strength of the associations was greater among these 9 genes than among all other significant genes (β = 0.789, SE = 0.28, p = .005). This result was largely driven by the five most significant candidate genes - NOTCH4, DRD2, KCNN3, GRM3, and TNF. Results were attenuated but remained significant when we dropped MHC genes from both sets (β = 0.738, SE = 0.32, p = .02) and when we compared the 7 significant non-MHC candidate genes to all other significantly related non-MHC synaptic genes (β = 0.896, SE = 0.42, p = .03). Conclusions regarding the significantly related candidate genes among the broader set of less-studied 86 candidate genes were similar. Thus, there is evidence that some of the schizophrenia candidate genes are more strongly related to schizophrenia than expected by chance.
Figure 1. Quantile-quantile plot of the −log10 p-values from the 25 most-studied candidate genes.
Observed gene-level −log10 p-values from MAGMA are plotted on the y-axis, with expected −log10 p-values plotted on the x-axis. Points are heat map colored according to the number of times each gene has been studied, and the vertical green lines are bootstrapped 95% confidence intervals.
DISCUSSION
The overarching goal of this study was to examine whether the results from a highly powered GWAS support the hypothesis that the most-studied schizophrenia candidate genes are particularly relevant to schizophrenia. The scientific community has invested enormous time, talent, and effort in candidate gene studies over the years. It has been estimated that at least 250 million USD have been invested in candidate gene studies in the 1990s and 2000s (7). These studies have contributed to an improved characterization and understanding of the biological functions of many of these candidate genes. However, we found little evidence that common SNPs within these genes are any more relevant to schizophrenia than SNPs within control sets of non-candidate genes. The set of top 25 candidate genes showed no evidence of being more associated with schizophrenia compared to all other genes, or relative to genes involved with type 2 diabetes and height, which are not expected to harbor a disproportionate number of risk alleles for schizophrenia. When we expanded the gene set to include all candidate genes that had been studied more than five times, there was marginal evidence that these 86 genes were more associated with schizophrenia when compared to all other genes and relative to genes associated with Type 2 diabetes, but they were no more significantly associated than genes involved in height or the ~1000 genes in a functional category (synaptic processes) we hypothesized a priori might be related to schizophrenia. Furthermore, only two of the 16 gene set results were significant when we measured strength of association using the minimum p-value per gene.
Although our results suggest that, taken as a group, schizophrenia candidate genes are no more associated with schizophrenia than random sets of control genes, they do not imply that this is true of all the candidate genes. Indeed, we present evidence that several of the most-studied candidate genes—particularly NOTCH4, DRD2, KCNN3, GRM3, and TNF—are more strongly related to schizophrenia than expected by chance. It is important to put this evidence in perspective. First, two of these five genes (NOTCH4 and TNF) are in the MHC, and given the long-range and complex nature of LD in this region, it is unclear whether it is variants in these two genes or variants in some of the other ~240 genes in the MHC that are relevant, and indeed recent evidence suggests the signal driving the NOTCH4 association comes not from NOTCH4 but from the nearby complement component 4 (C4A and C4B) MHC genes (25). For this reason, we hereafter restrict discussion of the most significantly associated candidate genes to those not in the MHC. Second, while variants in DRD2, KCNN3, and GRM3 all appear related to schizophrenia above chance, the specific polymorphisms most studied in these genes were not particularly related to schizophrenia and were not close to being among the 108 genome-wide significant SNPs discovered in the PGC study (12) (p = .22, 3.3 × 10−5, and .58 respectively). Thus, our results do not agree with most previous positive findings on these candidate genes. Finally, although there can be alternative motivations for further study of these candidate genes (e.g., the fact that they are well studied and well characterized in animal studies), our results suggest that the statistical rationale for further prioritization of these genes is weak. There were 128, 300, and 434 genes, respectively, more related to schizophrenia than DRD2, KCNN3, and GRM3. In Supplementary Table S7, we present results from the 100 top-ranked non-MHC genes, any one of which is arguably a better target of future studies than any previously studied candidate gene.
There are some important limitations to acknowledge in this study. First, we tested for enrichment of lower GWAS p-values from imputed, common SNPs. Thus we cannot rule out the possibility that these candidate genes contain rare variants important to the etiology of schizophrenia. Similarly, our study focused on variants +/−25 kb of gene boundaries, and ignored effects of trans-regulatory elements that can occur at great distances (26). However, for all of the top 25 candidate genes except DISC1 (27), the focus in the literature has been on variants within candidate gene boundaries, and typically on a specific common polymorphism within those genes. Understanding the role that rare variants might play on schizophrenia risk in these candidate genes awaits sequence or more accurate imputation data, and understanding the role of trans-regulatory elements awaits better understanding of long-range gene regulation and the incorporation of patterns of chromatin binding (28) or gene expression data with GWAS (29). Nevertheless, our results do not support the original hypotheses involving the most-studied candidate genes, and thus provide no reason to believe that rare variants in these genes, or the trans-elements that regulate them, will be particularly relevant to schizophrenia.
Additionally, the imputed data used by the PGC GWAS analysis did not adequately capture common polymorphisms within all the candidate genes. For example, the most frequently studied candidate polymorphisms for DRD4 and NOTCH4 were neither genotyped nor successfully imputed in the PGC GWAS. The most commonly studied polymorphism in DRD4 is a 13-bp indel, which is not well captured in the kind of sequencing done by the 1000 Genomes(21) project, the reference panel used to impute in the PGC data. The most commonly studied polymorphism in NOTCH4, rs367398, was also missing from the 1000 Genomes phase 1 reference panel, and the PGC data included no SNPs in LD with that polymorphism at R2 > 0.3. As already noted, this SNP is located in the MHC region, which has made genotyping and analyzing this section of the genome difficult, although a recent study suggests that a large proportion of the genetic risk to schizophrenia in the MHC is specific to a particular locus in the C4A gene (25).
In the field of genetics, candidate gene analyses have largely fallen out of favor due to concerns about low power, false positives, low replication rates (30,31), insufficient biological knowledge to correctly identify plausible candidate genes, and the increasingly low cost of whole-genome array data. Yet candidate gene research continues in other fields, despite these issues. For example, a Google Scholar search performed on July 31, 2016 for “COMT” generated three thousand search results from 2016 alone, many of which are classic candidate gene association studies; the first five phenotypes from this search were circadian preferences(32), affective well-being across the lifespan(33), cognitive outcomes after electroconvulsive therapy(34), effect of opioid treatment on pain relief(35), and second-language learning in adults(36). It is of course possible that these and other traits are exceptions to what we now know about complex traits studied to date by GWAS, for which thousands of risk variants exist, each which explain a very small amount of variation (37). Nevertheless, even if a given trait’s genetic architecture is simple, our findings, as well as those of our colleagues(2,5,8,11), call into question the notion that scientists have been able to guess, a priori, which genes, much less which polymorphisms within those genes, will be relevant to any given trait. Given our inchoate understanding of the biological mechanisms underlying most complex traits, we suggest that future candidate gene studies should base gene choice not on historical precedent or proposed biological underpinnings, but on rigorous statistical evidence from the same or related traits, and that such studies should be sufficiently powered to detect effects on the order of those typically observed in modern GWAS.
Supplementary Material
Acknowledgments
This work was funded by the following grants: R01 AA017889, R01 MH100141, T32 HD007289, T32 MH06880, T32 AA013525. We thank the investigators who were part of the PGC who put this data together, as well as Michelle Farris, Robin Hacker-Cary, Chris Klene, Valerie Slegesky, and Nicole Thatcher for helping with initial data collection, and the many participants in these studies, all of whom helped make this study possible.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
FINANCIAL DISCLOSURES
The authors report no biomedical financial interests or potential conflicts of interest.
References
- 1.Sullivan PF, Kendler KS, Neale MC. Schizophrenia as a Complex Trait. Arch Gen Psychiatry. 2003;60:1187–92. doi: 10.1001/archpsyc.60.12.1187. [DOI] [PubMed] [Google Scholar]
- 2.Allen NC, Bagade S, McQueen MB, Ioannidis JPa, Kavvoura FK, Khoury MJ, et al. Systematic meta-analyses and field synopsis of genetic association studies in schizophrenia: the SzGene database. Nat Genet. 2008;40(7):827–34. doi: 10.1038/ng.171. [DOI] [PubMed] [Google Scholar]
- 3.Cichon S, Craddock N, Daly M, Faraone SV, Gejman PV, Kelsoe J, et al. Genomewide association studies: history, rationale, and prospects for psychiatric disorders. Am J Psychiatry. 2009;166(5):540–56. doi: 10.1176/appi.ajp.2008.08091354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wei J, Hemmings GP. Nat Genet. 4. Vol. 25. Nature Publishing Group; 2000. The NOTCH4 locus is associated with susceptibility to schizophrenia; p. 376. [DOI] [PubMed] [Google Scholar]
- 5.Ioannidis JP, Ntzani EE, Trikalinos TA, Contopoulos-Ioannidis DG. Replication validity of genetic association studies. Nat Genet. 2001;29(3):306–9. doi: 10.1038/ng749. [DOI] [PubMed] [Google Scholar]
- 6.Mutsuddi M, Morris DW, Waggoner SG, Daly MJ, Scolnick EM, Sklar P. Analysis of high-resolution HapMap of DTNBP1 (Dysbindin) suggests no consistency between reported common variant associations and schizophrenia. Am J Hum Genet. 2006;79(5):903–9. doi: 10.1086/508942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Visscher PM, Brown MA, McCarthy MI, Yang J. Five Years of GWAS Discovery. Am J Hum Genet. 2012 Jan 13;90(1):7–24. doi: 10.1016/j.ajhg.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collins AL, Kim Y, Sklar P, O’Donovan MC, Sullivan PF. Psychol Med. 03. Vol. 42. Cambridge Univ Press; 2012. Hypothesis-driven candidate genes for schizophrenia compared to genome-wide association results; pp. 607–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Purcell SSM, Wray NRN, Stone JJL, Visscher PM, O’Donovan MC, Sullivan PF, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009:748–52. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi J, Levinson DF, Duan J, Sanders AR, Zheng Y, Pe’er I, et al. Common variants on chromosome 6p22.1 are associated with schizophrenia. Nature. 2009;460(7256):753–7. doi: 10.1038/nature08192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farrell MS, Werge T, Sklar P, Owen MJ, Ophoff RA, O’Donovan MC, et al. Evaluating historical candidate genes for schizophrenia. Mol Psychiatry. 2015 May;20(5):555–62. doi: 10.1038/mp.2015.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ripke S, Neale BM, Corvin A, Walters JTR, Farh K-H, Holmans Pa, et al. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–7. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lohmueller KE, Pearce CL, Pike M, Lander ES, Hirschhorn JN. Meta-analysis of genetic association studies supports a contribution of common variants to susceptibility to common disease. Nat Genet. 2003;33(2):177–82. doi: 10.1038/ng1071. [DOI] [PubMed] [Google Scholar]
- 14.Owen MJ, Craddock N, O’Donovan MC. Schizophrenia: Genes at last? Trends in Genetics. 2005:518–25. doi: 10.1016/j.tig.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 15.Sullivan PF. The genetics of schizophrenia. PLoS Med. 2005;2(7):0614–8. doi: 10.1371/journal.pmed.0020212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harrison PJ, Weinberger DR. Schizophrenia genes, gene expression, and neuropathology: on the matter of their convergence. Mol Psychiatry. 2005;10(1):40–68. doi: 10.1038/sj.mp.4001558. [DOI] [PubMed] [Google Scholar]
- 17.Zheng J, Erzurumluoglu AM, Elsworth BL, Kemp JP, Howe L, Haycock PC, et al. Bioinformatics. Oxford Univ Press; 2016. LD Hub: a centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Welter D, MacArthur J, Morales J, Burdett T, Hall P, Junkins H, et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42(D1) doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ruano D, Abecasis GR, Glaser B, Lips ES, Cornelisse LN, de Jong APH, et al. Functional Gene Group Analysis Reveals a Role of Synaptic Heterotrimeric G Proteins in Cognitive Ability. Am J Hum Genet. 2010;86(2):113–25. doi: 10.1016/j.ajhg.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lips ES, Cornelisse LN, Toonen RF, Min JL, Hultman CM, Holmans PA, et al. Functional gene group analysis identifies synaptic gene groups as risk factor for schizophrenia. Mol Psychiatry. 2012;17(10):996–1006. doi: 10.1038/mp.2011.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.The 1000 Genomes Project Consortium. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012 Nov 1;491(7422):56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: Generalized Gene-Set Analysis of GWAS Data. PLoS Comput Biol. 2015;11(4) doi: 10.1371/journal.pcbi.1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mishra A, Macgregor S, Amos CI, Wang L-E, Lee JE, Gershenwald JE, et al. VEGAS2: Software for More Flexible Gene-Based Testing. Twin Res Hum Genet. 2015;18(01):86–91. doi: 10.1017/thg.2014.79. [DOI] [PubMed] [Google Scholar]
- 24.De Leeuw CA, Neale BM, Heskes T, Posthuma D. The statistical properties of gene-set analysis. Nat Rev Genet. 2016;17(6):353–64. doi: 10.1038/nrg.2016.29. [DOI] [PubMed] [Google Scholar]
- 25.Sekar A, Bialas AR, de Rivera H, Davis A, Hammond TR, Kamitaki N, et al. Schizophrenia risk from complex variation of complement component 4. Nature. 2016;530(7589):177–83. doi: 10.1038/nature16549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Consortium TGte. Ardlie KG, Deluca DS, Segrè AV, Sullivan TJ, Young TR, et al. The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science. 2015;348(6235):648–60. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Millar JK, Wilson-Annan JC, Anderson S, Christie S, Taylor MS, Semple Ca, et al. Disruption of two novel genes by a translocation co-segregating with schizophrenia. Hum Mol Genet. 2000;9(9):1415–23. doi: 10.1093/hmg/9.9.1415. [DOI] [PubMed] [Google Scholar]
- 28.Won H, de la Torre-Ubieta L, Stein JL, Parikshak NN, Huang J, Opland CK, et al. Chromosome conformation elucidates regulatory relationships in developing human brain. Nature. 2016;538(7626):523–7. doi: 10.1038/nature19847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gusev A, Mancuso N, Finucane HK, Reshef Y, Song L, Safi A, et al. Transcriptome-wide association study of schizophrenia and chromatin activity yields mechanistic disease insights. bioRxiv. 2016 Aug 2; doi: 10.1038/s41588-018-0092-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ioannidis JPA, Tarone R, McLaughlin JK. The false-positive to false-negative ratio in epidemiologic studies. Epidemiology. 2011;22(4):450–6. doi: 10.1097/EDE.0b013e31821b506e. [DOI] [PubMed] [Google Scholar]
- 31.Koenen KC, Duncan LE, Liberzon I, Ressler KJ. From candidate genes to genome-wide association: The challenges and promise of posttraumatic stress disorder genetic studies. Biological Psychiatry. 2013:634–6. doi: 10.1016/j.biopsych.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jawinski P, Tegelkamp S, Sander C, Häntzsch M, Huang J, Mauche N, et al. Time to wake up: No impact of COMT Val158Met gene variation on circadian preferences, arousal regulation and sleep. Chronobiol Int. 2016;33(7):893–905. doi: 10.1080/07420528.2016.1178275. [DOI] [PubMed] [Google Scholar]
- 33.Turan B, Sims T, Best SE, Carstensen LL. Psychol Aging. 3. Vol. 31. American Psychological Association; 2016. Older age may offset genetic influence on affect: The COMT polymorphism and affective well-being across the life span; p. 287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bennett DM, Currie J, Fernie G, Perrin JS, Reid IC. Differences in Cognitive Outcomes After ECT Depending on BDNF and COMT Polymorphisms. J ECT. 2016 doi: 10.1097/YCT.0000000000000325. [DOI] [PubMed] [Google Scholar]
- 35.Elens L, Elisabeth N, Maja M, Rane A, Fellman V, van Schaik RH. Genetic predisposition to poor opioid response in preterm infants: Impact of KCNJ6 and COMT polymorphisms on pain relief after endotracheal intubation. Ther Drug Monit. 2016 doi: 10.1097/FTD.0000000000000301. [DOI] [PubMed] [Google Scholar]
- 36.Mamiya PC, Richards TL, Coe BP, Eichler EE, Kuhl PK, Geschwind DH, et al. Brain white matter structure and COMT gene are linked to second-language learning in adults. Proc Natl Acad Sci. 2016;113(26):7249–54. doi: 10.1073/pnas.1606602113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sullivan PF, Daly MJ, O’Donovan M. Genetic architectures of psychiatric disorders: the emerging picture and its implications. Nat Rev Genet. 2012;13(8):537–51. doi: 10.1038/nrg3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.