Abstract
We review the development and evolution of the ear neurosensory cells, the aggregation of neurosensory cells into an otic placode, the evolution of novel neurosensory structures dedicated to hearing and the evolution of novel nuclei and their input dedicated to processing those novel auditory stimuli. The evolution of the apparently novel auditory system lies in duplication and diversification of cell fate transcription regulation that allows variation at the cellular level [transforming a single neurosensory cell into a sensory cell connected to its targets by a sensory neuron as well as diversifying hair cells], organ level [duplication of organ development followed by diversification and novel stimulus acquisition] and brain nuclear level [multiplication of transcription factors to regulate various neuron and neuron aggregate fate to transform the spinal cord into the unique hindbrain organization]. Tying cell fate changes driven by bHLH and other transcription factors into cell and organ changes is at the moment tentative as not all relevant factors are known and their gene regulatory network is only rudimentary understood. Future research can use the blueprint proposed here to provide both the deeper molecular evolutionary understanding as well as a more detailed appreciation of developmental networks. This understanding can reveal how an auditory system evolved through transformation of existing cell fate determining networks and thus how neurosensory evolution occurred through molecular changes affecting cell fate decision processes. Appreciating the evolutionary cascade of developmental program changes could allow identifying essential steps needed to restore cells and organs in the future.
Keywords: cell type evolution, transcription factor multiplication, differential expression regulation
Introduction
Ever since Ohno (Ohno, 2013) first proposed in 1972 that gene duplication might be a driving force for evolutionary change, multiple examples of evolutionary innovation have been associated with gene duplication: neo-functionalization (changes in the protein sequence) and sub-functionalization [alteration in the expression of duplicated genes (Lynch and Force, 2000; Peter and Davidson, 2015; Wagner, 2011)]. Many genes and enhancers originally discovered in multicellular organism have now been detected in single-celled outgroups suggesting that gene regulation evolution combined with gene multiplications (Sebé-Pedrós et al., 2016) might drive derived traits of a particular cell type [aka apomeres (Arendt et al., 2016)]. This process forms the basis of multicellular evolution that, in turn, is the basis for organ and thus organismal evolution . Among the many genes that show duplication or multiplication in vertebrates relative to the hypothetical single cell ancestor of all metazoans are cell fate decision making genes such as those encoding basic Helix-Loop-Helix (bHLH) transcription factors (Pan et al., 2012b). Specifically, current data suggest multiplication of these factors occurred in evolution, resulting in the formation of entire new classes showing properties not found in single celled ancestors such as Class A bHLH transcription factors (Fritzsch et al., 2015a). Such cellular decision making molecular cascades were eventually modified by the evolution of novel signal cascades such as the Wnt signaling (Booth and King, 2016) and supplemented by the recruitment of other transcription factors co-opted from other organ development to generate unique multicellular body parts such as sensory organs (Arendt et al., 2016) with dedicated modifications of gene regulatory networks (Peter and Davidson, 2015).
Building on this general principle, we explore here how molecular diversification might have provided the material for major steps in inner ear cellular development and evolution and how sensory organ evolution might connect to the central nervous system evolution through matched specific changes in developmental gene regulatory networks:
The ancestral metazoan neurosensory cell likely was polarized into a base and an apex similar to a vertebrate olfactory neurosensory cell: one end it was specialized to receive a stimulus and the other end specialized to send the information decoded into an action potential along its own axon to the central nervous system for processing. An ancestral split generated two cell types out of this unicellular precursor: i). a sensory cell type specialized for sensory stimulus reception but without an axon and ii). a sensory neuron type specialized for conducting the information from the sensory cell to the central nervous system (Fritzsch et al., 2007; Gasparini et al., 2013; Patthey et al., 2014).
Aggregation of what might have initially been individually distributed neurosensory cells and neurons into placodes led to the formation of major cranial sensory organ formation such as the ear, lateral line, electroreceptors, olfaction and taste system (Maier et al., 2014; O'Neill et al., 2012; Schlosser et al., 2014). Some of these simple organ systems eventually split into multiple organs such as distinct lateral line neuromasts or distinct inner ear organs (Fritzsch et al., 2002). Splitting of such organs, in combination with the recruitment of novel genes, led to novel specializations that enable different mechanosensory organs to be tuned to specific sensory stimuli (Fritzsch et al., 2013).
Analyzing various sensory information provided by the different kind of placodally-derived receptor organs requires that central nervous system projections reach discrete areas of the brain (Grothe et al., 2004) for segregated information processing enabled by unique brain gene regulatory networks (GRN)-guided neuronal development (Nothwang, 2016). Here we will develop a concept of molecular transformation of a spinal cord like organization of nuclei into a hindbrain organization (Hernandez-Miranda et al., 2016) able to receive and process distinct information provided by placodally-derived sensory cells (Elliott et al., 2017; Fritzsch et al., 2005a; Fritzsch et al., 2006b).
A) Evolution and development of vertebrate mechanosensory hair cells and sensory neurons
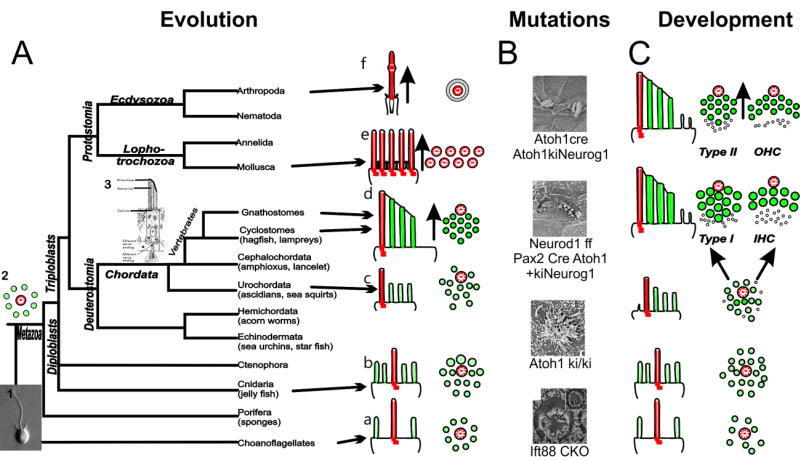
Neurosensory cells, like all other cells of multicellular organisms, evolved from a single celled ancestor, a choanoflagellate-like organism (Booth and King, 2016; Fritzsch and Straka, 2014). These single celled organisms were polarized into an apex and a base: they display on one end a kinocilium surrounded by interconnected microvilli that function as a basket to filter food propelled by the flagellum; the opposite end provides attachment to the substrate. Choanoflagellates thus show a cellular polarity in line with the basal-apical cellular polarity in epithelia (Boenigk, 2015; Leadbeater, 2015) as is so obvious in sponges (Ueda et al., 2016). Development of mechanosensory cells apparently recapitulates this process in that mechanosensory cells of vertebrates develop first an apical kinocilium surrounded by microvilli. The kinocilium moves into an acentric position as mechanosensory cells mature (Fig.1). Eliminating kinocilia mediated apical reorganization results in circular polarized hair cells (Jones et al., 2008). Beyond the kinocilium as a signaling center, the apical acentric polarity development relies on several regulatory factors, such as LGN and Gαi (Tarchini et al., 2016) and Bbs8 and Ift20 (May-Simera et al., 2015) to eliminate microvilli and move the kinocilium into an acentric position and regulate the height and distribution of stereocilia to properly polarize hair cells. The end product of this incompletely understood process (Lu and Sipe, 2016; Schüler et al., 2013) is a highly polarized cell that allows mechanical stimuli to exert changes on the cell’s resting potential depending on the direction of the stimulation (Fig. 1) through opening of channels attached to tip links [putative candidates are TMC1/2 (Shibata et al., 2016; Wu and Müller, 2016)]. Many mechano-sensitive cells display a permutation of this common scheme of kinocilia/microvilli. However, the variation of some non-vertebrate lineages can be arranged into a graded set of changes toward the vertebrate mechanosensory cell: a polarized organ pipe of stereocilia with or without a kinocilium (Fig. 1). Future work needs to determine how various proteins known to play part in hair cell polarity formation were recruited. During development hair cells go through a similar rearrangement (Fig.1a) and certain mutations ‘freeze’ hair cells at earlier developmental or evolutionary stages (Fig. 1b).
Figure 1. Evolution and development of the apex from centric to acentric organization.
Evolution(A): The kinocilia (red) shifts from a centric position (2) surrounded by microvilli/stereocilia (green) found in choanoflagellates to an acentric position (a–d) in Deuterostomia, establishing cell polarity and allowing directional stimulation in vertebrates (3). Protostomia show divergent specializations for mechanosensation (e,f). Movement direction is indicated by the arrow. Mutation (B): Various mutations that affect the stereocilia. Atoh1-cre, Atoh1kiNeurog1, Neurod1f/f Pax2-cre, Atoh1+kiNeurog1 and Atoh1ki/ki mice develop sterocilia aberrations suggesting that planar cell polarity is influenced by Atoh1 levels (Jahan et al., 2015a; Jahan et al., 2015b; Pan et al., 2012a). Itf88 CKO mice can develop circular stereocilia in the absence of a kinocilia (Jones et al., 2008). Development(C): Hair cells develop first a central kinocilium surrounded by microvilli followed by shift into an acentric position and alteration of stereocilia diameters (bottom to top). Only organ of Corti hair cells lose secondarily the kinocilium (top). Modified after (Fritzsch et al., 2007)
In addition to TMC1/2 and TMIE, Piezo channels, frequently found in proprioceptors and Merkel cells, are also associated with normal function of some hair cells, but are apparently not directly involved in mechanotransduction (Wu et al., 2016). In addition, multiple functionally important links between stereocilia and mechanotransduction defects can be found in various mutations. For example, mutations in stereocilin (Strc) cause the DFNb16 hearing loss, the most frequent hair cell-related hearing loss (Francey et al., 2012; Moteki et al., 2016). It would be important to establish how the stereocilin-mediated connections of stereocilia molecularly relate to the microvilli connections in choanoflagellates and sponges. This could help to reveal the evolutionary origin of the leading candidates for the mechanosensory transduction channel, TMC1/2 (Shibata et al., 2016) and TMHS/TMIE (Wu et al., 2016; Wu and Müller, 2016).
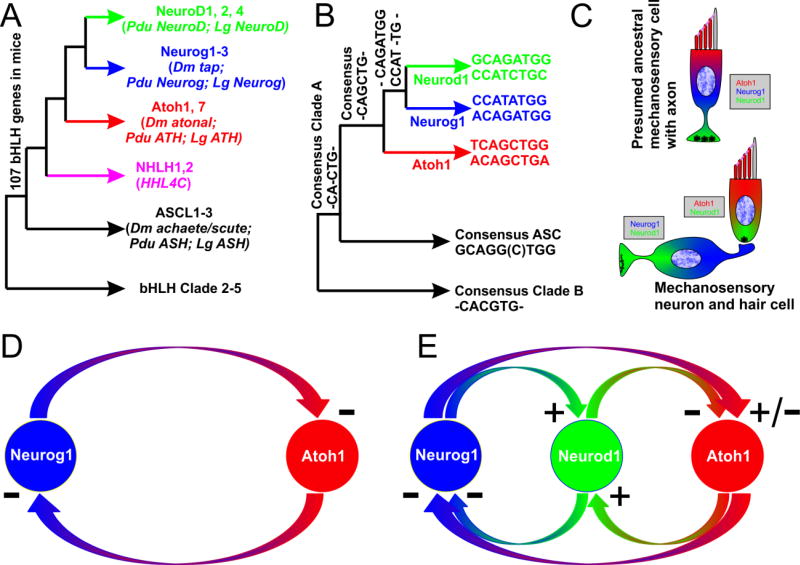
Superimposed on these transformations in the apex of mechanosensory cells that rearranges kinocilia and microvilli from a symmetric non-sensory organization with a motile kinocilium in the center to an asymmetric sensory system specialized for mechanotransduction, is a partially overlapping set of changes that lead to the segregation of mechanosensory cells into a sensory cell and a sensory neuron. This transformation correlates with the multiplication of certain cell fate determining transcription factors, the basic Helix-Loop-Helix transcription factors (bHLH TFs; Fig. 2) and their binding sites (Fritzsch et al., 2010; Fritzsch and Straka, 2014). Based on selective loss of mechanosensory hair cell differentiation in mice null for Atoh1 (Bermingham et al., 1999) and of all sensory neurons but also some hair cells in Neurog1 null mice (Ma et al., 2000) it was hypothesized that some lineage relationship exists between sensory neurons and mechanosensory hair cells (Fritzsch et al., 2000). Later, a third bHLH TF, Neurod1, was identified that was originally claimed to also lead to complete neuronal loss (Liu et al., 2000) but was later found to lead only to partial loss, mostly of spiral ganglion neurons, due to other partially co-expressed bHLH genes (Jahan et al., 2010a; Kim et al., 2001). Subsequent work showed that there is a transient expression of bHLH transcription factor Atoh1 in delaminating neurons and that expression of markers common to hair cell and neuronal precursors show a transient increase of hair cell precursor population, implying a possible transformation of neurons into hair cells (Matei et al., 2005). Further validation of this idea of a lineage relationship was provided using a Neurog1-cre line for some sensory epithelia (Raft et al., 2007) claiming a direct Neurog1-Atoh1 interaction (Fig. 2). Additional analysis centered on the fate of different cell types in the ear using a combination of knock-out and knock-in approaches has modified this model, mainly by adding the function of Neurod1 as a partially understood mediator of Neurog1 and Atoh1 interactions (Fig. 2).
Figure 2. Relationship of the bHLH transcription factors and their E-boxes indicates likely multiplication and diversification at molecular and cellular levels.
(A) Relationship between the bHLH transcription factors. (B) Relationship between the E-box binding sites of the various bHLH transcription factors. Note co-evolutionary changes in the bHLH transcription factors (A) and the E-box sequences (B). (C) The presumed ancestral mechanosensory cell with an axon may have contained three bHLH transcription factors: Atoh1, Neurod1, and Neurog1. Incomplete segregation of these three transcription factors result in a derived condition with separate hair cells expressing Atoh1 and Neurod1 and sensory neurons expressing Neurod1 and Neurog1. Some experiments suggest that Neurog1 and Atoh1 are in a negative feedback loop [D (Raft et al., 2007)]. Knockin of Neurog1 into Atoh1 and effects of Neurod1 deletion suggest that this feedback loop is more complicated and at least in part mediated by Neurod1 (E). Modified after (Fritzsch et al., 2010; Pan et al., 2012b)
Neurog1 clearly forms such a negative feedback loop to Atoh1 in the CNS but the role of Neurod1 was not clarified in this process (Gowan et al., 2001). Changes in neuronal or hair cell populations after Neurog1 deletion (Matei et al., 2005) was also claimed to indicate such a feedback loop in neurosensory precursors (Raft and Groves, 2015). However, a more complicated scenario of Neurog1 and Atoh1 being both variably cross-regulated by Neurod1 remains an alternative possibility (Fig. 2) in light of more recent evidence of the existence of such a complicated feed-forward and feed-back interactions between these three bHLH TFs (Pan et al., 2012b). For example, a conditional deletion model for Neurod1 showed that this transcription factor suppresses Atoh1 expression in several systems such as the cerebellum (Pan et al., 2009) and the ear (Jahan et al., 2010b) suggesting a negative feedback loop for Atoh1 and Neurod1. Consistent with such a role for Neurod1 is that loss of Neurod1 results in continued expression of an otherwise transient Atoh1 expression in sensory neurons and their transdifferentiation into intraganglionic hair cells (Jahan et al., 2010b). Such a potential negative feedback loop was also found in hair cells that show a premature and more profound expression of Atoh1 in the apex of Neurod1 CKO mice, resulting in a partial conversion of hair cell types as a consequence of premature and elevated expression of Atoh1 (Jahan et al., 2015a; Jahan et al., 2010b, 2013). To formally test a more direct interaction between Atoh1 and Neurog1 required replacement of Atoh1 by Neurog1. In contrast to the effective transformation of neurons into hair cells in Neurod1 null mice, Neurog1 proved to be unable to substitute for Atoh1 in hair cell precursors leading to the rapid degeneration of such hair cell precursors (Jahan et al., 2012) in a pattern reminiscent of complete (Fritzsch et al., 2005b) or conditional deletion of Atoh1 (Pan et al., 2011). These data clearly demonstrated that Neurog1 cannot signal alone effectively in postmitotic hair cell precursors to initiate a differentiation as hair cells (Jahan et al., 2012). This contrasts with the flexibility of some neuronal precursors that easily convert to intraganglionic hair cells upon elimination of Neurod1 (Jahan et al., 2010b). Importantly, mice heterozygotic for the Atoh1tgNeurog1 knockin construct coexpressed both Atoh1 and Neurog1 under Atoh1 promoter control with little negative effects on hair cell development or expression level of either gene. This suggested that in differentiating hair cells there is no direct feedback loop of Neurog1 on Atoh1 as previously claimed but a feedback loop of Atoh1 on Neurog1 remained a possibility (Raft and Groves, 2015).
It remained unclear whether this inability to rescue hair cell development or to convert hair cells into neurons was related to Neurog1 functional differences in activating Atoh1 specific e-boxes (Fritzsch et al., 2010) or expression level differences. The latter remained a possibility given the positive feedback loop Atoh1 protein exerts on its own regulation via specific enhancer binding (Fritzsch et al., 2006a; Helms et al., 2000). To distinguish between these two possibilities, a mouse line was made that combined a delayed deletion of Atoh1 (Pan et al., 2012a) with the misexpression of Neurog1 in the second allele (Jahan et al., 2015b). These mice displayed a surprisingly robust development of nearly all hair cells but in a disorganized organ of Corti. Most importantly, the stereocilia phenotypes resembled those observed in mutants of LGN and Gαi (Tarchini et al., 2016) and various Myosin mutants (Quint and Steel, 2003). Combined these data suggest that possibly the concentration and/or binding specificity of Atoh1 is an essential aspect of normal stereocilia development. Further support for the influence of Atoh1 on stereocilia development of hair cells comes from mice in which Atoh1 expression level was manipulated (Fig. 1) through Neurod1 mutations (Jahan et al., 2010b) or Sox2 manipulations (Dvorakova et al., 2016) or alterations in miRs (Soukup et al., 2009).
The mechanosensory cell specializations to allow sensory cells to develop this exquisite mechanosensation require a number of factors for their normal development such as the basic Helix-Loop-Helix transcription factors Atoh1 (Jahan et al., 2015b; Pan et al., 2012a) or Neurod1 (Jahan et al., 2015a) that is tightly regulated in addition through several other transcription factors such as Pou4f3 (Ikeda et al., 2015; Xiang et al., 2003). Gfi1 (Hertzano et al., 2004) and Barlh1 (Chellappa et al., 2008) to name a few. Unfortunately, further work is needed to narrow down how these multiple factors cooperate to regulate the coordinated transformation of circular polarized cellular apex found in choanoflagellates (Fritzsch and Straka, 2014) and certain hair cell mutations (Jones et al., 2008) into a highly polarized hair cell (Lu and Sipe, 2016). Additional information is needed to understand the development of different types of hair cells. Hair cells show variable stereociliary diameters that clearly are affected by bHLH genes (Jahan et al., 2013, 2015b), but also show the unique stereocilia arrangements of inner and outer hair cells that apparently evolved out of the much simpler stereocilia bundle found in ancestral hair cells of the vestibular type (Lewis et al., 1985). When and how factors that coordinate polarity of hair cells across sensory epithelia (May-Simera et al., 2015) evolved is unclear. Thus, while some connections between Atoh1 level of expression and some of the downstream factors needed for the apical development of the stereocilia bundle are likely (Taura et al., 2016), we do not yet understand how vestibular and cochlear hair cells molecularly evolved their unique stereocilia organization and loss of a kinocilium. Atoh1 and other factors are also expressed in and needed for Merkel cell development (Bermingham et al., 2001) and Merkel cells share a mechanotransduction channel [Piezo2 (Woo et al., 2014)] with some hair cells (Wu et al., 2016) indicating the possibility that this highly conserved channel is directly tied into Atoh1 signaling, possibly derived from an ancestral function in epidermal cells (Arendt et al., 2016). Other channels associated with mechanosensory hair cells have recently been identified as being part of the derived non-mechanosensory ‘hair cells’ of the electroreceptive organs, yet another permutation of receptor cell development driven by a nearly identical set of transcription factors (Bellono et al., 2017; Pierce et al., 2008).
In summary, we begin to understand how mechanosensory transduction evolved in terms of the molecular machinery necessary to develop the stereocilia arrangement and regulate mechanotransduction channels, but details of this process still require additional work to reveal the whole gene regulatory network.
Without doubt, the complete absence of any neuronal development in the ear of mice null for Neurog1 demonstrates the essential nature of this transcription factor for ear neurogenesis (Ma et al., 2000; Ma et al., 1998). However, while there is an obvious interaction of neuronal precursor formation with hair cell development (Matei et al., 2005), the detailed nature of it remains unclear at a molecular and cellular level (Jahan et al., 2015b; Raft and Groves, 2015). What is clear is the sequence of activations of transcription factors that result in proliferation of neuronal precursors and their differentiation (Goodrich, 2016). Integral to this emerging perspective are several transcription factors such as Neurod1 for normal differentiation and targeted projection to the brain and the periphery (Jahan et al., 2010a), possibly through regulation of the Pou-domain factor Brn3a (Pou4f1) and both regulate expression of neurotrophin receptors (Huang et al., 2001; Kim et al., 2001). The inner ear neurons are unique in that they co-express at various levels the two neurotrophin receptors TrkB and TrkC whereas hair cells and supporting cells express the two ligands, Bdnf and Ntf-3 (Fariñas et al., 2001; Fritzsch et al., 2016). These receptors and ligands are essential for the viability of the neurons and participate in certain aspects of guidance of neurons toward hair cells in response to the neurotrophins released by hair cells and supporting cells (Yang et al., 2011).
In essence, a nearly parallel cascade of isoforms of transcription factors that represent a permutation of a common gene regulatory network lead to differentiation of two distinct neurosensory cell types. Instead of a single neurosensory cell, vertebrates have two cell types that are locked into a mutually dependent system of short term support to maintain neurons via hair cells/supporting cells (Fritzsch et al., 2016) and long term support of hair cells via unknown factors provided by innervation (Kersigo and Fritzsch, 2015). It appears that the evolutionary split of single neurosensory cells into sensory neurons and sensory cells was possible by duplicating specific aspects of gene activation sequence (Atoh1 and Pou4f3 for hair cells, Neurog1 and Pou4f1 for neurons) with Neurod1 being expressed in both and providing a negative feedback loop (Fig. 2) for both Neurog1 and Atoh1 (Jahan et al., 2010b). The embryonic early expression of neurotrophins allows to actually trace the fate of both delaminating neuronal and hair cell precursors (Matei et al., 2005) as they are expressed in both cell types. However, Neurod1 and Pou4f1 regulate neurotrophin receptor expression whereas neurotrophin expression is apparently regulated by factors expressed in precursors, such as Sox2, that can regulate expression of neurotrophins in supporting cells and undifferentiated hair cells of Atoh1 null mice, and in addition by Atoh1 (Fritzsch et al., 2005b; Pan et al., 2011). Past work suggested that neurotrophins and their receptors evolved only in vertebrates that also had an ear (Hallböök et al., 2006). However, recent work suggests neurotrophin and neurotrophin receptor presence in other deuterostomes (Benito-Gutiérrez et al., 2006; Burke et al., 2006) and bilaterians (Zhu et al., 2008). Given the likely ancestry of neurotrophins, it is possible that the segregation of hair cell/neuronal precursors and the neurotrophin expression in the common precursor for both is associated with evolution of proliferative neurosensory precursor system as an integral part of the evolution of a vertebrate placode (Fritzsch and Beisel, 2003; Schlosser et al., 2014). Obviously, the trophically interacting hair cell/neuronal system could only evolve after neurotrophins and their receptors had evolved, much like efferents reaching hair cells could evolve after the cholinergic receptor evolution (Fritzsch and Elliott, 2017). Further work is needed to identify the enhancer modifications that allowed the duplicated GRN to be expressed selectively in either neuronal or sensory cell and how the GRN changes were tied into the mechanotransduction and axonal pathfinding of hair cells and neurons, respectively.
In summary, data presented here are in line with hypothesis of cellular diversification through transformation of entire cascades of early cell fate defining transcription factor regulatory networks to generate parallel networks of sister genes that alter radically the cell type of at least partially clonally related cells (Fritzsch et al., 2006a). In essence, through duplication and modification of an entire cascade of transcription factors, evolution transformed a single, neurosensory cell into two derived cells, in agreement with a recently proposed idea of apomeric relationships of diversified cells (Arendt et al., 2016). Further work is needed to detail how trans- and cis element variations enabled the distinct functions associated with regulating sensory neurons and hair cell differentiation. We have detailed here some aspects of this segregation and will highlight the consequences for neuronal projections in part C further below.
B) Evolution and development of a mechanosensory ear and the transformation into hearing organs
Statocysts as complex sensory organs are widespread among metazoans (Fritzsch and Beisel, 2003; Fritzsch and Straka, 2014). Most of these mechanosensors have unique sensory cells that perceive the translational forces such as gravity acting on a denser structure such as otoconia or a mobile body part through different specializations of the apical sensory structures (Fig. 3). This widespread distribution of mechanosensory organs able to monitor position in the earth gravity field, even in animals without a fully developed central nervous system such as diploblastic coelenterates and comb jellies, indicates the essential need for any mobile animal to monitor its position in space (Fritzsch and Straka, 2014). Among the transcription factors associated with development of these mechanosensory organs are the Pax2/5/8 factors (Jacobs et al., 2007; Matus et al., 2007). While Pax2/8 are clearly necessary for normal ear development in mice (Bouchard et al., 2010), using this transcription factor as an evolutionary marker is complicated by the fact that it is also essential for kidney development. In fact, the pronephros and otocyst are immediately adjacent in the Pax 2/8 expression domains in developing amphibians, albeit in mesoderm and ectoderm (Buisson et al., 2015). Interestingly enough, several other transcription factors that are essential for vertebrate ear development, such as Eya1/Six1 (Xu et al., 1999; Zou et al., 2008), Gata3 (Duncan and Fritzsch, 2013; Karis et al., 2001) are also essential for both kidney and ear development. In addition, several other transcription factors are needed for ear development (Foxi3, Fgf3/10, Fgfr2, Sox9, Tfap2a (Alsina and Streit, 2016; Birol et al., 2016; Khatri et al., 2014; McMahon, 2016; Papadopoulos et al., 2016; Singh and Groves, 2016). Based on the evolution of statocysts in diploblasts, one could argue that evolution of the proneural gene regulatory network (GRN), involving possibly Eya1/Six1, Foxi3, Pax2/8 and Gata3 network of interacting factors and its dependence on Fgfr2 signaling (Pauley et al., 2003; Pirvola et al., 2000), evolved in ectodermal sensory placodes and was co-opted for mesodermal kidney development in triploblasts. Further evolutionary GRN analysis in diploblast statocyst development is needed to validate or refute this suggestion.
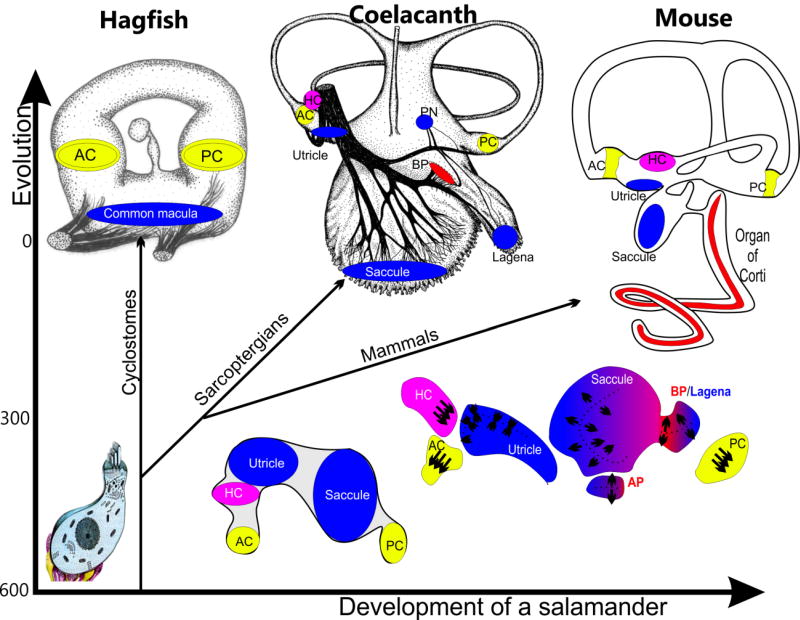
Figure 3. Inner ear evolution and development of sensory epithelia show multiplication and diversification.
Hagfish have a simple torus as a labyrinth with only three epithelia: a common macula for gravistatic orientation and 2 canal cristae for angular acceleration. Latimeria have multiplied and diversified these epithelia into three gravistatic organs (saccule, utricle and lagena) three canal cristae (anterior, horizontal and poster crista; AC, HC, PC) and two organs of unknown function the neglected papilla (PN) and the basilar papilla (BP). Mice have reduced the gravity sensing epithelia through the loss of the lagena to two (utricle, saccule), have three canals and associated cristae (AC,HC, PC) and have elongated the lagena duct into a coiled cochlea containing the organ of Corti for hearing. The splitting of sensory epithelia can be traced during development in salamanders as a progressive segregation of sensory primordia. Modified after (Fritzsch et al., 2002)
Beyond these early transcription factors and their expression in kidney and otocyst, the developing otic placode, but not the developing kidney, more highly or exclusively expresses several genes that ensure neurosensory development of the otic placode/ectoderm. Clearly, ear neurosensory development depends on several Fgfs (3,8,10) and Fgfr2 for overall neurosensory and Fgfr3 for supporting cell development (Pirvola et al., 2000; Puligilla et al., 2007). Kidney development also depends on Fgf 7/10 ligands signaling mostly via the Fgfr1 receptor (Bates, 2007; Rossini et al., 2005), but also Fgfr2 (McMahon, 2016). BMP4 downregulation combined with Fgf expression is an essential early step in neural induction (Fritzsch et al., 2006a; Meinhardt, 2015) that results in an unclear proportion of BMP4/Fgf signaling (Singh and Groves, 2016). This essential step of Bmp4 suppression and Fgf expression is not only essential for placode induction but also suffices to induce stem cells into differentiation as ear organoids in culture (Liu et al., 2016). How much of the developmental cascade of otic placode/otocyst development is recapitulated in these in vitro conditions and how closely the level of BMP4/Fgf protein signaling needs to be regulated for this development to occur remains to be determined. During neuronal development (most likely including the ear), the expression of early transcription factors leads to the expression of pro-proliferative and neural-fate stabilizing transcription factors, such as Otx2, Gbx2, Geminin, and Sox2 (Janesick et al., 2013; Singh and Groves, 2016; Yellajoshyula et al., 2011) and the Baf complex (Seo et al., 2005a; Seo et al., 2005b), that ultimately regulates chromatin remodeling and proneural bHLH gene expression. Level of bHLH gene expression in turn depends on the action of the Baf complex variably supported by Eya1/Six1, Pax2/8, Sox2, Foxi3 and Gata3. How all of these early transcription factors interact to define the level and topology of bHLH gene activation and how Wnt signaling fits in to regulate the size of the otic placode (Ohyama et al., 2007) remains to be determined experimentally. Loss of several factors can result in either incomplete invagination of the otic placode in mice mutant for Gata3 (Karis et al., 2001) or Pax2/8 (Bouchard et al., 2010), complete suppression of ear placode invagination such as in Foxi3 mutants (Birol et al., 2016; Singh and Groves, 2016) or in frogs exposed to RA (Fritzsch et al., 1998) or even incomplete formation of the ear following loss of Fgfr2 (Pirvola et al., 2000), indicating that several interactions are needed to move an otic placode forward to form an otocyst.
Eya1/Six1 play not only a role in preplacodal specification but also in later bHLH gene regulation such as Atoh1, Neurog1 (Ahmed et al., 2012a; Ahmed et al., 2012b) and several other transcriptional regulators of neuronal development of the otic placode (Riddiford and Schlosser, 2016). Pax2/8 as well as Foxi1/3 are chromatin remodeling factors that could enable expression of many other genes (Sharma et al., 2015; Singh and Groves, 2016). While these transcription factors can already be assembled into a rudimentary GRN for early neurosensory fate determination in developing otic region (Riddiford and Schlosser, 2016) and as initial aspects or otic placode enhancers begin to surface (Chen and Streit, 2015), the details of this network require more work to ensure guidance of otic development out of stem cells. Without a doubt, some of the factors needed for neurosensory development in the ear are not expressed or are very limitedly expressed in the developing kidney but can later be associated with kidney tumors due to their pro-proliferative signaling, such as Geminin (Dudderidge et al., 2005). Assuming that a neurosensory ectodermal ‘placode’ to generate statocysts evolved first in diploblasts, the lack of mesoderm and thus kidney formation in these animals suggests that a minimal network of Eya1, Pax2/8, Gata3 and Foxi3 was complemented in the otocyst to modify pro-proliferative placodal development toward neurogenesis through Sox and bHLH gene expression regulation that are also tied into the sensory organ development.
In essence, this new developmental GRN network (Riddiford and Schlosser, 2016; Schlosser et al., 2014), that possibly evolved with early statocysts already in diploblastic animals such as jelly fish, may have co-opted already existing GRNs dedicated to differentiate sensory cells (bHLH and Pou genes). Given that some of these factors are also participating in other developmental GRNs, their evolution specifically for neurosensory development regulation is less likely. For example, Atoh1 is not only regulating hair cell development but many other cells as well (Fritzsch et al., 2015a; Mulvaney and Dabdoub, 2012). This co-option of a cellular differentiation program ensured that existing single cell programs aggregate and integrate into a highly localized neurosensory development program leading to otic placode and otocyst/statocyst development. Some early stages of otic development in chicken are accompanied by cellular migration to coalesce disperse neurogenic pre-placodal cells into placodes (Steventon et al., 2016). Unfortunately, the various morphogenetic movements in early placode to otocyst formation (Alsina and Streit, 2016; Fritzsch et al., 1998) make it difficult to generalize from chicken development to otocyst evolution. Molecular developmental programs of more diploblast statocysts need to be analyzed to reveal the presence or absence of the basic regulator network of localized transformation of ectoderm into neurosensory ectoderm. It should be noted that several of the genes of this putative network are conserved in eukaryotes or bilaterians (Eya1, Six1, Pax2/8; Gata3, Sox2) and thus could indeed have evolved into a GRN that was coopted for its current function in otic development through altered downstream regulations and interactions with neurosensory transcription factors.
Independent of these actively investigated issues on the evolution of the molecular development of GRNs to make a placode, there is documented evolutionary change in the otic placode-derived ear (Lewis et al., 1985) which we like to explore next. For example, there is the simple addition of sensory epithelia, some of which acquire novel functions through association with specific structures to provide novel mechanical stimulation to these multiplied sensory epithelia (Fritzsch et al., 2002). In order to highlight this principle of duplication and diversification we will only discuss the molecular basis of the jawed vertebrate lateral/horizontal canal evolution and the molecular basis of the auditory organ evolution.
Lampreys and hagfish are jawless vertebrates that have no lateral (horizontal) canal with a uniquely associated lateral canal crista (Lewis et al., 1985). However, while the ear of hagfish is a simple torus with two canal cristae and an elongated gravity sensing common macula (Fig. 3), lampreys have evolved a complicated labyrinth with several unique features including two medial canals that together seem to serve the function of the single lateral canal of jawed vertebrates (Maklad et al., 2014). It appears that several genes drive lateral canal development, as a canal is missing in null mutants of these genes, Otx1 (Fritzsch et al., 2001; Morsli et al., 1999), N-Myc (Domínguez-Frutos et al., 2011; Kopecky et al., 2011) and Lmx1a (Koo et al., 2009; Nichols et al., 2008). In contrast to vertical canals where the Fgf10/BMP4 positive sensory epithelium drives canal formation (Chang et al., 2004; Pauley et al., 2003), a horizontal canal forms even in the very transient presence of a canal crista primordium in Foxg1 null mice (Hwang et al., 2009; Pauley et al., 2006). Notably, truncating ear proliferation through elimination of N-Myc seems to suffice to eliminate this evolutionary late addition to the jawed vertebrate ear development (Domínguez-Frutos et al., 2011; Kopecky et al., 2011) and proliferative precursors segregation is apparently dependent on repressive interactions between the notch signaling pathway (Daudet et al., 2007; Daudet and Lewis, 2005) and Lmx1a (Nichols et al., 2008).
The apparent independence of canal formation from canal crista formation in development of the horizontal canal suggests that GRNs driving development of either were paired through independent events. Comparison of lamprey ears with the sensory epithelia and their innervation in Otx1 null mutants suggest that a sensory patch of unknown function in lampreys may be the precursor for the horizontal canal crista recruited to function in the novel angular acceleration sensing system enabled through the development of a horizontal canal (Fritzsch et al., 2002). If further work supports this, it would indicate that formation of a separate sensory patch allowed implementation of a novel level of angular acceleration sensing once the molecular alterations in jawed vertebrates generated a horizontal canal. A similar predisposition seem to be the case for the segregated patches of neglected papilla that can under certain conditions evolve into the amphibian papilla for middle frequency hearing (Fritzsch and Wake, 1988). Enough flexibility in the boundary definitions of sensory epithelia is needed to allow such segregation of distinct patches of cells to occur and proliferation and Lmx1 seem to play a part in this flexibility (Fritzsch et al., 2013).
More complicated and contentious is the evolution of the basilar papilla and its transformation into the organ of Corti of mammals. There is agreement that a novel sensory patch had to form first to be later transformed in several steps into the auditory basilar papilla of most tetrapods that eventually was transformed into the coiled mammalian cochlea housing the organ of Corti (Fritzsch et al., 2013). Part of the contention derives from the finding that some aquatic sarcopterygian fish have such a unique sensory epithelium but have no known sound pressure communication (Fritzsch, 1987; Fritzsch, 1992). Furthermore, the changing taxonomic positions of basal sarcopterygians necessitating to invoke a loss of this sensory patch in lungfish (assuming the sensory organ in Latimeria is ancestral to the same named organ in tetrapods), or the sensory epithelium in Latimeria being a convergent evolution associated with an equally convergent evolution of a lagena and a lagenar recess (Fritzsch et al., 2013). A second area of contention is the structural dissimilarity of the organ of Corti of all therians (Jahan et al., 2015a) and its evolution already in monotremes in terms of inner and outer hair cells (Chen and Anderson, 1985; Ladhams and Pickles, 1996) and their prestin motor (Okoruwa et al., 2008) separated by pillar cells with unusual molecular and structural features not found in other supporting cell type of any vertebrate (Jahan et al., 2015a). Importantly, in monotremes, the basilar papilla/organ of Corti resides in the lagena recess that has a lagena sensory epithelium at its tip that is entirely separated from the sound conducting perilymphatic space connected to the round and oval window (Schultz et al., 2016). Obviously, eutherian mammals lack a lagena, have a coiled cochlea with the organ of Corti extending to the apex, and have a continuous perilymphatic space (scala tympani, scala vestibuli) that provides the means for sound pressure propagation from the oval to the round window. Whether or not the lagena was lost or incorporated into the expanding organ of Corti hinges currently on evidence provided by mutations that truncate proliferation of hair cell precursors (Fritzsch et al., 2013). In N-Myc mutant mice, the apex of the continuous organ of Corti develops a vestibular type of epithelium (Fig. 4) without any specialization of inner and outer hair cells reminiscent of an organ of Corti (Kopecky et al., 2011). In addition, much like in monotremes, the perilymphatic space loops around this unusual apical epithelium that is in a shortened cochlea duct resembling a monotreme lagena recess. Another similarity with monotremes of the N-Myc mutants is the lack of a ductus reuniens allowing a broad communication of the base of the cochlear/lagena recess with the saccule. Most interesting is that many supposedly early evolutionary markers for the ear such as Pax2 and Gata3 cause selective loss of the organ of Corti in their absence (Bouchard et al., 2010; Duncan and Fritzsch, 2013), indicating a much tighter control of the organ of Corti development, compared to vestibular organs, by apparently ubiquitously-expressed transcription factors. How early in organ of Corti/basilar papilla evolution those transcription factors got involved in its development remains to be shown across sarcopterygians.
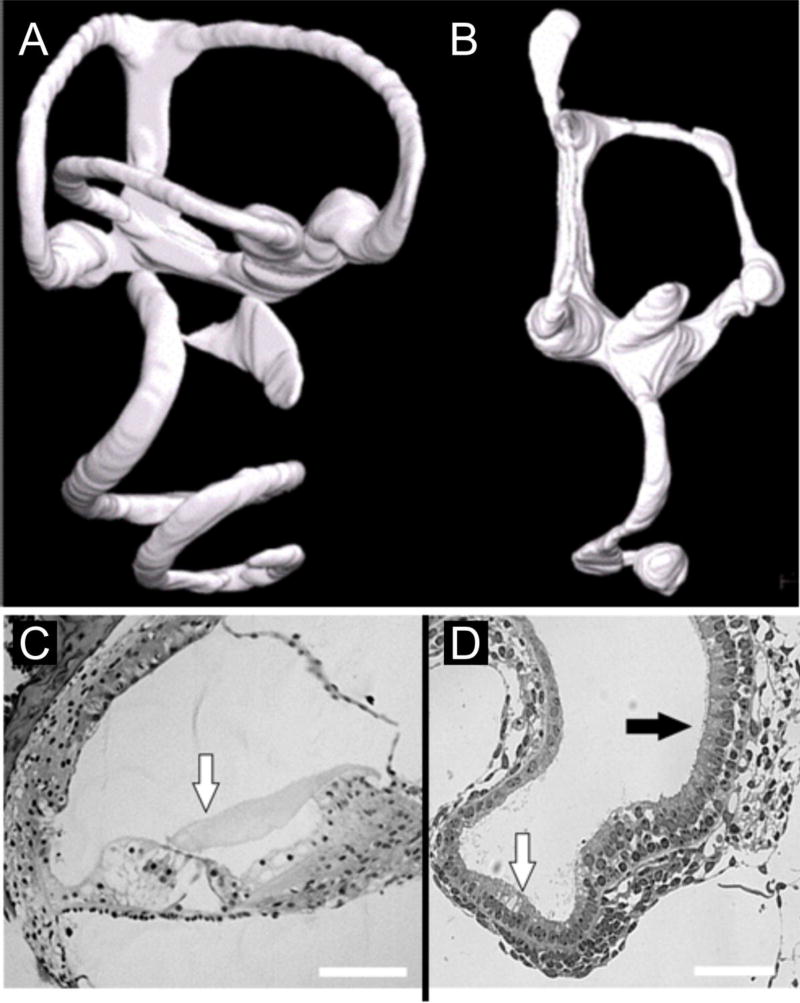
Figure 4. Loss of N-Myc results in loss of the horizontal canal and the formation of a “lagena” at the apex of the cochlea.
Compared with a control inner ear (A), loss of N-myc results in a truncated cochlea (B) that is in broad continuation with the saccule. Furthermore, there is loss of the horizontal canal and at the enlarged apex of the cochlea (B), there is a structure reminiscent of the lagena found in monotremes, including a vestibular-like organization of hair cells (black arrow in D) instead of one row if inner and three rows of outer hair cells found in the control organ of Corti (white arrow in C,D). Modified after (Kopecky et al., 2011)
The organ of Corti is the most sophisticatedly patterned sensory organ with a highly conserved and extremely stable cellular organization in all eutherian mammals (Jahan et al., 2015a). It evolved out of a less organized organ found in monotremes through reorganization of already specified inner and outer hair cells as a consequence of coiling related elongation that rearranged multiple rows of inner and outer hair cells of monotremes (Fritzsch et al., 2013; Schultz et al., 2016) into the single row of inner and three rows of outer hair cells of eutherian mammals (Lewis et al., 1985). Eliminating a single gene in mouse development, Foxg1, can alter the organ of Corti development to mimic that of monotremes in terms of shortening and rearrangement of hair cells into multiple rows (Fritzsch et al., 2013; Pauley et al., 2006) and similar effects can be achieved by altering convergent extension in PCP mutants (Jones et al., 2008).
Only the therian organ of Corti evolved pillar cells that allow a rocking movement of the lateral part to enhance endolymph flow across the inner hair cells for stimulation (Richter et al., 2007). Molecularly, this sophisticated arrangement into discrete rows of structurally and functionally different hair cells and supporting cells seems to depend on the segregated expression of transcription factors that are co-expressed in vestibular sensory epithelia but flank the medial and lateral aspect of the organ of Corti, Fgfs and BMP4 (Groves and Fekete, 2012; Pan et al., 2011). In addition, Wnt signaling plays a role in this cellular assembly (Munnamalai and Fekete, 2016; Sienknecht et al., 2014). Integrated into the diffusible factor signaling is the cell-cell interaction mediated by lateral inhibition via the Delta-Notch signal pathway (Korrapati et al., 2013) and differential effects of IHC and OHCs to mutations in the PCP pathway (Jones et al., 2008). This dual diffusion/cell-cell interaction program ensures the highly stereotyped organ of Corti development that requires both cell-cell interaction and diffusible signal changes. If both are disrupted the Organ of Corti shows a scrambled assembly of hair cells/supporting cells with limited specification of normally very distinct features (Jahan et al., 2015b) and mice are deaf. Molecular analysis of embryonic development in monotremes is needed to show how differentiation of two distinct hair cell types with unique numerical ratios in the medial and lateral aspect of the organ of Corti is guided. In essence, evolution of the organ of Corti could be an excellent case of sub-functionalization where differential expression regulation of diffusible factors (BMPs, Fgfs, Wnts) drives an innovative cellular arrangement largely specified by Delta/Notch interactions into a novel pattern enabling sound analysis over a vast range of frequencies.
In summary, once the first step of aggregation of neurosensory cell development into a discrete region was accomplished through either development of or co-option of a cellular GRN, further evolution followed the principles established for molecular and cellular evolution: multiply and diversify. Even such structurally and functionally different organs, such as the organ of Corti and the saccule, seem to form from a single anlage that needs to segregate properly (Daudet et al., 2007; Kopecky et al., 2011; Nichols et al., 2008) to enable normal regulation of expression of diffusible factors that drive the unusual cellular distribution and specializations of this most complicated cellular assembly in the vertebrate body.
C) Evolution of central projections and auditory nuclei
Processing of novel information requires that sensory organs not only are tuned to such novel stimuli but also transmit this information separately to the brain where it needs to be processed by a distinct set of neurons as a discrete sensory information. A prime example is the evolution of the basilar papilla/organ of Corti (Fig. 5) that projects in all tetrapods with a sound amplification capable middle ear (Manley and Sienknecht, 2013) to recognizable auditory nuclei of the brainstem (Grothe et al., 2004) using a dedicated set of sensory neurons (spiral ganglion neurons in eutherian mammals). In principle, there are three possible scenarios how this interconnected system of sound reception, information transmission and information processing could have evolved:
Outside-in with sensory organ evolution followed by sensory neuron evolution followed by auditory nuclei evolution
Inside-out starting from cochlear nuclei going backward to the auditory periphery.
Starting with the evolution of sensory neurons followed by sequential or simultaneous evolution of the sensory organ in the ear and the sensory nuclei in the brain.
Figure 5. Scheme of differences in vestibular, auditory, mechanoreception, and electroreception organ projections in the hindbrain as revealed by tracing experiments.
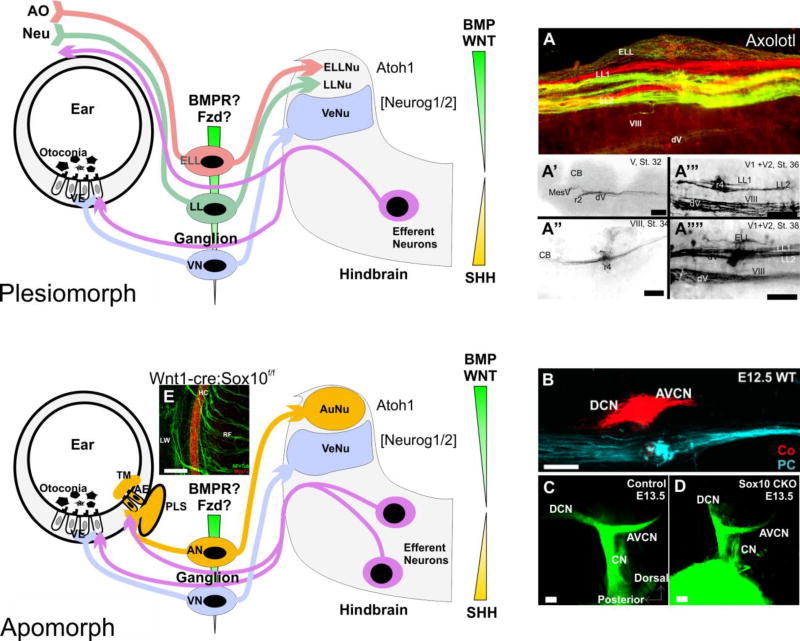
The various peripheral sensory organs [vestibular (VN), auditory (AN), mechanosensory lateral line (LL), or electrosensory ampullary organs (ELL)] each have unique central projections that terminate in distinct, non-overlapping regions in the hindbrain. The transition from aquatic, anamniotic vertebrates to terrestrial amniotic vertebrates was accompanied by the loss of all lateral line organs and the gain of auditory sound pressure receivers projecting via newly evolved auditory sensory neurons of the ear (AN) to apparently new auditory nuclei. In addition, efferent cells lose connections to mechanosensory lateral line and evolve new connections of a segregated population to auditory epithelia. Formation of central nuclei is mediated in part by bHLH transcription factors that also are important in ear neurosensory development (Atoh1, Neurog1) and their expression is specified by the dorso-ventral patterning morphogen gradients in the hindbrain (BMPs, Wnts and Shh). Afferent projection of different ganglia related to lateral line organs or other sensory modalities (A) develop progressively over time (A’, A”, A’”, A””). Likewise, different inner ear organs (B) project directly to their distinct target nuclei, possibly guided by morphogen gradients (indicated as colored wedges representing ligands in the brain and receptors in the ganglia). Elimination of Schwann cells through conditional deletion of Sox10 does not alter central projections (C,D) but partially disables afferents from finding the organ of Corti (E). Tectorial membrane, TM; auditory epithelia, AE; vestibular epithelia, AN, auditory neurons; VE, vestibular epithelia; VN, vestibular neurons; PLS, perilymphatic space. Modified after (Fritzsch et al., 2005a; Fritzsch et al., 2015b; Fritzsch et al., 2006b; Mao et al., 2014; Sienknecht et al., 2014).
Current information of trophic dependency strongly supports assumption 1. Sensory neurons depend on the peripheral target for viability (Fritzsch et al., 2016) and cochlear nuclei neurons depend on innervation for viability and differentiation much like other brainstem sensory neurons (Elliott et al., 2015; Rubel and Fritzsch, 2002). Conversely, lack of parts of auditory nuclei has only a limited effect on sensory neuron viability (Maricich et al., 2009) and hair cells depend on innervation only with a long delay (Kersigo and Fritzsch, 2015). During development, spiral ganglion neurons exit the cell cycle before hair cells and cochlear nucleus neurons (Altman and Bayer, 1980; Matei et al., 2005) and project both centrally and peripherally before their target cells start to differentiate (Fritzsch et al., 2005a; Fritzsch et al., 2005b). In fact, correct targeting of auditory afferents is possible without either hair cells or auditory nuclei indicating that other mechanisms guide afferents (Elliott et al., 2017). This aspect of development implies support for scenario 3, but might also represent a heterochronic shift in the developmental timetable.
Irrespective to this question, afferents need to respond to inner ear and CNS guidance cues to allow segregated projections to a unique inner ear sensory organ dedicated to sound perception as well as a unique central target able to process sound without vestibular stimulus mixing (Fig. 5). Salamanders lack a tympanic ear and show limited sensitivity to sound (Zeyl and Johnston, 2016) but have a basilar papilla with separate sensory projection within the CNS though they lack morphologically-distinct auditory nuclei (Herrick, 1948; Nieuwenhuys et al., 1998; Will and Fritzsch, 1988). Similar segregated projections in the absence of nuclei were found in the visual system (Fritzsch, 1980) and might indicate a more general evolutionary principle, segregated neuropils appear before segregated nuclei (Herrick, 1948). Details of the molecular basis of afferent segregation in the ear and to the CNS remain obscure but may be related to several known pathfinding molecules such Ephrins (Cramer and Gabriele, 2014; Siddiqui and Cramer, 2005), semaphorins, and other diffusible factors released from the hindbrain such as Wnts and Bmps (Henríquez and Osses, 2016; Seiradake et al., 2016) as well as cellular interactions. Clearly, Gata3 and Neurod1 are critical for this segregated projection from the ear to the cochlear nuclei (Fritzsch et al., 2006b; Goodrich, 2016; Jahan et al., 2010a) but exactly when the relevant genes were first expressed in the auditory inner ear neurons and how they tied into the novel pathfinding selection process to connect a novel periphery with a novel central target remains to be shown (Fritzsch et al., 2015b). Consistent with the ancestral split of a neurosensory cell with an axon into a hair cell and its sensory neuron is that central projections of sensory neurons do not depend on neural crest derived Schwann cells (Mao et al., 2014) whereas the peripheral projection is highly disorganized and many afferents miss the hair cells and sensory epithelia (Fig. 5). Moreover, at least in frogs, transplanted ears near the hindbrain can project directly to or reroute to the vestibular nuclei (Elliott et al., 2014), strongly supporting the notion that diffusible factors released from the hindbrain (Fig. 5) play a role in this process as substrate interactions are out of the question in these transplants. In contrast, inner ear afferents of ears transplanted to the orbit, enter randomly the midbrain (Elliott et al., 2013).
Irrespective to the sequence of formation of central projections, the spinal cord-like organization of the hindbrain has to be transformed into a hindbrain nuclear organization, necessitating alterations in functional longitudinal columns that extend from the spinal cord to the midbrain/hindbrain boundary (Albuixech-Crespo et al., 2017; Fritzsch and Glover, 2006; Nieuwenhuys and Puelles, 2015). Since vestibular, cochlear, mechanosensory, and electrosensory lateral line afferents are in all vertebrates restricted to the hindbrain (Fig. 5), the basic patterning of the spinal cord needs to be altered through the addition of those nuclei and/or transformation of existing spinal cord nuclei into nuclei able to receive and process this information distinctly. As it turns out, some of the transcription factors ensuring differentiation of the most dorsal spinal cord and hindbrain second order sensory nuclei development appear to be conserved from spinal cord to the cerebellum. Most dorsal in either is the expression of Atoh1, Neurog1/2 and Ascl1 (Bermingham et al., 2001; Fritzsch et al., 2006b) with Neurog1/2 being only transiently expressed. However, more recent data show subdivisions in these populations through the nested expression of various transcription factors that likely are driven by the intricate pattern generated by ventralizing Shh signal emitted from the floor plate and the dorsalizing Wnt/BMP signal emitted from the roof plate (Fritzsch et al., 2006b; Lai et al., 2016). The basic pattern of bHLH genes is supplemented by addition of other bHLH and homeobox factors (Hernandez-Miranda et al., 2016) and shows local, rhombomere-specific variation in the hindbrain. Most importantly, bHLH genes such as Ptf1a are both highly conserved in their expression (Sugahara et al., 2016) but also elicit a local rearrangement of nuclear development when mutated (Iskusnykh et al., 2016). How the GRNs of spinal cord cellular population are both systematically and rhombomere-specifically influenced to give rise to the four above mentioned nuclei dedicated to processing the information from the ear in vestibular and cochlear nuclei as well as the mechanosensory and electrosensory lateral line nuclei remains to be determined in the light of an apparently rather stable Hox code (Krumlauf, 2016; Murakami et al., 2004; Parker et al., 2016). What is beginning to emerge is that specific transcription factors can define specific subsets of brainstem nuclei such as the solitary tract (Qian et al., 2001) or auditory nuclei (Fritzsch et al., 2006b; Wang et al., 2005).
More recent data suggest that bHLH gene interactions and cross-regulation may be essential for different compartment specific alar plate differentiation. Chief among those is Ptf1a, a bHLH transcription factor that is essential for cerebellar (Pascual et al., 2007) (Millen et al., 2014) and dorsal cochlear nucleus development (Fujiyama et al., 2009) but other bHLH genes play a role as well (Cai et al., 2016). In addition, to be relevant for the formation of nuclei or brain areas, Ptf1a also plays an essential role in cell fate determination within a given nucleus defining excitatory and inhibitory neurons (Iskusnykh et al., 2016) Beyond formation of entire nuclei, several genes are now emerging that interact to define different cell types in different brainstem nuclei important for their normal function (Nothwang, 2016). Above we outlined how multiplication of existing cell fate determining bHLH genes provides the molecular start of distinct cell fate evolution (Arendt et al., 2016; Fritzsch et al., 2000) for neurosensory evolution of the ear. Here we suggest that the evolution of various rhombomere-specific hindbrain nuclei is a consequence of bHLH transcription factors that are variably tied into the Hox code regulation to achieve local cell fate changes and, by logical extension, formation of novel nuclei.
While cell fate switching during metamorphosis from lateral line to auditory (Herrick, 1948) was a reasonable idea in analogy to the well-established Reichert-Gaupp hypothesis of middle ear ossicle transformation (Maier and Ruf, 2016), it rather appears now that these nuclei actually die by apoptosis in frogs losing the lateral line (Fritzsch et al., 1988; Wahnschaffe et al., 1987) or co-exist in those retaining the lateral line. Formation of new nuclei is more likely to reflect assembly of novel interactions of the increasingly complex transcriptional landscape of the hindbrain that drives both expansion of cell populations through proliferation regulation and acquisition of novel phenotypes by regulatory changes of already complicated GRNs that evolved in the spinal cord to guide sophisticated, yet different, cell assemblies (Lai et al., 2016). In this context it is remarkable that some molecular conservation exists in peripheral pain receptors in skin and ear [Piezo1,2 (Wu et al., 2016)] and that loud sound causes pain sensation comparable to the pain perceived in the spinal cord.
In summary, it appears that gene regulatory network evolution is underlying both auditory neuron and auditory nuclei evolution. How the already established gene regulatory networks for inner ear neuron formation and hindbrain nuclear formation was nearly simultaneously modified to yield novel projections to process a novel stimulus acquired by a modified sensory epithelium dedicated for sound processing to be processed by a highly modified spinal cord neuronal network forming apparently novel auditory nuclei remains to be worked out.
Conclusion and outlook
This review summarizes evolutionary changes in molecular developmental gene regulatory networks that highlights specific, likely steps taken in the assembly of mechanosensory systems starting from a single celled organism ending with an integrated circuit consisting of an interactive neuronal connectome for auditory signal processing. While the broad outline of such changes appear to be highly supported by descriptive and experimental data, details in particular of the gene expression regulations underlying the functional diversification of multiplied GRNS remain obscure for the time being and require more work.
Highlights.
Molecular evolution predates cellular evolution predates organ and system evolution
Multiplication of transcription factors enables multiplications of cell types, expansion of cell types and segregation into new organ types.
Molecular evolution of the PNS and CNS are linked into morphogen diffusible factors.
Acknowledgments
This work was supported by NIH R01 DC005590 to BF and R03 DC015333 to KE.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahmed M, Wong EY, Sun J, Xu J, Wang F, Xu P-X. Eya1-Six1 interaction is sufficient to induce hair cell fate in the cochlea by activating Atoh1 expression in cooperation with Sox2. Developmental cell. 2012a;22:377–390. doi: 10.1016/j.devcel.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed M, Xu J, Xu P-X. EYA1 and SIX1 drive the neuronal developmental program in cooperation with the SWI/SNF chromatin-remodeling complex and SOX2 in the mammalian inner ear. Development. 2012b;139:1965–1977. doi: 10.1242/dev.071670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albuixech-Crespo B, López-Blanch L, Burguera D, Maeso I, Sánchez-Arrones L, Moreno-Bravo JA, Somorjai I, Pascual-Anaya J, Puelles E, Bovolenta P. Molecular regionalization of the developing amphioxus neural tube challenges major partitions of the vertebrate brain. PLoS biology. 2017;15:e2001573. doi: 10.1371/journal.pbio.2001573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsina B, Streit A. Morphogenetic Mechanisms of Inner Ear Development, Organogenetic Gene Networks. Springer; 2016. pp. 235–258. [Google Scholar]
- Altman J, Bayer SA. Development of the brain stem in the rat. III. Thymidine-radiographic study of the time of origin of neurons of the vestibular and auditory nuclei of the upper medulla. Journal of Comparative Neurology. 1980;194:877–904. doi: 10.1002/cne.901940410. [DOI] [PubMed] [Google Scholar]
- Arendt D, Musser JM, Baker CV, Bergman A, Cepko C, Erwin DH, Pavlicev M, Schlosser G, Widder S, Laubichler MD, Wagner GP. The origin and evolution of cell types. Nature reviews. Genetics. 2016;17:744–757. doi: 10.1038/nrg.2016.127. [DOI] [PubMed] [Google Scholar]
- Bates CM. Role of fibroblast growth factor receptor signaling in kidney development. Pediatric Nephrology. 2007;22:343–349. doi: 10.1007/s00467-006-0239-7. [DOI] [PubMed] [Google Scholar]
- Bellono NW, Leitch DB, Julius D. Molecular basis of ancestral vertebrate electroreception. Nature. 2017 doi: 10.1038/nature21401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benito-Gutiérrez È, Garcia-Fernàndez J, Comella JX. Origin and evolution of the Trk family of neurotrophic receptors. Molecular and Cellular Neuroscience. 2006;31:179–192. doi: 10.1016/j.mcn.2005.09.007. [DOI] [PubMed] [Google Scholar]
- Bermingham NA, Hassan BA, Price SD, Vollrath MA, Ben-Arie N, Eatock RA, Bellen HJ, Lysakowski A, Zoghbi HY. Math1: an essential gene for the generation of inner ear hair cells. Science. 1999;284:1837–1841. doi: 10.1126/science.284.5421.1837. [DOI] [PubMed] [Google Scholar]
- Bermingham NA, Hassan BA, Wang VY, Fernandez M, Banfi S, Bellen HJ, Fritzsch B, Zoghbi HY. Proprioceptor pathway development is dependent on Math1. Neuron. 2001;30:411–422. doi: 10.1016/s0896-6273(01)00305-1. [DOI] [PubMed] [Google Scholar]
- Birol O, Ohyama T, Edlund RK, Drakou K, Georgiades P, Groves AK. The mouse Foxi3 transcription factor is necessary for the development of posterior placodes. Developmental biology. 2016;409:139–151. doi: 10.1016/j.ydbio.2015.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boenigk J. The Choanoflagellates: Evolution, Biology and Ecology. Marine Biology Research. 2015;11:1118–1119. [Google Scholar]
- Booth DS, King N. Evolution: Gene regulation in transition. Nature. 2016 doi: 10.1038/nature18447. [DOI] [PubMed] [Google Scholar]
- Bouchard M, de Caprona D, Busslinger M, Xu P, Fritzsch B. Pax2 and Pax8 cooperate in mouse inner ear morphogenesis and innervation. BMC developmental biology. 2010;10:1. doi: 10.1186/1471-213X-10-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buisson I, Le Bouffant R, Futel M, Riou J-F, Umbhauer M. Pax8 and Pax2 are specifically required at different steps of Xenopus pronephros development. Developmental biology. 2015;397:175–190. doi: 10.1016/j.ydbio.2014.10.022. [DOI] [PubMed] [Google Scholar]
- Burke R, Angerer L, Elphick M, Humphrey G, Yaguchi S, Kiyama T, Liang S, Mu X, Agca C, Klein W. A genomic view of the sea urchin nervous system. Developmental biology. 2006;300:434–460. doi: 10.1016/j.ydbio.2006.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Kardon AP, Snyder LM, Kuzirian MS, Minestro S, de Souza L, Rubio ME, Maricich SM, Ross SE. Bhlhb5:: flpo allele uncovers a requirement for Bhlhb5 for the development of the dorsal cochlear nucleus. Developmental biology. 2016;414:149–160. doi: 10.1016/j.ydbio.2016.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang W, Brigande JV, Fekete DM, Wu DK. The development of semicircular canals in the inner ear: role of FGFs in sensory cristae. Development. 2004;131:4201–4211. doi: 10.1242/dev.01292. [DOI] [PubMed] [Google Scholar]
- Chellappa R, Li S, Pauley S, Jahan I, Jin K, Xiang M. Barhl1 regulatory sequences required for cell-specific gene expression and autoregulation in the inner ear and central nervous system. Molecular and cellular biology. 2008;28:1905–914. doi: 10.1128/MCB.01454-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Anderson L. The inner ear structures of the echidna—an SEM study. Experientia. 1985;41:1324–1326. [Google Scholar]
- Chen J, Streit A. A medium-scale assay for enhancer validation in amniotes. Developmental Dynamics. 2015;244:1291–1299. doi: 10.1002/dvdy.24306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer KS, Gabriele ML. Axon guidance in the auditory system: Multiple functions of Eph receptors. Neuroscience. 2014;277:152–162. doi: 10.1016/j.neuroscience.2014.06.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daudet N, Ariza-McNaughton L, Lewis J. Notch signalling is needed to maintain, but not to initiate, the formation of prosensory patches in the chick inner ear. Development. 2007;134:2369–2378. doi: 10.1242/dev.001842. [DOI] [PubMed] [Google Scholar]
- Daudet N, Lewis J. Two contrasting roles for Notch activity in chick inner ear development: specification of prosensory patches and lateral inhibition of hair-cell differentiation. Development. 2005;132:541–551. doi: 10.1242/dev.01589. [DOI] [PubMed] [Google Scholar]
- Domínguez-Frutos E, López-Hernández I, Vendrell V, Neves J, Gallozzi M, Gutsche K, Quintana L, Sharpe J, Knoepfler PS, Eisenman RN. N-myc controls proliferation, morphogenesis, and patterning of the inner ear. The Journal of Neuroscience. 2011;31:7178–7189. doi: 10.1523/JNEUROSCI.0785-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudderidge TJ, Stoeber K, Loddo M, Atkinson G, Fanshawe T, Griffiths DF, Williams GH. Mcm2, Geminin, and KI67 define proliferative state and are prognostic markers in renal cell carcinoma. Clinical cancer research. 2005;11:2510–2517. doi: 10.1158/1078-0432.CCR-04-1776. [DOI] [PubMed] [Google Scholar]
- Duncan JS, Fritzsch B. Continued expression of GATA3 is necessary for cochlear neurosensory development. PloS one. 2013;8:e62046. doi: 10.1371/journal.pone.0062046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvorakova M, Jahan I, Macova I, Chumak T, Bohuslavova R, Syka J, Fritzsch B, Pavlinkova G. Incomplete and delayed Sox2 deletion defines residual ear neurosensory development and maintenance. Sci Rep. 2016;6:38253. doi: 10.1038/srep38253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott K, Kersigo J, Pan N, Jahan I, Fritzsch B. Spiral Ganglion Neuron Projection Development to the hindbrain in mice lacking peripheral and/or central target differentiation. Frontiers in Neural Circuits. 2017:11. doi: 10.3389/fncir.2017.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott KL, Houston DW, DeCook R, Fritzsch B. Ear manipulations reveal a critical period for survival and dendritic development at the single-cell level in Mauthner neurons. Developmental neurobiology. 2015;75:1339–1351. doi: 10.1002/dneu.22287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott KL, Houston DW, Fritzsch B. Transplantation of Xenopus laevis tissues to determine the ability of motor neurons to acquire a novel target. PloS one. 2013;8:e55541. doi: 10.1371/journal.pone.0055541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott KL, Houston DW, Fritzsch B. Sensory afferent segregation in three-eared frogs resemble the dominance columns observed in three-eyed frogs. Scientific reports. 2014;5:8338–8338. doi: 10.1038/srep08338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fariñas I, Jones KR, Tessarollo L, Vigers AJ, Huang E, Kirstein M, De Caprona DC, Coppola V, Backus C, Reichardt LF. Spatial shaping of cochlear innervation by temporally regulated neurotrophin expression. The Journal of Neuroscience. 2001;21:6170–6180. doi: 10.1523/JNEUROSCI.21-16-06170.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francey LJ, Conlin LK, Kadesch HE, Clark D, Berrodin D, Sun Y, Glessner J, Hakonarson H, Jalas C, Landau C, Spinner NB, Kenna M, Sagi M, Rehm HL, Krantz ID. Genome-wide SNP genotyping identifies the Stereocilin (STRC) gene as a major contributor to pediatric bilateral sensorineural hearing impairment. Am J Med Genet A. 2012;158a:298–308. doi: 10.1002/ajmg.a.34391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzsch B. Retinal projections in European salamandridae. Cell and tissue research. 1980;213:325–341. doi: 10.1007/BF00234791. [DOI] [PubMed] [Google Scholar]
- Fritzsch B. Inner ear of the coelacanth fish Latimeria has tetrapod affinities. Nature. 1987;327:153–154. doi: 10.1038/327153a0. [DOI] [PubMed] [Google Scholar]
- Fritzsch B. The water-to-land transition: evolution of the tetrapod basilar papilla, middle ear, and auditory nuclei, The evolutionary biology of hearing. Springer; 1992. pp. 351–375. [Google Scholar]
- Fritzsch B, Barald KF, Lomax MI. Early embryology of the vertebrate ear, Development of the auditory system. Springer; 1998. pp. 80–145. [Google Scholar]
- Fritzsch B, Beisel K. Molecular conservation and novelties in vertebrate ear development. Current topics in developmental biology. 2003;57:1–44. doi: 10.1016/s0070-2153(03)57001-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzsch B, Beisel K, Jones K, Farinas I, Maklad A, Lee J, Reichardt L. Development and evolution of inner ear sensory epithelia and their innervation. Journal of neurobiology. 2002;53:143–156. doi: 10.1002/neu.10098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzsch B, Beisel KW, Bermingham NA. Developmental evolutionary biology of the vertebrate ear: conserving mechanoelectric transduction and developmental pathways in diverging morphologies. Neuroreport. 2000;11:R35–R44. doi: 10.1097/00001756-200011270-00013. [DOI] [PubMed] [Google Scholar]
- Fritzsch B, Beisel KW, Hansen LA. The molecular basis of neurosensory cell formation in ear development: a blueprint for hair cell and sensory neuron regeneration? Bioessays. 2006a;28:1181–1193. doi: 10.1002/bies.20502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzsch B, Beisel KW, Pauley S, Soukup G. Molecular evolution of the vertebrate mechanosensory cell and ear. The International journal of developmental biology. 2007;51:663. doi: 10.1387/ijdb.072367bf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzsch B, Eberl DF, Beisel KW. The role of bHLH genes in ear development and evolution: revisiting a 10-year-old hypothesis. Cellular and molecular life sciences. 2010;67:3089–3099. doi: 10.1007/s00018-010-0403-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzsch B, Elliott KL. Evolution and Development of the Inner Ear Efferent System: Transforming a Motor Neuron Population to Connect to the Most Unusual Motor Protein via Ancient Nicotinic Receptors. Frontiers in Cellular Neuroscience. 2017;11:114. doi: 10.3389/fncel.2017.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzsch B, Glover J. Evolution of the Deuterostome Central Nervous System: An Intercalation of Developmental Patterning Processes with Cellular Specification Processes-2.01 2006 [Google Scholar]
- Fritzsch B, Gregory D, Rosa-Molinar E. The development of the hindbrain afferent projections in the axolotl: evidence for timing as a specific mechanism of afferent fiber sorting. Zoology. 2005a;108:297–306. doi: 10.1016/j.zool.2005.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzsch B, Jahan I, Pan N, Elliott KL. Evolving gene regulatory networks into cellular networks guiding adaptive behavior: an outline how single cells could have evolved into a centralized neurosensory system. Cell and tissue research. 2015a;359:295–313. doi: 10.1007/s00441-014-2043-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzsch B, Kersigo J, Yang T, Jahan I, Pan N. Neurotrophic Factor Function During Ear Development: Expression Changes Define Critical Phases for Neuronal Viability, The Primary Auditory Neurons of the Mammalian Cochlea. Springer; New York: 2016. pp. 49–84. [Google Scholar]
- Fritzsch B, Matei VA, Nichols DH, Bermingham N, Jones K, Beisel KW, Wang VY. Atoh1 null mice show directed afferent fiber growth to undifferentiated ear sensory epithelia followed by incomplete fiber retention. Dev Dyn. 2005b;233:570–583. doi: 10.1002/dvdy.20370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzsch B, Pan N, Jahan I, Duncan JS, Kopecky BJ, Elliott KL, Kersigo J, Yang T. Evolution and development of the tetrapod auditory system: an organ of Corti-centric perspective. Evolution & development. 2013;15:63–79. doi: 10.1111/ede.12015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzsch B, Pan N, Jahan I, Elliott KL. Inner ear development: building a spiral ganglion and an organ of Corti out of unspecified ectoderm. Cell and tissue research. 2015b;361:7–24. doi: 10.1007/s00441-014-2031-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzsch B, Pauley S, Feng F, Matei V, Nichols D. The molecular and developmental basis of the evolution of the vertebrate auditory system. International Journal of Comparative Psychology. 2006b:19. [Google Scholar]
- Fritzsch B, Signore M, Simeone A. Otx1 null mutant mice show partial segregation of sensory epithelia comparable to lamprey ears. Development genes and evolution. 2001;211:388–396. doi: 10.1007/s004270100166. [DOI] [PubMed] [Google Scholar]
- Fritzsch B, Straka H. Evolution of vertebrate mechanosensory hair cells and inner ears: toward identifying stimuli that select mutation driven altered morphologies. J Comp Physiol A. 2014;200:5–18. doi: 10.1007/s00359-013-0865-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzsch B, Wahnschaffe U, Bartsch U. The evolution of the amphibian auditory system. Wiley; New York: 1988. Metamorphic changes in the octavolateralis system of amphibians; pp. 359–376. [Google Scholar]
- Fritzsch B, Wake M. The inner ear of gymnophione amphibians and its nerve supply: a comparative study of regressive events in a complex sensory system (Amphibia, Gymnophiona) Zoomorphology. 1988;108:201–217. [Google Scholar]
- Fujiyama T, Yamada M, Terao M, Terashima T, Hioki H, Inoue YU, Inoue T, Masuyama N, Obata K, Yanagawa Y. Inhibitory and excitatory subtypes of cochlear nucleus neurons are defined by distinct bHLH transcription factors, Ptf1a and Atoh1. Development. 2009;136:2049–2058. doi: 10.1242/dev.033480. [DOI] [PubMed] [Google Scholar]
- Gasparini F, Degasperi V, Shimeld SM, Burighel P, Manni L. Evolutionary conservation of the placodal transcriptional network during sexual and asexual development in chordates. Dev Dynam. 2013;242:752–766. doi: 10.1002/dvdy.23957. [DOI] [PubMed] [Google Scholar]
- Goodrich LV. Early Development of the Spiral Ganglion, The Primary Auditory Neurons of the Mammalian Cochlea. Springer; 2016. pp. 11–48. [Google Scholar]
- Gowan K, Helms AW, Hunsaker TL, Collisson T, Ebert PJ, Odom R, Johnson JE. Crossinhibitory activities of Ngn1 and Math1 allow specification of distinct dorsal interneurons. Neuron. 2001;31:219–232. doi: 10.1016/s0896-6273(01)00367-1. [DOI] [PubMed] [Google Scholar]
- Grothe B, Carr CE, Casseday JH, Fritzsch B, Köppl C. The evolution of central pathways and their neural processing patterns, Evolution of the vertebrate auditory system. Springer; 2004. pp. 289–359. [Google Scholar]
- Groves AK, Fekete DM. Shaping sound in space: the regulation of inner ear patterning. Development. 2012;139:245–257. doi: 10.1242/dev.067074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallböök F, Wilson K, Thorndyke M, Olinski RP. Formation and evolution of the chordate neurotrophin and Trk receptor genes. Brain, behavior and evolution. 2006;68:133–144. doi: 10.1159/000094083. [DOI] [PubMed] [Google Scholar]
- Helms AW, Abney AL, Ben-Arie N, Zoghbi HY, Johnson JE. Autoregulation and multiple enhancers control Math1 expression in the developing nervous system. Development. 2000;127:1185–1196. doi: 10.1242/dev.127.6.1185. [DOI] [PubMed] [Google Scholar]
- Henríquez JP, Osses N. Editorial: Morphogens in the Wiring of the Nervous System. Frontiers in cellular neuroscience. 2016;9:502. doi: 10.3389/fncel.2015.00502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Miranda LR, Müller T, Birchmeier C. The dorsal spinal cord and hindbrain: from developmental mechanisms to functional circuits. Developmental Biology. 2016 doi: 10.1016/j.ydbio.2016.10.008. [DOI] [PubMed] [Google Scholar]
- Herrick CJ. The brain of the tiger salamander. University of Chicago Press; Chicago: 1948. [Google Scholar]
- Hertzano R, Montcouquiol M, Rashi-Elkeles S, Elkon R, Yücel R, Frankel WN, Rechavi G, Möröy T, Friedman TB, Kelley MW. Transcription profiling of inner ears from Pou4f3ddl/ddl identifies Gfi1 as a target of the Pou4f3 deafness gene. Human molecular genetics. 2004;13:2143–2153. doi: 10.1093/hmg/ddh218. [DOI] [PubMed] [Google Scholar]
- Huang EJ, Liu W, Fritzsch B, Bianchi LM, Reichardt LF, Xiang M. Brn3a is a transcriptional regulator of soma size, target field innervation and axon pathfinding of inner ear sensory neurons. Development. 2001;128:2421–2432. doi: 10.1242/dev.128.13.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang CH, Simeone A, Lai E, Wu DK. Foxg1 is required for proper separation and formation of sensory cristae during inner ear development. Developmental Dynamics. 2009;238:2725–2734. doi: 10.1002/dvdy.22111. [DOI] [PubMed] [Google Scholar]
- Ikeda R, Pak K, Chavez E, Ryan AF. Transcription factors with conserved binding sites near ATOH1 on the POU4F3 gene enhance the induction of cochlear hair cells. Molecular neurobiology. 2015;51:672–684. doi: 10.1007/s12035-014-8801-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iskusnykh IY, Steshina EY, Chizhikov VV. Loss of Ptf1a Leads to a Widespread Cell-Fate Misspecification in the Brainstem, Affecting the Development of Somatosensory and Viscerosensory Nuclei. The Journal of Neuroscience. 2016;36:2691–2710. doi: 10.1523/JNEUROSCI.2526-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs DK, Nakanishi N, Yuan D, Camara A, Nichols SA, Hartenstein V. Evolution of sensory structures in basal metazoa. Integrative and Comparative Biology. 2007;47:712–723. doi: 10.1093/icb/icm094. [DOI] [PubMed] [Google Scholar]
- Jahan I, Kersigo J, Pan N, Fritzsch B. Neurod1 regulates survival and formation of connections in mouse ear and brain. Cell and tissue research. 2010a;341:95–110. doi: 10.1007/s00441-010-0984-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahan I, Pan N, Elliott KL, Fritzsch B. The quest for restoring hearing: understanding ear development more completely. Bioessays. 2015a;37:1016–1027. doi: 10.1002/bies.201500044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahan I, Pan N, Kersigo J, Calisto LE, Morris KA, Kopecky B, Duncan JS, Beisel KW, Fritzsch B. Expression of Neurog1 instead of Atoh1 can partially rescue organ of Corti cell survival. PLoS One. 2012;7:e30853. doi: 10.1371/journal.pone.0030853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahan I, Pan N, Kersigo J, Fritzsch B. Neurod1 suppresses hair cell differentiation in ear ganglia and regulates hair cell subtype development in the cochlea. PloS one. 2010b;5:e11661. doi: 10.1371/journal.pone.0011661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahan I, Pan N, Kersigo J, Fritzsch B. Beyond generalized hair cells: molecular cues for hair cell types. Hearing research. 2013;297:30–41. doi: 10.1016/j.heares.2012.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahan I, Pan N, Kersigo J, Fritzsch B. Neurog1 can partially substitute for Atoh1 function in hair cell differentiation and maintenance during organ of Corti development. Development. 2015b;142:2810–2821. doi: 10.1242/dev.123091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janesick A, Abbey R, Chung C, Liu S, Taketani M, Blumberg B. ERF and ETV3L are retinoic acid-inducible repressors required for primary neurogenesis. Development. 2013;140:3095–3106. doi: 10.1242/dev.093716. [DOI] [PubMed] [Google Scholar]
- Jones C, Roper VC, Foucher I, Qian D, Banizs B, Petit C, Yoder BK, Chen P. Ciliary proteins link basal body polarization to planar cell polarity regulation. Nat Genet. 2008;40:69–77. doi: 10.1038/ng.2007.54. [DOI] [PubMed] [Google Scholar]
- Karis A, Pata I, van Doorninck JH, Grosveld F, de Zeeuw CI, de Caprona D, Fritzsch B. Transcription factor GATA-3 alters pathway selection of olivocochlear neurons and affects morphogenesis of the ear. Journal of Comparative Neurology. 2001;429:615–630. doi: 10.1002/1096-9861(20010122)429:4<615::aid-cne8>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- Kersigo J, Fritzsch B. Inner ear hair cells deteriorate in mice engineered to have no or diminished innervation. Aging, neurogenesis and neuroinflammation in hearing loss and protection. 2015:22. doi: 10.3389/fnagi.2015.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatri SB, Edlund RK, Groves AK. Foxi3 is necessary for the induction of the chick otic placode in response to FGF signaling. Developmental biology. 2014;391:158–169. doi: 10.1016/j.ydbio.2014.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W-Y, Fritzsch B, Serls A, Bakel LA, Huang EJ, Reichardt LF, Barth DS, Lee JE. NeuroD-null mice are deaf due to a severe loss of the inner ear sensory neurons during development. Development. 2001;128:417–426. doi: 10.1242/dev.128.3.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo SK, Hill JK, Hwang CH, Lin ZS, Millen KJ, Wu DK. Lmx1a maintains proper neurogenic, sensory, and non-sensory domains in the mammalian inner ear. Developmental biology. 2009;333:14–25. doi: 10.1016/j.ydbio.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopecky B, Santi P, Johnson S, Schmitz H, Fritzsch B. Conditional deletion of N-Myc disrupts neurosensory and non-sensory development of the ear. Developmental Dynamics. 2011;240:1373–1390. doi: 10.1002/dvdy.22620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korrapati S, Roux I, Glowatzki E, Doetzlhofer A. Notch signaling limits supporting cell plasticity in the hair cell-damaged early postnatal murine cochlea. PLoS One. 2013;8:e73276. doi: 10.1371/journal.pone.0073276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumlauf R. Chapter Thirty-Four-Hox Genes and the Hindbrain: A Study in Segments. Current topics in developmental biology. 2016;116:581–596. doi: 10.1016/bs.ctdb.2015.12.011. [DOI] [PubMed] [Google Scholar]
- Ladhams A, Pickles J. Morphology of the monotreme organ of Corti and macula lagena. Journal of Comparative Neurology. 1996;366:335–347. doi: 10.1002/(SICI)1096-9861(19960304)366:2<335::AID-CNE11>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Lai HC, Seal RP, Johnson JE. Making sense out of spinal cord somatosensory development. Development. 2016;143:3434–3448. doi: 10.1242/dev.139592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leadbeater BS. The Choanoflagellates. Cambridge University Press; 2015. [Google Scholar]
- Lewis ER, Leverenz EL, Bialek WS. The vertebrate inner ear. CRC PressI Llc; 1985. [Google Scholar]
- Liu M, Pereira FA, Price SD, Chu M-j, Shope C, Himes D, Eatock RA, Brownell WE, Lysakowski A, Tsai M-J. Essential role of BETA2/NeuroD1 in development of the vestibular and auditory systems. Genes & development. 2000;14:2839–2854. doi: 10.1101/gad.840500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X-P, Koehler KR, Mikosz AM, Hashino E, Holt JR. Functional development of mechanosensitive hair cells in stem cell-derived organoids parallels native vestibular hair cells. Nature communications. 2016:7. doi: 10.1038/ncomms11508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Sipe CW. Developmental regulation of planar cell polarity and hair-bundle morphogenesis in auditory hair cells: lessons from human and mouse genetics. Wiley Interdisciplinary Reviews: Developmental Biology. 2016;5:85–101. doi: 10.1002/wdev.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Force A. The probability of duplicate gene preservation by subfunctionalization. Genetics. 2000;154:459–473. doi: 10.1093/genetics/154.1.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q, Anderson DJ, Fritzsch B. Neurogenin 1 null mutant ears develop fewer, morphologically normal hair cells in smaller sensory epithelia devoid of innervation. Journal of the Association for Research in Otolaryngology. 2000;1:129–143. doi: 10.1007/s101620010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q, Chen Z, del Barco Barrantes I, de la Pompa JL, Anderson DJ. neurogenin1 is essential for the determination of neuronal precursors for proximal cranial sensory ganglia. Neuron. 1998;20:469–482. doi: 10.1016/s0896-6273(00)80988-5. [DOI] [PubMed] [Google Scholar]
- Maier EC, Saxena A, Alsina B, Bronner ME, Whitfield TT. Sensational placodes: Neurogenesis in the otic and olfactory systems. Developmental biology. 2014;389:50–67. doi: 10.1016/j.ydbio.2014.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier W, Ruf I. Evolution of the mammalian middle ear: a historical review. Journal of anatomy. 2016;228:270–283. doi: 10.1111/joa.12379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maklad A, Reed C, Johnson NS, Fritzsch B. Anatomy of the lamprey ear: morphological evidence for occurrence of horizontal semicircular ducts in the labyrinth of Petromyzon marinus. Journal of anatomy. 2014;224:432–446. doi: 10.1111/joa.12159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manley GA, Sienknecht UJ. The evolution and development of middle ears in land vertebrates, The Middle Ear. Springer; 2013. pp. 7–30. [Google Scholar]
- Mao Y, Reiprich S, Wegner M, Fritzsch B. Targeted deletion of Sox10 by Wnt1-cre defects neuronal migration and projection in the mouse inner ear. PloS one. 2014;9:e94580. doi: 10.1371/journal.pone.0094580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maricich SM, Xia A, Mathes EL, Wang VY, Oghalai JS, Fritzsch B, Zoghbi HY. Atoh1-lineal neurons are required for hearing and for the survival of neurons in the spiral ganglion and brainstem accessory auditory nuclei. The Journal of Neuroscience. 2009;29:11123–11133. doi: 10.1523/JNEUROSCI.2232-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matei V, Pauley S, Kaing S, Rowitch D, Beisel K, Morris K, Feng F, Jones K, Lee J, Fritzsch B. Smaller inner ear sensory epithelia in Neurog1 null mice are related to earlier hair cell cycle exit. Developmental Dynamics. 2005;234:633–650. doi: 10.1002/dvdy.20551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matus DQ, Pang K, Daly M, Martindale MQ. Expression of Pax gene family members in the anthozoan cnidarian, Nematostella vectensis. Evolution & development. 2007;9:25–38. doi: 10.1111/j.1525-142X.2006.00135.x. [DOI] [PubMed] [Google Scholar]
- May-Simera HL, Petralia RS, Montcouquiol M, Wang Y-X, Szarama KB, Liu Y, Lin W, Deans MR, Pazour GJ, Kelley MW. Ciliary proteins Bbs8 and Ift20 promote planar cell polarity in the cochlea. Development. 2015;142:555–566. doi: 10.1242/dev.113696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon AP. Chapter Three-Development of the Mammalian Kidney. Current topics in developmental biology. 2016;117:31–64. doi: 10.1016/bs.ctdb.2015.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinhardt H. Models for patterning primary embryonic body axes: the role of space and time, Seminars in cell & developmental biology. Elsevier; 2015. pp. 103–117. [DOI] [PubMed] [Google Scholar]
- Millen KJ, Steshina EY, Iskusnykh IY, Chizhikov VV. Transformation of the cerebellum into more ventral brainstem fates causes cerebellar agenesis in the absence of Ptf1a function. Proceedings of the National Academy of Sciences. 2014;111:E1777–E1786. doi: 10.1073/pnas.1315024111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morsli H, Tuorto F, Choo D, Postiglione MP, Simeone A, Wu DK. Otx1 and Otx2 activities are required for the normal development of the mouse inner ear. Development. 1999;126:2335–2343. doi: 10.1242/dev.126.11.2335. [DOI] [PubMed] [Google Scholar]
- Moteki H, Azaiez H, Sloan-Heggen CM, Booth K, Nishio SY, Wakui K, Yamaguchi T, Kolbe DL, Iwasa YI, Shearer AE, Fukushima Y, Smith RJ, Usami SI. Detection and Confirmation of Deafness-Causing Copy Number Variations in the STRC Gene by Massively Parallel Sequencing and Comparative Genomic Hybridization. Ann Otol Rhinol Laryngol. 2016;125:918–923. doi: 10.1177/0003489416661345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulvaney J, Dabdoub A. Atoh1, an essential transcription factor in neurogenesis and intestinal and inner ear development: function, regulation, and context dependency. Journal of the Association for Research in Otolaryngology. 2012;13:281–293. doi: 10.1007/s10162-012-0317-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munnamalai V, Fekete DM. Notch-Wnt-Bmp crosstalk regulates radial patterning in the mouse cochlea in a spatiotemporal manner. Development. 2016;143:4003–4015. doi: 10.1242/dev.139469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami Y, Pasqualetti M, Takio Y, Hirano S, Rijli FM, Kuratani S. Segmental development of reticulospinal and branchiomotor neurons in lamprey: insights into the evolution of the vertebrate hindbrain. Development. 2004;131:983–995. doi: 10.1242/dev.00986. [DOI] [PubMed] [Google Scholar]
- Nichols DH, Pauley S, Jahan I, Beisel KW, Millen KJ, Fritzsch B. Lmx1a is required for segregation of sensory epithelia and normal ear histogenesis and morphogenesis. Cell and tissue research. 2008;334:339–358. doi: 10.1007/s00441-008-0709-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwenhuys R, Puelles L. Towards a new neuromorphology. Springer; 2015. [Google Scholar]
- Nieuwenhuys R, Ten Donkelaar H, Nicholson C. The Central Nervous System of Vertebrates. Vol. 2. Springer; London: 1998. [Google Scholar]
- Nothwang HG. Evolution of mammalian sound localization circuits: A developmental perspective. Prog Neurobiol. 2016;141:1–24. doi: 10.1016/j.pneurobio.2016.02.003. [DOI] [PubMed] [Google Scholar]
- O’Neill P, Mak S-S, Fritzsch B, Ladher RK, Baker CV. The amniote paratympanic organ develops from a previously undiscovered sensory placode. Nature communications. 2012;3:1041. doi: 10.1038/ncomms2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. Evolution by gene duplication. Springer Science & Business Media; 2013. [Google Scholar]
- Ohyama T, Groves AK, Martin K. The first steps towards hearing: mechanisms of otic placode induction. International Journal of Developmental Biology. 2007;51:463–472. doi: 10.1387/ijdb.072320to. [DOI] [PubMed] [Google Scholar]
- Okoruwa OE, Weston MD, Sanjeevi DC, Millemon AR, Fritzsch B, Hallworth R, Beisel KW. Evolutionary insights into the unique electromotility motor of mammalian outer hair cells. Evolution & development. 2008;10:300–315. doi: 10.1111/j.1525-142X.2008.00239.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan N, Jahan I, Kersigo J, Duncan JS, Kopecky B, Fritzsch B. A novel Atoh1 “self- terminating” mouse model reveals the necessity of proper Atoh1 level and duration for hair cell differentiation and viability. PLoS One. 2012a;7:e30358. doi: 10.1371/journal.pone.0030358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan N, Jahan I, Kersigo J, Kopecky B, Santi P, Johnson S, Schmitz H, Fritzsch B. Conditional deletion of Atoh1 using Pax2-Cre results in viable mice without differentiated cochlear hair cells that have lost most of the organ of Corti. Hear Res. 2011;275:66–80. doi: 10.1016/j.heares.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan N, Jahan I, Lee JE, Fritzsch B. Defects in the cerebella of conditional Neurod1 null mice correlate with effective Tg (Atoh1-cre) recombination and granule cell requirements for Neurod1 for differentiation. Cell and tissue research. 2009;337:407–428. doi: 10.1007/s00441-009-0826-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan N, Kopecky B, Jahan I, Fritzsch B. Understanding the evolution and development of neurosensory transcription factors of the ear to enhance therapeutic translation. Cell and tissue research. 2012b;349:415–432. doi: 10.1007/s00441-012-1454-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos T, Krochmal M, Cisek K, Fernandes M, Husi H, Stevens R, Bascands J-L, Schanstra JP, Klein J. Omics databases on kidney disease: where they can be found and how to benefit from them. Clinical kidney journal. 2016;9:343–352. doi: 10.1093/ckj/sfv155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker HJ, Bronner ME, Krumlauf R. The vertebrate Hox gene regulatory network for hindbrain segmentation: Evolution and diversification. Bioessays. 2016;38:526–538. doi: 10.1002/bies.201600010. [DOI] [PubMed] [Google Scholar]
- Pascual M, Abasolo I, Mingorance-Le Meur A, Martínez A, Del Rio JA, Wright CV, Real FX, Soriano E. Cerebellar GABAergic progenitors adopt an external granule cell-like phenotype in the absence of Ptf1a transcription factor expression. Proceedings of the National Academy of Sciences. 2007;104:5193–5198. doi: 10.1073/pnas.0605699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patthey C, Schlosser G, Shimeld SM. The evolutionary history of vertebrate cranial placodes-I: cell type evolution. Developmental biology. 2014;389:82–97. doi: 10.1016/j.ydbio.2014.01.017. [DOI] [PubMed] [Google Scholar]
- Pauley S, Lai E, Fritzsch B. Foxg1 is required for morphogenesis and histogenesis of the mammalian inner ear. Developmental Dynamics. 2006;235:2470–2482. doi: 10.1002/dvdy.20839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauley S, Wright TJ, Pirvola U, Ornitz D, Beisel K, Fritzsch B. Expression and function of FGF10 in mammalian inner ear development. Developmental dynamics. 2003;227:203–215. doi: 10.1002/dvdy.10297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter I, Davidson EH. Genomic control process: development and evolution. Academic Press; 2015. [Google Scholar]
- Pierce ML, Weston MD, Fritzsch B, Gabel HW, Ruvkun G, Soukup GA. MicroRNA-183 family conservation and ciliated neurosensory organ expression. Evolution & development. 2008;10:106–113. doi: 10.1111/j.1525-142X.2007.00217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirvola U, Spencer-Dene B, Xing-Qun L, Kettunen P, Thesleff I, Fritzsch B, Dickson C, Ylikoski J. FGF/FGFR-2 (IIIb) signaling is essential for inner ear morphogenesis. The Journal of Neuroscience. 2000;20:6125–6134. doi: 10.1523/JNEUROSCI.20-16-06125.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puligilla C, Feng F, Ishikawa K, Bertuzzi S, Dabdoub A, Griffith AJ, Fritzsch B, Kelley MW. Disruption of fibroblast growth factor receptor 3 signaling results in defects in cellular differentiation, neuronal patterning, and hearing impairment. Developmental Dynamics. 2007;236:1905–1917. doi: 10.1002/dvdy.21192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Y, Fritzsch B, Shirasawa S, Chen C-L, Choi Y, Ma Q. Formation of brainstem (nor) adrenergic centers and first-order relay visceral sensory neurons is dependent on homeodomain protein Rnx/Tlx3. Genes & development. 2001;15:2533–2545. doi: 10.1101/gad.921501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quint E, Steel KP. Use of mouse genetics for studying inner ear development. Current topics in developmental biology. 2003;57:45–83. doi: 10.1016/s0070-2153(03)57002-8. [DOI] [PubMed] [Google Scholar]
- Raft S, Groves AK. Segregating neural and mechanosensory fates in the developing ear: patterning, signaling, and transcriptional control. Cell and tissue research. 2015;359:315–332. doi: 10.1007/s00441-014-1917-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raft S, Koundakjian EJ, Quinones H, Jayasena CS, Goodrich LV, Johnson JE, Segil N, Groves AK. Cross-regulation of Ngn1 and Math1 coordinates the production of neurons and sensory hair cells during inner ear development. Development. 2007;134:4405–4415. doi: 10.1242/dev.009118. [DOI] [PubMed] [Google Scholar]
- Richter C-P, Emadi G, Getnick G, Quesnel A, Dallos P. Tectorial membrane stiffness gradients. Biophysical journal. 2007;93:2265–2276. doi: 10.1529/biophysj.106.094474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddiford N, Schlosser G. Dissecting the pre-placodal transcriptome to reveal presumptive direct targets of Six1 and Eya1 in cranial placodes. eLife. 2016;5:e17666. doi: 10.7554/eLife.17666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossini M, Cheunsuchon B, Donnert E, Ma L-J, Thomas JW, Neilson EG, Fogo AB. Immunolocalization of fibroblast growth factor-1 (FGF-1), its receptor (FGFR-1), and fibroblast-specific protein-1 (FSP-1) in inflammatory renal disease. Kidney international. 2005;68:2621–2628. doi: 10.1111/j.1523-1755.2005.00734.x. [DOI] [PubMed] [Google Scholar]
- Rubel EW, Fritzsch B. Auditory system development: primary auditory neurons and their targets. Annual review of neuroscience. 2002;25:51–101. doi: 10.1146/annurev.neuro.25.112701.142849. [DOI] [PubMed] [Google Scholar]
- Schlosser G, Patthey C, Shimeld SM. The evolutionary history of vertebrate cranial placodes II. Evolution of ectodermal patterning. Developmental biology. 2014;389:98–119. doi: 10.1016/j.ydbio.2014.01.019. [DOI] [PubMed] [Google Scholar]
- Schüler S, Hauptmann J, Perner B, Kessels MM, Englert C, Qualmann B. Ciliated sensory hair cell formation and function require the F-BAR protein syndapin I and the WH2 domain-based actin nucleator Cobl. J Cell Sci. 2013;126:196–208. doi: 10.1242/jcs.111674. [DOI] [PubMed] [Google Scholar]
- Schultz JA, Zeller U, Luo ZX. Inner ear labyrinth anatomy of monotremes and implications for mammalian inner ear evolution. Journal of Morphology. 2016 doi: 10.1002/jmor.20632. [DOI] [PubMed] [Google Scholar]
- Sebé-Pedrós A, Ballaré C, Parra-Acero H, Chiva C, Tena JJ, Sabidó E, Gómez-Skarmeta JL, Di Croce L, Ruiz-Trillo I. The Dynamic Regulatory Genome of Capsaspora and the Origin of Animal Multicellularity. Cell. 2016;165:1224–1237. doi: 10.1016/j.cell.2016.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiradake E, Jones EY, Klein R. Structural Perspectives on Axon Guidance. Annual Review of Cell and Developmental Biology. 2016;32:577–608. doi: 10.1146/annurev-cellbio-111315-125008. [DOI] [PubMed] [Google Scholar]
- Seo S, Herr A, Lim J-W, Richardson GA, Richardson H, Kroll KL. Geminin regulates neuronal differentiation by antagonizing Brg1 activity. Genes & development. 2005a;19:1723–1734. doi: 10.1101/gad.1319105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo S, Richardson GA, Kroll KL. The SWI/SNF chromatin remodeling protein Brg1 is required for vertebrate neurogenesis and mediates transactivation of Ngn and NeuroD. Development. 2005b;132:105–115. doi: 10.1242/dev.01548. [DOI] [PubMed] [Google Scholar]
- Sharma R, Sanchez-Ferras O, Bouchard M. Pax genes in renal development, disease and regeneration. Seminars in cell & developmental biology; Elsevier; 2015. pp. 97–106. [DOI] [PubMed] [Google Scholar]
- Shibata SB, Ranum PT, Moteki H, Pan B, Goodwin AT, Goodman SS, Abbas PJ, Holt JR, Smith RJ. RNA Interference Prevents Autosomal-Dominant Hearing Loss. Am J Hum Genet. 2016;98:1101–1113. doi: 10.1016/j.ajhg.2016.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui SA, Cramer KS. Differential expression of Eph receptors and ephrins in the cochlear ganglion and eighth cranial nerve of the chick embryo. J Comp Neurol. 2005;482:309–319. doi: 10.1002/cne.20396. [DOI] [PubMed] [Google Scholar]
- Sienknecht UJ, Köppl C, Fritzsch B. Evolution and development of hair cell polarity and efferent function in the inner ear. Brain, behavior and evolution. 2014;83:150–161. doi: 10.1159/000357752. [DOI] [PubMed] [Google Scholar]
- Singh S, Groves AK. The molecular basis of craniofacial placode development. Wiley Interdiscip Rev Dev Biol. 2016;5:363–376. doi: 10.1002/wdev.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soukup GA, Fritzsch B, Pierce ML, Weston MD, Jahan I, McManus MT, Harfe BD. Residual microRNA expression dictates the extent of inner ear development in conditional Dicer knockout mice. Developmental biology. 2009;328:328–341. doi: 10.1016/j.ydbio.2009.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steventon B, Mayor R, Streit A. Directional cell movements downstream of Gbx2 and Otx2 control the assembly of sensory placodes. Biology Open. 2016;5:1620–1624. doi: 10.1242/bio.020966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugahara F, Pascual-Anaya J, Oisi Y, Kuraku S, Aota S-i, Adachi N, Takagi W, Hirai T, Sato N, Murakami Y. Evidence from cyclostomes for complex regionalization of the ancestral vertebrate brain. Nature. 2016 doi: 10.1038/nature16518. [DOI] [PubMed] [Google Scholar]
- Tarchini B, Tadenev AL, Devanney N, Cayouette M. A link between planar polarity and staircase-like bundle architecture in hair cells. Development. 2016;143:3926–3932. doi: 10.1242/dev.139089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taura A, Taura K, Koyama Y, Yamamoto N, Nakagawa T, Ito J, Ryan A. Hair cell stereociliary bundle regeneration by espin gene transduction after aminoglycoside damage and hair cell induction by Notch inhibition. Gene therapy. 2016 doi: 10.1038/gt.2016.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda N, Richards GS, Degnan BM, Kranz A, Adamska M, Croll RP, Degnan SM. An ancient role for nitric oxide in regulating the animal pelagobenthic life cycle: evidence from a marine sponge. Sci Rep. 2016;6:37546. doi: 10.1038/srep37546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A. The origins of evolutionary innovations: a theory of transformative change in living systems. OUP Oxford: 2011. [Google Scholar]
- Wahnschaffe U, Bartsch U, Fritzsch B. Metamorphic changes within the lateral-line system of Anura. Anatomy and embryology. 1987;175:431–442. doi: 10.1007/BF00309679. [DOI] [PubMed] [Google Scholar]
- Wang VY, Rose MF, Zoghbi HY. Math1 expression redefines the rhombic lip derivatives and reveals novel lineages within the brainstem and cerebellum. Neuron. 2005;48:31–43. doi: 10.1016/j.neuron.2005.08.024. [DOI] [PubMed] [Google Scholar]
- Will U, Fritzsch B. The evolution of the amphibian auditory system. Wiley; New York: 1988. The eighth nerve of amphibians: peripheral and central distribution; pp. 159–183. [Google Scholar]
- Woo S-H, Ranade S, Weyer AD, Dubin AE, Baba Y, Qiu Z, Petrus M, Miyamoto T, Reddy K, Lumpkin EA. Piezo2 is required for Merkel-cell mechanotransduction. Nature. 2014;509:622–626. doi: 10.1038/nature13251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Grillet N, Zhao B, Cunningham C, Harkins-Perry S, Coste B, Ranade S, Zebarjadi N, Beurg M, Fettiplace R, Patapoutian A, Muller U. Mechanosensory hair cells express two molecularly distinct mechanotransduction channels. Nat Neurosci. 2016 doi: 10.1038/nn.4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Müller U. Molecular Identity of the Mechanotransduction Channel in Hair Cells: Not Quiet There Yet. Journal of Neuroscience. 2016;36:10927–10934. doi: 10.1523/JNEUROSCI.1149-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang M, Maklad A, Pirvola U, Fritzsch B. Brn3c null mutant mice show long-term, incomplete retention of some afferent inner ear innervation. BMC neuroscience. 2003;4:1. doi: 10.1186/1471-2202-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P-X, Adams J, Peters H, Brown MC, Heaney S, Maas R. Eya1-deficient mice lack ears and kidneys and show abnormal apoptosis of organ primordia. Nature genetics. 1999;23:113–117. doi: 10.1038/12722. [DOI] [PubMed] [Google Scholar]
- Yang T, Kersigo J, Jahan I, Pan N, Fritzsch B. The molecular basis of making spiral ganglion neurons and connecting them to hair cells of the organ of Corti. Hearing research. 2011;278:21–33. doi: 10.1016/j.heares.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yellajoshyula D, Patterson ES, Elitt MS, Kroll KL. Geminin promotes neural fate acquisition of embryonic stem cells by maintaining chromatin in an accessible and hyperacetylated state. Proceedings of the National Academy of Sciences. 2011;108:3294–3299. doi: 10.1073/pnas.1012053108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeyl JN, Johnston CE. Comparative and developmental patterns of amphibious auditory function in salamanders. Journal of Comparative Physiology A. 2016;202:879–894. doi: 10.1007/s00359-016-1128-6. [DOI] [PubMed] [Google Scholar]
- Zhu B, Pennack JA, McQuilton P, Forero MG, Mizuguchi K, Sutcliffe B, Gu C-J, Fenton JC, Hidalgo A. Drosophila neurotrophins reveal a common mechanism for nervous system formation. PLoS Biol. 2008;6:e284. doi: 10.1371/journal.pbio.0060284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou D, Erickson C, Kim E-H, Jin D, Fritzsch B, Xu P-X. Eya1 gene dosage critically affects the development of sensory epithelia in the mammalian inner ear. Human molecular genetics. 2008;17:3340–3356. doi: 10.1093/hmg/ddn229. [DOI] [PMC free article] [PubMed] [Google Scholar]