Abstract
If unrepaired, damage to genomic DNA can cause mutations and/or be cytotoxic. Single base lesions are repaired via the base excision repair (BER) pathway. The first step in BER is the recognition and removal of the nucleobase lesion by a glycosylase enzyme. For example, human oxoguanine glycosylase 1 (hOGG1) is responsible for removal of the prototypic oxidatively damaged nucleobase, 8-oxo-7,8-dihydroguanine (8-oxoG). To date, most studies of glycosylases have used free duplex DNA substrates. However, cellular DNA is packaged as repeating nucleosome units, with 145 base pair segments of DNA wrapped around histone protein octamers. Previous studies revealed inhibition of hOGG1 at the nucleosome dyad axis and in the absence of chromatin remodelers. In this study, we reveal that even in the absence of chromatin remodelers or external cofactors, hOGG1 can initiate BER at positions off the dyad axis and that this activity is facilitated by spontaneous and transient unwrapping of DNA from the histones. Additionally, we find that solution accessibility as determined by hydroxyl radical footprinting is not fully predictive of glycosylase activity and that histone tails can suppress hOGG1 activity. We therefore suggest that local nuances in the nucleosome environment and histone-DNA interactions can impact glycosylase activity.
Keywords: human oxoguanine glycosylase 1, base excision repair, glycosylase, nucleosome
1. Introduction
Genomic DNA is under constant threat of damage from free radicals, radiation, and environmental toxins1. Persistent chemical modification of a nucleobase can be cytotoxic and/or mutagenic, and is the underlying cause of aging and cancer1. Therefore, cells have developed a variety of essential repair pathways to remove lesions from DNA. Single base lesions are rectified via the base excision repair (BER) pathway, which is initiated by a glycosylase enzyme specific to the lesion. For example, the oxidation of guanine to 8-oxo-7,8-dihydroguanine (8-oxoG) is repaired in humans by oxoguanine glycosylase 1 (hOGG1). Glycosylases bind the DNA and flip the lesion out of the base stack into the enzyme’s active site. Glycosylases share a common SN1 type mechanism for cleavage of the glycosidic bond at the lesion, ultimately resulting in an abasic site2–4.
The majority of studies to examine glycosylase activity have used free duplex DNA as the substrate. While these studies provide essential mechanistic information about glycosylases, cellular DNA packaged as chromatin presents a more complex substrate context for repair in vivo.5 The primary structural repeating unit of chromatin is the nucleosome. Individual nucleosomes, or nucleosome core particles (NCP), serve as a model system for the lowest-order element of DNA packaging. An NCP consists of 145–147 base pairs of duplex DNA wrapped around a protein core. The octameric protein core contains two copies each of the four histone proteins: H2A, H2B, H3, and H4. The histone proteins have unstructured lysine-rich tails which are subject to post-translational modification, such as acetylation6. The NCP contains a dyad axis of symmetry aligned with the central base pair of the wrapped DNA. The translational position of a base refers to its distance from the central base pair. Further, the rotational position of a base describes its orientation relative to the histone core. Outward facing bases are solution accessible while bases facing inward to the histone core are sequestered.
Several factors have been proposed to impact the initiation of repair by glycosylases in packaged DNA substrates, including the steric bulk of the histone proteins and the specific position of the lesion. A recent review by Odell et al. summarized the studies to date and proposed a set of general tenets for glycosylase activity in NCPs5. First, spontaneous unwrapping of nucleosomal DNA transiently exposes sites that are normally occluded. Ensemble FRET measurements7–9 and single molecule FRET techniques10, 11 have quantified the rates and extent of DNA end unwrapping. This dynamic motion ultimately results in increased accessibility of bases closer to the ends of DNA compared to bases at the dyad axis. Early work using restriction enzymes showed increased accessibility of restriction sites closer to ends of nucleosomal DNA compared to restriction sites on the dyad axis12, 13. This trend was also observed with the human glycosylase endonuclease III (hNTH1), which had increased excision of thymine glycol at the ends of the DNA compared to on the dyad axis;14 interestingly, it was recently reported that human cells contain a factor that facilitates hNTH1 activity in NCP substrates15. Further, previous studies have revealed significant inhibition of several other glycosylases, including hOGG1, acting on lesions positioned at the dyad axis14, 16, 17. Notably, the abilities of uracil DNA glycosylase (UDG) and alkyladenine glycosylase to excise an outward facing lesion at the dyad axis were observed as exceptions16. Finally, it has been proposed that lesions with outward rotational position are more easily repaired than inward facing lesions, though exceptions have been observed14, 16, 18–24. Most importantly, the general applicability of these principles to all glycosylases and lesions has yet to be fully explored.
In the case of hOGG1, inhibition has been observed in NCP substrates when the lesion is located at the dyad axis16, 17, 25. Activity of hOGG1 in NCP substrates has been observed upon addition of chromatin remodeling complexes17, 25 and when the lesion is located in the linker region between two NCPs of a dinucleosome substrate25. Given the known importance of transient DNA unwrapping, hOGG1 activity with lesions of varied translational position warrants further exploration.
To investigate the role of DNA unwrapping on hOGG1 activity, we created NCP substrates with single base lesions incorporated off the dyad axis. The lesions were positioned approximately 20 base pairs from the end of the DNA within the region that is known to show dynamic unwrapping. To further investigate how the rotational position of a lesion impacts glycosylase activity, we incorporated lesions in positions of varying solution accessibility. Finally, we investigated the impact the local histone environment on hOGG1 activity via acetylation and omission of the histone H2B tail.
2. Materials and Methods
2.1 Oligonucleotide Synthesis and Purification
Oligomers were synthesized on a MerMade 4 (BioAutomation) DNA synthesizer using phosphoramidite chemistry. All synthesis reagents and phosphoramidites were purchased from Glen Research. The final trityl group was retained for an initial round of HPLC purification (Dynamax Microsorb C18 column, 10 × 250 mm; A = acetonitrile, B = 30 mM ammonium acetate; 5:95 to 35:65 A:B over 30 min at 3.5 mL/min). Following this first round of purification, oligomers were incubated with 20% v/v aqueous acetic acid for 60 min at room temperature to remove the trityl group. For oligomers containing only canonical bases or uracil (U), HPLC at 90°C was used for a second round of purification (Agilent PLRP-S column, 250 mm × 4.6 mm; A = 100 mM triethylammonium acetate [TEAA] in 5% aqueous MeCN, B = 100 mM TEAA in MeCN; 0:100 to 35:65 A:B over 35 min at 1 mL/min). For oligomers containing 8-oxoG, incorporation of 8-oxoG was carried out according to Glen Research protocols. The trityl group was removed on the synthesizer before purification and 2-mercaptoethanol was included during cleavage of the oligomer from the bead. Anion-exchange HPLC was used to purify the 8-oxoG-containing oligomers (Dionex DNAPac PA100 anion-exchange column; A = 10% acetonitrile, B = 0.8 M ammonium acetate in 10% acetonitrile; 70:30 to 0:100 A:B over 35 min at 1 mL/min). Collected peaks were desalted by buffer exchange using centrifugal concentrators (Sartorius Vivaspin Turbo 15, 5 kDa MWCO).
2.2 Ligation of 145mer Oligonucleotides
We synthesized the DNA strands in shorter pieces for assembly into full length 145mer via enzymatic ligation (Scheme S1). The short oligomers were phosphorylated with T4 polynucleotide kinase (New England Biolabs) with an additional 2 mM ATP in the reaction buffer. Phosphorylated oligomers were annealed to scaffold oligomers in a 1:1 ratio by heating to 90 °C for 5 min followed by cooling to 25 °C at a rate of 1°C/min. Next, the oligomers were ligated for 3 h at ambient temperature using T4 Ligase (New England Biolabs). Full-length ligation products were purified by 8% denaturing PAGE (0.8 mm thickness). Electrospray ionization mass spectrometry was used to confirm the identity of the purified 145mers. Oligonucleotide concentrations were determined by their absorbance at 260 nm using molar extinction coefficients calculated with OligoAnalyzer 3.1 (www.idtdna.com).
2.3 Glycosylase Expression and Purification
His6-tagged hOGG1 was recombinantly expressed in E. coli and purified as previously described16, 26. SDS PAGE analysis showed hOGG1 purity to be >98%. UDG was purchased from New England Biolabs. The total concentration of each enzyme was determined by Bradford assay with γ-globulin standards (Bio-Rad Laboratories).
2.4 Histone Proteins and Octamer Assembly
Expression and purification of canonical X. laevis histone proteins and assembly of the histone octamer were performed according to the published method of Luger, et al27, 28. Chemically acetylated H2B protein was prepared based on previously published methods29, 30, which provides solution-accessible lysine residues as potential substrates. Briefly, 0.3 µmol of purified H2B was dissolved in 1 mL of freshly prepared 7 M urea solution with 500 mM ammonium acetate and 50 mM ammonium bicarbonate (pH 8.5). The mixture was kept on ice, and 1.5 µL of freshly prepared 10.6 M acetic anhydride was added every 15 min. Concurrently, the pH of the reaction was constantly adjusted to 8.0–8.5 with ammonium hydroxide. After 60 min, the reaction was quenched with 100 µL of 1M Tris-HCl before transferring to a dialysis device for overnight dialysis into water with 2 mM 2-mercaptoethanol. Electrospray ionization mass spectrometry revealed that between 4–14 acetyl groups were added to H2B (Figure S1). The acetylated H2B protein was lyophilized and stored at −20 °C until used for octamer assembly. The tailless X. laevis H2B protein (residues 24–122) was purchased from The Histone Source (Colorado State University).
2.5 Nucleosome Core Particle Reconstitution
NCPs were prepared by stepwise dialysis as reported previously16, 31. Briefly, a Slide-a-Lyzer MINI dialysis device was equilibrated in buffer (10 mM Tris-HCl [pH 7.5], 1 mM EDTA, 1 mM dithiothreitol [DTT], 2 M NaCl) at 4 °C. Radiolabeled duplex (50 µL of 1 µM) was added to the dialysis device and allowed to incubate for 30 min before addition of histone octamer in a 1:1.05 molar ratio. The concentration of NaCl in the dialysis buffer was progressively lowered at 60 min intervals (1.2 M, 1.0 M, 0.6 M, 0 M). The final dialysis step was carried out for 3 h. Samples were filtered to remove precipitate and NCP formation was confirmed by 7 % native polyacrylamide (60:1 acrylamide:bisacrylamide; 0.25× TBE) gel electrophoresis (3 h at 150 V, 4 °C). A representative native gel is shown in Figure S2. Only NCP preparations with less than 5 % free duplex DNA (as determined by native gel) were used in the experiments.
2.6 Hydroxyl Radical Footprinting
Hydroxyl Radical Footprinting procedures were based on previously published methods32, 33. Briefly, 5 pmol of NCPs containing 32P radiolabeled DNA were suspended in 52.5 µL buffer (10 mM Tris-HCl [pH 7.5], 1 mM EDTA) and mixed with 7.5 µL each 10 mM Fe(II)-EDTA, 10 mM sodium ascorbate, and 0.12% w/v aqueous hydrogen peroxide. The reaction was incubated at ambient temperature for 10 min before quenching with the addition of 50 µL 1 mM EDTA in 25% w/v glycerol. The sample was immediately loaded on a pre-running 7% native polyacrylamide (60:1 acrylamide:bisacrylamide, 0.25 × TBE) and electrophoresed for 3 h at 150 V at 4 °C. The band containing NCPs was excised and NCPs were eluted into buffer (300 mM sodium acetate [pH 8.0], 1 mM EDTA) overnight. The resulting eluent was concentrated (Sartorius Vivaspin Turbo 15, 5 kDa MWCO) and extracted twice against 25:24:1 phenol:chloroform:isoamyl alcohol. An ethanol precipitation was performed with the addition of 20 µL co-precipitation reagent (0.5 mg/mL tRNA in 300 mM sodium acetate [pH 8.0], 1 mM EDTA). Samples were dissolved in formamide and run on an 8 % denaturing PAGE. The gel was dried and exposed before phosphorimaging. The bands were quantitated using SAFA gel analysis software34.
2.7 Glycosylase Kinetics Experiments
Kinetics experiments were based on previously published protocols16. Briefly, radiolabeled, lesion-containing substrate (either duplex or NCP-incorporated) and glycosylase were prepared at 2 × experimental concentration (final concentration 20 nM and 0.64 µM, respectively) in reaction buffer (20 mM Tris-HCl [pH 7.6], 25 mM NaCl, 75 mM KCl, 1 mM EDTA, 1 mM DTT, 200 µg/ml BSA). Following a temperature pre-equilibration at 37 °C for 2 min, equal volumes of substrate (8 µL) and glycosylase (8 µL) preparations were mixed for varying amounts of time before addition of 1M NaOH quench (16 µL, final concentration 500 mM). For each time course a negative control sample (QC) was prepared by adding 1 M NaOH quench (16 µL) to substrate (8 µL) followed by addition of glycosylase (8 µL) before incubation at 37 °C for the duration of the longest time point. This control serves to reveal any pre-existing damage or incidental damage due to heating or sample work-up (generally less than 10 %). Samples were heated to 90 °C for 2 min after addition of quench to induce a strand break at abasic sites. DNA was isolated from proteins by extraction with 25:24:1 phenol:chloroform:isoamyl alcohol before desalting by ethanol precipitation. Samples were electrophoresed on an 8 % denaturing PAGE, imaged by phosphorimagery (BioRad Pharos FX) and quantitated by densitometry.
The fraction of product at each time point, (t), was determined by the formula
where δS (t) and δP (t) are the densities of the substrate and product bands, respectively, at time t. The product yield, (t), was corrected for pre-existing and incidental substrate damage using the following formula:
where FP(0) is the fraction of product observed in the QC sample. The mean product yield from four independent trials (two separate NCP preparations with two trials each) was determined at each time point. The averaged data were fit to the modified first-order integrated rate law:
or
where [P(t)] is the concentration of product at time t, [P(∞) is the maximum concentration of product, and [P(0)] is the amount of product at time t = 0(generally close to zero), using a nonlinear least squares regression (Kaleidagraph). Reaction rates, kobs, were reported from the fits. A two-tailed student’s t test was performed to obtain the p values for the tailless H2B NCPs and acetylated H2B NCPs in comparison to the canonical NCPs. We considered p < 0.05 to be significant.
2.8 Molecular Modeling
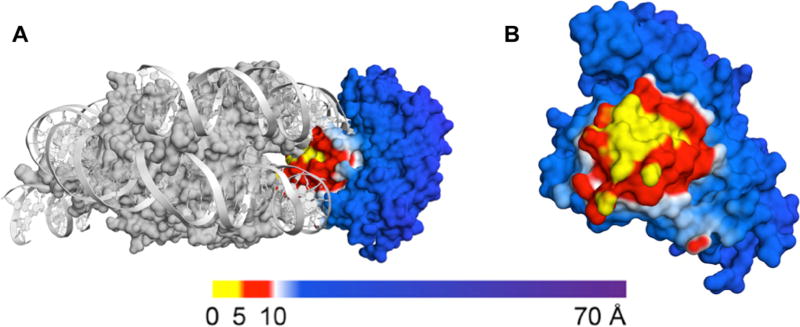
Molecular models were created in PyMOL (The PyMOL Molecular Graphics System, Version 1.74 Schrödinger, LLC.) to visualize hOGG1 binding to the 601 NCP. Structures of hOGG1-bound-NCPs were created by aligning the crystal structure of hOGG1 bound to DNA (PDB ID: 1ebm) with the 601 NCP crystal structure (PDB ID: 3lz0) as described previously16. The glycosylase surface was colored in PyMol according to proximity to the histone core (regions within 5 Å of the octamer are yellow; regions within 5–10 Å of the octamer are red; regions greater than 10 Å from the octamer are blue).
3. Results
3.1 Formation of Homogenous NCPs
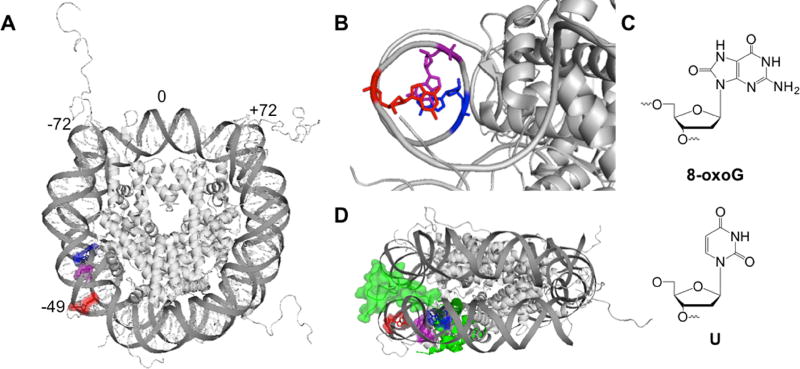
In this study, we evaluated the impact of lesion position on BER in packaged DNA. Our DNA substrates were based on the Widom 601 positioning sequence, which binds in a single translational and rotational position around the histone octamer35. We used the Widom 601 NCP crystal structure36 to guide our placement of DNA lesions off the dyad axis and in one of three rotational positions: out toward solution (OUT), approximately 90° from solution (MID), or in toward the histone core (IN) (Figure 1A, 1B). These OUT, MID, and IN positions correspond to locations −49, −52, and −54, respectively, which indicate the distance from the center of the DNA strand. A single 8-oxoG or U was incorporated at these sites during DNA synthesis (Figure 1C). The complementary 601 strand was modified such that 8-oxoG was paired with C and U was paired with G to mimic a deaminated C:G base pair. X. laevis histone proteins were individually expressed in E. coli, purified, assembled into an octamer, and used to form NCPs following the salt dialysis method31. Histone octamers containing either a tailless or acetylated H2B protein were also used to assemble NCPs.
Figure 1.
Representation of the NCP structure and lesions used in this study. (A) Merged crystal structure of an NCP containing Widom 601 DNA and histone octamer with tails (PDB codes 3lz0 and 1kx5, respectively) with off-dyad lesion locations highlighted using PyMol. The DNA strands are numbered such that 0 corresponds to center of the 145mer DNA, and the 5′ end is base −72. The OUT, MID, and IN facing lesion positioned at base −49, −52, and −54, respectively. (B) Rotational positions of lesions (one DNA strand shown for simplicity). The location of the OUT facing lesion is highlighted in red, the MID facing lesion in purple, and the IN lesion in blue. (C) Lesions used in this study. (D) Side view of NCP. The H2B tail is highlighted in green and represented in surface mode.
3.2 Hydroxyl Radical Footprinting Defines Rotational Position of Lesions
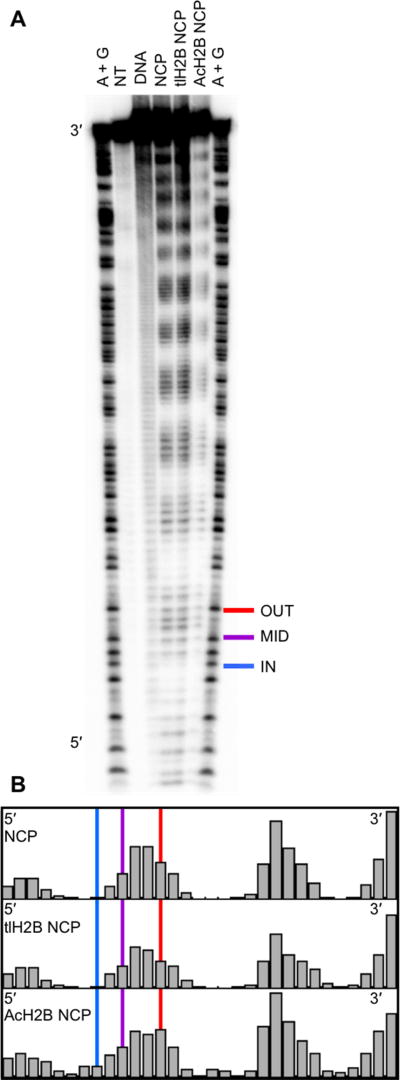
Hydroxyl radical footprinting was used to establish the solution accessibility of each lesion position. The hydroxyl radical footprinting technique uses Fenton chemistry to generate hydroxyl radicals, which abstract a hydrogen atom from the DNA backbone to create a strand break33. A sample of free duplex Widom 601 DNA treated with hydroxyl radicals shows an unbiased pattern of reactivity throughout the length of the DNA strand (Figure 2A, DNA lane). In comparison, DNA incorporated into an NCP shows an oscillating pattern of damage (Figure 2A, NCP lane). There are regions of protection and damage corresponding to the DNA wrapping in toward the histone core and out toward solution, respectively. Notably, the oscillatory pattern persists both upstream and downstream of the lesion positions, indicating that while this region is known to be more dynamic11, it is still in contact with the histone core and does not react like free duplex DNA with hydroxyl radical treatment. The integrated band area at each base position, and the relative reactivity of lesion positions is shown in Figure 2B34. Indeed, the relative hydroxyl radical reactivity at the selected lesion positions correlates with predicted solution accessibility based on the Widom 601 NCP crystal structure (ie. OUT > MID > IN).
Fig. 2.
Hydroxyl Radical Footprinting of DNA in an NCP. (A) The Hydroxyl Radical Footprinting experiment was analyzed by denaturing PAGE. The flanking “A+G” lanes display a ladder of the 601 DNA sequence created using Maxam Gilbert reactions. The “NT” lane shows a no treatment control. The “DNA” lane shows unbiased damage of hydroxyl radical treatment on free duplex DNA. Hydroxyl radical treatment of canonical (“NCP”), tailless H2B octamer (“tlH2B NCP”), and acetylated H2B octamer (“AcH2B NCP”) NCPs reveals an oscillating pattern of damage. The location of OUT (red), MID (purple), and IN (blue) lesions in this study are highlighted. (B) Quantitation of band density of the footprinting using SAFA software to show relative solvent accessibility. The vertical lines highlight the location of each lesion position.
Further, hydroxyl radical footprinting was used to evaluate the impact of changes to histone proteins on the solution accessibility of the DNA bases. NCPs prepared using a histone octamer containing either tailless H2B or acetylated H2B histone proteins showed no systematic differences in the oscillating pattern or the relative solution accessibility of the studied OUT, MID, and IN positions when compared to the canonical histone octamer (Figure 2A, 2B).
3.3 Glycosylase Activity in NCPs with Off-Dyad Lesions
Kinetics experiments were performed to test the activity of glycosylase enzymes on NCP substrates. NCP substrates containing a single 8-oxoG or U lesion were incubated at 37 °C with hOGG1 or UDG before quenching with NaOH. Formation of product over time was monitored on denaturing polyacrylamide gels (Figure S3). In these experiments, a 32-fold excess of enzyme over substrate was used to ensure single-turnover conditions. We have previously shown that further increases in the excess of enzyme do not yield an increased rate of glycosylase activity in NCP substrates16. The observed rate (kobs) therefore reflects the slowest step up to or including the chemistry step of glycosidic bond cleavage.
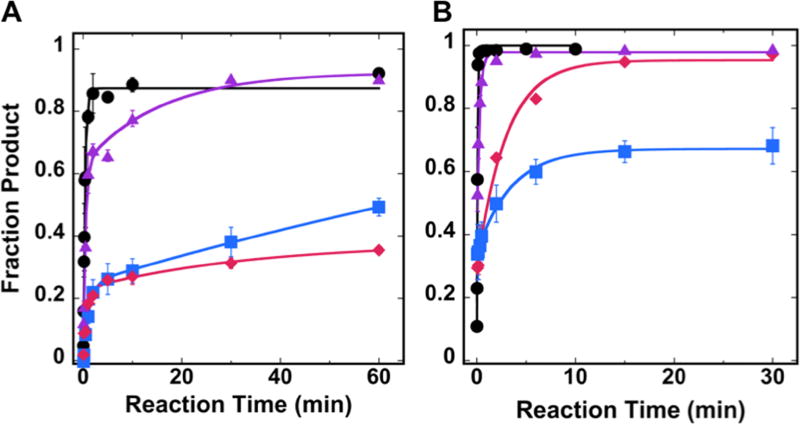
Control experiments were performed in which free duplex DNA substrates containing a single 8-oxoG or U lesion (the OUT facing lesion position is shown as a representative example) were converted to 87 and 99 % product, respectively (Figure 3). The data were fit to a single exponential, and the differences in kobs and product formation for free duplex DNA substrates of each lesion/glycosylase pair varied minimally with lesion position (data not shown).
Fig. 3.
Single-turnover kinetic results for hOGG1 and UDG in NCP substrates. Kinetic time courses were performed to evaluate the activity of (A) hOGG1 or (B) UDG on either free duplex DNA (●) or NCP substrates with off-dyad lesions. The NCP substrates contained lesions of varying rotational position: OUT (
 ), MID (
), MID (
 ), or IN (
), or IN (
 ). Experiments were conducted using 20 nM NCP substrate and 0.64 µM glycosylase in 20 mM Tris-HCl (pH 7.6), 25 mM NaCl, 75 mM KCl, 1 mM EDTA, 1 mM DTT, 200 µg/ml BSA. Data for free duplex DNA substrates were fit to a single exponential equation; all NCP data were fit to a double exponential equation. Error bars represent the standard error (n=4).
). Experiments were conducted using 20 nM NCP substrate and 0.64 µM glycosylase in 20 mM Tris-HCl (pH 7.6), 25 mM NaCl, 75 mM KCl, 1 mM EDTA, 1 mM DTT, 200 µg/ml BSA. Data for free duplex DNA substrates were fit to a single exponential equation; all NCP data were fit to a double exponential equation. Error bars represent the standard error (n=4).
For NCP substrates, we previously showed complete inhibition of hOGG1 activity on lesions located at the dyad axis, regardless of rotational position16. Interestingly, in the current work, we observe that activity of hOGG1 is restored on NCP substrates when the lesions are located off the dyad axis (Figure 3A). Surprisingly, however, the product yield of these kinetic time courses does not correlate with the solvent accessibility of the lesions as determined by hydroxyl radical footprinting. The most product formation (93%) was observed at the MID facing lesion site and 49% product formation was observed for the IN facing lesion site. The most unanticipated result was the relative inhibition of hOGG1 at the most solvent accessible lesion tested in this experiment (OUT), which reached a maximum of 38% product (Figure 3A, Table 1). In contrast to the free duplex DNA substrates, the data for all NCP substrates were best fit to a double exponential model, reflecting a faster kinetic phase and a slower kinetic phase (Table 1). While the product yield varies between different rotationally positioned substrates, the kobs values are the same within error.
Table 1.
kobs Values Determined for Free Duplex DNA and NCP Substrates
| enzyme | NCP | substrate | kobs/min (% product) |
|---|---|---|---|
| hOGG1 | N/A | 8-oxoG free duplex | 2.6 ± 0.2 (87%) |
| canonical | 8-oxoGOUT | 1.4 ± 0.4 (23%) 0.03 ± 0.03 (15%) | |
| canonical | 8-oxoGMID | 2.0 ± 0.4 (63%), 0.06 ± 0.04 (30%) | |
| canonical | 8-oxoGIN | 0.8 ± 0.1 (25%), N.R.1 (≥ 24%) | |
| tailless H2B | 8-oxoGOUT | 2.3 ± 0.4 (28%), 0.05 ± 0.02 (18%) | |
| acetylated H2B | 8-oxoGOUT | 4.7 ± 1.4 (23%), 0.11 ± 0.03 (33%) | |
| UDG | N/A | U free duplex | 12.6 ± 1.6 (99%) |
| canonical | UOUT | 37.7 ± 33.4 (29%), 0.3 ± 0.05 (67%) | |
| canonical | UMID | 20.1 ± 5.3 (52%), 3.3 ± 0.6 (46%) | |
| canonical | UIN | 42.3 ± 19.9 (34%), 0.3 ± 0.04 (33%) |
N.R. indicates an undetermined kobs value for this kinetic phase.
The unexpected pattern of glycosylase activity and solvent accessibility was also manifested for another glycosylase, UDG, acting on the off-dyad lesions. However, for UDG, rather than observing differences in the amount of product, we observed differences in kobs; while full product conversion was achieved for both the OUT and MID positions, U at the MID position was excised approximately 10-fold faster than at the OUT position (Figure 3B, Table 1).
3.4 Glycosylase Activity in NCPs with H2B-Modified Octamers
Upon observing the disaccord between the solvent accessibility of the lesions and the resulting glycosylase activity, we modeled the Widom 601 DNA wrapped around a histone core containing histone tails (PDB ID 1kx5), which are present in our in vitro experiments. In this model we observed that our off-dyad lesions are near the H2B tail that protrudes between the superhelices (Figure 1D). We hypothesized that interactions between the histone tail and the incoming glycosylase could explain the suppressed product yield of hOGG1 at the OUT lesion position as well as the slower kobs for UDG.
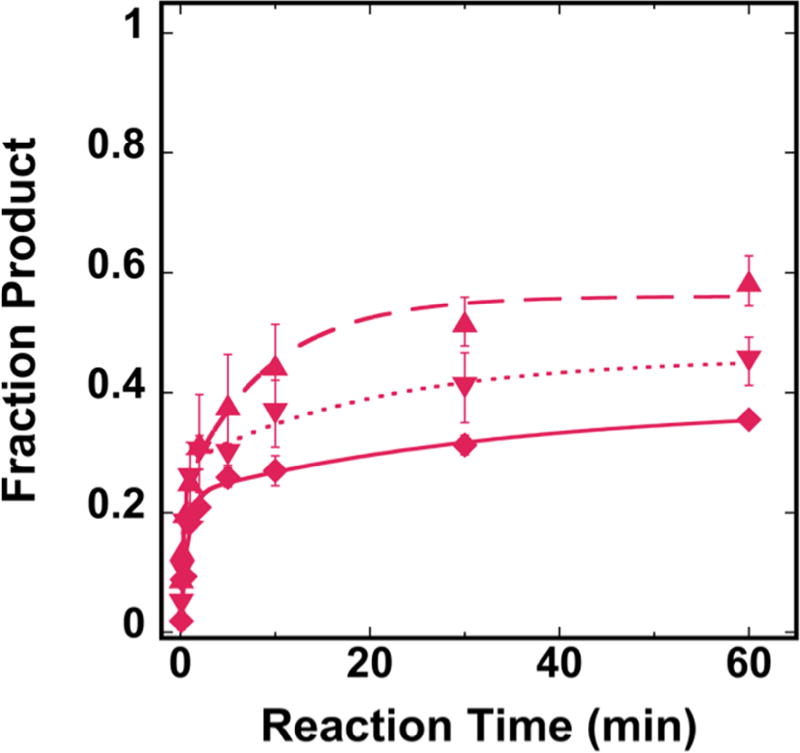
To test the impact of the nearby H2B tail on hOGG1 activity, we created two modified histone octamers. First, we created NCPs using an octamer with chemically acetylated H2B proteins, where addition of an acetyl group neutralizes the positive charge of lysine. In order to further test the steric impact of the H2B tail, we created NCPs using an octamer with tailless H2B proteins. We tested the activity of hOGG1 on these H2B-modified NCPs with an OUT facing 8-oxoG and compared product formation to the canonical NCP substrate. While the tailless H2B NCP does not show a statistically significant increase in product formation, there is an increase in product with the acetylated H2B substrate (p = 0.0016) (Figure 4).
Fig. 4.
Single-turnover kinetic results for hOGG1 on 8-oxoG OUT NCP substrates with H2B modified NCPs. The octamer used to create the NCPs was varied to include a canonical octamer (
 , solid line), tailless H2B octamer (
, solid line), tailless H2B octamer (
 , short dashed line) or acetylated H2B octamer (
, short dashed line) or acetylated H2B octamer (
 , long dashed line). Experiments were conducted using 20 nM NCP substrate and 0.64 µM total hOGG1 in 20 mM Tris-HCl (pH 7.6), 25 mM NaCl, 75 mM KCl, 1 mM EDTA, 1 mM DTT, 200 µg/ml BSA. All data were fit to a double exponential equation. Error bars represent the standard error (n=4).
, long dashed line). Experiments were conducted using 20 nM NCP substrate and 0.64 µM total hOGG1 in 20 mM Tris-HCl (pH 7.6), 25 mM NaCl, 75 mM KCl, 1 mM EDTA, 1 mM DTT, 200 µg/ml BSA. All data were fit to a double exponential equation. Error bars represent the standard error (n=4).
4. Discussion
To investigate the role of DNA unwrapping on glycosylase activity, we created NCP substrates with single base lesions located approximately 20 base pairs from the end of the DNA. In general, activity of the tested enzymes increases when the lesions are moved closer to the DNA ends, relative to when the lesions are at the dyad. While hOGG1 was completely inhibited at the dyad axis regardless of the rotational position of the lesion16, product formation is observed at all of the off dyad positions studied here.
While the data for free duplex DNA substrates fit well to a single exponential model, the kinetic data for the NCP substrates are more complex. For the off-dyad lesions, double exponential models were required to fit the data for all lesion positions and glycosylases. We attribute the biphasic kinetics to the existence of two distinct substrate populations. Each NCP substrate shows a fast phase, which we attribute to a configuration that can be processed directly by the glycosylase. The slow kinetic phase for NCP substrates is generally two orders of magnitude slower than the fast phase. We attribute the slower phase to a population of NCPs that require a rate-limiting conformational change to adopt a structure permissive for repair. This conformational change is consistent with the transient unwrapping of DNA to expose the off-dyad lesion sites, and is not observed in experiments with lesions positioned on the dyad16. Previously, the Stivers group reported multiphasic kinetics in experiments with UDG excising U at sites off the dyad31.
As no crystal structures of glycosylase-bound NCPs have been reported, we used molecular modeling to analyze the interaction of the glycosylase and the NCP (Figure 5). To generate these models, we merged the crystal structure of the 601 NCP with the crystal structure of DNA-bound hOGG1. We chose to model the glycosylase-bound NCP using the 601 NCP crystal structure lacking histone tails36 in order to avoid assigning static positions to the unstructured histone tails. The surface of the glycosylase was colored according to its proximity to the histone core as follows: regions within 5 Å of the octamer are yellow; regions within 5–10 Å of the octamer are red; regions greater than 10 Å away from the octamer are blue16.
Fig. 5.
Molecular model of NCP-bound hOGG1. The NCP binding surface of hOGG1 was generated by merging the 601 NCP PDB (3lz0, gray) and hOGG1 bound to DNA (1ebm). (A) A side view of hOGG1 bound to the NCP is shown. (B) A front view of the binding surface of hOGG1 to NCPs with off dyad 8-oxoG OUT facing (base position −49) is shown and the NCP is hidden for clarity. The hOGG1 surface model is colored according to the distance to the histone core (regions less than 5 Å away from the octamer are yellow; regions within 5–10 Å of the octamer are red; regions beyond 10 Å away from the octamer are blue).
Upon modeling hOGG1 bound to the NCP at the outward facing off-dyad lesion, we found the region near the enzyme’s active site to have several amino acid residues within 5 Å of the octamer and an even larger region between 5–10 Å of the octamer. This region represents a significant steric clash between the histone core and hOGG1, which we might predict would prevent enzyme binding and, ultimately, excision of the lesion. However, 38% product formation is observed for hOGG1 at this off-dyad lesion position. The formation of product despite the potential for steric clash in a static model at this position emphasizes the contribution of DNA unwrapping and dynamics to hOGG1 accessibility in an NCP.
There are conflicting reports on the correlation between solution accessibility of a lesion and enzyme activity in an NCP substrate14, 16, 18–24. We found that for hOGG1, the product yield for NCP substrates did not correlate with the solution accessibility of the lesions as determined by hydroxyl radical footprinting. Furthermore, while full product conversion was observed for both the OUT and MID position with UDG, the rate of excision at the MID position was ~10 times faster than at the OUT position. It is clear that solution accessibility as determined by hydroxyl radical footprinting is not able to predict all chemistry within an NCP. We therefore suggest that local nuances in the NCP environment and histone-DNA interaction have a significant impact on glycosylase activity and may have a more influential role than solution accessibility at certain positions in the NCP. Other groups who have observed complex kinetics of UDG in NCP substrates have come to similar conclusions31.
We created a tailless H2B version of the histone octamer to test the influence of completely removing the histone tail on glycosylase activity. We found that for hOGG1, removing the H2B tails does not lead to a statistically significant increase in product formation. While we therefore conclude that the inhibition of hOGG1 is not completely derived from steric interference of the histone tail, the phenomenon of histone tail clipping in vivo by endopeptidases is an area of recent interest and has been observed in all histone proteins in several organisms37. It has been shown that histones lacking N-terminal tails allow for increased accessibility of nucleosomal DNA to transcription factors via increased unwrapping of DNA from the NCP38–40. Other BER enzymes have also shown varying levels of inhibition in NCP substrates due to the presence of histone tails41, 42.
Histone proteins are also targets for post-translational modification, particularly acetylation at the positively charged lysine residues in the N-terminal tails. In this study, NCPs with acetylated H2B proteins showed an increase in product formation when compared to unmodified NCPs. While the H2B tail associates with the nearby DNA in unmodified NCPs, it has been shown that acetylation causes release of H2B tails from the DNA43. Further, FRET studies have shown that acetylation of histones facilitates DNA unwrapping44, 45. We therefore attribute the significant increase in product formation to the neutralization of the lysine residues and resulting relaxation of the NCP structure. However, the incomplete recovery of product formation in acetylated H2B NCPs suggests that electrostatic interference of the lysine residues on H2B is not the only contributing factor to hOGG1 inhibition at the OUT lesion position.
Conclusion
In summary, we demonstrate that hOGG1 can excise 8-oxoG from nucleosomal DNA in the absence of external cofactors or chromatin remodelers and that this activity is modulated by transient DNA unwrapping. We further demonstrate the complexity of the NCP environment, as we observe a disparity between solution accessibility and enzyme activity. Increased activity of hOGG1 in response to acetylation of H2B reveals intricacies in the NCP environment due to the electrostatic influence of the histone tails. A more complete understanding of BER in NCP substrates will require additional studies on the impact of histone modifications, chromatin remodeling complexes, and other cellular factors.
Supplementary Material
Highlights.
Packaged DNA presents a complex substrate for DNA glycosylases
We found that hOGG1 is able to excise lesions near the ends of nucleosomal DNA
Transient DNA unwrapping promotes excision of otherwise occluded lesions
Solution accessibility of DNA bases is not fully predictive of hOGG1 activity
Acetylation of H2B increases hOGG1 product formation at the studied lesion site
Acknowledgments
This work was supported by the National Institutes of Health R01ES019296. EEK was supported by the National Institutes of Health T32GM007601. We thank Paul Caffrey and Dr. Shashank Gupta for helpful discussion.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest statement
The authors declare that there are no conflicts of interest.
References
- 1.Dizdaroglu M. Oxidatively induced DNA damage and its repair in cancer. Mutation Research/Reviews in Mutation Research. 2015;763:212–245. doi: 10.1016/j.mrrev.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 2.Werner RM, Stivers JT. Kinetic isotope effect studies of the reaction catalyzed by uracil DNA glycosylase: Evidence for an oxocarbenium ion– uracil anion intermediate. Biochemistry. 2000;39:14054–14064. doi: 10.1021/bi0018178. [DOI] [PubMed] [Google Scholar]
- 3.McCann JA, Berti PJ. Transition-state analysis of the DNA repair enzyme MutY. Journal of the American Chemical Society. 2008;130:5789–5797. doi: 10.1021/ja711363s. [DOI] [PubMed] [Google Scholar]
- 4.Drohat AC, Maiti A. Mechanisms for enzymatic cleavage of the N-glycosidic bond in DNA. Organic & biomolecular chemistry. 2014;12:8367–8378. doi: 10.1039/c4ob01063a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Odell ID, Wallace SS, Pederson DS. Rules of engagement for base excision repair in chromatin. Journal of cellular physiology. 2013;228:258–266. doi: 10.1002/jcp.24134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Allfrey V, Faulkner R, Mirsky A. Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proceedings of the National Academy of Sciences. 1964;51:786–794. doi: 10.1073/pnas.51.5.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li G, Widom J. Nucleosomes facilitate their own invasion. Nature structural & molecular biology. 2004;11:763–769. doi: 10.1038/nsmb801. [DOI] [PubMed] [Google Scholar]
- 8.Li G, Levitus M, Bustamante C, Widom J. Rapid spontaneous accessibility of nucleosomal DNA. Nature structural & molecular biology. 2005;12:46–53. doi: 10.1038/nsmb869. [DOI] [PubMed] [Google Scholar]
- 9.Tims HS, Gurunathan K, Levitus M, Widom J. Dynamics of Nucleosome Invasion by DNA Binding Proteins. Journal of Molecular Biology. 2011;411:430–448. doi: 10.1016/j.jmb.2011.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tomschik M, Zheng H, van Holde K, Zlatanova J, Leuba SH. Fast, long-range, reversible conformational fluctuations in nucleosomes revealed by single-pair fluorescence resonance energy transfer. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:3278–3283. doi: 10.1073/pnas.0500189102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wei S, Falk SJ, Black BE, Lee T-H. A novel hybrid single molecule approach reveals spontaneous DNA motion in the nucleosome. Nucleic acids research. 2015;43:e111–e111. doi: 10.1093/nar/gkv549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Linxweiler W, Hörz W. Reconstitution of mononucleosomes: characterization of distinct particles that differ in the position of the histone core. Nucleic acids research. 1984;12:9395–9413. doi: 10.1093/nar/12.24.9395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Polach K, Widom J. Mechanism of protein access to specific DNA sequences in chromatin: a dynamic equilibrium model for gene regulation. Journal of molecular biology. 1995;254:130–149. doi: 10.1006/jmbi.1995.0606. [DOI] [PubMed] [Google Scholar]
- 14.Prasad A, Wallace SS, Pederson DS. Initiation of base excision repair of oxidative lesions in nucleosomes by the human, bifunctional DNA glycosylase NTH1. Molecular and cellular biology. 2007;27:8442–8453. doi: 10.1128/MCB.00791-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maher R, Marsden C, Averil A, Wallace S, Sweasy J, Pederson D. Human cells contain a factor that facilitates the DNA glycosylase-mediated excision of oxidized bases from occluded sites in nucleosomes. DNA Repair. 2017 doi: 10.1016/j.dnarep.2017.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Olmon ED, Delaney S. Differential Ability of Five DNA Glycosylases to Recognize and Repair Damage on Nucleosomal DNA. ACS chemical biology. 2017;12:692–701. doi: 10.1021/acschembio.6b00921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Menoni H, Gasparutto D, Hamiche A, Cadet J, Dimitrov S, Bouvet P, Angelov D. ATP-dependent chromatin remodeling is required for base excision repair in conventional but not in variant H2A. Bbd nucleosomes. Molecular and cellular biology. 2007;27:5949–5956. doi: 10.1128/MCB.00376-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hinz JM, Rodriguez Y, Smerdon MJ. Rotational dynamics of DNA on the nucleosome surface markedly impact accessibility to a DNA repair enzyme. Proceedings of the National Academy of Sciences. 2010;107:4646–4651. doi: 10.1073/pnas.0914443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nilsen H, Lindahl T, Verreault A. DNA base excision repair of uracil residues in reconstituted nucleosome core particles. The EMBO journal. 2002;21:5943–5952. doi: 10.1093/emboj/cdf581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beard BC, Wilson SH, Smerdon MJ. Suppressed catalytic activity of base excision repair enzymes on rotationally positioned uracil in nucleosomes. Proceedings of the National Academy of Sciences. 2003;100:7465–7470. doi: 10.1073/pnas.1330328100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Odell ID, Barbour J-E, Murphy DL, Della-Maria JA, Sweasy JB, Tomkinson AE, Wallace SS, Pederson DS. Nucleosome disruption by DNA ligase III-XRCC1 promotes efficient base excision repair. Molecular and cellular biology. 2011;31:4623–4632. doi: 10.1128/MCB.05715-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Odell ID, Newick K, Heintz NH, Wallace SS, Pederson DS. Non-specific DNA binding interferes with the efficient excision of oxidative lesions from chromatin by the human DNA glycosylase, NEIL1. DNA repair. 2010;9:134–143. doi: 10.1016/j.dnarep.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cole HA, Tabor-Godwin JM, Hayes JJ. Uracil DNA glycosylase activity on nucleosomal DNA depends on rotational orientation of targets. Journal of Biological Chemistry. 2010;285:2876–2885. doi: 10.1074/jbc.M109.073544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rodriguez Y, Smerdon MJ. The structural location of DNA lesions in nucleosome core particles determines accessibility by base excision repair enzymes. Journal of Biological Chemistry. 2013;288:13863–13875. doi: 10.1074/jbc.M112.441444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Menoni H, Shukla MS, Gerson V, Dimitrov S, Angelov D. Base excision repair of 8-oxoG in dinucleosomes. Nucleic acids research. 2011;40:692–700. doi: 10.1093/nar/gkr761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jarem DA, Wilson NR, Delaney S. Structure-dependent DNA damage and repair in a trinucleotide repeat sequence. Biochemistry. 2009;48:6655–6663. doi: 10.1021/bi9007403. [DOI] [PubMed] [Google Scholar]
- 27.Luger K, Rechsteiner TJ, Richmond TJ. Preparation of nucleosome core particle from recombinant histones. Methods in enzymology. 1999;304:3–19. doi: 10.1016/s0076-6879(99)04003-3. [DOI] [PubMed] [Google Scholar]
- 28.Luger K, Rechsteiner TJ, Richmond TJ. Expression and purification of recombinant histones and nucleosome reconstitution. Chromatin Protocols. 1999:1–16. doi: 10.1385/1-59259-681-9:1. [DOI] [PubMed] [Google Scholar]
- 29.Bheda P, Swatkoski S, Fiedler KL, Boeke JD, Cotter RJ, Wolberger C. Biotinylation of lysine method identifies acetylated histone H3 lysine 79 in Saccharomyces cerevisiae as a substrate for Sir2. Proceedings of the National Academy of Sciences. 2012;109:E916–E925. doi: 10.1073/pnas.1121471109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parsons XH, Garcia SN, Pillus L, Kadonaga JT. Histone deacetylation by Sir2 generates a transcriptionally repressed nucleoprotein complex. Proceedings of the National Academy of Sciences. 2003;100:1609–1614. doi: 10.1073/pnas.0434064100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ye Y, Stahley MR, Xu J, Friedman JI, Sun Y, McKnight JN, Gray JJ, Bowman GD, Stivers JT. Enzymatic Excision of Uracil Residues in Nucleosomes Depends on the Local DNA Structure and Dynamics. Biochemistry. 2012;51:6028–6038. doi: 10.1021/bi3006412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hayes JJ, Tullius TD, Wolffe AP. The structure of DNA in a nucleosome. Proceedings of the National Academy of Sciences. 1990;87:7405–7409. doi: 10.1073/pnas.87.19.7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jain SS, Tullius TD. Footprinting protein–DNA complexes using the hydroxyl radical. Nature protocols. 2008;3:1092–1100. doi: 10.1038/nprot.2008.72. [DOI] [PubMed] [Google Scholar]
- 34.Das R, Laederach A, Pearlman SM, Herschlag D, Altman RB. SAFA: semi-automated footprinting analysis software for high-throughput quantification of nucleic acid footprinting experiments. Rna. 2005;11:344–354. doi: 10.1261/rna.7214405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chua EYD, Vasudevan D, Davey GE, Wu B, Davey CA. The mechanics behind DNA sequence-dependent properties of the nucleosome. Nucleic Acids Research. 2012;40:6338–6352. doi: 10.1093/nar/gks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vasudevan D, Chua EYD, Davey CA. Crystal Structures of Nucleosome Core Particles Containing the ‘601’ Strong Positioning Sequence. Journal of Molecular Biology. 2010;403:1–10. doi: 10.1016/j.jmb.2010.08.039. [DOI] [PubMed] [Google Scholar]
- 37.Zhou P, Wu E, Alam HB, Li Y. Histone cleavage as a mechanism for epigenetic regulation: current insights and perspectives. Current molecular medicine. 2014;14:1164. doi: 10.2174/1566524014666141015155630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vitolo JM, Thiriet C, Hayes JJ. The H3-H4 N-terminal tail domains are the primary mediators of transcription factor IIIA access to 5S DNA within a nucleosome. Molecular and Cellular Biology. 2000;20:2167–2175. doi: 10.1128/mcb.20.6.2167-2175.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vettese-Dadey M, Walter P, Chen H, Juan L-J, Workman JL. Role of the histone amino termini in facilitated binding of a transcription factor, GAL4-AH, to nucleosome cores. Molecular and Cellular Biology. 1994;14:970–981. doi: 10.1128/mcb.14.2.970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Godde JS, Nakatani Y, Wolffe AP. The amino-terminal tails of the core histones and the translational position of the TATA box determine TBP/TFIIA association with nucleosomal DNA. Nucleic Acids Research. 1995;23:4557–4564. doi: 10.1093/nar/23.22.4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beard BC, Stevenson JJ, Wilson SH, Smerdon MJ. Base excision repair in nucleosomes lacking histone tails. DNA Repair. 2005;4:203–209. doi: 10.1016/j.dnarep.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 42.Chafin DR, Vitolo JM, Henricksen LA, Bambara RA, Hayes JJ. Human DNA ligase I efficiently seals nicks in nucleosomes. The EMBO journal. 2000;19:5492–5501. doi: 10.1093/emboj/19.20.5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fu I, Cai Y, Geacintov NE, Zhang Y, Broyde S. Nucleosome Histone Tail Conformation and Dynamics: Impacts of Lysine Acetylation and a Nearby Minor Groove Benzo[a]pyrene-Derived Lesion. Biochemistry. 2017;56:1963–1973. doi: 10.1021/acs.biochem.6b01208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim J, Lee J, Lee T-H. Lysine acetylation facilitates spontaneous dna dynamics in the nucleosome. The Journal of Physical Chemistry B. 2015;119:15001–15005. doi: 10.1021/acs.jpcb.5b09734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim J, Wei S, Lee J, Yue H, Lee T-H. Single-Molecule Observation Reveals Spontaneous Protein Dynamics in the Nucleosome. The Journal of Physical Chemistry B. 2016;120:8925–8931. doi: 10.1021/acs.jpcb.6b06235. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.