Abstract
Asymmetrical division can be a reason for microbial populations heterogeneity. In particular, budding yeast daughter cells are more vulnerable to stresses than the mothers. It was suggested that yeast mother cells could also differ from each other depending on their replicative age. To test this, we measured the levels of Idh1-GFP, Idh2-GFP, Trx2-GFP, Pdr5-GFP and Can1-GFP proteins in cells of the few first, most represented, age cohorts. Pdr5p and Can1p were selected because of the pronounced mother-bud asymmetry for these proteins distributions, Trx2p as indicator of oxidative stress. Isocitrate dehydrogenase subunits Idh1p and Idh2p were assessed because their levels are regulated by mitochondria. We found a small negative correlation between yeast replicative age and Idh1-GFP or Idh2-GFP but not Trx2-GFP levels. Mitochondrial network fragmentation was also confirmed as an early event of replicative aging. No significant difference in the membrane proteins levels Pdr5p and Can1p was found. Moreover, the elder mother cells showed lower coefficient of variation for Pdr5p levels compared to the younger ones and the daughters. Our data suggest that the levels of stress-response proteins Pdr5p and Trx2p in the mother cells are stable during the first few cell cycles regardless of their mother-bud asymmetry.
Introduction
Some microorganisms show significant levels of individual cells heterogeneity1–3. This could be due to DNA replication errors and subsequent proliferation of the new subclones within the population4. However, there can also be non-genetic sources of the heterogeneity. Some factors that are present in the cells at very low copy numbers5 can be unevenly distributed during the cell divisions. For instance, an inequality in the inherited mitochondria can render differences between the individual cells6. A switch of aggregation-prone proteins to prion states was also shown to be a source of phenotypic heterogeneity in yeast populations7. Another source of cell-to-cell variability are cycling processes. The low-frequency localization pulsing of stress-response transcription factors was reported in yeast8. Finally, the concentrations of many mRNAs and proteins are also fluctuating due to subsequent activation of the factors specific to the cell cycle stages9,10.
Additionally, asymmetrical cell division is an important mechanism of mother and daughter cells diversification. For instance, it was shown that in stationary phase Saccharomyces cerevisiae mother and daughter cells segregate into the populations of ‘nonquiescent’ and ‘quiescent’ cells3,11. Moreover, baker’s yeast cells have the limited capacity for budding12. This phenomenon is usually referred to as “replicative aging”. It was shown that after 15–20 divisions mother cells reach a senescence entry point and then die13,14. However, due to the dilution by the newborn cells the proportion of such replicatively old ones is extremely low (see for instance15). At the same time, some changes were reported for relatively young yeast ‘cohorts’, which are the mostly represented ones in yeast populations. First, Kale & Jazwinski16 have shown that the resistance to genotoxic stresses is different between yeast mother cells of relatively young ‘cohorts’. Next, the change of mitochondrial morphology was reported for yeast cells of replicative age 5–917. We have also shown recently that the resistance to acetic acid stresses and heat shock is higher in yeast mother cells within replicative age 2–418 compared to that of the daughter cells or the older mothers. We found that the percentage of cells with distinguishably different mitochondrial populations is enriched in yeast cells with age ≥519. Finally, it was shown that aggregates containing Hsp104 accumulate in yeast cells which have already produced 3–6 buds, while being absent in the newborn cells20. According to these observations, it was suggested that the replicative age provides additional source for cell variability21.
If this is indeed the case, yeast mother cells of highly represented age ‘cohorts’ (i.e. with 1–9 bud scars) are expected to carry significantly different concentration of several proteins. We reasoned that, as the level of stress-induced damage increases with the replicative age, the abundances of stress-response proteins are also likely to correlate with the age. To test this hypothesis we selected three stress-response proteins: (1) mitochondrial isocitrate dehydrogenases Idh1p, Idh2p which levels are regulated by mitochondria-to-nucleus signaling pathway, activated by dysfunctional mitochondria22–24 and (2) cytoplasmic thioredoxin Trx2p. TRX2 gene is regulated by H2O2-sensor transcription factor Yap1p25, thus Trx2p concentration in individual cell reflects the level of oxidative stress response activation.
We have also selected two other proteins, Pdr5p and Can1p, mostly because they are supposed to show a significant asymmetry in mother-daughter distribution. Pdr5p is a plasma membrane pleiotropic drug resistance ABC-transporter, its expression is regulated by transcription factors PDR1 and PDR3 26,27. These transcription factors are activated, among others, in response to mitochondrial dysfunction28. Changes of mitochondria functioning are one of the earliest manifestations of yeast aging17,19, so we reasoned that this protein is likely to be accumulated in mid-aged mother cells. Can1p is arginine permease, it has been recently found to be one of the key regulators of yeast replicative lifespan29.
Thus, we measured the levels of Trx2-GFP, Idh1-GFP, Idh2-GFP, Pdr5-GFP and Can1-GFP in the yeast cells of different replicative age cohorts. As a positive control for our method, we have also tested the heterologously expressed fragment of human huntingtin 103Q-CFP. This protein is known to form aggregates, which in yeast tend to remain in the mother cells30. We found that while Trx2-GFP, Pdr5-GFP, Can1-GFP did not show any significant variation with mother cells age, the levels of Idh1-GFP\Idh2-GFP and 103Q-CFP showed negative and positive correlations correspondingly.
Results
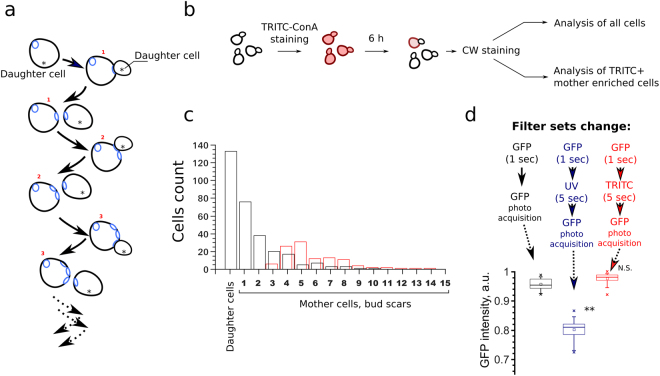
Asymmetrical division of yeast cells allow to attribute a replicative age (the number of produced buds) to each cell (Fig. 1a). The number of cells in each replicative age cohort is approximately a half of the number of previous (younger) replicative age cohort. In addition to the biological significance of this fact, it makes it technically difficult to find in yeast suspension the cells of replicative age higher than five. To increase sample size of mothers cells with age 5–9 we stained yeast cells with TRITC-ConA and allowed them to perform 4–5 divisions afterwards (Fig. 1b, see materials and methods). TRITC-ConA positive cells were substantially enriched by mother cells with 4–8 bud scars (Fig. 1c). Therefore, in each experiment in addition to whole population of yeast cells we analyzed a fraction of TRITC-ConA positive cells. The age of yeast cells was analyzed by the number of bud scars visualized by Calcofluor White (CW), while the levels of proteins were estimated by the average intensity of GFP in cells (see methods section for details). Importantly, we calculated bud scars of the cells only after they were photographed in GFP-channel. Indeed, even short illumination of yeast cells with UV-light (U-MNU2 filter set, required for CW staining) bleached GFP, while that was not the case for green-light (U-MNIBA3 filter set, required for TRITC-ConA staining, see Fig. 1d).
Figure 1.
Experimental approach. (a) Yeast cell division cycles result in the formation of the mother cells with different numbers of chitin enriched bud scars. Asterisks indicate the buds (daughter cells), digits - the replicative age (number of bud scars) of mother cells. (b) An outline of the performed experimental procedures. Exponentially growing yeast cells were stained with TRITC-ConA, then grown for 6 hours, stained with CW (Calcofluor white) and analyzed. In each biological replicate the first 40–60 randomly selected cells were photographed in GFP- and then CW-channel (analysis of all cells), afterwards 40–60 TRITC-positive cells from the same sample were selected and photographed in the same channels. (c) Distribution of yeast cells in the replicative aging cohorts (black bars). The distribution of TRITC-ConA enriched mother cells (red bars). (d) The ratio of Trx2-GFP fluorescence intensity before and after the illumination with U-MNU2 (UV, excitation wavelength λ = 360–370 nm) and U-MNG2 (TRITC, excitation wavelength λ = 530–550 nm) filter sets. **P value < 0.01 according to Wilcoxon rank-sum unpaired test.
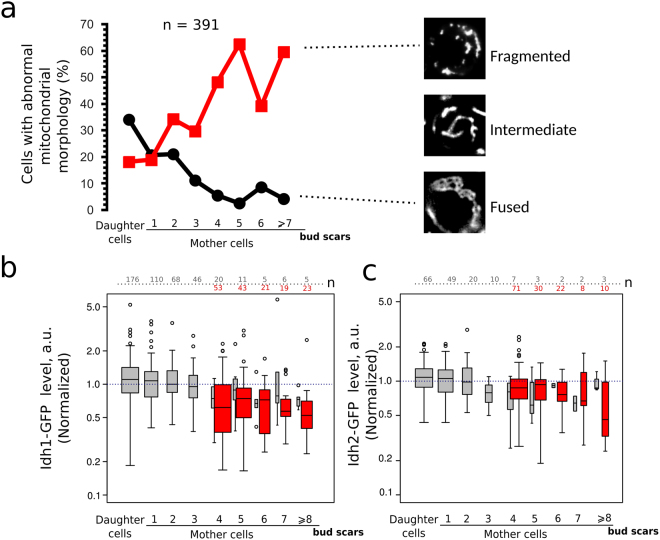
Is it possible to explain heterogeneity of yeast clonal cultures by the replicative age dependent accumulation of some proteins in mother cells? To test this we measured the levels of the proteins chosen for the aforementioned reasons in the yeast age cohorts. We suggested earlier that accumulation of dysfunctional mitochondria can be the reason for the early decline of the stress resistance in yeast cells19. Therefore, as the first step we compared the mitochondrial morphology and the levels of Idh1-GFP protein in the different replicative age cohorts. IDH1 is regulated by retrograde mitochondria-to-nucleus signaling pathway31. We noticed that in accordance with the results of Lam et al.17, there is a correlation between the yeast age and mitochondrial reticulum morphology (Fig. 2a). While the age positively correlated with the percentage of the cells with fragmented mitochondria the proportion of yeast cells with the hyperfused morphology decreased with age (Fig. 2a). At the same time we found a significant negative correlation between yeast mother cell replicative age and the level of Idh1-GFP (Table 1, Fig. 2b). Same correlation of age and protein level was found for another isoform of isocitrate dehydrogenase Idh2 (Table 1, Fig. 2c).
Figure 2.
The levels of Idh1-GFP and mitochondrial morphology in yeast cells correlate with the age of the mother cells. (a) The percentage of yeast cells with fragmented or fused mitochondrial network morphology. The data (391 cells) were pooled from four separate day experiments. Correlation coefficient (Kendall’s tau) for yeast age and percentage of yeast cells with fragmented mitochondria is equal to 0.714 (P = 0.014). The difference between the age cohorts of the mother cells is significant, p-value = 0.00018 according to Pearson’s Chi-squared test with simulated p-value. The representative photographs of different types mitochondrial morphology networks visualized by mitochondrial protein Idh1-GFP are shown in the right. Idh1-GFP (b) and Idh2-GFP (c) levels in individual cells of different replicative age cohorts. Grey boxplots represent the random samples of yeast cells. There is a significant negative correlation between yeast mother cells age and Idh1-GFP level (see Table 1). Red box plot represents TRITC+ (age > 4) enriched mother cells. The numbers of analyzed cells for each age class are shown at the top of the boxplot. The bin size for TRITC+ and TRITC− cells are designated separately.
Table 1.
Mother-bud asymmetry and the correlation of protein levels with replicative age of the mother cells.
| Protein and conditions | Correlation of protein level with replicative age of mothers. Kendall’s tau (NS — non significant) | Mother/bud asymmetry (ratio of the means, p-value according to Wilcoxon Mann Whitney two-tailed test) |
|---|---|---|
| Idh1-GFP | −0.149* (p = 0.000976) | 1.01 p = 0.206 |
| Idh2-GFP | −0.197* (p = 0.01082) | 0.88 p =0.009253 |
| Trx2-GFP | 0.078NS | 1.18 p = 3.5 × 10−6 |
| Trx2-GFP, H2O2 | 0.01NS | 1.28 p = 0.048 |
| Pdr5-GFP | 0.07NS | 1.38 p = 2 × 10−5 |
| Pdr5-GFP, clotrimazole | 0.076NS | 0.97 p = 0.66 |
| Can1-GFP | 0.049NS | 1.49 p = 10−16 |
| 103Q-CFP | 0.11* (p = 0.02656) | 1.76 p = 10−16 |
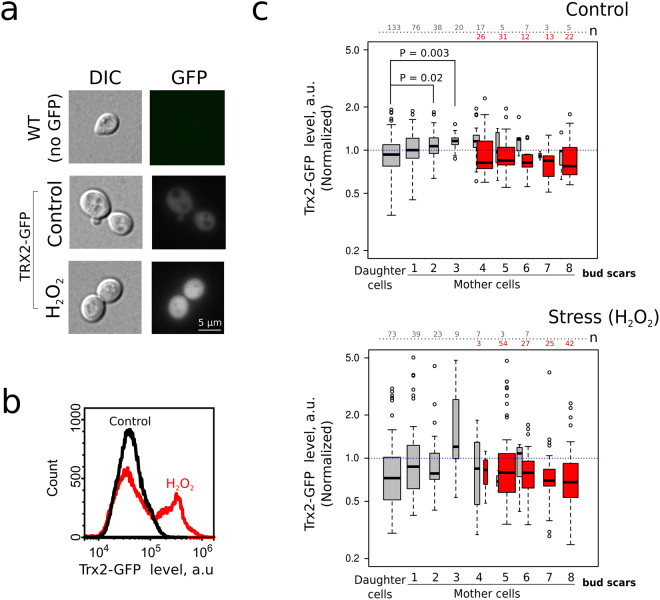
The mitochondrial dysfunction could be a reason for redox imbalance and oxidative stress32. Increased hydrogen peroxide levels in yeast cytoplasm induce relocalization of transcription factor Yap1p and subsequent activation of the target genes, in particular, TRX2 33,34. To assess Yap1p activation we measured Trx2-GFP levels in yeast cells (Fig. 3a). We expected that replicative age-dependent mitochondrial dysfunction can be manifested in differential activation of Yap1p depending on the replicative age of a cohort. Indeed, we found that only a portion of cells increased Trx2-GFP levels in response to exogenous hydrogen peroxide (Fig. 3b). However, the analysis of the different age cohorts showed that there was no statistically significant difference in Trx2-GFP levels, with the exception of mother-to-bud asymmetry in the absence of hydrogen peroxide (Fig. 3c, Table 1).
Figure 3.
Trx2-GFP level displays mother-daughter asymmetry but does not differ between the mother cells of different ages. (a) Representative photograph of control yeast cells and yeast cells expressing Trx2-GFP. Treatment of yeast cells with H2O2 (2 mM) induces an increase of Trx2-GFP levels; (b) Analysis of Trx2-GFP levels by flow cytometry under the control conditions and in the presence of 2 mM H2O2; (c) Quantification of Trx2-GFP in yeast cells of different age cohorts under the control conditions (upper panel) or treated with H2O2. Red box plots indicate Trx2-GFP levels in TRITC-ConA positive (age > 4 enriched) mother cells. The numbers of analyzed cells for each age class are shown at the top of the boxplot. The bin size for TRITC+ and TRITC− cells are designated separately. The only detected difference was between the untreated daughter and the mother cells (*P < 0.05; **P < 0.005 according to Nemenyi test).
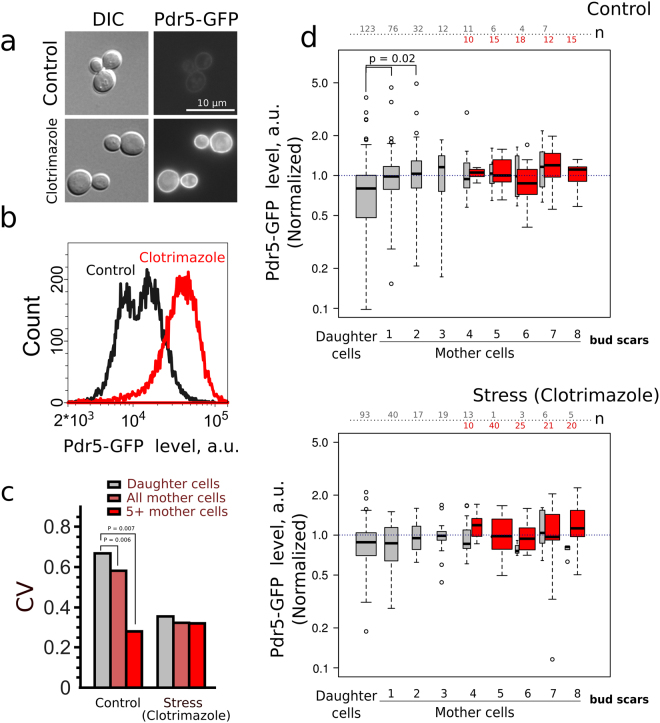
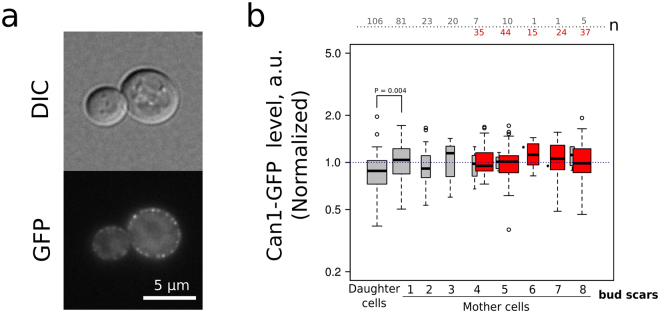
Next, we tested the levels of major pleiotropic drug resistance pump Pdr5p. Functional Pdr5p is localized in plasma membrane, therefore we analyzed only the border area of the cell (see Figure S1). Clotrimazole is an inducer of PDR5 gene35. Accordingly, we have shown that clotrimazole supplementation induces accumulation of Pdr5-GFP in yeast cells (Fig. 4a,b). To test the ability of yeast cells belonging to different age cohorts to activate mechanisms of antibiotic resistance we compared Pdr5-GFP levels and variance in the absence and in the presence of clotrimazole (Fig. 4c,d). Surprisingly, despite pronounced mother-to-bud asymmetry of Pdr5-GFP levels the Kruskal–Wallis H test did not show significant difference among cohorts of mother cells (see Table 1). Moreover, there was no significant correlation between mother cells age and Pdr5-GFP levels (see Table 1). Interestingly, we found that there is a significantly lower coefficient of variation of Pdr5-GFP levels in ‘mature’ (TRITC+) yeast mother cells than in overall mother cell population which is enriched with ‘young’ mother cells (Fig. 4c). Clotrimazole treatment did not induce age-specific accumulation of Pdr5-GFP (Fig. 4d, lower panel; Table 1). Similar results were obtained when we compared the levels of arginine permease Can1-GFP, the plasma membrane protein, in yeast of different age cohort. CAN1 was recently identified in screening for most noisy S.cerevisiae genes36. However, despite the significant mother-to-bud asymmetry (Fig. 5, Table 1) we did not find any age-dependent variation of Can1-GFP levels.
Figure 4.
The level of Pdr5-GFP in yeast cells does not differ in the mother cells aging cohorts. (a) A representative photograph of the control and clotrimazole treated (20 μM) cells. (b) Quantification of Pdr5-GFP levels increase induced by clotrimazole. (c) Cell-to-cell coefficient of variation (CV) of Pdr5-GFP levels for daughter cells, all mother cells and TRITC+ (age > 4 enriched) mother cells with replicative age above five. P-value was calculated with Fligner-Killeen (median) test. (d) Pdr5-GFP levels in individual cells of different yeast cells replicative age cohorts. Grey boxplots represent the random samples of yeast cells, the red box plot represents TRITC+ cells. The control and the stress conditions (20 μM clotrimazole) were analyzed. The numbers of analyzed cells for each age class are shown at the top of the boxplot. The bin size for TRITC+ and TRITC− cells are designated separately. P value was calculated by Nemenyi-Test.
Figure 5.
The level of Can1-GFP does not differ in the mother cells aging cohorts. (a) Representative photograph of the control yeast cells expressing Can1-GFP. (b) Can1-GFP levels in the individual cells of different replicative age cohorts (n = 410 cells). Grey boxplots represent the random samples of yeast cells, the red box plot represents TRITC+ (age > 4 enriched) mother cells. The numbers of analyzed cells for each age class are shown at the top of the boxplot. The bin size for TRITC+ and TRITC− cells are designated separately. P value was calculated according Nemenyi-Test.
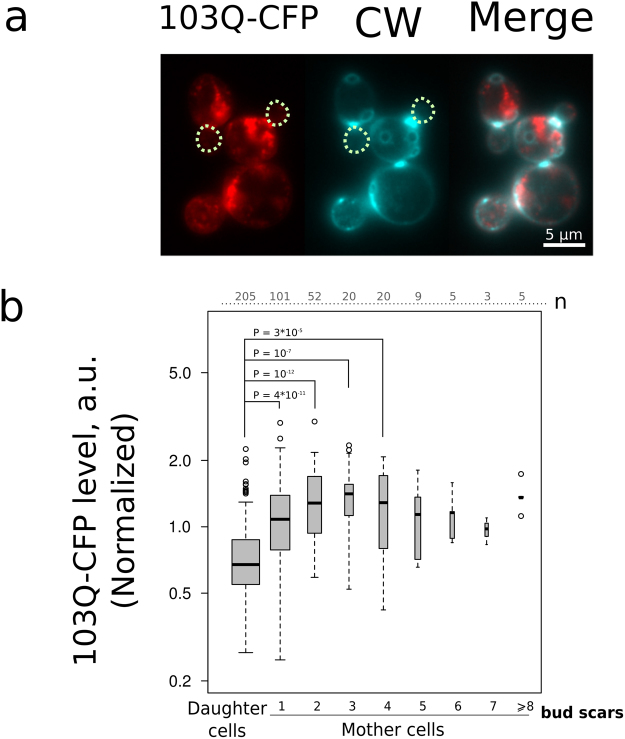
Apart from plasma membrane proteins, protein aggregates can be asymmetrically distributed between the mother cell and the bud. This asymmetry is a result of either constrained diffusion across the bud neck37 or an active directed transport of such aggregates38. We expected that the continuous protein synthesis can supply aggregates with additional components eventually making the old mother cells enriched by such aggregates if compared to ‘young’ mother cells. To test this possibility we used polyglutamine (103Q) fragment of human mutant huntingtin gene fused with cyan fluorescent protein (CFP)39,40. The gene was set under the regulation of PGAL promoter. 103Q-CFP was expressed for 5 hours and assessed using fluorescent microscopy. The daughter cells were found to be mainly devoid of CFP aggregates (Fig. 6a, Table 1). As expected, we found a small positive correlation (Kendall’s Tau 0.11, P = 0.02) between replicative age and average intensity of CFP in cells (Table 1). Notably, this correlation arose mostly from the differences within the several first age cohorts of the mother cells (Fig. 1b). As 103Q-CFP was expressed for 5 hours (i.e. 3–4 cell divisions), there was no increase in CFP levels within mother cells of replicative age above four.
Figure 6.
103Q-CFP level displays mother-daughter asymmetry. (a) Representative photograph of the group of cells expressing 103Q-CFP (red) stained with calcofluor white (blue). Image was generated by pseudo colouring: CFP-channel black and white image was copied to R channel, CW-channel image — to G and B channels of the RGB file. The dotted circles mark daughter cells. (b) Quantification of the average 103Q-CFP levels for replicative age cohorts. The numbers of analyzed cells for each age class are shown at the top of the boxplot. P value was calculated by Nemenyi-Test.
Discussion
The age-dependent differentiation of asymmetrically divided microorganisms might contribute to optimization of their performance under stressful conditions. Indeed, it is suggested that heterogeneity of some traits can benefit microorganisms upon stress36,41. In the case of pathogens, the inhibition of such mechanisms could be a promising strategy to prevent the survival of the relatively resistant subpopulation. It was shown that old mother cells of some pathogenic yeast species are more resistant to antimycotics or host immune system. This leads to an increased proportion of the old cells in the host42–44. Understanding the mechanisms of yeast replicative aging is important not only because it is a model of mammalian aging but also because it can help to reveal the bet hedging strategies acting to increase the survival of the clonal population.
Baker’s yeast S.cerevisiae is a widely used model system for aging studies. It was found that some genetic modifications can change average replicative lifespan in the range of 20–30 generations45–47. However, yeast mother cells of replicative age above 20 are diluted by more than million of their descendants and thus are excluded from the selection pressure. Accordingly, few first replicative age cohorts represent the majority of the cells (see Fig. 1c). We suggested that the active mechanisms of cell components partitioning could cause significant differences between these ‘major’ mother cell cohorts. If so, the genesis of such differences could be an important mechanism of phenotypic heterogeneity in yeast clonal populations. In our work we compared the levels of several selected proteins to test this possibility. We found that the levels of mitochondrial protein Idh1 negatively correlated with replicative age of yeast exponentially growing population (Fig. 2). The increasing replicative age was also associated with the changes of mitochondrial morphology. The latter observation is in line with the work of Lam et al.17 who showed that the changes of mitochondrial morphology are one of the first manifestations of replicative aging. In the same work the authors found another early hallmark of aging — increased reactive oxygen species (ROS) levels (as detected by dihydroethidium bromide staining). An increase in the reactive oxygen species (ROS) production rate and the reshaping of mitochondrial network can induce each other. Indeed, on the one hand, dysfunctional mitochondria promote ROS accumulation in the cells32. On the other hand, ROS can induce depolarization of mitochondria48,49 in this way inhibiting mitochondrial fusion and promoting fragmentation of mitochondrial network50,51. We did not find any indications of early age-associated ROS increase — there was no correlation between Trx2-GFP levels and replicative age in our experiments (Fig. 3). Importantly, the Trx2-GFP levels were responsive to exogenously added hydrogen peroxide (Fig. 3a,b). Therefore, during the course of yeast replicative aging the reshaping of mitochondrial network takes place earlier than ROS accumulation.
The inhibition of mitochondrial fission increases the lifespan of yeast and filamentous fungi52. The latter is not surprising because mitochondrial fission and fusion cycles are closely interconnected with cell metabolism and stress resistance53. In our work we tested the possible connections between age-dependent mitochondrial network reshaping and the accumulation of major pleiotropic drug resistance ABC-transporter Pdr5p in the plasma membrane. The possible regulatory connection between mitochondrial dynamics and PDR5 expression has been recently found54. Earlier, it was found that mitochondrial dysfunction activates pleiotropic drug resistance28,55. Importantly, in the screening based on the method of transposon mutagenesis for PDR5 activators, three out of five insertions were found in yeast mitofusin gene FZO1 55. The product of this gene is responsible for mitochondria fusion. However, we found that in the case of membrane proteins Pdr5p and Can1p the significant mother-to-bud asymmetry did not lead to a significant trends of the protein levels changes in the mother cells (Figs 4 and 5). We explain these results by the orchestrated proteostasis of these plasma membrane proteins which compensates the possible variability of PDR5 expression that can be induced by disorganized mitochondrial network. For instance, it was shown that Pdr5p and Can1p can be degraded via ubiquitin-dependent endocytosis56,57. Otherwise, the age-dependent difference of mitochondrial network during early phases of yeast replicative aging could be an epiphenomenon, which does not affect any parameters determining cell fitness.
Newborn daughter cells emerge with the reset replicative lifespan, while deleterious processes are expected to randomly affect the cell systems with age. Therefore, we expected an increased variation of stress-response proteins levels in replicative age cohorts of elder mother cells compared to daughter cells or young mothers. We also hypothesized that bet hedging of PDR5 expression level could be a reasonable strategy for cells to survive possible environmental stresses. However, we did not find any drift in Pdr5-GFP levels in yeast mother cells with age. Moreover, we found that the coefficient of variation of Pdr5-GFP levels was significantly lower in the mother cells with replicative age above five if compared to the daughter cells (Fig. 4c). This data indicates that individual variation of Pdr5 levels is achieved only at the level of mother-bud asymmetry, whereas in mother cells the levels are stable.
To summarize, we compared the level of five yeast proteins and one heterologous protein in different replicative age cohorts of yeast cells. Our results showed that cell-to-cell variations in the expression of these proteins are mostly due to the mother-bud difference. We, however, confirmed the previous reports showing that mitochondrial network morphology decreases its interconnectivity already during early replicative aging. Also a small age-dependent decrease in Idh1p and Idh2p levels was found for mother cells. This result suggests that a decrease in isocitrate dehydrogenase subunits levels may cause the early age-dependent decline in TCA cycle capacity. At the same time, the coefficients of variation of Idh1-GFP for mother and daughter cells were not significantly different (according to Fligner-Killeen test). Therefore, our results did not provide any strong evidence for the hypothesis of mother cell age-specific differentiation. Probably a study of less abundant proteins will show higher correlations with mother cell age. We suggest that changes related to mitochondria — organelles which have some autonomy from the nuclear regulation — can initiate the differentiation in asymmetrically divided eukaryotes.
Material and Methods
Yeast strains and growth conditions
In the study we used TRX2-GFP, PDR5-GFP, IDH2-GFP and CAN1-GFP yeast strains from yeast GFP clone collection58. IDH1-GFP (IDH1-GFP LEU2 TRP1 W303-1A) and 103Q-CFP (PGAL-103Q-CFP HIS3 W303-1A) were constructed earlier in our laboratory in W303 mat a genetic background40,59. Yeast strains were grown in YPD (yeast peptone d-glucose) liquid medium with the exception of 103Q-CFP strain. 103Q-CFP was grown in YPRaf (yeast peptone raffinose), the expression of PGAL-103Q-CFP was induced by replacement of YPRaf with YPRafGal (yeast peptone raffinose and galactose) medium for five hours. Yeast media (YPD, YPRaf, YPRafGal) were prepared as described by Sherman60.
Microscopy
For microscopy yeast cells were grown overnight on solid YPD medium, then resuspended in 200 µl PBS buffer, stained with with TRITC-ConA (tetramethylrhodamine conjugate of concanavalin A, 50 µg/ml) and incubated for 10 minutes at room temperature, then washed twice with PBS buffer, added to 5 ml liquid YPD medium and cultivated for 6 hours (unless indicated otherwise) to allow 4–5 rounds of budding. Importantly, TRITC-ConA does not affect GFP fluorescence (Fig. S4). One could argue that the observed decrease of Idh1-GFP with age is a result of the transition from solid to liquid medium. However, we obtained the similar results in the experiments with prolonged preincubation in liquid YPD medium (Figure S5). For stress induction, 2 mM H2O2 or 20 µM clotrimazole were added and the cells were incubated for additional 2–3 hours. Thereafter cells were stained with 5 µM calcofluor white (CW) in order to count the bud scars. Then the cells were washed to remove the medium and resuspended in PBS buffer. Fluorescence microscopy was carried out using Olympus BX51 upright microscope, UPLANFLN 100x lens (N.A. 1.3) with the set of filters presented in Table S1. Images were taken with Olympus DP30BW CCD camera using Olympus CellF software. Color photographs of the cells were generated by pseudocoloring: GFP-channel black and white image was copied into green channel of RGB file.
We imaged cells in GFP-channel and afterwards were switching to CW-channel to count the bud scars. Imaged cells were ranged by their replicative age as follows: a single cell without any bud scars or a bud larger than half of the volume of it’s mother cell were counted as ‘daughter cells’, a single cell with one bud scar or a cell without a bud scar but with a bud - as age 1, etc. During each experiment we imaged 50 cells at random (i.e. it can be considered as random sampling from yeast population) and 50 TRITC-positive cells to enrich data with the old mother cells.
GFP levels in cells were measured using ImageJ software. Average signal intensity was measured for each cell, background signal intensity was subtracted. For each experiment the individual cell signal intensity was normalized to the population mean. The latter step was necessary due to the moderate differences between separate day experiments that were probably due to the instability in the mercury lamp excitation intensity.
In case of Trx2-GFP, Idh1-GFP, Idh2-GFP and 103Q-CFP we analyzed the whole cell fluorescence signals. To determine Pdr5-GFP and Can1-GFP levels, we measured the signal only within the plasma membrane area (0.25 µm [4 px] wide cell perimeter, Fig. S1). We used GIMP and Inkscape to crop and arrange the figure panels.
Mitochondrial morphology assay
Cells expressing Idh1-GFP were photographed in GFP-channel (see Table S1) and the number of bud scars was counted and recorded in CW-channel. Next, photograph of each individual cell was attributed to one of three mitochondrial morphology classes: fused, intermediate or fragmented. Attribution was made by visual similarity between sample photographs of each aging class (Fig. 2a inset, Figure S3). Importantly, attribution was made by experimentator who did not know the replicative age of each cell. As a result, the table of individual cells with age and mitochondrial morphology attribute was generated.
Flow cytometry
For FACS experiments yeast cells were grown overnight in liquid YPD medium to 4 × 107 cells/ml, then the stress was induced by 2 mM H2O2 or 20 µM clotrimazole followed by 2–3 hour incubation. Yeast cells were washed and resuspended in PBS buffer. To quantify GFP fluorescence we performed flow cytometric analysis using CytoFLEX flow cytometer (Beckman Coulter). All flow cytometry data were analyzed with CytExpert software (Beckman Coulter). At least 10000 cells were analyzed per experiment.
Data Analysis and visualization
Each data set (i.e. strain, conditions) was analyzed first by Kruskall-Wallis test. Each age cohort of yeast cells was considered as separate experimental condition for this test. None of the tested conditions showed any significance if only mother cells were taken for analysis. If a result of Kruskall-Wallis test (including daughter cells) was significant (P < 0.05), than Nemenyi test was applied and all P values below cut-off threshold (P < 0.05) were indicated on the figures. For Nemenyi test mother cells with replicative age above 8 were considered as a single condition. Chi-squared contingency table test with Monte Carlo simulation (50000 replicates) was used to analyze difference in mitochondrial morphology between different age cohorts. Fligner-Killeen Test of Homogeneity of Variances was used to test of the null hypothesis that the variances in each of the age cohorts are the same. Cells that were found using TRITC-ConA visual enrichment were analyzed separately from the cells that were photographed without visual selection procedures. Data is plotted as boxplots with boxes widths proportional to the square-roots of the number of observations in the groups. Tests were conducted using R. Data were visualized with R or QtiPlot.
Data Availability
The datasets generated and/or analysed during the current study are available from the corresponding author on reasonable request.
Electronic supplementary material
Acknowledgements
We are grateful to Dr. Eugene Rikhvanov and Dr. Boris Feniouk for useful comments on our manuscript. The experiments on mitochondrial dynamics (Figure 2) were supported by Russian Foundation for Basic Research grant 16-04-01381; The rest of the work (Figures 1, 3–6) were conducted with the support of Russian Science Foundation grant 14-50-00029.
Author Contributions
D.K. and F.S. designed the experiments. A.A., K.G. and M.S. performed the experiments. A.A., K.G., M.S. and D.K. analyzed the data. A.A., K.G., and D.K. prepared the figures. D.K. drafted the manuscript. All authors contributed to manuscript editing and reviewed the final manuscript. D.K. and F.S. supervised the project and acquired funding.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-13576-w.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Dubnau D, Losick R. Bistability in bacteria. Mol. Microbiol. 2006;61:564–572. doi: 10.1111/j.1365-2958.2006.05249.x. [DOI] [PubMed] [Google Scholar]
- 2.Veening J-W, et al. Bet-hedging and epigenetic inheritance in bacterial cell development. Proc. Natl. Acad. Sci. USA. 2008;105:4393–4398. doi: 10.1073/pnas.0700463105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aragon AD, et al. Characterization of differentiated quiescent and nonquiescent cells in yeast stationary-phase cultures. Mol. Biol. Cell. 2008;19:1271–1280. doi: 10.1091/mbc.E07-07-0666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Halme A, Bumgarner S, Styles C, Fink GR. Genetic and epigenetic regulation of the FLO gene family generates cell-surface variation in yeast. Cell. 2004;116:405–415. doi: 10.1016/S0092-8674(04)00118-7. [DOI] [PubMed] [Google Scholar]
- 5.Ghaemmaghami S, et al. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 6.Guantes R, Díaz-Colunga J, Iborra FJ. Mitochondria and the non-genetic origins of cell-to-cell variability: More is different. Bioessays. 2016;38:64–76. doi: 10.1002/bies.201500082. [DOI] [PubMed] [Google Scholar]
- 7.Halfmann R, et al. Prions are a common mechanism for phenotypic inheritance in wild yeasts. Nature. 2012;482:363–368. doi: 10.1038/nature10875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dalal CK, Cai L, Lin Y, Rahbar K, Elowitz MB. Pulsatile dynamics in the yeast proteome. Curr. Biol. 2014;24:2189–2194. doi: 10.1016/j.cub.2014.07.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spellman PT, et al. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cho RJ, et al. A genome-wide transcriptional analysis of the mitotic cell cycle. Mol. Cell. 1998;2:65–73. doi: 10.1016/S1097-2765(00)80114-8. [DOI] [PubMed] [Google Scholar]
- 11.Allen C, et al. Isolation of quiescent and nonquiescent cells from yeast stationary-phase cultures. J. Cell Biol. 2006;174:89–100. doi: 10.1083/jcb.200604072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mortimer RK, Johnston JR. Life span of individual yeast cells. Nature. 1959;183:1751–1752. doi: 10.1038/1831751a0. [DOI] [PubMed] [Google Scholar]
- 13.Fehrmann S, et al. Aging yeast cells undergo a sharp entry into senescence unrelated to the loss of mitochondrial membrane potential. Cell Rep. 2013;5:1589–1599. doi: 10.1016/j.celrep.2013.11.013. [DOI] [PubMed] [Google Scholar]
- 14.Janssens GE, Veenhoff LM. The Natural Variation in Lifespans of Single Yeast Cells Is Related to Variation in Cell Size, Ribosomal Protein, and Division Time. PLoS One. 2016;11:e0167394. doi: 10.1371/journal.pone.0167394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maxwell, C. S. & Magwene, P. M. The Quick and the Dead: Microbial Demography at the Yeast Thermal Limit. Mol. Ecol. 10.1111/mec.13955 (2016). [DOI] [PubMed]
- 16.Kale SP, Jazwinski SM. Differential response to UV stress and DNA damage during the yeast replicative life span. Dev. Genet. 1996;18:154–160. doi: 10.1002/(SICI)1520-6408(1996)18:2<154::AID-DVG8>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 17.Lam YT, Aung-Htut MT, Lim YL, Yang H, Dawes IW. Changes in reactive oxygen species begin early during replicative aging of Saccharomyces cerevisiae cells. Free Radic. Biol. Med. 2011;50:963–970. doi: 10.1016/j.freeradbiomed.2011.01.013. [DOI] [PubMed] [Google Scholar]
- 18.Knorre DA, et al. Sir2-dependent daughter-to-mother transport of the damaged proteins in yeast is required to prevent high stress sensitivity of the daughters. Cell Cycle. 2010;9:4501–4505. doi: 10.4161/cc.9.22.13683. [DOI] [PubMed] [Google Scholar]
- 19.Sorokin MI, Knorre DA, Severin FF. Early manifestations of replicative aging in the yeast Saccharomyces cerevisiae. Microbial Cell. 2014;1:37–42. doi: 10.15698/mic2014.01.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saarikangas, J. & Barral, Y. Protein aggregates are associated with replicative aging without compromising protein quality control. Elife4, (2015). [DOI] [PMC free article] [PubMed]
- 21.Jazwinski SM, Egilmez NK, Chen JB. Replication control and cellular life span. Exp. Gerontol. 1989;24:423–436. doi: 10.1016/0531-5565(89)90049-1. [DOI] [PubMed] [Google Scholar]
- 22.Liu Z, Butow RA. A transcriptional switch in the expression of yeast tricarboxylic acid cycle genes in response to a reduction or loss of respiratory function. Mol. Cell. Biol. 1999;19:6720–6728. doi: 10.1128/MCB.19.10.6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Z, Butow RA. Mitochondrial retrograde signaling. Annu. Rev. Genet. 2006;40:159–185. doi: 10.1146/annurev.genet.40.110405.090613. [DOI] [PubMed] [Google Scholar]
- 24.Zyrina, A. N., Smirnova, E. A., Markova, O. V., Severin, F. F. & Knorre, D. A. Mitochondrial Superoxide Dismutase and Yap1p Act as a Signaling Module Contributing to Ethanol Tolerance of the Yeast Saccharomyces cerevisiae. Appl. Environ. Microbiol. 83 (2017). [DOI] [PMC free article] [PubMed]
- 25.Kuge, S. & Jones, N. YAP1 dependent activation of TRX2 is essential for the response of Saccharomyces cerevisiae to oxidative stress by hydroperoxides. EMBO J (1994). [DOI] [PMC free article] [PubMed]
- 26.Balzi E, Wang M, Leterme S, Van Dyck L, Goffeau A. PDR5, a novel yeast multidrug resistance conferring transporter controlled by the transcription regulator PDR1. J. Biol. Chem. 1994;269:2206–2214. [PubMed] [Google Scholar]
- 27.Dexter D, Moye-Rowley WS, Wu AL, Golin J. Mutations in the yeast PDR3, PDR4, PDR7 and PDR9 pleiotropic (multiple) drug resistance loci affect the transcript level of an ATP binding cassette transporter encoding gene, PDR5. Genetics. 1994;136:505–515. doi: 10.1093/genetics/136.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Panwar SL, Moye-Rowley WS. Long chain base tolerance in Saccharomyces cerevisiae is induced by retrograde signals from the mitochondria. J. Biol. Chem. 2006;281:6376–6384. doi: 10.1074/jbc.M512115200. [DOI] [PubMed] [Google Scholar]
- 29.Beaupere C, et al. CAN1 Arginine Permease Deficiency Extends Yeast Replicative Lifespan via Translational Activation of Stress Response Genes. Cell Rep. 2017;18:1884–1892. doi: 10.1016/j.celrep.2017.01.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu B, et al. Segregation of protein aggregates involves actin and the polarity machinery. Cell. 2011;147:959–961. doi: 10.1016/j.cell.2011.11.018. [DOI] [PubMed] [Google Scholar]
- 31.Liao X, Butow RA. RTG1 and RTG2: two yeast genes required for a novel path of communication from mitochondria to the nucleus. Cell. 1993;72:61–71. doi: 10.1016/0092-8674(93)90050-Z. [DOI] [PubMed] [Google Scholar]
- 32.Barros MH, Netto LES, Kowaltowski AJ. H(2)O(2) generation in Saccharomyces cerevisiae respiratory pet mutants: effect of cytochrome c. Free Radic. Biol. Med. 2003;35:179–188. doi: 10.1016/S0891-5849(03)00307-1. [DOI] [PubMed] [Google Scholar]
- 33.Stephen DW, Rivers SL, Jamieson DJ. The role of the YAP1 and YAP2 genes in the regulation of the adaptive oxidative stress responses of Saccharomyces cerevisiae. Mol. Microbiol. 1995;16:415–423. doi: 10.1111/j.1365-2958.1995.tb02407.x. [DOI] [PubMed] [Google Scholar]
- 34.Coleman ST, Epping EA, Steggerda SM, Moye-Rowley WS. Yap1p activates gene transcription in an oxidant-specific fashion. Mol. Cell. Biol. 1999;19:8302–8313. doi: 10.1128/MCB.19.12.8302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Golin J, et al. Studies with novel Pdr5p substrates demonstrate a strong size dependence for xenobiotic efflux. J. Biol. Chem. 2003;278:5963–5969. doi: 10.1074/jbc.M210908200. [DOI] [PubMed] [Google Scholar]
- 36.Liu J, et al. Natural yeast promoter variants reveal epistasis in the generation of transcriptional-mediated noise and its potential benefit in stressful conditions. Genome Biol. Evol. 2015;7:969–984. doi: 10.1093/gbe/evv047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou C, et al. Motility and segregation of Hsp104-associated protein aggregates in budding yeast. Cell. 2011;147:1186–1196. doi: 10.1016/j.cell.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu B, et al. The polarisome is required for segregation and retrograde transport of protein aggregates. Cell. 2010;140:257–267. doi: 10.1016/j.cell.2009.12.031. [DOI] [PubMed] [Google Scholar]
- 39.Meriin AB, et al. Huntington toxicity in yeast model depends on polyglutamine aggregation mediated by a prion-like protein Rnq1. J. Cell Biol. 2002;157:997–1004. doi: 10.1083/jcb.200112104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sokolov S, Pozniakovsky A, Bocharova N, Knorre D, Severin F. Expression of an expanded polyglutamine domain in yeast causes death with apoptotic markers. Biochimica et Biophysica Acta - Bioenergetics. 2006;1757:660–666. doi: 10.1016/j.bbabio.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 41.Holland SL, Reader T, Dyer PS, Avery SV. Phenotypic heterogeneity is a selected trait in natural yeast populations subject to environmental stress. Environ. Microbiol. 2014;16:1729–1740. doi: 10.1111/1462-2920.12243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bouklas T, et al. Generational distribution of a Candida glabrata population: Resilient old cells prevail, while younger cells dominate in the vulnerable host. PLoS Pathog. 2017;13:e1006355. doi: 10.1371/journal.ppat.1006355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bouklas T, Fries BC. Aging as an emergent factor that contributes to phenotypic variation in Cryptococcus neoformans. Fungal Genet. Biol. 2015;78:59–64. doi: 10.1016/j.fgb.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bouklas, T. et al. Old Cryptococcus neoformans cells contribute to virulence in chronic cryptococcosis. MBio4 (2013). [DOI] [PMC free article] [PubMed]
- 45.Kim S, Benguria A, Lai CY, Jazwinski SM. Modulation of life-span by histone deacetylase genes in Saccharomyces cerevisiae. Mol. Biol. Cell. 1999;10:3125–3136. doi: 10.1091/mbc.10.10.3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McCormick MA, et al. A Comprehensive Analysis of Replicative Lifespan in 4,698 Single-Gene Deletion Strains Uncovers Conserved Mechanisms of Aging. Cell Metab. 2015;22:895–906. doi: 10.1016/j.cmet.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kang, W. K., Kim, Y. H., Kang, H. A., Kwon, K.-S. & Kim, J.-Y. Sir2 phosphorylation through cAMP-PKA and CK2 signaling inhibits the lifespan extension activity of Sir2 in yeast. Elife4 (2015). [DOI] [PMC free article] [PubMed]
- 48.Trendeleva T, Sukhanova E, Ural’skaya L, Saris N-E, Zvyagilskaya R. Effect of prooxidants on yeast mitochondria. J. Bioenerg. Biomembr. 2011;43:633–644. doi: 10.1007/s10863-011-9403-2. [DOI] [PubMed] [Google Scholar]
- 49.Zorov DB, Juhaszova M, Sollott SJ. Mitochondrial reactive oxygen species (ROS) and ROS-induced ROS release. Physiol. Rev. 2014;94:909–950. doi: 10.1152/physrev.00026.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu S, Zhou F, Zhang Z, Xing D. Mitochondrial oxidative stress causes mitochondrial fragmentation via differential modulation of mitochondrial fission-fusion proteins. FEBS J. 2011;278:941–954. doi: 10.1111/j.1742-4658.2011.08010.x. [DOI] [PubMed] [Google Scholar]
- 51.Rogov, A. G., Ovchenkova, A. P., Goleva, T. N., Kireev, I. I. & Zvyagilskaya, R. A. New yeast models for studying mitochondrial morphology as affected by oxidative stress and other factors. Anal. Biochem. 10.1016/j.ab.2017.04.003 (2017). [DOI] [PubMed]
- 52.Scheckhuber CQ, et al. Reducing mitochondrial fission results in increased life span and fitness of two fungal ageing models. Nat. Cell Biol. 2007;9:99–105. doi: 10.1038/ncb1524. [DOI] [PubMed] [Google Scholar]
- 53.Mishra P, Chan DC. Metabolic regulation of mitochondrial dynamics. J. Cell Biol. 2016;212:379–387. doi: 10.1083/jcb.201511036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mutlu N, Garipler G, Akdoğan E, Dunn CD. Activation of the pleiotropic drug resistance pathway can promote mitochondrial DNA retention by fusion-defective mitochondria in Saccharomyces cerevisiae. G3. 2014;4:1247–1258. doi: 10.1534/g3.114.010330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hallstrom TC, Moye-Rowley WS. Multiple signals from dysfunctional mitochondria activate the pleiotropic drug resistance pathway in Saccharomyces cerevisiae. J. Biol. Chem. 2000;275:37347–37356. doi: 10.1074/jbc.M007338200. [DOI] [PubMed] [Google Scholar]
- 56.Egner R, Mahé Y, Pandjaitan R, Kuchler K. Endocytosis and vacuolar degradation of the plasma membrane-localized Pdr5 ATP-binding cassette multidrug transporter in Saccharomyces cerevisiae. Mol. Cell. Biol. 1995;15:5879–5887. doi: 10.1128/MCB.15.11.5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ghaddar K, et al. Substrate-induced ubiquitylation and endocytosis of yeast amino acid permeases. Mol. Cell. Biol. 2014;34:4447–4463. doi: 10.1128/MCB.00699-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huh W-K, et al. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 59.Karavaeva IE, et al. Mitochondrial depolarization in yeast zygotes inhibits clonal expansion of selfish mtDNA. J. Cell Sci. 2017;130:1274–1284. doi: 10.1242/jcs.197269. [DOI] [PubMed] [Google Scholar]
- 60.Sherman F. Getting started with yeast. Methods Enzymol. 2002;350:3–41. doi: 10.1016/S0076-6879(02)50954-X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analysed during the current study are available from the corresponding author on reasonable request.