Abstract
Since the report of its discovery in E. coli in late 2015, the plasmid-mediated colistin resistance gene, mcr-1, has been detected in various bacterial species in clinical setting and various environmental niches. However, the transmission mechanisms of this gene in Salmonella is less defined. In this study, we conducted a comprehensive study to characterize the genetic features of mcr-1-positive Salmonella strains isolated from animals and foods. Our data revealed that Salmonella recovered from animals and food specimens exhibited highly different PFGE patterns, and acquired mcr-1-encoding plasmids via different mechanism. Plasmids harboring mcr-1 in Salmonella food isolates were all conjugative and similar as plasmids reported in other species of Enterobacteriaceae, whereas mcr-1-bearing plasmids from animal Salmonella isolates were not conjugative, and belonged to the IncHI2 type. The lack of a region carrying the tra genes was found to account for the inability to undergo conjugation for various sizes of IncHI2 plasmids harbored by animal strains. These data suggest that transmission of mcr-1-positive Salmonella from animal to food might not be a common event and food isolates may have acquired mcr-1-bearing plasmids from other mcr-1-positive bacteria such as E. coli, which co-exist in food samples.
Introduction
Since the discovery of the mobile colistin resistance determinant mcr-1 in E. coli, it has been reported in almost all parts of the world and in many different species of bacteria1–7. It was speculated that widespread dissemination of this resistance element may be due to the extensive use of polymyxin as growth promoter in animals, imposing an enormous selection pressure on this genetic element in Enterobacteriaceae in particular E. coli. It is important to know the prevalence and transmission of mcr-1 in key Enterobacteriaceae species such as Salmonella, which is an important human pathogen prevalent in animal GI tract in particular chicken and pigs. The mcr-1 gene was first reported in Salmonella through analysis of WGS sequence available in GenBank, in which mcr-1-bearing plasmids were identified in 10 clinical S. enterica isolates submitted between 2012 and 2015, including 8S. Typhimurium, 1S. Paratyphi B var Java and 1S. Virchow strains8. The mcr-1 gene was subsequently reported to be recoverable from Salmonella strains isolated from food, animals and clinical specimens in Europe, the US and China9–15. Three types of mcr-1-carrying plasmids including IncX4, IncI1and IncHI2 types of plasmids of various sizes have been reported in Salmonella 9,11,12. However, these data remain scattered, and do not provide a comprehensive view on the prevalence of the mcr-1 gene in Salmonella or the transmission kinetics of the different types of mobile elements that harbor such resistance determinant. To address these important issues, we investigated the prevalence of the mcr-1 gene in Salmonella strains recovered from animals and food collected nationwide in China and investigated the transmission potential of the mcr-1 gene recovered from two different categories of Salmonella isolates. Our data provided a comprehensive view of the extent of mcr-1 contamination in Salmonella in animals and food in China, and essential insights into the transmission mechanism of mcr-1 in Salmonella.
Results
Prevalence of mcr-1 in Salmonella isolated from animals and food samples
A total of 1312 non-repeated Salmonella isolates were recovered from animal fecal samples in various regions of China during a four-years Nationwide Salmonella Antimicrobial Resistance Surveillance Program (2012–2015) conducted in the China Institute of Veterinary Drug Control. A total of 175 Salmonella isolated were obtained from 1914 animal fecal samples in 2012; 290 isolates were obtained from 3205 animal fecal samples in 2013; 333 isolates were obtained from 2980 fecal samples in 2014 and 514 were obtained from 3586 fecal samples in 2015. The animal fecal samples were collected from Shanghai, Sichuan, Shandong, Guangdong, Inner Mongolia, Liaoning, Henan, Hunan, Hubei, Yunnan, Chongqing, Shaanxi, Shanxi, Jiangxi, Jilin, Hebei, Guangxi, Fujian, Beijing and Anhui provinces or municipalities. Among these 1312 Salmonella isolates, 569 isolates were from chicken fecal samples and 743 were from pig fecal samples. The mcr-1 gene screening for these 1312 Salmonella isolates showed that 26 (2%) Salmonella isolates were positive to mcr-1, among which 19 were from 2013 and 7 were from 2015. No mcr-1-positive Salmonella isolate was detected in Salmonella isolates collected in 2012 and 2014. A total of 830 Salmonella strains isolated from various of food samples including chicken, pork, beef and seafood in Shenzhen in 2015, and 230 food-borne Salmonella isolates collected from various provinces and submitted to the China CDC during the period 2012~2015 were tested. Twelve out of 830 (1.4%) Salmonella isolates collected from Shenzhen were positive for mcr-1; the corresponding positive rate for the CDC samples was 1.3% (4 out of 230).
Antimicrobial susceptibility and serotype distribution of mcr-1-positive Salmonella
All mcr-1 positive Salmonella isolates were subjected to determination of their susceptibilities to 14 antibiotics, the results of which were shown in Table 1. These isolates generally displayed a high rate of resistance to other antibiotics, in addition to colistin, yet substantial variations between strains recovered from animal and food were observed. Animal isolates exhibited a very high rate of resistance to most of test agents, while none of the Salmonella strains in this category were resistant to ceftriaxone. In contrast, 13% of Salmonella isolates from food were resistant to ceftriaxone, while the resistance rate to other antibiotics tested was relatively low compared to organisms recovered from animals (Table 1).
Table 1.
Antimicrobial susceptibility of mcr-1-positive Salmonella strains isolated from different sources.
| Antibiotics | Break-point (µg/ml) | % resistance | ||
|---|---|---|---|---|
| Food isolates | ||||
| WT (n = 15) | TC (n = 15) | Animal isolates (n = 26) | ||
| Ampicillin | ≥32 | 94 | 94 | 100 |
| Amoxicillin/Clavulanic acid | ≥32/16 | 13 | 13 | 62 |
| Ceftriaxone | ≥4 | 13 | 13 | 0 |
| Ceftazidime | ≥4 | 13 | 13 | 0 |
| Cefotaxime | ≥4 | 13 | 13 | 0 |
| Meropenem | ≥4 | 13 | 13 | 0 |
| Chloramphenicol | ≥32 | 75 | 13 | 96 |
| Gentamicin | ≥16 | 31 | 26 | 100 |
| Nalidixic acid | 32 | 38 | 0 | 96 |
| Ciprofloxacin | ≥1 | 25 | 0 | 100 |
| Trimethoprim/ Sulfamethoxazole | ≥4/76 | 44 | 17 | 96 |
| Tetracycline | ≥16 | 100 | 0 | 100 |
| *Colistin | ≥4 | 100 | 100 | 100 |
| Azithromycin | ≥16 | 0 | 0 | 0 |
WT, wild type; TC, transconjugants;
*there is no CLSI breakpoint for colistin in Enterobacteriaceae. The forthcoming 2017 CLSI guideline proposed it to be 4 µg/ml.
Serotypes and genetic relatedness of mcr-1-positive Salmonella strains
Most of the mcr-1-positive Salmonella strains were found to be S. Typhimurium. The 26 animal isolates included 21S. Typhimurium, and 5S. Newport. Among the 21S. Typhimurium strains, 19 were isolated in different locations in Henan province in 2013 and were found to exhibit almost identical PFGE pattern, suggesting that clonal dissemination of this specific clone in animals in Henan province had occurred; 2 were isolated from Shandong province in 2015 and exhibited identical PFGE pattern. Another 5 animal isolates belonged to S. Newport that were isolated from Guangdong and Guangxi provinces with two different PFGE patterns. Among the food-borne Salmonella isolates, 9 were S. Derby that belonged to the same clone and were isolated from different locations in Shenzhen on the same day in 2015; five were S. Typhimurium, among which three were isolated from Shenzhen with very similar PFGE pattern and two were from other parts of China with different PFGE patterns, and one was S. Welteweden. Interestingly, mcr-1-positive Salmonella isolated from animal and food showed very different serotypes and PFGE pattern. None of the food Salmonella isolate showed identical PFGE pattern as animal Salmonella isolate, which might suggest that the transmission of mcr-1-positve animal Salmonella to food products is not a common event (Fig. 1).
Figure 1.
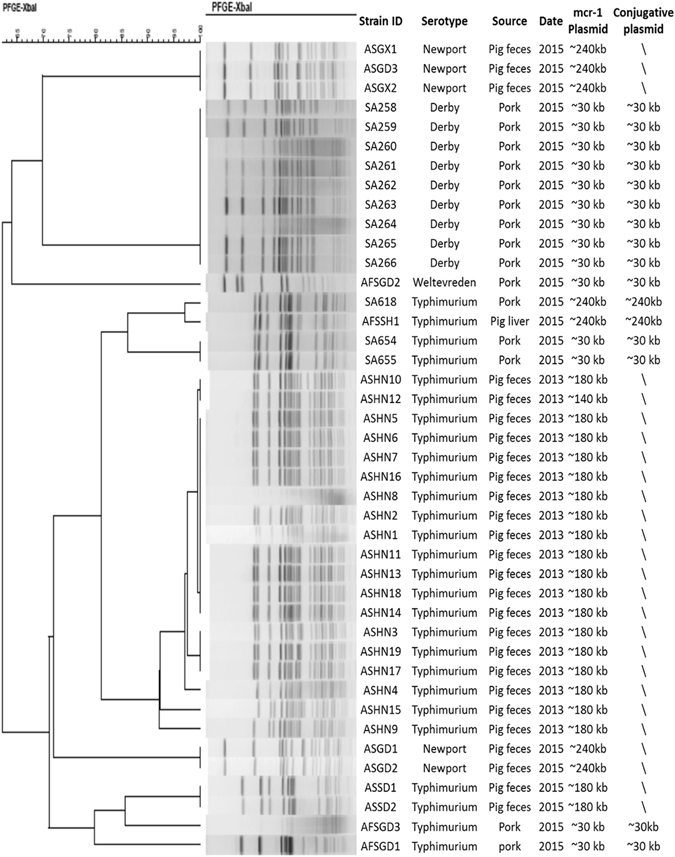
Summary of genetic characteristics of mcr-1-bearing Salmonella strains isolated from different sources. The isolation location and sources of the Salmonella isolates were designed as follow: ASHN1-ASHN19, Salmonella strains isolated from animal fecal samples from Henan province; ASSD1-ASSD2; Salmonella strains isolated from animal fecal samples from Shandong province; ASGD1-ASGD3, Salmonella strains isolated from animal fecal samples from Guangdong province; ASGX1-ASGX2, Salmonella strains isolated from animal fecal samples from Guangxi province; AFSGD1-AFSGD3, Salmonella strains isolated from food samples from Guangdong province; AFSSH1, Salmonella strains isolated from food samples from Shanghai province; SA258-SA266, Salmonella strains isolated from food samples in Shenzhen.
Mechanisms of transmission of mcr-1 among Salmonella of different sources
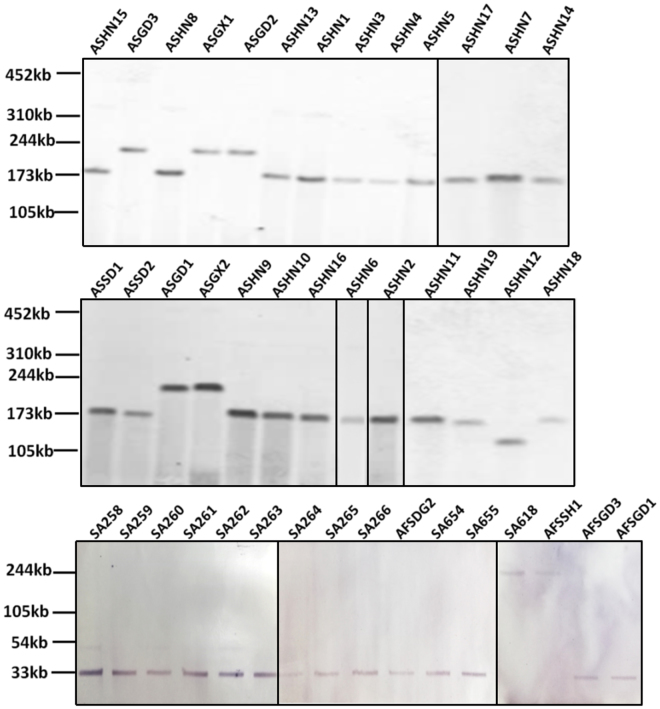
To investigate the transmission potential of mcr-1 among Salmonella strains isolated from animal and food, all mcr-1-positive strains were tested for their ability to undergo conjugation. Surprisingly, strains isolated from different sources exhibited highly different conjugation rate: the mcr-1 gene in all the animal isolates tested were not conjugative, while the gene in all food isolates were conjugative. S1-PFGE and Southern hybridization were then performed all animal Salmonella isolates and both parental strains and transconjugants of food isolates to investigate the genetic features of plasmids recoverable from these Salmonella strains. Among the 26 animal isolates tested, 20 were found to contain one ~180 kb mcr-1-bearing plasmid, 5 contained a ~240 kb plasmid and one contained a ~140 kb plasmid. Among the 16 Salmonella food isolates tested, conjugative plasmids of 30 kb and 240 kb in size were recoverable from 14 and 2 strains respectively (Figs 1, 2).
Figure 2.
S1-PFGE and Southern Hybridization of all Salmonella isolates from animal and food. The results of hybridization to mcr-1 probe was shown.
Genetic features of mcr-1-encoding plasmids recovered from Salmonella strains isolated from different sources
To further delineate the genetic features of different conjugative and non-conjugative plasmids harboring the mcr-1 element, representative plasmids were selected from different strains including two ~30 kb and one ~250 kb plasmid from food isolates, one each of ~140 kb and ~240 kb and two ~180 kb plasmids from animal isolates, for sequencing using the Illumina platforms. One ~180 kb plasmid recovered from strain ASSD2 was further sequenced by the PacBio platform to obtain the complete map (Table 2). Illumina contiges of two ~30 kb plasmids harbored by Salmonella strains, S. Weltevreden AFSGD2 and S. Derby SA258, were obtained and aligned to several previously reported plasmids, pOW3E1(KX129783.1) and pECJP-B65-33 (KX084392.1). These two plasmids were shown to belong to IncX4 type and could be aligned very well to pOW3E1(KX129783.1) ( > 99% in both identity and coverage) (Data not shown).
Table 2.
Origin and genetic features of mcr-1-bearing plasmids in Salmonella strains subjected to sequence analysis in this study.
| Strain ID | Serotype | source | Year of Isolation | S1-PFGE | CN | Plasmid types | Complete sequences/ contigs |
|---|---|---|---|---|---|---|---|
| AFSGD2 | Weltervredn | PK | 2014 | ~30 kb | C | IncX4 | contigs |
| SA258 | Derby | PK | 2015 | ~30 kb | C | IncX4 | contigs |
| ASGD2 | Newport | PS | 2015 | ~240 kb | NC | IncHI2 | contigs |
| ASSD2 | Typhimurium | PS | 2015 | ~180 kb | NC | IncHI2 | pASSD2-MCR1 |
| ASHN8 | Typhimurium | PS | 2013 | ~180 kb | NC | IncHI2 | contigs |
| ASHN12 | Typhimurium | PS | 2013 | ~140 kb | NC | IncHI2 | contigs |
| SA618 | Typhimurium | PS | 2015 | ~250 kb | C | IncHI2 | contigs |
HS, human stool; PK, pork; PS, pig feces; CN, conjugative nature; C, conjugative; NC, non-conjugative.
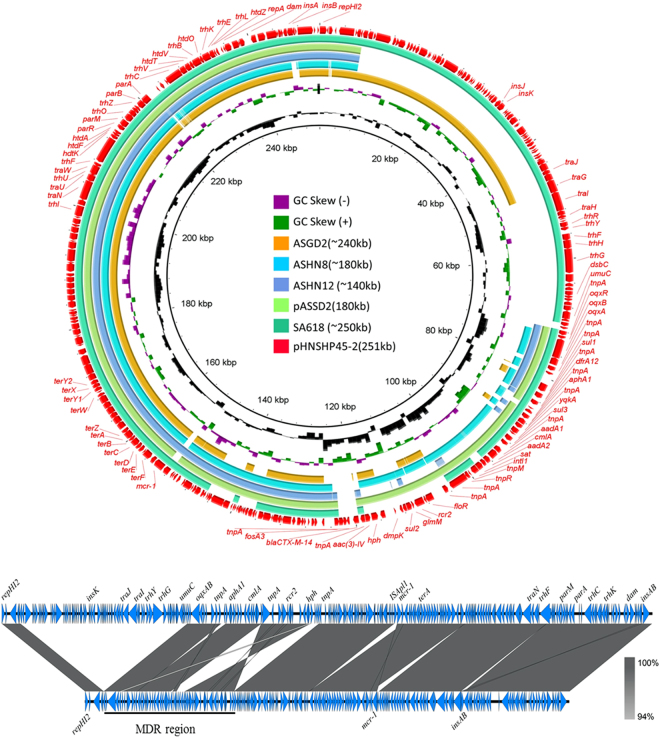
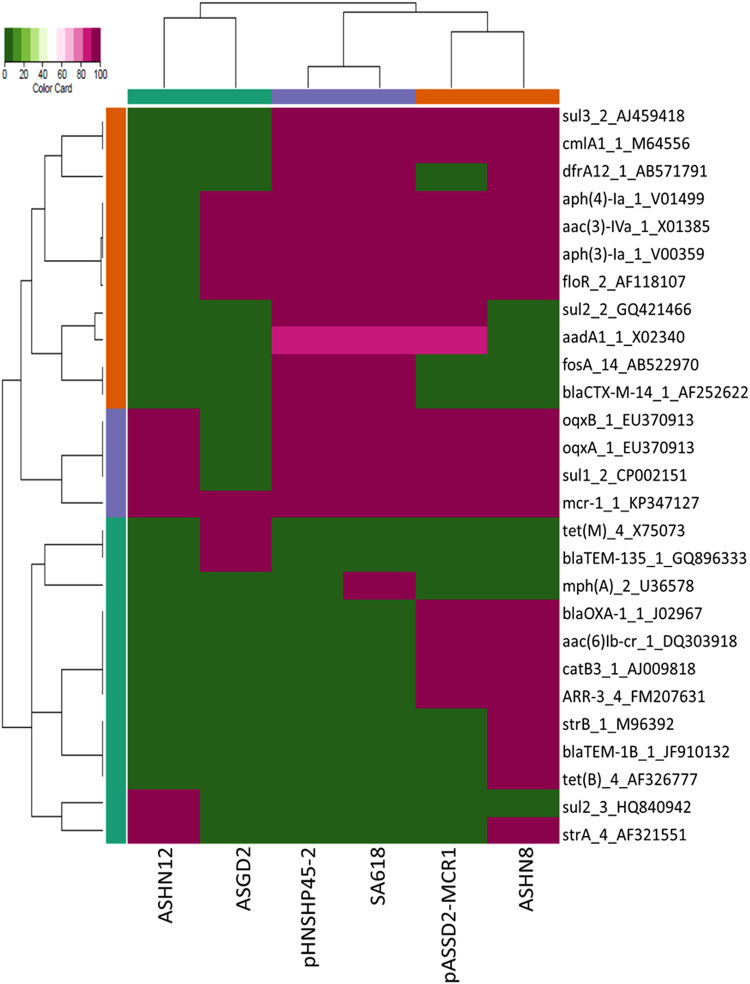
The Illumina sequencing data showed that both conjugative or non-conjugative plasmids with sizes of ~140 kb, ~180 kb, ~240 kb and ~250 kb belonged to the IncHI2 type, which contained all or part of the previously reported ~251 kb plasmid pHNSHP45-2 (KU341381.1) (Fig. 3). The illumina sequence of a conjugative ~250 kb plasmid recovered from S. Typhimurium SA618 was obtained and compared to pHNSHP45-2, with results showing that this conjugative plasmid displayed a high degree of sequence homology to pHNSHP45-2 (Fig. 3). Antimicrobial resistance gene analysis showed that conjugative plasmid from SA618 contained one more mphA(2) gene (Fig. 4). Interestingly, alignment of Illumina reads of the non-conjugative ~240 kb plasmids recovered from the S. Newport strains ASGD2 to pHNSHP45-2 revealed that this plasmid aligned very well with pHNSHP45-2 except that a region containing the tra genes, which were responsible for conjugation, was absent in the plasmid of strain ASGD2, as well as some mobile elements in the Multidrug Resistance (MDR) Region was absent (Fig. 3). Antimicrobial resistance gene analysis showed that the plasmid from S. Newport strains ASGD2 lacked the mobile elements carrying bla CTX-M-14, oqxAB, fosA3, aadA1 and other antimicrobial resistance genes such as dfrA12 and cmlA1, but gained mobile elements carrying other antimicrobial resistance genes and transposase genes such as tetM and bla TEM135. Interestingly, different from pHNSHP45-2 that carried an intact Tn6330, the mobile element carrying mcr-1 on this plasmid lacked the downstream ISApl1. The lack of a tra region may explain why the ~240 kb plasmids of Salmonella animal isolates were non-conjugative (Figs 3, 4).
Figure 3.
Alignment of conjugative and non-conjugative IncHI2 plasmids/contigs against pHNSHP45-2. (a) the circular map was created by BRIG tools; the linear map was generated by EasyFig. Genes in the reference plasmid, pHNSHP45-2, which was reported previously, are labeled by red arrows. Two other plasmids, pHSHLJ1-MCR1, pASSD2-MCR1 and contigs of other plasmids labeled with different colors were aligned to the reference plasmid. The gaps in the plasmid sequences represent the missing sequences when compared to the reference plasmid; (b) alignment of pASSD2-MCR1 to pHNSHP45-2 using Easyfig. The major different between these two plasmids is the absence of a gene fragment encoding tra genes that are responsible for plasmid conjugation was detected in pASSD2-MCR1 accounting for its smaller size compared to pHNSHP45-2. In addition, some variations were also detected in the MCR regions of these two plasmids. The other backbone regions in these two plasmids were almost identical.
Figure 4.
Antimicrobial resistance-related gene analysis for all IncHI2 plasmids with various sizes. Plasmid information was shown in Table 2.
The complete sequence of one ~180 kb, non-conjugative plasmid recovered from the S. Typhimurium strain ASSD2, namely pASSD2-MCR1, was obtained and shown to be 187, 257 bp in size. This plasmid was found to belong to IncHI2 and exhibit high level homology to pHNSHP45-2. Detailed alignment between pHNSHP45-2 and the non-conjugative plasmid pASSD2-MCR1 revealed several different genetic rearrangements including: (i) the mcr-1 gene without ISApl1 in the upstream in pASSD2-MCR1 was located in different sites compared to plasmid pHNSHP45-2; (ii) the mobile element insAB was repeatedly inserted in different locations in pASSD2-MCR1; (iii) the MDR region differed by one inversion; (iv) a large region of ca. 70 kb harboring the tra regions was absent in pASSD2-MCR1; (v) loss of mobile element carrying bla CTX-M-14 and fosA3 and gaining a mobile element carrying bla OXA-1, aac(6)-Ib-cr and catB3 (Figs 3, 4). These different genetic features, detectable among the two plasmids, depicted the evolution routes of the IncI2 class of mcr-1-carrying plasmids, and indicated that the loss of the ca. 70 kb tra region in pASSD2-MCR1 could contribute to the loss of transferability of this plasmid. The illumina sequence data of the other non-conjugative plasmids with sizes of ~180 kb recovered from Salmonella ASHN8 was shown to be highly homologous to pASSD2-MCR1(187 kb) and belong to the IncHI2 type plasmid, and that it obtained some more mobile elements carrying different antimicrobial resistance genes such as StrAB, bla TEM and tetB (Fig. 4). Similar as the ~240 kb plasmid in this study, the mobile element carrying mcr-1 on the ~180 kb plasmids also lacked the downstream ISApl1. The ~140 kb plasmid recovered from S. Typhimurium strain ASHN12 was very similar to pASSD2-MCR1(187 kb) in the plasmid backbone region, but with further deletions in the MDR region. It lacked almost all the mobile elements carrying antimicrobial resistance gene presence in pHNSHP45-2, but only remained the mobile element carrying oqxAB gene. It also acquired one more antimicrobial resistance genes, StrA gene compared to pHNSHP45-2 (Figs 3, 4). Interesting, plasmid from ASHN12 was found to carry the intact Tn6330 as reported in plasmid, pHNSHP45-2, with two copies of ISApl1 located up- and down-stream of the mcr-1 gene.
Genetic features of mcr-1-positive plasmids obtained from various sources were consistent with the antimicrobial resistance phenotypes of Salmonella isolates. All animal Salmonella isolates carried mcr-1 plasmids without the bla CTX-M-14 and fosA3 genes, therefore none of them was resistant to cephalosporins and fosfomycin. Two Salmonella isolates from food carried conjugative ~250 kb plasmids harboring bla CTX-M-14 and fosA3 mobile elements, therefore these strains were resistant to cephalosporins and fosfomycin.
Discussion
Compared to other bacterial species of Enterobacteriaceae, the prevalence rate of mcr-1 in Salmonella was found to be much lower in each of these surveys, regardless of the site of recovery16–20. The low prevalence of mcr-1 was also reported in a study from China, England and Wales3,21. However, another study in China reported the high prevalence of Salmonella from animals9. In addition, our data showed that most of the Salmonella strains that harbored mcr-1-bearing plasmids were S. Typhimurium, suggesting that mcr-1-bearing plasmids might have strong association with specific serotypes of Salmonella. The close association between S. Typhimurium and mcr-1-bearing plasmids entails further investigation. In animal and food Salmonella isolates, clonal spread of mcr-1-positive Salmonella was common. S. Derby strains isolated from food samples in Shenzhen, China were genetically identical, and were therefore likely the result of clonal dissemination of a single mcr-1-bearing strain. All the animal isolates could be grouped into three clusters: 19S. Typhimurium isolated in 2013 in Henan province exhibited similar PFGE patterns, five S. Newport strains isolated from 2015 belonged to two types of PFGE patterns and two S. Typhimurium isolated from 2015 were genetically identical. These data suggested that the mcr-1-positive Salmonella is similar from same location, while diverse between different part of China. However, it is interesting that mcr-1-positive Salmonella from food and animal did not share similar PFGE pattern, suggesting the less common event of transmission of mcr-1-positive Salmonella from animals to food samples.
This phenomenon was further supported by data on genetic analysis of mcr-1-bearing plasmids carried by Salmonella strains isolated from animals and food specimens. All mcr-1-bearing plasmids recovered from animal Salmonella isolates were non-conjugative with sizes of ~140 kb, ~180 kb or ~240 kb, the representative of which were shown to be IncHI2. However, all mcr-1-bearing plasmids from food Salmonella isolates were conjugative and belonged to two types, ~33 kb IncX4 and ~250 kb IncHI2, which exhibited identical sequences to plasmids of similar types recoverable from other bacterial species in the family of Enterobacteriaceae. These two types of plasmids were not detected in Salmonella isolates of animal origin, suggesting that they may be transmitted to Salmonella from other mcr-1-positive bacteria such as E. coli which co-exist in food but not in animal GI tract.
In conclusion, results of this nationwide surveillance of mcr-1 and its transmission mechanisms in Salmonella provide comprehensive understanding of the features and mechanisms of transmission of mcr-1 in Salmonella recovered from different settings, and hence lay the foundation for future development of strategies to control the transmission of this colistin resistance determinant among Gram negative bacterial pathogens.
Materials and Methods
Salmonella strains
Salmonella strains were collected from animals and food nationwide in China. All test strains were isolated in CHROMagar Salmonella agar (CHROMagar Company, Paris, France) and XLT4 agar (Oxoid). Suspected Salmonella colonies were selected for biochemical confirmation using the API 20 E system (bioMérieux, Marcy l’ Etoile, France), as well as via molecular identification by PCR assay targeting the invA gene, followed by sequencing. Salmonella serotyping was conducted by performing the slide agglutination test, using Salmonella antisera (S & A Reagents Lab Ltd., Bangkok, Thailand) according to the Kaufman-White scheme. All animal fecal samples were collected in accordance with relevant guidelines and regulations of the China Institute of Veterinary Drug Control, Beijing, P. R. China. All experimental protocols were approved by the China Institute of Veterinary Drug Control, Beijing, P. R. China.
Screening of the mcr-1 gene in Salmonella
Salmonella genomic DNA was prepared using the boiling method. PCR was performed using the primers targeting mcr-1 as reported previously16. The genetic identity of all amplification products was confirmed by nucleotide sequencing.
Antimicrobial susceptibility tests
All Salmonella isolates were subjected to antimicrobial susceptibility tests by the standard agar dilution method as described by the Clinical and Laboratory Standards Institute22,23. Fourteen antimicrobials as shown in Table 1 were tested. Escherichia coli strain ATCC 25922 was used as the quality control.
Conjugation, PFGE, S1-PFGE and Southern Hybridization
Conjugation experiments were carried out using the mixed broth method as previously described24. PFGE, S1-PFGE and Southern Hybridization were performed as previously described25.
Plasmid sequencing
Representative plasmids with sizes of ~33 kb, ~60 kb, 140 kb ~180 kb and ~240 kb, recovered from Salmonella parental strains and transconjugants, were subjected to plasmid sequencing using the Illumina and PacBio platforms and analysed as previously described26. The complete nucleotide sequences of the ~180 kb plasmid, pASSD2-MCR1(KX856065), were submitted to GenBank.
Acknowledgements
This work was supported by the Chinese National Key Basic Research and Development Program (2013CB127200) and National Science Foundation of China (31302142).
Author Contributions
C.M.W., S.C. designed the research, analyzed the data and wrote the manuscript, M.Q.C., J.H.Z. performed the research, analyzed the data and drafted the manuscript, C.P.Z., C.B.W. collected part of the strains, R.C.L. analyzed the sequencing data, E.W.C. analyzed the data and edited the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Mingquan Cui and Jinfei Zhang contributed equally to this work.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Congming Wu, Email: wucm@cau.edu.cn.
Sheng Chen, Email: sheng.chen@polyu.edu.hk.
References
- 1.Gao R, et al. Dissemination and Mechanism for the MCR-1 Colistin Resistance. PLoS Pathog. 2016;12:e1005957. doi: 10.1371/journal.ppat.1005957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gao R, Wang Q, Li P, Li Z, Feng Y. Genome sequence and characteristics of plasmid pWH12, a variant of the mcr-1-harbouring plasmid pHNSHP45, from the multi-drug resistant E. coli. Virulence. 2016;7:732–735. doi: 10.1080/21505594.2016.1193279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li XP, et al. Clonal spread of mcr-1 in PMQR-carrying ST34 Salmonella isolates from animals in China. Sci Rep. 2016;6:38511. doi: 10.1038/srep38511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun J, et al. Co-transfer of blaNDM-5 and mcr-1 by an IncX3-X4 hybrid plasmid in Escherichia coli. Nat Microbiol. 2016;1:16176. doi: 10.1038/nmicrobiol.2016.176. [DOI] [PubMed] [Google Scholar]
- 5.Wang Q, et al. Complex dissemination of the diversified mcr-1-harbouring plasmids in Escherichia coli of different sequence types. Oncotarget. 2016;7:82112–82122. doi: 10.18632/oncotarget.12621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang RS, et al. Emergence of NDM-5- and MCR-1-Producing Escherichia coli Clones ST648 and ST156 from a Single Muscovy Duck (Cairina moschata) Antimicrob Agents Chemother. 2016;60:6899–6902. doi: 10.1128/AAC.01365-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ye H, et al. Diversified mcr-1-Harbouring Plasmid Reservoirs Confer Resistance to Colistin in Human Gut Microbiota. MBio. 2016;7:e00177. doi: 10.1128/mBio.00177-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doumith, M. et al. Detection of the plasmid-mediated mcr-1 gene conferring colistin resistance in human and food isolates of Salmonella enterica and Escherichia coli in England and Wales. J Antimicrob Chemother, doi:10.1093/jac/dkw093 (2016). [DOI] [PubMed]
- 9.Yi L, et al. mcr-1-Harboring Salmonella enterica Serovar Typhimurium Sequence Type 34 in Pigs, China. Emerg Infect Dis. 2017;23:291–295. doi: 10.3201/eid2302.161543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anjum, M. F. et al. Colistin resistance in Salmonella and Escherichia coli isolates from a pig farm in Great Britain. J Antimicrob Chemother, doi:10.1093/jac/dkw149 (2016). [DOI] [PubMed]
- 11.Campos, J., Cristino, L., Peixe, L. & Antunes, P. MCR-1 in multidrug-resistant and copper-tolerant clinically relevant Salmonella 1,4,[5],12:i:- and S. Rissen clones in Portugal, 2011 to 2015. Euro Surveill21, doi:10.2807/1560-7917.ES.2016.21.26.30270 (2016). [DOI] [PubMed]
- 12.Yang, Y. Q. et al. Co-occurrence of mcr-1 and ESBL on a single plasmid in Salmonella enterica. J Antimicrob Chemother, doi:10.1093/jac/dkw243 (2016). [DOI] [PubMed]
- 13.Figueiredo, R. et al. Detection of an mcr-1-encoding plasmid mediating colistin resistance in Salmonella enterica from retail meat in Portugal. J Antimicrob Chemother, doi:10.1093/jac/dkw240 (2016). [DOI] [PubMed]
- 14.Quesada A, et al. Detection of plasmid mediated colistin resistance (MCR-1) in Escherichia coli and Salmonella enterica isolated from poultry and swine in Spain. Res Vet Sci. 2016;105:134–135. doi: 10.1016/j.rvsc.2016.02.003. [DOI] [PubMed] [Google Scholar]
- 15.Vinueza-Burgos C, Cevallos M, Ron-Garrido L, Bertrand S, De Zutter L. Prevalence and Diversity of Salmonella Serotypes in Ecuadorian Broilers at Slaughter Age. PLoS One. 2016;11:e0159567. doi: 10.1371/journal.pone.0159567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu YY, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 17.Cannatelli, A. et al. First detection of the mcr-1 colistin resistance gene in Escherichia coli, Italy. Antimicrob Agents Chemother, doi:10.1128/AAC.00246-16 (2016). [DOI] [PMC free article] [PubMed]
- 18.Li R, et al. Genetic characterization of mcr-1-bearing plasmids to depict molecular mechanisms underlying dissemination of the colistin resistance determinant. J Antimicrob Chemother. 2017;72:393–401. doi: 10.1093/jac/dkw411. [DOI] [PubMed] [Google Scholar]
- 19.Liu, Y. Y. et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis, doi:10.1016/S1473-3099(15)00424-7 (2015). [DOI] [PubMed]
- 20.Malhotra-Kumar, S. et al. Colistin resistance gene mcr-1 harboured on a multidrug resistant plasmid. Lancet Infect Dis, doi:10.1016/S1473-3099(16)00012-8 (2016). [DOI] [PubMed]
- 21.Doumith M, et al. Detection of the plasmid-mediated mcr-1 gene conferring colistin resistance in human and food isolates of Salmonella enterica and Escherichia coli in England and Wales. J Antimicrob Chemother. 2016;71:2300–2305. doi: 10.1093/jac/dkw093. [DOI] [PubMed] [Google Scholar]
- 22.CLSI. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-fifth informational supplement. CLSI document M100-S25, Wayne, PA: Clinical and Laboratory Standards Institute (2015).
- 23.CLSI. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-sixth informational supplement. CLSI document M100-S26, Wayne, PA: Clinical and Laboratory Standards Institute (2016).
- 24.Borgia S, et al. Outbreak of carbapenem-resistant enterobacteriaceae containing blaNDM-1, Ontario, Canada. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 2012;55:e109–117. doi: 10.1093/cid/cis737. [DOI] [PubMed] [Google Scholar]
- 25.Zhang, R. et al. Emergence of carbapenem-resistant Serotype K1 hypervirulent Klebsiella pneumoniae (hvKP) strains in China. Antimicrobial agents and chemotherapy, doi:10.1128/AAC.02173-15 (2015). [DOI] [PMC free article] [PubMed]
- 26.Ye, L. et al. Characterization of an IncA/C Multidrug Resistance Plasmid in Vibrio alginolyticus. Antimicrob Agents Chemother, doi:10.1128/AAC.00300-16 (2016). [DOI] [PMC free article] [PubMed]