Abstract
Background:
With one-third of the world’s population infected, tuberculosis (TB) is one of the most common infectious diseases and a major public health problem, especially in developing countries. The efficacy of the BCG vaccine for controlling the disease in adults is poor. The development of an effective TB vaccine is a global objective. An effective tuberculosis vaccine should stimulate cellular immunity. DNA vaccines are a new generation of vaccines with the potential to achieve this goal. The aim of this study was to produce a DNA vaccine of Mtb72F.
Methods:
mtb32C, mtb39, and mtb32N were cloned into pcDNA3.1 using restriction enzyme digestion and T4 DNA ligase. Colony-PCR and restriction enzyme digestion were performed to detect transformed bacteria. DNA sequencing confirmed the desired gene insertion into the vector. A Chinese hamster ovary (CHO) cell line was transfected with the recombinant plasmid and RT-PCR was performed to assess gene expression.
Results:
Gel electrophoresis showed the expected amplified gene fragments of 429, 614, and 1200 base pairs (bps) for mtb32C, mtb32N, and mtb39, respectively. Enzyme digestion and gel electrophoresis showed the expected fragments, indicating the desired gene position and orientation in the recombinant plasmid. This finding was verified by DNA sequencing, and RT-PCR demonstrated gene expression in the CHO cell line.
Conclusion:
An Mtb72F DNA plasmid was successfully constructed. This plasmid may be a candidate for animal immunizations.
Key Words: Cloning, DNA vaccine, mtb32C, mtb32N, mtb39, Mycobacterium tuberculosis
Introduction
According to the World Health Organization’s (WHO) report approximately 10 million people are infected with tuberculosis (TB) annually. Currently, BCG is the only TB vaccine in use. BCG can prevent tuberculosis meningitis and disseminated disease (2), but it cannot protect adults from latent infections, nor can it be used in immunosuppressed patients (3).
An effective vaccine is desperately needed to control TB worldwide (4). Numerous TB vaccines, including recombinant protein vaccines, live attenuated vaccines, and DNA vaccines, have been produced. A DNA vaccine could be effective in controlling the disease. DNA vaccines, which often stimulate cellular immunity and both TCD8+ and TCD4+ responses, are good candidates for induction of immunity against intracellular pathogens such as Mycobacterium tuberculosis (M.tuberculosis). Several studies have shown that TB DNA vaccines can protect animals against primary infections, and can also be used as boosters after to enhance immune responses after BCG vaccination (5). In addition, DNA vaccines may be useful as therapeutic vaccines to treat the disease. It has been demonstrated that simultaneous use of DNA vaccines and anti-TB drugs in infected animals reduced recovery response times compared with anti-TB drugs alone.
Mtb72F is a recombinant protein vaccine against M. tuberculosis that provided protective immunity in mice, rabbits, guinea pigs, and monkeys (7-9). In response to immunization cellular immune responses were stimulated; CD4+ T cells proliferated and IFNγ production increased. Also, no serious adverse events were reported in a human phase I clinical trial (10).
Mtb72F consists of the C- and N-terminal open reading frames (ORFs) of Mtb32 (Rv0125) with the mtb39 gene (Rv1196) between them. The resulting recombinant vaccine, Mtb72F, provided greater protection against TB in animals than Mtb32 or Mtb39 separately or mixed (11). Mtb39 is an Mtb 39-kDa protein that induces T cell proliferation and also IFNγ production in mononuclear cells of PPD-positive patients (12). Mtb32 is a secreted serine protease that can also stimulate cellular immunity (13).
The aim of this study was to construct a DNA vaccine candidate encoding Mtb72F by cloning the mtb39 gene between the C- and N-terminal ORFs of mtb32.
Materials and Methods
Cloning of mtb32C into pCDNA3.1
In this study the pET21b/Mtb72F vector, which was previously constructed (14), was used as the DNA template to PCR amplify the desired products.
Amplification of mtb32C
Sense and anti-sense primers, respectively, were as follows: (Fu1) 5' CAATTAAAGCTTGCCACCATGGGAACGGCCGCGTCCGAT 3´ and (R1) 5´ CTAATCGGATCCGGCCGGGGGTCCCTCGGCCAA 3´ (15). A Kozak sequence was placed 3´ of the HindIII enzyme restriction site in Fu1 primer (bold sequence). Underline letter show HindIII and BamHI restriction site in Fu1 and R1 primers respectively.
The PCR mixture consisted of 2.5 μl of 10X pfu buffer with MgSO4, 2.5 units of pfu enzyme (Thermo Scientific, South Korea), 0.5 μl each of sense and antisense primers (10 pmol), 0.5 μl of dNTPs (10mM), and 10 ng of DNA template in a final volume of 25 μl.
The PCR program was initial denaturation at 95 °C for 10 min followed by 35 cycles of amplification at 95 °C for 30 seconds, 56 °C for 30 seconds, and 72 °C for 60 seconds in a thermocycler (BIO-RAD MyCyclerTM). The PCR product was purified on a silica column (Bioneer, (EU) Lithuania) according to the manufacturer's instructions.
Then pcDNA3.1(+) (Invitrogen, USA) and the purified mtb32C PCR product were digested with BamHI and HindIII (Roche, Switzerland) at 37 °C for 2 hours. The enzymes were inactivated by heating at 80 °C for 15 minutes and the digested products were electrophoresed on a 1.5% agarose gel and purified.
Insertion and cloning of mtb32C
The ligation mixture contained 2 units of T4 ligase (Thermo Scientific, South Korea), 2 µl of T4 ligase 10X buffer, 0.5 μl (15 ng/µl) of mtb32C, and 2 µl (50 ng/µl) of pcDNA3.1(+) in a final volume of 20 µl. The mixture was incubated at room temperature for 18 hours. E.coli DH5α strain competent cells were prepared with 50 mM CaCl2. Ten µl of the ligation mixture were added to the competent bacteria and incubated on ice for 20 minutes. Bacteria were transformed using the heat shock protocol and then incubated at 37 °C for 1 hr in antibiotic-free Luria-Bertani (LB) broth. The transformed E. coli were cultured on plates containing LB agar with 100 µg/ml ampicillin at 37 °C for 16 hrss.
Insertion of mtb32C into pcDNA3.1(+) was analyzed by colony PCR and restriction digests.
Cloning of mtb32N into pcDNA3.1/mtb32C
mtb32N was amplified with the sense and antisense primers: (F3) 5' CTAATCGAATTCGCCCCGCCGGCCTTGTCGCAGGAC 3' (14) and (Ru3) 5' CTAATCTCTAGACTATCAGGACGCGGCCGTGTTCAT 3', respectively.
A stop codon was placed 3' of the XbaI restriction site in the reverse primer, show by bold letters. Underlined letters show XbaI and EcoRI enzeymes restriction site in F3 and Ru3 primers, respectively.
The PCR mixture and cycling program for mtb32N were the same as that used to amplify mtb32C. The purified mtb32N PCR product and pcDNA3.1/Mtb32C were digested with XbaI and EcoRI (Roche Company). The ligation mixture contained 2 units of T4 DNA ligase, 2 µl of T4 DNA ligase buffer, 3 μl of mtb32N (11 ng/µl), 5 µl of pcDNA3.1/Mtb32C (19 ng/µl), and 8 μl of H2O. The mixture was incubated at room temperature overnight and the bacteria were transformed as above.
Cloning of Mtb39 into pcDNA3.1/Mtb32/cMtb32N
pET21b/Mtb72F, obtained previously (14), and pcDNA3.1/Mtb32C/Mtb32N were digested with EcoRI and BamHI and mtb39 was purified. The ligation mixture contained 2 units of T4 DNA ligase, 2 µl of T4 DNA ligase buffer, 1.1 µl (84 ng/µl) of pcDNA3.1/Mtb32C/Mtb32N, and 6.5 µl of mtb39 (3.4 ng/µl).
Bacteria were transformed as above. The cloning of mtb39 was confirmed by colony-PCR and digestion with HindIII and EcoRI.
The recombinant construct was confirmed by sequencing (Bioneer, (EU) Lithuania).
Transfection
The CHO cells were grown in RPMI medium containing 10% fetal bovine serum (FBS) in a 95% humidified incubator with 5% CO2. One day before transfection, the cells were seeded in a 6-well cell culture plate.
One µg of pcDNA3.1/Mtb32C/Mtb32N was added to Superfect® (301305 Qiagen) and mixed gently at room temperature for 20 min. The volume was then adjusted to 500 µl with RPMI, cells were washed with PBS, the transfection medium was added to the cells, and the plate was incubated at 37 °C with 5% CO2 for 2 hours. The transfection medium was replaced with RPMI containing 10% FBS and the plate was incubated at 37 °C in a humidified atmosphere containing 5% CO2 as above for 2 days.
RT-PCR
The transfected cells were washed with PBS, scraped from the plates, and resuspended in 200 μl of PBS. RNA was extracted with a High Pure RNA Isolation kit (Roche cat. no. 11828665001). CDNA was synthesized with a First Strand cDNA Synthesis kit (Thermo Scientific cat. no. K1611) using oligo (dT) and R1 primers.
Results
Cloning of mtb32C into pCDNA3.1
Mtb32C was amplified successfully by PCR. Fig. 1 shows a 429 bp nucleic acid fragment of mtb32C. Four bacteria colonies were grown on ampicillin LB agar 16 hours after transformation. Colony-PCR was performed for all colonies. One colony was positive for mtb32C fragment and showed 429 bp fragment (Fig. 2, lane 6). In addition, the mtb32C insertion into recombinant plasmid was confirmed by double enzyme digestion method (Fig. 3). A high molecular mass band representing the plasmid, and a band of 429 bp, representing mtb32C, were obtained.
Fig. 1.
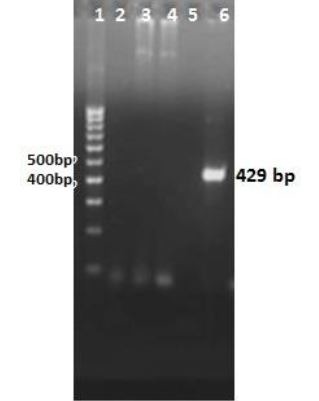
Agarose gel of colony-PCR products to assess insertion of mtb32C into pcDNA3.1 using mtb32C specific primers F3 and Ru3. Lane 1: DNA size marker (100 bp); lane 2: PCR negative control; lanes 3, 4, 5, 6: colony-PCR products from colonies grown on LB agar after transformation. PCR of one colony (lane 6), amplified a 429 bp fragment, indicating insertion of mtb32C.
Fig. 2.
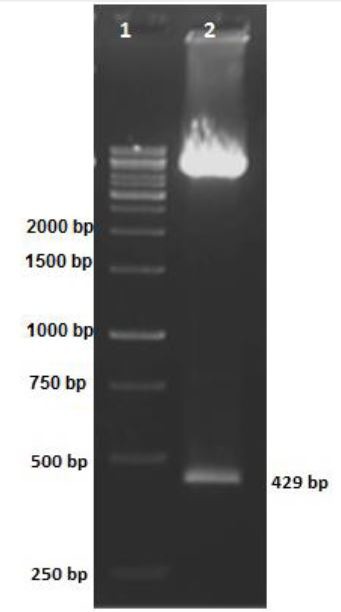
Agarose gel of pcDNA3.1 containing mtb32C after digestion with BamHI and HindIII. Lane 1: DNA size marker (1000 bp); lane 2: BamHI and HindIII-digested plasmid; the 429 bp fragment indicates the presence of mtb32C
Fig. 3.
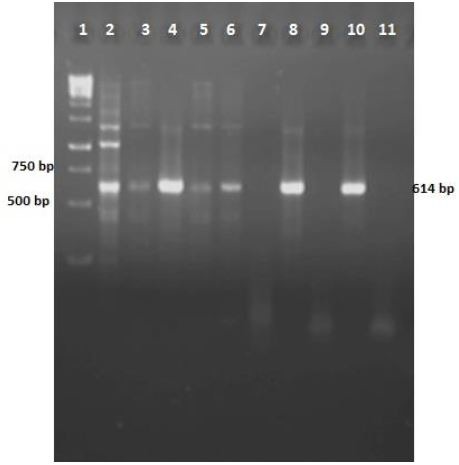
Agarose gel of colony-PCR results for mtb32N in the pcDNA3.1/Mtb32C. Lane 1: DNA size marker (1000 bp); lanes 2-10: colony PCR results from bacterial clones grown on LB agar after transformation; lane 11: PCR negative control.
Cloning of mtb32N into pcDNA3.1/mtb32C
PCR of mtb32N with primers F3 and Ru3 amplified a 614-bp fragment on agarose gel electrophoresis Fig. 4. Nine bacteria colonies were developed following bacteria transformation. Colony-PCR was done for colonies of growing bacteria and represented seven positive mtb32N insertion (Figure 5). Double Digestion of pcDNA3.1/Mtb32C/MTB32N recombinant plasmid showed a 1045-bp fragment containing mtb32C and mtb32N (Fig. 4, lane 1).
Fig. 4.
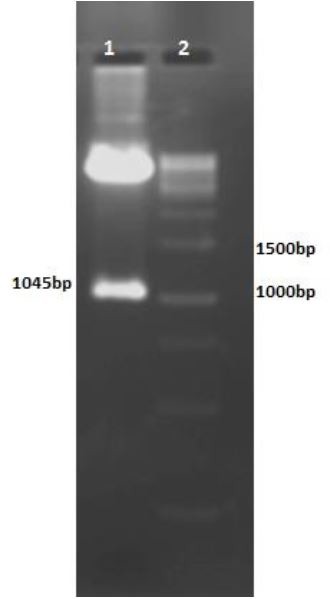
Agarose gel of HindIII and XbaI-digested pcDNA3.1/Mtb32C/Mtb32N. Lane 1: HindIII and XbaIdigested digested vector; the 1045 bp fragment consist of mtb32C and mtb32N; lane 2: DNA size marker (1000 bp).
Cloning of Mtb39 into pcDNA3.1/Mtb32/cMtb32N
Colony-PCR of transformants using mtb39C primers were amplified a 1200-bp fragment, confirming the presence of mtb39C in all examined colonies (Fig. 5, lanes 1-10). Dual Digestion of pcDNA3.1/Mtb32C/Mtb39/MTB32N with HindIII and EcoRI resulted in a high molecular weight band representing the plasmid and a 1630-bp fragment corresponding to mtb32C plus mtb39 (Fig. 6, lane 2).
Fig. 5.
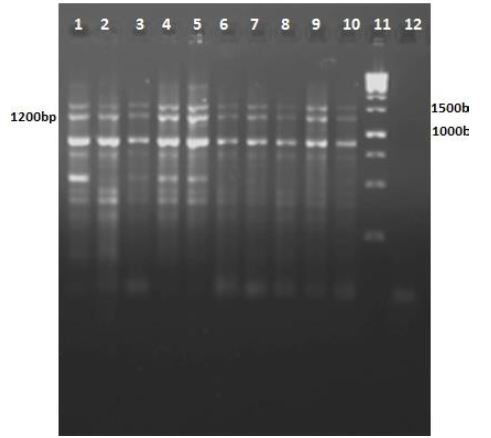
Agarose gel of colony-PCR products using mtb39-specific primers: Fu1 and R1. Lanes 1- 10: mtb39 PCR products from colonies grown on agar plates following transformation. The 1200 bp fragments represent mtb39; lane 11: DNA size marker (1000bp); lane 12: PCR negative control.
Fig. 6.
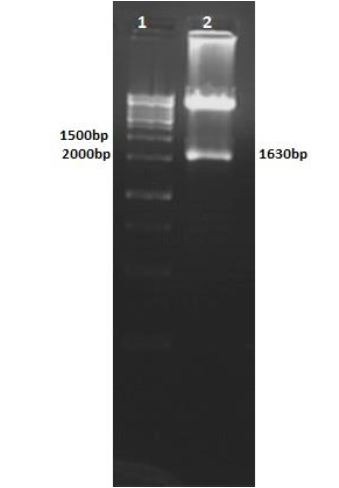
Agarose gel of HindIII- and EcoRI-digested pcDNA3.1/Mtb32c/Mtb39/Mtb32N. Lane 1: DNA size marker (1000 bp); lane 2: digested vector: the 1630 bp fragment contains mtb32C and mtb39.
Correct gene insertion into the final recombinant plasmid was confirmed by DNA sequencing. Alignment of the sequence showed 100% homology with M. tuberculosis H37Rv strain and confirmed the correct orientation of gene insertion into pcDNA3.1(+).
Expression of recombinant protein
CHO cells were transfected with the final construct using Superfect®. Recombinant protein expression was examined by RT-PCR using mtb32C-specific primers. The presence of desired 429 bp fragment in agarose gel electrophoresis was shown in Fig. 7, lane 2, confirmed the expression of recombinant protein in CHO cells.
Fig. 7.
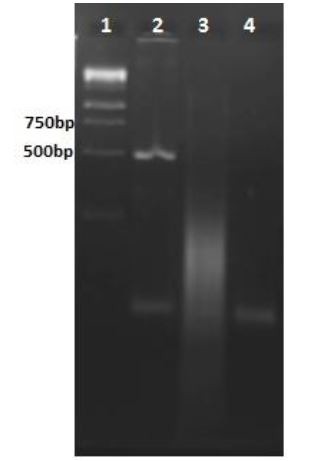
Agarose gel electrophoresis of mtb32C RT-PCR products. Lane 1: DNA size marker (1000 bp); lane 2: mtb32C RT-PCR product; lane 3: PCR product after amplification using isolated RNA as the template in the reaction to check for DNA contamination; lane 4: PCR negative control.
Discussion
Although the exact number is not known, it is estimated that 2-3 million people die annually from TB infections. Therefore, making a new vaccine that offers better protection than BCG is a global goal.
MTB32A was discovered in 1999 (13). The hydrophobicity of this protein makes it a potential candidate for stimulating T cell immunity (16). The advantage of MTB32A as a vaccine candidate is its rapid secretion after translation (13), because posttranslational modifications produce different protein isoforms with different immunogenic properties (16). As MTB32A do not affected by post translational modifications therefore expression in another host could not alter its immunogenic properties. MTB39 has more than 10 epitopes that are recognized by T cells; therefore, a low amount of Downloaded from the antigen can produce strong immune responses. Several hydrophobic regions in MTB39 that can stimulate T cell responses (12).
Tuberculosis is a zoonotic disease that infects several animal species and is transmissible from animals to humans; consequently, TB cannot be completely eliminated until it is eradicated in animals (17).
Protein subunit vaccines are inactive at high temperatures and expensive therefore they could not distribute into wildlife habitat. These characteristics make their application difficult in undomesticated animals. DNA vaccines and attenuated vaccines are less expensive to produce than subunit vaccines, making them more economical for animal applications.
Many studies have determined that DNA vaccines can control disease progression and prevent relapse in TB-infected animals (18). DNA vaccines
have also been reported to be useful in the treatment of multidrug-resistant TB-infected mice (19); however, DNA vaccines have also been shown to induce immunopathology when used to treat TB-infected animals (18). The effect of the Mtb72F DNA vaccine candidate in animal models needs further study.
The prevalence of M. tuberculosis in Iran is 13 cases per 100,000. Consequently, we need an appropriate program to control and reduce the incidence of tuberculosis in our country (20).
In this study, Mtb72F was successfully cloned and expressed in pcDNA3.1. This vector can be used for animal vaccination in subsequent studies.
Acknowledgements
This work was supported by a grant (No: 89078) from Mashhad University of Medical Sciences, Mashhad, IR. Iran. The authors declare that there is no conflict of interest.
References
- 1.Organization WH. Global tuberculosis control:epidemiology, strategy, financing: WHO report 2009. World Health Organization. 2009 [Google Scholar]
- 2.Colditz GA, Brewer TF, Berkey CS, Wilson ME, Burdick E, Fineberg HV, et al. Efficacy of BCG vaccine in the prevention of tuberculosis: meta-analysis of the published literature. Jama. 1994;271(9):698–702. [PubMed] [Google Scholar]
- 3.Andersen P, Doherty TM. The success and failure of BCG—implications for a novel tuberculosis vaccine. Nature Reviews Microbiology. 2005;3(8):656–62. doi: 10.1038/nrmicro1211. [DOI] [PubMed] [Google Scholar]
- 4.Andersen P. Tuberculosis vaccines—an update. Nature Reviews Microbiology. 2007;5(7):484–7. doi: 10.1038/nrmicro1703. [DOI] [PubMed] [Google Scholar]
- 5.Lowrie DB, Silva CL, Tascon RE. DNA vaccines against tuberculosis. Immunology and cell biology. 1997;75(6):591–4. doi: 10.1038/icb.1997.93. [DOI] [PubMed] [Google Scholar]
- 6.Lowrie DB, Tascon RE, Bonato VL, Lima VM, Faccioli LH, Stavropoulos E, et al. Therapy of tuberculosis in mice by DNA vaccination. Nature. 1999;400(6741):269–71. doi: 10.1038/22326. [DOI] [PubMed] [Google Scholar]
- 7.Brandt L, Skeiky YA, Alderson MR, Lobet Y, Dalemans W, Turner OC, et al. The protective effect of the Mycobacterium bovis BCG vaccine is increased by coadministration with the Mycobacterium tuberculosis 72-kilodalton fusion polyprotein Mtb72F in M.tuberculosis-infected guinea pigs. Infection and immunity. 2004;72(11):6622–32. doi: 10.1128/IAI.72.11.6622-6632.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsenova L, Harbacheuski R, Moreira AL, Ellison E, Dalemans W, Alderson MR, et al. Evaluation of the Mtb72F polyprotein vaccine in a rabbit model of tuberculous meningitis. Infection and immunity. 2006;74(4):2392–401. doi: 10.1128/IAI.74.4.2392-2401.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reed SG, Coler RN, Dalemans W, Tan EV, Cruz ECD, Basaraba RJ, et al. Defined tuberculosis vaccine, Mtb72F/AS02A, evidence of protection in cynomolgus monkeys. Proceedings of the National Academy of Sciences. 2009;106(7):2301–6. doi: 10.1073/pnas.0712077106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spertini F, Audran R, Lurati F, Ofori-Anyinam O, Zysset F, Vandepapelière P, et al. The candidate tuberculosis vaccine Mtb72F/AS02 in PPD positive adults: A randomized controlled phase I/II study. Tuberculosis. 2013;93(2):179–88. doi: 10.1016/j.tube.2012.10.011. [DOI] [PubMed] [Google Scholar]
- 11.Skeiky YA AM, Ovendale PJ, Guderian JA, Brandt L, Dillon DC, et al. Differential immune responses and protective efficacy induced by components of a tuberculosis polyprotein vaccine, Mtb72F, delivered as naked DNA or recombinant protein. J Immunol. 2004;172(12):7618–28. doi: 10.4049/jimmunol.172.12.7618. [DOI] [PubMed] [Google Scholar]
- 12.Dillon DC, Alderson MR, Day CH, Lewinsohn DM, Coler R, Bement T, et al. Molecular Characterization and Human T-Cell Responses to a Member of a Novel Mycobacterium tuberculosis mtb39 Gene Family. Infection and immunity. 1999;67(6):2941–50. doi: 10.1128/iai.67.6.2941-2950.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Skeiky YA, Lodes MJ, Guderian JA, Mohamath R, Bement T, Alderson MR, et al. Cloning, Expression, and Immunological Evaluation of Two Putative Secreted Serine Protease Antigens ofMycobacterium tuberculosis. Infection and immunity. 1999;67(8):3998–4007. doi: 10.1128/iai.67.8.3998-4007.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nabavinia MS, Naderi Nasab M, Meshkat Z, Derakhshan M, Khaje-Karamadini M. Construction of an Expression Vector Containing Mtb72F of Mycobacterium tuberculosis. Cell J. 2012;14(1):61–6. Epub 2013/04/30. [PMC free article] [PubMed] [Google Scholar]
- 15.Nabavinia MS, Nasab MN, Meshkat Z, Derakhshan M, Khaje-Karamadini M. Construction and Evaluation of an Expression Vector Containing Mtb32C (Rv0125) of Mycobacterium tuberculosis. Avicenna J Med Biotechnol. 2011;3(4):207–10. Epub 2011/10/01. [PMC free article] [PubMed] [Google Scholar]
- 16.Sable SB, Plikaytis BB, Shinnick TM. Tuberculosis subunit vaccine development: impact of physicochemical properties of mycobacterial test antigens. Vaccine. 2007;25(9):1553–66. doi: 10.1016/j.vaccine.2006.11.014. [DOI] [PubMed] [Google Scholar]
- 17.Thoen C, LoBue P, de Kantor I. The importance of Mycobacterium bovis as a zoonosis. Veterinary Microbiology. 2006;112(2-4):339–45. doi: 10.1016/j.vetmic.2005.11.047. [DOI] [PubMed] [Google Scholar]
- 18.Lowrie DB. DNA vaccines for therapy of tuberculosis: where are we now? Vaccine. 2006;24(12):1983–9. doi: 10.1016/j.vaccine.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 19.Liang Y, Wu X, Zhang J, Li N, Yu Q, Yang Y, et al. The treatment of mice infected with multidrug-resistant Mycobacterium tuberculosis using DNA vaccines or in combination with rifampin. Vaccine. 2008;26(35):4536–40. doi: 10.1016/j.vaccine.2008.06.066. [DOI] [PubMed] [Google Scholar]
- 20.Derakhshan M, Safdari H, Sadeghi A, Ghazvini K, Mohammadi S. Prevalence of Mycobacterium tuberculosis in the samples referred to the tuberculosis research laboratory in Mashhad Ghaem Hospital during 2005-2006. Iranian Journal of Microbiology. 2009;1(3):20–2. [Google Scholar]


