Abstract
Background:
The prevalence of hepatitis C virus (HCV) infection is increasing worldwide. Cytotoxic Tlymphocyte-associated protein 4 (CTLA-4) may play a role in the intensity of the disease. The aim of this study was to evaluate the association between genetic variants of the CTLA-4 and HCV infection.
Methods:
Restriction fragment length polymorphism-polymerase chain reaction (RFLP-PCR) was performed as the genotyping assay at four different positions (+49 A>G, -318 C>T, -1722 T>C, and - 1661 A>G). Haplotypes were analyzed using PHASE software. Sixty-five HCV patients and 65 healthy individuals as controls who were referred to the hepatitis clinic in Mashhad, Iran, were recruited. Genomic DNA was extracted from whole blood of participants
Results:
In a dominant analysis model of the -1661 position (GG vs. AA+AG), the AA genotype was more common in controls than in patients (adjusted P = 0.0003; OR = 0.15, 95% CI = 0.051 -0.42). The GCAT haplotype was also more prevalent in controls than in patients (adjusted P = 0.01; OR = 0.40, 95% CI = 0.20-0.81). Furthermore, the ACGT/ACGT diplotype was more common in controls than in patients (P = 0.0037; OR = 0.15, 95% CI = 0.04-0.54). In addition, the ACGT/ACAT diplotype was more frequent in patients than controls (adjusted P =0.003; OR = 2.48, 95% CI = 1.37- 4.50).
Conclusion:
Our results indicated that polymorphisms in CTLA-4 and certain haplotypes may affect the risk of HCV infection in our population, although a larger sample size may be required to confirm this association.
Key Words: CTLA-4 gene Polymorphism, Haplotype analysis, Hepatitis C virus
Introduction
Cytotoxic T-lymphocyte-associated protein 4 (CTLA-4) plays a key role in the negative regulation of T cell immune responses, and is mostly found on the surface of cells that are responsible for activation and regulation of T cells. Also, previous studies have shown that CTLA-4 has a critical role in the hemostasis of T cells (1-5). The CTLA-4 gene (CTLA-4) is located on chromosome 2q33.3 and contains four exons. The CTLA-4 protein (CTLA-4) is a T cell molecule expressed on the cell surface after T helper cell activation (6,7). Due to the importance of CTLA-4 in T cell activities, any changes in expression or malfunctions of CTLA-4 may lead to autoimmune diseases such as multiple sclerosis (8). Studies have shown that molecular variants of CTLA-4 are involved in T cell-mediated inflammation and autoimmune diseases including type 1 diabetes, autoimmune thyroid disease, multiple sclerosis, Addison’s disease, celiac disease, and systemic lupus erythematosus (SLE) (9-12).
Hepatitis C virus (HCV) infection, with almost 170 million infected people worldwide, is an important disease that is affected by immune cell responses. In addition, T lymphocyte response has an important role in the spontaneous clearance of HCV. Studies in similar fields can help to determine some factors that may influence hepatitis infections (4, 13). An "A to G" nucleotide change at position +49 of exon 1 in CTLA-4 has a key role in susceptibility to autoimmune type 1 hepatitis among different races. This polymorphism affects CTLA-4 gene regulation; thus, CTLA-4 as an inhibitory receptor may influence disease susceptibility and inhibit autoimmunity (14, 15). In this study, we analyzed the genotype and haplotype distributions of the CTLA-4 single-nucleotide polymorphisms (SNPs) +49 A>G, -318 C>T, -1722 T>C, and -1661 A>G, in HCV patients and controls.
Materials and Methods
Sample collection and DNA extraction
The study was approved by the Ethical Committee of Mashhad University of Medical Sciences and informed written consent was obtained from all participants. Ten ml of blood were collected from 65 HCV patients and 65 healthy controls who tested negative for HCV. Peripheral blood mononuclear cells (PBMCs) were isolated and their genomic DNA was extracted using a genomic DNA extraction Kit (GeNet Bio, Korea). Real-time PCR was used for the HCV viral load test. Risk factors for HCV infection, such as drug and alcohol addiction, transfusion, HIV infection, and tattoos were considered in the final analysis when results were adjusted for confounders (Table 1).
Table 1.
Demographic and risk factors feature in HCV patients and healthy groups.
| Characteristics | Patients | Controls | P-Value |
|---|---|---|---|
| M: F | 56:9 | 30:36 | <0.05 |
| Age (years; mean ± SD) | 41.86±10.29 | 36.77±12.28 | <0.05 |
| Addiction | 31 (47.69%) | - | <0.05 |
| Alcohol | 27 (41.53%) | 1 (1.53%) | <0.05 |
| Transfusion | 26 (40%) | 4 (6.15%) | <0.05 |
| Tattoo | 22 (33.84%) | - | <0.05 |
| ALT (U.L-1, mean ± SD) | 35.82±30.2 | 26.80±21.38 | <0.1 |
| Increased ALT* | 17 (26.15) | 11 (16.92) | <0.05 |
Alanine aminotransferase assay
Alanine aminotransferase (ALT) was assessed using a kit (Pars Azmoon, Iran) in HCV patients and controls. According to the kit instruction methods, the ALT normal range was < 31 U.L-1 in women and < 41 U.L-1 in men.
Genotyping assay
CTLA-4 polymorphisms were genotyped by the polymerase chain reaction-restriction fragment length polymorphism method (PCR-RFLP). Two different primers were designed for four positions of CTLA-4. The sequence at each position is shown in Table 2. The PCR (96 well thermal cycler Applied Biosystems) conditions for the +49 A>G position were the following: initial denaturation for 4 minutes at 94°C followed by 30 cycles of denaturation for 40 seconds at 94°C, annealing for 45 seconds at 57°C, and extension for 30 seconds at 72°C, with a final extension for 10 min at 72°C. PCR amplifications were performed in 30 μl reaction volumes containing 100 ng of DNA, 1 μl of 10 pmol of each primer, 1X reaction buffer, 2.5 mM MgCl2, 200 mM each of dNTPs, and 1 unit of Taq polymerase (GeNet Bio, Korea). The 162 base pair (bp) PCR product was electrophoresed in a 3% agarose gel. The PCR product was digested with BbvI (Bse XI, 100u, ER1451) and incubated at 65°C for 8 hours in a 20 μl volume.To genotype the CTLA-4 promoter at position -318 C>T, the PCR volume and program were similar to that of the +49 G<A position, but the annealing condition was 40 seconds at 60°C. The 247 bp product was digested with MseI. The PCR conditions for theCTLA-4 polymorphisms at positions −1661 and −1722 were as follows: initial denaturation for 5 minutes at 94°C, then thirty cycles of 15 seconds at 94°C, 30 seconds at 60°C, and 45 seconds at 72°C, with a final extension for 5 min at 72°C. PCR products were incubated for 3 h at 65°C with MseI and BbvI to genotype -1661 and -1722 SNPs, respectively. All amplified products were analysed on 3% agarose gels.
Table 2.
Primer sequences of CTLA-4 gene (+49 A>G, -318 C>T, -1722 T>C, and -1661 A>G).
| CTLA-4 gene | Forward primer | Revers primer |
|---|---|---|
| +49 A>G | 5'-GCTCTACTTCCTGAAGACCT-3' | AGTCTCACTCACCTTTGCAG-3''-5 |
| -318 C>T | -3' AAATGAATTGGACTGGATGGT'-5 | TTACGAGAAAGGAAGCCGTG-3''-5 |
| -1722 T>C | 5'-CTAAGAGCATCCGCTTGCACCT-3' | TGGGTGTGATGCACAGAAGCCTTTT-3''-5 |
| -1661 A>G |
HCV genotyping
HCV was genotyped using methods described by Afshari et al., and Ohno et al. (16, 17).
Statistical analysis
Statistical analyses were carried out using SPSS version 21 (SPSS Inc, Chicago, Illinois) software. For determination of normality, data was assessed with Kolmogorov-Smirnov test. Allele frequencies and genotype distributions at the four positions of CTLA-4 of patients and controls were compared using the χ2 test. For haplotype and diplotype analyses, PHASE (18) software (v2.1) was used. P values less than 0.05 were considered statistically significant.
Results
The patient groups’ ages ranged from 16 to 61 years and 86.1% were males. The control groups’ ages ranged from 15 to 63 years and 46.1% were males. The mean ages of patients and controls were 41.86 ± 10.29 years (mean ± SD) and 36.77 + 12.28 years (mean ± SD), respectively (Table 1). For genotype results, Hardy-Weinberg equilibrium was also tested for all participates at the four positions of CTLA-4. A deviation (P < 0.05) was observed in patients at positions -318 and -1772 and the frequency of alleles in these two positions was not common in two groups; therefore, to rule out any genotyping errors, 5% of samples were re-genotyped and no technical issues were found. The genotypes and allele frequencies for the four SNPs are shown in Table 2.
Polymorphisms analyses
If an A allele (of +49 A>G) was present in CTLA-4, the digested product was 162 bps, whereas a G allele yielded 90 and 72 bp fragments. With T at position -318, three fragments of 130, 96, and 21 bp were seen, whereas with C at position -318, only two fragments of 21 bp and 226 bp were seen. In CTLA-4 at position +49 A>G, we found the AA genotype frequency was greater in patients than controls (78.46% vs. 60%). The frequency of an A allele was also greater in patients than controls (78.46% vs. 73.07%); however, the difference was not significant (P > 0.05). In a recessive model analysis (AA vs. AG+GG), the AA genotype was significantly different between patients and controls (P = 0.024; OR=2.43, 95% CI= 1.12-5.26). After adjusting for confounders, including drug and alcohol addiction, transfusion, and tattoos, which might influence the risk of HCV infection, these differences between patients and controls were not significantly different (adjusted P = 0.1; OR = 2.76, 95% CI = 0.81-9.40).
No significant differences between patients and controls were found at CTLA-4 positions -318 C>T or -1772 T>C for any of the genotype distributions or allele frequencies (P > 0.05).
Allele A (homozygous subjects) at -1661 was revealed as 347- and 139-bp bands, whereas the G allele at this site showed a 486-bp fragment. Three bands were observed in those who were heterozygous for CTLA-4. In the -1722 (T>C) polymorphism, the digested amplicon from homozygous subjects (allele C) resulted in two bands of 270 and 216 bps, whereas digested amplicons from homozygous subjects with T alleles showed one band of 486 bp. At this position, digested amplicons from heterozygous subjects displayed three bands, similar to the -1661 position.
The genotype distribution analysis of CTLA-4 at position -1661 A>G showed that the AG genotype was more frequent in patients than controls (85.07% vs. 58.46%); however, this difference was not significant after adjusting for confounders. In a recessive model analysis (AA vs. AG+GG), the AA genotype was more common in controls than in patients (adjusted P = 0.0003; OR = 0.15, 95% CI = 0.05-0.42) (Table 3).
Table 3.
Distribution of genotypes frequency in three positions of CTLA-4 gene polymorphisms.
| CTLA-4 locus +49 A/G | Control (n=65) N (%) | Patients (n=65) N (%) | P value Crude | OR Crude (95% C.I.) | P value adjusted* | OR adjusted* (95% C.I.) |
|---|---|---|---|---|---|---|
| AA | 39 (60) | 51 (78.46) | Ref | |||
| AG | 17 (26.15) | 0 (0) | 0. 99 | - | - | - |
| GG | 9 (13.84) | 14 (21.53) | 0.84 | 0.90 | 0.84 | 0.90 |
| (0.35-2.33 ) | (0.35-2.33 ) | |||||
| AA | 39 (60) | 51 (78.46) | 0.1 | 2.76 | ||
| AG/GG | 26 (39.99 | 14 (21.53) | 0.024 | 2.43 | (0.81-9.40) | |
| (1.12-5.26) | ||||||
| Allele Frequency | ||||||
| A | 95 (73.07) | 102 (78.46) | 0.31 | 1.34 | 0.31 | 1.34 |
| G | 35 (26.92) | 28 (21.53) | (0.76-2.37) | (0.76-2.37) | ||
| CTLA-4 locus -318 T/C | Control (n = 65) N (%) | Patients (n = 65 ) N (%) | P value Crude | OR Crude (95% C.I.) | P value adjusted* | OR adjusted* (95% C.I.) |
| CC | 55 (48.61) | 55 (48.61) | Ref | |||
| CT | 9 (13.84) | 10 (15.38) | 0.83 | 0.90 | 0.83 | 0.90 |
| (0.34-2.39) | (0.34-2.39) | |||||
| TT | 1 (1.53) | 0 (0) | 1 | - | - | - |
| CC | 55 (84.61) | 55 (84.61) | 1 | 1 | 1 | 1 |
| CT/TT | 10 (15.37) | 10 915.37) | (0.39-2.59) | (0.39-2.59) | ||
| Allele Frequency | ||||||
| C | 119 (91.53) | 120 (92.30) | 0.82 | 1.11 | 0.31 | 1.34 |
| T | 11 (8.46) | 10 (7.69) | (0.45-2.71) | (0.76-2.34) | ||
| CTLA-4 locus -1661 A/G | Control (n = 65) N (%) | Patients (n = 65 ) N (%) | P value Crude | OR Crude (95% C.I.) | P value adjusted* | OR adjusted* (95% C.I.) |
| AA | 23 (35.38) | 5 (7.46) | Ref | |||
| AG | 38 (58.46) | 57 (85.07) | 0.00031 | 0.14 | 0.00031 | 0.14 |
| (0.05-0.41) | (0.05-0.41) | |||||
| GG | 4 (6.15) | 5 (7.46) | 0.04 | 0.17 | 0.04 | 0.17 |
| (0.03-0.89) | (0.03-0.89) | |||||
| AA | 23 (35.38) | 5 (7.46) | 0.0003 | 0.15 | 0.0003 | 0.15 |
| AG/GG | 42 (64.61) | 62 (92.53) | (0.05-0.42) | (0.05-0.42) | ||
| Allele Frequency | ||||||
| A | 84 (66.61) | 67 (0.5) | 0.096 | 0.67 | 0.096 | 0.67 |
| (0.14-1.08) | (0.14-1.08) | |||||
| G | 56 (35.38) | 67 (0.5) | ||||
| CTLA-4 locus -1772 T/C | Control (n = 65) N (%) | Patients (n = 65 ) N (%) | P value Crude | OR Crude (95% C.I.) | P value adjusted* | OR adjusted* (95% C.I.) |
| TT | 64 (95.52) | 56 (86.15) | Ref | |||
| CT | 2 (2.98) | 8 (12.30) | 0.06 | 0.22 | 0.06 | 0.22 |
| (0.04-1.07) | (0.04-1.07) | |||||
| CC | 1 (1.49) | 1 (1.53) | 0.92 | 0.87 | 0.92 | 0.87 |
| (0.05-14.31) | (0.05-14.31) | |||||
| CC | 1 (1.49) | 1 (1.53) | 0.07 | 0.29 | 0.07 | 0.29 |
| CT/TT | 66 (98.5) | 64 (98.45) | (0.08-1.13) | (0.08-1.13) | ||
| Allele Frequency | ||||||
| C | 130 (97.01) | 120 (92.30) | 0.099 | 0.37 (0.11-1.21) | 0.099 | 0.37 (0.11-1.21) |
| T | 4 (2.98) | 10 (7.69) | ||||
Adjusted based on transfusion, tattoo and addiction
C.I. = Confidence interval
Haplotype and diplotype distributions
At CTLA-4 positions +49 A>G, -318 C>T, -1722 T>C, and -1661 A>G, haplotypes and diplotypes were analyzed using PHASE software (Table 4). The GCAT haplotype was significantly greater in the controls than in patients (15.1% vs. 6.7%) (adjusted P = 0.01; OR = 0.40, 95% CI = 0.20-0.81). The ACTA/ACTA diplotype distribution was also more common in controls than patients, and this difference remained significant after adjusting for confounders (P = 0.0037; OR = 0.15, 95% CI = 0.04-0.54). In addition, the ACTA/ACGT diplotype was significantly greater in patients than in controls (adjusted P = 0.003; OR = 2.48, 95% CI = 1.37-4.50).
Table 4.
Distribution of haplotype and diplotype frequencies in patient and control groups.
| Locus | Controls 65 | Frequency (%) | Patients 65 | Frequency (%) | P value | OR | 95% C.I. |
|---|---|---|---|---|---|---|---|
| Haplotype | |||||||
| ACGT | 76 | 44.2 | 73 | 37.6 | 0.20 | 0.76 | 0.50-1.15 |
| ACGC | 3 | 1.7 | 11 | 5.7 | 0.065 | 3.39 | 0.93-12.35 |
| ACAT | 47 | 27.3 | 75 | 38.7 | 0.22 | 1.68 | 1.08-2.61 |
| ATGT | 2 | 1.2 | 1 | 0.5 | 0.50 | 0.44 | 0.04-4.90 |
| ATAT | 9 | 5.2 | 7 | 3.6 | 0.45 | 0.68 | 0.25-1.86 |
| GCGT | 26 | 15.1 | 13 | 6.7 | 0.01 | 0.40 | 0.20-0.81 |
| GCGC | 6 | 3.5 | 12 | 6.2 | 0.24 | 1.82 | 0.67-4.97 |
| Diplotype | |||||||
| ACGT / ACGT | 15 | 17.44 | 3 | 3.09 | 0.0037 | 0.15 | 0.04-0.54 |
| ACGT /ACAT | 34 | 39.54 | 60 | 61.85 | 0.003 | 2.48 | 1.37-4.50 |
| ACGT/ ATGT | 2 | 2.32 | 1 | 1.03 | 0.50 | 0.43 | 0.04-4.91 |
| ACGT/ ATAT | 4 | 4.65 | 5 | 5.15 | 0.87 | 1.11 | 0.29-4.29 |
| ACAT/ACAT | 2 | 2.32 | 3 | 3.03 | 0.75 | 1.34 | 0.22-8.22 |
| ACAT/ ATAT | 1 | 1.16 | 1 | 1.03 | 0.93 | 0.88 | 0.05-14.37 |
| GCGT/ GCGT | 2 | 2.32 | 1 | 1.03 | 0.50 | 0.44 | 0.04-4.91 |
| GCGT/ GCGC | 6 | 6.97 | 10 | 10.3 | 0.43 | 1.53 | 0.53-4.40 |
HCV genotypes
All participants in the study tested negative for HIV. As shown in Table 1, 40% of patients and 6.15% of controls had histories of blood transfusions, and this difference was significant (P<0.05). In the patient group, 47.69% of participants were drug addicted, while no controls were drug addicted. In the patient group, 41.53% of participants were alcohol addicted while only one control subject (1.5%) was alcohol addicted (P< .05). Other risk factors are listed in Table 1. The HCV was genotyped for 65% of patients as follows: 3a: 53.02%, 1a: 15.41%, 1a/1b: 33.34 % and type 2: 2.22 %. The relationship between genotypes of hepatitis virus and some risk factors is shown in Figure 1(Fig. 1).
Fig. 1.
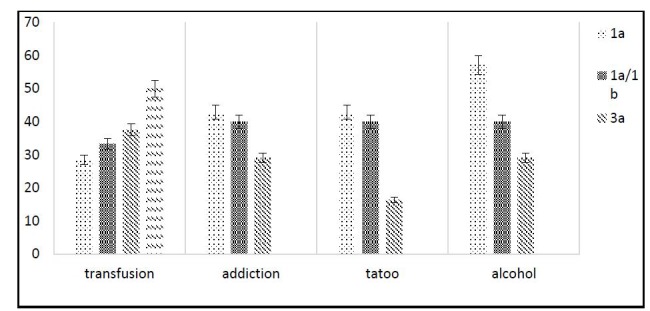
Relationship between common risk factors (alcohol, drug addiction, transfusion and tattoo) and virus genotype in HCV patients.
Biochemical results (ALT assay
For the biochemical test, ALT levels were normal in 73.85% of patients and 83.08% of controls. In patients, an elevated level of ALT up to 35.82 ± 30.2 (U.L-1) was noticed, while in controls it was 26.80 ± 21.38; however the ALP level difference in two groups was not statistically significant.
Discussion
Genetic variations are shown to be associated with susceptibility to various viral and non-viral infections. Hepatitis C infection is influenced by many environmental factors and variations in genetic materials (19). We previously showed that genetic variation of IL-10 (-1082 G>A) can affect susceptibility to hepatitis infection (20). More studies on HCV infection have also been carried out. These studies showed that the genetic polymorphism diversity in different populations have different patterns and these differences can be related to the constitutional differences in populations (21,22). The present study also shows a possible association between these genetic variations and susceptibility to HCV infection, as the ACTA/ACGT diplotype distribution was more common in patients than in controls. However, a larger sample size may be needed to confirm our results.
The association between CTLA-4 promoter polymorphisms (SNPs) and susceptibility to HCV infection has been widely investigated. Yee et al. reported that after treatment with a combination interferon and ribavirin, the GG genotype of CTLA-4 at position +49 A>G was more common in patients than in controls (23).
In another study, Djilali-Saiah et al. studied children from families with autoimmune hepatitis and evaluated the relationship between CD28 and a CTLA-4 exon 1 polymorphism in French and Canadian populations. They found that the ″A” allele of CTLA-4 at position +49 was more frequent in patients than controls (83.5% vs. 50%) and associated with type 1 autoimmune hepatitis (24). We also showed the “A” allele was more common in patients than controls (78.46% vs. 73.03%). Fan et al. also investigated the relationship between autoimmune hepatitis (AIH), primary biliary cirrhosis (PBC), and CTLA-4 polymorphisms at positions +49 and -318 in a Chinese population. Their results indicated that the G allele at +49 A>G was more common in PBC patients than controls (62.9% vs. 56.6%), and the C allele at -318 C>T was more common in AIH patients than controls (93.6% vs. 85.6%) (25). However, our results were not significantly different and the frequency of CT genotype was not common in our patients.
In 2009, Xing Gu et al. examined SNPs at position +49 A>G in CTLA-4 and hepatitis B virus-related hepatocellular carcinoma (HCC) susceptibility in 1003 Chinese subjects. Their results showed that the AA genotype was more common in patients than in controls (14.4% vs 9.5%) and also the “G” allele had a protective effect against hepatitis B virus- (HBV) related HCC (26). The result of their study was similar to our results in which the AA genotype was more common in our patients than in controls. Thio et al. investigated the association between CTLA-4 gene polymorphisms (+49 A>G, -318 C>T, -1722 T>C, and -1661 A>G) and HBV infection. They suggested that the CACCG and TACCG haplotypes were associated with viral clearance (27). In contrast to the Thio et al. study; we found the GCAT haplotype to be the most frequent one in patients with the disease.
Although none of these studies reported diplotype frequencies, our study suggested the ACTA/ACGT diplotype distribution could be related to susceptibility to HCV infection (61.85% and 39.54% in patients and controls, respectively) (Adjusted P = 0.003). Because of the higher frequency of the GCAT haplotype (15.1% vs. 6.7%) (+49 A>G, -318 C>T, -1722 T>C, and -1661 A>G) and the ACTA/ACTA diplotype in the control group vs. patients (17.44% vs. 3.095%), these variants may be protective against HCV infection. Our results remained significant even after adjusting for confounders (tattoo, transfusion, and alcohol and drug addiction). On the other hand, using a dominant model, we found that the “A” allele was significantly more common in controls than patients at position -1661 of CTLA-4 (adjusted P = 0.0003) (Table 2).
Increasing sample size would be helpful to confirm the association between genetic variations in this gene and susceptibility to HCV infection. We showed that transfusion, addiction, and alcohol had similar frequencies of 40, 47, and 41%, respectively, in our patients when we adjusted our results for these confounders. We also found that 57.14% of alcoholaddicted patients were infected with HCV genotype 1a. This study was conducted on a limited number of patients based on estimation of the minimum required sample size using standard statistical formulas; therefore, further studies with larger sample sizes are warranted.
Acknowledgements
The authors declare no conflicts of interest. This study was supported financially by Mashhad University of Medical Sciences, Mashhad, Iran (Grant number: 901012).
References
- 1.Ariyan C, Salvalaggio P, Fecteau S, Deng S, Rogozinski L, Mandelbrot D, et al. Cutting edge: transplantation tolerance through enhanced CTLA-4 expression. J Immunol. 2003;171(11):5673–7. doi: 10.4049/jimmunol.171.11.5673. Epub 2003/11/25. [DOI] [PubMed] [Google Scholar]
- 2.Ueda H, Howson JM, Esposito L, Heward J, Snook H, Chamberlain G, et al. Association of the Tcell regulatory gene CTLA4 with susceptibility to autoimmune disease. Nature. 2003;423(6939):506–11. doi: 10.1038/nature01621. Epub 2003/05/02. [DOI] [PubMed] [Google Scholar]
- 3.Sahin M, Erdogan MF, Erdogan G. Cytotoxic T lymphocyte-associated molecule-4 polymorphisms in Turkish Graves' disease patients and association with probability of remission after antithyroid therapy. European journal of internal medicine. 2005;16(5):352–5. doi: 10.1016/j.ejim.2005.06.007. Epub 2005/09/03. [DOI] [PubMed] [Google Scholar]
- 4.Schott E, Witt H, Hinrichsen H, Neumann K, Weich V, Bergk A, et al. Gender-dependent association of CTLA4 polymorphisms with resolution of hepatitis C virus infection. Journal of hepatology. 2007;46(3):372–80. doi: 10.1016/j.jhep.2006.09.011. Epub 2006/12/08. [DOI] [PubMed] [Google Scholar]
- 5.Lei C, Dongqing Z, Yeqing S, Oaks MK, Lishan C, Jianzhong J, et al. Association of the CTLA-4 gene with rheumatoid arthritis in Chinese Han population. European journal of human genetics. 2005;13(7):823–8. doi: 10.1038/sj.ejhg.5201423. Epub 2005/04/21. [DOI] [PubMed] [Google Scholar]
- 6.Ei Wafai RJ, Chmaisse HN, Makki RF, Fakhoury H. Association of HLA class II alleles and CTLA-4 polymorphism with type 1 diabetes. Saudi journal of kidney diseases and transplantation : an official publication of the Saudi Center for Organ Transplantation, Saudi Arabia. 2011;22(2):273–81. Epub 2011/03/23. [PubMed] [Google Scholar]
- 7.Suppiah V, Alloza I, Heggarty S, Goris A, Dubois B, Carton H, et al. The CTLA4 +49 A/G*G-CT60*G haplotype is associated with susceptibility to multiple sclerosis in Flanders. Journal of neuroimmunology. 2005;164(1-2):148–53. doi: 10.1016/j.jneuroim.2005.04.003. Epub 2005/05/21. [DOI] [PubMed] [Google Scholar]
- 8.Fukazawa T, Yanagawa T, Kikuchi S, Yabe I, Sasaki H, Hamada T, et al. CTLA-4 gene polymorphism may modulate disease in Japanese multiple sclerosis patients. Journal of the neurological sciences. 1999;171(1):49–55. doi: 10.1016/s0022-510x(99)00251-8. Epub 1999/11/24. [DOI] [PubMed] [Google Scholar]
- 9.Chistiakov DA, Savost'anov KV, Turakulov RI, Efremov IA, Demurov LM. Genetic analysis and functional evaluation of the C/T(-318) and A/G(-1661) polymorphisms of the CTLA-4 gene in patients affected with Graves' disease. Clin Immunol. 2006;118(2-3):233–42. doi: 10.1016/j.clim.2005.09.017. Epub 2005/11/22. [DOI] [PubMed] [Google Scholar]
- 10.Chen Z, Fei M, Fu D, Zhang L, Ma Y, Wang Y, et al. Association between cytotoxic T lymphocyte antigen-4 polymorphism and type 1 diabetes: A metaanalysis. Gene. 2013;516(2):263–70. doi: 10.1016/j.gene.2012.12.030. [DOI] [PubMed] [Google Scholar]
- 11.Erfani N, Razmkhah M, Talei AR, Pezeshki AM, Doroudchi M, Monabati A, et al. Cytotoxic T lymphocyte antigen-4 promoter variants in breast cancer. Cancer genetics and cytogenetics. 2006;165(2):114–20. doi: 10.1016/j.cancergencyto.2005.07.020. Epub 2006/03/11. [DOI] [PubMed] [Google Scholar]
- 12.Taha Khalaf A L, Song JQ L, Gao TT L, Yu XP L, Lei TC L. CTLA-4 gene polymorphism and the risk of systemic lupus erythematosus in the Chinese population. Journal of biomedicine & biotechnology. 2011;2011:167395. doi: 10.1155/2011/167395. Epub 2011/09/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Agarwal K, Czaja AJ, Jones DE, Donaldson PT. Cytotoxic T lymphocyte antigen-4 (CTLA-4) gene polymorphisms and susceptibility to type 1 autoimmune hepatitis. Hepatology. 2000;31(1):49–53. doi: 10.1002/hep.510310110. Epub 1999/12/29. [DOI] [PubMed] [Google Scholar]
- 14.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells . Nucleic acids research . 1988;16(3):1215. doi: 10.1093/nar/16.3.1215. Epub 1988/02/11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kouki T, Sawai Y, Gardine CA, Fisfalen ME, Alegre ML, DeGroot LJ. CTLA-4 gene polymorphism at position 49 in exon 1 reduces the inhibitory function of CTLA-4 and contributes to the pathogenesis of Graves' disease. J Immunol. 2000;165(11):6606–11. doi: 10.4049/jimmunol.165.11.6606. Epub 2000/11/22. [DOI] [PubMed] [Google Scholar]
- 16.Ohno O, Mizokami M, Wu RR, Saleh MG, Ohba K, Orito E, et al. New hepatitis C virus (HCV) genotyping system that allows for identification of HCV genotypes 1a, 1b, 2a, 2b, 3a, 3b, 4, 5a, and 6a. Journal of clinical microbiology. 1997;35(1):201–7. doi: 10.1128/jcm.35.1.201-207.1997. Epub 1997/01/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Afshari R, Nomani H, Zaniani FR, Nabavinia MS, Mirbagheri Z, Meshkat M, et al. Genotype distribution of hepatitis C virus in Khorasan Razavi Province, Iran. Turk J Med Sci. 2014;44(4):656–60. doi: 10.3906/sag-1305-64. [DOI] [PubMed] [Google Scholar]
- 18.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. American journal of human genetics. 2001;68(4):978–89. doi: 10.1086/319501. Epub 2001/03/20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bataller R, North KE, Brenner DA. Genetic polymorphisms and the progression of liver fibrosis: a critical appraisal. Hepatology. 2003;37(3):493–503. doi: 10.1053/jhep.2003.50127. [DOI] [PubMed] [Google Scholar]
- 20.Sepahi S, Pasdar A, Ahadi M, Gerayli S, Rostami S, Meshkat Z. Haplotype Analysis of Interleukin-10 Gene Promoter Polymorphisms in Chronic Hepatitis C Infection: A Case Control Study. Viral immunology. 2014;27(8):398–403. doi: 10.1089/vim.2014.0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morgan TR, Lambrecht RW, Bonkovsky HL, Chung RT, Naishadham D, Sterling RK, et al. DNA polymorphisms and response to treatment in patients with chronic hepatitis C: results from the HALT-C trial. Journal of hepatology. 2008;49(4):548–56. doi: 10.1016/j.jhep.2008.05.011. Epub 2008/07/16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rehermann B, Nascimbeni M. Immunology of hepatitis B virus and hepatitis C virus infection. Nature reviews Immunology. 2005;5(3):215–29. doi: 10.1038/nri1573. Epub 2005/03/02. [DOI] [PubMed] [Google Scholar]
- 23.Yee LJ, Perez KA, Tang J, van Leeuwen DJ, Kaslow RA. Association of CTLA4 polymorphisms with sustained response to interferon and ribavirin therapy for chronic hepatitis C virus infection. The Journal of infectious diseases. 2003;187(8):1264–71. doi: 10.1086/374561. Epub 2003/04/16. [DOI] [PubMed] [Google Scholar]
- 24.Djilali-Saiah I, Ouellette P, Caillat-Zucman S, Debray D, Kohn JI, Alvarez F. CTLA-4/CD 28 region polymorphisms in children from families with autoimmune hepatitis. Human immunology. 2001;62(12):1356–62. doi: 10.1016/s0198-8859(01)00344-5. Epub 2002/01/05. [DOI] [PubMed] [Google Scholar]
- 25.Fan LY, Tu XQ, Cheng QB, Zhu Y, Feltens R, Pfeiffer T, et al. Cytotoxic T lymphocyte associated antigen-4 gene polymorphisms confer susceptibility to primary biliary cirrhosis and autoimmune hepatitis in Chinese population. World journal of gastroenterology: WJG. 2004;10(20):3056–9. doi: 10.3748/wjg.v10.i20.3056. Epub 2004/09/21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gu X, Qi P, Zhou F, Ji Q, Wang H, Dou T, et al. +49G > A polymorphism in the cytotoxic Tlymphocyte antigen-4 gene increases susceptibility to hepatitis B-related hepatocellular carcinoma in a male Chinese population. Human immunology. 2010;71(1):83–7. doi: 10.1016/j.humimm.2009.09.353. Epub 2009/09/26. [DOI] [PubMed] [Google Scholar]
- 27.Thio CL, Mosbruger TL, Kaslow RA, Karp CL, Strathdee SA, Vlahov D. Cytotoxic T-lymphocyte antigen 4 gene and recovery from hepatitis B virus infection. Journal of virology. 2004;78(20):11258–62. doi: 10.1128/JVI.78.20.11258-11262.2004. Epub 2004/09/29. [DOI] [PMC free article] [PubMed] [Google Scholar]


