Abstract
Examples of bioactive peptides derived from internal sequences of proteins are known for decades. The great majority of these findings appear to be fortuitous rather than the result of a deliberate and methodological-based enterprise. In the present work, we describe the identification and the biological activities of novel antimicrobial peptides unveiled as internal fragments of various plant proteins founded on our hypothesis-driven search strategy. All putative encrypted antimicrobial peptides were selected based upon their physicochemical properties that were iteratively selected by an in-house computer program named Kamal. The selected peptides were chemically synthesized and evaluated for their interaction with model membranes. Sixteen of these peptides showed antimicrobial activity against human and/or plant pathogens, some with a wide spectrum of activity presenting similar or superior inhibition efficacy when compared to classical antimicrobial peptides (AMPs). These original and previously unforeseen molecules constitute a broader and undisputable set of evidences produced by our group that illustrate how the intragenic concept is a workable reality and should be carefully explored not only for microbicidal agents but also for many other biological functions.
Introduction
In Greek Philosophy but also under the historical knowledge accumulated in Physics, Aristotle introduced the concept of first cause or prime mover, meaning the first of all substances or the commencement of everything that exists in nature. Assuming that complex and superior beings are not logically supposed to be the unique cause (origin, explanation) of themselves, under the framework proposed by the thinker, things in the universe should be caused by previous and original ones. Therefore, these causes exist in succession, as a chain of events distending regressively.
The paradigmatic cosmological theory of the Big Bang, initially suggested in Georges Lemaître’s pivotal work, “Un univers homogène de masse constante et de rayon croissant rendant compte de la vitesse radiale des nébuleuses extra-galactiques”1 and his subsequent Primeval-Atom hypothesis2,3; the Universal Common Ancestry theory4, strengthened by the recent advances in genome sequencing; or the present models of development and differentiation of stem cells5,6, seem to share the same archetypical structure as proposed by the Aristotelian framework. It means that all these theories and models conceive a fundamental starting point of its own substance that during a certain period of time moves towards an increasing structural and functional complexity until the inevitable finitude. By analogy, the complete genome of any living form could also be considered as the primordial starting point in a chain of events. It means that a linearly organized genetic material undergoes metabolic processing yielding structures with forms and functions that, in many ways, as a direct function of time and environmental conditions, appear to be different or substantially apart from its origin. Considering a broader and interspecific scenario, the actual differences in shapes, sizes and behavior our senses perceive in all living creatures, may also be seen as successful products of the internal possibilities (potentialities) of the prime genetic material exposed to a number of evolutionary factors in the time line.
In our previous work7, inspired by this theoretical reasoning, protein sequences were perceived as the prime source from where antimicrobial peptides (AMPs) could be located in one or more regions of the complete polypeptide chain of some mature and expressed proteins. In strict tandem with Aristotle’s thought, the primary structures of certain proteins were assumed to be the material cause of our system, i.e. the raw material (potentiality) from which antimicrobial peptides (actuality) could be physically attained in the same manner that in the cytoplasm newly translated precursor polypeptides (pre-pro-proteins) from correspondent mRNA’s are enzymatically processed into mature proteins and/or peptides. The practical differences of our approach rest on what Aristotle’s analytical method defined as formal, final and efficient causes. In short, an in-house made computer program, called Kamal, generates interactive routines of in silico processing of protein sequences oriented by specific physicochemical parameters capable of identifying putative antimicrobial peptide fragments inside larger polypeptide chains7. These physicochemical parameters, e.g. net charge, average hydrophobicity, and hydrophobic moment, were originally derived from native AMPs described in the literature and used as archetypal parameters to define antimicrobial activity, or in Aristotelian terms, the formal cause. Protein fragments with matching physicochemical properties to the above-described antimicrobial archetype were filtered, chemically synthesized, and tested accordingly to evaluate their microbicidal function (final cause). Evidences of “encrypted” protein fragments showing dissimilar biological activities to their original parent-molecules have been available for some time8–13. However, to our best knowledge, a systematic hypothesis-driven initiative to produce analytical means able to rationally decipher unseen bioactive potentialities inside protein primary structures into biologically relevant functional actualities in the form of microbicidal molecules or “intragenic” antimicrobial peptides (IAPs) was first demonstrated by our group7,14,15.
Our methodology relies in two reinforcing steps: the filtering of genomes/protein collections using Kamal, and a classification tool to identify the IAPs that induce comparable disturbances in model phospholipid membranes. The physicochemical properties of peptide subgroups associated with potent antimicrobial activity are fed back to Kamal and used for successive rounds of peptide filtering/evaluation7. In fact, our system mimics an evolutionary process, with the generation of diversity followed by the selection of the fittest. The present work reports a substantial quantitative and qualitative improvement of our methodology on bioactive peptide screening by showing a new generation of IAPs with antibacterial and antifungal activities obtained from various protein sequences belonging to different plants. The biological activities of two frog skin antimicrobial peptides, DS01 from Phyllomedusa oreades and Ascaphin-8 from Ascaphus truei, are also evaluated as references of native AMPs16,17. Our work demonstrates that intragenic antimicrobial peptides can be as active as known AMPs, with similar selectivity and breadth of antimicrobial spectrum. Furthermore, it provides additional evidence that organisms expressing intragenic peptides could be a feasible alternative to transgenic technology using conventional AMPs18,19 in the constant need for novel agents for the control of pathogens.
Results
Plants genomes as sources of IAPs
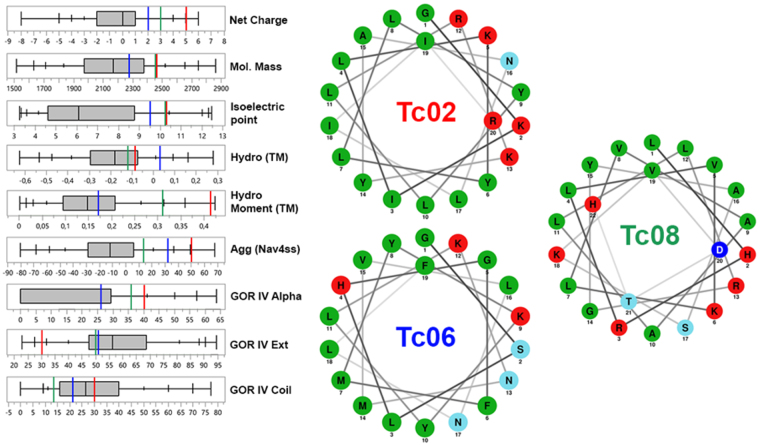
Publicly available protein collections from Theobroma cacao, Arabidopsis thaliana, Citrus sinensis and Gossypium. raimondii were submitted to the software Kamal v1.0 alpha for the filtering of putative IAPs encrypted within proteins. Kamal uses calculated physicochemical properties derived from protein primary structure as descriptors of peptide activity, as previously demonstrated by our group7. Nine filters were sequentially applied to plant genomes to uncover putative IAPs, as described in Table 1. Kamal generates a list of protein fragments with higher probability of antimicrobial activity, which should be curated by the user aided by auxiliary tools available online20. By extensive user examination of the putative IAPs generated by Kamal, twenty-one peptides were selected for solid phase peptide synthesis (Table 2). Tc02, Tc06 and Tc08 have their helical wheel projections and their physicochemical properties depicted in Fig. 1 as an illustration. The distribution of amino acid residues in these IAPs in theoretical helical segments emulates the characteristic amphiphilic character of AMPs20. Furthermore, to investigate the physicochemical properties of putative IAPs prospected from the T. cacao genome in relation to randomly selected peptides, quantile plots were generated. Tc02, Tc06 and Tc08 have a net charge, isoelectric point21 and an aggregation tendency (Na4vSS)22 higher than the 3rd quartile of a sample of ten thousand peptides from 16 to 22 amino acid residues prospected randomly from the same T. cacao protein collection. Moreover, average hydrophobicity23 and hydrophobic moment24 are in the third quartile of the reference set (Fig. 1). These three peptides are representative of the putative IAPs derived from plant genomes selected for solid phase peptide synthesis (Supplementary Material 01, Table S1 and Figure S1). Therefore, putative IAPs selected for synthesis tend to be more cationic, more hydrophobic, and more prone to aggregation than the average T. cacao random protein fragment of the same number of residues.
Table 1.
User-defined physicochemical properties used for the search of IAPs.
| Physicochemical Parameters | Window Range | |
|---|---|---|
| Peptide size | 16–22 aa | |
| Residue filter | C and P* | |
| Net Charge | +1 up to +5 | |
| Molecular Mass (Da) | 1400 up to 4000 | |
| Hydrophobicity Tm scale23 | −0.485 up to +0.516 | |
| Hydrophobic Moment Tm scale23 | +0.516 up to +1.267 | |
| Aggregation22 | −10 up to +55 | |
| Isoelectric Point21 | 9 up to 13 | |
| Windowed Hydrophobicity54 | Max 1 up to 3 | Min −2 up to −0.5 |
*Exclude peptides with cysteine (C) or proline (P) residues.
Table 2.
Peptide IDs, source proteins, scanned organisms, peptide sequences and number of residues of the current generation of IAPs.
| Peptide ID | Source protein | Organism (s) | Peptide Sequence (*) | N.r. |
|---|---|---|---|---|
| Tc01 | Embryo defective 1381 isoform 2 | Theobroma cacao | VALRLAKEVIKVQQGW | 16 |
| Tc02 | ARM repeat superfamily protein, putative isoform 1 | Theobroma cacao | GKILKYLLYLLRKYANLIIR | 20 |
| Tc03 | Tc05_g014340 (12–32)** | Theobroma cacao | IKLRNVLKYLFRIDVIKEDIL | 21 |
| Tc04 | Purple acid phosphatase 3 | Theobroma cacao | RVLKDVESALRESVANWKIVIG | 22 |
| Tc05 | Gamma interferon responsive lysosomal thiol reductase family protein, putative | Theobroma cacao | IVNHLVKLFDKGLNSIVNLR | 20 |
| Tc06 | Cytochrome P450, family 87, subfamily A, polypeptide 2 | Theobroma cacao | GSLHGFMYKYLKNMVLNLF | 19 |
| Tc07 | Laccase 17 | Theobroma cacao | LIKVVNHVQYNVTLHWHGIR | 20 |
| Tc08 | Uncharacterized protein TCM_026695 | Theobroma cacao | LHRLVKLVAALLRGYASKVDTH | 22 |
| Tc09 | NB-ARC domain-containing disease resistance protein, putative | Theobroma cacao | GIVLKDLFSEKLRRYKIVIG | 20 |
| Tc10 | NB-ARC domain-containing disease resistance protein, putative | Theobroma cacao | GLLFKELQKLIRYQIFIGK | 19 |
| Tc11 | LRR and NB-ARC domains-containing disease resistance protein, putative isoform 1 | Theobroma cacao | LLDKLKRTLLSIEAVLI | 17 |
| At01 | Cytochrome P450, family 87, subfamily A, polypeptide 2 | Arabidopsis thaliana | GSLHGFMYKYLKNMVLTLF | 19 |
| At02 | SIP121-0-PQ4 (45-60)*** | Arabidopsis thaliana | KVLSKVHTLLKAVLAL | 16 |
| At03 | YELLOW STRIPE like | Arabidopsis thaliana | GAKLAKKQVRALGKFFSF | 18 |
| At04 | YELLOW STRIPE like | Arabidopsis thaliana | GLYNFIKVLGRTVFGLYKQF | 20 |
| Cs01 | PREDICTED: cytochrome P450 87A3 | Citrus sinensis | GSLHGFMYRYLKNMVLNLF | 19 |
| Zm01 | PREDICTED: cytochrome P450 87A3-like | Zea mays | GSLHGFMYKYLKTLVLRLY | 19 |
| Cs02 | photosystem II CP47 chlorophyll apoprotein (chloroplast) | Citrus sinensis | FFGHIWHGARTLFRDVFA | 18 |
| Cs03 | hypothetical protein CISIN_1g015590mg | Citrus sinensis | FFYNVIKIYGNMAGRISK | 18 |
| Gr01 | PREDICTED: uncharacterized protein LOC105765114 | Gossypium raimondii | GFKLGRKLVKVFKWII | 16 |
| Gr02 | hypothetical protein B456_004G254100 | Gossypium raimondii | ANRLLEAYKMLLKFLGNLR | 19 |
*Carboxyamidated peptides. **Protein not available on NCBInr database. This protein was obtained from the predicted proteins downloaded from cocoagendb.cirad.fr. ***Protein nor available on NCBInr database. This protein was obtained from a list of oxidative-stress from A. thaliana 55.
Figure 1.
Physicochemical parameters and helical wheel plot of the IAPs Tc02, Tc06 and Tc08. Quantile box plots of the net charge, molecular mass (Da), isoelectric point21, average hydrophobicity (TM scale23), Hydrophobic moment (TM scale23), Aggregation (Nav4SS parameter from Aggrescan22), GOR IV53 alpha helix, extended configuration and coil structures was generated for 10.000 randomly selected peptides ranging from 16–22 amino acids from T. cacao genome. Color bars in each quantile plot represent the physicochemical parameters for the IAPs Tc02 (red), Tc06 (blue) and Tc08 (green). Helical wheel plots for the same IAPs are also represented, demonstrating that once structured as α-helical segments, these peptides show an amphiphilic character similar to classical AMPs. Hydrophobic residues are represented in green, positively charged residues are represented in red, negatively charged residues are represented in blue and polar uncharged residues in light blue. Tc02, Tc06 and Tc08 were selected based on their tested antimicrobial activities (Tables 4 and 5) to illustrate physicochemical parameters of the filtered IAPs when compared to randomly selected peptides from T. cacao. Physicochemical properties and helical wheel plots of the remaining IAPs are shown on Supplementary Material 01.
The interaction of IAPs with model membranes
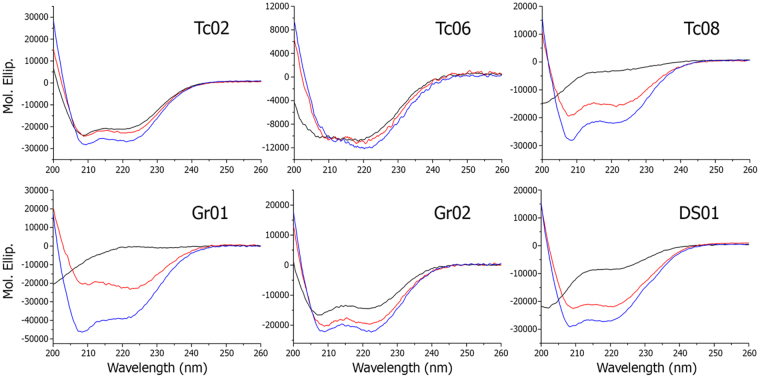
The secondary structure of synthetic IAPs was investigated in buffer and after the addition of dimyristoilphophatidilcoline (DMPC) and 2:1 dimyristoilphophatidilcoline: dimyristoilphophatidilglycerol (DMPC:DMPG) large unilamelar vesicules (LUVs) at 50-fold molar excess. In buffer, most peptides presented far Ultraviolet-Circular Dichroism (UV-CD) spectra compatible with random coil structures (Table 3). Nevertheless, some were already structured as α-helices or β-sheets even before the addition of vesicles. Peptides Tc02 and Gr02 are structured as stable α-helical segments in buffer, and membrane addition accentuated their helical content (Fig. 2). IAPs, such as At02, Gr01 and Tc08, transitioned from random coil structures in buffer to α-helical segments upon membrane interaction, similarly to the classical AMP DS017,16,25 (Fig. 2). Other molecules presented Far-UV CD spectra compatible with β-sheets and were considered separately. The helical percentages of the 21 IAPs evaluated herein in buffer and following the addition of LUVs are described in Table 3. This data corroborates our previous results that IAPs behaves as most membrane-active α-helical AMPs undergoing structure change in the presence of membranes.
Table 3.
Helical percentages of IAPs with or without LUVs.
| Peptide ID | %Hel buffer | %Hel 2:1 DMPC/DMPG | %Hel DMPC |
|---|---|---|---|
| Tc01 | 5.0 | 64.8 | 4.5 |
| Tc02 | 60.4 | 77.0 | 65.6 |
| Tc03 | * | 50.7 | 36.8 |
| Tc04 | 14.3 | 36.3 | 15.8 |
| Tc05 | 8.3 | 68.8 | 34.9 |
| Tc06 | 29.4 | 34.9 | 31.5 |
| Tc07 | 23.5 | 19.9 | 17.1 |
| Tc08 | 9.0 | 62.1 | 40.4 |
| Tc09 | 12.3 | 0.0 | 19.3 |
| Tc10 | 20.3 | 49.6 | 5.1 |
| Tc11 | 45.8 | 56.7 | 57.0 |
| At01 | 32.4 | 28.4 | 26.4 |
| At02 | 13.3 | 79.8 | 32.6 |
| At03 | 0.0 | 30.8 | 0.0 |
| At04 | * | * | * |
| Cs01 | 26.3 | 27.5 | 37.8 |
| Zm01 | 31.0 | 65.5 | 42.2 |
| Cs02 | 4.3 | 52.9 | 52.8 |
| Cs03 | * | * | * |
| Gr01 | 0.9 | 115.0 | 57.7 |
| Gr02 | 56.7 | 64.2 | 50.5 |
| Asc-8 | 2.7 | 88.5 | 74.2 |
| DS01 | 23.2 | 72.4 | 59.4 |
*β-sheet structure.
Figure 2.
Circular Dichroism (CD) spectra of IAPs. Far-UV CD scans were performed to evaluate the secondary structure of IAPs in buffer alone (40 μM solution peptide in phosphate buffer), represented as a black line, and after the addition of 2 mM DMPC, represented as a red line, or in the presence of 2 mM 2:1 DMPC:DMPG, blue line. Tc02 and Gr02 dichroic bands indicate that both peptides are already partially structured as α-helical segments in buffer, and the addition of LUVs induces further structuration. Tc08, Gr01 and DS01 are randomly structured in buffer, only showing α-helix dichroic bands in the presence of LUVs.
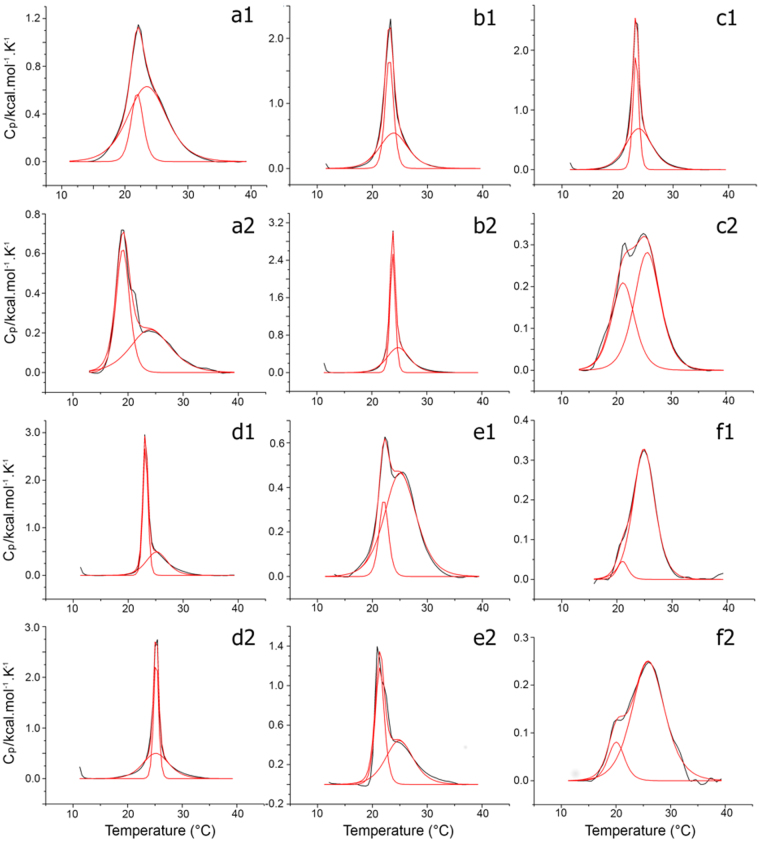
We have further investigated the thermotropic behavior of phospholipid membranes enriched with 4 mol% IAPs by differential scanning calorimetry (Fig. 3). The putative IAPs varied in their ability to alter the P′β→Lα main phase transition of DMPC and 2:1 DMPC:DMPG LUVs. Peptides Tc02 and Gr02 disturbed the main phase transition of vesicles similarly to DS01 (Fig. 3). These two IAPs induced a two-component main phase transition with a more cooperative endotherm at a lower temperature (sharp component) superimposed by another less-cooperative endotherm at a higher temperature (broad component). These alterations are characteristic of deep peptide penetration in the acyl interior of model membranes, a feature previously associated with antimicrobial activity7. Other peptides, such as Gr01, disturbed the main phase transition of LUVs to a lesser extent, indicating that although this peptide structures as an α-helical segment, it interacts more superficially with model membranes (Fig. 3).
Figure 3.
Heating thermal scans of DMPC and 2:1 DMPC:DMPG LUVs enriched with 4 mol% IAPs. Non two-state model fitting of P′β → Lα phase transition of a solution of 0.5 mM of (1) DMPC or (2) 2:1 DMPC:DMPG LUVs enriched with 4 mol% of Tc02 (a), Tc06 (b), Tc08 (c), Gr01 (d), Gr02 (e) or DS01 (f). Black line corresponds to experimental data. Red line corresponds to model fitting.
IAPs are potent and broad antimicrobial agents with low hemolytic activities
The antimicrobial activity of the 21 putative IAPs versus Gram-positive and Gram-negative bacteria, yeasts and filamentous fungi were evaluated. A panel of plant and human pathogenic microorganisms were assayed. Sixteen IAPs, or approximately 76% of the synthesized IAPs, showed inhibitory activity at concentrations up to 256 µM against at least one of the evaluated microorganisms (Tables 4 and 5). Five of them, Tc02, Tc08, At02, Gr01 and Gr02 were as potent as DS0116 and Asc-817, with a similar amplitude of spectrum of activity. Moreover, some specificity towards particular groups of microorganisms were observed. Tc02 inhibited the growth of yeasts and filamentous fungi more efficiently than Gram-positive and -negative bacteria, while Tc08 presented an opposite pattern of activity. The same is true for At04, which showed almost negligible activity to most microorganisms, except for Staphylococcus aureus. Tc06, At01, Cs01 and Zm01 were also as potent as AMPs in inhibiting the growth of yeasts. Other peptides, such as At02, Gr01 and Gr02, inhibited the growth of microorganisms without regard to specific membrane structures or phospholipid compositions.
Table 4.
Antimicrobial activity of IAPs against yeasts and filamentous fungi.
| Peptides | Minimum inhibitory concentration (µM ± SD)* | |||||||
|---|---|---|---|---|---|---|---|---|
| Candida albicans | Cryptococcus neoformans | Fusarium solani | Fusarium oxysporum | Fusarium oxysporum f.sp. vasinfectum | Ramularia areola | Rhizoctonia solani | Trichoderma asperellum | |
| Tc02 | 1.0 | 1.0 | 1.0 | 2.0 | 0.83 ± 0.29 | 0.83 ± 0.29 | 1.0 | 1.0 |
| Tc03 | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| Tc05 | 128.0 | 128.0 | N/A | N/A | N/A | N/A | N/A | N/A |
| Tc06 | 4.0 | 10.67 ± 4.61 | 32.0 | 16.0 | 16.0 | 4.0 | N/A | 6.67 ± 2.31 |
| Tc08 | 4.0 | 8.0 | 32.0 | 32.0 | 2.0 | 4.0 | 16.0 | 16.0 |
| Tc10 | 64.0 | 64.0 | N/A | N/A | N/A | 64.0 | N/A | N/A |
| At01 | 16.0 ± 0.0 | 16.0 ± 0.0 | 32.0 ± 0.0 | 16.0 ± 0.0 | 8.0 ± 0.0 | 4.0 ± 0.0 | N/A | 8.0 ± 0.0 |
| At02 | 8.0 ± 0.0 | 4.0 ± 0.0 | 16.0 ± 0.0 | 4.0 ± 0.0 | 4.0 ± 0.0 | 2.0 ± 0.0 | 16.0 ± 0.0 | 16.0 ± 0.0 |
| At03 | 128.0 ± 0.0 | 64.0 ± 0.0 | N/A | 256.0 ± 0.0 | 64.0 ± 0.0 | 16.0 ± 0.0 | N/A | 128.0 ± 0.0 |
| At04 | N/A | 64,0 ± 0,0 | N/A | N/A | N/A | N/A | N/A | N/A |
| Cs01 | 2.67 ± 1.15 | 8.0 | 16.0 | 32.0 | 8.0 | 4.0 | N/A | 32.0 |
| Zm01 | 6.67 ± 2.31 | 4.0 | 4.0 | 16.0 | 10.67 ± 4.61 | 2.0 | 32.0 | 16.0 |
| Cs02 | 32.0 | 32.0 | 8.0 | 16.0 | 13.33 ± 4.61 | 16.0 | 32.0 | 8.0 |
| Cs03 | 128.0 | 32.0 | 128.0 | 128.0 | 128.0 | 64.0 | N/A | 256.0 |
| Gr01 | 1.0 | 1.0 | 0.5 | 0.5 | 0.5 | 0.5 | 1.0 | 1.0 |
| Gr02 | 4.0 | 2.67 ± 1.15 | 2.0 | 4.0 | 1.67 ± 0.57 | 2.0 | 4.0 | 4.0 |
| Asc-8 | 8.0 | 4.0 | 4.0 | 8.0 | 2.0 | 4.0 | 16.0 | 3.33 ± 1.15 |
| DS01 | 4.0 | 8.0 | 16.0 | 16.0 | 4.0 | 8.0 | 64.0 | 8.0 |
| Fluconazole | 1.63 | 1.63 | N/A | N/A | 26.12 | N/A | N/A | N/A |
| Amphotericin B | 0.27 | 0.13 | 1.08 | 0.54 | 1.08 | 0.54 | 1.08 | 2.16 |
*Standard deviation equals 0 is not displayed in the table. N/A – No antimicrobial activity observed. Peptides Tc01, Tc04, Tc07, Tc09 and Tc11 did not show antimicrobial activity against any of the tested microorganisms.
Table 5.
Antimicrobial activity of IAPs against Gram–positive and –negative bacteria.
| Peptides | Minimum inhibitory concentration (µM ± SD) | |||||||
|---|---|---|---|---|---|---|---|---|
| Escherichia coli | Pseudomonas aeruginosa | Bacillus subtilis | Bacillus cereus | Staphylococcus aureus | Erwinia carotovora | Pseudomonas syringae pv. tabaci | Xanthomonas campestris | |
| Tc02 | 10.67 ± 4.61 | 16.0 | 4.0 | 13.33 ± 4.61 | 8.0 | 16.0 | 4.0 | 16.0 |
| Tc03 | 64.0 | N/A | 16.0 | 128.0 | N/A | 32.0 | 32.0 | 64.0 |
| Tc05 | N/A | N/A | 64.0 | N/A | N/A | N/A | N/A | N/A |
| Tc06 | 32.0 | 26.67 ± 9.24 | 16.0 | 64.0 | 13.33 ± 4.61 | 128.0 | 16.0 | 64.0 |
| Tc08 | 2.0 | 4.0 | 2.0 | 6.67 ± 2.65 | 2.0 | 3.33 ± 1.15 | 4.0 | 4.0 |
| Tc10 | 26.67 ± 9.24 | 128.0 | 16.0 | N/A | N/A | N/A | N/A | 128.0 |
| At01 | 64.0 ± 0.0 | 32.0 ± 0.0 | 16.0 ± 0.0 | 128.0 ± 0.0 | 16.0 ± 0.0 | 256.0 ± 0.0 | 16.0 ± 0.0 | 128.0 ± 0.0 |
| At02 | 2.0 ± 0.0 | 5.33 ± 2.31 | 1.0 ± 0.0 | 3.33 ± 1.15 | 4.0 ± 0.0 | 1.0 ± 0.0 | 4.0 ± 0.0 | 1.0 ± 0.0 |
| At03 | 16.0 ± 0.0 | N/A | 8.0 ± 0.0 | N/A | N/A | 32.0 ± 0.0 | 8.0 ± 0.0 | 32.0 ± 0.0 |
| At04 | N/A | N/A | 256.0 ± 0.0 | N/A | 16.0 ± 0.0 | N/A | N/A | N/A |
| Cs01 | 32.0 | 32.0 | 8.0 | 64.0 | 4.0 | 128.0 | 16.0 | 64.0 |
| Zm01 | 13.33 ± 4.61 | 16.0 | 8.0 | 64.0 | 16 | 16.0 | 32.0 | 16.0 |
| Cs02 | 8.0 | N/A | 3.33 ± 1.15 | 64.0 | 8.0 | 32.0 | 4.0 | 32.0 |
| Cs03 | 128.0 | N/A | 64.0 | N/A | 128.0 | 16.0 | 128.0 | N/A |
| Gr01 | 0.5 | 3.33 ± 1.15 | 1.0 | 1.0 | 0.83 ± 0.29 | 0.5 | 1.0 | 1.0 |
| Gr02 | 4.0 | 32.0 | 1.0 | 4.0 | 1.0 | 1.0 | 2.0 | 4.0 |
| Asc-8 | 2.0 | 4.0 | 1.0 | 4.0 | 2.0 | 3.33 ± 1.15 | 2.0 | 2.0 |
| DS01 | 1.0 | 2.0 | 1.0 | 16.0 | 8.0 | 0.5 | 1.0 | 0.5 |
| Ampicilin | 10.77 | N/A | 0.08 | N/A | 5.38 | 1.35 | 2.69 | 1.35 |
| Gentamicin | 0.52 | 0.13 | 0.033 | 8.37 | 0.26 | 0.53 | 0.52 | 0.53 |
*Standard deviation equals 0 is not displayed in the table. N/A – No antimicrobial activity observed. Peptides Tc01, Tc04, Tc07, Tc09 and Tc11 did not show antimicrobial activity against any of the tested microorganisms.
The microbicidal activities of these IAPs were also evaluated by standard dilution assays and are presented as Supplementary Material (Supplementary Material 02, Tables S2 and S3). Inhibitory and microbicidal concentrations were very similar for the most active peptides, except for Tc06, At01, Cs01 and Zm01, which presented non-detectable microbicide concentrations to various microorganisms, while they inhibited their growth at concentrations lower than 32 µM, indicating a microbistatic activity rather than microbicidal.
The sixteen synthetic IAPs that displayed antimicrobial activity were tested for their cytotoxicity using human erythrocytes as model. Although the IAPs described herein were filtered to hinder the growth of plant pathogens (Table 1), erythrocyte lysis is used as a standard to evaluate peptide selectivity16,17,26. IAPs induced erythrocyte lysis at variable concentrations similar to the AMPs Asc-817 and DS0116 (Table 6 and Supplementary Material 03, Table S4). While Asc-8 is a known hemolytic molecule, DS01 did not induce significant alterations in red or white blood cells at the tested concentrations up to 45 μM16. IAPs induced 50% hemolysis at intermediate concentrations to these two AMPs. Tc02, Gr01 and Gr02, three of the peptides with highest antimicrobial activities, induced 50% hemolysis at 64 μM, a concentration approximately 32 times higher than that required for fungicidal activity, or 12 times higher than that necessary for antibacterial activity.
Table 6.
Hemolytic activity.
| Peptides | Hemolytic activity of human red blood cells (%) |
|---|---|
| SC50** | |
| Tc02 | 64 µM |
| Tc03 | >128 µM |
| Tc05 | >128 µM |
| Tc06 | >128 µM |
| Tc08 | >128 µM |
| Tc10 | >128 µM |
| At01 | >128 µM |
| At02 | >128 µM |
| At03 | >128 µM |
| At04 | >128 µM |
| Cs01 | >128 µM |
| Zm01 | >128 µM |
| Cs02 | >128 µM |
| Cs03 | >128 µM |
| Gr01 | 64 µM |
| Gr02 | 64 µM |
| Asc-8 | 16 µM |
| Ds01 | >128 µM |
**IAP concentration that at least 50% of red blood cells remain intact.
IAPs from T. cacao inhibit the germination of Moniliophthora perniciosa basidiospores
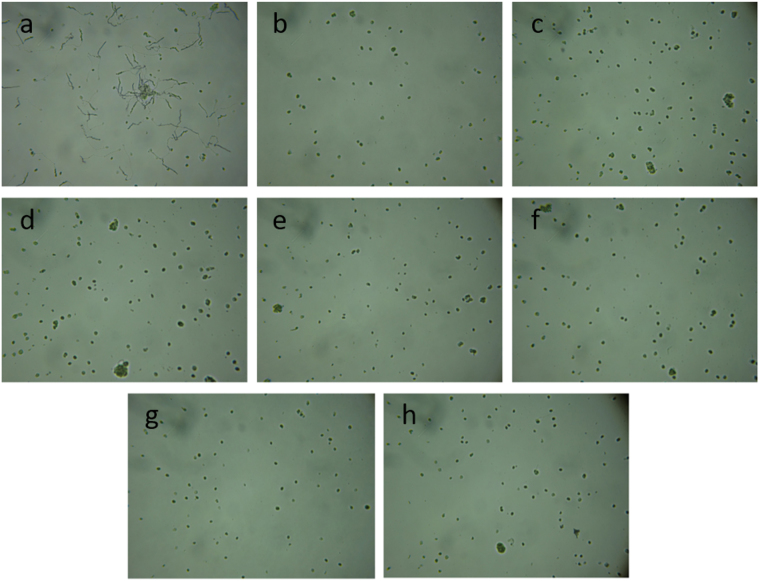
The eleven IAPs filtered from the T. cacao genome were evaluated for their potential to inhibit the germination of M. perniciosa basidiospores, the causal agent of witches’ broom disease27. M. perniciosa basidiospores germinate and infect cocoa meristems 4 hours after contact with cocoa meristems28. Five IAPs and the amphibian peptides DS01 and Asc-8 inhibited the germination of 1 × 106 basidiospores after 4 hours of incubation in the evaluated concentrations, ranging from 256–0.5 µM (Fig. 4). No basidiospores germination were detected even with incubation for longer time periods (24 h and 48 h, Supplementary Material 04, Figure S2). Tc02, a fragment of the ARM repeat superfamily protein, putative isoform 1 from T. cacao, inhibited at least 95% basidiospore germination (IC95) of M. perniciosa basidiospores at 16 µM. Tc06 and Tc10 also inhibited significantly the growth of fungal spores, with an IC95 of 64 µM (Table 7).
Figure 4.
Moniliophthora perniciosa basidiospores germination test. After 4 hours of growth, the basidiospores were visualized. (a) Without peptides; (b) Tc02, 16 μM; (c) Tc05, 256 μM; (d) Tc06, 64 μM; (e). Tc08, 128 μM; (f) Tc10, 64 μM; (g). Asc-8, 16 μM; (h). DS01, 256 μM. The concentrations described for each peptide correspond to the minimum concentrations required to inhibit at least 95% basidiospore germination.
Table 7.
Inhibitory concentration of T. cacao IAPs on M. perniciosa basidiospores germination.
| Peptides | Inhibitory concentration (µM) | |
|---|---|---|
| IC95* | IC50** | |
| Tc02 | 16 | 8 |
| Tc05 | 256 | 128 |
| Tc06 | 64 | 32 |
| Tc08 | 128 | 64 |
| Tc10 | 64 | 32 |
| Asc-8 | 16 | 8 |
| DS01 | 256 | 128 |
*Inhibitory concentration that at least 95% of basidiospores germination was inhibited. **Inhibitory concentration that at least 50% of basidiospores germination was inhibited.
IAPs from G. raimondii inhibit areolate mildew development on cotton leaves
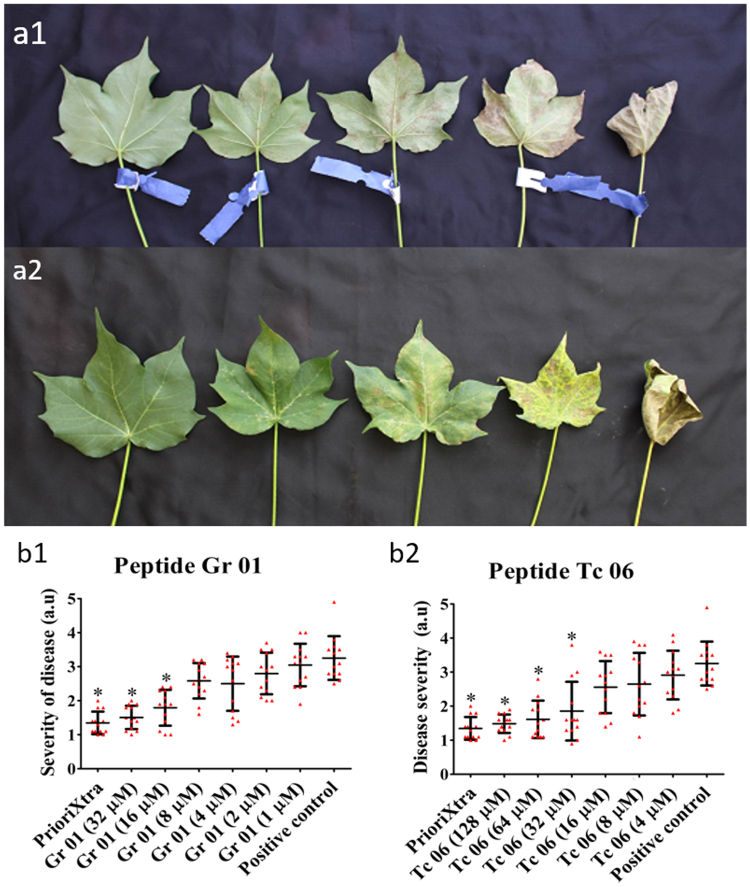
IAPs were also able to inhibit the cotton pathogen Ramularia areola 29 growth in vitro (Table 4). The IAPs Gr01 and Gr02, prospected from G. raimondii proteins, along with other peptides such as Tc06, At03 and Cs02, originally prospected for other plants but found with 100% identity in the cotton genome, were evaluated for their potential to inhibit disease development on cotton leaves infected with R. areola spores. Disease severity was evaluated in a scale from 1 (no symptoms) to 5 (over 50% of leaf area showing disease symptoms) (Fig. 5a). IAPs and 2 × 106 spores were incubated for 20 minutes and this solution was applied in plant leaves. Gr01 and Tc06 showed significant results (p < 0.05, One-way ANOVA) on the control of areolate mildew down to 16 and 32 µM, respectively (Fig. 5b). Moreover, IAPs showed similar qualitative results to the commercial fungicide used as control. The number of spores collected from the leaves was also significantly decreased in the presence of the IAPs (Supplementary Material 05, Figure S3). Surprisingly, Gr02 did not show a significant reduction in areolate mildew severity and spores concentration. This demonstrates that in vitro biological activities might not be observed in vivo, and novel assays are necessary to accurately address these results.
Figure 5.
Ramularia areola infection test. Disease severity index ranging from 1 (without symptoms) and 5 (over 50% of leaf area showing symptoms) in both abaxial (a1) and adaxial surface (a2). Areolate mildew severity in cotton leaves after treatment with a commercial fungicide (PrioriXtra) and different concentrations of IAPs Gr01 (b1) and Tc06 (b2). Gr01 and Tc06 showed comparable results to fungicide at concentrations starting at 16 µM and 32 µM, respectively.
Discussion
We are persuaded that mature proteins represent a feasible and copious source from where smaller bioactive polypeptide chains may be earned, particularly antimicrobials. In the present report, we confirm our previous findings7 by demonstrating additional proofs of this concept exploring plant protein sequences as sources of their derived peptide primary structures and, evaluating their respective biological activities against varied pathogens. The IAPs searched in plant genomes described herein have equivalent potency, selectivity, and display a similar activity spectrum towards microorganisms as AMPs from various sources15,30–32. Moreover, these IAPs fold into α-helical segments upon membrane interaction producing a range of membrane disturbances, in the same manner as mature antimicrobial peptides from frog skin secretions7,33 (Figs 2 and 3). The disturbances introduced by the IAPs in model membranes, as depicted in Fig. 3, are typical of molecules that are localized at the polar/nonpolar interface of the phospholipid bilayer, with a tendency to disturb the acyl interior of membranes, similar to DS01 and other AMPs in the literature34. Furthermore, membrane disturbance is directly associated with peptide helicity, and the same is observed for the IAPs (Fig. 2). Our data demonstrates that classical antimicrobial peptides, selected for their membrane disruptive potential by millions of years of evolution35, can be mimicked in several aspects by internal protein segments with comparable physicochemical properties, but with no identifiable adaptive pressure for such activity when encrypted in parent proteins. Once identified and characterized, these IAPs can be used to confer resistance to a variety of pathogens, by transforming plants with fragments of their own genetic material, providing an alternative to the transgenic technology. This was demonstrated by our group7, where soybean IAPs were able to decrease symptons of Asian rust (Phakopsora pachyrhizi) on soybean leaves in vitro and in vivo.
To the best of our knowledge, this is the first time that molecules with significant antimicrobial activity are obtained from T. cacao and G. raimondii. The IAPs Tc02, Tc06 and Tc10 inhibited the germination of M. perniciosa basidiospores and can be considered promising agents against other T. cacao pathogens, such as Phytophthora spp. and Moniliophthora roreri 36. Despite the encouraging results, cocoa transformation is challenging due to its long growth period to yield its first fruits37. Micro-Tom tomatoes transformation represents an alternative to evaluate the intragenic approach in the control of M. perniciosa infection38. For cotton plants, the inhibition of areolate mildew symptoms (Fig. 5b1 and b2) and the decrease in spore concentration (Supplementary Material 05, Figure S3) with comparable results to a commercial fungicide indicates that peptides Gr01 and Tc06 are suitable candidates for cotton plant transformation.
The evidences we have encountered so far by applying our methodology, alongside most of the native bioactive peptide data available in the literature7,16,31,39, compel us to infer that other sources of proteins in animals, algae and microorganisms should yield similar results to the ones we have evaluated here. Putative IAPs can be prospected from both clade-specific proteins or from evolutionarily conserved homologous genes that span different taxa. The latter seems to be the case of the IAPs Tc06 and Cs02. Tc06 is derived from cytochrome P450 family 87, found with 100% identity in many eudicots and with more than 75% identity in some monocots such as Oryza sativa and Zea mays (Table 2), while the IAP Cs02 is a fragment of the photosystem II CP47 chlorophyll apoprotein, found identically in Bacteria and Eukarya kingdoms (Supplementary Material 06, Figure S4). However, at the present phase of our investigations it appears that more important than a most needed systematic search for novel bioactive peptides in other protein sources that would reinforce our findings, some fundamental questions seem to emerge from the current scenario that deserve to be addressed with higher priority: 1. Are these facts pointing out to a general phenomenon? 2. If so, what would be the biological and physiological implications of that? 3. Could protein degradation (into bioactive peptides) be considered an overlooked step of specific protein post-processing and/or function in nature and not only an earlier stage of proteolysis destined to supply amino acids to various metabolic pathways? 4. What would be the implications of this concept in biotechnology, agriculture and human health?
From our perspective, these questions have no easy answers. Each one of them appears to open such a wide range of theoretical and experimental research possibilities that, at present, are beyond our best work force and probably lifetime effort.
It is our understanding that although the empirical results on microbicidal activities presented here constitute a robust and assorted set of material evidences on which our prime hypothesis is anchored, they seem to represent just the tip of a much larger aspect of the reality that need to be investigated rigorously not only for different types of antimicrobial properties and modes of action, but also for other important biological functions such as analgesic, anti-inflammatory, hypotensive, for example. In fact, our group recently identified encrypted opioid peptides on dehydrin protein family from Coffea spp., showing similar results to Leu-Enkephalin40,41. We also identified a conserved Met-Enkephalin in a F-box protein from T. cacao (NCBI EOY06230.1 – Residues Lys 176 to Lys 184) with similar cleavage sites for serine proteases as found on human pro-enkephalins (unpublished data). Such results highlight how abundant encrypted peptides can be, suggest another facet to genome plasticity and therefore offer new extrinsic principles to biodiversity42–44. Mechanistic features and physicochemical studies on the antimicrobial peptides evaluated in this work are available as a second part of the current paper, entitled “Towards an experimental classification system for membrane active peptides”.
Material and Methods
IAPs search criteria and peptide synthesis
The predicted proteins from Theobroma cacao were downloaded from Cacao Genome Database (www.cacaogenomedb.org) on October 2013. Arabidopsis thaliana, Gossypium raimondii and Citrus sinensis genomes were downloaded from the Phytozome portal (https://phytozome.jgi.doe.gov) on February, July and August 2014, respectively. All genomes were searched for putative IAPs. IAPs were searched through a set of defined physicochemical parameters using Kamal alpha v.1.0 software7. Physicochemical descriptors were updated from our previous report and are listed in Table 1. A total of 21 peptides were chemically synthesized using Fmoc/t-butyl strategy45. Peptide chain elongation was performed on Rink Amide resin, yielding C-terminal amidated peptides. Two known AMPs from frog skin secretions, Dermaseptin DS0116 and Asc-817, were also synthesized. Helical wheel plots were created using an Internet Tool (http://lbqp.unb.br/NetWheels/).
Mass spectrometry analyses
All crude synthetic peptides were analyzed by mass spectrometry to confirm peptide mass and amino acid sequence. Experiments were carried out in an UltrafleXtreme MALDI-TOF/TOF (Bruker Daltonics), controlled by FlexControl 3.0 software (Bruker Daltonics) using α-cyano-4-hydroxycinnamic acid matrix (Fluka) for ionization. Peptides monoisotopic mass were obtained in reflector mode over a range of 700–3500 m/z with external calibration using Peptide Calibration Standard II (Bruker Daltonics). Peptide MS/MS spectra were obtained by means of LIFT fragmentation after analyzing the obtained MS spectra and selection of precursor ions for fragmentation. The software FlexAnalysis 3.0 (Bruker Daltonics) was used for mass spectrometric data analysis. Peptide primary structures were inferred by means of manual interpretation of fragmentation spectra.
HPLC purification and peptide quantification
Reverse phase HPLC (RP-HPLC) of the synthetic peptides were performed in two different scales: analytical and preparative, both using C-18 columns, Grace Vydac 218TP54 and Grace Vydac 218TP1022, respectively. Ultrapure H2O + 0.1% (v/v) TFA (J.T. Baker) and acetonitrile acetonitrile (J.T. Baker) +0.1% (v/v) TFA (J.T. Baker) were used as solvent A and B, respectively. The chromatography method was set as a 50 minutes protocol composed of three steps: 1- 0–5 min: 5% (v/v) solvent B; 2- 5–45 min: gradient of solvent B up to 95% (v/v); 3- 45–50 min: 95% (v/v) of solvent B). Fractions were manually collected and analyzed by mass spectrometry to confirm the elution time of each synthetic peptide. Peptides were then purified in a preparative scale using the same parameters described above. Fractions of interest were collected and analyzed by mass spectrometry to confirm purity. Purified synthetic peptides containing Trp or, at least, three Tyr residues were quantified using calculated molar absorption coefficients46. The remaining peptides were quantified using the UV absorbance of the peptide bond according to the literature47.
Large unilamelar vesicles (LUVs) preparation
DMPC and 2:1 DMPC:DMPG (w/w) were dissolved in chloroform and methanol (3:1 v/v) at 10 mg/mL, dried using a rotary evaporator and left 3 hours under high vacuum in a freeze dryer. Phospholipids were then dissolved in 20 mM Sodium phosphate – NaOH, 150 mM NaCl, pH 7.4 and hand-shaken until the formation of a cloudy solution, which was passed 19 times through a 100 nm polycarbonate membrane at 30 °C for the formation of large unilamelar vesicles (LUVs) using a mini-extruder (Avanti Polar Lipids). Phospholipid concentration was estimated according to the ammonium ferrothiocyanate method48.
Differential scanning calorimetry (DSC)
Thermograms were obtained using a VP-DSC (GE Healthcare) at a temperature range from 10 to 40 °C at a scanning rate of 0.5 °C/min. Blank thermograms using buffer alone and 0.5 mM DMPC or 2:1 (w/w) DMPC:DMPG LUVs in buffer were acquired as reference. Peptides were added to fresh samples of 0.5 mM LUVs at a concentration of 20 μM (0.04 mol/mol peptide/phospholipids) at room temperature, immediately followed by DSC data acquisition. Each sample was subjected to various thermal scans until there were no distinguishable changes in the thermal profile of the main phase transition (P′β→Lα) of phospholipids between scans. Data was concentration normalized, baseline subtracted (linear connect), and fitted to a non two-state transition with two peaks determined by the user applying the MicroCal Origin software v7.0. Re-scans for selected cases were acquired using fresh peptide and LUVs solutions to check the reproducibility of the data.
Circular Dichroism (CD)
Experiments were conducted on a Jasco-J815 spectropolarimeter (Jasco International Co.). Spectra were acquired at room temperature from 200 to 260 nm as an average of 4 readings using a 0.1 cm path length cell, data pitch of 0.2 nm and a response time of 0.5 s. Data Scans of buffer and 2 mM DMPC and 2:1 DMPC:DMPG LUVs solutions were acquired and subtracted from each peptide data. Peptides were scanned at a concentration of 40 μM in buffer and then 50 fold excess of DMPC and 2:1 DMPC:DMPG LUVs were added, resulting in a molar ratio of 0.02 peptide/phospholipid. The spectra were converted to mean residue ellipticity and readings at [θ]222 nm were used to estimate helix percentages49.
Evaluation of antimicrobial activity of IAPs
The susceptibility test of microorganisms was performed to evaluate the inhibitory activity of the IAPs. Protocols M7-A1050, M27-A351 and M38-A252 from Clinical & Laboratory Standards Institute (CLSI) were used for bacteria, yeasts and filamentous fungi tests, respectively. Briefly, different concentration of IAPs (256-0.5 µM) were tested against 16 microorganisms, in a final volume of 100 µL. Media conditions, final cell concentration and incubation conditions are presented in Supplementary Material 07, Table S5. Three biological repetitions, with 2 technical replicates each, were performed for each test, on polystyrene flat-bottom 96 wells microplates. The minimum inhibitory concentration (MIC) was defined as the concentration that no cells/hyphae were detected when visualized by optical microscopy.
The microbicidal activity of the IAPS was also evaluated. After the growing period showed on Supplementary Material 07 for each organism, 10 µL of the minimum inhibitory concentration and raising concentrations up to 256 µM were transferred to agar plates, and incubated at the same conditions (Table 2). YPD agar and Mueller-Hinton agar were used for fungi/yeasts and bacteria, respectively. Meanwhile, susceptibility microplates were incubated again to evaluate if there would be any growing after a longer period. For both approaches, the minimal killing concentration (MKC) was defined as the concentration that no growth was observed after incubation. Three biological replicates were performed for each test.
Evaluation of cytotoxicity of IAPs
The cytotoxicity of the selected IAPS was performed using human red blood cells in strict accordance with relevant guidelines and regulations (Ethical committee – UnB # 1.939.989). Different concentrations of IAPs (128 µM–0.5 µM) were incubated with a final concentration of 2.5 × 105 red blood cells.mL−1. Briefly, 100 µL of red blood cells was incubated with 100 µL of different concentrations of each IAP, prepared in PBS, for 30 minutes at 37 °C, with constant shaking at 120 rpm. PBS and Triton X-100 0.1% (v/v) were used as negative and positive controls, respectively. After incubation, the solution was centrifuged at 900 × g for five minutes followed by transferring 50 µL of the supernatant to flat-bottom polystyrene 96-well micro plates containing 50 µL of PBS. Microplate reading was performed at a BioTek Multireader, using a wavelength of 540 nm. All tests were performed in 3 biological replicates, composed of 3 biological repetitions each. The obtained data was subtracted by the negative control and relative to the positive control, defined as 100% lysis.
Inhibition of M. perniciosa basidiospore germination by T. cacao IAPs
The antimicrobial potential of T. cacao IAPs for inhibiting M. perniciosa basidiospores was evaluated. Briefly, 1 × 106 basidiospores mL−1 were incubated for one hour with different concentrations of T. cacao IAPs (256-0.5 µM), at 24 °C, in a final volume of 100 µL. Autoclaved dH20 was used as a control. After incubation, 3 droplets of 10 µL each were placed in a slide coated with agar (1.2% (w/v)). Slides were incubated for 4, 24 and 48 hours at 24 °C. Slides were visualized using a DMRXA light microscope (Leica). Approximately 150–200 basidiospores per droplet from each time point were counted and compared to the control condition, which was defined as 100% germination. All experiments were performed in biological triplicates, with 3 technical repetitions each.
IAP areolate mildew inhibition on cotton leaves
Different concentration of IAPs Tc06 (128-4 µM) and Gr01 (32-1 µM) were mixed with Ramularia areola spore solution (final concentration 2 × 106 spores.mL−1) for 20 minutes prior to application on both surfaces of cotton leaves (cultivar IMA5675 B2RF). R. areola spores was also mixed with the commercial fungicide PrioriXtra (Syngenta®) (1,5 mL.L−1) as negative control for disease development, and with water, as positive control.
Experimental groups were composed of 13 pots, with two plants each. From each plant, two leaves from sixty one days old cotton plants were selected for treatment. Sixteen days after application, disease severity and number of spores per leaf per experimental group were evaluated. For severity analysis, a score from 1 (no symptoms) to 5 (symptoms over 50% of foliar area) was attributed for each leave. The final score for each treatment was an average from the four leaves from each pot. Spore counting was performed using a Neubauer chamber. Briefly, four leaves from each pot were scrapped and counted, yielding a final spore count per pot. Data and statistical analysis were performed using Excel, GraphdPad Prism and Past softwares.
Electronic supplementary material
Acknowledgements
The authors wish to express their gratitude to Dr. Eliseu R. Alves and Mr Álvaro Salles for their support throughout this project and are also grateful to EMBRAPA, CEPLAC, IMA-MT, FINEP (Rede de Cacau do Renorbio, Vassoura-de-bruxa) and CNPq (Bilateral Project Brazil-Belgium) for providing the infrastructure and indispensable funding.
Author Contributions
Intragenic antimicrobial peptides identification was performed by M.H.S.R., G.D.B. and C.B.Jr. Synthesis and analytical steps were performed by M.H.S.R. and M.V.P. Antimicrobial and haemolytic activities were designed and performed by M.H.S.R., F.Y.A., M.O and K.P.G. Peptide-membrane interactions were performed by M.H.S.R. R. areola infection tests were designed and performed by J.L.C.F. and R.G. The manuscript was written by M.H.S.R., G.D.B. and C.B.Jr. All authors have read and approved the final manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-13685-6.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lemaître G. Un Univers homogène de masse constante et de rayon croissant rendant compte de la vitesse radiale des nébuleuses extra-galactiques. Ann. la Société Sci. Bruxelles. 1927;47:49–59. [Google Scholar]
- 2.Lemaître G. The Beginning of the World from the Point of View of Quantum Theory. Nature. 1931;127:706–706. doi: 10.1038/127706b0. [DOI] [Google Scholar]
- 3.Kragh HS, Lambert D. The Context of Discovery: Lemaître and the Origin of the Primeval-Atom Universe. Ann. Sci. 2007;64:445–470. doi: 10.1080/00033790701317692. [DOI] [Google Scholar]
- 4.Theobald DL. A formal test of the theory of universal common ancestry. Nature. 2010;465:219–222. doi: 10.1038/nature09014. [DOI] [PubMed] [Google Scholar]
- 5.David B, Derrick JR, Weissman IL. Hematopoietic Stem Cells: The Paradigmatic Tissue-Specific Stem Cell. Am. J. Pathol. 2006;169:338–346. doi: 10.2353/ajpath.2006.060312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fu, N., Lindeman, G. J. & Visvader, J. E. The Mammary Stem Cell Hierarchy. Current Topics in Developmental Biology107 (Elsevier Inc., 2014). [DOI] [PubMed]
- 7.Brand GD, et al. Probing Protein Sequences as Sources for Encrypted Antimicrobial Peptides. PLoS One. 2012;7:e45848. doi: 10.1371/journal.pone.0045848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meisel H, Bockelmann W. Bioactive peptides encrypted in milk proteins: Proteolytic activation and thropho-functional properties. Antonie van Leeuwenhoek, Int. J. Gen. Mol. Microbiol. 1999;76:207–215. doi: 10.1023/A:1002063805780. [DOI] [PubMed] [Google Scholar]
- 9.Dores RM, Lecaudé S, Bauer D, Danielson PB. Analyzing the evolution of the opioid/orphanin gene family. Mass Spectrom. Rev. 2002;21:220–243. doi: 10.1002/mas.10029. [DOI] [PubMed] [Google Scholar]
- 10.Demidova-Rice TN, Wolf L, Deckenback J, Hamblin MR, Herman IM. Human platelet-rich plasma- and extracellular matrix-derived peptides promote impaired cutaneous wound healing in vivo. PLoS One. 2012;7:e32146. doi: 10.1371/journal.pone.0032146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dziuba B, Dziuba M. New milk protein-derived peptides with potential antimicrobial activity: An approach based on bioinformatic studies. Int. J. Mol. Sci. 2014;15:14531–14545. doi: 10.3390/ijms150814531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saikia K, Sravani YD, Ramakrishnan V, Chaudhary N. Highly potent antimicrobial peptides from N-terminal membrane-binding region of E. coli MreB. Sci. Rep. 2017;7:42994. doi: 10.1038/srep42994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumar R, Jangir PK, Das J, Taneja B, Sharma R. Genome Analysis of Staphylococcus capitis TE8 Reveals Repertoire of Antimicrobial Peptides and Adaptation Strategies for Growth on Human Skin. Sci. Rep. 2017;7:10447. doi: 10.1038/s41598-017-11020-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bloch, C. Jr., Silva, L. P. & Magalhães, B. S. Peptides and polypeptides identification comprises detection of potential functions DNA fragments, for genetic engineering in organisms. Patent Number(s): BR200506317-A; Derwent Primary Accession Number: 2007-709053 [67]; International Patent Classification: C12N-015/09; C12Q-001/68; G01N-033/68; G01N-037/00 (2007).
- 15.Brand, G. D. Estratégias para prospecção e predição de peptídeos bioativos. 217p. (2007).
- 16.Brand GD, et al. Dermaseptins from Phyllomedusa oreades and Phyllomedusa distincta: Anti-Trypanosoma cruzi activity without cytotoxicity to mammalian cells. J. Biol. Chem. 2002;277:49332–49340. doi: 10.1074/jbc.M209289200. [DOI] [PubMed] [Google Scholar]
- 17.Conlon JM, Sonnevend A, Davidson C, David Smith D, Nielsen PF. The ascaphins: A family of antimicrobial peptides from the skin secretions of the most primitive extant frog, Ascaphus truei. Biochem. Biophys. Res. Commun. 2004;320:170–175. doi: 10.1016/j.bbrc.2004.05.141. [DOI] [PubMed] [Google Scholar]
- 18.Ponti D, Mangoni ML, Mignogna G, Simmaco M, Barra D. An amphibian antimicrobial peptide variant expressed in Nicotiana tabacum confers resistance to phytopathogens. Biochem. J. 2003;370:121–7. doi: 10.1042/bj20021444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rivero M, et al. Stacking of antimicrobial genes in potato transgenic plants confers increased resistance to bacterial and fungal pathogens. J. Biotechnol. 2012;157:334–343. doi: 10.1016/j.jbiotec.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 20.Wang G, Li X, Wang Z. APD2: The updated antimicrobial peptide database and its application in peptide design. Nucleic Acids Res. 2009;37:933–937. doi: 10.1093/nar/gkn823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bjellqvist B, et al. The focusing positions of polypeptides in immobilized pH gradients can be predicted from their amino acid sequences. Electrophoresis. 1993;14:1023–1031. doi: 10.1002/elps.11501401163. [DOI] [PubMed] [Google Scholar]
- 22.Conchillo-Solé O, et al. AGGRESCAN: a server for the prediction and evaluation of ‘hot spots’ of aggregation in polypeptides. BMC Bioinformatics. 2007;8:65. doi: 10.1186/1471-2105-8-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao G, London E. An amino acid ‘transmembrane tendency’ scale that approaches the theoretical limit to accuracy for prediction of transmembrane helices: relationship to biological hydrophobicity. Protein Sci. 2006;15:1987–2001. doi: 10.1110/ps.062286306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eisenberg D, Weiss RM, Terwilliger TC. The helical hydrophobic moment: a measure of the amphiphilicity of a helix. Nature. 1982;299:371–374. doi: 10.1038/299371a0. [DOI] [PubMed] [Google Scholar]
- 25.Chen Y, et al. Role of peptide hydrophobicity in the mechanism of action of α-helical antimicrobial peptides. Antimicrob. Agents Chemother. 2007;51:1398–1406. doi: 10.1128/AAC.00925-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsuzaki K. Control of cell selectivity of antimicrobial peptides. Biochim. Biophys. Acta - Biomembr. 2009;1788:1687–1692. doi: 10.1016/j.bbamem.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 27.Evans HC. Pleomorphism in Crinipellis perniciosa, causal agent of witches’ broom disease of cocoa. Trans. Br. Mycol. Soc. 1980;74:515–523. doi: 10.1016/S0007-1536(80)80051-9. [DOI] [Google Scholar]
- 28.Sena K, Alemanno L, Gramacho KP. The infection process of Moniliophthora perniciosa in cacao. Plant Pathol. 2014;63:1272–1281. doi: 10.1111/ppa.12224. [DOI] [Google Scholar]
- 29.Galbieri R, et al. Ramularia areola sporulation potential in Brazilian cotton. Summa Phytopathol. 2015;41:233–235. doi: 10.1590/0100-5405/2008. [DOI] [Google Scholar]
- 30.Yeaman MR, Yount NY. Mechanisms of antimicrobial peptide action and resistance. Pharmacol. Rev. 2003;55:27–55. doi: 10.1124/pr.55.1.2. [DOI] [PubMed] [Google Scholar]
- 31.Melo MN, Ferre R, Castanho MARB. Antimicrobial peptides: linking partition, activity and high membrane-bound concentrations. Nat. Rev. Microbiol. 2009;7:245–250. doi: 10.1038/nrmicro2095. [DOI] [PubMed] [Google Scholar]
- 32.Wang G, Li X, Wang Z. APD3: The antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 2016;44:D1087–D1093. doi: 10.1093/nar/gkv1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McElhaney RN. The use of differential scanning calorimetry and differential thermal analysis in studies of model and biological membranes. Chem. Phys. Lipids. 1982;30:229–259. doi: 10.1016/0009-3084(82)90053-6. [DOI] [PubMed] [Google Scholar]
- 34.Seto GWJ, et al. Interactions of the Australian tree frog antimicrobial peptides aurein 1.2, citropin 1.1 and maculatin 1.1 with lipid model membranes: Differential scanning calorimetric and Fourier transform infrared spectroscopic studies. Biochim. Biophys. Acta - Biomembr. 2007;1768:2787–2800. doi: 10.1016/j.bbamem.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 35.Vanhoye D, Bruston F, Nicolas P, Amiche M. Antimicrobial peptides from hylid and ranin frogs originated from a 150-million-year-old ancestral precursor with a conserved signal peptide but a hypermutable antimicrobial domain. Eur. J. Biochem. 2003;270:2068–2081. doi: 10.1046/j.1432-1033.2003.03584.x. [DOI] [PubMed] [Google Scholar]
- 36.Evans HC. Cacao Diseases—The Trilogy Revisited. Phytopathology. 2007;97:1640–1643. doi: 10.1094/PHYTO-97-12-1640. [DOI] [PubMed] [Google Scholar]
- 37.Argout X, et al. The genome of Theobroma cacao. Nat. Genet. 2011;43:101–8. doi: 10.1038/ng.736. [DOI] [PubMed] [Google Scholar]
- 38.Deganello J, Leal GA, Rossi ML, Peres LEP, Figueira A. Interaction of Moniliophthora perniciosa biotypes with micro-tom tomato: A model system to investigate the witches’ broom disease of Theobroma cacao. Plant Pathol. 2014;63:1251–1263. doi: 10.1111/ppa.12206. [DOI] [Google Scholar]
- 39.Zosloff M. Magainlns, a Class of Antimicrobial Peptldes from Xenopus Skin: Isolation, Characterization of Two Active Forms, and Partial cDNA Sequence of a Precursor. J. Occup. Environ. Med. 1988;30:470. doi: 10.1097/00043764-198806000-00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vinecky, F. Identificação e caracterização de peptídeos opioides presentes na fração proteica de grãos de Coffea arabica. 72p. (2015).
- 41.Vinecky, F., Moreira, K. G., Mortari, M. R. & Bloch, C. Jr. Opioid peptide. Patent number: BR2015/050120. US Patent number: 15/504,385 (2016).
- 42.Bennett PM. Genome Plasticity. Genomics, Proteomics, Clin. Bacteriol. 2004;266:71–113. doi: 10.1385/1-59259-763-7:071. [DOI] [Google Scholar]
- 43.Dubcovsky J, Dvorak J. Genome plasticity a key factor in the success of polyploid wheat under domestication. Science. 2007;316:1862–6. doi: 10.1126/science.1143986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vandecraen J, Chandler M, Aertsen A, Van Houdt R. The impact of insertion sequences on bacterial genome plasticity and adaptability. Crit. Rev. Microbiol. 2017;13:1–22. doi: 10.1080/1040841X.2017.1303661. [DOI] [PubMed] [Google Scholar]
- 45.Chan, W. C. & White, P. D. Fmoc Solid Phase Peptide Synthesis: A Practical Approach. (Oxford University Press, 2000).
- 46.Kirschenbaum DM. Molar absorptivity and A1cm1% values for proteins at selected wavelengths of the ultraviolet and visible regions-XXI. J. Quant. Spectrosc. Radiat. Transf. 1982;27:39–53. doi: 10.1016/0022-4073(82)90042-5. [DOI] [Google Scholar]
- 47.Waddell W. A simple UV spectrophotometric method for the determination of protein. J. Lab. Clin. Med. 1956;48:311–314. [PubMed] [Google Scholar]
- 48.Stewart JC. Colorimetric determination of phospholipids with ammonium ferrothiocyanate. Anal. Biochem. 1980;104:10–4. doi: 10.1016/0003-2697(80)90269-9. [DOI] [PubMed] [Google Scholar]
- 49.Chen YH, Yang JT, Chau KH. Determination of the helix and beta form of proteins in aqueous solution by circular dichroism. Biochemistry. 1974;13:3350–9. doi: 10.1021/bi00713a027. [DOI] [PubMed] [Google Scholar]
- 50.CLSI. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobicaly; Approved Standard - Tenth Edition. CLSI DocumentM07–A10, (Clinical and Laboratory Standards Institute, 2012).
- 51.CLSI. Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts: Approved Atandard - Third Edition. CLSI DocumentM27-A3, (Clinical and Laboratory Standards Institute, 2008).
- 52.CLSI. Reference Method for Broth Dilution Antifungal Susceptibility Testing of Filamentous Fungi; Approved Standard—Second Edition. CLSI DocumentM38-A2, (Clinical and Laboratory Standards Institute, 2008).
- 53.Garnier J, Gibrat JF, Robson B. GOR IV GOR secondary structure prediction method version IV. Meth Enzym. 1996;266:540–553. doi: 10.1016/S0076-6879(96)66034-0. [DOI] [PubMed] [Google Scholar]
- 54.Kyte J, Doolittle RF. A simple method for displaying the hudropathic character of a protein. J. Mol. Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 55.De Coninck B, et al. Mining the genome of Arabidopsis thaliana as a basis for the identification of novel bioactive peptides involved in oxidative stress tolerance. J. Exp. Bot. 2013;64:5297–5307. doi: 10.1093/jxb/ert295. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.