Abstract
Bacterial pathogens must sense and respond to newly encountered host environments to regulate the expression of critical virulence factors that allow for niche adaptation and successful colonization. Among bacterial pathogens, non-typhoidal serovars of Salmonella enterica, such as serovar Typhimurium (S. Tm), are a primary cause of foodborne illnesses that lead to hospitalizations and deaths worldwide. S. Tm causes acute inflammatory diarrhea that can progress to invasive systemic disease in susceptible patients. The gastrointestinal tract and intramacrophage environments are two critically important niches during S. Tm infection, and each presents unique challenges to limit S. Tm growth. The intestinal tract is home to billions of commensal microbes, termed the microbiota, which limits the amount of available nutrients for invading pathogens such as S. Tm. Therefore, S. Tm encodes strategies to manipulate the commensal population and side-step this nutritional competition. During subsequent stages of disease, S. Tm resists host immune cell mechanisms of killing. Host cells use antimicrobial peptides, acidification of vacuoles, and nutrient limitation to kill phagocytosed microbes, and yet S. Tm is able to subvert these defense systems. In this review, we discuss recently described molecular mechanisms that S. Tm uses to outcompete the resident microbiota within the gastrointestinal tract. S. Tm directly eliminates close competitors via bacterial cell-to-cell contact as well as by stimulating a host immune response to eliminate specific members of the microbiota. Additionally, S. Tm tightly regulates the expression of key virulence factors that enable S. Tm to withstand host immune defenses within macrophages. Additionally, we highlight the chemical and physical signals that S. Tm senses as cues to adapt to each of these environments. These strategies ultimately allow S. Tm to successfully adapt to these two disparate host environments. It is critical to better understand bacterial adaptation strategies because disruption of these pathways and mechanisms, especially those shared by multiple pathogens, may provide novel therapeutic intervention strategies.
Keywords: Salmonella, macrophages, signaling pathways, infection, microbiota
Salmonella enterica Serovar Typhimurium Infection
Non-typhoidal serovars of Salmonella enterica (NTS) are leading causes of foodborne illness and diarrheal disease worldwide (Graham et al., 2000; Vojdani et al., 2008; Scallan et al., 2011; Ansari et al., 2012; Kabir et al., 2012; Kozak et al., 2013). In the United States, NTS infections result in more hospitalizations and deaths compared to infections caused by any other foodborne pathogen (Scallan et al., 2011). Among NTS, serovar Typhimurium (S. Tm) is one of the most commonly isolated from patients around the globe (Galanis et al., 2006). NTS infections typically present as a self-limiting diarrheal disease (Acheson and Hohmann, 2001; Gordon, 2008); however, NTS gastrointestinal infections can develop into systemic disease in immunocompromised patients, as well as a small subset of immunocompetent patients (Acheson and Hohmann, 2001; Gordon, 2008). Currently, there are no effective vaccines against gastrointestinal infections. Additionally, treatment options are limited because antibiotics may lead to increased levels of S. Tm shedding and also because S. Tm is developing resistance to many antibiotics (Wiström et al., 1992; Martin, 2012; Diard et al., 2014; Gopinath et al., 2014; Strugnell et al., 2014). Accordingly, alternative therapeutic intervention strategies are needed.
S. Tm establishes infection in the gastrointestinal tract and causes acute gastroenteritis. A common feature of S. Tm disease is inflammatory diarrhea indicated by the presence of neutrophils in patient stool samples (Harris et al., 1972). Type III secretion systems (T3SSs) are molecular syringe-like structures that allow Gram-negative organisms to directly inject effector proteins into the cytosol of host cells (Deng et al., 2017). S. Tm uses a T3SS encoded within Salmonella Pathogenicity Island (SPI) 1 (T3SS-1) to actively invade epithelial cells, induce inflammation, and breach the epithelial barrier (Galán and Curtiss, 1989; Tsolis et al., 1999). After exiting the intestinal tract, S. Tm is phagocytosed by resident and recruited immune cells, including macrophages. S. Tm utilizes the SPI-2 encoded T3SS (T3SS-2) to survive and replicate within these phagocytes (Hensel et al., 1995; Ochman et al., 1996; Shea et al., 1996). The cumulative effects of T3SS-2 cause unchecked bacterial replication during systemic infection and lethal disease (Yoon et al., 2009).
Decades of research has identified a vast repertoire of S. Tm virulence determinants (extensively reviewed in Fàbrega and Vila, 2013). Recent studies have expanded our understanding of factors that influence virulence gene expression, including growth phase and environmental signals (Kröger et al., 2013; Srikumar et al., 2015); however, less is known about the signal transduction pathways that link environmental signals to virulence gene expression. In this review, we discuss recent findings concerning strategies that S. Tm uses to overcome microbiota- and host-derived obstacles within the intestinal and intramacrophage environments. Additionally, we highlight signals that S. Tm uses to coordinate expression of virulence genes required for adaptation to these distinct environments.
Making Room Within the Crowded Intestinal Tract
The gastrointestinal tract is home to billions of microbes termed the microbiota. Interactions between the host, the microbiota, and pathogens have profound impacts on infection (Yurist-Doutsch et al., 2014; McKenney and Pamer, 2015; Bäumler and Sperandio, 2016; Gart et al., 2016; Kendall and Sperandio, 2016; McKenney et al., 2016). The microbiota function as a barrier to limit pathogen colonization and shedding (Endt et al., 2010), an ability collectively referred to as colonization resistance. Colonization resistance is largely attributed to the ability of the microbiota to outcompete invading pathogens for nutrients; however, the microbiota can also modulate host mucosal immune responses important for clearing infection (Endt et al., 2010; Thiemann et al., 2017). Dysbiosis, or alterations to the microbiota, creates a non-competitive niche in which S. Tm is able to establish infection and rapidly replicate in the intestine. Antibiotic use results in disruptions to the microbiota and is a key risk factor associated with the Salmonella-associated diarrhea (Acheson and Hohmann, 2001; Gordon, 2008).
Although antibiotic-related dysbiosis provides an opening for S. Tm to establish infection, S. Tm also directly perturbs the microbiota to enhance and prolong infection (Figure 1A). S. Tm relies primarily on the T3SS-1, and to an extent the T3SS-2, to induce host inflammation, and the resulting innate immune response non-specifically targets the microbiota along with S. Tm (Barthel et al., 2003; Coburn et al., 2005; Stecher et al., 2007; Barman et al., 2008; Lawley et al., 2008; Sekirov et al., 2008; Juricova et al., 2013; Lam and Monack, 2014; Drumo et al., 2016; Rivera-Chávez et al., 2016b). As a result of T3SS-induced inflammation, a proportion of infecting S. Tm cells succumb to host immune responses; however, a sufficient amount of S. Tm cells survive to successfully establish infection (Raffatellu et al., 2009; Liu et al., 2012; Bogomolnaya et al., 2013; Maier et al., 2014; Diaz-Ochoa et al., 2016). Several environmental conditions contribute to T3SS-1 expression (Golubeva et al., 2012). For example, oxygen limitation and high salt concentrations enhance SPI-1 expression and epithelial cell invasion (Galán and Curtiss, 1990; Lee and Falkow, 1990; Bajaj et al., 1996; Mizusaki et al., 2008), whereas bile and some long and short chain fatty acids, such as a butyrate, oleate, myristate, and palmitate, repress T3SS-1 expression (Prouty and Gunn, 2000; Lawhon et al., 2002; Gantois et al., 2006; Eade et al., 2016; Golubeva et al., 2016). The balance of activating and repressing signals is thought to enrich S. Tm invasion in the ileum in vivo (Lawhon et al., 2002; Gantois et al., 2006; Eade et al., 2016). Expression of the T3SS-1 is regulated by a feed-forward loop in which the regulatory proteins HilD, HilC, and RtsA positively control expression of the master transcription factor HilA (Bajaj et al., 1995; Ellermeier et al., 2005). HilD, HilC, and RtsA are transcription factors that bind to the hilA promoter to induce hilA expression (Schechter and Lee, 2001; Olekhnovich and Kadner, 2002; Ellermeier and Slauch, 2004). HilA then activates the expression of the remaining transcription factors and structural components of the T3SS-1, as well as non-SPI-1-encoded effectors (Phoebe Lostroh and Lee, 2001). Expression of the T3SS-1 core regulatory system is in turn regulated by accessory factors that are presumed to respond to environmental signals (Golubeva et al., 2012), but how these signals are incorporated into the SPI-1 regulatory pathway has not been fully elucidated.
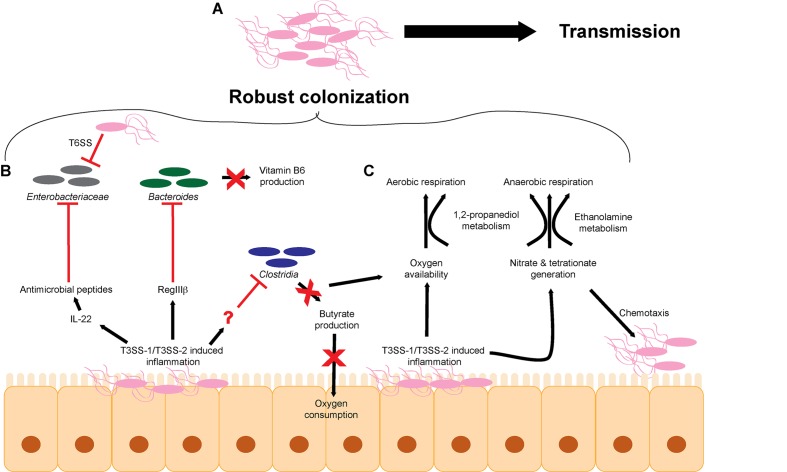
FIGURE 1.
S. Tm overcomes microbiota-mediated colonization resistance in the intestinal tract. (A) The cumulative effects of T3SS-1/T3SS-2-induced inflammation lead to robust colonization of the intestinal tract and efficient transmission to a new host. (B) S. Tm utilizes the T3SS-1 and T3SS-2 to generate a robust host immune response within the intestinal tract to eliminate members of the microbiota. (C) T3SS-1/T3SS-2-induced inflammation leads to the production of metabolites such as electron acceptors including oxygen, nitrate, and tetrathionate that promote aerobic and anaerobic respiration. T3SS, type III secretion system; T6SS, type VI secretion system.
Besides causing unspecific disruption to the microbiota as a whole, S. Tm-induced inflammation impacts particular intestinal microbes that alter concentrations of metabolites and/or host responses that would otherwise limit S. Tm infection (Figure 1B). For example, S. Tm infection induces RegIIIβ expression, and RegIIIβ is directly bactericidal against Bacteroides sp. and Eubacterium rectale (Miki et al., 2017). RegIII lectin family proteins are expressed in the intestinal tract and are important for maintaining intestinal homeostasis and combating pathogens (Cash et al., 2006; Vaishnava et al., 2008; Miki et al., 2012). Suppression of Bacteroides sp. is associated with changes in metabolite availability in the gut, most notably a decrease in vitamin B6 concentrations (Miki et al., 2017). Significantly, reconstitution of Bacteroides or supplementation of vitamin B6 contributes to the resolution of S. Tm infection, further underscoring the complex interplay of host, bacteria, and metabolites (Miki et al., 2017). S. Tm T3SS-1 and T3SS-2-induced inflammation also leads to depletion of Clostridia sp. in the intestine, which enhances S. Tm colonization (Rivera-Chávez et al., 2016b) (detailed in the next section). The host factor that directly depletes Clostridia sp. has not been identified.
Additionally, commensal Escherichia coli represents a minor component of the microbiota during homeostasis, but during general dysbiosis, the proportion of E. coli within the bacterial community increases (Winter et al., 2013). E. coli and S. Tm compete for metabolites and deploy molecules to limit growth of each other (Raffatellu et al., 2009; Deriu et al., 2013; Sassone-Corsi et al., 2016). For example, iron is an essential nutrient for most microbes and is typically limited within the host; therefore, microbes have evolved mechanisms to scavenge iron (Behnsen and Raffatellu, 2016). E. coli and S. Tm produce and secrete siderophores, which are small molecules that chelate iron (Crouch et al., 2008; Behnsen and Raffatellu, 2016). E. coli siderophores can be conjugated to microcins, small antibacterial peptides that kill bacterial cells through an unknown mechanism (Rebuffat, 2012; Sassone-Corsi et al., 2016). To fight back, the S. Tm T3SS-1 and T3SS-2 promote expression of the chemokine IL-22, which results in the production of antimicrobials that kill E. coli but are ineffective against S. Tm (Godinez et al., 2008; Raffatellu et al., 2009; Stelter et al., 2011; Liu et al., 2012; Behnsen et al., 2014). By inducing IL-22, S. Tm eliminates Enterobacteriaceae species and thus direct nutritional competitors.
S. Tm also takes direct action against competitors using a type VI secretion system (T6SS). T6SSs are commonly found in the Proteobacteria and Bacteroidetes phyla and are structurally homologous to bacteriophage tail complexes (Bönemann et al., 2009; Russell et al., 2014; Hood et al., 2017). These dynamic structures contract to directly inject effector proteins into target cells (Basler et al., 2012). Although T6SS can inject effector proteins into host cells (Ma and Mekalanos, 2010; Schwarz et al., 2010), increasing evidence suggest that T6SSs primarily target and cause subsequent death of bacterial cells (Mougous et al., 2006; Pukatzki et al., 2006; Bingle et al., 2008; Cianfanelli et al., 2016). The T6SS encoded by S. Tm has selective bactericidal efficacy against commensal organisms in vitro, including other members of the Enterobacteriaceae, such as E. coli W3110, Klebsiella oxytoca, and Klebsiella variicola (Brunet et al., 2015; Sana et al., 2016). However, Enterobacter cloacae and E. coli JB2 are resistant to S. Tm T6SS attack (Sana et al., 2016). It is currently unclear why different species are susceptible or resistant. The efficacy of the S. Tm T6SS against K. oxytoca data were validated in vivo during intestinal co-infection studies (Sana et al., 2016). These data support an important role for the S. Tm T6SS against other members of the microbiota; however, the benefits of selective killing remain to be defined. A better understanding of the commensal organisms that are directly targeted by S. Tm via the T6SS in vivo may reveal essential metabolites that are being differentially regulated that may impact S. Tm growth or virulence gene expression.
Collectively, these findings reveal that a central strategy S. Tm uses to colonize the host is to actively displace members of the microbiota. S. Tm achieves this through manipulation and exploitation of host responses (Stecher et al., 2007; Mooney et al., 2015). Moreover, S. Tm is capable of directly eliminating particular bacterial species. This transforms the intestinal tract into an environment that S. Tm is optimized to survive in and ultimately lowers the barrier of colonization resistance.
Metabolism in the Face of Inflammation
Dysbiosis not only results in changes in the bacterial populations but also changes the chemistry of the intestine (Zeng et al., 2017). Hence, a second aspect of S. Tm infection includes exploiting nutrients generated specifically during infection. The majority of bacteria that comprise the microbiota are obligate anaerobes that rely on fermentation for growth (Gibson and Roberfroid, 1995). Enterobacteriaceae, including S. Tm, are able to gain energy by respiration. Respiration generates higher amounts of ATP compared to fermentation and thereby enables S. Tm to outgrow many members of the microbiota (Wiles et al., 2006; Brochier-Armanet et al., 2009; Marteyn et al., 2010; Maier et al., 2013; Ng et al., 2013; Winter et al., 2013; Zeng et al., 2017) (Figure 1C). During homeostasis, host colonocytes consume oxygen, yielding a localized environment characterized by low oxygen partial pressure (Carreau et al., 2011; Espey, 2013). Colonocytes preferentially oxidize short-chain fatty acids, such as butyrate, to respire oxygen (Hamer et al., 2008; Donohoe et al., 2012; Kelly et al., 2015). Because Clostridia are the major producers of butyrate in the intestine, depletion of Clostridia with antibiotics or during S. Tm infection leads to a concomitant decrease in oxygen consumption by colonocytes (Sekirov et al., 2008; Louis and Flint, 2009; Gill et al., 2012; Vital et al., 2014; Rivera-Chávez et al., 2016b). S. Tm capitalizes on newly available oxygen to rapidly grow within the intestine (Rivera-Chávez et al., 2016b). These findings reveal a complex role for oxygen during infection. On the one hand, low levels of oxygen enhance SPI-1 expression, leading to increased inflammation and subsequent restriction of commensal organisms such as Clostridia. On the other hand, newly available oxygen promotes growth within the intestine. Oxygen limitation might therefore allow S. Tm to regulate the kinetics of SPI-1 expression within the intestine. In this proposed model, oxygen limitation early on during infection would promote SPI-1 expression, which would then in turn deplete commensal Clostridia via the host immune response, which would then result in an increase in oxygen availability leading to S. Tm outgrowth and subsequent reduction in SPI-1 expression. Additionally, the decrease in butyrate availability may also restrict the abundance of butyrate metabolizing members of the microbiota leading to even greater levels of dysbiosis.
The electron acceptors nitrate and tetrathionate also support S. Tm growth during infection (Winter et al., 2010; Lopez et al., 2012, 2015). The T3SS-1-secreted effector SopE is a bacteriophage-encoded activator of host Rho GTPases that results in host cytoskeletal rearrangements and activation of immune signaling pathways (Hardt et al., 1998). Additionally, SopE induces expression of host inducible nitric oxide synthase (iNOS) and generates nitrate (Lopez et al., 2012). Tetrathionate generation is a two-step process. Microbiota-produced hydrogen sulfide is converted by host colonocytes to thiosulfate, which reacts with oxygen radicals produced by host NADPH oxidase to generate tetrathionate (Winter et al., 2010). Tetrathionate reduction is coupled to ethanolamine, 1,2-propanediol, or fructose-asparagine oxidation (Price-Carter et al., 2001; Thiennimitr et al., 2011; Ali et al., 2014; Sabag-Daigle et al., 2016; Faber et al., 2017). Ethanolamine is a component of phosphatidylethanolamine, one of the most abundant phospholipids in host and microbial membranes (Randle et al., 1969; Dawaliby et al., 2015). The turnover of enterocytes and microbial cells as well as the diet provide a continuously replenished source of ethanolamine in the intestine (Cotton, 1972; Kawai et al., 1974; Dowhan, 1997, 2003; Snoeck et al., 2005; Bakovic et al., 2007). The accumulation of 1,2-propanediol depends primarily on the microbiota. It is thought that the ability of Bacteroides sp. to breakdown complex carbohydrates allows for the production of 1,2-propanediol as a byproduct of fermentation of methyl-pentoses (Faber et al., 2017), although experiments are needed to demonstrate this during the course of infection. This model is supported by the findings that 1,2-propanediol is nearly undetectable in germ-free mice; however, the presence of either Bacteroides fragilis or Bacteroides thetaiotaomicron is associated with 1,2-propanediol accumulation (Faber et al., 2017). Additionally, expression of S. Tm genes coding for 1,2-propanediol metabolism are induced in the presence of B. thetaiotaomicron in vivo (Ng et al., 2013; Faber et al., 2017). Furthermore, the presence of B. thetaiotaomicron within the intestinal tract enhances S. Tm expression of additional carbohydrate metabolism and transport genes, including sialic acid and fucose catabolic pathways (Ng et al., 2013). The B. thetaiotaomicron-encoded sialidase liberates sialic acid, which promotes S. Tm sialic acid metabolism and intestinal growth (Ng et al., 2013). These data indicate a somewhat paradoxical relationship between S. Tm and Bacteroides sp. As discussed above, S. Tm-induced inflammation restricts the levels of Bacteroides sp. (Miki et al., 2017) and yet Bacteroides sp. are critical for the accumulation of multiple metabolites that S. Tm utilizes during infection (Ng et al., 2013; Faber et al., 2017). These seemingly conflicting ideas suggest an even more complex relationship between pathogen and microbiota. Rather than presence or absence of commensals, S. Tm may require a fine-tuned abundance of microbes to generate beneficial metabolites without these organisms directly competing with S. Tm.
S. Tm also exploits electron acceptors as spatiotemporal cues for colonization and tissue invasion. Nitrate and tetrathionate are indirect signals for S. Tm chemotaxis and thereby influence subsequent stages in S. Tm infection (Rivera-Chavez et al., 2013). The chemotaxis proteins Aer and Tsr sense changes in redox and proton motive force during tetrathionate and nitrate respiration, respectively (Edwards et al., 2006; Rivera-Chávez et al., 2016a). Both Aer and Tsr promote T3SS-1/T3SS-2-dependent intestinal colonization (Rivera-Chavez et al., 2013). Tsr-dependent chemotaxis correlates with localized host production of iNOS and enables S. Tm to invade ileal Peyer’s patches (Rivera-Chávez et al., 2016a). Surprisingly, no additive effect is seen with Aer and Tsr-dependent chemotaxis (Rivera-Chavez et al., 2013). This suggests that these two sensing pathways may be interconnected and perhaps functionally redundant. Connectivity of these two pathways is further supported by the observation that the presence of nitrate reduces the expression of genes involved in tetrathionate respiration as well as the growth advantage conferred by tetrathionate respiration (Lopez et al., 2012). It is currently unclear how S. Tm balances the sensing and utilization of two signals that may restrict one another.
Much of our understanding of the host–microbiota–pathogen interplay has been generated through genetic manipulation of the pathogen, and alternative experimental approaches are shedding new light on this interaction. For example, defined commensal communities have been used to reconstitute the microbiota of germ-free mice and study the contributions of specific members of the microbiota necessary for effective colonization resistance (Dewhirst et al., 1999; Stecher et al., 2010; Brugiroux et al., 2016). Studies using defined microbial communities will also contribute to understanding how differences in microbial species between susceptible and resistant intestinal environments impact S. Tm virulence gene expression. Additionally, a recent RNA-seq study assessed gene expression in S. Tm grown under 22 different in vitro conditions that mimicked aspects of infection, which could reveal new signals important for controlling the expression of key metabolic and virulence determinants (Kröger et al., 2013). Future in vivo metabolomics studies may identify specific microbiota or diet-dependent molecules that impact S. Tm virulence gene expression and/or host response. In line with this, a high salt diet was recently identified as significantly altering the host response during S. Tm infection (Tubbs et al., 2017). Thus, the ability to manipulate commensal microbial communities and intestinal metabolite concentrations offers an exciting tool to further understand how microbiota and metabolites not only impact S. Tm growth but also impact S. Tm virulence.
Collectively, these findings highlight that S. Tm thrives during host dysbiosis, benefits from the host immune response, and utilizes virulence factors to directly and indirectly suppress members of the microbiota. However, S. Tm relies on members of the indigenous microbial community to expand its metabolic capabilities during infection that in turn promote outgrowth and transmission (Maier et al., 2013). Although some host factors and cytokines that are involved in the development of inflammation during infection are known (Behnsen et al., 2015), additional components are likely to contribute to S. Tm infection. A more comprehensive understanding of immune responses that contribute to dysbiosis and/or generate a nutritional niche for S. Tm is necessary to fully understand S. Tm infection strategies, and indeed, this remains an active area of research. Host inflammatory components could be enhancing intestinal pathology, restricting microbiota reconstitution, molding a new nutrient niche, or enhancing all aspects of infection.
Intramacrophage Adaptation
After benefiting from components of the host immune response during intestinal colonization, S. Tm must withstand the bactericidal efforts of host phagocytes during systemic infection. S. Tm dissemination from the intestinal tract to systemic sites of infection largely depends on phagocytic cells (Vazquez-Torres et al., 1999). Of the host phagocytes, macrophages frequently interact with S. Tm during dissemination and within systemic sites, including the spleen and liver (Salcedo et al., 2001). Macrophages utilize several strategies such as acidification of phagosomes, generation of reactive oxygen and nitrogen species, and production of antimicrobial proteins and peptides to kill internalized pathogens (Flannagan et al., 2009); however, S. Tm is able to withstand these defense mechanisms through multiple molecular mechanisms. The T3SS-2 and associated secreted effectors create a replicative niche within macrophages termed the Salmonella containing vacuole (SCV) by modulating diverse host processes (Hensel et al., 1995; Ochman et al., 1996; Shea et al., 1996; Figueira and Holden, 2012). For example, these effectors inhibit SCV–lysosome fusion, modify host vesicle trafficking, localize the SCV within the cell, and evade the host autophagic response (Uchiya et al., 1999; Shotland et al., 2003; Guignot et al., 2004; Boucrot et al., 2005; Brumell and Scidmore, 2007; Jackson et al., 2008; Owen et al., 2014, 2016). Multiple environmental signals and bacterial regulators influence this critical adaptation step (discussed below).
Counteracting Host Defense Mechanisms
Pathogen recognition is the first line of defense for phagocytic cells such as macrophages. Host Toll-like receptors (TLRs) are transmembrane proteins located on the plasma membrane and endosomal membranes that recognize conserved molecular patterns associated with pathogens such as lipids, proteins, and nucleic acids (Kawai and Akira, 2010). TLRs have profound impacts on adaptive immunity and are thus broadly essential for host defense (Iwasaki and Medzhitov, 2004). Additionally, TLRs are critical for control of S. Tm replication as well as host survival during infection (Weiss et al., 2004). TLR activation and downstream signaling through the adapter proteins MyD88 and TRIF is linked to initial SCV acidification (Arpaia et al., 2011). The SCV acidifies rapidly following phagocytosis, dropping to a pH between 4.0 and 5.0 within 60 min of formation (Rathman et al., 1996). Acidification of the SCV is essential for S. Tm expression and functional formation of the T3SS-2 as well as secretion of effectors (Rathman et al., 1996; Beuzón et al., 1999; Hansen-Wester et al., 2002; Rappl et al., 2003; Arpaia et al., 2011; Chakraborty et al., 2015). Thus, S. Tm co-opts part of the macrophage defense system to serve as an initiating signal that promotes bacterial survival.
In the host, concentrations of free iron are extremely low in part to limit growth of invading pathogens (Kühn, 2015; Behnsen and Raffatellu, 2016). For example, the host expresses iron regulatory proteins (IRPs) and lipocalin-2 to limit the amount of iron available within macrophages (Kühn, 2015; Nairz et al., 2015a,b). Host-mediated iron limitation significantly restricts S. Tm replication within macrophages and reduces lethal disease (Nairz et al., 2015a). Additionally, IRP and lipocalin-2 influence the immune response during S. Tm infection (Nairz et al., 2015a,b). Indeed, modulating the immune response, specifically interleukin 10, can rescue the bactericidal defects of lipocalin-2 deficient cells and mice (Nairz et al., 2015b). These results suggest that host iron concentrations indirectly affect S. Tm survival through alteration of the host immune response rather than by directly starving S. Tm of iron. Nonetheless, modulation of iron concentrations does influence S. Tm expression of virulence genes. For example, SPI-2 expression is reduced when S. Tm is grown in the presence of iron (Choi and Groisman, 2013; Choi et al., 2014), and the iron-sensing transcription factors Fur and PmrA limit SPI-2 expression during macrophage infection (Wösten et al., 2000; Choi and Groisman, 2013; Choi et al., 2014). However, SPI-2 expression has also been shown to be reduced when iron is chelated from cultures and during infection of macrophages that have low iron concentrations (Zaharik et al., 2002; Nairz et al., 2009). These contrasting findings require further investigation. Similarly, it is unclear what the iron availability is within the SCV with and without host iron acquisition mediators IRP and lipocalin-2.
Altogether, these findings highlight that S. Tm senses and responds to macrophage defense mechanisms, which impact S. Tm virulence and survival. By utilizing host defenses as signals, S. Tm incorporates antimicrobial processes into a bacterial signal transduction pathway that creates a suitable replication niche in an otherwise inhospitable environment.
Intramacrophage Sensing and Signaling
In addition to host defense-linked signals, S. Tm responds to concentrations of cations, nutrients, and ATP to activate expression of SPI-2 and other virulence factors to ensure survival (Valdivia and Falkow, 1997; Cirillo et al., 1998; Deiwick et al., 1999; Kim and Falkow, 2004; Löber et al., 2006; Osborne and Coombes, 2011; Lee and Groisman, 2012; Blair et al., 2013) (Figure 2). To sense these environmental signals and regulate SPI-2 expression, S. Tm uses several two-component systems (TCS) (Fass and Groisman, 2009). TCS are typically comprised of a sensor kinase that autophosphorylates upon sensing of a stimulus and then phosphorylates its paired response regulator, which in turn binds DNA to activate transcription of target genes (Stock et al., 2000). The PhoPQ TCS senses acidic pH, divalent cations, antimicrobial peptides, and potentially other signals to activate SPI-2 expression and modify components of the bacterial outer membrane (Bader et al., 2005; Prost et al., 2007; Dalebroux et al., 2014; Hicks et al., 2015). In addition to SPI-2, PhoP regulates other virulence factors, including the mgtCBR operon. The mgtCBR operon encodes an inner membrane protein, a Mg2+ transporter, and a regulator and is critical for intramacrophage survival (Soncini et al., 1996; Blanc-Potard and Groisman, 1997; Alix and Blanc-Potard, 2007; Lee and Groisman, 2010). It is still unclear what host or bacterial factors contribute to the presence of these SPI-2 activating signals within the SCV.
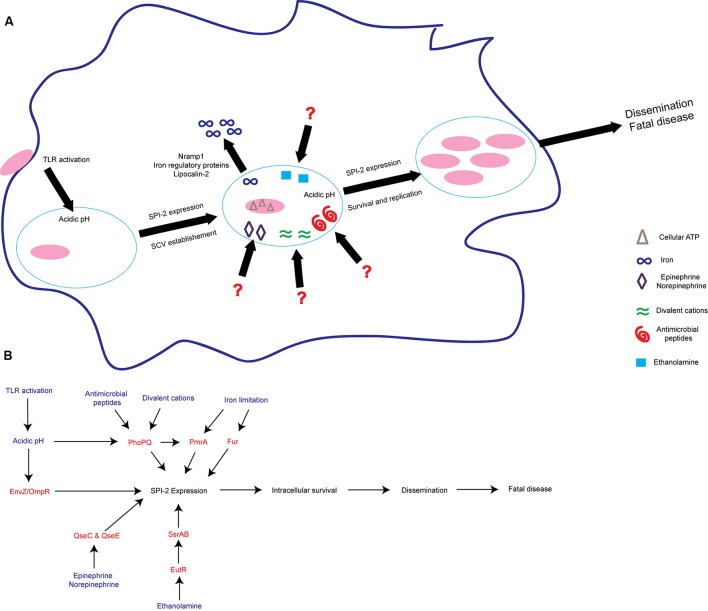
FIGURE 2.
S. Tm senses environmental cues to adapt to the intramacrophage environment. (A) S. Tm recognizes diverse signals within the macrophage. These include iron limitation, ethanolamine, acidic pH, antimicrobial peptides, divalent cations, epinephrine/norepinephrine, and cellular ATP. These signals act to induce SPI-2 expression and establish the SCV as a replicative niche. (B) Schematic of some of the regulatory proteins and associated signals important for activating SPI-2 expression. Signals are written blue; regulatory proteins and TCS are written in red. TLRs, Toll-like receptors; SCV, Salmonella containing vacuole.
S. Tm also responds to signals that are not specific to the SCV, but rather are found in multiple environments throughout the host. For example, the host hormones epinephrine and norepinephrine (epi/NE) are ubiquitous throughout the body (Boyanova, 2017). The bacterial TCS QseBC and QseEF sense and respond to epi/NE during infection (Clarke et al., 2006; Reading et al., 2009). The sensor kinase QseC regulates the expression of genes encoded within SPI-1 and enhances epithelial cell invasion under conditions that promote SPI-1 expression (Moreira et al., 2010). Additionally, during macrophage infection, QseC activates SPI-2 expression to enhance intramacrophage survival (Moreira et al., 2010). Moreover, both epi/NE-responsive histidine sensor kinases, QseC and QseE, are required for systemic infection (Rasko et al., 2008; Moreira et al., 2010; Moreira and Sperandio, 2012). NE induces expression of both SPI-1 and SPI-2 associated genes, however, only expression of SPI-2 associated genes is QseC dependent (Moreira et al., 2010). These findings reveal that the same signal (epi or NE) enhances S. Tm virulence gene expression depending on the surrounding environment. The additional components of the intramacrophage environment that allow these epi/NE-dependent signaling pathways to distinguish between a SPI-1 or SPI-2 inducing condition warrant further studies.
Ethanolamine is another signal that plays environment-dependent roles in expression of S. Tm virulence traits (Anderson and Kendall, 2016). Ethanolamine is present in serum and is maintained intracellularly by host cells in part to recycle and produce phosphatidylethanolamine (Nikawa et al., 1986; Lipton et al., 1988, 1990; Sandra and Cai, 1991; Shiao and Vance, 1995). In the Enterobacteriaceae, including S. Tm, the transcription factor EutR directly senses ethanolamine (Roof and Roth, 1992; Luzader et al., 2013). EutR-dependent signaling promotes ethanolamine metabolism during intestinal infection (Anderson et al., 2015). As infection progresses, ethanolamine promotes S. Tm dissemination to systemic sites independently of metabolism (Anderson et al., 2015). Although ethanolamine metabolism does not provide a growth benefit for S. Tm during systemic infection (Stojiljkovic et al., 1995; Thiennimitr et al., 2011; Steeb et al., 2013; Anderson et al., 2015), EutR directly activates expression of SPI-2 within macrophages leading to increased survival and early dissemination (Anderson et al., 2015). It is currently unclear how ethanolamine can signal through the same receptor, EutR, to promote niche adaptation in distinct host environments. However, it is clear that ethanolamine plays a dual role in S. Tm infection by supporting growth in the inflamed intestine as well as enhancing subsequent stages of S. Tm disease.
The production of Salmonella-induced filaments (SIFs) is a critical component of S. Tm adaptation to intracellular environments (Garcia-del Portillo et al., 1993; Stein et al., 1996). This SIF network remodels the host endosomal network, allowing S. Tm to gain access to endocytosed molecules (Ohlson et al., 2008; Liss et al., 2017). The SCV/SIF continuum is required for efficient intracellular metabolism and promotes S. Tm replication (Liss et al., 2017). While the focus of this study was on access to metabolites (Liss et al., 2017), the same principle of incorporating extracellular molecules into the SCV may be true for signaling molecules that promote virulence. In such a model, signaling molecules present within host endosomes, or recently endocytosed molecules from the extracellular environment, would be shuttled through the SCV/SIF continuum and potentially impact S. Tm virulence gene expression. This revelation opens the possibility that large sets of molecules within various host tissues are able to reach the SCV within macrophages in vivo. Additionally, genes that have previously been identified as not being induced during macrophage infection in vitro may be the result of a relevant signal being absent from the culture conditions. Determining in vivo signals that S. Tm recognizes remains a daunting challenge but is necessary for a thorough understanding of the events that trigger intracellular adaptation.
The majority of work on environmental signals and sensing within macrophages has focused on transcriptional regulation; however, recent proteomic and RNA-seq approaches have shown that regulation of gene expression is more complex. Post-translational modifications as well as small regulatory RNA (sRNA)-induced post-transcriptional changes regulate virulence during macrophage infection (Ansong et al., 2013; Westermann et al., 2016). sRNAs are non-coding 50–500 nucleotide transcripts that utilize base-pair interactions to post-transcriptionally regulate the expression of target mRNAs (Hébrard et al., 2012). S. Tm encodes approximately 300 unique sRNAs, and the expression of a subset of sRNAs is highly sensitive to signals encountered within the macrophage environment, such as nutrient starvation, and are controlled by key components of the SPI-2 regulatory system (Kröger et al., 2012, 2013; Amin et al., 2016; Colgan et al., 2016). Specifically, the sRNA PinT regulates SPI-2 expression and is critical for host adaptation in vivo (Chaudhuri et al., 2013; Westermann et al., 2016). It is possible that post-transcriptional regulation helps S. Tm incorporate environmental signals sensed within the SCV to modulate survival.
In addition to active intracellular replication, entering a non-replicating yet viable state is an alternative adaptation strategy. A portion of the infecting S. Tm population enters a persistent state within macrophages that is independent of SPI-2 (Helaine et al., 2010). These non-replicating S. Tm remain viable and are not killed by macrophage antimicrobial defenses (Helaine et al., 2010). This phenomenon has also been demonstrated within non-phagocytic cells in vivo (Núñez-Hernández et al., 2013). It is unclear how these persister cells impact disease progression in vivo, but perhaps persister cells promote asymptomatic carriage and transmission. Future studies are required to determine if there is transcriptional overlap between these persister cells and actively replicating S. Tm or if perhaps post-transcriptional regulation contributes to this phenotypic switch. Advancements in single cell expression techniques will allow replicating and dormant cells to be distinguished from one another. Similarly, it remains unclear what signals present within the SCV trigger the shift from replication to persistence. Although distinct signals may be responsible for transitioning to a persistent state, it is also possible that loss of the signals important for replication leads to dormancy.
Altogether, these studies reveal complex and dynamic regulatory circuits important for S. Tm survival within macrophages. S. Tm must appropriately repress and activate virulence gene expression to ensure adaptation to the SCV (Kato et al., 2003; Choi and Groisman, 2013; Westermann et al., 2016). Additionally, S. Tm must be able to initiate distinct regulatory pathways at different time points during infection (Moreira et al., 2010; Moreira and Sperandio, 2012; Anderson et al., 2015). Further understanding of these activating and suppressing cues, and how they are balanced with one another, will enhance our understanding of S. Tm pathogenesis.
Host Cell Death
The replication-suitable SCV within macrophages is a temporary niche, as host cells die. The way in which cells die has a tremendous impact on host physiology and inflammation (Pasparakis and Vandenabeele, 2015; Medina and Ravichandran, 2016). Programmed host cell death can be classified based on the effector proteins required and the state of inflammation each type of death induces (Kroemer et al., 2009). Three of the major programmed cell death pathways are apoptosis, necroptosis, and pyroptosis (Kroemer et al., 2009). S. Tm infection induces apoptosis-like features in infected macrophages including chromatin condensation and caspase-3 and caspase-8 activity (Chen et al., 1996; Lindgren et al., 1996; Monack et al., 1996; van der Velden et al., 2000; Man et al., 2013; Günster et al., 2017). Additionally, S. Tm infection can induce RIPK3 and MLKL-dependent necroptosis (Robinson et al., 2012; Günster et al., 2017). The best-characterized form of S. Tm-induced cell death is caspase-1/caspase-11-dependent pyroptosis (Brennan and Cookson, 2000; Monack et al., 2001; Fink and Cookson, 2006; Fink et al., 2008). Host cells employ several means to recognize S. Tm virulence proteins and initiate regulated pyroptosis. For example, macrophage NLRC4 (Ipaf) recognizes S. Tm flagellin and components of T3SS-1 to activate caspase-1 (Franchi et al., 2006; Miao et al., 2006, 2010b; Zhao et al., 2011). Additionally, macrophage NAIP1 also recognizes components of T3SS-1 to initiate a pyroptotic death (Rayamajhi et al., 2013; Yang et al., 2013). Caspase-1 and caspase-11, as well as their activators NLRP3 and NLRC4, impact S. Tm pathogenesis and bacterial burden in vivo (Lara-Tejero et al., 2006; Raupach et al., 2006; Broz et al., 2010, 2012). Interestingly, the host is able to enhance clearance of S. Tm infection when pyroptosis is experimentally induced in vivo (Miao et al., 2010a; Stewart et al., 2011; Aachoui et al., 2013; Jorgensen et al., 2016). These findings suggest that S. Tm might try to evade inducing host cell death during some stages of infection. Although most commonly studied in macrophages, S. Tm-induced cell death also occurs in dendritic and epithelial cells (van der Velden et al., 2003; Sellin et al., 2014; Rauch et al., 2017). It remains uncertain if infected cells are simply overwhelmed by S. Tm over time in vitro, if S. Tm actively regulates virulence to promote or evade host cell death in vivo, if cell death is occurring throughout infection, and what the consequences of host cell death are during systemic disease.
Conclusion
S. Tm is able to recognize, adapt to, and survive within the intestinal tract and the intramacrophage environment during infection. These two environments present very different obstacles to infection, and S. Tm utilizes diverse strategies to overcome these distinct barriers. A better understanding of the signaling molecules, the signal transduction pathways, and the cross talk between signal transduction pathways that promote niche recognition may provide novel opportunities for therapeutic intervention. Importantly, several of the strategies used by S. Tm to adapt to host environments are conserved among several organisms such as pathogenic E. coli, Listeria monocytogenes, Brucella abortus, and Staphylococcus aureus. For example, pathogens adapt to the host or regulate virulence by utilizing and/or sensing nitrate (Spees et al., 2013; Winter et al., 2013), epi/NE (Curtis et al., 2014; Halang et al., 2015; Moreira et al., 2016; Rooks et al., 2017), ethanolamine (Maadani et al., 2007; Kendall et al., 2012; DebRoy et al., 2014; Gonyar and Kendall, 2014; Mellin et al., 2014; Subashchandrabose et al., 2014), iron (Almirón et al., 2001; Zimbler et al., 2012; Hammer et al., 2013, 2014; Mashruwala et al., 2015; Palmer and Skaar, 2016), and pH (Heinzen et al., 1996; Sturgill-Koszycki and Swanson, 2000; Singh et al., 2008; Vandal et al., 2008). Additionally, T6SS are widely conserved, particularly within Proteobacteria and Bacteroidetes (Hood et al., 2017), and promote colonization of intestinal pathogens (Fu et al., 2013; Sana et al., 2017). Perhaps the fact that S. Tm utilizes all of these strategies, rather than a select few, makes S. Tm so successful and such a major burden on global healthcare (Scallan et al., 2011). Therefore, further study of these environmental adaptation strategies, using S. Tm as a model organism, will enhance our understanding of the host–microbiota–pathogen interface.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. Work in the MMK lab is supported by the National Institutes of Health (NIH) grant AI118732. CA was supported through NIH training grant 5T32AI007046 and University of Virginia School of Medicine Wagner Fellowship.
References
- Aachoui Y., Leaf I. A., Hagar J. A., Fontana M. F., Campos C. G., Zak D. E., et al. (2013). Caspase-11 protects against bacteria that escape the vacuole. Science 339 975–978. 10.1126/science.1230751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acheson D., Hohmann E. L. (2001). Nontyphoidal salmonellosis. Clin. Infect. Dis. 32 263–269. 10.1086/318457 [DOI] [PubMed] [Google Scholar]
- Ali M. M., Newsom D. L., Gonzalez J. F., Sabag-Daigle A., Stahl C., Steidley B., et al. (2014). Fructose-asparagine is a primary nutrient during growth of Salmonella in the inflamed intestine. PLOS Pathog. 10:e1004209. 10.1371/journal.ppat.1004209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alix E., Blanc-Potard A.-B. (2007). MgtC: a key player in intramacrophage survival. Trends Microbiol. 15 252–256. 10.1016/j.tim.2007.03.007 [DOI] [PubMed] [Google Scholar]
- Almirón M., Martínez M., Sanjuan N., Ugalde R. A. (2001). Ferrochelatase is present in Brucella abortus and is critical for its intracellular survival and virulence. Infect. Immun. 69 6225–6230. 10.1128/IAI.69.10.6225-6230.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin S. V., Roberts J. T., Patterson D. G., Coley A. B., Allred J. A., Denner J. M., et al. (2016). Novel small RNA (sRNA) landscape of the starvation-stress response transcriptome of Salmonella enterica serovar typhimurium. RNA Biol. 13 331–342. 10.1080/15476286.2016.1144010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson C. J., Kendall M. M. (2016). Location, location, location. Salmonella senses ethanolamine to gauge distinct host environments and coordinate gene expression. Microb. Cell 3 89–91. 10.15698/mic2016.02.479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson C. J., Clark D. E., Adli M., Kendall M. M. (2015). Ethanolamine signaling promotes Salmonella niche recognition and adaptation during infection. PLOS Pathog. 11:e1005278. 10.1371/journal.ppat.1005278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari S., Sherchand J. B., Parajuli K., Mishra S. K., Dahal R. K., Shrestha S., et al. (2012). Bacterial etiology of acute diarrhea in children under five years of age. J. Nepal Health Res. Counc. 10 218–223. [PubMed] [Google Scholar]
- Ansong C., Wu S., Meng D., Liu X., Brewer H. M., Kaiser B. L. D., et al. (2013). Top-down proteomics reveals a unique protein S-thiolation switch in Salmonella Typhimurium in response to infection-like conditions. Proc. Natl. Acad. Sci. U.S.A. 110 10153–10158. 10.1073/pnas.1221210110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arpaia N., Godec J., Lau L., Sivick K. E., McLaughlin L. M., Jones M. B., et al. (2011). TLR signaling is required for Salmonella typhimurium virulence. Cell 144 675–688. 10.1016/j.cell.2011.01.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader M. W., Sanowar S., Daley M. E., Schneider A. R., Cho U., Xu W., et al. (2005). Recognition of antimicrobial peptides by a bacterial sensor kinase. Cell 122 461–472. 10.1016/j.cell.2005.05.030 [DOI] [PubMed] [Google Scholar]
- Bajaj V., Hwang C., Lee C. A. (1995). hilA is a novel ompR/toxR family member that activates the expression of Salmonella typhimurium invasion genes. Mol. Microbiol. 18 715–727. 10.1111/j.1365-2958.1995.mmi_18040715.x [DOI] [PubMed] [Google Scholar]
- Bajaj V., Lucas R. L., Hwang C., Lee C. A. (1996). Co-ordinate regulation of Salmonella typhimurium invasion genes by environmental and regulatory factors is mediated by control of HilA expression. Mol. Microbiol. 22 703–714. 10.1046/j.1365-2958.1996.d01-1718.x [DOI] [PubMed] [Google Scholar]
- Bakovic M., Fullerton M. D., Michel V. (2007). Metabolic and molecular aspects of ethanolamine phospholipid biosynthesis: the role of ctp:phosphoethanolamine cytidylyltransferase (pcyt2). Biochem. Cell Biol. 85 283–300. 10.1139/O07-006 [DOI] [PubMed] [Google Scholar]
- Barman M., Unold D., Shifley K., Amir E., Hung K., Bos N., et al. (2008). Enteric salmonellosis disrupts the microbial ecology of the murine gastrointestinal tract. Infect. Immun. 76 907–915. 10.1128/IAI.01432-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthel M., Hapfelmeier S., Quintanilla-Martinez L., Kremer M., Rohde M., Hogardt M., et al. (2003). Pretreatment of mice with streptomycin provides a Salmonella enterica serovar typhimurium colitis model that allows analysis of both pathogen and host. Infect. Immun. 71 2839–2858. 10.1128/IAI.71.5.2839-2858.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basler M., Pilhofer M., Henderson P. G., Jensen J. G., Mekalanos J. (2012). Type VI secretion requires a dynamic contractile phage tail-like structure. Nature 483 182–186. 10.1038/nature10846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäumler A. J., Sperandio V. (2016). Interactions between the microbiota and pathogenic bacteria in the gut. Nature 535 85–93. 10.1038/nature18849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnsen J., Jellbauer S., Wong C. P., Edwards R. A., George M. D., Ouyang W., et al. (2014). The cytokine Il-22 promotes pathogen colonization by suppressing related commensal bacteria. Immunity 40 262–273. 10.1016/j.immuni.2014.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnsen J., Perez-Lopez A., Nuccio S.-P., Raffatellu M. (2015). Exploiting host immunity: the Salmonella paradigm. Trends Immunol. 36 112–120. 10.1016/j.it.2014.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnsen J., Raffatellu M. (2016). Siderophores: more than stealing iron. mBio 7:e01906–16. 10.1128/mBio.01906-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beuzón C. R., Banks G., Deiwick J., Hensel M., Holden D. W. (1999). pH-dependent secretion of SseB, a product of the Spi-2 type III secretion system of Salmonella typhimurium. Mol. Microbiol. 33 806–816. 10.1046/j.1365-2958.1999.01527.x [DOI] [PubMed] [Google Scholar]
- Bingle L. E., Bailey C. M., Pallen M. J. (2008). Type VI secretion: a beginner’s guide. Curr. Opin. Microbiol. 11 3–8. 10.1016/j.mib.2008.01.006 [DOI] [PubMed] [Google Scholar]
- Blair J. M. A., Richmond G. E., Bailey A. M., Ivens A., Piddock L. J. V. (2013). Choice of bacterial growth medium alters the transcriptome and phenotype of Salmonella enterica serovar typhimurium. PLOS ONE 8:e63912. 10.1371/journal.pone.0063912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc-Potard A. B., Groisman E. A. (1997). The Salmonella selC locus contains a pathogenicity island mediating intramacrophage survival. EMBO J. 16 5376–5385. 10.1093/emboj/16.17.5376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogomolnaya L. M., Andrews K. D., Talamantes M., Maple A., Ragoza Y., Vazquez-Torres A., et al. (2013). The ABC-type efflux pump MacAB protects Salmonella enterica serovar typhimurium from oxidative stress. mBio 4:e00630–13. 10.1128/mBio.00630-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bönemann G., Pietrosiuk A., Diemand A., Zentgraf H., Mogk A. (2009). Remodelling of VipA/VipB tubules by Clpv-mediated threading is crucial for type VI protein secretion. EMBO J. 28 315–325. 10.1038/emboj.2008.269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucrot E., Henry T., Borg J.-P., Gorvel J.-P., Méresse S. (2005). The intracellular fate of Salmonella depends on the recruitment of kinesin. Science 308 1174–1178. 10.1126/science.1110225 [DOI] [PubMed] [Google Scholar]
- Boyanova L. (2017). Stress hormone epinephrine (adrenaline) and Norepinephrine (noradrenaline) effects on the anaerobic bacteria. Anaerobe 44 13–19. 10.1016/j.anaerobe.2017.01.003 [DOI] [PubMed] [Google Scholar]
- Brennan M. A., Cookson B. T. (2000). Salmonella induces macrophage death by caspase-1-dependent necrosis. Mol. Microbiol. 38 31–40. 10.1046/j.1365-2958.2000.02103.x [DOI] [PubMed] [Google Scholar]
- Brochier-Armanet C., Talla E., Gribaldo S. (2009). The multiple evolutionary histories of dioxygen reductases: implications for the origin and evolution of aerobic respiration. Mol. Biol. Evol. 26 285–297. 10.1093/molbev/msn246 [DOI] [PubMed] [Google Scholar]
- Broz P., Newton K., Lamkanfi M., Mariathasan S., Dixit V. M., Monack D. M. (2010). Redundant roles for inflammasome receptors NLRP3 and NLRC4 in Host defense against Salmonella. J. Exp. Med. 207 1745–1755. 10.1084/jem.20100257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broz P., Ruby T., Belhocine K., Bouley D. M., Kayagaki N., Dixit V. M., et al. (2012). Caspase-11 increases susceptibility to Salmonella infection in the absence of caspase-1. Nature 490 288–291. 10.1038/nature11419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugiroux S., Beutler M., Pfann C., Garzetti D., Ruscheweyh H.-J., Ring D., et al. (2016). Genome-guided design of a defined mouse microbiota that confers colonization resistance against Salmonella enterica serovar typhimurium. Nat. Microbiol. 2:16215. 10.1038/nmicrobiol.2016.215 [DOI] [PubMed] [Google Scholar]
- Brumell J. H., Scidmore M. A. (2007). Manipulation of Rab GTPase function by intracellular bacterial pathogens. Microbiol. Mol. Biol. Rev. 71 636–652. 10.1128/MMBR.00023-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet Y. R., Khodr A., Logger L., Aussel L., Mignot T., Rimsky S., et al. (2015). H-NS silencing of the Salmonella pathogenicity island 6-encoded type VI secretion system limits Salmonella enterica serovar typhimurium interbacterial killing. Infect. Immun. 83 2738–2750. 10.1128/IAI.00198-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreau A., Hafny-Rahbi B. E., Matejuk A., Grillon C., Kieda C. (2011). Why is the partial oxygen pressure of human tissues a crucial parameter? Small molecules and hypoxia. J. Cell. Mol. Med. 15 1239–1253. 10.1111/j.1582-4934.2011.01258.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cash H. L., Whitham C. V., Behrendt C. L., Hooper L. V. (2006). Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science 313 1126–1130. 10.1126/science.1127119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty S., Mizusaki H., Kenney L. J. (2015). A FRET-based DNA biosensor tracks OmpR-dependent acidification of Salmonella during macrophage infection. PLOS Biol. 13:e1002116. 10.1371/journal.pbio.1002116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri R. R., Morgan E., Peters S. E., Pleasance S. J., Hudson D. L., Davies H. M., et al. (2013). Comprehensive assignment of roles for Salmonella typhimurium genes in intestinal colonization of food-producing animals. PLOS Genet. 9:e1003456. 10.1371/journal.pgen.1003456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L. M., Kaniga K., Galán J. E. (1996). Salmonella spp. are cytotoxic for cultured macrophages. Mol. Microbiol. 21 1101–1115. 10.1046/j.1365-2958.1996.471410.x [DOI] [PubMed] [Google Scholar]
- Choi E., Kim H., Lee H., Nam D., Choi J., Shin D. (2014). The iron-sensing fur regulator controls expression timing and levels of Salmonella pathogenicity island 2 genes in the course of environmental acidification. Infect. Immun. 82 2203–2210. 10.1128/IAI.01625-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J., Groisman E. A. (2013). The lipopolysaccharide modification regulator PmrA limits Salmonella virulence by repressing the type three-secretion system Spi/Ssa. Proc. Natl. Acad. Sci. U.S.A. 110 9499–9504. 10.1073/pnas.1303420110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cianfanelli F. R., Monlezun L., Coulthurst S. J. (2016). Aim, load, fire: the type VI secretion system, a bacterial nanoweapon. Trends Microbiol. 24 51–62. 10.1016/j.tim.2015.10.005 [DOI] [PubMed] [Google Scholar]
- Cirillo D. M., Valdivia R. H., Monack D. M., Falkow S. (1998). Macrophage-dependent induction of the Salmonella pathogenicity island 2 type III secretion system and its role in intracellular survival. Mol. Microbiol. 30 175–188. 10.1046/j.1365-2958.1998.01048.x [DOI] [PubMed] [Google Scholar]
- Clarke M. B., Hughes D. T., Zhu C., Boedeker E. C., Sperandio V. (2006). The QseC sensor kinase: a bacterial adrenergic receptor. Proc. Natl. Acad. Sci. U.S.A. 103 10420–10425. 10.1073/pnas.0604343103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coburn B., Li Y., Owen D., Vallance B. A., Finlay B. B. (2005). Salmonella enterica serovar typhimurium pathogenicity island 2 is necessary for complete virulence in a mouse model of infectious enterocolitis. Infect. Immun. 73 3219–3227. 10.1128/IAI.73.6.3219-3227.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colgan A. M., Kröger C., Diard M., Hardt W.-D., Puente J. L., Sivasankaran S. K., et al. (2016). The impact of 18 ancestral and horizontally-acquired regulatory proteins upon the transcriptome and sRNA landscape of Salmonella enterica serovar typhimurium. PLOS Genet. 12:e1006258. 10.1371/journal.pgen.1006258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotton P. B. (1972). Non-dietary lipid in the intestinal lumen. Gut 13 675–681. 10.1136/gut.13.9.675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crouch M.-L. V., Castor M., Karlinsey J. E., Kalhorn T., Fang F. C. (2008). Biosynthesis and IroC-dependent export of the siderophore salmochelin are essential for virulence of Salmonella enterica serovar typhimurium. Mol. Microbiol. 67 971–983. 10.1111/j.1365-2958.2007.06089.x [DOI] [PubMed] [Google Scholar]
- Curtis M. M., Russell R., Moreira C. G., Adebesin A. M., Wang C., Williams N. S., et al. (2014). QseC inhibitors as an antivirulence approach for gram-negative pathogens. mBio 5:e02165–14. 10.1128/mBio.02165-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalebroux Z. D., Matamouros S., Whittington D., Bishop R. E., Miller S. I. (2014). PhoPQ regulates acidic glycerophospholipid content of the Salmonella Typhimurium outer membrane. Proc. Natl. Acad. Sci. U.S.A. 111 1963–1968. 10.1073/pnas.1316901111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawaliby R., Trubbia C., Delporte C., Noyon C., Ruysschaert J.-M., Antwerpen P. V., et al. (2015). Phosphatidylethanolamine is a key regulator of membrane fluidity in eukaryotic cells. J. Biol. Chem. 291 3658–3667. 10.1074/jbc.M115.706523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DebRoy S., Gebbie M., Ramesh A., Goodson J. R., Cruz M. R., van Hoof A., et al. (2014). Riboswitches. a riboswitch-containing sRNA controls gene expression by sequestration of a response regulator. Science 345 937–940. 10.1126/science.1255091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deiwick J., Nikolaus T., Erdogan S., Hensel M. (1999). Environmental regulation of Salmonella pathogenicity island 2 gene expression. Mol. Microbiol. 31 1759–1773. 10.1046/j.1365-2958.1999.01312.x [DOI] [PubMed] [Google Scholar]
- Deng W., Marshall N. C., Rowland J. L., McCoy J. M., Worrall L. J., Santos A. S., et al. (2017). Assembly, structure, function and regulation of type III secretion systems. Nat. Rev. Microbiol. 15 323–337. 10.1038/nrmicro.2017.20 [DOI] [PubMed] [Google Scholar]
- Deriu E., Liu J. Z., Pezeshki M., Edwards R. A., Ochoa R. J., Contreras H., et al. (2013). Probiotic bacteria reduce Salmonella typhimurium intestinal colonization by competing for iron. Cell Host Microbe 14 26–37. 10.1016/j.chom.2013.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewhirst F. E., Chien C.-C., Paster B. J., Ericson R. L., Orcutt R. P., Schauer D. B., et al. (1999). Phylogeny of the defined murine microbiota: altered schaedler flora. Appl. Environ. Microbiol. 65 3287–3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diard M., Sellin M. E., Dolowschiak T., Arnoldini M., Ackermann M., Hardt W.-D. (2014). Antibiotic treatment selects for cooperative virulence of Salmonella Typhimurium. Curr. Biol. 24 2000–2005. 10.1016/j.cub.2014.07.028 [DOI] [PubMed] [Google Scholar]
- Diaz-Ochoa V. E., Lam D., Lee C. S., Klaus S., Behnsen J., Liu J. Z., et al. (2016). Salmonella mitigates oxidative stress and thrives in the inflamed gut by evading calprotectin-mediated manganese sequestration. Cell Host Microbe 19 814–825. 10.1016/j.chom.2016.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donohoe D. R., Wali A., Brylawski B. P., Bultman S. J. (2012). Microbial regulation of glucose metabolism and cell-cycle progression in mammalian colonocytes. PLOS ONE 7:e46589. 10.1371/journal.pone.0046589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowhan W. (1997). Phosphatidylserine decarboxylases: pyruvoyl-dependent enzymes from bacteria to mammals. Methods Enzymol. 280 81–88. 10.1016/S0076-6879(97)80104-8 [DOI] [PubMed] [Google Scholar]
- Dowhan W. (2003). Molecular basis for membrane phospholipid diversity: why are there so many lipids? Annu. Rev. Biochem. 66 199–232. 10.1146/annurev.biochem.66.1.199 [DOI] [PubMed] [Google Scholar]
- Drumo R., Pesciaroli M., Ruggeri J., Tarantino M., Chirullo B., Pistoia C., et al. (2016). Salmonella enterica serovar typhimurium exploits inflammation to modify swine intestinal microbiota. Front. Cell. Infect. Microbiol. 5:106. 10.3389/fcimb.2015.00106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eade C. R., Hung C.-C., Bullard B., Gonzalez-Escobedo G., Gunn J. S., Altier C. (2016). Bile acids function synergistically to repress invasion gene expression in Salmonella by destabilizing the invasion regulator HilD. Infect. Immun. 84 2198–2208. 10.1128/IAI.00177-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards J. C., Johnson M. S., Taylor B. L. (2006). Differentiation between electron transport sensing and proton motive force sensing by the Aer and Tsr receptors for aerotaxis. Mol. Microbiol. 62 823–837. 10.1111/j.1365-2958.2006.05411.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellermeier C. D., Ellermeier J. R., Slauch J. M. (2005). HilD, HilC and RtsA constitute a feed forward loop that controls expression of the SPI1 type three secretion system regulator HilA in Salmonella enterica serovar typhimurium. Mol. Microbiol. 57 691–705. 10.1111/j.1365-2958.2005.04737.x [DOI] [PubMed] [Google Scholar]
- Ellermeier C. D., Slauch J. M. (2004). RtsA coordinately regulates DsbA and the Salmonella pathogenicity island 1 type III secretion system. J. Bacteriol. 186 68–79. 10.1128/JB.186.1.68-79.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endt K., Stecher B., Chaffron S., Slack E., Tchitchek N., Benecke A., et al. (2010). The microbiota mediates pathogen clearance from the gut lumen after non-typhoidal Salmonella diarrhea. PLOS Pathog. 6:e1001097. 10.1371/journal.ppat.1001097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espey M. G. (2013). Role of oxygen gradients in shaping redox relationships between the human intestine and its microbiota. Free Radic. Biol. Med. 55 130–140. 10.1016/j.freeradbiomed.2012.10.554 [DOI] [PubMed] [Google Scholar]
- Faber F., Thiennimitr P., Spiga L., Byndloss M. X., Litvak Y., Lawhon S., et al. (2017). Respiration of microbiota-derived 1,2-propanediol drives Salmonella expansion during colitis. PLOS Pathog. 13:e1006129. 10.1371/journal.ppat.1006129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fàbrega A., Vila J. (2013). Salmonella enterica serovar typhimurium skills to succeed in the host: virulence and regulation. Clin. Microbiol. Rev. 26 308–341. 10.1128/CMR.00066-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fass E., Groisman E. A. (2009). Control of Salmonella pathogenicity island-2 gene expression. Curr. Opin. Microbiol. 12 199–204. 10.1016/j.mib.2009.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueira R., Holden D. W. (2012). Functions of the Salmonella pathogenicity island 2 (SPI-2) type III secretion system effectors. Microbiology 158 1147–1161. 10.1099/mic.0.058115-0 [DOI] [PubMed] [Google Scholar]
- Fink S. L., Bergsbaken T., Cookson B. T. (2008). Anthrax lethal toxin and Salmonella elicit the common cell death pathway of caspase-1-dependent pyroptosis via distinct mechanisms. Proc. Natl. Acad. Sci. U.S.A. 105 4312–4317. 10.1073/pnas.0707370105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink S. L., Cookson B. T. (2006). Caspase-1-dependent pore formation during pyroptosis leads to osmotic lysis of infected host macrophages. Cell. Microbiol. 8 1812–1825. 10.1111/j.1462-5822.2006.00751.x [DOI] [PubMed] [Google Scholar]
- Flannagan R. S., Cosío G., Grinstein S. (2009). Antimicrobial mechanisms of phagocytes and bacterial evasion strategies. Nat. Rev. Microbiol. 7 355–366. 10.1038/nrmicro2128 [DOI] [PubMed] [Google Scholar]
- Franchi L., Amer A., Body-Malapel M., Kanneganti T.-D., Özören N., Jagirdar R., et al. (2006). Cytosolic flagellin requires Ipaf for activation of caspase-1 and interleukin 1β in Salmonella-infected macrophages. Nat. Immunol. 7 576–582. 10.1038/ni1346 [DOI] [PubMed] [Google Scholar]
- Fu Y., Waldor M. K., Mekalanos J. J. (2013). Tn-Seq analysis of Vibrio cholerae intestinal colonization reveals a role for T6SS-mediated antibacterial activity in the host. Cell Host Microbe 14 652–663. 10.1016/j.chom.2013.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galán J. E., Curtiss R. (1989). Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc. Natl. Acad. Sci. U.S.A. 86 6383–6387. 10.1073/pnas.86.16.6383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galán J. E., Curtiss R. (1990). Expression of Salmonella typhimurium genes required for invasion is regulated by changes in DNA supercoiling. Infect. Immun. 58 1879–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galanis E., Lo Fo Wong D. M., Patrick M. E., Binsztein N., Cieslik A., Chalermchaikit T., et al. (2006). Web-based surveillance and global Salmonella distribution, 2000–2002. Emerg. Infect. Dis. 12 381–388. 10.3201/eid1203.050854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gantois I., Ducatelle R., Pasmans F., Haesebrouck F., Hautefort I., Thompson A., et al. (2006). Butyrate specifically down-regulates Salmonella pathogenicity island 1 gene expression. Appl. Environ. Microbiol. 72 946–949. 10.1128/AEM.72.1.946-949.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-del Portillo F., Zwick M. B., Leung K. Y., Finlay B. B. (1993). Salmonella induces the formation of filamentous structures containing lysosomal membrane glycoproteins in epithelial cells. Proc. Natl. Acad. Sci. U.S.A. 90 10544–10548. 10.1073/pnas.90.22.10544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gart E. V., Suchodolski J. S., Welsh T. H., Alaniz R. C., Randel R. D., Lawhon S. D. (2016). Salmonella typhimurium and multidirectional communication in the gut. Front. Microbiol. 7:1827. 10.3389/fmicb.2016.01827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson G. R., Roberfroid M. B. (1995). Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. J. Nutr. 125 1401–1412. [DOI] [PubMed] [Google Scholar]
- Gill N., Ferreira R. B. R., Antunes L. C. M., Willing B. P., Sekirov I., Al-Zahrani F., et al. (2012). Neutrophil elastase alters the murine gut microbiota resulting in enhanced Salmonella colonization. PLOS ONE 7:e49646. 10.1371/journal.pone.0049646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godinez I., Haneda T., Raffatellu M., George M. D., Paixão T. A., Rolán H. G., et al. (2008). T Cells help to amplify inflammatory responses induced by Salmonella enterica serotype typhimurium in the intestinal mucosa. Infect. Immun. 76 2008–2017. 10.1128/IAI.01691-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golubeva Y. A., Ellermeier J. R., Cott Chubiz J. E., Slauch J. M. (2016). Intestinal long-chain fatty acids act as a direct signal to modulate expression of the Salmonella pathogenicity island 1 type III secretion system. mBio 7:e02170–15. 10.1128/mBio.02170-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golubeva Y. A., Sadik A. Y., Ellermeier J. R., Slauch J. M. (2012). Integrating global regulatory input into the Salmonella pathogenicity island 1 type III secretion system. Genetics 190 79–90. 10.1534/genetics.111.132779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonyar L. A., Kendall M. M. (2014). Ethanolamine and choline promote expression of putative and characterized fimbriae in enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 82 193–201. 10.1128/IAI.00980-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopinath S., Lichtman J. S., Bouley D. M., Elias J. E., Monack D. M. (2014). Role of disease-associated tolerance in infectious superspreaders. Proc. Natl. Acad. Sci. U.S.A. 111 15780–15785. 10.1073/pnas.1409968111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon M. A. (2008). Salmonella infections in immunocompromised adults. J. Infect. 56 413–422. 10.1016/j.jinf.2008.03.012 [DOI] [PubMed] [Google Scholar]
- Graham S. M., Molyneux E. M., Walsh A. L., Cheesbrough J. S., Molyneux M. E., Hart C. A. (2000). Nontyphoidal Salmonella infections of children in tropical Africa. Pediatr. Infect. Dis. J. 19 1189–1196. 10.1097/00006454-200012000-00016 [DOI] [PubMed] [Google Scholar]
- Guignot J., Caron E., Beuzón C., Bucci C., Kagan J., Roy C., et al. (2004). Microtubule motors control membrane dynamics of Salmonella-containing vacuoles. J. Cell Sci. 117 1033–1045. 10.1242/jcs.00949 [DOI] [PubMed] [Google Scholar]
- Günster R. A., Matthews S. A., Holden D. W., Thurston T. L. M. (2017). SseK1 and SseK3 type III secretion system effectors inhibit NF-κB signaling and necroptotic cell death in Salmonella-infected macrophages. Infect. Immun. 85:e00010–17. 10.1128/IAI.00010-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halang P., Toulouse C., Geißel B., Michel B., Flauger B., Müller M., et al. (2015). Response of Vibrio cholerae to the catecholamine hormones epinephrine and norepinephrine. J. Bacteriol. 197 3769–3778. 10.1128/JB.00345-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamer H. M., Jonkers D., Venema K., Vanhoutvin S., Troost F. J., Brummer R.-J. (2008). Review article: the role of butyrate on colonic function. Aliment. Pharmacol. Ther. 27 104–119. 10.1111/j.1365-2036.2007.03562.x [DOI] [PubMed] [Google Scholar]
- Hammer N. D., Cassat J. E., Noto M. J., Lojek L. J., Chadha A. D., Schmitz J. E., et al. (2014). Inter- and intraspecies metabolite exchange promotes virulence of antibiotic-resistant Staphylococcus aureus. Cell Host Microbe 16 531–537. 10.1016/j.chom.2014.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer N. D., Reniere M. L., Cassat J. E., Zhang Y., Hirsch A. O., Hood M. I., et al. (2013). Two heme-dependent terminal oxidases power Staphylococcus aureus organ-specific colonization of the vertebrate host. mBio 4:e00241–13. 10.1128/mBio.00241-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen-Wester I., Stecher B., Hensel M. (2002). Type III secretion of Salmonella enterica serovar typhimurium translocated effectors and SseFG. Infect. Immun. 70 1403–1409. 10.1128/IAI.70.3.1403-1409.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardt W.-D., Chen L.-M., Schuebel K. E., Bustelo X. R., Galán J. E. (1998). S. typhimurium encodes an activator of Rho GTPases that induces membrane ruffling and nuclear responses in host cells. Cell 93 815–826. 10.1016/S0092-8674(00)81442-7 [DOI] [PubMed] [Google Scholar]
- Harris J. C., Dupont H. L., Hornick R. B. (1972). Fecal leukocytes in diarrheal illness. Ann. Intern. Med. 76 697–703. 10.7326/0003-4819-76-5-697 [DOI] [PubMed] [Google Scholar]
- Hébrard M., Kröger C., Srikumar S., Colgan A., Händler K., Hinton J. C. D. (2012). sRNAs and the virulence of Salmonella enterica serovar typhimurium. RNA Biol. 9 437–445. 10.4161/rna.20480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinzen R. A., Scidmore M. A., Rockey D. D., Hackstadt T. (1996). Differential interaction with endocytic and exocytic pathways distinguish parasitophorous vacuoles of Coxiella burnetii and Chlamydia trachomatis. Infect. Immun. 64 796–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helaine S., Thompson J. A., Watson K. G., Liu M., Boyle C., Holden D. W. (2010). Dynamics of intracellular bacterial replication at the single cell level. Proc. Natl. Acad. Sci. U.S.A. 107 3746–3751. 10.1073/pnas.1000041107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensel M., Shea J. E., Gleeson C., Jones M. D., Dalton E., Holden D. W. (1995). Simultaneous identification of bacterial virulence genes by negative selection. Science 269 400–403. 10.1126/science.7618105 [DOI] [PubMed] [Google Scholar]
- Hicks K. G., Delbecq S. P., Sancho-Vaello E., Blanc M.-P., Dove K. K., Prost L. R., et al. (2015). Acidic pH and divalent cation sensing by PhoQ are dispensable for systemic salmonellae virulence. eLife 4:e06792. 10.7554/eLife.06792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood R. D., Peterson S. B., Mougous J. D. (2017). From striking out to striking gold: discovering that type VI secretion targets bacteria. Cell Host Microbe 21 286–289. 10.1016/j.chom.2017.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki A., Medzhitov R. (2004). Toll-like receptor control of the adaptive immune responses. Nat. Immunol. 5 987–995. 10.1038/ni1112 [DOI] [PubMed] [Google Scholar]
- Jackson L. K., Nawabi P., Hentea C., Roark E. A., Haldar K. (2008). The Salmonella virulence protein SifA is a G protein antagonist. Proc. Natl. Acad. Sci. U.S.A. 105 14141–14146. 10.1073/pnas.0801872105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen I., Zhang Y., Krantz B. A., Miao E. A. (2016). Pyroptosis triggers pore-induced intracellular traps (PITs) that capture bacteria and lead to their clearance by efferocytosis. J. Exp. Med. 213 2113–2128. 10.1084/jem.20151613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juricova H., Videnska P., Lukac M., Faldynova M., Babak V., Havlickova H., et al. (2013). Influence of Salmonella enterica serovar enteritidis infection on the development of the cecum microbiota in newly hatched chicks. Appl. Environ. Microbiol. 79 745–747. 10.1128/AEM.02628-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabir M. R., Hossain M. A., Paul S. K., Mahmud C., Ahmad S., Mahmud N. U., et al. (2012). Enteropathogens associated with acute diarrhea in a tertiary hospital of Bangladesh. Mymensingh Med. J. 21 618–623. [PubMed] [Google Scholar]
- Kato A., Latifi T., Groisman E. A. (2003). Closing the loop: the PmrA/PmrB two-component system negatively controls expression of its posttranscriptional activator PmrD. Proc. Natl. Acad. Sci. U.S.A. 100 4706–4711. 10.1073/pnas.0836837100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai K., Fujita M., Nakao M. (1974). Lipid components of two different regions of an intestinal epithelial cell membrane of mouse. Biochim. Biophys. Acta 369 222–233. 10.1016/0005-2760(74)90253-7 [DOI] [PubMed] [Google Scholar]
- Kawai T., Akira S. (2010). The role of pattern-recognition receptors in innate immunity: update on toll-like receptors. Nat. Immunol. 11 373–384. 10.1038/ni.1863 [DOI] [PubMed] [Google Scholar]
- Kelly C. J., Zheng L., Campbell E. L., Saeedi B., Scholz C. C., Bayless A. J., et al. (2015). Crosstalk between microbiota-derived short-chain fatty acids and intestinal epithelial HIF augments tissue barrier function. Cell Host Microbe 17 662–671. 10.1016/j.chom.2015.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendall M. M., Gruber C. C., Parker C. T., Sperandio V. (2012). Ethanolamine controls expression of genes encoding components involved in interkingdom signaling and virulence in enterohemorrhagic Escherichia coli O157:H7. mBio 3:e00050–12. 10.1128/mBio.00050-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendall M. M., Sperandio V. (2016). What a dinner party! Mechanisms and functions of interkingdom signaling in host-pathogen associations. mBio 7:e01748–15. 10.1128/mBio.01748-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C. C., Falkow S. (2004). Delineation of upstream signaling events in the Salmonella pathogenicity island 2 transcriptional activation pathway. J. Bacteriol. 186 4694–4704. 10.1128/JB.186.14.4694-4704.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak G. K., MacDonald D., Landry L., Farber J. M. (2013). Foodborne outbreaks in Canada linked to produce: 2001 through 2009. J. Food Prot. 76 173–183. 10.4315/0362-028X.JFP-12-126 [DOI] [PubMed] [Google Scholar]
- Kroemer G., Galluzzi L., Vandenabeele P., Abrams J., Alnemri E., Baehrecke E., et al. (2009). Classification of cell death. Cell Death Differ. 16 3–11. 10.1038/cdd.2008.150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kröger C., Colgan A., Srikumar S., Händler K., Sivasankaran S. K., Hammarlöf D. L., et al. (2013). An infection-relevant transcriptomic compendium for Salmonella enterica serovar typhimurium. Cell Host Microbe 14 683–695. 10.1016/j.chom.2013.11.010 [DOI] [PubMed] [Google Scholar]
- Kröger C., Dillon S. C., Cameron A. D. S., Papenfort K., Sivasankaran S. K., Hokamp K., et al. (2012). The transcriptional landscape and small RNAs of Salmonella enterica serovar typhimurium. Proc. Natl. Acad. Sci. U.S.A. 109 E1277–E1286. 10.1073/pnas.1201061109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühn L. C. (2015). Iron regulatory proteins and their role in controlling iron metabolism. Metallomics 7 232–243. 10.1039/C4MT00164H [DOI] [PubMed] [Google Scholar]
- Lam L. H., Monack D. M. (2014). Intraspecies competition for niches in the distal gut dictate transmission during persistent Salmonella infection. PLOS Pathog. 10:e1004527. 10.1371/journal.ppat.1004527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara-Tejero M., Sutterwala F. S., Ogura Y., Grant E. P., Bertin J., Coyle A. J., et al. (2006). Role of the caspase-1 inflammasome in Salmonella typhimurium pathogenesis. J. Exp. Med. 203 1407–1412. 10.1084/jem.20060206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawhon S. D., Maurer R., Suyemoto M., Altier C. (2002). Intestinal short-chain fatty acids alter Salmonella typhimurium invasion gene expression and virulence through BarA/SirA. Mol. Microbiol. 46 1451–1464. 10.1046/j.1365-2958.2002.03268.x [DOI] [PubMed] [Google Scholar]
- Lawley T. D., Bouley D. M., Hoy Y. E., Gerke C., Relman D. A., Monack D. M. (2008). Host transmission of Salmonella enterica serovar typhimurium is controlled by virulence factors and indigenous intestinal microbiota. Infect. Immun. 76 403–416. 10.1128/IAI.01189-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. A., Falkow S. (1990). The ability of Salmonella to enter mammalian cells is affected by bacterial growth state. Proc. Natl. Acad. Sci. U.S.A. 87 4304–4308. 10.1073/pnas.87.11.4304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E.-J., Groisman E. A. (2010). An antisense RNA that governs the expression kinetics of a multifunctional virulence gene. Mol. Microbiol. 76 1020–1033. 10.1111/j.1365-2958.2010.07161.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E.-J., Groisman E. A. (2012). Control of a Salmonella virulence locus by an ATP-sensing leader mRNA. Nature 486 271–275. 10.1038/nature11090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindgren S. W., Stojiljkovic I., Heffron F. (1996). Macrophage killing is an essential virulence mechanism of Salmonella typhimurium. Proc. Natl. Acad. Sci. U.S.A. 93 4197–4201. 10.1073/pnas.93.9.4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipton B. A., Davidson E. P., Ginsberg B. H., Yorek M. A. (1990). Ethanolamine metabolism in cultured bovine aortic endothelial cells. J. Biol. Chem. 265 7195–7201. [PubMed] [Google Scholar]
- Lipton B. A., Yorek M. A., Ginsberg B. H. (1988). Ethanolamine and choline transport in cultured bovine aortic endothelial cells. J. Cell. Physiol. 137 571–576. 10.1002/jcp.1041370325 [DOI] [PubMed] [Google Scholar]
- Liss V., Swart A. L., Kehl A., Hermanns N., Zhang Y., Chikkaballi D., et al. (2017). Salmonella enterica remodels the host cell endosomal system for efficient intravacuolar nutrition. Cell Host Microbe 21 390–402. 10.1016/j.chom.2017.02.005 [DOI] [PubMed] [Google Scholar]
- Liu J. Z., Jellbauer S., Poe A. J., Ton V., Pesciaroli M., Kehl-Fie T. E., et al. (2012). Zinc sequestration by the neutrophil protein calprotectin enhances Salmonella growth in the inflamed gut. Cell Host Microbe 11 227–239. 10.1016/j.chom.2012.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löber S., Jäckel D., Kaiser N., Hensel M. (2006). Regulation of Salmonella pathogenicity island 2 genes by independent environmental signals. Int. J. Med. Microbiol. 296 435–447. 10.1016/j.ijmm.2006.05.001 [DOI] [PubMed] [Google Scholar]
- Lopez C. A., Rivera-Chávez F., Byndloss M. X., Bäumler A. J. (2015). The periplasmic nitrate reductase NapABC supports luminal growth of Salmonella enterica serovar typhimurium during colitis. Infect. Immun. 83 3470–3478. 10.1128/IAI.00351-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez C. A., Winter S. E., Rivera-Chávez F., Xavier M. N., Poon V., Nuccio S.-P., et al. (2012). Phage-mediated acquisition of a type III secreted effector protein boosts growth of Salmonella by nitrate respiration. mBio 3:e00143–12. 10.1128/mBio.00143-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis P., Flint H. J. (2009). Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol. Lett. 294 1–8. 10.1111/j.1574-6968.2009.01514.x [DOI] [PubMed] [Google Scholar]
- Luzader D. H., Clark D. E., Gonyar L. A., Kendall M. M. (2013). EutR is a direct regulator of genes that contribute to metabolism and virulence in enterohemorrhagic Escherichia coli O157:H7. J. Bacteriol. 195 4947–4953. 10.1128/JB.00937-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma A. T., Mekalanos J. J. (2010). In vivo actin cross-linking induced by Vibrio cholerae type VI secretion system is associated with intestinal inflammation. Proc. Natl. Acad. Sci. U.S.A. 107 4365–4370. 10.1073/pnas.0915156107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maadani A., Fox K. A., Mylonakis E., Garsin D. A. (2007). Enterococcus faecalis mutations affecting virulence in the Caenorhabditis elegans model host. Infect. Immun. 75 2634–2637. 10.1128/IAI.01372-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier L., Diard M., Sellin M. E., Chouffane E.-S., Trautwein-Weidner K., Periaswamy B., et al. (2014). Granulocytes impose a tight bottleneck upon the gut luminal pathogen population during Salmonella typhimurium colitis. PLOS Pathog. 10:e1004557. 10.1371/journal.ppat.1004557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier L., Vyas R., Cordova C. D., Lindsay H., Schmidt T. S. B., Brugiroux S., et al. (2013). Microbiota-derived hydrogen fuels Salmonella typhimurium invasion of the gut ecosystem. Cell Host Microbe 14 641–651. 10.1016/j.chom.2013.11.002 [DOI] [PubMed] [Google Scholar]
- Man S. M., Tourlomousis P., Hopkins L., Monie T. P., Fitzgerald K. A., Bryant C. E. (2013). Salmonella infection induces recruitment of caspase-8 to the inflammasome to modulate interleukin-1β production. J. Immunol. 191 5239–5246. 10.4049/jimmunol.1301581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marteyn B., West N., Browning D., Cole J., Shaw J., Palm F., et al. (2010). Modulation of Shigella virulence in response to available oxygen in vivo. Nature 465 355–358. 10.1038/nature08970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin L. B. (2012). Vaccines for typhoid fever and other salmonelloses. Curr. Opin. Infect. Dis. 25 489–499. 10.1097/QCO.0b013e328356ffeb [DOI] [PubMed] [Google Scholar]
- Mashruwala A. A., Pang Y. Y., Rosario-Cruz Z., Chahal H. K., Benson M. A., Mike L. A., et al. (2015). Nfu facilitates the maturation of iron-sulfur proteins and participates in virulence in Staphylococcus aureus. Mol. Microbiol. 95 383–409. 10.1111/mmi.12860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenney E. S., Kendall M. M., Napier B. (2016). Microbiota and pathogen ‘pas de deux’: setting up and breaking down barriers to intestinal infection. Pathog. Dis. 74:ftw051. 10.1093/femspd/ftw051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenney P. T., Pamer E. G. (2015). From hype to hope: the gut microbiota in enteric infectious disease. Cell 163 1326–1332. 10.1016/j.cell.2015.11.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina C. B., Ravichandran K. S. (2016). Do not let death do us part: ‘find-me’ signals in communication between dying cells and the phagocytes. Cell Death Differ. 23 979–989. 10.1038/cdd.2016.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellin J. R., Koutero M., Dar D., Nahori M.-A., Sorek R., Cossart P. (2014). Sequestration of a two-component response regulator by a riboswitch-regulated noncoding RNA. Science 345 940–943. 10.1126/science.1255083 [DOI] [PubMed] [Google Scholar]
- Miao E. A., Alpuche-Aranda C. M., Dors M., Clark A. E., Bader M. W., Miller S. I., et al. (2006). Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1β via Ipaf. Nat. Immunol. 7 569–575. 10.1038/ni1344 [DOI] [PubMed] [Google Scholar]
- Miao E. A., Leaf I. A., Treuting P. M., Mao D. P., Dors M., Sarkar A., et al. (2010a). Caspase-1-induced pyroptosis is an innate immune effector mechanism against intracellular bacteria. Nat. Immunol. 11 1136–1142. 10.1038/ni.1960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao E. A., Mao D. P., Yudkovsky N., Bonneau R., Lorang C. G., Warren S. E., et al. (2010b). Innate immune detection of the type III secretion apparatus through the NLRC4 inflammasome. Proc. Natl. Acad. Sci. U.S.A. 107 3076–3080. 10.1073/pnas.0913087107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki T., Goto R., Fujimoto M., Okada N., Hardt W.-D. (2017). The bactericidal lectin RegIIIβ prolongs gut colonization and enteropathy in the streptomycin mouse model for Salmonella diarrhea. Cell Host Microbe 21 195–207. 10.1016/j.chom.2016.12.008 [DOI] [PubMed] [Google Scholar]
- Miki T., Holst O., Hardt W.-D. (2012). The bactericidal activity of the C-type lectin RegIIIβ against gram-negative bacteria involves binding to lipid A. J. Biol. Chem. 287 34844–34855. 10.1074/jbc.M112.399998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizusaki H., Takaya A., Yamamoto T., Aizawa S.-I. (2008). Signal pathway in salt-activated expression of the Salmonella pathogenicity island 1 type III secretion system in Salmonella enterica serovar typhimurium. J. Bacteriol. 190 4624–4631. 10.1128/JB.01957-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monack D. M., Detweiler C. S., Falkow S. (2001). Salmonella pathogenicity island 2-dependent macrophage death is mediated in part by the host cysteine protease caspase-1. Cell. Microbiol. 3 825–837. 10.1046/j.1462-5822.2001.00162.x [DOI] [PubMed] [Google Scholar]
- Monack D. M., Raupach B., Hromockyj A. E., Falkow S. (1996). Salmonella typhimurium invasion induces apoptosis in infected macrophages. Proc. Natl. Acad. Sci. U.S.A. 93 9833–9838. 10.1073/pnas.93.18.9833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooney J. P., Lokken K. L., Byndloss M. X., George M. D., Velazquez E. M., Faber F., et al. (2015). Inflammation-associated alterations to the intestinal microbiota reduce colonization resistance against non-typhoidal Salmonella during concurrent malaria parasite infection. Sci. Rep. 5:14603. 10.1038/srep14603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira C. G., Russell R., Mishra A. A., Narayanan S., Ritchie J. M., Waldor M. K., et al. (2016). Bacterial adrenergic sensors regulate virulence of enteric pathogens in the gut. mBio 7:e00826–16. 10.1128/mBio.00826-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira C. G., Sperandio V. (2012). Interplay between the QseC and QseE bacterial adrenergic sensor kinases in Salmonella enterica serovar typhimurium pathogenesis. Infect. Immun. 80 4344–4353. 10.1128/IAI.00803-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira C. G., Weinshenker D., Sperandio V. (2010). QseC mediates Salmonella enterica serovar typhimurium virulence in vitro and in vivo. Infect. Immun. 78 914–926. 10.1128/IAI.01038-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mougous J. D., Cuff M. E., Raunser S., Shen A., Zhou M., Gifford C. A., et al. (2006). A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 312 1526–1530. 10.1126/science.1128393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nairz M., Ferring-Appel D., Casarrubea D., Sonnweber T., Viatte L., Schroll A., et al. (2015a). Iron regulatory proteins mediate host resistance to Salmonella infection. Cell Host Microbe 18 254–261. 10.1016/j.chom.2015.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nairz M., Schroll A., Haschka D., Dichtl S., Sonnweber T., Theurl I., et al. (2015b). Lipocalin-2 ensures host defense against Salmonella typhimurium by controlling macrophage iron homeostasis and immune response. Eur. J. Immunol. 45 3073–3086. 10.1002/eji.201545569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nairz M., Fritsche G., Crouch M.-L. V., Barton H. C., Fang F. C., Weiss G. (2009). Slc11a1 limits intracellular growth of Salmonella enterica sv. Typhimurium by promoting macrophage immune effector functions and impairing bacterial iron acquisition. Cell. Microbiol. 11 1365–1381. 10.1111/j.1462-5822.2009.01337.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng K. M., Ferreyra J. A., Higginbottom S. K., Lynch J. B., Kashyap P. C., Gopinath S., et al. (2013). Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature 502 96–99. 10.1038/nature12503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikawa J., Tsukagoshi Y., Yamashita S. (1986). Cloning of a gene encoding choline transport in Saccharomyces cerevisiae. J. Bacteriol. 166 328–330. 10.1128/jb.166.1.328-330.1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Núñez-Hernández C., Tierrez A., Ortega Á. D., Pucciarelli M. G., Godoy M., Eisman B., et al. (2013). Genome expression analysis of nonproliferating intracellular Salmonella enterica serovar typhimurium unravels an acid pH-dependent PhoP-PhoQ response essential for dormancy. Infect. Immun. 81 154–165. 10.1128/IAI.01080-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H., Soncini F. C., Solomon F., Groisman E. A. (1996). Identification of a pathogenicity island required for Salmonella survival in host cells. Proc. Natl. Acad. Sci. U.S.A. 93 7800–7804. 10.1073/pnas.93.15.7800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlson M. B., Huang Z., Alto N. M., Blanc M.-P., Dixon J. E., Chai J., et al. (2008). Structure and function of Salmonella SifA indicate that its interactions with SKIP, SseJ, and RhoA family GTPases induce endosomal tubulation. Cell Host Microbe 4 434–446. 10.1016/j.chom.2008.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olekhnovich I. N., Kadner R. J. (2002). DNA-binding activities of the HilC and HilD virulence regulatory proteins of Salmonella enterica serovar typhimurium. J. Bacteriol. 184 4148–4160. 10.1128/JB.184.15.4148-4160.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne S. E., Coombes B. K. (2011). Transcriptional priming of Salmonella pathogenicity island-2 precedes cellular invasion. PLOS ONE 6:e21648. 10.1371/journal.pone.0021648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen K. A., Anderson C. J., Casanova J. E. (2016). Salmonella suppresses the TRIF-dependent type I interferon response in macrophages. mBio 7:e02051–15. 10.1128/mBio.02051-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen K. A., Meyer C. B., Bouton A. H., Casanova J. E. (2014). Activation of focal adhesion kinase by Salmonella suppresses autophagy via an Akt/mTOR signaling pathway and promotes bacterial survival in macrophages. PLOS Pathog. 10:e1004159. 10.1371/journal.ppat.1004159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer L. D., Skaar E. P. (2016). Transition metals and virulence in bacteria. Annu. Rev. Genet. 50 67–91. 10.1146/annurev-genet-120215-035146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasparakis M., Vandenabeele P. (2015). Necroptosis and its role in inflammation. Nature 517 311–320. 10.1038/nature14191 [DOI] [PubMed] [Google Scholar]
- Phoebe Lostroh C., Lee C. A. (2001). The Salmonella pathogenicity island-1 type III secretion system. Microbes Infect. 3 1281–1291. 10.1016/S1286-4579(01)01488-5 [DOI] [PubMed] [Google Scholar]
- Price-Carter M., Tingey J., Bobik T. A., Roth J. R. (2001). The alternative electron acceptor tetrathionate supports B12-dependent anaerobic growth of Salmonella enterica serovar typhimurium on ethanolamine or 1,2-propanediol. J. Bacteriol. 183 2463–2475. 10.1128/JB.183.8.2463-2475.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prost L. R., Daley M. E., Le Sage V., Bader M. W., Le Moual H., Klevit R. E., et al. (2007). Activation of the bacterial sensor kinase PhoQ by acidic pH. Mol. Cell 26 165–174. 10.1016/j.molcel.2007.03.008 [DOI] [PubMed] [Google Scholar]
- Prouty A. M., Gunn J. S. (2000). Salmonella enterica serovar typhimurium invasion is repressed in the presence of bile. Infect. Immun. 68 6763–6769. 10.1128/IAI.68.12.6763-6769.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pukatzki S., Ma A. T., Sturtevant D., Krastins B., Sarracino D., Nelson W. C., et al. (2006). Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc. Natl. Acad. Sci. U.S.A. 103 1528–1533. 10.1073/pnas.0510322103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffatellu M., George M. D., Akiyama Y., Hornsby M. J., Nuccio S.-P., Paixao T. A., et al. (2009). Lipocalin-2 resistance confers an advantage to Salmonella enterica serotype typhimurium for growth and survival in the inflamed intestine. Cell Host Microbe 5 476–486. 10.1016/j.chom.2009.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randle C. L., Albro P. W., Dittmer J. C. (1969). The phosphoglyceride composition of gram-negative bacteria and the changes in composition during growth. Biochim. Biophys. Acta 187 214–220. 10.1016/0005-2760(69)90030-7 [DOI] [PubMed] [Google Scholar]
- Rappl C., Deiwick J., Hensel M. (2003). Acidic pH is required for the functional assembly of the type III secretion system encoded by Salmonella pathogenicity island 2. FEMS Microbiol. Lett. 226 363–372. 10.1016/S0378-1097(03)00638-4 [DOI] [PubMed] [Google Scholar]
- Rasko D. A., Moreira C. G., Li D. R., Reading N. C., Ritchie J. M., Waldor M. K., et al. (2008). Targeting QseC signaling and virulence for antibiotic development. Science 321 1078–1080. 10.1126/science.1160354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathman M., Sjaastad M. D., Falkow S. (1996). Acidification of phagosomes containing Salmonella typhimurium in murine macrophages. Infect. Immun. 64 2765–2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch I., Deets K. A., Ji D. X., von Moltke J., Tenthorey J. L., Lee A. Y., et al. (2017). NAIP-NLRC4 inflammasomes coordinate intestinal epithelial cell expulsion with eicosanoid and IL-18 release via activation of caspase-1 and -8. Immunity 46 649–659. 10.1016/j.immuni.2017.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raupach B., Peuschel S.-K., Monack D. M., Zychlinsky A. (2006). Caspase-1-mediated activation of interleukin-1β (IL-1β) and IL-18 contributes to innate immune defenses against Salmonella enterica serovar typhimurium infection. Infect. Immun. 74 4922–4926. 10.1128/IAI.00417-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayamajhi M., Zak D. E., Chavarria-Smith J., Vance R. E., Miao E. A. (2013). Cutting edge: mouse NAIP1 detects the type III secretion system needle protein. J. Immunol. 191 3986–3989. 10.4049/jimmunol.1301549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reading N. C., Rasko D. A., Torres A. G., Sperandio V. (2009). The two-component system QseEF and the membrane protein QseG link adrenergic and stress sensing to bacterial pathogenesis. Proc. Natl. Acad. Sci. U.S.A. 106 5889–5894. 10.1073/pnas.0811409106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebuffat S. (2012). Microcins in action: amazing defence strategies of enterobacteria. Biochem. Soc. Trans. 40 1456–1462. 10.1042/BST20120183 [DOI] [PubMed] [Google Scholar]
- Rivera-Chávez F., Lopez C. A., Zhang L. F., García-Pastor L., Chávez-Arroyo A., Lokken K. L., et al. (2016a). Energy taxis toward host-derived nitrate supports a Salmonella pathogenicity island 1-independent mechanism of invasion. mBio 7:e00960–16. 10.1128/mBio.00960-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Chávez F., Zhang L. F., Faber F., Lopez C. A., Byndloss M. X., Olsan E. E., et al. (2016b). Depletion of butyrate-producing Clostridia from the gut microbiota drives an aerobic luminal expansion of Salmonella. Cell Host Microbe 19 443–454. 10.1016/j.chom.2016.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Chavez F., Winter S. E., Lopez C. A., Xavier M. N., Winter M. G., Nuccio S.-P., et al. (2013). Salmonella uses energy taxis to benefit from intestinal inflammation. PLOS Pathog. 9:e1003267. 10.1371/journal.ppat.1003267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson N., McComb S., Mulligan R., Dudani R., Krishnan L., Sad S. (2012). Type I interferon induces necroptosis in macrophages during infection with Salmonella enterica serovar typhimurium. Nat. Immunol. 13 954–962. 10.1038/ni.2397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roof D. M., Roth J. R. (1992). Autogenous regulation of ethanolamine utilization by a transcriptional activator of the eut operon in Salmonella typhimurium. J. Bacteriol. 174 6634–6643. 10.1128/jb.174.20.6634-6643.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooks M. G., Veiga P., Reeves A. Z., Lavoie S., Yasuda K., Asano Y., et al. (2017). QseC inhibition as an antivirulence approach for colitis-associated bacteria. Proc. Natl. Acad. Sci. U.S.A. 114 142–147. 10.1073/pnas.1612836114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell A. B., Wexler A. G., Harding B. N., Whitney J. C., Bohn A. J., Goo Y. A., et al. (2014). A type VI secretion-related pathway in Bacteroidetes mediates interbacterial antagonism. Cell Host Microbe 16 227–236. 10.1016/j.chom.2014.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabag-Daigle A., Blunk H. M., Sengupta A., Wu J., Bogard A. J., Ali M. M., et al. (2016). A metabolic intermediate of the fructose-asparagine utilization pathway inhibits growth of a Salmonella FraB mutant. Sci. Rep. 6:28117. 10.1038/srep28117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salcedo S. P., Noursadeghi M., Cohen J., Holden D. W. (2001). Intracellular replication of Salmonella typhimurium strains in specific subsets of splenic macrophages in vivo. Cell. Microbiol. 3 587–597. 10.1046/j.1462-5822.2001.00137.x [DOI] [PubMed] [Google Scholar]
- Sana T. G., Flaugnatti N., Lugo K. A., Lam L. H., Jacobson A., Baylot V., et al. (2016). Salmonella typhimurium utilizes a T6SS-mediated antibacterial weapon to establish in the host gut. Proc. Natl. Acad. Sci. U.S.A. 113 E5044–E5051. 10.1073/pnas.1608858113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sana T. G., Lugo K. A., Monack D. M. (2017). T6SS: the bacterial “fight club” in the host gut. PLOS Pathog. 13:e1006325. 10.1371/journal.ppat.1006325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandra A., Cai J. (1991). Plasma membrane appearance of phosphatidylethanolamine in stimulated macrophages. J. Leukoc. Biol. 50 19–27. [DOI] [PubMed] [Google Scholar]
- Sassone-Corsi M., Nuccio S.-P., Liu H., Hernandez D., Vu C. T., Takahashi A. A., et al. (2016). Microcins mediate competition among Enterobacteriaceae in the inflamed gut. Nature 540 280–283. 10.1038/nature20557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scallan E., Hoekstra R. M., Angulo F. J., Tauxe R. V., Widdowson M.-A., Roy S. L., et al. (2011). Foodborne illness acquired in the United States—major pathogens. Emerg. Infect. Dis. 17 7–15. 10.3201/eid1701.P11101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schechter L. M., Lee C. A. (2001). AraC/XylS family members, HilC and HilD, directly bind and derepress the Salmonella typhimurium hilA promoter. Mol. Microbiol. 40 1289–1299. 10.1046/j.1365-2958.2001.02462.x [DOI] [PubMed] [Google Scholar]
- Schwarz S., Hood R. D., Mougous J. D. (2010). What is type VI secretion doing in all those bugs? Trends Microbiol. 18 531–537. 10.1016/j.tim.2010.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekirov I., Tam N. M., Jogova M., Robertson M. L., Li Y., Lupp C., et al. (2008). Antibiotic-induced perturbations of the intestinal microbiota alter host susceptibility to enteric infection. Infect. Immun. 76 4726–4736. 10.1128/IAI.00319-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellin M. E., Müller A. A., Felmy B., Dolowschiak T., Diard M., Tardivel A., et al. (2014). Epithelium-intrinsic NAIP/NLRC4 inflammasome drives infected enterocyte expulsion to restrict Salmonella replication in the intestinal mucosa. Cell Host Microbe 16 237–248. 10.1016/j.chom.2014.07.001 [DOI] [PubMed] [Google Scholar]
- Shea J. E., Hensel M., Gleeson C., Holden D. W. (1996). Identification of a virulence locus encoding a second type III secretion system in Salmonella typhimurium. Proc. Natl. Acad. Sci. U.S.A. 93 2593–2597. 10.1073/pnas.93.6.2593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiao Y. J., Vance J. E. (1995). Evidence for an ethanolamine cycle: differential recycling of the ethanolamine moiety of phosphatidylethanolamine derived from phosphatidylserine and ethanolamine. Biochem. J. 310 673–679. 10.1042/bj3100673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shotland Y., Krämer H., Groisman E. A. (2003). The Salmonella SpiC protein targets the mammalian Hook3 protein function to alter cellular trafficking. Mol. Microbiol. 49 1565–1576. 10.1046/j.1365-2958.2003.03668.x [DOI] [PubMed] [Google Scholar]
- Singh R., Jamieson A., Cresswell P. (2008). GILT is a critical host factor for Listeria monocytogenes infection. Nature 455 1244–1247. 10.1038/nature07344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snoeck V., Goddeeris B., Cox E. (2005). The role of enterocytes in the intestinal barrier function and antigen uptake. Microbes Infect. 7 997–1004. 10.1016/j.micinf.2005.04.003 [DOI] [PubMed] [Google Scholar]
- Soncini F. C., García Véscovi E., Solomon F., Groisman E. A. (1996). Molecular basis of the magnesium deprivation response in Salmonella typhimurium: identification of PhoP-regulated genes. J. Bacteriol. 178 5092–5099. 10.1128/jb.178.17.5092-5099.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spees A. M., Wangdi T., Lopez C. A., Kingsbury D. D., Xavier M. N., Winter S. E., et al. (2013). Streptomycin-induced inflammation enhances Escherichia coli gut colonization through nitrate respiration. mBio 4:e00430–13. 10.1128/mBio.00430-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikumar S., Kröger C., Hébrard M., Colgan A., Owen S. V., Sivasankaran S. K., et al. (2015). RNA-seq brings new insights to the intra-macrophage transcriptome of Salmonella typhimurium. PLOS Pathog. 11:e1005262. 10.1371/journal.ppat.1005262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stecher B., Chaffron S., Käppeli R., Hapfelmeier S., Freedrich S., Weber T. C., et al. (2010). Like will to like: abundances of closely related species can predict susceptibility to intestinal colonization by pathogenic and commensal bacteria. PLOS Pathog. 6:e1000711. 10.1371/journal.ppat.1000711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stecher B., Robbiani R., Walker A. W., Westendorf A. M., Barthel M., Kremer M., et al. (2007). Salmonella enterica serovar typhimurium exploits inflammation to compete with the intestinal microbiota. PLOS Biol. 5:e244. 10.1371/journal.pbio.0050244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steeb B., Claudi B., Burton N. A., Tienz P., Schmidt A., Farhan H., et al. (2013). Parallel exploitation of diverse host nutrients enhances Salmonella virulence. PLOS Pathog. 9:e1003301. 10.1371/journal.ppat.1003301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein M. A., Leung K. Y., Zwick M., Portillo F. G., Finlay B. B. (1996). Identification of a Salmonella virulence gene required for formation of filamentous structures containing lysosomal membrane glycoproteins within epithelial cells. Mol. Microbiol. 20 151–164. 10.1111/j.1365-2958.1996.tb02497.x [DOI] [PubMed] [Google Scholar]
- Stelter C., Käppeli R., König C., Krah A., Hardt W.-D., Stecher B., et al. (2011). Salmonella-induced mucosal lectin RegIIIβ kills competing gut microbiota. PLOS ONE 6:e20749. 10.1371/journal.pone.0020749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart M. K., Cummings L. A., Johnson M. L., Berezow A. B., Cookson B. T. (2011). Regulation of phenotypic heterogeneity permits Salmonella evasion of the host caspase-1 inflammatory response. Proc. Natl. Acad. Sci. U.S.A. 108 20742–20747. 10.1073/pnas.1108963108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock A. M., Robinson V. L., Goudreau P. N. (2000). Two-component signal transduction. Annu. Rev. Biochem. 69 183–215. 10.1146/annurev.biochem.69.1.183 [DOI] [PubMed] [Google Scholar]
- Stojiljkovic I., Bäumler A. J., Heffron F. (1995). Ethanolamine utilization in Salmonella typhimurium: nucleotide sequence, protein expression, and mutational analysis of the cchA cchB eutE eutJ eutG eutH gene cluster. J. Bacteriol. 177 1357–1366. 10.1128/jb.177.5.1357-1366.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strugnell R. A., Scott T. A., Wang N., Yang C., Peres N., Bedoui S., et al. (2014). Salmonella vaccines: lessons from the mouse model or bad teaching? Curr. Opin. Microbiol. 17 99–105. 10.1016/j.mib.2013.12.004 [DOI] [PubMed] [Google Scholar]
- Sturgill-Koszycki S., Swanson M. S. (2000). Legionella pneumophila replication vacuoles mature into acidic, endocytic organelles. J. Exp. Med. 192 1261–1272. 10.1084/jem.192.9.1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subashchandrabose S., Hazen T. H., Brumbaugh A. R., Himpsl S. D., Smith S. N., Ernst R. D., et al. (2014). Host-specific induction of Escherichia coli fitness genes during human urinary tract infection. Proc. Natl. Acad. Sci. U.S.A. 111 18327–18332. 10.1073/pnas.1415959112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiemann S., Smit N., Roy U., Lesker T. R., Gálvez E. J. C., Helmecke J., et al. (2017). Enhancement of IFNγ production by distinct commensals ameliorates Salmonella-induced disease. Cell Host Microbe 21 682.e5–694.e5. 10.1016/j.chom.2017.05.005 [DOI] [PubMed] [Google Scholar]
- Thiennimitr P., Winter S. E., Winter M. G., Xavier M. N., Tolstikov V., Huseby D. L., et al. (2011). Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc. Natl. Acad. Sci. U.S.A. 108 17480–17485. 10.1073/pnas.1107857108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsolis R. M., Adams L. G., Ficht T. A., Bäumler A. J. (1999). Contribution of Salmonella typhimurium virulence factors to diarrheal disease in calves. Infect. Immun. 67 4879–4885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tubbs A. L., Liu B., Rogers T. D., Sartor R. B., Miao E. A. (2017). Dietary salt exacerbates experimental colitis. J. Immunol. 199 1051–1059. 10.4049/jimmunol.1700356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiya K., Barbieri M. A., Funato K., Shah A. H., Stahl P. D., Groisman E. A. (1999). A Salmonella virulence protein that inhibits cellular trafficking. EMBO J. 18 3924–3933. 10.1093/emboj/18.14.3924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaishnava S., Behrendt C. L., Ismail A. S., Eckmann L., Hooper L. V. (2008). Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc. Natl. Acad. Sci. U.S.A. 105 20858–20863. 10.1073/pnas.0808723105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdivia R. H., Falkow S. (1997). Fluorescence-based isolation of bacterial genes expressed within host cells. Science 277 2007–2011. 10.1126/science.277.5334.2007 [DOI] [PubMed] [Google Scholar]
- van der Velden A. W. M., Lindgren S. W., Worley M. J., Heffron F. (2000). Salmonella pathogenicity island 1-independent induction of apoptosis in infected macrophages by Salmonella enterica serotype typhimurium. Infect. Immun. 68 5702–5709. 10.1128/IAI.68.10.5702-5709.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Velden A. W. M., Velasquez M., Starnbach M. N. (2003). Salmonella rapidly kill dendritic cells via a caspase-1- dependent mechanism. J. Immunol. 171 6742–6749. 10.4049/jimmunol.171.12.6742 [DOI] [PubMed] [Google Scholar]
- Vandal O. H., Pierini L. M., Schnappinger D., Nathan C. F., Ehrt S. (2008). A membrane protein preserves intrabacterial pH in intraphagosomal Mycobacterium tuberculosis. Nat. Med. 14 849–854. 10.1038/nm.1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez-Torres A., Jones-Carson J., Bäumler A. J., Falkow S., Valdivia R., Brown W., et al. (1999). Extraintestinal dissemination of Salmonella by Cd18-expressing phagocytes. Nature 401 804–808. 10.1038/44593 [DOI] [PubMed] [Google Scholar]
- Vital M., Howe A. C., Tiedje J. M. (2014). Revealing the bacterial butyrate synthesis pathways by analyzing (Meta)genomic Data. mBio 5:e00889–14. 10.1128/mBio.00889-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vojdani J. D., Beuchat L. R., Tauxe R. V. (2008). Juice-associated outbreaks of human illness in the United States, 1995 through 2005. J. Food Prot. 71 356–364. 10.4315/0362-028X-71.2.356 [DOI] [PubMed] [Google Scholar]
- Weiss D. S., Raupach B., Takeda K., Akira S., Zychlinsky A. (2004). Toll-like receptors are temporally involved in host defense. J. Immunol. 172 4463–4469. 10.4049/jimmunol.172.7.4463 [DOI] [PubMed] [Google Scholar]
- Westermann A. J., Förstner K. U., Amman F., Barquist L., Chao Y., Schulte L. N., et al. (2016). Dual RNA-Seq unveils noncoding RNA functions in host–pathogen interactions. Nature 529 496–501. 10.1038/nature16547 [DOI] [PubMed] [Google Scholar]
- Wiles S., Pickard K. M., Peng K., MacDonald T. T., Frankel G. (2006). In vivo bioluminescence imaging of the murine pathogen Citrobacter rodentium. Infect. Immun. 74 5391–5396. 10.1128/IAI.00848-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter S. E., Thiennimitr P., Winter M. G., Butler B. P., Huseby D. L., Crawford R. W., et al. (2010). Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467 426–429. 10.1038/nature09415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter S. E., Winter M. G., Xavier M. N., Thiennimitr P., Poon V., Keestra A. M., et al. (2013). Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science 339 708–711. 10.1126/science.1232467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiström J., Jertborn M., Ekwall E., Norlin K., Söderquist B., Strömberg A., et al. (1992). Empiric treatment of acute diarrheal disease with norfloxacin. A randomized, placebo-controlled study. Swedish study group. Ann. Intern. Med. 117 202–208. 10.7326/0003-4819-117-3-202 [DOI] [PubMed] [Google Scholar]
- Wösten M. M., Kox L. F., Chamnongpol S., Soncini F. C., Groisman E. A. (2000). A signal transduction system that responds to extracellular iron. Cell 103 113–125. 10.1016/S0092-8674(00)00092-1 [DOI] [PubMed] [Google Scholar]
- Yang J., Zhao Y., Shi J., Shao F. (2013). Human NAIP and mouse NAIP1 recognize bacterial type III secretion needle protein for inflammasome activation. Proc. Natl. Acad. Sci. U.S.A. 110 14408–14413. 10.1073/pnas.1306376110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon H., McDermott J. E., Porwollik S., McClelland M., Heffron F. (2009). Coordinated regulation of virulence during systemic infection of Salmonella enterica serovar typhimurium. PLOS Pathog. 5:e1000306. 10.1371/journal.ppat.1000306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurist-Doutsch S., Arrieta M.-C., Vogt S. L., Finlay B. B. (2014). Gastrointestinal microbiota–mediated control of enteric pathogens. Annu. Rev. Genet. 48 361–382. 10.1146/annurev-genet-120213-092421 [DOI] [PubMed] [Google Scholar]
- Zaharik M. L., Vallance B. A., Puente J. L., Gros P., Finlay B. B. (2002). Host–pathogen interactions: host resistance factor nramp1 up-regulates the expression of Salmonella pathogenicity island-2 virulence genes. Proc. Natl. Acad. Sci. U.S.A. 99 15705–15710. 10.1073/pnas.252415599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng M. Y., Inohara N., Nuñez G. (2017). Mechanisms of inflammation-driven bacterial dysbiosis in the gut. Mucosal Immunol. 10 18–26. 10.1038/mi.2016.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Yang J., Shi J., Gong Y.-N., Lu Q., Xu H., et al. (2011). The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus. Nature 477 596–600. 10.1038/nature10510 [DOI] [PubMed] [Google Scholar]
- Zimbler D. L., Park T. M., Arivett B. A., Penwell W. F., Greer S. M., Woodruff T. M., et al. (2012). Stress response and virulence functions of the Acinetobacter baumannii NfuA Fe-S scaffold protein. J. Bacteriol. 194 2884–2893. 10.1128/JB.00213-12 [DOI] [PMC free article] [PubMed] [Google Scholar]