Abstract
We aimed to investigate the effect of hotspot variations of XRCC2 gene on the risk of head and neck cancer (HNC) in 400 patients and 400 controls. Five polymorphisms of XRCC2 gene G4234C (rs3218384), G4088T (rs3218373), G3063A (rs2040639), R188H (rs3218536) and rs7802034 were analyzed using Allele- specific polymerase chain reaction (ARMS-PCR) followed by sequence analysis. For rs3218373, the GG genotype indicated a statistically significant 3-fold increased risk of HNC (P < 0.001) after multivariate adjustment. For rs7802034, the GG genotype suggested statistically significant 2-fold increased risk of HNC (P < 0.001). For SNP of rs3218536, the AA genotype indicated a significant 3-fold increased risk of HNC (P < 0.001). Additionally, haplotype analysis revealed that TACAG, TGGAG, TACGG and TAGGA haplotypes of XRCC2 polymorphisms are associated with HNC risk. Two SNPs in XRCC2 (rs2040639 and rs3218384) were found increased in strong linkage disequilibrium. Furthermore, joint effect model showed 20 fold (OR = 19.89; 95% CI = 2.65–149.36, P = 0.003) increased HNC risk in patients carrying four homozygous risk alleles of selected polymorphisms. These results show that allele distributions and genotypes of XRCC2 SNPs are significantly associated with increased HNC risk and could be a genetic adjuster for the said disease.
Introduction
Various damaging agents such as chemicals, radiations and some endogenous elements affect DNA integrity which ultimately result in single strand breaks (SSBs). Unrepaired SSBs may lead to double strand breaks (DSBs) during the S phase of the cell cycle1. Accumulation of these unrepaired DSBs can cause cell death and initiate malignancies2. There are several mechanisms which repair these DSB. Homologous recombination repair (HRR) is the key pathway for this DNA repair, functioning in S phase of somatic mammalian cell cycle2. Defective HRR has been reported to be closely related to different human cancers3. A wide range of crucial molecules have been identified to participate in HRR process such as RAD51 paralogs (RAD51B, RAD51C, RAD51D, XRCC2, XRCC3) and can serve as central proteins during HRR process4,5. X-ray repair cross complementing group 2 (XRCC2) gene, XRCC2 protein, together with other proteins of RAD51 6, forms a complex which plays a critical role in chromosomal segregation and apoptotic response to DSBs7. This crucial function of the XRCC2 protein for the HRR process has been demonstrated in earlier studies where over 100 fold HRR reduction in XRCC2 deficient hamster cells was observed compared to parental cells8.
Many earlier studies have found that single nucleotide polymorphisms (SNPs) in the DNA repair gene might modify DNA repair capacity and subsequently influence the susceptibility of cancer9. Common genetic variants in XRCC2, particularly a coding SNP in exon 3 (R188H, dbSNP ID rs3218536), have been identified as potential cancer susceptibility loci recently, though the association results were controversial6,10. The XRCC2 R188H polymorphism has been proposed to be a genetic modifier for smoking related pancreatic cancer10, pharyngeal cancer11, oral cancer12 and ovarian cancer13, though validation studies could not provide confirmation14,15. Some of the studies have also implicated XRCC2 R188H in breast cancer16–19, however, the Breast Cancer Association Consortium20 and other subsequent studies found no association between R188H and breast cancer risk21,22, or evidence of a modest protective association23,24.
Since XRCC2 genomic sequence is highly polymorphic, it is of interest to identify genetic defects which have a functional potential to affect the final repairing efficiency of XRCC2, and subsequently the development of head and neck cancer. On the basis of these observations, it was planned to study the role of XRCC2 gene as a candidate involved in the underlying cause of head and neck carcinogenesis.
Materials and Methods
Study subjects and ethical approval
The subjects included in this study consisted of 400 diagnosed head and neck cancer patients and an equal number of age- and sex-matched controls. These subjects were collected during 2011 to 2015. The diagnosis of the head and neck cancer patients was made histologically at Nuclear Oncology Radiation Institute (NORI) Islamabad and Pakistan institute of medicine Sciences (PIMS). Controls were selected from individuals receiving routine medical examinations in these hospitals. The selection procedure for patients included confirmed histological diagnosis of HNC, no preoperative therapy and availability of complete follow-up data. The inclusion criterion for the controls was age and sex matched healthy individuals with absence of prior history of cancerous or precancerous lesions. Patients and controls suffering from any other familial disease (diabetes, blood pressure, and cardiovascular, renal, or hepatic impairment) were excluded from this study. A written informed consent was obtained from all subjects. Additionally, a structured questionnaire, including information about demographic factors, smoking habits and dietary habits was also used to interview all subjects who provided written informed consent. Peripheral blood samples were collected from all study subjects. This study was approved by the institutional Ethical Review Boards of COMSATS Institute of Information Technology, Islamabad and both collaborating hospitals. Additionally, all experiments performed were in accordance with relevant guidelines and regulations.
DNA extraction
Approximately 3–4 ml blood sample was collected in vacutainer tubes from enrolled subjects in this study. DNA was extracted from whole blood by Phenol chloroform method with some modifications25. The extracted DNA was quantified by 2% ethidium bromide gels and spectrophotometrically using Nano Drop (Thermoscientifiv, USA) and stored at −20 °C until used.
SNPs selection
Five functional polymorphisms in DNA repair gene XRCC2 were selected using a set of web-based SNP selection tools (http://snpinfo.niehs.nih.gov/snpinfo/snpfunc.htm). Potential functional SNPs were included to meet the following criteria: (1) Validated SNPs with minor allele frequency > 5% in the Asian population; (2) SNPs present in the promoter of XRCC2 gene such as G4234C (rs3218384), G4088T (rs3218373) G3063A (rs2040639); (3) SNPs present in exonic region of XRCC2 gene such as R188H (rs3218536); (4) SNPs present in intronic region of XRCC2 gene such as (rs7802034).
Genotyping
Genotyping was performed by Allele- specific polymerase chain reaction (ARMS-PCR). Primers for PCR amplification were designed by WASP (web based allele specific primer designing tool)26. Primers specific for each polymorphism are given in Supplemetary Table S1. PCR reaction was carried out in a reaction volume of 10 µl containing 50–100 ng genomic DNA, 100 µ M of each primers and Solis BioDyne master mix. Thermal cycling protocol used was: 94 °C for 30 sec, optimized annealing temperature for 45 sec, 72 °C for 1 min and final extension for 7 minutes. PCR products were visualized on a 2% agarose gel electrophoresis (100 V, 300 A for 45 min). Products by the presence or absence of bands specific for wild or mutant primers, in each well, were evaluated using UV trans illuminator (Gel Doc BioRad, USA). Internal control β-Actin was used in each reaction as a positive control for PCR. PCR products of thirty patients with homozygous wild, homozygous mutant and heterozygous mutant genotype were further confirmed by sequence analysis. Thirty control (normal) PCR product with different genotypic patterns were also sequenced along with cancerous samples to compare the sequencing results. DNA sequencing was carried out by MC lab (USA) by automated fluorescent sequencing to verify both nucleotide sequence and presence of specific SNPs. Results of DNA sequencing were analyzed using BioEdit software version7.0.5.
Statistical analyses
Statistical analysis was performed using GraphPad prism software v 6.0. The chi-squared test and one sample t-test was performed to assess difference of collected data of age, gender, family history, smoking status, histological type of HNC and different treatment modalities for HNC between the control and case group. Hardy-Weinberg equilibrium test was used to compare the actual genotypes with the expected number based on Hardy-Weinberg equilibrium theory (p = allele frequency, q = 1-p, p2 + q2 = 1) in controls. The difference in allele frequencies and genotypes between the control and case group was analyzed by Chi-squared tests. Logistic regression, with the adjustment of age and gender, was applied to calculate the odds ratios (ORs) and 95% confidence intervals (CIs). Three logistic regression models (additive, dominant, and recessive) were also used to analyze the SNPs. For SNP-SNP interactions, we used a adjusted logistic regression model to estimate the multiplicative interaction effect of the SNPs. P < 0.05 was considered to be statistically significant.
Haplotypes were generated from the genotyped data. The linkage disequilibrium (LD) and haplotype analysis were performed using Haploview 4.2, which uses the expectation maximization (EM) algorithm. Bonferroni correction was used to account for multiple testing and a two-tailed p value < 0.01 (=0.05/5 SNPs) was considered statistically significant.
Results
Case-Control Study
400 head and neck cancer patients and 400 control subjects were tested for five selected SNPs of XRCC2 gene (rs3218373, rs2040639, rs3218384, rs7802034 and rs3218536). Demographic data of these head and neck cancer patients and control individuals is given in Table 1.
Table 1.
Frequency distribution analysis of selected demographic and risk factors in head and neck cancer cases and controls.
| Variables | Cases (N = 400) | Controls (N = 400) | OR (95%CI) | P-value |
|---|---|---|---|---|
| Age (years) | ||||
| Median (Range) | 45 (17–68) | 45 (22–65) | ||
| Gender | ||||
| Males | 243 (60.7%) | 272 (68%) | ||
| Females | 157 (39.3%) | 128 (32%) | 0.06a | |
| Age | ||||
| ≤45 | 289 (72.3%) | 251 (62.7%) | ||
| >45 | 111 (27.7%) | 149 (37.3%) | 0.09a | |
| Family History of Cancer | ||||
| Yes | 153 (38.3%) | 21 (5.3%) | 11.17(6.89 to 18.1) | <0.0001b |
| No | 247 (61.7%) | 379 (94.7%) | ||
| Smoking History (cigarette, paan, bidi, betel quid, moist sniff) | ||||
| Smokers | 259 (64.7%) | 227 (56.7%) | 1.39 (1.05 to 1.86) | 0.02b |
| Non-Smokers | 141 (35.3%) | 173 (43.3%) | ||
| Types of head and neck cancer | ||||
| Oral Cavity | 182(45.5%) | — | ||
| Nasal Cavity | 88 (22%) | — | 0.03a | |
| Pharynx | 78 (19.5%) | — | ||
| Larynx | 52 (13%) | — | ||
| Types of treatment | ||||
| Radiotherapy | 88 (22%) | — | ||
| Chemotherapy | 94 (23.5%) | — | 0.09a | |
| Surgery | 218 (54.5%) | — | ||
Abbreviations: N, number of samples; OR, odds ratio (crude); CI, confidence interval; level of significance p-value ≤ 0.05 calculated by one samples t-testa and χ² -testb.
Distribution of the XRCC2 SNPs in head and neck cancer
The distribution of the genotypes and the allele frequencies of all of the studied polymorphisms are shown in Table 2. A significant association was observed between XRCC2 and head and neck cancer. In case of first selected SNP (rs3218373) of XRCC2 gene, frequency of heterozygous mutant (TG) and homozygous mutant (GG) genotypes was observed significantly higher in HNC patients compared to healthy controls (OR: 2.14, 95% CI: 1.47 to 3.10, P = 0.0001; OR: 2.58, 95% CI: 1.77 to 3.74, P < 0.0001 respectively). Frequency of G allele of respective polymorphism (rs3218373) was also found statistically higher in the patient group (OR: 2.73, 95% CI: 2.18 to 3.4, P < 0.0001). Genotyping of second selected SNP (rs2040639) of XRCC2 showed that the frequency of G allele of respective SNP was significantly higher in the control group (OR: 0.64, 95% CI: 0.49 to 0.81, P = 0.0003) compared to patients as shown in Table 1.
Table 2.
Distribution of five selected SNPs in XRCC2 gene in head and neck cancer.
| Genotype/Allele | Cases n (%) | Controls n (%) | OR (95% CI) | P- value |
|---|---|---|---|---|
| rs3218373 | ||||
| TT | 197 (49.2%) | 299 (74.7%) | 1 | 1 |
| TG | 97 (24.3%) | 52 (13%) | 2.14 (1.47 to 3.10) | P < 0.0001 |
| GG | 106 (26.5%) | 49 (12.3%) | 2.58 (1.77 to 3.74) | P < 0.0001 |
| T allele frequency | 491 (61.3%) | 650 (8%) | 1 | 1 |
| G allele frequency | 309 (38.7%) | 150 (19%) | 2.73 (2.18 to 3.4) | P < 0.0001 |
| rs2040639 | ||||
| AA | 301 (75.3%) | 259 (64.7%) | 1 | 1 |
| AG | 60 (15%) | 85 (21.3%) | 0.65 (0.45 to 0.94) | P = 0.0223 |
| GG | 39 (9.7%) | 56 (14%) | 0.66 (0.42 to 1.02) | P = 0.0645 |
| A allele frequency | 662 (82.7%) | 603 (75%) | 1 | 1 |
| G allele frequency | 138 (17.3%) | 197 (25%) | 0.64(0.49–0.81) | P = 0.0003 |
| rs3218384 | ||||
| GG | 331 (82.7%) | 269 (67.3%) | 1 | 1 |
| GC | 48 (12%) | 79 (19.7%) | 0.55 (0.37 to 0.81) | P = 0.003 |
| CC | 21 (5.3%) | 52 (13%) | 0.37 (0.21 to 0.62) | P = 0.0002 |
| G allele frequency | 710 (88.7%) | 617 (77%) | 1 | 1 |
| C allele frequency | 90 (11.3%) | 183 (23%) | 0.42 (0.32 to 0.56) | P < 0.0001 |
| rs7802034 | ||||
| AA | 232 (58%) | 199 (49.7%) | 1 | 1 |
| AG | 101 (25.3%) | 170 (42.5%) | 0.45 (0.33 to 0.61) | P < 0.0001 |
| GG | 67 (16.7%) | 31 (7.8%) | 2.39 (1.52 to 3.75) | P < 0.0001 |
| A allele frequency | 565 (70.6%) | 568 (71%) | 1 | 1 |
| G allele frequency | 235 (29.4%) | 232 (29%) | 0.98 (0.79 to 1.22) | P = 0.699 |
| rs3218536 | ||||
| GG | 240 (60%) | 336 (84%) | 1 | 1 |
| GA | 64 (16%) | 26 (6.5%) | 2.73 (1.69 to 4.42) | P < 0.0001 |
| AA | 96 (24%) | 38 (9.5%) | 3.00 (2.00 to 4.51) | P < 0.0001 |
| G allele frequency | 544 (68%) | 698 (87%) | 1 | 1 |
| A allele frequency | 256 (32%) | 102 (13%) | 3.22 (2.49 to 4.16) | P < 0.0001 |
Abbreviations: n, Number of samples; OR, odds ratio; CI, confidence interval; p-value ≤ 0.05 considered as statistically significant. P-values in bold have still maintained their significance after Bonferroni correction (0.05/25 = 0.002). ORs were adjusted for age, sex and smoking status of cancer in logistic regression model.
Genotyping of third selected SNP (rs3218384) of XRCC21 revealed that the frequencies of heterozygous mutant (GC) (OR: 0.55, 95% CI: 0.37 to 0.81, P = 0.003) and homozygous mutant (CC) (OR: 0.37, 95% CI: 0.21 to 0.62, P = 0.0002) genotype along with C allele frequency (OR: 0.42, 95% CI: 0.32 to 0.56, P < 0.0001) were significantly higher in the control group compared to patients. In case of fourth selected SNP (rs7802034), frequency of heterozygous mutant (AG) genotype was significantly higher in control group compared to patient group (OR: 0.45, 95% CI: 0.33 to 0.61, P < 0.0001). However, the frequency of homozygous mutant (GG) genotype was statistically significantly higher in patients compared to control group (OR: 2.39, 95% CI: 1.52 to 3.75, P < 0.0001). Genotyping of fifth SNP (rs3218536) showed that frequency of heterozygous mutant (GA) (OR: 2.73, 95% CI: 1.69 to 4.42, P < 0.0001) and homozygous mutant (AA) genotypes (OR: 3.00, 95% CI: 2.00 to 4.51, P < 0.0001) was significantly higher in patients compared with the controls. Moreover, the frequency of the A allele of respective SNP (rs3218536) was also statistically higher in the patients (OR: 3.22, 95% CI: 2.49 to 4.19, P < 0.0001) as shown in Table 1. Additionally hospital specific analysis (i.e. cases from Hospital 1 only; and then cases from Hospital 2 only) was also performed and no significant difference in frequency of selected polymorphisms was observed in patients from hospital 1 compared to patients from hospital 2 (Supplementary Table S2).
In genetic association studies, statistical power to detect disease susceptibility loci depended on the genetic models tested. Therefore, the genotype frequencies were further analyzed by three genetic models: additive, dominant, and recessive model. For rs3218373, a significant association between this polymorphism and increased HNC risk was found in dominant model (OR = 3.05, 95% CI = 2.26–4.11, P < 0.0001), recessive model (OR = 2.58, 95% CI = 1.77–3.74, P < 0.0001) and additive model (OR = 2.73, 95% CI = 2.20–3.48, P < 0.0001). Similarly, an increased risk of HNC risk was also found in recessive model (OR = 2.39, 95% CI = 1.52–3.75, P = 0.0001) of polymorphism rs7802034. Moreover, significant positive correlations between rs3218536 and HNC risk were also identified in dominant (OR: 3.50, 95% CI = 2.51–4.88, P < 0.0001), recessive (OR: 3.00, 95% CI = 2.00–4.51, P < 0.0001) and additive model (OR: 3.22, 95% CI = 2.49–4.15, P < 0.0001) as shown in Table 3.
Table 3.
Analysis of the five selected SNPs based on three genetic models.
| XRCC2 Genotype/Allele | Model | OR (95% CI) | p- value |
|---|---|---|---|
| rs3218373 | |||
| TT vs TG + GG | Dominant | 3.05(2.26–4.11) | <0.0001 |
| G/G vs TT + TG | Recessive | 2.58(1.77–3.74) | <0.0001 |
| G vs T | Additive | 2.73 (2.20–3.48) | <0.0001 |
| rs2040639 | |||
| AA vs AG + GG | Dominant | 0.60(0.44–0.82) | 0.001 |
| GG vs AA + AG | Recessive | 0.66(0.42–1.02) | 0.06 |
| G vs A | Additive | 0.64(0.49–0.81) | 0.0003 |
| rs3218384 | |||
| GG vs GC + CC | Dominant | 0.42(0.31–5.97) | <0.0001 |
| CC vs GG + GC | Recessive | 0.37(0.21–0.62) | 0.0002 |
| C vs G | Additive | 0.43(0.32–0.56) | <0.0001 |
| rs7802034 | |||
| AA vs AG + GG | Dominant | 1.39(1.05–1.84) | 0.02 |
| GG vs AA + AG | Recessive | 2.39(1.52–3.75) | <0.0001 |
| G vs A | Additive | 1.02(0.82–1.26) | 0.87 |
| rs3218536 | |||
| GG vs GA + AA | Dominant | 3.50(2.51–4.88) | <0.0001 |
| AA vs GA + GG | Recessive | 3.00(2.00–4.51) | <0.0001 |
| A vs G | Additive | 3.22(2.49–4.15) | <0.0001 |
Abbreviations: OR, odds ratio; CI, confidence interval; p-value ≤ 0.05 considered as statistically significant. P-values in bold have still maintained their significance after Bonferroni correction (0.05/15 = 0.003). ORs were adjusted for age, sex and smoking status of study cohort in logistic regression model.
Association of the XRCC2 SNPs with different histological subtype and smoking status of HNC patients
Frequencies of selected polymorphisms were calculated in different histological subtypes of head and neck cancer such as oral, nasal, pharyngeal and laryngeal carcinoma. In case of oral carcinoma, mutant genotype of polymorphism rs3218373 (OR = 1.94; 95% CI; 1.24–3.04; P = 0.003), rs2040639 (OR = 0.31; 95% CI; 0.16–0.59; P = 0.0004), rs3218384 (OR = 0.07; 95% CI; 0.01–0.30; P = 0.0003) and rs3218536 (OR = 1.93; 95% CI; 1.16–3.22; P = 0.01) were observed significantly associated with oral carcinoma. In case of nasal carcinoma, mutant genotype of polymorphisms rs3218373 (OR = 2.38; 95% CI; 1.35–4.2; P = 0.0002), rs7802034 (OR = 4.21; 95% CI; 2.31–7.6; p = 0.0001) and rs3218536 (OR = 4.44; 95% CI; 2.54–7.7; P < 0.0001) were observed significantly associated with the said histological type. For pharyngeal carcinoma, mutant genotype of polymorphisms rs3218373 (OR = 2.47; 95% CI; 1.36–4.45; P = 0.002), rs7802034 (OR = 3.31; 95% CI; 1.73–6.35; P = 0.0003) and rs3218536 (OR = 2.65; 95% CI; 1.40–4.99; p = 0.002) were observed significantly associated with the pharyngeal carcinoma. For laryngeal carcinoma, mutant genotype of polymorphism rs3218373 (OR = 3.18; 95% CI = 1.64–6.16; P = 0.0006) and rs3218536 (OR = 5.04; 95% CI = 2.60–9.77; P < 0.0001) were observed significantly associated with said histological type of head and neck cancer, as shown in Table 4.
Table 4.
Distribution of genotypes and odds ratios (OR) for different histological subtypes of HNC patients
| Genotypes XRCC2 | Controls (n = 400) | n | Oral cavity (n = 182) OR (95% CI) | P- value | n | Nasal Cavity (n = 88) OR (95% CI) | P- value | n | Pharynx (n = 78) OR (95% CI) | P- value | n | Larynx (n = 52) OR (95% CI) | P- value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs3218373 | |||||||||||||
| TT | 299 | 104 | 1 | 30 | 1 | 41 | 1 | 22 | 1 | ||||
| TG | 52 | 30 | 0.94(0.57–1.54) | 0.80 | 36 | 4.63(2.76–7.7) | <0.0001 | 17 | 1.86(1.01–3.43) | 0.04 | 14 | 2.46(1.25–4.85) | 0.009 |
| GG | 49 | 48 | 1.94(1.24–3.04) | 0.003 | 22 | 2.38(1.35–4.2) | 0.0002 | 20 | 2.47(1.36–4.45) | 0.002 | 16 | 3.18(1.64–6.16) | 0.0006 |
| rs2040639 | |||||||||||||
| AA | 259 | 149 | 1 | 60 | 1 | 60 | 1 | 32 | 1 | ||||
| AG | 85 | 20 | 0.34(0.20–0.58) | <0.0001 | 16 | 0.82(0.45–1.4) | 0.52 | 11 | 0.60(0.30–1.20) | 0.15 | 13 | 1.23(0.63–2.41) | 0.53 |
| GG | 56 | 13 | 0.31(0.16–0.59) | 0.0004 | 12 | 0.96(0.49–1.8) | 0.92 | 7 | 0.6090.26–1.38) | 0.23 | 7 | 0.96(0.41–2.22) | 0.91 |
| rs3218384 | |||||||||||||
| GG | 269 | 159 | 1 | 65 | 1 | 63 | 1 | 44 | 1 | ||||
| GC | 79 | 21 | 0.39(0.23–0.66) | 0.0005 | 13 | 0.70(0.37–1.3) | 0.28 | 9 | 0.53(0.25–1.10) | 0.09 | 5 | 0.43(1.16–1.12) | 0.08 |
| CC | 52 | 2 | 0.07(0.01–0.30) | 0.0003 | 10 | 0.85(0.41–1.7) | 0.67 | 6 | 0.55(0.23–1.34) | 0.19 | 3 | ||
| rs7802034 | |||||||||||||
| AA | 199 | 126 | 1 | 45 | 1 | 42 | 1 | 19 | 1 | ||||
| AG | 170 | 37 | 0.04(0.01–0.19) | <0.0001 | 20 | 0.39(0.23–0.6) | 0.0008 | 19 | 0.43(0.25–0.75) | 0.003 | 25 | 0.40(1.12–1.36) | 0.14 |
| GG | 31 | 19 | 0.89(0.48–1.63) | 0.70 | 23 | 4.21(2.31–7.6) | <0.0001 | 17 | 3.31(1.73–6.35) | 0.0003 | 8 | 1.25(0.70–2.23) | 0.44 |
| rs3218536 | |||||||||||||
| GG | 336 | 129 | 1 | 44 | 1 | 48 | 1 | 19 | 1 | ||||
| GA | 26 | 20 | 1.13(0.61–2.09) | 0.69 | 16 | 3.19(1.63–6.2) | 0.0007 | 13 | 2.87(1.40–5.88) | 0.003 | 15 | 5.83(2.83–11.9) | <0.0001 |
| AA | 38 | 33 | 1.93(1.16–3.22) | 0.01 | 28 | 4.44(2.54–7.7) | <0.0001 | 17 | 2.65(1.40–4.99) | 0.002 | 18 | 5.04(2.60–9.77) | <0.0001 |
OR, odds ratio; CI, confidence interval; p-value ≤ 0.05 considered as statistically significant. ORs were adjusted for age, sex and smoking status of study cohort in logistic regression model.
In case of genotype frequency correlation and smoking status of head and neck cancer, logistic regression model analysis was conducted using SNPs genotypes as a dependent variable and demographic parameters such as age, sex and smoking status as independent variable (Table 5). The results showed that smoking risk factor was associated with increased frequency of mutant genotype of rs3218373 (OR = 2.33; 95% CI = 1.003–5.42; P < 0.04) and rs3218536 (OR = 4.39; 95% CI = 1.007–19.10; P < 0.04) in head and neck cancer as shown in Table 5.
Table 5.
Association of selected polymorphisms of XRCC2 gene with smoking status.
| Polymorphisms | B | S.E | Wald | Sig | OR | 95% CI |
|---|---|---|---|---|---|---|
| rs3218373 | 0.826 | 0.428 | 3.720 | 0.04 | 2.33 | 1.003–5.42 |
| rs2040639 | −0.264 | 0.428 | 0.38 | 0.53 | 0.768 | 0.332–1.77 |
| rs3218384 | 0.248 | 0.527 | 0.191 | 0.62 | 1.281 | 0.42–3.893 |
| rs7802034 | −0.606 | 0.562 | 1.164 | 0.281 | 0.545 | 0.181–1.641 |
| rs3218536 | 1.478 | 0.751 | 3.877 | 0.04 | 4.386 | 1.007–19.10 |
Abbreviations: OR, odds ratio; CI, confidence interval; p-value ≤ 0.05 considered as statistically significant. ORs were adjusted for age and sex status of study cohort in logistic regression model.
Haplotype analysis of the XRCC2 SNPs
It was also investigated whether the five SNPs were in linkage disequilibrium. Any common haplotypes associated with the disease and rare haplotypes (with frequency < 5%) were excluded from the association analysis. The most common haplotypes of the five polymorphisms, calculated by Haploview 4.2, are summarized in Table 6.
Table 6.
The distribution of XRCC2 haplotypes in HNC patients and controls.
| rs3218373 | XRCC2 haplotypes (SNPs) | rs3218536 | Frequency | x² | P-value | |||
|---|---|---|---|---|---|---|---|---|
| rs2040639 | rs3218384 | rs7802034 | Cases | Controls | ||||
| G | A | C | A | G | 0.004 | 0.000 | — | — |
| G | A | C | A | G | 0.000 | 0.001 | — | — |
| G | A | C | G | G | 0.000 | 0.016 | 12.9 | 0.00 |
| G | A | G | A | A | 0.060 | 0.011 | 27.4 | 1.6e–007 |
| G | A | G | A | G | 0.139 | 0.048 | 39.9 | 2.7e–010 |
| G | A | G | G | G | 0.054 | 0.030 | 5.88 | 0.015 |
| G | G | C | A | A | 0.009 | 0.002 | — | — |
| G | G | C | A | G | 0.019 | 0.010 | 2.31 | 0.128 |
| G | G | C | G | A | 0.010 | 0.002 | 3.85 | 0.049 |
| G | G | C | G | G | 0.014 | 0.010 | 0.63 | 0.424 |
| G | G | G | A | G | 0.016 | 0.026 | 1.88 | 0.169 |
| G | G | G | G | G | 0.008 | 0.027 | 8.27 | 0.004 |
| T | A | C | A | A | 0.003 | 0.007 | — | — |
| T | A | C | A | G | 0.000 | 0.088 | 73.1 | 1.2e–017 |
| T | A | C | G | A | 0.000 | 0.006 | — | — |
| T | A | C | G | G | 0.000 | 0.057 | 46.9 | 7.6e–012 |
| T | A | G | A | A | 0.102 | 0.072 | 4.56 | 0.032 |
| T | A | G | A | G | 0.288 | 0.332 | 3.35 | 0.066 |
| T | A | G | G | A | 0.054 | 0.005 | 33.3 | 8.1e–009 |
| T | A | G | G | G | 0.087 | 0.081 | 0.22 | 0.637 |
| T | G | C | A | A | 0.014 | 0.002 | 6.18 | 0.012 |
| T | G | C | A | G | 0.025 | 0.009 | 5.83 | 0.015 |
| T | G | C | G | A | 0.011 | 0.002 | 4.38 | 0.036 |
| T | G | C | G | G | 0.003 | 0.010 | 3.53 | 0.060 |
| T | G | G | A | A | 0.000 | 0.012 | 8.02 | 0.004 |
| T | G | G | A | G | 0.010 | 0.095 | 57.2 | 4.0e–014 |
| T | G | G | G | G | 0.012 | 0.035 | 9.91 | 0.001 |
| G | A | G | G | A | 0.035 | 0.000 | 29.0 | 7.2e–008 |
| G | G | G | A | A | 0.016 | 0.000 | 13.3 | 0.000 |
| T | G | G | G | A | 0.005 | 0.000 | — | — |
Abbreviations: SNP, single nucleotide polymorphism; OR, odds ratio; CI, confidence interval; *p-value ≤ 0.05 considered statistical significant. P values in bold have still maintained their significance after Bonferroni correction.
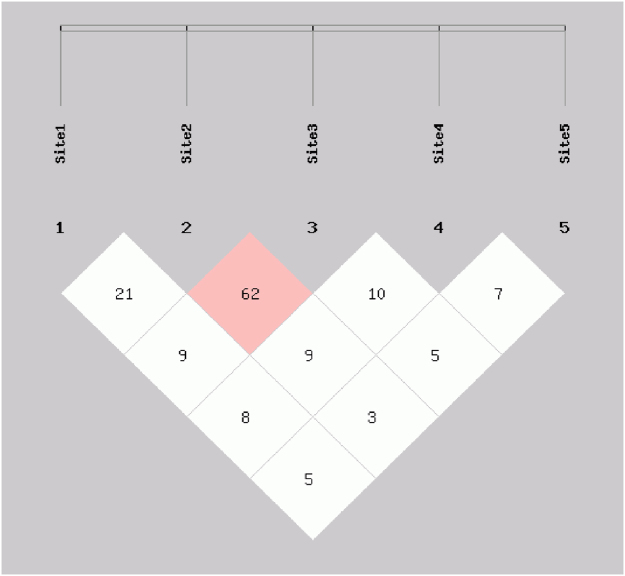
The haplotypes were generated using the five XRCC2 intragenic SNPs (rs3218373, rs2040639, rs3218384, rs7802034 and rs3218536) among the HNC cases and controls, and thirty different haplotypes were generated which accounted for most of the haplotypes in the cancer patients and control groups (with frequency < 5%). For commonly observed haplotypes, GAGAA haplotype (P = 1.6e-007), GAGAG haplotype (P = 2.7e-010), GAGGG haplotype (P = 0.02), GGCGA haplotype (P = 0.04), TAGAA haplotype (P = 0.03), TAGGA haplotype (P = 8.10e-009), TGCAA haplotype (P = 0.01), TGCAG haplotype (P = 0.01) and TGCGA haplotype (P = 0.03) were found linked with significant increase in head and neck cancer risk while GGGGG (P = 0.0004), TGGAA (P = 0.004), TGGAG (P = 4.06e-014) and TGGGG (P = 0.001) were observed associated with a significant reduction in head and cancer risk. The other seventeen common haplotypes including GACAG, GACGA, GACGG, GGCAA, GGCAG, GGCGG, GGGAG, TACAA, TACAG, TACGA, TACGG, TAGAG, TAGGG,TGCGG, GAGGA, GGGAA and TGGGA were observed not associated with the risk of head and neck cancer as shown in Table 6. Since, this study was based on a relatively small sample size, we applied a Bonferroni correction to decrease the type I error. Following the Bonferroni correction the haplotypes GAGAA, GAGAG, TACAG, TACGG, TAGGA, TGGAG and GAGGA still maintained their significance. Furthermore, two of the SNPs in XRCC2 (rs2040639 and rs3218384) were in strong LD (Fig. 1).
Figure 1.
Pairwise linkage disequilibrium plot for examined XRCC2 SNPs. Site 1 is for rs3218373, site 2 is for rs2040639, site 3 for rs3218384, site 4 for rs7802034 and site 5 for rs3218536. The darker region shows higher r2-value.
Combined genotype analysis of XRCC2 SNPs
Table 7 summarizes the association studies among the combined genotypes of the four SNPs and overall risk for head and neck cancer using conditional logistic regression model. The analysis revealed that rs3218373 and rs2040639 had a positive correlation with increased risk of HNC (OR = 2.89; 95% CI = 0.12–0.72; P < 0.001). A significant positive correlation was also observed between rs3218373 vs rs3218384 (OR = 2.44; 95% CI = 0.99–6.033; P < 0.05) and between rs2040639 vs rs3218536 (OR = 2.51; 95% CI = 0.99–6.031; P < 0.05) with increased risk of HNC as shown in Table 7.
Table 7.
Logistic regression model of SNP-SNP interactions and HNC risk.
| SNP-SNP interactions | B | S.E | Wald | Sig | OR | 95% CI |
|---|---|---|---|---|---|---|
| rs3218373 vs rs2040639 | 1.241 | 0.462 | 7.216 | 0.001 | 2.89 | 0.117–0.715 |
| rs3218373 vs rs3218384 | 0.895 | 0.460 | 3.775 | 0.05 | 2.447 | 0.992–6.033 |
| rs3218373 vs rs7802034 | −0.199 | 0.455 | 0.192 | 0.66 | 0.819 | 0.336–1.998 |
| rs3218373 vs rs3218536 | 0.629 | 0.463 | 1.841 | 0.175 | 1.875 | 0.756–4.648 |
| rs2040639 vs rs3218384 | 0.488 | 0.475 | 1.053 | 0.305 | 1.628 | 0.642–4.133 |
| rs2040639 vs rs7802034 | 0.414 | 0.448 | 0.852 | 0.386 | 1.512 | 0.628–3.642 |
| rs2040639 vs rs3218536 | 0.919 | 0.470 | 3.822 | 0.05 | 2.508 | 0.998–6.305 |
| rs3218384 vs rs7802034 | 0.225 | 0.458 | 0.242 | 0.623 | 1.252 | 0.510–3.073 |
| rs3218384 vs rs3218536 | −0.470 | 0.463 | 1.033 | 0.310 | 0.625 | 0.252–1.548 |
| rs7802034 vs rs3218536 | 0.699 | 0.452 | 2.393 | 0.122 | 2.013 | 0.830–4.882 |
Abbreviations: OR, odds ratio; CI, confidence interval; p-value ≤ 0.05 considered as statistically significant. ORs were adjusted for age, sex and smoking status of study cohort in logistic regression model.
Discussion
Many earlier studies have reported that the genes involved in DNA repair and in the maintenance of genome integrity plays a crucial role in protection against mutations. Although single nucleotide polymorphisms have been identified in these DNA repair genes, such as XRCC2, but the influence of specific genetic variants on repair phenotype and cancer risk has not yet been identified27–31. Thus, an attempt was undertaken in this study to determine whether single nucleotide polymorphisms (SNPs) in XRCC2 gene are associated with head and neck cancer. In this study, we successfully genotyped a total of five SNPs in different regions of XRCC2 gene such as promoter region, exonic region and intronic region and examined their possible association with head and neck cancer risk. We also investigated that whether or not these five polymorphisms are in linkage disequilibrium and common haplotypes of these SNPs are associated with head and neck carcinogenesis. Finally, we estimated the association among the combined genotypes of five selected SNPs and overall risk of head and neck cancer.
The majority of earlier studies on cancer susceptibility have focused only on XRCC2 rs3218536 SNP. In present study five polymorphisms, present in different regions of XRCC2 gene, such as promoter region, exonic region and intronic region were screened in HNC cancer patients and controls. Among selected promoter polymorphisms, frequency of risk allele of first promoter polymorphism rs3218373 was observed significantly higher in patients compared to controls. However, in the case of other two promoter polymorphisms (rs2040639 and rs3218384), frequency of risk allele was observed significantly higher in controls compared to patients. Similar results have earlier been reported where significant association was observed between XRCC2 promoter polymorphism and oral cancer risk32, breast cancer risk33, thyroid cancer risk34 and bladder cancer risk35. Although the functional consequences of these polymorphisms are unknown, their location in important domain(s) of XRCC2 may control translation and mRNA decay and are also sites for RNA interference34.
In this study, fourth selected polymorphism of XRCC2 gene, rs7802034, was located in the non-coding region and mutant allele frequency was observed associated with increased risk of HNC. Even though intronic SNPs are unlikely to have a direct functional role, still several studies have provided evidence that SNPs located in non-coding DNA, especially in intronic gene regions near the exon/intron boundaries, can inactivate pre-mRNA splice sites consequently affecting gene expression36,37, or can activate cryptic splice sites leading to exonization38. Furthermore, the presence of SNPs in 3‘-UTR of selected genes can modify the binding with specific microRNAs (miRNAs)39. For fifth selected polymorphism of XRCC2 gene, rs3218536, frequency of risk allele was observed significantly higher in patients compared to controls. Similar results have earlier been reported where significant association was observed between XRCC2 polymorphism rs3218536 with laryngeal and pharyngeal cancer risk40, thyroid cancer risk41, ovarian cancer42, gastric cancer risk43, oral cancer44 and head and neck cancer45. Nevertheless, some of the studies have also reported that there is no significant association between XRCC2- rs3218536 polymorphism and thyroid cancer46,47. In some of the studies rs3218536 polymorphism has been considered a genetic adjuster for ovarian and colorectal cancer patients48,49. The influence of these specific genetic variants on repair phenotype and cancer risk is yet not clear. However, amino acid 188 is conserved in humans, mice and rat XRCC2 proteins as well as human RAD51C, suggesting a potential functional role in DNA repair activity50. Romanowicz-Makowska et al., (2016) reported that rs3218536 polymorphism has shown a functional significance and may be responsible for a low DNA repair capacity phenotype characteristic of cancer patients including larynx carcinoma31. Future determination of functional and active sites in human XRCC2 protein may clarify the biological importance of this amino acid residue.
It is believed that haplotype analysis can provide more information than single-locus analysis19. In second step of study, we successfully established haplotypes for the XRCC2 gene from different combinations of five SNPs. GAGAA, GAGAG, GAGGG, GGCGA, TAGAA, TAGGA, TGCAA, TGCAG and TGCGA haplotypes of selected polymorphism were observed linked with significant increase in head and neck cancer risk. These haplotypes possess risky alleles which are consistently over represented in head and neck cancer patients relative to healthy controls, suggesting a leading role of selected polymorphisms in risk determination. To produce more information, linkage disequilibrium was calculated for five SNPs of XRCC2 gene and rs2040639 was found in complete linkage disequilibrium with rs3218384. It is possible that SNPs of this gene may have a collective effect on DNA repair outcomes. It has earlier been reported that interaction of polymorphisms, of the same gene or other genes, by linkage disequilibrium may be important in modulating the overall repair activity. This might explain the influence of genetic variations in the carcinogenic process51.
After this step, SNP-SNP interaction was calculated which showed that rs3218373-rs2040639 and rs2040639-rs3218536 combinations were associated with an increased head and neck cancer risk. Similar results have earlier been reported where it was suggested that mutations in XRCC2 gene may contribute to decreased or lost DNA repair capacity12,46. Furthermore, SNPs of XRCC2 may also increase the risk of several types of cancer including thyroid, brain and breast cancer52,53. In previously reported studies, cumulative meta-analyses have suggested no such significant association. Theoretically, genetic variants in XRCC2 gene can change the regular function of this gene, disturb the DNA repair and subsequently increase the cancer risk54. Nevertheless, some of the previous studies have reported that the variant alleles of this polymorphism can increase resistance to DNA damage induced by cisplatin50,55 which enlightens protective function of this polymorphism under certain conditions.
In conclusion, current evidence suggest that analyzed XRCC2 polymorphisms are directly associated with HNC risk. Nevertheless, several potential limitations of this study need to be considered before making a final conclusion. Firstly, studies should incorporate a larger sample size with various ethnic groups to further confirm the association between SNPs of XRCC2 and head and neck susceptibility. Secondly, HNC is a polygenic disease, therefore other genetic and environmental factors should also be assessed. Thirdly, few well-known risk factors for HNC such as human papilloma virus (HPV) has been discovered. In such cases correlation between sexual behavior of the subjects and head and neck carcinogenesis should be considered. Fourthly, subjects in this case-control study came from two hospitals and this may cause selection bias that can have substantial impact on the overall conclusions. As a result, large-scale studies adjusting for a wide range of factors are recommended to validate these findings. In conclusion, our results indicate that these five polymorphisms of XRCC2 gene may be related to individual susceptibility to head and neck risk in Pakistani population.
Electronic supplementary material
Acknowledgements
Authors acknowledge financial and infrastructural help from Higher Education Commission of Pakistan (HEC) and COMSATS Institute of Information Technology (CIIT), Islamabad. Authors are thankful to patients and staff of Nuclear Medicine Oncology & Radiotherapy Institute (NORI), Islamabad, for contribution in this research.
Author Contributions
All of the authors read and approved the final version of manuscript. S.S., R.S. and K.B. collected and isolated the DNA samples. S.S. performed the genotyping and drafted the manuscript. I.M. performed the genotyping studies, statistical analyses of the data and draft of the manuscript. M.A.K. supervised the project, helped to analyze the statistical data and provided critical revisions. All of the authors discussed the results and commented on the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Soma Saeed and Ishrat Mahjabeen have contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-13461-6.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Khanna KK, Jackson SP. DNA double-strand breaks: signaling, repair and the cancer connection. Nat. Genet. 2001;27:247–254. doi: 10.1038/85798. [DOI] [PubMed] [Google Scholar]
- 2.Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nat. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 3.Cerbinskaite A, Mukhopadhyay A, Plummer ER, Curtin NJ, Edmondson RJ. Defective homologous recombination in human cancers. Cancer. Treat. Rev. 2010;38(2):89–100. doi: 10.1016/j.ctrv.2011.04.015. [DOI] [PubMed] [Google Scholar]
- 4.West SC. Molecular views of recombination proteins and their control. Nat. Rev. Mol. Cell Biol. 2003;4:435–445. doi: 10.1038/nrm1127. [DOI] [PubMed] [Google Scholar]
- 5.Suwaki N, Klare K, Tarsounas M. RAD51 paralogs: Roles in DNA damage signalling, recombinational repair and tumorigenesis. Sem. Cell Dev. Biol. 2001;22(8):898–905. doi: 10.1016/j.semcdb.2011.07.019. [DOI] [PubMed] [Google Scholar]
- 6.Braybrooke JP, Spink KG, Thacker J, Hickson ID. The RAD51 family member, RAD51L3, is a DNA-stimulated ATPase that forms a complex with XRCC2. J. Biol. Chem. 2000;275:29100–29106. doi: 10.1074/jbc.M002075200. [DOI] [PubMed] [Google Scholar]
- 7.Griffin CS, Simpson PJ, Wilson CR, Thacker J. Mammalian recombination-repair genes XRCC2 and XRCC3 promote correct chromosome segregation. Nat. Cell Boil. 2000;2:757–761. doi: 10.1038/35036399. [DOI] [PubMed] [Google Scholar]
- 8.Johnson RD, Liu N, Jasin M. Mammalian XRCC2 promotes the repair of DNA double-strand breaks by homologous recombination. Nat. 1999;401:397–399. doi: 10.1038/43932. [DOI] [PubMed] [Google Scholar]
- 9.Clarkson SG, Wood RD. Polymorphisms in the human XPD (ERCC2) gene, DNA repair capacity and cancer susceptibility: an appraisal. DNA Repair (Amst) 2005;4:1068–1074. doi: 10.1016/j.dnarep.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 10.Jiao L, et al. XRCC2 and XRCC3 gene polymorphism and risk of pancreatic cancer. Am. J. Gastroenterol. 2008;103:360–7. doi: 10.1111/j.1572-0241.2007.01615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Benhamou S, et al. DNA repair gene XRCC2 and XRCC3 polymorphisms and susceptibility to cancers of the upper aerodigestive tract. Int. J. Cancer. 2004;112:901–4. doi: 10.1002/ijc.20474. [DOI] [PubMed] [Google Scholar]
- 12.Yen CY, et al. Combinational polymorphisms of four DNA repair genes XRCC1, XRCC2, XRCC3, and XRCC4 and their association with oral cancer in Taiwan. J. Oral. Pathol. Med. 2008;37:271–7. doi: 10.1111/j.1600-0714.2007.00608.x. [DOI] [PubMed] [Google Scholar]
- 13.Auranen A, et al. Polymorphisms in DNA repair genes and epithelial ovarian cancer risk. Int. J. Cancer. 2005;117:611–8. doi: 10.1002/ijc.21047. [DOI] [PubMed] [Google Scholar]
- 14.Beesley J, et al. Association between single nucleotide polymorphisms in hormone metabolism and DNA repair genes and epithelial ovarian cancer: results from two Australian studies and an additional validation set. Cancer Epidemiol. Biomarkers Prev. 2007;16:2557–65. doi: 10.1158/1055-9965.EPI-07-0542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pearce CL, et al. Validating genetic risk associations for ovarian cancer through the international Ovarian Cancer Association Consortium. Br. J. Cancer. 2009;100:412–20. doi: 10.1038/sj.bjc.6604820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han J, et al. Polymorphisms in DNA double-strand break repair genes and breast cancer risk in the Nurses’ Health Study. Carcinogen. 2004;25:189–95. doi: 10.1093/carcin/bgh002. [DOI] [PubMed] [Google Scholar]
- 17.Han J, et al. Interaction between genetic variations in DNA repair genes and plasma folate on breast cancer risk. Cancer Epidemiol. Biomarkers Prev. 2004;13:520–4. doi: 10.1158/1055-9965.EPI-03-0332. [DOI] [PubMed] [Google Scholar]
- 18.Kuschel B, et al. Variants in DNA double-strand break repair genes and breast cancer susceptibility. Hum. Mol. Genet. 2002;11:1399–407. doi: 10.1093/hmg/11.12.1399. [DOI] [PubMed] [Google Scholar]
- 19.Rafii S, et al. A potential role for the XRCC2 R188H polymorphic site in DNA-damage repair and breast cancer. Hum. Mol. Genet. 2002;11:1433–8. doi: 10.1093/hmg/11.12.1433. [DOI] [PubMed] [Google Scholar]
- 20.Commonly studied single-nucleotide polymorphisms and breast cancer: results from the Breast Cancer Association Consortium. J. Natl. Cancer Inst. 98, 1382–96 (2006). [DOI] [PubMed]
- 21.Garcia-Closas M, et al. Polymorphisms in DNA double-strand break repair genes and risk of breast cancer: two population-based studies in USA and Poland, and meta-analyses. Hum. Genet. 2006;119:376–88. doi: 10.1007/s00439-006-0135-z. [DOI] [PubMed] [Google Scholar]
- 22.Brooks J, et al. Polymorphisms in RAD51, XRCC2, and XRCC3 are not related to breast cancer risk. Cancer Epidemiol. Biomarkers Prev. 2008;17:1016–9. doi: 10.1158/1055-9965.EPI-08-0065. [DOI] [PubMed] [Google Scholar]
- 23.Loizidou MA, et al. Genetic polymorphisms in the DNA repair genes XRCC1, XRCC2 and XRCC3 and risk of breast cancer in Cyprus. Breast Cancer Res. Treat. 2009;115:623–7. doi: 10.1007/s10549-008-0084-4. [DOI] [PubMed] [Google Scholar]
- 24.Pooley KA, et al. Common single nucleotide polymorphisms in DNA double-strand break repair genes and breast cancer risk. Cancer Epidemiol. Biomarkers Prev. 2008;17:3482–9. doi: 10.1158/1055-9965.EPI-08-0594. [DOI] [PubMed] [Google Scholar]
- 25.Mahjabeen I, et al. Genetic variations in XRCC1 gene in sporadic head and neck cancer (HNC) patients. Pathology & Oncology Research. 2013;19(2):183–188. doi: 10.1007/s12253-012-9567-z. [DOI] [PubMed] [Google Scholar]
- 26.Liu J, et al. An improved allele-specific PCR primer design method for SNP marker analysis and its application. Plant Methods. 2012;8(1):34. doi: 10.1186/1746-4811-8-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang R, Wang W, Zhang JW. Correlation between XRCC2 and XRCC5 single nucleotide polymorphisms and drug-sensitivity of human lung cancer cells. Zhonghua. Yi. Xue. Za. Zhi. 2008;88:3059–3062. [PubMed] [Google Scholar]
- 28.Romanowicz-Makowska H, Smolarz B, Połać I, Sporny S. Single nucleotide polymorphisms of RAD51 G135C, XRCC2 Arg188His and XRCC3 Thr241Met homologous recombination repair genes and the risk of sporadic endometrial cancer in Polish women. J. Obstet. Gynaecol. Res. 2012;38:918–924. doi: 10.1111/j.1447-0756.2011.01811.x. [DOI] [PubMed] [Google Scholar]
- 29.Han J, Hankinson SE, Hunter DJ, De Vivo I. Genetic variations in XRCC2 and XRCC3 are not associated with endometrial cancer risk. Cancer Epidemiol. Biomarkers Prev. 2004;13:330–331. doi: 10.1158/1055-9965.EPI-03-0332. [DOI] [PubMed] [Google Scholar]
- 30.Lin WY, Camp NJ, Cannon-Albright LA. A role for XRCC2 gene polymorphisms in breast cancer risk and survival. J. Med. Genet. 2011;48:477–484. doi: 10.1136/jmedgenet-2011-100018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Romanowicz-Makowska H, et al. Polymorphism of the DNA repair genes RAD51 and XRCC2 in smoking- and drinking-related laryngeal cancer in a Polish population. Arch. Med. Sci. 2012;8(6):1065–1075. doi: 10.5114/aoms.2012.32417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang CH, et al. Single nucleotide polymorphism barcoding to evaluate oral cancer risk using odds ratio-based genetic algorithms. The Kaohsiung J. of Med. Sci. 2012;28(7):362–368. doi: 10.1016/j.kjms.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 33.Pelttari LM, et al. RAD51, XRCC3, and XRCC2 mutation screening in Finnish breast cancer families. SpringerPlus. 2015;4(1):92. doi: 10.1186/s40064-015-0880-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sarwar R, et al. Association of Promoter Polymorphisms in XRCC2 Gene Involved in DNA Double Strand Break Repair and Increased Susceptibility to Thyroid Cancer Risk in Pakistani Population. J Carcinog Mutagen. 2016;7:265. [Google Scholar]
- 35.Figueroa JD, et al. Evaluation of genetic variation in the double-strand break repair pathway and bladder cancer risk. Carcinogenesis. 2007;28(8):1788–1793. doi: 10.1093/carcin/bgm132. [DOI] [PubMed] [Google Scholar]
- 36.Lomelin D, Jorgenson E, Risch N. Human genetic variation recognizes functional elements in noncoding sequence. Genome Research. 2010;20(3):311–319. doi: 10.1101/gr.094151.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vogelstein B, et al. Cancer Genome Landscapes. Science. 2013;339(6127):1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang GS, Cooper TA. Splicing in disease: disruption of the splicing code and the decoding machinery. Nature Reviews Genetics. 2007;8(10):749–761. doi: 10.1038/nrg2164. [DOI] [PubMed] [Google Scholar]
- 39.Naccarati, A. et al. Polymorphisms in miRNA binding sites of nucleotide excision repair genes and colorectal cancer risk. Carcinogenesis. bgs172 (2012). [DOI] [PubMed]
- 40.He Y, et al. Impact of XRCC2 Arg188His polymorphism on cancer susceptibility: a meta-analysis. PLoS one. 2014;9(3):e91202. doi: 10.1371/journal.pone.0091202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yan L, et al. Association Studies between XRCC1, XRCC2, XRCC3 Polymorphisms and Differentiated Thyroid Carcinoma. Cellular Physiology and Biochem. 2016;38(3):1075–1084. doi: 10.1159/000443058. [DOI] [PubMed] [Google Scholar]
- 42.Yu X, et al. Association between ERCC2 rs13181 and XRCC2 rs3218536 polymorphisms and the risk of ovarian cancer: a meta-analysis. Int. J. Clin. Exp. Med. 2016;9(2):1558–1566. [Google Scholar]
- 43.Gok İ, et al. Polymorphisms in DNA repair genes XRCC2 and XRCC3 risk of gastric cancer in Turkey. Bosnian. J. of Basic Med. Sci. 2014;14(4):214–218. doi: 10.17305/bjbms.2014.4.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang CH, et al. A Systematic Gene–Gene and Gene–Environment Interaction Analysis of DNA Repair Genes XRCC1, XRCC2, XRCC3, XRCC4, and Oral Cancer Risk. Omics: A J. of Integrative Bio. 2015;19(4):238–247. doi: 10.1089/omi.2014.0121. [DOI] [PubMed] [Google Scholar]
- 45.Choudhury JH, Choudhury B, Kundu S, Ghosh SK. Combined effect of tobacco and DNA repair genes polymorphisms of XRCC1 and XRCC2 influence high risk of head and neck squamous cell carcinoma in northeast Indian population. Medical Onco. 2014;31(8):1–9. doi: 10.1007/s12032-014-0067-8. [DOI] [PubMed] [Google Scholar]
- 46.Bastos HN, et al. Association of polymorphisms in genes of the homologous recombination DNA repair pathway and thyroid cancer risk. Thyroid. 2009;19:1067–1075. doi: 10.1089/thy.2009.0099. [DOI] [PubMed] [Google Scholar]
- 47.Garcia-Quispes WA, et al. Association studies of OGG1, XRCC1, XRCC2 and XRCC3 polymorphisms with differentiated thyroid cancer. Mutat. Res. 2011;709-710:67–72. doi: 10.1016/j.mrfmmm.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 48.Curtin K, Lin WY, George R. Genetic variants in XRCC2: new insights into colorectal cancer tumorigenesis. Cancer Epidemiol. Biomarkers Prev. 2009;18:2476–2484. doi: 10.1158/1055-9965.EPI-09-0187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Krupa R, Sliwinski T, Wisniewska-Jarosinska M. Polymorphisms in RAD51, XRCC2 and XRCC3 genes of the homologous recombination repair in colorectal cancer - a case control study. Mol. Biol. Rep. 2011;38:2849–2854. doi: 10.1007/s11033-010-0430-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Danoy P, et al. A naturally occurring genetic variant of human XRCC2 (R188H) confers increased resistance to cisplatin-induced DNA damage. Biochem. Biophys. Res. Commun. 2007;352:763–768. doi: 10.1016/j.bbrc.2006.11.083. [DOI] [PubMed] [Google Scholar]
- 51.Kiyohara, C., Takayama, K. & Nakanishi, Y. Lung Cancer Risk and Genetic Polymorphisms in DNA Repair Pathways: AMeta-Analysis. J. of Nucleic Acids. 701760 (2010). [DOI] [PMC free article] [PubMed]
- 52.Cetinkunar S, et al. The effect of polymorphism in DNA repair genes RAD51 and XRCC2 in colorectal cancer in Turkish population. Int. J. Clin. Exp. Med. 2015;8:2649–2655. [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang Y, et al. The Arg188His polymorphism in the XRCC2 gene and the risk of cancer. Tumour Biol. 2014;35:3541–3549. doi: 10.1007/s13277-013-1468-6. [DOI] [PubMed] [Google Scholar]
- 54.He Y, et al. Impact of XRCC2 Arg188His Polymorphism on Cancer Susceptibility: A Meta-Analysis. PLoS. ONE. 2014;9:3. doi: 10.1371/journal.pone.0091202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Romanowicz H, Brys M, Forma E, Smolarz B. Lack of Association between the 4234G/C X-Ray Repair Cross-Complementing 2 (XRCC2) Gene Polymorphism and the Risk of Endometrial Cancer among Polish Population. J. Gynecol. Res. Obstet. 2016;2(1):047–050. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.