Abstract
Objective
The aim of this study was to reveal the exact changes during the occurrence of breast cancer to explore significant new and promising genes or factors related to this disease.
Methods
We compared the gene expression profiles of breast cancer tissues with its uninvolved normal breast tissues as controls using the cDNA microarray analysis in seven breast cancer patients. Further, one representative gene, named IFI30, was quantitatively analyzed by real-time PCR to confirm the result of the cDNA microarray analysis.
Results
A total of 427 genes were identified with significantly differential expression, 221 genes were up-regulated and 206 genes were down-regulated. And the result of cDNA microarray analysis was validated by detection of IFI30 mRNA level changes by real-time PCR. Genes for cell proliferation, cell cycle, cell division, mitosis, apoptosis, and immune response were enriched in the up-regulated genes, while genes for cell adhesion, proteolysis, and transport were significantly enriched in the down-regulated genes in breast cancer tissues compared with normal breast tissues by a gene ontology analysis.
Conclusion
Our present study revealed a range of differentially expressed genes between breast cancer tissues and normal breast tissues, and provide candidate genes for further study focusing on the pathogenesis and new biomarkers for breast cancer.
Keywords: Breast neoplasms, Candidate genes, Microarray
Introduction
Breast cancer is the most frequent cancer among women worldwide. As the latest WHO statistics show that there were 1.67 million new cases diagnosed in 2012, which was 25% of all cancers.1 And breast cancer is still the leading cause of cancer-related death in women in less developed countries, although preventive approaches and treatments have greatly improved in the past decades.2 The activation and overexpression of oncogenes, as well as the decreased expression or deletion of cancer suppressor genes, have been proven to play important roles in the development of breast cancer.3, 4, 5, 6 Although, a large number of studies on molecular genetics and molecular biology have revealed that multiple genes and factors take part in the pathogenesis of breast cancer, the exact mechanism is still unclear. Therefore, there is a huge need to identify new and promising genes or factors which can be used as biomarkers to predict the prognosis or sensitivity as well as the effect of treatments for breast cancer.
Currently, transcriptional profiling of gene expression achieved by cDNA microarrays has greatly increased our knowledge of the pathomechanisms involved in various types of cancers. This high-through put technique identifying a large number of promising genes has been successfully used in distinguishing subclasses of caner as well as predicting the clinical outcome and the response to particular treatments.7, 8 Thus, we choose to use the cDNA microarray technology to discover the exact changes during the occurrence of breast cancer to look for new promising genes or factors.
In the present study, we compared the gene expression profiles of breast cancer tissue with its uninvolved normal breast tissue as the control using the cDNA microarray analysis. We found that differences existed between the two groups in gene expression profiles. And a large number of genes were identified that were significantly changed during the occurrence of the disease. Furthermore, we confirmed the cDNA microarray results by detecting one selected gene's change of expression by the use of quantitative real-time PCR. Thus, the cDNA microarray analysis here provide plenty of candidate genes for further study focusing on the pathogenesis and biomarkers of breast cancer.
Material and methods
Ethics statement
This study was approved by the Institutional Review Board of the Second Hospital of Shandong University, and written informed consent was obtained from each patient.
Patients
The enrolled specimens were collected from seven women patients with pathologically confirmed invasive breast cancer in the Second Hospital of Shandong University from May 1, 2009 to Feb 28, 2010. All the patients enrolled followed strict eligibility and exclusion criteria as follows: eligibility criteria were female, histologically confirmed invasive breast ductal carcinoma without prior treatment before surgery, patients underwent modified radical mastectomy, having available breast cancer tissue samples as well as normal breast tissue samples, complete basic clinical and pathological information, and a signed informed consent. The exclusion criteria included prior invasive malignancy within five years, pregnancy, receiving neoadjuvant therapy for breast cancer, bilateral breast cancer, and having a family history of breast cancer. The general characteristics of the seven patients enrolled are shown in Table 1.
Table 1.
The general characteristics of the seven patients enrolled.
| No | Age | Pathological type | Histological grade | Tumor size | ER | PR | Her-2 | Ki67(%) |
|---|---|---|---|---|---|---|---|---|
| 1 | 55 | IDC | II | 1.5*0.5 | +++ | ± | − | 10 |
| 2 | 41 | IDC | II | 3.0*2.0 | + | +++ | − | 5 |
| 3 | 59 | IDC | II | 2.0*2.0 | +++ | +++ | − | 10 |
| 4 | 53 | IDC | II | 1.8*1.0 | +++ | +++ | − | 10 |
| 5 | 39 | IDC | II | 1.0*0.8 | +++ | ++ | − | 30 |
| 6 | 57 | IDC | II | 1.8*1.5 | +++ | +++ | − | 30 |
| 7 | 35 | IDC | II | 2.3*1.8 | +++ | +++ | − | 50 |
Clinical tissue sample collections
All tumor tissues and adjacent normal tissues were collected into liquid nitrogen within 30 min after the resection of the mammary tissue. Pathological review of hematoxylin and eosin-stained sections was used for evaluation of the histological characteristics of all specimens. All the adjacent normal breast tissues were obtained from breast cancer patients who underwent modified radical mastectomy, and the uninvolved parts referred to the region where more than 5 cm away from the tumor sites. All samples were reevaluated independently by two pathologists to confirm the diagnosis and to estimate the tumor cell content. All these enrolled cancer specimens contained at least 60 percent breast cancer cells, and all normal specimens were pathologically confirmed as normal ductal epithelium without atypical hyperplasia.
Gene expression microarray analyses
Total RNA was extracted from those frozen tissues with Trizol Reagent (Invitrogen, Gaithersburg, MD, USA) following the manufacturer's instructions. And the RNA concentration and purity were determined by A260 and A260/280 ratio through ultraviolet spectrophotometer (Nanodrop, ND-1000), and further checked by electrophoresis. The human long oligonucleotide microarray used in the present study was constructed by CapitalBio Corporation (Beijing, China). The microarray genechip consists of 5′-amino-modified 70-mer probes represented 35,035 well-characterized human genes. The P value for significance cutoff was 0.05. This experimental protocol has been described previously.9
Real-time RT-PCR assay
Total RNA was extracted by TRIzol according to the manufacturer's protocol. Total RNA was dissolved in nuclease-free water. All RNAs were verified for purity as well as concentration by NanoDrop analysis. After genomic DNA was digested by DNase I and the residual RNA was purified, all the RNA was reverse-transcribed in a final volume of 20 μl using Superscript II (Invitrogen, USA) according to the manufacturer's protocol. Quantitative analysis of IFI30 mRNA expression was performed in paired breast cancer tissues and normal breast tissues by using the 7900 HT Fast RealTime PCR system (Applied Biosystems, USA). IFI30 was amplified with the following primers: 5′-ATGTCACGCTGGTGCCCTAC-3′ (forward primer) and 5′-GTCAAGTTCATCCAACACGCA-3′ (reverse primer). ACTB was used as an endogenous control with the following primers: 5′-CATGTACGTTGCTATCCAGGC-3′ (forward primer) and 5′-CTCCTTAATGTCACGCACGAT-3′ (reverse primer). The results were evaluated by the comparative threshold cycle value method (2−ΔΔct). Each RT-qPCR experiment was repeated three times.
Results
Genes differentially expressed between breast cancer tissues and normal breast tissues
To identify candidate genes which may contribute to the pathogenesis of breast cancer, gene expression profiles of breast cancer tissues and normal breast tissues were comparatively analyzed by the cDNA microarray technique. A total of 427 genes were screened as differently expressed in breast cancer tissues compared with normal breast tissues at significant levels (P < 0.05). They included 221 up-regulated genes and 206 down-regulated genes. Among these differentially expressed genes, 36 genes were up-regulated >5-fold; such as PTTG1, CDC2, KIAA0101, DLG7, NUF2, S100P, TPX2, and IFI30.
Different gene expression profiles between breast cancer tissues and normal breast tissues
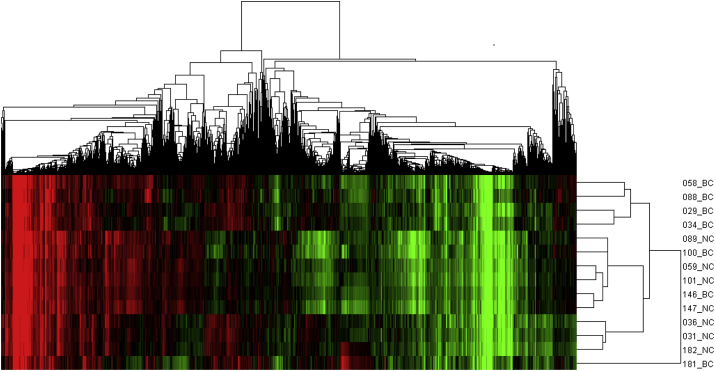
In order to clarify the different gene expression profiles between breast cancer tissues and normal breast tissues, we conducted cluster analysis of 14 tissue samples based on the detected genes. The result is shown in Fig. 1. The gene expression profiles of breast cancer samples were partly distinguished from the normal breast tissues, indicating that differences existed between the two groups.
Fig. 1.
Cluster analysis results for gene expression in breast cancer tissues and normal breast tissues. The expression values clustered in the red-shaded areas indicate up-regulation, and the green-shaded areas indicate down-regulation. The abbreviations in the figure were as follows: BC, for breast cancer tissues and NC, for normal breast tissues.
Functional classification of the differentially expressed genes
To explore candidate biological pathways which may contribute to the pathogenesis of breast cancer, we performed a hierarchical clustering followed by a gene ontology (GO) analysis on the differentially expressed genes. The GO analysis of the differentially expressed genes was based on molecular function as well as biological processes and revealed a number of distinct functional groups. Cell proliferation, cell cycle, cell division, mitosis, apoptosis, and immune response genes were enriched in the up-regulated genes, while cell adhesion, proteolysis, and transport genes were significantly enriched in the down-regulated genes in breast cancer tissues compared with normal breast tissues. Several representative and important biological processes, including a large number of related differentially expressed genes shown in Table 2, Table 3, Table 4, Table 5, are involved in immune function, cell proliferation, cell adhesion, and apoptosis.
Table 2.
Differential expressed genes related to immune function.
| Accession | Gene symbol | Description | Change | Fold change | P value |
|---|---|---|---|---|---|
| NM_006332 | IFI30 | Gamma-interferon-inducible lysosomal thiol reductase precursor | ↑ | 5.4016 | <0.001 |
| NM_130398 | EXO1 | Exonuclease 1 | ↑ | 3.3573 | <0.001 |
| NM_005101 | ISG15 | Interferon-induced 17 kDa protein precursor | ↑ | 7.4359 | <0.001 |
| NM_002351 | SH2D1A | SH2 domain-containing protein 1A | ↑ | 6.3158 | <0.001 |
| NM_005514 | HLA-B | HLA class I histocompatibility antigen | ↑ | 2.2504 | 0.0057 |
| NM_003467 | CXCR4 | C-X-C chemokine receptor type 4 | ↑ | 2.3894 | 0.0057 |
| NM_022873 | IFI6 | Interferon-induced protein 6-16 precursor | ↑ | 2.6298 | 0.0069 |
| NM_000593 | TAP1 | Antigen peptide transporter 1 | ↑ | 3.6752 | 0.0125 |
| NM_002117 | HLA-C | HLA class I histocompatibility antigen | ↑ | 2.0259 | 0.0167 |
| NM_004335 | BST2 | Bone marrow stromal antigen 2 precursor | ↑ | 4.3109 | 0.0200 |
| NM_015932 | POMP | Proteasome maturation protein | ↑ | 2.1079 | 0.0209 |
| NM_000569 | FCGR3A | Low affinity immunoglobulin gamma Fc | ↑ | 2.2588 | 0.0267 |
| NM_148170 | CTSC | Cathepsin C | ↑ | 2.7163 | 0.0267 |
| NM_006847 | LILRB4 | Leukocyte immunoglobulin-like receptor subfamily B member 4 precursor | ↑ | 2.2951 | 0.0301 |
| NM_005533 | IFI35 | Interferon-induced 35 kDa protein | ↑ | 3.5268 | 0.0328 |
| NM_004048 | B2M | Beta-2-microglobulin precursor | ↑ | 2.0588 | 0.0328 |
| NM_004031 | IRF7 | Interferon regulatory factor 7 | ↑ | 2.3360 | 0.0414 |
| NM_002996 | CX3CL1 | Fractalkine precursor | ↓ | 0.3260 | 0.0141 |
| NM_181524 | PIK3R1 | Phosphatidylinositol 3-kinase regulatory subunit alpha | ↓ | 0.4631 | 0.0167 |
| NM_199168 | CXCL12 | chemokine (C-X-C motif) ligand 12 | ↓ | 0.4500 | 0.0210 |
Table 3.
Differential expressed genes related to cell proliferation.
| Accession | Gene symbol | Description | Change | Fold change | P value |
|---|---|---|---|---|---|
| NM_005192 | CDKN3 | Cyclin-dependent kinase inhibitor 3 | ↑ | 5.1270 | <0.001 |
| NM_001311 | CRIP1 | Cysteine-rich protein | ↑ | 3.7188 | <0.001 |
| NM_012112 | TPX2 | Targeting protein for Xklp2 | ↑ | 9.6235 | <0.001 |
| NM_002351 | CKS2 | Cyclin-dependent kinases regulatory subunit 2 | ↑ | 5.0202 | <0.001 |
| NM_006845 | KIF2C | Kinesin-like protein KIF2C | ↑ | 3.9517 | <0.001 |
| NM_003467 | CXCR4 | C-X-C chemokine receptor type 4 | ↑ | 2.3894 | 0.0057 |
| NM_005030 | PLK1 | Serine/threonine-protein kinase PLK1 | ↑ | 2.6298 | 0.0069 |
| NM_021173 | POLD4 | DNA polymerase subunit delta 4 | ↑ | 2.1277 | 0.0110 |
| NM_003318 | TTK | Dual specificity protein kinase TTK | ↑ | 4.9328 | 0.0110 |
| NM_012346 | NUP62 | Nucleoporin 62 kDa | ↑ | 2.0221 | 0.0125 |
| NM_004336 | BUB1 | Mitotic checkpoint serine/threonine-protein kinase BUB1 | ↑ | 7.5387 | 0.0125 |
| NM_007045 | FGFR1OP | FGFR1 oncogene partner | ↑ | 2.4699 | 0.0125 |
| NM_033285 | TP53INP1 | Tumor protein p53-inducible nuclear protein 1 | ↑ | 2.2895 | 0.0125 |
| NM_016343 | CENPF | Centromere protein F | ↑ | 5.0906 | 0.0166 |
| NM_004335 | BST2 | Bone marrow stromal antigen 2 precursor | ↑ | 4.3109 | 0.0200 |
| NM_005727 | TSPAN1 | Tetraspanin-1 | ↑ | 3.3838 | 0.0210 |
| NM_003225 | TFF1 | Trefoil factor 1 precursor | ↑ | 6.4284 | 0.0210 |
| NM_004494 | HDGF | Hepatoma-derived growth facto | ↑ | 2.0478 | 0.0301 |
| NM_014750 | DLG7 | Discs large homolog 7 | ↑ | 9.9318 | 0.0327 |
| NM_003503 | CDC7 | Cell division cycle 7-related protein kinase | ↑ | 2.3667 | 0.0327 |
| NM_017548 | CDV3 | CDV3 homolog | ↑ | 2.3609 | 0.0414 |
| NM_004585 | RARRES3 | Retinoic acid receptor responder protein 3 | ↑ | 2.2753 | 0.0498 |
| NM_006533 | MIA | Melanoma-derived growth regulatory protein precursor | ↓ | 0.3528 | 0.0141 |
| NM_003888 | ALDH1A2 | Retinal dehydrogenase 2 | ↓ | 0.4265 | 0.0141 |
| NM_002178 | IGFBP6 | Insulin-like growth factor-binding protein 6 precursor | ↓ | 0.3603 | 0.0142 |
| NM_001145 | ANG | Ribonuclease 4 precursor | ↓ | 0.4966 | 0.0200 |
| NM_000612 | IGF2 | Insulin-like growth factor II precursor | ↓ | 0.4022 | 0.0210 |
| NM_005228 | EGFR | Epidermal growth factor receptor precursor | ↓ | 0.4726 | 0.0448 |
Table 4.
Differential expressed genes related to cell adhesion.
| Accession | Gene symbol | Description | Change | Fold change | P value |
|---|---|---|---|---|---|
| NM_002026 | FN1 | Fibronectin precursor | ↑ | 4.5627 | <0.001 |
| NM_130398 | LPXN | Leupaxin | ↑ | 2.1309 | 0.0210 |
| NM_005727 | TSPAN1 | Tetraspanin-1 | ↑ | 3.3838 | 0.0210 |
| NM_016639 | TNFRSF12A | Tumor necrosis factor receptor superfamily member 12A precursor | ↑ | 2.5041 | 0.0267 |
| NM_006288 | THY1 | Thy-1 membrane glycoprotein precursor | ↑ | 2.4001 | 0.0448 |
| NM_001670 | ARVCF | Armadillo repeat protein deleted in velo-cardio-facial syndrome | ↓ | 0.4794 | 0.0141 |
| NM_002996 | CX3CL1 | Fractalkine precursor | ↓ | 0.3260 | 0.0141 |
| NM_005168 | RND3 | Rho-related GTP-binding protein RhoE | ↓ | 0.3721 | 0.0142 |
| NM_006329 | FBLN5 | Fibulin-5 precursor | ↓ | 0.4262 | 0.0200 |
| NM_000228 | LAMB3 | Laminin subunit beta-3 precursor | ↓ | 0.4043 | 0.0200 |
| NM_199168 | CXCL12 | Stromal cell-derived factor 1 precursor | ↓ | 0.4500 | 0.0210 |
| NM_002404 | MFAP4 | Microfibril-associated glycoprotein 4 precursor | ↓ | 0.4093 | 0.0301 |
| NM_033254 | BOC | Brother of CDO precursor | ↓ | 0.4480 | 0.0301 |
| NM_001937 | DPT | Dermatopontin precursor | ↓ | 0.4719 | 0.0301 |
Table 5.
Differential expressed genes related to apoptosis.
| Accession | Gene symbol | Description | Change | Fold change | P value |
|---|---|---|---|---|---|
| NM_013258 | PYCARD | Apoptosis-associated speck-like protein containing a CARD | ↑ | 2.6746 | <0.001 |
| NM_004324 | BAX | BAX protein | ↑ | 2.1953 | <0.001 |
| NM_001908 | CTSB | Cathepsin B precursor | ↑ | 2.0506 | 0.0069 |
| NM_021127 | PMAIP1 | Phorbol-12-myristate-13-acetate-induced protein 1 | ↑ | 4.3945 | 0.0069 |
| NM_014397 | NEK6 | Serine/threonine-protein kinase Nek6 | ↑ | 2.0326 | 0.0069 |
| NM_001168 | BIRC5 | Baculoviral IAP repeat-containing protein 5 | ↑ | 6.7460 | 0.0069 |
| NM_002192 | INHBA | Inhibin beta A chain precursor | ↑ | 3.3210 | 0.0125 |
| NM_012346 | NUP62 | l-amino-acid oxidase precursor | ↑ | 2.0221 | 0.0125 |
| NM_033379 | CDC2 | Cell division control protein 2 homolog | ↑ | 13.5476 | 0.0125 |
| NM_033285 | TP53INP1 | Tumor protein p53-inducible nuclear protein 1 | ↑ | 2.2895 | 0.0125 |
| NM_002466 | MYBL2 | Myb-related protein B | ↑ | 7.5657 | 0.0142 |
| NM_022073 | EGLN3 | Egl nine homolog 3 | ↑ | 2.6077 | 0.0200 |
| NM_005356 | LCK | Proto-oncogene tyrosine-protein kinase LCK | ↑ | 4.8360 | 0.0210 |
| NM_016639 | TNFRSF12A | Tumor necrosis factor receptor superfamily member 12A precursor | ↑ | 2.5041 | 0.0267 |
| NM_006904 | PRKDC | DNA-dependent protein kinase catalytic subunit | ↑ | 2.0768 | 0.0301 |
| NM_000041 | APOE | Apolipoprotein E precursor | ↑ | 2.2342 | 0.0301 |
| NM_016640 | MRPS30 | Mitochondrial 28S ribosomal protein S30 | ↑ | 6.1910 | 0.0414 |
| NM_006288 | THY1 | Thy-1 membrane glycoprotein precursor | ↑ | 2.4001 | 0.0448 |
| NM_003012 | SFRP1 | Secreted frizzled-related protein 1 precursor | ↓ | 0.2201 | 0.0141 |
| NM_181524 | PIK3R1 | Phosphatidylinositol 3-kinase regulatory subunit alpha | ↓ | 0.4631 | 0.0166 |
| NM_016109 | ANGPTL4 | Angiopoietin-related protein 4 precursor | ↓ | 0.4228 | 0.0210 |
Validation of differentially expressed genes
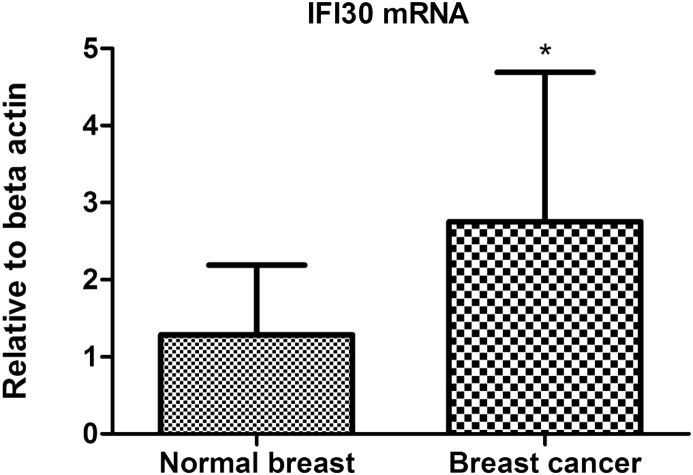
To confirm the result of the cDNA microarray analysis, one representative gene, IFI30, was quantitatively analyzed by real-time PCR. This gene was selected for three reasons as follows: 1) it was up-regulated with a high fold-change at a significant level, 2) it appeared in both functional classification and clustering analysis, 3) it presented a promising function by the current research findings in cancer pathogenesis as it is related to cancer immunity, in which we were interested. As shown in Fig. 2, real-time PCR results confirmed IFI30 mRNA was increased significantly in breast cancer tissues compared with the corresponding adjacent normal breast tissues (P = 0.009), which was in accordance with the result from cDNA microarray analysis.
Fig. 2.
Validation of the differentially expressed genes by IFI30 detected through quantitative real-time PCR. Relative means ± SD for IFI30 mRNA. IFI30 mRNA increased significantly in breast cancer tissues compared with the corresponding adjacent normal breast tissues (*P = 0.009), which was in accordance with the result from cDNA microarray analysis.
Discussion
In the present study, we have revealed differentially expressed genes in breast cancer compared with uninvolved normal control tissue by using the cDNA microarray analysis of the gene expression profiles. A total of 427 genes were identified with significantly differential expression, 221 genes were up-regulated and 206 genes were down-regulated. Multiple biological processes were found by GO analysis in which these differentially expressed genes took part in. Cell proliferation, cell cycle, cell division, mitosis, apoptosis, and immune response genes were enriched in the up-regulated genes, while cell adhesion, proteolysis, and transport genes were significantly enriched in the down-regulated genes in breast cancer tissues compared with normal breast tissues.
Cell proliferation, cell cycle, cell division, apoptosis, immune response, and cell adhesion have all been demonstrated to be closely associated with cancers.10, 11 Many oncogenes and tumor suppressors involved in these key processes have been found to contribute to the occurrence as well as development of malignant diseases. CXCR4, a 352-amino acid rhodopsin-like G protein-coupled receptor (GPCR), which can selectively bind to the CXC chemokine stromal cell-derived factor 1 (SDF-1), has been identified in a large number of studies to play a central role in cancer proliferation, invasion, and dissemination in a majority of cancers.12 It has been established that CXCR4 plays a significant role in tumorigenesis as well as the progression of malignant diseases,13 especially breast cancer.14 As reported, at least 23 different cancers express CXCR4,15 including breast cancer,16 prostate cancer, ovarian cancer, and melanoma.17 In addition, the CXCR4 is highly expressed in breast cancer, while present at low levels or absent in normal breast tissues.16 This is consistent with the results of our cDNA microarray analysis. Additionally, the ligand of CXCR4, CXCL12, also known as SDF-1, was found to be significantly down-regulated in breast cancer tissues compared with normal breast samples. Our result is in accordance with what has been identified by many studies. The reduced expression of CXCL12 in breast cancer has been associated with aggressive behavior of breast cancer, which shows that it plays critical roles in progression, metastasis, and prognosis of the disease.18, 19 In conclusion, our findings in the present study do identify some significant factors related to breast cancer which have been demonstrated in previous studies.
Additionally, IFI30, one of the most significantly increased genes in breast cancer tissues in our study, is a unique member of the thiol reductase family because of its optimal pH being 4.5–5.5,20, 21, 22 and it is the only one localized in lysosomes and phagosomes.23 It has been demonstrated that the IFI30 protein is constitutively expressed in professional APC and plays important roles in the process of exogenous antigen processing as well as presentation,24 moreover affects the anti-tumor immunity.25 This gene is a newly found tumor suppressor gene with a promising future as a target in malignant disease treatment and the effects of this gene have been identified in several cancers, including melanomas,26 diffuse large B-Cell lymphoma,27 prostate cancer,28 and glioblastoma.29 Furthermore, we have confirmed that the absence of GILT expression in primary breast cancer was independently associated with poor disease-free survival of patients.9
However, there are some deficiencies in our study. The samples we used in this cDNA microarray analysis were breast cancer tissues and uninvolved normal breast tissues. They contain a mixture of different cell types, which may influence the exact changes found in our work. But, we have previously demonstrated the changes in IFI30 expression by laser-capture microdissection (LCM) to obtain a relatively pure population of epithelial cells and neoplastic cells from uninvolved breast tissues and breast cancer tissues for mRNA evaluation.9 The validation of differentially expressed genes by quantitative real-time PCR was done with only one gene in our study, and the number of genes needs to be extended to include more genes in the future. Altogether, we think that most of the differentially expressed genes found in our present work worthy of further study to uncover their potential effects in breast cancer.
Conclusions
Our present study revealed a range of differentially expressed genes by comparing the gene expression profiles of breast cancer tissues with uninvolved normal control breast tissue. Most of these genes have the potential to play critical roles in the pathogenesis or development of breast cancer according to the functional analysis. Thus, we provide candidate genes for further research to explore potential biomarkers or therapeutic targets in this disease.
Conflict of interest
The authors have no potential conflicts of interest, including financial interests and relationships and affiliations relevant to the subject of their manuscript.
Acknowledgments
This study is supported by the Foundation of the Minister-affiliated Hospital Key Project of the Ministry of Health of the People's Republic of China, China (07090122). And we are grateful to the Central Research Laboratory, the Second Hospital of Shandong University, for technical assistance and generous support.
Footnotes
Peer review under responsibility of Chinese Medical Association.
References
- 1.Siegel R., Ma J., Zou Z., Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Hutchinson L. Breast cancer: challenges, controversies, breakthroughs. Nat Rev Clin Oncol. 2010;7:669–670. doi: 10.1038/nrclinonc.2010.192. [DOI] [PubMed] [Google Scholar]
- 3.Moelans C.B., van der Groep P., Hoefnagel L.D. Genomic evolution from primary breast carcinoma to distant metastasis: few copy number changes of breast cancer related genes. Cancer Lett. 2014;344:138–146. doi: 10.1016/j.canlet.2013.10.025. [DOI] [PubMed] [Google Scholar]
- 4.Gru A.A., Allred D.C. FGFR1 amplification and the progression of non-invasive to invasive breast cancer. Breast Cancer Res. 2012;14:116. doi: 10.1186/bcr3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morgan R., Boxall A., Harrington K.J. Targeting the HOX/PBX dimer in breast cancer. Breast Cancer Res Treat. 2012;136:389–398. doi: 10.1007/s10549-012-2259-2. [DOI] [PubMed] [Google Scholar]
- 6.Douglass S., Ali S., Meeson A.P., Browell D., Kirby J.A. The role of FOXP3 in the development and metastatic spread of breast cancer. Cancer Metastasis Rev. 2012;31:843–854. doi: 10.1007/s10555-012-9395-3. [DOI] [PubMed] [Google Scholar]
- 7.Lin D.W., Nelson P.S. Microarray analysis and tumor classification. N Engl J Med. 2006;355:960. doi: 10.1056/NEJMc061813. [DOI] [PubMed] [Google Scholar]
- 8.Quackenbush J. Microarray analysis and tumor classification. N Engl J Med. 2006;354:2463–2472. doi: 10.1056/NEJMra042342. [DOI] [PubMed] [Google Scholar]
- 9.Morgan R., Boxall A., Harrington K.J. Absence of gamma-interferon-inducible lysosomal thiol reductase (GILT) is associated with poor disease-free survival in breast cancer patients. PLoS One. 2014;9:e109449. doi: 10.1371/journal.pone.0109449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jin Y., Da W. Screening of key genes in gastric cancer with DNA microarray analysis. Eur J Med Res. 2013;18:37. doi: 10.1186/2047-783X-18-37. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Fendri K., Patten S.A., Kaufman G.N. Microarray expression profiling identifies genes with altered expression in Adolescent Idiopathic Scoliosis. Eur Spine J. 2013;22:1300–1311. doi: 10.1007/s00586-013-2728-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cojoc M., Peitzsch C., Trautmann F., Polishchuk L., Telegeev G.D., Dubrovska A. Emerging targets in cancer management: role of the CXCL12/CXCR4 axis. Onco Targets Ther. 2013;6:1347–1361. doi: 10.2147/OTT.S36109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He H., Wang C., Shen Z. Upregulated expression of C-X-C chemokine receptor 4 is an independent prognostic predictor for patients with gastric cancer. PLoS One. 2013;8:e71864. doi: 10.1371/journal.pone.0071864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Z., Ni C., Chen W. Expression of CXCR4 and breast cancer prognosis: a systematic review and meta-analysis. BMC Cancer. 2014;14:49. doi: 10.1186/1471-2407-14-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Balkwill F.R. The chemokine system and cancer. J Pathol. 2012;226:148–157. doi: 10.1002/path.3029. [DOI] [PubMed] [Google Scholar]
- 16.Müller A., Homey B., Soto H. Involvement of chemokine receptors in breast cancer metastasis. Nature. 2001;410:50–56. doi: 10.1038/35065016. [DOI] [PubMed] [Google Scholar]
- 17.Balkwill F. The significance of cancer cell expression of the chemokine receptor CXCR4. Semin Cancer Biol. 2004;14:171–179. doi: 10.1016/j.semcancer.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 18.Lv Z.D., Kong B., Liu X.P. CXCL12 chemokine expression suppresses human breast cancer growth and metastasis in vitro and in vivo. Int J Clin Exp Pathol. 2014;7:6671–6678. [PMC free article] [PubMed] [Google Scholar]
- 19.Graham N.A., Graeber T.G. Complexity of metastasis-associated SDF-1 ligand signaling in breast cancer stem cells. Proc Natl Acad Sci USA. 2014;111:7503–7504. doi: 10.1073/pnas.1405991111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luster A.D., Weinshank R.L., Feinman R., Ravetch J.V. Molecular and biochemical characterization of a novel gamma-interferon-inducible protein. J Biol Chem. 1988;263:12036–12043. [PubMed] [Google Scholar]
- 21.Arunachalam B., Pan M., Cresswell P. Intracellular formation and cell surface expression of a complex of an intact lysosomal protein and MHC class II molecules. J Immunol (Baltimore, Md.: 1950) 1998;160:5797–5806. [PubMed] [Google Scholar]
- 22.Arunachalam B., Phan U.T., Geuze H.J., Cresswell P. Enzymatic reduction of disulfide bonds in lysosomes: characterization of a gamma-interferon-inducible lysosomal thiol reductase (GILT) Proc Natl Acad Sci USA. 2000;97:745–750. doi: 10.1073/pnas.97.2.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Singh R., Cresswell P. Defective cross-presentation of viral antigens in GILT-free mice. Science (New York, NY) 2010;328:1394–1398. doi: 10.1126/science.1189176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maric M., Arunachalam B., Phan U.T. Defective antigen processing in GILT-free mice. Science. 2001;294:1361–1365. doi: 10.1126/science.1065500. [DOI] [PubMed] [Google Scholar]
- 25.Haque M.A., Li P., Jackson S.K. Absence of gamma-interferon-inducible lysosomal thiol reductase in melanomas disrupts T cell recognition of select immunodominant epitopes. J Exp Med. 2002;195:1267–1277. doi: 10.1084/jem.20011853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goldstein O.G., Hajiaghamohseni L.M., Amria S., Sundaram K., Reddy S.V., Haque A. Gamma-IFN-inducible-lysosomal thiol reductase modulates acidic proteases and HLA class II antigen processing in melanoma. Cancer Immunol Immunother. 2008;57:1461–1470. doi: 10.1007/s00262-008-0483-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Phipps-Yonas H., Cui H., Sebastiao N. Low GILT expression is associated with poor patient survival in diffuse large B-cell lymphoma. Front Immunol. 2013;4:425. doi: 10.3389/fimmu.2013.00425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doonan B.P., Haque A. HLA class II antigen presentation in prostate Cancer cells: a novel approach to prostate tumor immunotherapy. Open Cancer Immunol J. 2010;3:1–7. doi: 10.2174/1876401001003010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haque A., Das A., Hajiaghamohseni L.M., Younger A., Banik N.L., Ray S.K. Induction of apoptosis and immune response by all-trans retinoic acid plus interferon-gamma in human malignant glioblastoma T98G and U87MG cells. Cancer Immunol Immunother. 2007;56:615–625. doi: 10.1007/s00262-006-0219-6. [DOI] [PMC free article] [PubMed] [Google Scholar]