Abstract
Long noncoding RNAs (lncRNAs) are emerging as powerful regulators of cardiac development and disease. However, our understanding of the importance of these molecules in cardiac fibrosis is limited. Using an integrated genomic screen, we identified Wisper (Wisp2 super-enhancer–associated RNA) as a cardiac fibroblast–enriched lncRNA that regulates cardiac fibrosis after injury. Wisper expression was correlated with cardiac fibrosis both in a murine model of myocardial infarction (MI) and in heart tissue from human patients suffering from aortic stenosis. Loss-of-function approaches in vitro using modified antisense oligonucleotides (ASOs) demonstrated that Wisper is a specific regulator of cardiac fibroblast proliferation, migration, and survival. Accordingly, ASO-mediated silencing of Wisper in vivo attenuated MI-induced fibrosis and cardiac dysfunction. Functionally, Wisper regulates cardiac fibroblast gene expression programs critical for cell identity, extracellular matrix deposition, proliferation, and survival. In addition, its association with TIA1-related protein allows it to control the expression of a profibrotic form of lysyl hydroxylase 2, implicated in collagen cross-linking and stabilization of the matrix. Together, our findings identify Wisper as a cardiac fibroblast–enriched super-enhancer–associated lncRNA that represents an attractive therapeutic target to reduce the pathological development of cardiac fibrosis in response to MI and prevent adverse remodeling in the damaged heart.
INTRODUCTION
Acute myocardial infarction (MI) due to coronary artery disease typically leads to maladaptive myocardial remodeling and heart failure (HF) (1, 2). HF places a major economic and clinical burden on the industrialized world, accounting for more than 400,000 deaths and more than 20 billion dollars in annual health care costs in the United States alone (3). Initial translational research has focused on the contracting cells of the heart, the cardiomyocytes (CMs), as a target in therapies aimed at restoring cardiac function. This was despite a wide appreciation that acute and chronic injuries trigger tissue remodeling, which invariably results in and is a consequence of the development of cardiac fibrosis (1). The destruction of the myocardium after infarction is compensated by the excessive production of extracellular matrix (ECM) and the formation of a collagen-rich fibrotic scar. Scar formation, tissue remodeling, and progressive interstitial fibrosis lead to a severe loss of function and ultimately HF (1, 2). Moreover, cross-linking enzymes and posttranslational modifications can alter collagen fibrils. This has important implications for matrix synthesis and degradation, which ultimately determine the onset of diastolic dysfunction (4). Despite this clinical importance, very few therapeutic modalities are available to prevent the development of HF. Antifibrotic drugs include blockers of the renin-angiotensin-aldosterone system and mineralocorticoid receptor antagonists but are inefficient in the vast majority of fibrotic diseases (5). Current medications typically slow the progression of the disease rather than prevent or reverse it, which could be achieved if cardiac fibroblasts (CFs) were the primary cell target (6). There is therefore an urgent need to develop alternative therapeutic strategies—for instance, targeting fibroblast differentiation into myofibroblasts or alteration of collagen cross-linking. To achieve this, a deeper characterization of the CF gene program and its associated cellular processes is required to identify specific regulatory molecules and targets (7, 8).
Activation and differentiation of CFs into myofibroblasts initiate the pathological process in the diseased heart. Myofibroblasts synthesize and secrete soluble procollagen I and III, which are processed by metalloproteinases, cross-linked by lysyl oxidases and hydroxylases, and assembled into dense fibers. The ability of myofibroblasts to resist apoptosis and secrete large quantities of profibrotic signaling molecules contributes to the overall pathogenesis of HF (1, 6). Like all differentiated cells, CF identity is hardwired by specific gene regulatory networks (GRNs) (7). These GRNs are controlled by core transcription factors (TFs), proteins that interact in a combinatorial manner at cis-regulatory sequences on DNA to regulate downstream programs dictating cell identity and behavior (9, 10). Enhancers, regions of DNA that can be bound by TFs, represent the key information processing units within the genome and integrate developmental, temporal, spatial, and environmental cues (11). In addition, enhancers may assemble together, generating large enhancer clusters named super-enhancers (SEs) (10, 12, 13). These SEs have important regulatory characteristics, including exquisite cell/tissue specificity, and appear to be crucial for the maintenance of cell identity. These elements are enriched in single-nucleotide polymorphisms linked to common traits and diseases specific to the tissues that harbor them (12). These findings have led many to speculate that SEs could hold therapeutic potential, provided that the means of modulating their activities could be tightly controlled (10, 14).
With the recognition that the mammalian genome is predominantly non–protein-coding (15), the classical protein-centric view of GRN regulation appears to have been premature. RNA-sequencing (RNA-seq) approaches have revealed that the majority of the noncoding genome is actively transcribed, generating thousands of small and long regulatory noncoding RNAs (ncRNAs) (15). Although the implication of microRNAs in the development of stress- and age-induced cardiac fibrosis is well documented (16–18), the more abundant and more diverse long ncRNA (lncRNA) group remains to be comprehensively characterized during heart remodeling. Despite increasing implications in CM hypertrophy and function (19–22), the involvement of lncRNAs in regulating cardiac fibrosis needs to be demonstrated (23). lncRNAs are able to regulate GRN activity via a disparate array of transcriptional and posttranscriptional mechanisms (24). Active enhancers are transcribed into ncRNAs, and SEs tend to produce more RNAs than typical enhancers (TEs) (25–27). Enhancer-associated lncRNAs are important for trapping TF proteins on DNA, modifying the local chromatin environment, and organizing nuclear three-dimensional topologies to ensure the correct activation of target gene programs (27, 28). Together, this suggests that targeting lncRNAs associated with CF-specific enhancers may represent a powerful means to modulate CF behavior.
In line with this hypothesis, we recently characterized the long noncoding transcriptome in a murine model of MI and identified hundreds of novel heart-enriched lncRNAs (29). The vast majority of these transcripts were associated with active heart-specific enhancers (29, 30), especially those dynamically modulated after MI. Some of these transcripts were conserved in humans and shown to be differentially expressed in cardiac disease including aortic stenosis (AOS) and dilated cardiomyopathy (29). These observations suggested that enhancer-associated lncRNAs were likely to represent interesting therapeutic targets for pathological fibrosis (9, 10). We therefore integrated previously generated transcriptomic and epigenomic data sets to identify Wisper (Wisp2 super-enhancer–associated RNA) as a CF-enriched lncRNA that could regulate cardiac fibrosis. Of crucial importance, WISPER is conserved in humans, and its expression in the human heart correlates with collagen content and the severity of cardiac fibrosis. These results highlight the potential for CF-specific SE-associated lncRNAs as therapeutic targets for the amelioration of cardiac fibrosis and ultimately HF.
RESULTS
Wisper is cardiac SE-associated lncRNAs
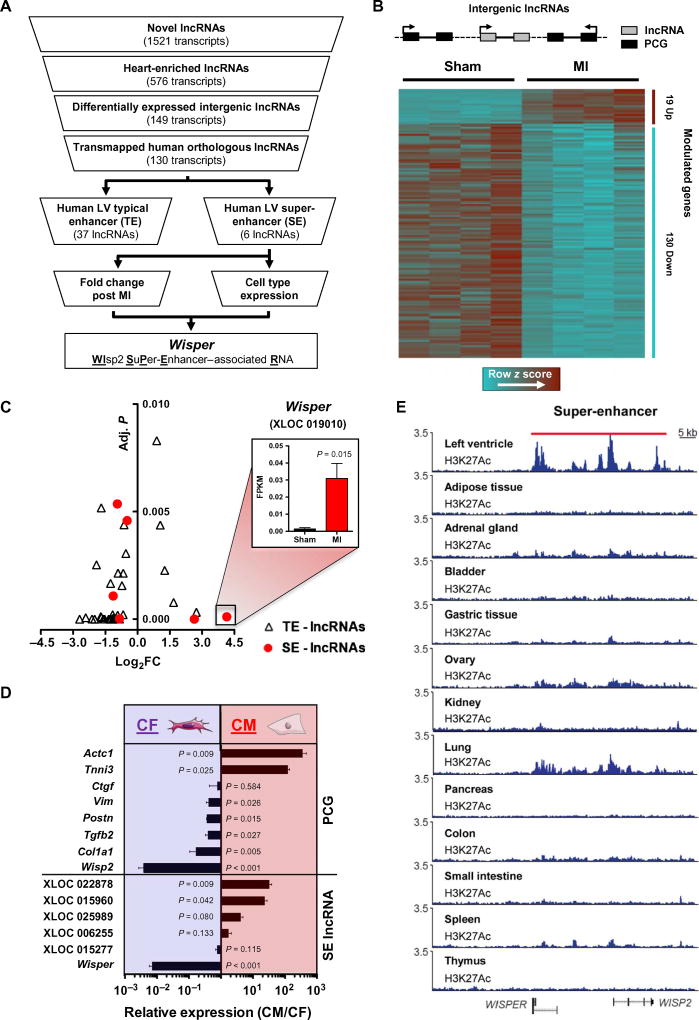
Emerging evidence suggests that SEs and the lncRNAs associated with them represent specific regulators of cell state and identity during development and disease (10). On the basis of this cogent rationale, we set out to identify SE-associated lncRNAs modulated in the damaged myocardium, which were conserved among mouse and human genomes (Fig. 1A). Previously, our laboratory identified 1521 novel lncRNAs in the murine heart (29). A large number of these novel lncRNAs were associated with active cardiac-specific enhancers. Using this transcriptomic data set, we first filtered for all novel lncRNAs that were classified as heart-enriched. To facilitate downstream functional assessment and alleviate any confounding interpretations based on overlapping protein-coding genes (PCGs), we further filtered lncRNAs for those that were intergenic and differentially expressed in the border zone (BZ) 14 days after MI, resulting in the inclusion of 149 lncRNAs (Fig. 1B). To identify putative human orthologs, we mapped these transcripts to the human genome using TransMap, a cross-species alignment tool. Globally, of these 149 mouse lncRNAs, 130 were predicted to have human orthologs. Considering that lncRNAs associated with TEs and SEs likely represent high-priority functional candidates (10), we next examined an enhancer catalog generated in 23 human tissues that used histone 3 lysine 27 acetylation (H3K27Ac) marks and the Rank Ordering of Super-Enhancers (ROSE) algorithm to identify tissue-specific TEs and SEs (12). We found that 37 (28%) of our lncRNAs map to TEs and 6 (5%) mapped to human heart–specific SEs. The most up-regulated lncRNA in the infarcted mouse heart was one of the six SE-associated lncRNAs, supporting the notion that these transcripts could play important roles in the transcriptional reprogramming that underpins cardiac remodeling (Fig. 1C). To dissect the cardiac cell specificity of the six SE-associated lncRNAs, we evaluated the expression in CFs and CMs isolated from the adult murine heart. Expression of these transcripts and canonical PCGs was determined via quantitative reverse transcription polymerase chain reaction (qRT-PCR) (Fig. 1D). The CM-specific PCGs, Tnni3 and Actc1, were highly enriched in CM preparations, whereas CF-specific PCGs, Col1a1, Tgfb2, Postn, and Vim, were specifically enriched in CFs. Among the SE-associated lncRNAs, two were significantly enriched in CMs (P = 0.009 and 0.42, respectively), whereas one lncRNA was highly enriched in CFs (P < 0.001). This transcript was more enriched in CFs than the CF-specific PCGs, suggesting that it could have important functions in this particular cardiac cell type and therefore could play a role in the fibrotic response of the heart after infarction. Proximal to this lncRNA was the gene encoding the matricellular protein Ccn5 (Wisp2), which had recently been implicated in pathological myocardial fibrosis (31). We therefore named this lncRNA Wisper and its human ortholog WISPER. This transcript was found to be substantially conserved in the two species with a 57.3% sequence identity between the two orthologs (fig. S1, A to C). The SE from which this lncRNA was derived was uniquely active in the adult human heart as compared to other tissues (Fig. 1E). Nevertheless, the SE overlapped with WISP2 sequences, suggesting that it contains several constituent enhancers, which may encode a different cis-regulatory potential (32). Considering the enrichment of this transcript in CFs, coupled to its up-regulation in the BZ after MI, we suspected that it could represent an interesting target molecule for the regulation of pathological fibrosis and was selected for further investigation.
Fig. 1. Identification of SE-associated lncRNAs.
(A) Selection strategy of novel SE-associated lncRNAs from a genome-wide profiling of the cardiac transcriptome after MI. (B) Schematic of an intergenic lncRNA located between two PCGs. Heat map showing clustering of heart-enriched intergenic lncRNAs differentially expressed after MI (adjusted P < 0.01). (C) Volcano plot of lncRNAs associated with TEs (triangle) or with SEs (circle). The x axis shows lncRNAs expression in infarcted versus sham-operated animals (log2 fold change) quantified from RNA-seq data; the y axis shows adjusted P value. Wisper expression in sham-operated (black bar) and infarcted heart (red bar) is expressed in FPKM (fragments per kilobase of exon per million fragments mapped). Bars represent mean ± SEM (n = 4). P value was determined by Student’s t test. (D) qRT-PCR analysis of SE-associated lncRNAs (SE-lncRNA) and PCG expression in CMs and fibro-blasts isolated from neonatal mouse hearts. Data represent fold change ratio (CM/CF) mean ± SEM (n = 3). P value was determined by Student’s t test. (E) H3K27Ac signature of the locus encompassing Wisper in different human tissues. The red bar highlights the SE region.
Wisper expression is enriched in CFs and associated with cardiac fibrosis
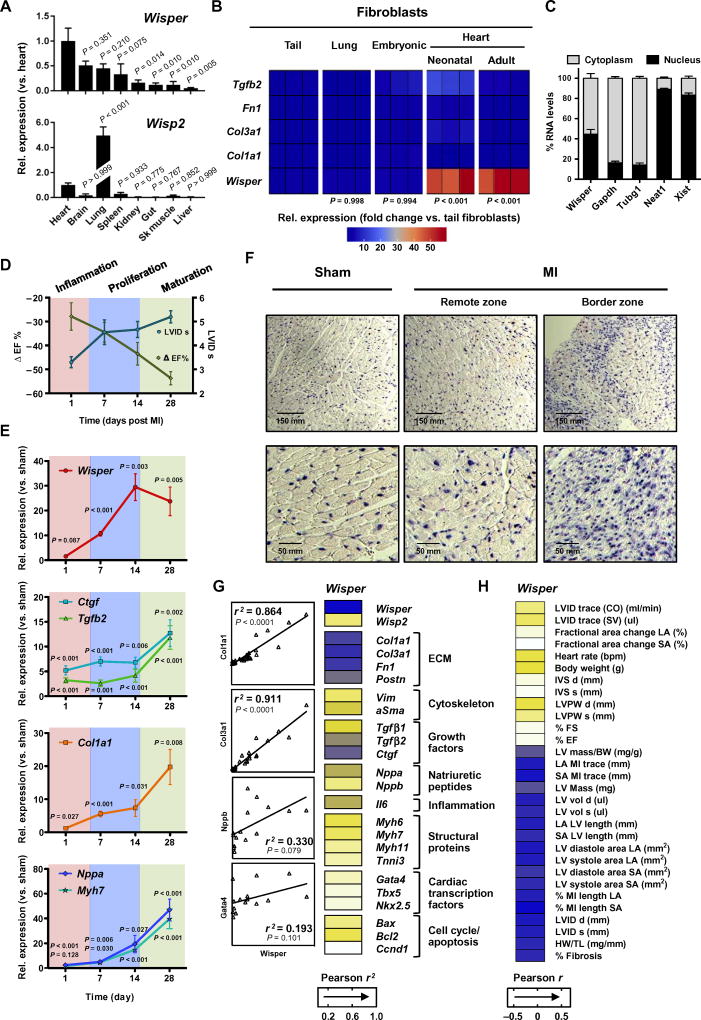
Integrative transcriptome analysis using publicly available RNA-seq data sets demonstrated that Wisper was a heart-enriched transcript (29). To validate this finding, we conducted qRT-PCR using RNA isolated from different adult mouse tissues. Wisper was more expressed in the heart than in all other tissues (Fig. 2A). On the other hand, Wips2 expression was not characterized by the same tissue distribution. We next quantified Wisper expression in fibroblasts of cardiac and noncardiac origins. In comparison to fibroblast-associated PCGs such as Tgfb2, Fn1, Col3a1, and Col1a1, which were similarly expressed in fibroblasts from different sources, Wisper was significantly enriched in fibroblasts isolated from neonatal and adult hearts (Fig. 2B; P < 0.001). Several binding sites for cardiac TFs such as GATA4 and NKX2–5 were identified in the SE element from which WISPER derives (fig. S1D). This was in contrast to WISP2, which was characterized by distinct TF binding sites at its promoter such as ATOH1 binding sites and E-box, which were known to regulate lung-specific expression (fig. S1E) (33). Finally, lncRNA functions are typically dependent on subcellular localization, with those present primarily in the nucleus involved in chromatin regulation, whereas cytoplasmic lncRNAs influence posttranscriptional processes (24). Wisper was found to be equally distributed between the two sub-cellular compartments, indicating that it could play roles in both transcriptional and posttranscriptional regulatory processes (Fig. 2C).
Fig. 2. Wisper is enriched in CFs and up-regulated in fibrotic myocardial tissue.
(A) Wisper and Wisp2 expression in different mouse tissues quantified by qRT-PCR. Bars represent mean expression relative to the heart ± SEM (n = 4). P value was determined by one-way analysis of variance (ANOVA) (Fisher’s test). (B) Heat map representation of Wisper and fibroblast gene expression (relative to tail fibroblasts) in fibroblasts isolated from different tissues. Three independent experiments are shown. P value was determined by one-way ANOVA (Fisher’s test). (C) Percentage of nuclear (black bar) and cytoplasmic (gray bar) RNA concentrations of Wisper, Gapdh, and Tubg1 (cytoplasmic markers), and Neat1 and Xist (nuclear markers) measured by qRT-PCR after subcellular fractionation in CFs. Data represent mean ± SEM (n = 4). (D) Ejection fraction (ΔEF; green line) and systolic left ventricular internal dimension (LVID s; blue line) after MI by echo-cardiography as compared to sham. Three different phases of remodeling are highlighted. (E) Expression kinetics of Wisper and canonical markers of mal-adaptive cardiac remodeling measured by qRT-PCR. Graphs show means normalized to sham ± SEM (n = 6 to 10 animals). P values were determined by two-way ANOVA (Fisher’s test). (F) Detection of Wisper expression in sham and infarcted heart sections by in situ hybridization. (G) Heat map of correlations of expression between Wisper and various genes implicated in postinfarction biological processes. Correlations between Wisper and Col1a1, Col3a1, Nppb, and Gata4 are shown as examples. Pearson’s correlation test [r2; 95% confidence interval (CI)]. (H) Heat map representation of the correlation between Wisper expression in the BZ and echocardiographic traits. Pearson’s correlation test (r; 95% CI).
After MI, the myocardium undergoes a remodeling process that is characterized by three distinct phases. These include an inflammatory response (days 1 to 3 after injury), proliferation and granulation tissue formation (days 7 to 14), and scar maturation (days 21 to 28) (34). Profibrotic pathways are typically associated with the proliferative phase, in which differentiation of myofibroblasts and subsequent proliferation, migration, and secretion of ECM components take place. To assign Wisper to either of these phases, MI was induced in the mouse heart by ligation of the left anterior descending (LAD) artery and Wisper expression was assessed over a 28-day period. Cardiac dimensions and function were assessed by echocardiography (Fig. 2D and table S1). Gene expression profiling demonstrated the expected expression kinetics for fibrotic and hypertrophy-associated PCGs (Fig. 2E). Wisper was maximally expressed 14 days after MI, corresponding to the proliferative phase in which myofibroblasts migrate to the BZ and actively secrete collagen and other ECM components. The temporal kinetics of Wisper induction implicated this transcript in cardiac fibrosis driving pathological remodeling. To support these findings, we perfomed RNA in situ hybridization using probes against Wisper on mouse hearts 14 days after infarction. In accordance with Wisper expression in activated fibroblasts, a marked signal was observed in the BZ of infarcted hearts (Fig. 2F and fig. S2A). Wisper expression was also detected, albeit less frequently, in the interstitial space of the viable muscle. We therefore evaluated whether Wisper expression correlated with gene programs linked to cardiac fibrosis. Wisper expression was highly correlated with PCGs relevant to ECM deposition (Fig. 2G) and also highly correlated with echocardiographic traits linked to remodeling of the injured myocardium (Fig. 2H). Moreover, to evaluate the tissue specificity of Wisper expression under stress conditions, we used a mouse model of renovascular hypertension, namely, the one-kidney, one-clip (1K1C) model (35). Cardiac hypertrophy and fibrosis develop in response to volume overload in this model (fig. S2, B to D). In addition, the single hypoxic kidney is also characterized by extensive fibrosis (fig. S2D). Strikingly, Wisper was induced in the stressed heart but not in the stressed kidney. In contrast, Wisp2 expression was up-regulated in both organs.
Wisper controls CF behavior and survival
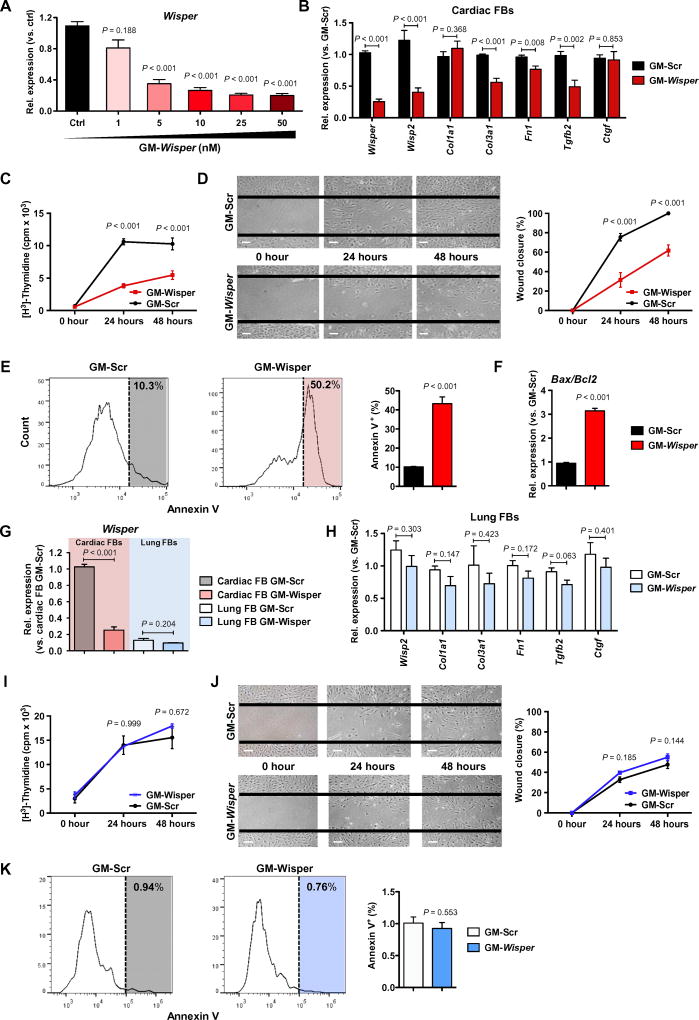
To characterize the functional role of Wisper, a loss-of-function approach was used in isolated adult murine CFs. Modified antisense oligonucleotides (ASOs) called GapmeRs were used to initiate nuclear ribonuclease H–mediated degradation of the transcript and to deplete Wisper in both the nucleus and cytoplasm (fig. S1A). Titration experiments showed that 10 nM GapmeRs targeting Wisper was sufficient to achieve maximal knockdown in adult CFs without affecting CF integrity (Fig. 3A). This concentration was therefore used for subsequent experiments. Silencing of Wisper expression resulted in a specific impact on ECM-associated PCG expression (Fig. 3B). Col3a1, Fn1, and Tgfb2 were down-regulated; however, Col1a1 and Ctgf were not affected. Wisper depletion also led to the down-regulation of the proximal PCG Wisp2, suggestive of a possible cis-regulatory role in adult CFs. Considering the effect on gene expression, we suspected that Wisper could be fundamentally involved in the transdifferentiation of CFs into myofibroblasts. We therefore isolated neonatal murine CFs, which can be induced to differentiate through cell passaging in vitro. Differentiation of these cells resulted in the up-regulation of myofibroblast-specific PCGs such as Tgfb2, Fn1, Col1a1, Col3a1, and aSma [also known as Acta2] (fig. S3A). Wisper and Wisp2 expression was closely associated with myofibroblast differentiation. Wisper depletion in differentiated myofibroblasts resulted in a significant down-regulation of Col3a1, Fn1, Tgfb2, and aSma expression (fig. S3B; P < 0.001), similar to what was observed in adult CFs (Fig. 3B). The myofibroblast-associated α-smooth muscle actin (α-SMA) protein was also significantly down-regulated (fig. S3C; P = 0.011). Again, Col1a1 was not affected by Wisper depletion, supporting a specific regulatory role on individual ECM genes.
Fig. 3. Wisper controls CF behavior and survival.
(A) Wisper expression in adult CFs after GapmeR transfection at increasing concentrations. Bars represent means normalized to control ± SEM (n ≥ 6). P values were determined by Student’s t test. (B and H) Gene expression measured by qRT-PCR in adult CFs (B) or lung fibroblasts (FBs) (H) after transfection with GapmeRs (10 nM, 48 hours; n ≥ 5) targeting Wisper (GM-Wisper) or scrambled GapmeRs (GM-Scr). P values were determined by two-way ANOVA (Fisher’s test). (C and I) Proliferation of adult CFs (C) or lung fibroblasts (I) after GapmeR transfection (10 nM) quantified by [H3]thymidine incorporation. Data are expressed as mean counts per million (cpm) ± SEM (n > 3). P values were determined by two-way ANOVA (Bonferroni’s test). (D and J) Migration of adult CFs (D) or lung fibroblasts (J) after GapmeR transfection (10 nM) measured using a wound closure assay. Representative pictures are shown. Scale bars, 100 µM. Graphs show the percentage of wound closure at each time point ± SEM (n ≥ 3). P values were determined by two-way ANOVA (Bonferroni’s test). (E and K) Representative fluorescence-activated cell sorting (FACS) analysis of annexin V-positive adult CFs (E) or lung fibroblasts (K) after GapmeR transfection (10 nM). Graphs show the percentage of apoptotic cells ± SEM (n = 3). P values were determined by Student’s t test. (F) Bax expression–to-Bcl2 expression ratio in adult CFs after GM-Wisper or GM-Scr transfection (10 nM, 48 hours). Bars represent means ± SEM (n = 3). P values were determined by Student’s t test. (G) Wisper expression in adult CFs and lung fibroblasts after GM-Wisper transfection (10 nM). Bars represent means normalized to GM-Scr ± SEM (n ≥ 5). P values were determined by Student’s t test.
Because Wisper depletion was found to have a large impact on fibroblast identity, we proceeded to evaluate cell behavior after Wisper knockdown in adult CFs. We found a significant decrease in proliferation in CFs 24 hours after Wisper knockdown (Fig. 3C; P < 0.001), suggesting that Wisper was important for the proliferative ability of CFs. Using a wound closure assay, we demonstrated a significant attenuation of the migratory ability of Wisper-depleted CFs (Fig. 3D; P < 0.001). Finally, apoptosis was assessed in Wisper-depleted CFs via testing annexin V positivity by flow cytometry analysis. At 24 hours after transfection, there was a fivefold increase in the percentage of apoptotic CFs treated with GapmeRs targeting Wisper (Fig. 3E). In addition, the ratio between Bax and Bcl2 expression indicated an increase in proapoptotic signaling in CFs upon Wisper depletion (Fig. 3F). Together, these data suggest that Wisper knockdown affects cell survival in CFs. To confirm the cardiac specificity of Wisper function, we used the same dose of Wisper-targeting GapmeRs in fibroblasts of noncardiac origin, that is, lung fibroblasts (Fig. 3G), and observed no impact on the expression of fibroblast-associated PCGs (Fig. 3H), proliferation (Fig. 3I), migration (Fig. 3J), and apoptosis (Fig. 3K). Finally, despite being largely CF-enriched, Wisper is also expressed at low concentrations in CMs. Therefore, we also tested the effects of GapmeRs in neonatal CMs to detect any unanticipated effects on these cells (fig. S3D). Although GapmeR treatment slightly decreased Wisper expression, it did not affect CM-specific gene expression, structure, cross-sectional area, or cell number. The number of CFs, which are typically present in neonatal CM cultures, was reduced after Wisper depletion, suggesting that decreased Wisper expression in CM cultures reflects modulation in contaminating CFs.
Wisper regulates specific CF gene programs
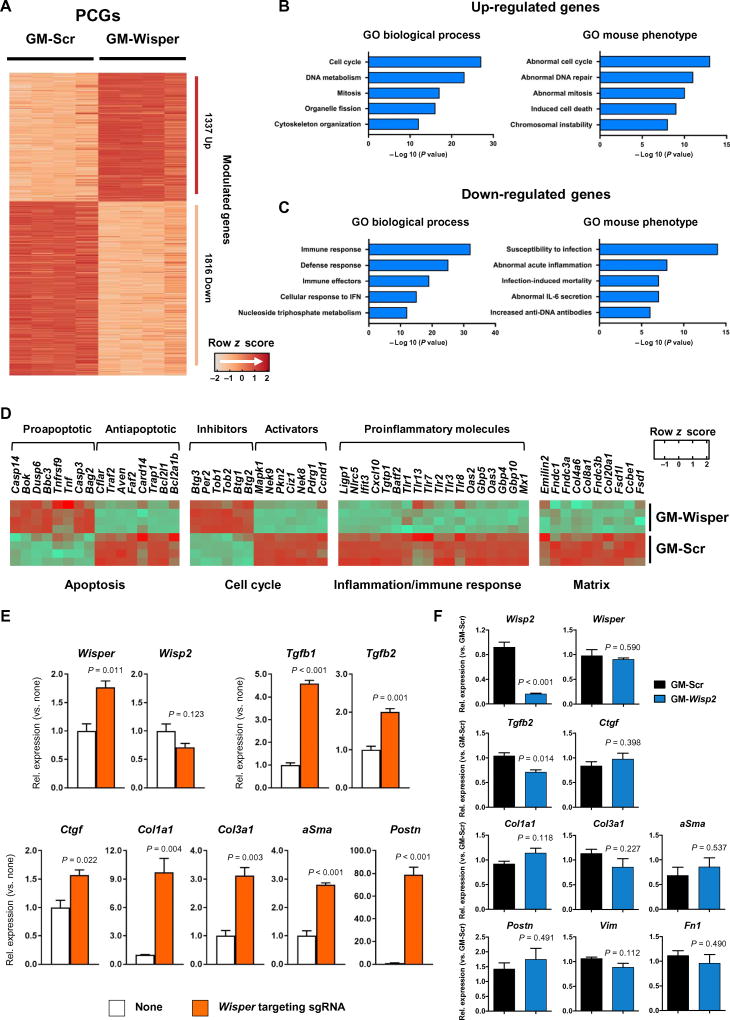
To determine whether Wisper played a global role in the regulation of specific CF gene programs, RNA-seq was performed on scrambled and Wisper-specific GapmeR-treated adult CFs. We identified 3153 differentially expressed PCGs (fold change, >2; adjusted P value, <0.05) in Wisper-depleted CFs, of which 1337 were up-regulated and 1816 were down-regulated (Fig. 4A). Using Gene Ontology (GO) analysis, we found that up-regulated PCGs were associated with biological processes linked to the control of cell cycle and mitosis (Fig. 4B). Furthermore, these PCGs were also associated with mouse phenotypes linked to abnormal control of cell cycle and induction of cell death, both processes observed in Wisper-depleted CFs. Down-regulated PCGs were associated with modulation of the immune response and inflammatory phenotypes in the mouse (Fig. 4C). These findings are consistent with the important roles that CFs play during acute inflammation after infarction (34). Up-regulated genes included many important proapoptotic molecules (for example, Dusp6 and Casp3), whereas antiapoptotic genes were down-regulated (for example, Bcl2l1 and Bcl2a1b) (Fig. 4D). Similarly, cell cycle inhibitors (for example, Btg1 and Tob1) were induced upon Wisper deletion, whereas cell cycle activators (for example, Ccnd1) were down-regulated. Many regulators of the immune response were also down-regulated (for example, Cxcl10). Key PCGs linked to the ECM (for example, Col4a6 and Col8a1) were depleted in Wisper GapmeR-treated cells. Furthermore, we also tested the capacity of Wisper to induce a fibroblastic gene program in nonfibroblastic cells when enacted at its own site of transcription. We used a CRISPR (clustered regularly interspaced short palindromic repeats)–based gain-of-function approach (CRISPR-on). P19CL6 cells were transfected with components of the synergistic activation mediator described by the Zhang laboratory in combination with a Wisper-targeting guide RNA engineered to contain two MS2 aptamers (36). Significant Wisper expression was measured 2 days after transfection (P < 0.011). In turn, prototypic fibroblast genes were induced. Wisp2 was not activated in these cells (Fig. 4E). Considering the cis-based regulatory roles linked to SE-associated lncRNAs, we proceeded to assess the impact of Wisper depletion on proximal PCGs embedded within its topologically associated domain (TAD). Cell type–invariant TADs are typically established during pluripotency and are critical for configuring the three-dimensional chromatin architecture that ensures correct temporal and spatial interactions between distal enhancers and their target promoters. We therefore used publicly available high-throughput confirmation capture data sets from mouse embryonic stem cells to interrogate the topological nature of this locus (fig. S4A). Of the 10 PCGs within the Wisper-harboring TAD, 5 were differentially expressed upon Wisper depletion in adult CFs, one of which is Wisp2. We therefore evaluated whether Wisper could exert its action via cis-regulation of Wisp2 expression. We examined the specificity of the gene expression programs in CFs after GapmeR-mediated Wisp2 depletion, which resulted in a significant loss of Wisp2 expression (P < 0.001) without affecting Wisper concentrations (Fig. 4F). The canonical ECM proteins previously examined, whose expression was modified by Wisper depletion, were not affected by Wisp2 knockdown. These data support a role for Wisper in dictating gene programs associated with cell identity and behavior in CFs, independent of Wisp2 expression. Finally, the RNA-seq data also allowed us to assess the impact of Wisper depletion on the annotated long noncoding transcriptome (fig. S4B). Among the 435 up-regulated IncRNAs, well-characterized IncRNAs such as Neat1 and Gas5 were included (fig. S4C). Conversely, 276 IncRNAs were down-regulated; among them, Malat1, Ftx, and Firre have all been implicated in cell cycle control and apoptosis in various cell types (37, 38).
Fig. 4. Regulation of CF gene programs by Wisper.
(A) Hierarchical clustering of PCGs differentially expressed in adult CFs after transfection with GapmeRs targeting Wisper (GM-Wisper) or scrambled GapmeRs (GM-Scr) as assessed by RNA-seq (fold change, >2; adjusted P < 0.05). (B and C) GO terms linked to biological processes and mouse phenotypes for the up-regulated (B) or down-regulated (C) PCGs in Wisper-depleted adult CFs. (D) Heat map representing functional sets of PCGs modulated by GapmeR-mediated Wisper depletion in CFs. PCGs are clustered based on biological and cellular functions. (E) Expression of relevant fibrosis-related genes measured by qRT-PCR in P19CL6 cells after CRISPR-on–mediated Wisper induction (Wisper targeting sgRNA; orange bar) as compared to control (none; white bar). Mean ± SEM (n = 3). P values were determined by Student’s t test. (F) GapmeR-induced depletion of Wisp2 in adult CFs (10 nM, 48 hours). Expression of relevant fibrosis-related PCGs. Bars show means normalized to control (GM-Scr) ± SEM (n ≥ 3). P values were determined by Student’s t test.
Wisper is associated with TIA1-related protein and regulates lysyl hydroxylase 2 expression
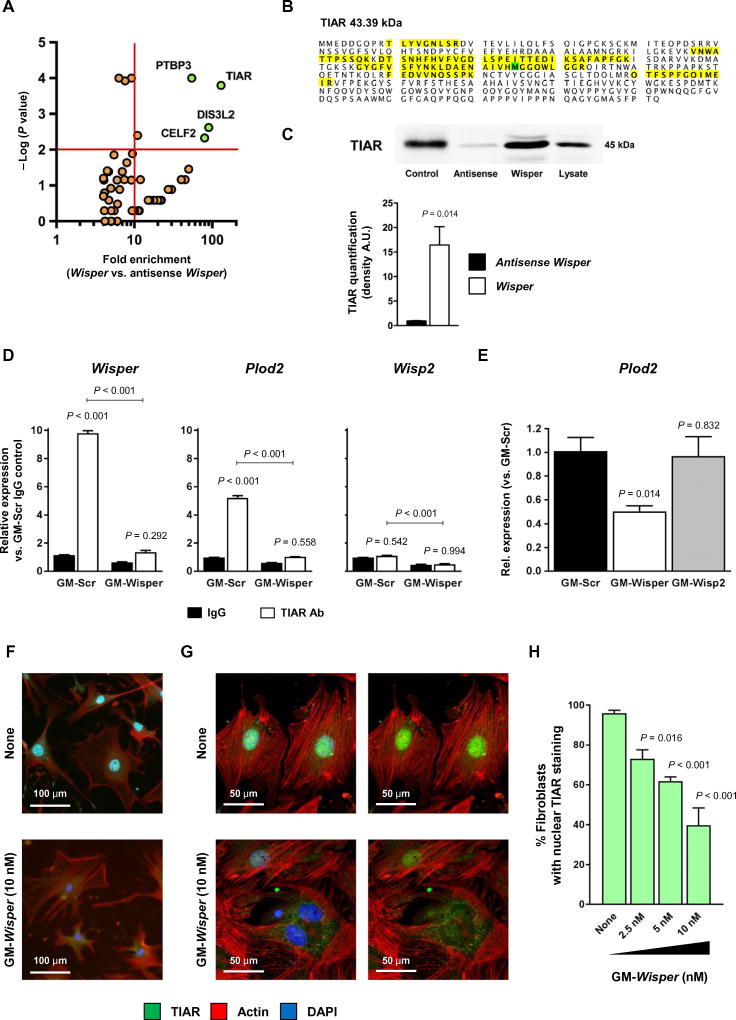
Many IncRNAs exert their function via interaction with proteins. To identify relevant Wisper-binding proteins, we performed an IncRNA pulldown assay. A biotinylated Wisper probe was therefore used as a bait to selectively extract putative Wisper protein partners from an adult CF lysate (fig. S4, D and E). An antisense Wisper transcript was used as control. Then, proteins were identified by shotgun mass spectrometry. Four proteins were detected as specifically associated with Wisper, namely, TIAR, PTB3, DIS3L2, and CELF2 (Fig. 5A). All four RNA binding proteins have been implicated in RNA processing. TIAR, PTBP3, and CELF2 are splicing factors, and DIS3L2 has been involved in target mRNA-mediated microRNA degradation as well as mRNA decay (39–43). These proteins demonstrate relevant functions as regulators of differentiation, proliferation, and apoptosis during development and in adulthood. Nevertheless, protein inference based on peptide analysis unambiguously identified TIAR [TIAl-related protein, also referred to as TIA1 cytotoxic granule-associated RNA binding protein-like 1 (TIAL1)] as a prime candidate with 26% amino acid coverage (102 of 392; Fig. 5B). More importantly, TIAR has been related to tissue fibrosis via its capacity to regulate expression of lysyl hydroxylase 2 [also known as procollagen lysine, 2-oxoglutarate 5-dioxygenase (Plod2)] (44) and thereby the extent of collagen cross-linking. We confirmed the strong association of TIAR with Wisper using Western blotting to detect TIAR after Wisper pulldown (Fig. 5C). Then, we performed an RNA immunoprecipitation assay to validate Wisper-TIAR interaction (Fig. 5D). RT-PCR after TIAR immunoprecipitation detected Wisper and Plod2 as TIAR-bound transcripts but not Wisp2. Wisper knockdown reduced the amounts of TIAR-associated Wisper as expected and, in addition, also affected the amounts of TIAR-bound Plod2 (Fig. 5D). Down-regulation of Wisper expression in CFs therefore resulted in decreased Plod2 expression, whereas Wisp2 depletion did not change Plod2 concentrations (Fig. 5E). TIAR has been demonstrated to shuttle between the cytoplasm and the nucleus (45). Because Wisper was also found in both the cytoplasm and the nucleus, we investigated whether TIAR nuclear translocation could depend on Wisper action. Primary CFs were therefore transfected with Wisper-targeting GapmeRs, and TIAR subcellular localization was determined by immunostaining (Fig. 5, F to H). Wisper knockdown resulted in a dose-dependent decrease in the number of fibroblasts with nuclear TIAR staining. Confocal microscopy confirmed the absence of TIAR in the nucleus and retention of the protein in the cytoplasm of treated cells.
Fig. 5. Wisper is associated with TIA1-related protein and regulates lysyl hydroxylase 2 expression.
(A) Proteins enriched after pulldown using biotinylated Wisper and identified by mass spectrometry. The x axis shows the protein enrichment in the Wisper group as compared to the control antisense Wisper group; the y axis shows the −log P value determined by Fisher’s test. (B) Amino acid composition of the protein TIAR. The detected amino acids by mass spectrometry are highlighted in yellow (103 of 392 amino acids; 26% coverage). (C) Protein quantification of TIAR by Western blotting in the pulled-down protein fraction. Purified TIAR is used as positive control. Graph shows means ± SEM (n = 3). P values were determined by Student’s t test. A.U., arbitrary units. (D) Quantification of RNA after immunoprecipitation using an immunoglobulin G (IgG) directed against TIAR or a control IgG. Protein lysate was produced from CFs after transfection with GapmeRs targeting Wisper (GM-Wisper) or scrambled GapmeRs (GM-Scr) (10 nM, 48 hours). Graph shows means ± SEM (n = 6). P values were determined by two-way ANOVA (Fisher’s test). Ab, antibody. (E) Plod2 expression in adult CFs after GM-Wisper, GM-Wisp2, or GM-Scr transfection (10 nM, 48 hours). Bars represent means ± SEM (n = 3). P values were determined by one-way ANOVA. (F) Immunohistochemistry analysis of TIAR expression in CFs transfected with GapmeRs targeting Wisper (GM-Wisper) or left untreated (none; 10 nM, 48 hours). TIAR, green; actin, red; 4’,6-diamidino-2-phenylindole (DAPI), blue. (G) Confocal microscopy analysis of TIAR subcellular localization in CFs treated as in (F). (H) Percentage of CFs with nuclear TIAR staining after GM-Wisper treatment at various doses. Bars represent means ± SEM (n > 6). P values were calculated by two-way ANOVA (Fisher’s test).
Preventive Wisper depletion in vivo inhibits cardiac fibrosis
To test whether the antifibrotic effects of Wisper in cultured CFs may hold therapeutic potential in cardiac fibrosis, we performed loss-of-function experiments in mice. We first completed a dose escalation study in adult untouched mice by administering Wisper GapmeRs (5, 10, and 15 mg/kg) (fig. S5A). Control mice received a scrambled GapmeR at a dose of 10 mg/kg. All mice injected with 15 mg kg died within the first week after GapmeR injection, whereas the 5 and 10 mg/kg doses had no impact on survival (fig. S5B). Echocardiography performed at 4,14, and 28 days after injection showed that mice receiving Wisper GapmeRs (10 mg/kg) demonstrated signs of cardiac remodeling, particularly a thickening of the interventricular septum. Animals injected with 5 mg/kg were not different from controls (fig. S5C and table S2). To determine the impact of acute and massive Wisper depletion on cardiac dimensions and function, we performed echocardiography in untouched mice receiving Wisper GapmeR (15 mg/kg) 4 days after injection. At this time point, the myocardium exhibited structural alterations, including an increased thickness of the ventricular septum and of the posterior wall, with a concomitant reduction of the left ventricular cavity. This peculiar situation was characterized by a paradoxical increase in EF, associated to a largely decreased stroke volume, creating a very detrimental situation (fig. S5D). Wisper was significantly depleted in isolated adult CFs (P < 0.007), and a robust down-regulation of key PCGs, including Collal, Col3al, Vim, and Postn, was also observed (fig. S5, E and F). Finally, a significant up-regulation of cardiac stress markers, including Ctgf and Myh7 (P = 0.012 and 0.011, respectively), was measured. Despite a dramatic decrease of collagen expression in the heart in animals receiving this toxic dose of Wisper GapmeRs, no effects on collagen expression were observed in the kidneys or the liver (fig. S5G). Signs of liver damage were, however, evident because plasma concentrations of both aspartate aminotransferase and alanine aminotransferase were elevated (fig. S5H). The amounts of these enzymes in the blood of mice receiving 5 mg/kg were not changed, indicating no toxic effects of Wisper GapmeR at this concentration. On the basis of these data, the 5 mg/kg dose was therefore selected for subsequent experiments in vivo.
We used GapmeRs to perturb the induction of Wisper in a preventive strategy by delivering scrambled and Wisper-targeting GapmeRs 3 days before the induction of MI and assessed cardiac function 7 and 14 days after MI (fig. S6A). RNA was isolated and histological sections were generated 14 days after infarction. Consistent with the observed effects in vitro, delivery of Wisper GapmeR resulted in the blunted expression of Wisper in the heart (fig. S6B) and of key ECM-associated PCGs (fig. S6C). Wisper was not affected by GapmeR treatment in sham-operated animals, suggesting that only stress-stimulated Wisper expression was sensitive to knockdown using a dose of 5 mg/kg. Gene expression was measured more than 2 weeks after GapmeR administration. Compared to scrambled GapmeR-treated animals, mice treated with Wisper GapmeR exhibited improved structural and functional parameters at 7 and 14 days after MI, as assessed by echocardiography (fig. S6, D and E, and table S3). Wisper-depleted mice demonstrated decreased remodeling, as indicated by decreased heart weight–to–tibial length ratio (fig. S6F). Infarct size was reduced and a significant decrease in myocardial fibrosis was observed (fig. S6G; P = 0.006). Considering the impact of Wisper depletion on CF proliferation in vitro, we also assessed nonmyocyte cell proliferation in vivo via 5-bromo-2’-deoxyuridine (BrdU) incorporation. The reduced numbers of BrdU-positive nonmyocyte cells in Wisper GapmeR-treated animals were indicative of decreased proliferation in this subpopulation (fig. S6H). However, despite these largely beneficial effects of Wisper depletion on maladaptive remodeling, decreased CF survival before injury may also be expected to affect the acute wound healing process. We observed that Wisper GapmeR-treated mice exhibited a higher rate of mortality during the acute phase after MI, primarily resulting from left ventricular wall rupture, suggesting that Wisper silencing might impair the acute would healing process that relies on CF activity (fig. S6I). This prompted us to test the therapeutic potential of Wisper depletion in a therapeutic protocol more relevant for a clinical setting.
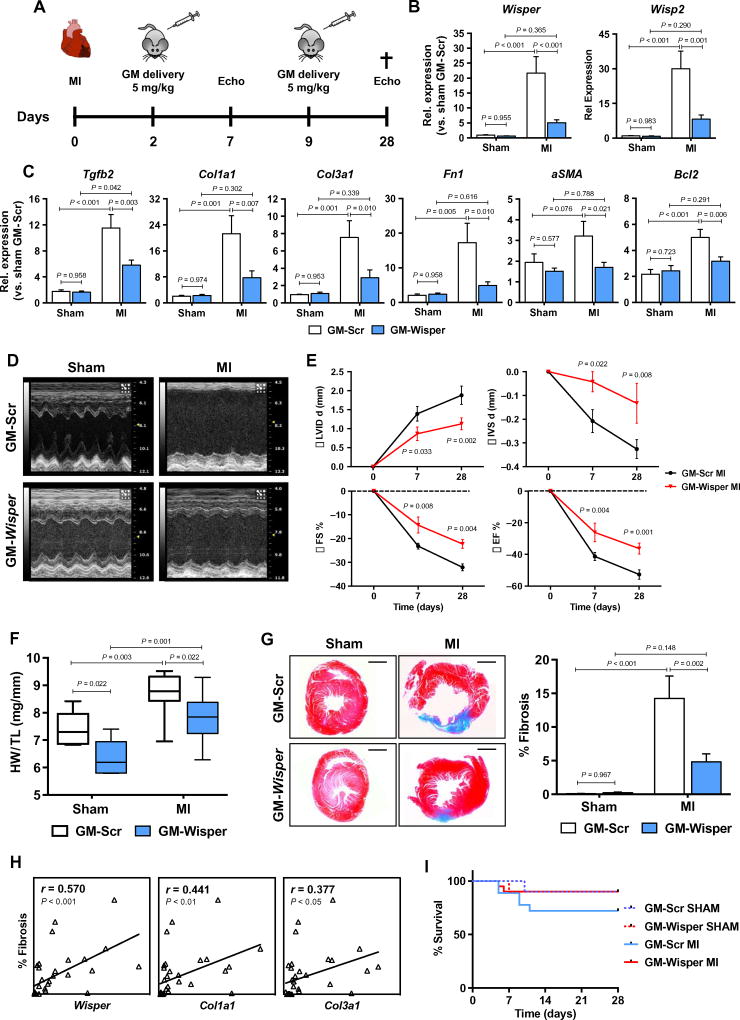
Therapeutic depletion of Wisper in vivo inhibits cardiac fibrosis and improves function
To evaluate the potential utility of Wisper targeting as an antifibrotic therapy, we used a therapeutic protocol in which Wisper was depleted after MI. MI was induced and Wisper GapmeRs were injected 2 and 9 days after injury to avoid affecting acute wound healing and to subsequently modulate the evolution of the pathological proliferative phase of the remodeling process (Fig. 6A). Cardiac dimensions and function were assessed by echocardiography at 7 and 28 days after MI. RNA was isolated upon sacrifice at 28 days, a temporal point coinciding with the evolution of the mature scar and the development of pathological remodeling. Both Wisper and its proximal PCG Wisp2 demonstrated blunted cardiac expression in Wisper GapmeR-treated mice (Fig. 6B). This profile was associated with a significant impact on the expression of key ECM and profibrotic PCGs including Tgfb2 (P = 0.003), Collal (P = 0.007), CoBal (P = 0.010), Fnl (P = 0.010), and aSma (P = 0.021) (Fig. 6C and fig. S7A). Additionally, expression of cardiac stress markers was also blunted upon Wisper silencing, suggestive of a beneficial impact on cardiac hypertrophy (fig. S7A). At both 7 and 28 days after MI, Wisper-depleted mice exhibited significantly improved cardiac function (Fig. 6E) (%FS: P = 0.008 and 0.004 at 7 and 28 days, respectively) and decreased remodeling (Fig. 6, D to F, and table S4). This response was associated with a reduction in infarct size and a significant perturbation of cardiac fibrosis (Fig. 6G; P = 0.002), resulting in preserved tissue architecture and reduced thinning of the myocardial wall after Wisper knockdown. In support of this, postinfarction fibrosis was found to be highly correlated with Wisper expression in treated mice and even more highly correlated with the expression of canonical ECM associated PCGs such as Collal and CoBal (Fig. 6H). Mortality rate in treated animals was not augmented during the acute phase, indicating that the wound healing process was not negatively affected (Fig. 6I). Wisper-depleted mice had a slightly increased survival rate after injury when compared to controls, suggestive of an overall beneficial effect on mortality rates. Collectively, these data support an important role for Wisper in pathological cardiac fibrosis and demonstrate that therapeutic targeting of Wisper after MI elicits clinically desirable effects.
Fig. 6. Therapeutic depletion of Wisper inhibits cardiac fibrosis and improves function.
(A) Overview of the experimental setup with injections of GapmeRs targeting Wisper (GM-Wisper; 5 mg/kg) or scrambled GapmeRs (GM-Scr) at 2 and 9 days after Ml. Echocardiography was performed 7 and 28 days after the surgery. The cross indicates time of sacrifice and tissue collection (sham, n = 6; MI, n = 9). (B and C) Expression of Wisper (B) and fibrotic/ stress genes (C) after GapmeR injection (white, GM-Scr; light blue, GM-Wisper) in sham-operated and MI mice 28 days after surgery. Bars represent means normalized to sham GM-Scr ± SEM. P values were determined by two-way ANOVA (Fisher’s test). (D) M-mode images of the left ventricle (LV) of GapmeR-injected mice 28 days after MI. (E) Echocardiographic assessment of cardiac dimension (LVID d, diastolic left ventricular internal dimension; IVS d, diastolic intraventricular septum) and function (FS%, fractional shortening). Graphs show means normalized to the average values of sham ± SEM. P values were determined by two-way ANOVA (Fisher’s test). (F) Ratio of heart weight (HW) to tibial length (TL) in GapmeR-injected sham and MI mice. P values were determined by two-way ANOVA (Fisher’s test). (G) Fibrotic tissue quantification by Masson’s trichrome staining on heart sections. Graph shows the percentage of cardiac fibrosis as measured by ImageJ (n ≥ 6). Bars represent means normalized to sham GM-Scr ± SEM. P values were determined by two-way ANOVA (Fisher’s test). (H) Correlation analysis between the percentage of cardiac fibrosis and Wisper, Colla1, and CoBa1 expression. Pearson’s correlation test (r, 95% CI). (I) Effect of GapmeR injection on survival after Ml.
WISPER expression correlates with the extent of fibrosis in the diseased human heart
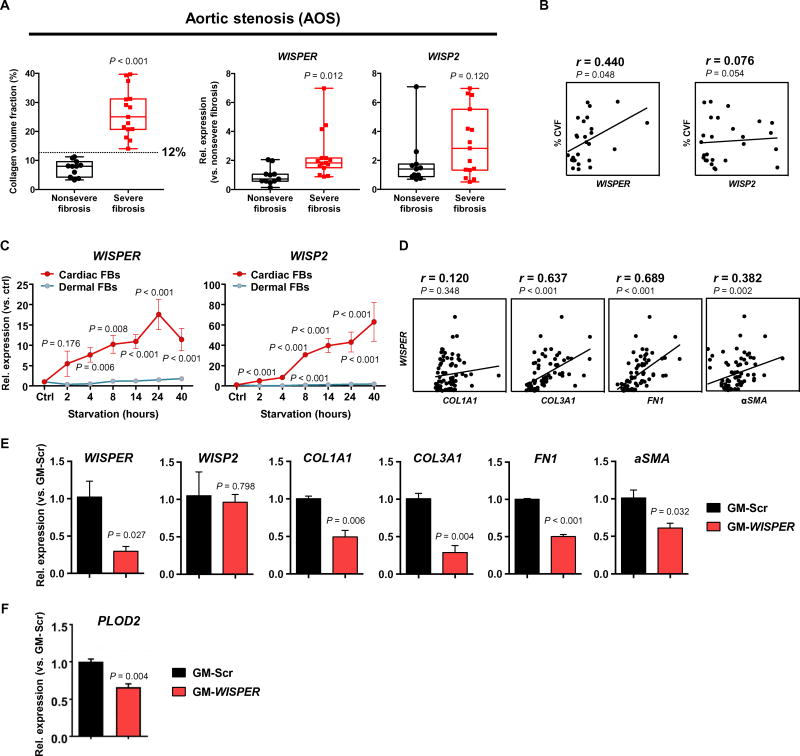
A putative ortholog of Wisper was identified in the human genome, mapping to an LV-specific SE (Fig. 1E and fig. S1, A to C). Primers were designed to amplify this transcript, which could be consistently detected in RNA isolated from human cardiac biopsies, formally demonstrating that WISPER was evolutionary conserved. We therefore proceeded to quantify WISPER expression in RNA isolated from the interventricular septum of patients suffering from AOS (table S5). AOS is associated with extensive myocardial fibrosis that directly contributes to LV dysfunction. In cardiac samples from all AOS patients, the collagen volume fraction (CVF) was higher than that in samples from healthy volunteers (46). After analyzing the distribution of CVF in the AOS cohort, two groups were identified: a group with nonsevere fibrosis (n = 11) with a CVF lower than 12% and a severe fibrosis group (n = 15) with a CVF greater than 12%. WISPER expression, and not the expression of its proximal PCG WISP2, was significantly increased in the severe fibrosis group (Fig. 7A; P = 0.012 and 0.120, respectively). Moreover, in a correlation analysis, WISPER expression and not WISP2 was found to be associated with the degree of CVF in all AOS patients (Fig. 7B). To evaluate the functional importance of WISPER in human fibroblasts, we used human dermal fibroblasts and human CFs. Both cell types can be induced to differentiate into myofibroblasts via serum starvation. This process is associated with the up-regulation of COL1A1, COL3A1, and aSMA in both fibroblast populations (fig. S7, B and C). WISPER was significantly induced by serum starvation in CFs but not in dermal fibroblasts (Fig. 7C; P < 0.001). WISP2 was also up-regulated in differentiating CFs. Supporting observations in the mouse, WISPER expression was correlated with COL3A1, FN1, and aSMA expression in differentiating human CFs (Fig. 7D). The functional importance of WISPER was tested in knockdown experiments. WISPER depletion using GapmeRs resulted in decreased expression of COL1A1, COL3A1, FN1, and aSMA in human CFs (Fig. 7E). Silencing of WISPER did not affect WISP2 expression, suggesting again that WISPER controls fibrotic gene expression independently of WISP2. Finally, PLOD2 expression was decreased in human CFs after GapmeR-mediated WISPER depletion (Fig. 7F). These findings demonstrate that WISPER is an IncRNA conserved in humans and highlight the translational relevance of WISPER as a therapeutic target in cardiac fibrosis.
Fig. 7. WISPER, a functionally conserved human ortholog of mouse Wisper.
(A) Box plot representation of the CVF in the heart of the nonsevere (n = 11) and severe fibrosis (n = 15) groups of patients affected by AOS. The line indicates the CVF cutoff value used to differentiate the two groups (12%). Box plots showing WISPER and WISP2 relative expression analyzed by qRT-PCR in cardiac biopsies from the two different fibrotic groups. Data show the mean, all the individual values, and the minimal to maximal variation. P values were determined by Student’s t test (compared to nonsevere fibrosis group). (B) Correlation analysis between the CVF and WISPER and WISP2 expression quantified by qRT-PCR. Pearson’s correlation test (r; 95% CI). (C) Time course of WISPER and WISP2 expression in differentiating human CFs [red, cardiac fibroblast (FBs)] and dermal FBs (green). Graphs show means normalized to control ± SEM (n = 6 to 14). P values versus control were determined by one-way ANOVA. (D) Correlation analysis between WISPER expression and COL1A1, COL3A1, FN1, and aSMA expression in differentiating human CFs. Spearman’s correlation test (r; 95% CI). (E) Effect of GapmeR-induced WISPER depletion (25 nM, 48 hours) on WISP2 and fibroblast gene expression in human CFs. Bars show mean ± SEM (n = 3). P values were determined by Student’s t test. (F) Effect of GapmeR-induced WISPER depletion (25 nM, 48 hours) on PLOD2 expression in human CFs. Bars show mean ± SEM (n = 3). P values were determined by Student’s t test.
DISCUSSION
After MI, activated CFs and associated ECM production assume crucial roles during the acute and chronic phases of the adaptive response of the heart to new hemodynamic conditions (1). However, maladaptive changes in ECM dynamics lead to the long-term disruption of myocardial architecture and function, eventually leading to HF. Excessive ECM deposition and its adverse effects are potentially modifiable (5, 34); a reduction in the development of fibrosis can be observed in humans treated with therapeutics aimed at limiting pathological remodeling of the diseased heart (5). Because of the nontargeted nature of these approaches, beneficial effects on fibrosis are relatively modest. Nonetheless, these agents improved clinical outcomes, providing a strong rationale for the development of highly specific therapeutics directly targeting the CF population.
The goal of this study was to identify fibrosis-associated therapeutic targets. We focused on the enhancer landscape and their associated transcripts and discovered the evolutionary conserved SE-associated IncRNA that we named Wisper, a polyadenylated and multiexonic CF-enriched transcript. Although the human genome encodes thousands of polyadenylated multiexonic IncRNAs, their functional and translational importance in controlling pathological remodeling and fibrosis remains largely unexplored. In contrast, the roles of IncRNAs in cardiogenesis and in maintaining identity in adult CMs are emerging, supporting their translational importance as therapeutic targets in the diseased adult heart (16, 47). We recently demonstrated that developmental enhancer–associated IncRNAs are reexpressed in stressed mouse and human hearts, potentially as part of a reactivation of a fetal gene program under pathological conditions (19, 30). The IncRNA Mhrt, which is associated with the Myh7 locus, was shown to protect the heart from hypertrophy and failure (20). Two other IncRNAs, Chast and Chaer, have recently been shown to play important roles during maladaptive cardiac remodeling (21, 48). Down-regulation of Chast after GapmeR administration can prevent tissue remodeling or even induce its regression after stress. Although extremely encouraging, these transcripts are primarily CM-enriched. Therefore, although their modulation produces beneficial effects on pathological fibrosis, they do not specifically control CF biology. Wisper, on the other hand, is a highly CF-enriched IncRNA. Furthermore, Wisper depletion leads to a global transcriptional reprogramming of the networks controlling proliferation, migration, apoptosis, and differentiation solely in CFs. Considering its distribution within both the cytoplasm and nucleus, Wisper could exert its action via both cis- and trans-regulatory functions, presumably via interaction with nuclear and cytoplasmic RNA binding proteins. In this context, we identified TIAR as a Wisper-associated protein. TIAR plays an important role in strengthening alternative 5′ splice sites (49). PLOD2 has two splice variants with the long form containing an additional exon, whose inclusion is controlled by TIAR. TIAR has been therefore implicated in tissue fibrosis via its capacity to promote production of the long profibrotic PLOD2 form (44). The short form is not expressed in the heart. Moreover, both Wisper and Plod2 bind TIAR, and down-regulation of Wisper in CFs decreases TlAB/Plod2 interaction and Plod2 expression, suggesting that Wisper regulates Plod2 mRNA posttranscriptional processing and its cellular concentrations via controlling Plod2 association with TIAR. PLOD2 has also been shown to induce collagen synthesis independently of its action on cross-linking and stabilization of the matrix (50), further supporting a central role for TIAR and PLOD2 in fibrosis. In addition, TIAR is a multifunctional protein that dictates many aspects of RNA metabolism. TIAR functionally interacts with ncRNAs, including IncRNAs (51, 52). IncRNA-mediated regulation of TIAR is therefore emerging as a common mechanism to confer context and cell specificity on an otherwise ubiquitously acting system. In particular, TIAR has been implicated in the control of cell proliferation and apoptosis. The GO terms associated with the transcriptional programs after Wisper knockdown are highly reminiscent of the GO terms associated with modulated genes in response to TIAR knockdown (53). In this context, Wisper functions, in part, by controlling TIAR shuttling into the nucleus (45). Blocking nuclear translocation of TIAR in Wisper-depleted cells is therefore expected to produce global effects on fibroblast gene programs. Three other proteins were identified by mass spectrometry after Wisper pulldown. PTBP3, DIS3L2, and CELF2 demonstrate relevant functions as regulators of differentiation, proliferation, and apoptosis (41–43). Their roles in CFs, and in particular in the development of fibrosis, have not been investigated. These candidates therefore warrant further characterization. It is important to note, however, that many IncRNAs are associated with RNA processing factors involved in maturation of the primary transcripts via posttranscriptional modification. Association of these proteins with a multiexonic IncRNA such as Wisper might also reflect the need for this transcript to be appropriately processed for ensuring its functions. In this regard, TIAR, while being a splicing factor, is the only protein of this small group with a demonstrated link to fibrosis.
The extent of gene reprogramming in fibroblast after Wisper knockdown demonstrates that Wisper exerts several important functions, some of which are probably independent of TIAR. We therefore examined the expression of genes topologically associated within the Wisper locus. Among those, Wisp2 is comodulated with Wisper during fibroblast differentiation and after Wisper knockdown, suggesting that Wisper could directly control its proximal coding gene in cis. However, several pieces of evidence indicate that concomitant expression of Wisper and Wisp2 does not necessarily reflect cis-regulation. Although Wisper knockdown in mouse CFs in vitro induces Wisp2 down-regulation, this effect is not observed in human CFs. In addition, the expression signature after Wisp2 loss of function in CFs is distinct from that observed after Wisper knockdown, suggesting that Wisper effects are not mediated via Wisp2. Moreover, CRISPR-on–mediated induction of Wisper expression at its site of transcription does not activate Wisp2 expression. Finally, Wisp2 is highly expressed in the lungs where modest Wisper expression is observed. The reason for Wisp2 being highly expressed in this organ is possibly related to the presence, in its promoter, of binding sites for TFs that are known to induce lung-specific gene programs. In the heart, Wisp2 has recently been implicated in the therapeutic regression of cardiac fibrosis (31). Wisp2 appears to act therefore more as a compensatory molecule that limits the extent of fibrosis rather than as a promoter of fibrosis. We therefore believe that Wisp2 down-regulation in the heart occurs secondary to Wisper depletion as a direct consequence of the induced beneficial impact on cardiac fibrosis.
Unlike most tissues in which myofibroblasts undergo apoptosis or revert to a quiescent state in the absence of pathological stress, this does not occur in the heart. However, GapmeR-mediated depletion of Wisper before MI attenuates pathological fibrosis and remodeling. Nevertheless, preventive Wisper depletion also negatively affects the acute wound healing process, resulting in cardiac rupture and increased mortality. Loss of CFs in the stressed heart affects the matrix that is key for the homeostatic maintenance of myocardial integrity (6). CFs produce the collagenous matrix that prevents myofiber slippage and sustains ventricular chamber geometry under normal conditions; interfering with this process results in weakening of the ventricular wall and susceptibility to rupture. Along the same lines, it is likely that massive doses of anti-Wisper GapmeRs destabilize the delicate architecture of the normal heart, leading to cardiac dysfunction. This has been observed in other situations in which important modulators of fibrosis, such as muscleblind-like1 (MBNL1), are down-regulated during the acute phase of the response to infarction (54). MBNL1 is an RNA binding protein that regulates transcripts that promote myofibroblast differentiation, and mice lacking MBNL1 develop cardiomyopathy with reduced function.
The detrimental effects of the Wisper GapmeR treatment during the acute phase could be overcome when using a therapeutic protocol in which GapmeRs were administered 2 days after infarction. Wisper knockdown during this clinically relevant window of time did not negatively affect acute wound healing while still blunting pathological fibrosis, reducing remodeling, and improving cardiac function at 7 and 28 days after infarction. These data coupled with the observation that human WISPER expression is correlated with fibrosis in AOS patients support translation into clinical scenarios. Clearance of activated CFs from the diseased heart appears to be a highly efficient process leading to progressive reduction of pathological fibrosis and diminished incidence of HF (5, 6). One could also envisage Wisper being targeted in the context of unchecked reactive fibrosis in the myocardium independently of the underlying disease etiology. This could be of high clinical relevance because myocardial fibrosis persists in patients with HF even when treated according to current guidelines and correlates with mortality. An important finding in our study is the apparent cardiac specificity of Wisper expression. Under basal conditions, Wisper is clearly a CF-enriched transcript. Cardiac TF binding sites in the SE element provide an explanation for this interesting feature. Along this vein, data obtained in the renovascular hypertensive model are quite informative. Both the heart and the kidney develop severe fibrosis, but Wisper is only induced in the heart. This observation suggests that, in the stressed kidney, fibrosis is not Wisper-dependent. Nevertheless, we cannot exclude the possibility that Wisper expression could be induced in other cell types and/or tissues. This certainly warrants further investigation. Finally, our data also demonstrate that Wisper is potentially a highly sensitive marker for pathological fibrosis. Considering the ability to detect IncRNAs circulating in human plasma (55), measurement of circulating WISPER concentrations might provide a noninvasive means to monitor cardiac remodeling. We have previously analyzed a series of novel IncRNAs including Wisper and correlated their expression with echocardiographic traits after infarction, supporting the notion that these transcripts may represent interesting marker candidates (29). Together, the identification of this CF-specific regulatory molecule therefore represents an important step toward the development of targeted antifibrotic therapeutic approaches and diagnostic tools.
MATERIALS AND METHODS
Study design
The main goal of our study was to evaluate cell-specific IncRNAs as therapeutic targets for cardiac fibrosis and heart disease. Using a stringent pipeline, we selected high-priority IncRNA candidates that are associated with cardiac-specific SEs. We focused our attention on one CF-enriched IncRNA named Wisper. Loss-of-function experiments were performed to assess the role of Wisper in the biology of CFs and in fibrosis. Modified ASOs (GapmeRs) were used to silence Wisper expression in mouse CFs. Knockdown experiments were performed in human CFs to verify the evolutionary conserved function of this transcript. For experiments in vivo, LAD artery ligation was chosen as a well-established animal model of MI. Experimental groups include at least six animals to robustly identify alteration in cardiac function and fibrosis. Mice were randomly assigned to treatment groups. To test whether Wisper may hold therapeutic potential in cardiac fibrosis, we performed loss-of-function experiments in vivo via GapmeR injection. In the preventive approach, injection was performed 3 days before MI. In the therapeutic approach, GapmeRs were injected 2 and 9 days after infarction. GapmeR injection and echocardiographic measurements were double-blinded until statistical analysis. Cardiac fibrosis was quantified by Masson’s trichrome staining on heart sections. All sample measurements were blinded. In addition, to evaluate the tissue specificity of Wisper expression, we took advantage of the 1K1C model of renovascular hypertension in the mouse. This model is characterized by the development of cardiac hypertrophy and fibrosis secondary to volume-dependent hypertension. Fibrosis also develops in the clipped kidney, allowing direct comparison of Wisper expression in two different fibrotic organs. WISPER human ortholog was identified and detected in RNA isolated from human cardiac biopsies of patients suffering from AOS (all primers are listed in table S6). This pathology is associated with extensive myocardial fibrosis that directly contributes to LV dysfunction and HF. Samples of cardiac tissue from AOS patients (n = 26) were divided into two groups after analyzing the distribution for CVF, as described in the text. No data were excluded in this analysis.
RNA-seq–based lncRNA profiling after MI
RNA-seq on infarcted and sham-operated hearts (14 days after infarction), ab initio transcript reconstruction, differential expression analysis of IncRNAs, heart specificity analysis, and human ortholog identification data sets were previously described (29).
SE mapping to human IncRNA orthologs
The locations of SEs were downloaded from (12). These regions were defined using H3K27ac chromatin immunoprecipitation sequencing from the Epigenome Roadmap (56). IncRNAs arising from SEs and TEs were determined by genomic overlap.
Animal experiments
Animal experiments were approved by the Government Veterinary Office (Lausanne, Switzerland) and performed according to the University of Lausanne Medical School institutional guidelines.
MI model
MI in mice (numbers of animals per group are in the figure legends) was induced as previously described (57). Briefly, male C57BL/6 mice (Charles River) at 12 weeks of age were anesthetized by intraperitoneal injection of ketamine/xylazine/acepromazine (65/15/2 mg/kg) and placed on artificial ventilation. After left thoracotomy, the pericardium was gently opened and a 7.0 silk ligature (Aesculap) was tied around the LAD artery near the insertion of the left auricular appendage. Occlusion of the artery was verified by the rapid blanching of the LV. In sham-operated mice, the ligature was placed in an identical location but not tied. After surgery, mice were gradually weaned from the respirator until spontaneous respiration was resumed and replaced in the cage.
1K1C renovascular hypertension in mice
A 1K1C renovascular hypertension in mice was produced as previously described (35). Briefly, a left lateral abdominal incision is used to expose the kidney. A clip (0.12 mm opening) is placed around the left renal artery to reduce renal blood flow. Another incision is made in the right lateral abdominal wall, and a right nephrectomy is performed. Animals develop volume overload–dependent cardiac hypertrophy and renal failure.
Echocardiograph
Transthoracic echocardiography was performed using a 30-MHz probe and the Vevo 770 Ultrasound machine (VisualSonics). Mice were lightly anesthetized with 1% isoflurane, maintaining heart rate at 400 to 500 beats per minute, and placed in dorsal recumbency on a heated 37°C platform. The heart was imaged in the two-dimensional mode in the parasternal long-axis view. From this view, an M-mode cursor was positioned perpendicular to the interventricular septum and the posterior wall of the LV at the level of the papillary muscles. LV free wall thickness in diastole (LVWT d) and in systole (LVWT s) as well as LV diameter in diastole (LVD d) and in systole (LVD s) were measured according to the American Society of Echocardiography guidelines. The measurements were taken in three separate M-mode images and averaged. EF was calculated using the formula %EF = [(LVVD – LVVS)/LVVD] × 100, where LVVD and LVVS are LV volume in diastole and systole, respectively.
GapmeR delivery in vivo
GapmeR-Wisper and GapmeR-Scrambled control A (Exiqon) were diluted in NaCl 0.9% isotonic solution to obtain the final exact concentration (5, 10, or 15 mg/kg) just before the injection. GapmeRs were delivered intraperitoneally using a U100 insulin 0.3-ml syringe and a 30-gauge, 8-mm needle.
Primary cell culture and transfection
Methods are described in detail in the Supplementary Materials.
Neonatal mouse CM and fibroblasts
Cells were obtained from neonatal C57BL/6 mice.
Adult mouse cardiac, lung, and tail fibroblasts
Cells were obtained from 12-week-old C57BL/6 mice.
Human fibroblasts
Adult human dermal fibroblasts were purchased from Gibco. Primary cell cultures of human CFs were obtained by tissue enzymatic digestion with collagenase and explant outgrowth from atrial samples, as previously described (58).
GapmeR transfection in mouse primary cell
Adult CFs, neonatal CF, lung fibroblasts, and CMs were transfected with 10 nM (if not differently specified in the text) of LNA IncRNA GapmeRs (GapmeR-Wisper or scrambled GapmeR or GapmeR-Wisp2; Exiqon).
GapmeR transfection in human CFs
Fibroblasts were transfected with 25 nM LNA IncRNA GapmeRs (GapmeR-Wisper or scrambled GapmeR; Exiqon).
CRISPR-on assay
CRISPR-based gain of function was used to activate Wisper expression in P19CL6 cells (RCB2318; RIKEN Cell Bank). Cells were cultured in Dulbecco’s modified Eagle’s medium with 10% fetal calf serum (FCS) and transfected with a component of the synergistic activation mediator described by the Zhang laboratory in combination with a Wisper-targeting guide RNA engineered to contain two MS2 aptamers [sgRNA(MS2); Addgene plasmid no. 61424] and containing the sequence GTCGACTCTGCTATACTCCA. Plasmids were transfected at a 1:1 ratio using Lipofectamine 2000 (Life Technologies) according to the manufacturer’s instructions. Total RNA was isolated 48 hours after transfection using the miRNeasy kit (Qiagen) and subjected to qRT-PCR.
Cytoplasmic and nuclear compartment fractionation
Adult CFs (2 ×106) were washed twice with cold phosphate-buffered saline (PBS). Nuclear and cytoplasmic RNA fractions were isolated using the Cytoplasmic and Nuclear RNA purification kit (Norgen Biotek Corp.) according to the manufacturer’s instructions. Total RNA was isolated from 1 × 106 adult CFs using the miRNeasy kit (Qiagen) and used as input.
Immunohistochemistry
Cardiomyocytes
CMs were transfected with 10 nM GapmeR-Wisper or GapmeR-Scrambled for 48 hours. Cells were then fixed for 10 min in 4% paraformaldehyde in PBS and permeabilized with 0.2% Triton X-100 in PBS. After treatment with blocking buffer (PBS containing 0.001% Triton X-100, 1% bovine serum albumin, and 1% FCS), cells were incubated overnight at 4°C with anti–α-sarcomeric actinin antibodies (1:400; Sigma). One day after, cells were washed three times and incubated for 1 hour at room temperature in the dark with conjugated anti-mouse donkey secondary antibodies (1:500, Alexa Fluor 488; Life Technology). Nuclei were stained with DAPI (Invitrogen). Unbound antibodies were washed out with PBS 0.1% Tween. Subsequently, coverslips were mounted with Fluoromount-G (SouthernBiotech) and analyzed with an inverted Axiovision Observer Zl fluorescence microscope (Carl Zeiss). Five pictures for each different condition were taken. The dimension of the cells was measured using ImageJ. CMs were counted as α-sarcomeric actinin–positive and DAPI-positive cells, whereas nonmyocyte cells were counted as α-sarcomeric actinin–negative and DAPI-positive cells.
Cardiac fibroblasts
Cells were transfected with 5 nM GapmeR-Wisper or GapmeR-Scrambled for 48 hours. Cells were fixed as described above and incubated overnight at 4°C with anti-TIAL antibody (1:200, AM29499; Abeam). One day thereafter, cells were washed three times and incubated for 1 hour at room temperature with conjugated anti-rabbit goat secondary antibodies (1:250, Alexa Fluor 488; Life Technology). Actin filaments were stained using Texas Red-X Phalloidin (1:100, T7471; Life Technologies), and nuclei were stained with DAPI (Invitrogen). Unbound antibodies were washed out with PBS 0.1% Tween. Subsequently, coverslips were mounted with Fluoromount-G (Southern Biotech) and TIAL localization was analyzed with an inverted Axiovision Observer Zl fluorescence microscope (Carl Zeiss).
Assessment of cardiac fibrosis
Hearts were harvested from sham-operated and MI mice, and atria and big vessels were eliminated. Cardiac tissues were fixed in 4% formalin overnight. Paraffin tissue sections were processed for Masson’s trichrome staining using standard histological procedures. The percentage of fibrotic tissue was determined by measuring collagen deposition (blue) on Masson’s trichrome-stained sections using ImageJ.
RNA pulldown
Biotinylated Wisper sense and Wisper antisense were in vitro transcribed using the T7 or T3 RNA polymerase (Promega) and Biotin RNA Labeling Mix (Roche) and then purified with Quick Spin columns (Roche) according to the manufacturers’ instructions. Biotinylated RNA (4 µg) was denaturated for 5 min at 65°C in RNA structure buffer (10 mM tris-HCl, 10 mM MgCl2, and 100 mM NH4C1) and cooled to room temperature. In brief, freshly harvested CFs were washed in ice-cold PBS. Cells were lysed in 1 ml lysis buffer [25 mM tris-HCl (pH 7.4), 150 mM NaCl, 5 mM MgCl2, 0.1 mM EDTA, 0.5% NP-40, 1 mM dithiothreitol (DTT), 1 mM phenylmethylsulfonyl fluoride, ribonuclease inhibitor (0.1 U/µ1; Promega), and l× protease inhibitor cocktail (Sigma)]. Streptavidin Dynabeads were washed in NT2 buffer [50 mM tris-HCl (pH 7.4), 150 mM NaCl, 1 mM MgCl2, 0.05% NP-40,1 mM DTT, 20 mM EDTA, 400 mM vanadyl-ribonucleoside, RNase inhibitor (0.1 U/µl; Promega), and l× protease inhibitor cocktail (Sigma)] and used to preclear the lysate (20 min at 4°C). The precleared cell lysate was incubated with biotinylated RNA along with RNase inhibitor (0.1 U/µ1; Promega) and yeast transfer RNA (20 µg/ml; Ambion) for 1.5 hours at room temperature followed by addition of 60 µ1 of Streptavidin Dynabeads for 1.5 hours. The beads containing the RNA protein complex were washed thrice in NT2 buffer and then were directly boiled in SDS-gel loading dye. Retrieved proteins were loaded on a 12% polyacrylamide gel in denaturating SDS–polyacrylamide gel electrophoresis (SDS-PAGE) buffer and visualized with Candiano colloidal Coomassie staining. Mass spectrometry was performed at the Protein Analysis Facility (University of Lausanne, Switzerland). Analysis of the pulled-down proteins was performed using Scaffold4. Protein enrichment was calculated by Fisher’s exact test.
Western blot of RNA pulldown
Four percent of the samples used for RNA pulldown were used as input. Proteins were resolved on 10% SDS-PAGE minigels and transferred to polyvinylidene difluoride membranes (Bio-Rad). The membranes were blocked and incubated with primary antibody against TIAR (Santa Cruz Biotechnology Inc.) at 4°C overnight. The primary antibody was detected using infrared IrDye antibody reagents, and membranes were scanned using the Odyssey Infrared Imaging System (LI-COR Biosciences). Protein quantification on the scanned Western blots was performed using the software provided with the scanner (LI-COR Biosciences). Mouse TIAR lysate (20 µg; Santa Cruz Biotechnology) was used as a control.
RNA immune precipitation
Cells were harvested in lysis buffer as described previously. Supernatant was precleared with Dynabeads G (Invitrogen). The precleared lysate was incubated with normal IgGor with anti-TIAR antibody (Santa Cruz Biotechnology) at 4°C overnight followed by addition of 50 µl of Dynabeads G for 1 hour at 4°C. The immunoprecipitated complexes were washed thrice in NT2 buffer and eluted in 500 µ1 of QIAzol. Total RNA extraction and complementary DNA synthesis were performed as described before. RT-PCR system (Applied Biosystems) was then used to measure expression of Wisper, Wisp2, and Plod2.
Human tissue sampling
The present study conforms to the principles of the Declaration of Helsinki. All subjects were duly informed and gave written consent. Samples were collected as previously described (46).
Collagen volume fraction
The fraction of myocardium occupied by collagen was quantified as previously described (46). A cluster analysis was performed according to the CVF values to define the nonsevere (CVF, <12%; n = 11) and the severe (CVF, >12%; n = 15) fibrosis groups (46).
Echocardiography analysis
LV mass was measured from M-mode recordings. LV mass index was calculated by dividing LV mass by body surface area. LV end-systolic and end-diastolic volume indices (LVESVI and LVEDVI, respectively), corresponding to the LV volumes corrected by body surface area, and the LVEF were determined in all patients. The following pulsed Doppler measurements were obtained: maximum early (VE) transmitral velocity in diastole, maximum late (VA) transmittal velocity in diastole, the deceleration time of the early mitral filling wave (DT), and the isovolumetric relaxation time (IVRT).
Sequencing of RNA isolated from GapmeR-transfected adult CFs
Total RNA was isolated from adult CF transfected (10 nM, 48 hours) with GapmeR-Wisper (n = 4) or GapmeR-Scrambled (n = 4) using the miRNeasy kit (Qiagen). Sequencing libraries were prepared according to the Illumina RNA Seq library kit instructions with PolyA selection. Libraries were sequenced with the Illumina HiSeq2000 (1 × 100 bp). Purity-filtered reads were adapter- and quality-trimmed with Cutadapt (v. 1.3) and filtered for low complexity with seq_crumbs (v. 0.1.8). Reads were aligned against the Mus musculus. GRCm38.82 genome using STAR (v. 2.4.2a) (59). The number of read counts per gene locus was summarized with HTSeq count (v. 0.6.1) (60) using Mus musculus. GRCm38.82 gene annotation. Quality of the RNA-seq data alignment was assessed using RSeQC (v. 2.3.7) (61). Reads were also aligned to the Mus musculus.GRCm38.82 transcriptome using STAR (v. 2.4.2a) (59), and the estimation of the isoform abundance was computed using RSEM (v. 1.2.19) (62). Statistical analysis was performed for genes and isoforms independently in R (R version 3.1.2). Genes/isoforms with low counts were filtered out according to the rule of 1 cpm in at least one sample. Library sizes were scaled using trimmed mean of M-value (TMM) normalization (EdgeR v 3.8.5) (63) and log-transformed with limma voom function (R version 3.22.4). Statistical quality controls were performed through pairwise sample correlations, clustering, and sample principal components analysis. Replicates cluster together and are well separated between conditions. Differential expression was computed with limma (64) by fitting data into a linear model, adding the factor for the batch effect, and comparing GapmeR versus control conditions. The P values were adjusted for multiple comparisons using the Benjamini-Hochberg method (65), controlling for false discovery rate or adjusted P value.
Statistical analysis
GraphPad Software (version 6 or 7) was used for statistical analysis. Data throughout the paper are expressed as mean ± SEM. Statistical significance between two columns was assessed by two-tailed unpaired Student’s t test; for more than two columns, one-way ANOVA [Fisher’s least significant difference (LSD) test] analysis was used. Two-way ANOVA (Fisher’s LSD test) was used to evaluate statistical significance between two or more groups. Correlation analysis was performed with Pearson (r or r2 values; 95% CI) or Spearman (r; 95% CI) test. Significance in percentage of animal survival was calculated with log-rank (Mantel-Cox) test. P values <0.05 were considered significant in all events. Individual-level data and exact P values are shown in table S7.
Supplementary Material
Fig. S1. Conservation between the mouse and human Wisper transcripts.
Fig. S2. Wisper is expressed in the stressed fibrotic heart but not in the stressed fibrotic kidney.
Fig. S3. Effects of Wisper knockdown in neonatal CFs and CMs.
Fig. S4. TAD centered on the murine Wisper locus; long noncoding transcriptome after Wisper knockdown in fibroblasts; Wisper pulldown assay.
Fig. S5. Dose-response analysis after injection of GapmeRs targeting Wisper in vivo.
Fig. S6. Preventative Wisper depletion inhibits cardiac fibrosis and improves function.
Fig. S7. Expression of cardiac markers of stress after therapeutic depletion of Wisper in the infarcted heart; expression of myofibroblast genes in differentiating cardiac and dermal human fibroblasts.
Table S1. Echocardiographic parameters measured 1, 7, 14, and 28 days after MI.
Table S2. Echocardiographic parameters measured 4, 14, and 28 days in untouched mice injected with 5, 10, or 15 mg/kg of GM-Scrambled (control) or GM-Wisper (Wisper).
Table S3. Echocardiographic parameters measured 7 and 14 days after MI in mice injected with GapmeRs 3 days before MI.
Table S4. Echocardiographic parameters measured 7 and 28 days after MI in mice injected with GapmeRs 2 days and 9 days after MI.
Table S5. Characteristics of patients suffering from AOS and developing cardiac fibrosis.
Table S6. List of the primers used in this study.
Table S7. Individual-level data and exact P values (Excel format).
Acknowledgments
We thank T. Beckmann (University of Lausanne Medical School, Switzerland) and W. Verhesen (Maastricht University, Netherlands) for their technical assistance. We are grateful to J. Diez (University of Navarra, Spain) for granting access to human tissue collections and to L. Martínez and A. González (University of Navarra) for their help in culturing human fibroblasts. We thank A.-C. Clerc (University of Lausanne) for generating the 1K1C model in mice.
Funding: This project was supported, in part, by grants from the Swiss National Science Foundation, Bern, Switzerland (406340-128129 and 31003A-163476 to T.P.); from the Ministry of Economy and Competitiveness, Spain (CB16/11/ 00483, RYC-2010-05797, and PI15/01909 to A.G); and from the European Union (FIBRO-TARGETS consortium, grant HEALTH-2013-602904). BJ.A. is a Hope Funds for Cancer Research Grillo-Marxuach Family Fellow. B.S. received funding from the Netherlands Organization for Scientific Research (Vidi grant no. 91714363) and the Netherlands Heart Foundation (Dekker 2014T105).
Footnotes
www.sciencetranslationalmedicine.org/cgi/content/full/9/395/eaai9118/DC1 Materials and Methods
Author contributions: T.P. and S.O. designed the experiments and wrote the manuscript. R.M. performed and analyzed the experiments and created the figures. BJ.A. and R.A.Y. identified cardiac SEs. A.S. performed animal experiments. B.S. performed in situ hybridization. C.-C.T., D. Maric, and M.N. contributed to primary cell isolation, cell culture, and GapmeR transfection experiments. M.A. performed the annexin V analysis by FACS. D. Maison and I.P. performed the RNA pulldown and RNA immunoprecipitation assays. A.G. performed analysis of human cardiac tissue and cells.
Competing interests: S.O. and T.P. filed a patent about therapeutic use of cardiac-enriched lncRNAs including Wisper (patent title: “Diagnostic, prognostic and therapeutic uses of lncRNAs for heart disease and regenerative medicine″; international application number: PCT/EP2014/078868; applicant: University of Lausanne).
Data and materials availability:
All the data and materials are available through the Gene Expression Omnibus using the following accession numbers: LV (GSM908951 and GSM906396), adipose tissue (GSM906394), adrenal gland (GSM1013126 and GSM896163), bladder (GSM1013133), gastric (GSM1013122, GSM1013128, and GSM910555), ovary (GSM956009), pancreas (GSM1013129 and GSM906397), colon (GSM915331 and GSM910559), small intestine (GSM1013131), spleen (GSM1013132 and GSM906398), and thymus (GSM1013125).
References
- 1.Prabhu SD, Frangogiannis NG. The biological basis for cardiac repair after myocardial infarction: From inflammation to fibrosis. Circ. Res. 2016;119:91–112. doi: 10.1161/CIRCRESAHA.116.303577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frangogiannis NG. Pathophysiology of myocardial infarction. Compr. Physiol. 2015;5:1841–1875. doi: 10.1002/cphy.c150006. [DOI] [PubMed] [Google Scholar]
- 3.Writing Group Members. Mozaffarian D, Benjamin EJ, Go AS, Arnett DK, Blaha MJ, Cushman M, Das SR, de Ferranti S, Despres J-P, Fullerton HJ, Howard VJ, Huffman MD, Isasi CR, Jimenez MC, Judd SE, Kissela BM, Lichtman JH, Lisabeth LD, Liu S, Mackey RH, Magid DJ, McGuire DK, Mohler ER, III, Moy CS, Muntner P, Mussolino ME, Nasir K, Neumar RW, Nichol G, Palaniappan L, Pandey DK, Reeves MJ, Rodriguez CJ, Rosamond W, Sorlie PD, Stein J, Towfighi A, Turan TN, Virani SS, Woo D, Yeh RW, B M. Turner; American Heart Association Statistics Committee; Stroke Statistics Subcommittee, Heart disease and stroke statistics— 2016 update: A report from the American Heart Association. Circulation. 2016;133:e38–e360. doi: 10.1161/CIR.0000000000000350. [DOI] [PubMed] [Google Scholar]
- 4.Spinale FG, Zile MR. Integrating the myocardial matrix into heart failure recognition and management. Cire. Res. 2013;113:725–738. doi: 10.1161/CIRCRESAHA.113.300309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schelbert EB, Fonarow GC, Bonow RO, Butler J, Gheorghiade M. Therapeutic targets in heart failure: Refocusing on the myocardial interstitium. J. Am. Coll. Cardiol. 2014;63:2188–2198. doi: 10.1016/j.jacc.2014.01.068. [DOI] [PubMed] [Google Scholar]
- 6.Weber KT, Sun Y, Bhattacharya SK, Ahokas RA, Gerling IC. Myofib rob last-mediated mechanisms of pathological remodelling of the heart. Nat. Rev. Cardiol. 2013;10:15–26. doi: 10.1038/nrcardio.2012.158. [DOI] [PubMed] [Google Scholar]
- 7.Lighthouse JK, Small EM. Transcriptional control of cardiac fibroblast plasticity. J. Mol. Cell. Cardiol. 2016;91:52–60. doi: 10.1016/j.yjmcc.2015.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schuetze KB, McKinsey TA, Long CS. Targeting cardiac fibroblasts to treat fibrosis of the heart: Focus on HDACs. J. Mol. Cell. Cardiol. 2014;70:100–107. doi: 10.1016/j.yjmcc.2014.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ounzain S, Pedrazzini T. The promise of enhancer-associated long noncoding RNAs in cardiac regeneration. Trends Cardiovasc. Med. 2015;25:592–602. doi: 10.1016/j.tcm.2015.01.014. [DOI] [PubMed] [Google Scholar]
- 10.Ounzain S, Pedrazzini T. Super-enhancer Incs to cardiovascular development and disease. Biochim. Biophys. Acta. 2016;1863:1953–1960. doi: 10.1016/j.bbamcr.2015.11.026. [DOI] [PubMed] [Google Scholar]
- 11.Wamstad JA, Wang X, Demuren OO, Boyer LA. Distal enhancers: New insights into heart development and disease. Trends Cell Biol. 2014;24:294–302. doi: 10.1016/j.tcb.2013.10.008. [DOI] [PubMed] [Google Scholar]
- 12.Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA, Hoke HA, Young RA. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Niederriter AR, Varshney A, Parker SCJ, Martin DM. Super enhancers in cancers, complex disease, and developmental disorders. Genes. 2015;6:1183–1200. doi: 10.3390/genes6041183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morris KV, Mattick JS. The rise of regulatory RNA. Nat. Rev. Genet. 2014;15:423–437. doi: 10.1038/nrg3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumarswamy R, Thum T. Non-coding RNAs in cardiac remodeling and heart failure. Cire. Res. 2013;113:676–689. doi: 10.1161/CIRCRESAHA.113.300226. [DOI] [PubMed] [Google Scholar]
- 17.Creemers EE, van Rooij E. Function and therapeutic potential of noncoding rnas in cardiac fibrosis. Cire. Res. 2016;118:108–118. doi: 10.1161/CIRCRESAHA.115.305242. [DOI] [PubMed] [Google Scholar]
- 18.Boon RA, Dimmeler S. MicroRNAs in myocardial infarction. Nat. Rev. Cardiol. 2015;12:135–142. doi: 10.1038/nrcardio.2014.207. [DOI] [PubMed] [Google Scholar]
- 19.Ounzain S, Micheletti R, Arnan C, Plaisance I, Cecchi D, Schroen B, Reverter F, Alexanian M, Gonzales C, Ng SY, Bussotti G, Pezzuto I, Notredame C, Heymans S, Guigo R, Johnson R, Pedrazzini T. CARMEN, a human super enhancer-associated long noncoding RNA controlling cardiac specification, differentiation and homeostasis. J. Mol. Cell. Cardiol. 2015;89:98–112. doi: 10.1016/j.yjmcc.2015.09.016. [DOI] [PubMed] [Google Scholar]
- 20.Han P, Li W, Lin C-H, Yang J, Shang C, Nurnberg ST, Jin KK, Xu W, Lin C-Y, Lin C-J, Xiong Y, Chien HC, Zhou B, Ashley E, Bernstein D, Chen P-S, Chen H-S, Quertermous T, Chang C-P. A long noncoding RNA protects the heart from pathological hypertrophy. Nature. 2014;514:102–106. doi: 10.1038/nature13596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Viereck J, Kumarswamy R, Foinquinos A, Xiao K, Avramopoulos P, Kunz M, Dittrich M, Maetzig T, Zimmer K, Remke J, Just A, Fendrich J, Scherf K, Bolesani E, Schambach A, Weidemann F, Zweigerdt R, de Windt LJ, Engelhardt S, Dandekar T, Batkai S, Thum T. Long noncoding RNA Chast promotes cardiac remodeling. Sci. Transl. Med. 2016;8:326ra322. doi: 10.1126/scitranslmed.aaf1475. [DOI] [PubMed] [Google Scholar]
- 22.Wang K, Liu F, Zhou L-Y, Long B, Yuan S-M, Wang Y, Liu C-Y, Sun T, Zhang X-J, Li P-F. The long noncoding RNA CHRF regulates cardiac hypertrophy by targeting miR-489. Cire. Res. 2014;114:1377–1388. doi: 10.1161/CIRCRESAHA.114.302476. [DOI] [PubMed] [Google Scholar]
- 23.Piccoli M-T, Bär C, Thum T. Non-coding RNAs as modulators of the cardiac fibroblast phenotype. J. Mol. Cell. Cardiol. 2016;92:75–81. doi: 10.1016/j.yjmcc.2015.12.023. [DOI] [PubMed] [Google Scholar]
- 24.Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat. Struct. Mol. Biol. 2013;20:300–307. doi: 10.1038/nsmb.2480. [DOI] [PubMed] [Google Scholar]
- 25.Li W, Notani D, Rosenfeld MG. Enhancers as non-coding RNA transcription units: Recent insights and future perspectives. Nat. Rev. Genet. 2016;17:207–223. doi: 10.1038/nrg.2016.4. [DOI] [PubMed] [Google Scholar]
- 26.Sigova AA, Mullen AC, Molinie B, Gupta S, Orlando DA, Guenther MG, Almada AE, Lin C, Sharp PA, Giallourakis CC, Young RA. Divergent transcription of long noncoding RNA/mRNA gene pairs in embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A. 2013;110:2876–2881. doi: 10.1073/pnas.1221904110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sigova AA, Abraham BJ, Ji X, Molinie B, Hannett NM, Guo YE, Jangi M, Giallourakis CC, Sharp PA, Young RA. Transcription factor trapping by RNA in gene regulatory elements. Science. 2015;350:978–981. doi: 10.1126/science.aad3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mele M, Rinn JL. “Cat’s Cradling” the 3D genome by the act of LncRNA transcription. Mol. Cell. 2016;62:657–664. doi: 10.1016/j.molcel.2016.05.011. [DOI] [PubMed] [Google Scholar]
- 29.Ounzain S, Micheletti R, Beckmann T, Schroen B, Alexanian M, Pezzuto I, Crippa S, Nemir M, Sarre A, Johnson R, Dauvillier J, Burdet F, Ibberson M, Guigo R, Xenarios I, Heymans S, Pedrazzini T. Genome-wide profiling of the cardiac transcriptome after myocardial infarction identifies novel heart-specific long non-coding RNAs. Eur. Heart J. 2015;36:353–368. doi: 10.1093/eurheartj/ehu180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ounzain S, Pezzuto I, Micheletti R, Burdet F, Sheta R, Nemir M, Gonzales C, Sarre A, Alexanian M, Blow MJ, May D, Johnson R, Dauvillier J, Pennacchio LA, Pedrazzini T. Functional importance of cardiac enhancer-associated noncoding RNAs in heart development and disease. J. Mol. Cell. Cardiol. 2014;76:55–70. doi: 10.1016/j.yjmcc.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jeong D, Lee M-A, Li Y, Yang DK, Kho C, Oh JG, Hong G, Lee A, Song MH, LaRocca TJ, Chen J, Liang L, Mitsuyama S, D’Escamard V, Kovacic JC, Kwak TH, Hajjar RJ, Park WJ. Matricellular protein CCN5 reverses established cardiac fibrosis. J. Am. Coll. Cardiol. 2016;67:1556–1568. doi: 10.1016/j.jacc.2016.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pott S, Lieb JD. What are super-enhancers? Nat. Genet. 2015;47:8–12. doi: 10.1038/ng.3167. [DOI] [PubMed] [Google Scholar]
- 33.Okubo T, Hogan BLM. Hyperactive Wnt signaling changes the developmental potential of embryonic lung endoderm. J. Biol. 2004;3:11. doi: 10.1186/jbiol3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frangogiannis NG. The inflammatory response in myocardial injury, repair,and remodelling. Nat. Rev. Cardiol. 2014;11:255–265. doi: 10.1038/nrcardio.2014.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wiesel P, Mazzolai L, Nussberger J, Pedrazzini T. Two-kidney, one clip and one-kidney, one clip hypertension in mice. Hypertension. 1997;29:1025–1030. doi: 10.1161/01.hyp.29.4.1025. [DOI] [PubMed] [Google Scholar]
- 36.Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C, Hsu PD, Habib N, Gootenberg JS, Nishimasu H, Nureki O, Zhang F. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517:583–588. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang F, Yi F, Han X, Du Q, Liang Z. MALAT-1 interacts with hnRNP C in cell cycle regulation. FEBS Lett. 2013;587:3175–3181. doi: 10.1016/j.febslet.2013.07.048. [DOI] [PubMed] [Google Scholar]
- 38.Hacisuleyman E, Goff LA, Trapnell C, Williams A, Henao-Mejia J, Sun L, McClanahan P, Hendrickson DG, Sauvageau M, Kelley DR, Morse M, Engreitz J, Lander ES, Guttman M, Lodish HF, Flavell R, Raj A, Rinn JL. Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat. Struct. Mol. Biol. 2014;21:198–206. doi: 10.1038/nsmb.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.C Le Guiner, Lejeune F, Galiana D, Kister L, Breathnach R, Stévenin J, Del Gatto-Konczak F. TIA-1 and TIAR activate splicing of alternative exons with weak 5’ splice sites followed by a U-rich stretch on their own pre-mRNAs. J. Biol. Chem. 2001;276:40638–40646. doi: 10.1074/jbc.M105642200. [DOI] [PubMed] [Google Scholar]
- 40.Waris S, Wilce MCJ, Wilce JA. RNA recognition and stress granule formation by TIA proteins. Int. J. Mol. Sci. 2014;15:23377–23388. doi: 10.3390/ijms151223377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tan L-Y, Whitfield P, Llorian M, Monzon-Casanova E, Diaz-Munoz MD, Turner M, Smith CWJ. Generation of functionally distinct isoforms of PTBP3 by alternative splicing and translation initiation. Nucleic Acids Res. 2015;43:5586–5600. doi: 10.1093/nar/gkv429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blech-Hermoni Y, Stillwagon SJ, Ladd AN. Diversity and conservation of CELF1 and CELF2 RNA and protein expression patterns during embryonic development. Dev. Dyn. 2013;242:767–777. doi: 10.1002/dvdy.23959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haas G, Cetin S, Messmer M, Chane-Woon-Ming B, Terenzi O, Chicher J, Kuhn L, Hammann P, Pfeffer S. Identification of factors involved in target RNA-directed microRNA degradation. Nucleic Acids Res. 2016;44:2873–2887. doi: 10.1093/nar/gkw040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yeowell HN, Walker LC, Mauger DM, Seth P, Garcia-Blanco MA. TIA nuclear proteins regulate the alternate splicing of lysyl hydroxylase 2. J. Invest. Dermatol. 2009;129:1402–1411. doi: 10.1038/jid.2008.386. [DOI] [PubMed] [Google Scholar]
- 45.Zhang T, Delestienne N, Huez G, Kruys V, Gueydan C. Identification of the sequence determinants mediating the nucleo-cytoplasmic shuttling of TIAR and TIA-1 RNA-binding proteins. J. Cell Sci. 2005;118:5453–5463. doi: 10.1242/jcs.02669. [DOI] [PubMed] [Google Scholar]
- 46.Beaumont J, López B, Hermida N, Schroen B, San José G, Heymans S, Valencia F, Gómez-Doblas JJ, De Teresa E, Díez J, González A. microRNA-122 down-regulation may play a role in severe myocardial fibrosis in human aortic stenosis through TGF-P1 up-regulation. Clin. Sci. 2014;126:497–506. doi: 10.1042/CS20130538. [DOI] [PubMed] [Google Scholar]
- 47.Rizki G, Boyer LA. Lncing epigenetic control of transcription to cardiovascular development and disease. Circ. Res. 2015;117:192–206. doi: 10.1161/CIRCRESAHA.117.304156. [DOI] [PubMed] [Google Scholar]
- 48.Wang Z, Zhang X-J, Ji Y-X, Zhang P, Deng K-Q, Gong J, Ren S, Wang X, Chen I, Wang H, Gao C, Yokota T, Ang YS, Li S, Cass A, Vondriska TM, Li G, Deb A, Srivastava D, Yang H-T, Xiao X, Li H, Wang Y. The long noncoding RNA Chaer defines an epigenetic checkpoint in cardiac hypertrophy. Nat. Med. 2016;22:1131–1139. doi: 10.1038/nm.4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sánchez-Jimenez C, Izquierdo JM. T-cell intracellular antigens in health and disease. Cell Cycle. 2015;14:2033–2043. doi: 10.1080/15384101.2015.1053668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu J, Reinhardt DP, Batmunkh C, Lindenmaier W, Far RK-K, Notbohm H, Hunzelmann N, Brinckmann J. Functional diversity of lysyl hydroxylase 2 in collagen synthesis of human dermal fibroblasts. Exp. Cell Res. 2006;312:3485–3494. doi: 10.1016/j.yexcr.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 51.Wang Z, Kayikci M, Briese M, Zarnack K, Luscombe NM, Rot G, Zupan B, Curk T, Ule J. iCLIP predicts the dual splicing effects of TIA-RNA interactions. PLOS Biol. 2010;8:e1000530. doi: 10.1371/journal.pbio.1000530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu L, Yue H, Liu Q, Yuan J, Li J, Wei G, Chen X, Lu Y, Guo M, Luo J, Chen R. LncRNA MT1JP functions as a tumor suppressor by interacting with TIAR to modulate the p53 pathway. Oncotarget. 2016;7:15787–15800. doi: 10.18632/oncotarget.7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sánchez-Jiménez C, Izquierdo JM. T-cell intracellular antigen (TIA)-proteins deficiency in murine embryonic fibroblasts alters cell cycle progression and induces autophagy. PLOS ONE. 2013;8:e75127. doi: 10.1371/journal.pone.0075127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Davis J, Salomonis N, Ghearing N, Lin S-CJ, Kwong JQ, Mohan A, Swanson MS, Molkentin JD. MBNL1-mediated regulation of differentiation RNAs promotes myofibroblast transformation and the fibrotic response. Nat. Commun. 2015;6:10084. doi: 10.1038/ncomms10084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kumarswamy R, Bauters C, Volkmann I, Maury F, Fetisch J, Holzmann A, Lemesle G, de Groote P, Pinet F, Thum T. Circulating long noncoding RNA, LIPCAR, predicts survival in patients with heart failure. Circ. Res. 2014;114:1569–1575. doi: 10.1161/CIRCRESAHA.114.303915. [DOI] [PubMed] [Google Scholar]
- 56.Bernstein BE, Stamatoyannopoulos JA, Costello JF, Ren B, Milosavljevic A, Meissner A, Kellis M, Marra MA, Beaudet AL, Ecker JR, Farnham PJ, Hirst M, Lander ES, Mikkelsen TS, Thomson JA. The NIH roadmap epigenomics mapping consortium. Nat. Biotechnol. 2010;28:1045–1048. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Michael LH, Entman ML, Hartley CJ, Youker KA, Zhu J, Hall SR, Hawkins HK, Berens K, Ballantyne CM. Myocardial ischemia and reperfusion: A murine model. Am. J. Physiol. 1995;269:H2147–H2154. doi: 10.1152/ajpheart.1995.269.6.H2147. [DOI] [PubMed] [Google Scholar]
- 58.López B, Querejeta R, González A, Larman M, Díez J. Collagen cross-linking but not collagen amount associates with elevated filling pressures in hypertensive patients with stage C heart failure: Potential role of lysyl oxidase. Hypertension. 2012;60:677–683. doi: 10.1161/HYPERTENSIONAHA.112.196113. [DOI] [PubMed] [Google Scholar]
- 59.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Anders S, Pyl PT, Huber W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang L, Wang S, Li W. RSeQC: Quality control of RNA-seq experiments. Bioinformatics. 2012;28:2184–2185. doi: 10.1093/bioinformatics/bts356. [DOI] [PubMed] [Google Scholar]
- 62.Li B, Dewey CN. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bionformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Stat. Methodol. 1995;57:289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Conservation between the mouse and human Wisper transcripts.
Fig. S2. Wisper is expressed in the stressed fibrotic heart but not in the stressed fibrotic kidney.
Fig. S3. Effects of Wisper knockdown in neonatal CFs and CMs.
Fig. S4. TAD centered on the murine Wisper locus; long noncoding transcriptome after Wisper knockdown in fibroblasts; Wisper pulldown assay.
Fig. S5. Dose-response analysis after injection of GapmeRs targeting Wisper in vivo.
Fig. S6. Preventative Wisper depletion inhibits cardiac fibrosis and improves function.
Fig. S7. Expression of cardiac markers of stress after therapeutic depletion of Wisper in the infarcted heart; expression of myofibroblast genes in differentiating cardiac and dermal human fibroblasts.
Table S1. Echocardiographic parameters measured 1, 7, 14, and 28 days after MI.
Table S2. Echocardiographic parameters measured 4, 14, and 28 days in untouched mice injected with 5, 10, or 15 mg/kg of GM-Scrambled (control) or GM-Wisper (Wisper).
Table S3. Echocardiographic parameters measured 7 and 14 days after MI in mice injected with GapmeRs 3 days before MI.
Table S4. Echocardiographic parameters measured 7 and 28 days after MI in mice injected with GapmeRs 2 days and 9 days after MI.
Table S5. Characteristics of patients suffering from AOS and developing cardiac fibrosis.
Table S6. List of the primers used in this study.
Table S7. Individual-level data and exact P values (Excel format).