Abstract
Objective
The aim of this study was to determine the etiology and prevalence of pediatric CAP in Beijing using a real-time polymerase chain reaction (PCR) technique.
Methods
Between February 15, 2011 and January 18, 2012, 371 pediatric patients with CAP were enrolled at Beijing Children's Hospital. Sixteen respiratory viruses and two bacteria were detected from tracheal aspirate specimens using commercially available multiplex real-time reverse transcription PCR (RT-PCR) kits.
Results
A single viral pathogen was detected in 35.3% of enrolled patients, multiple viruses in 11.6%, and virus/bacteria coinfection in 17.8%. In contrast, only 6.5% of patients had a single bacterial pathogen and 2.2% were infected with multiple bacteria. The etiological agent was unknown for 26.7% of patients. The most common viruses were respiratory syncytial virus (RSV) (43.9%), rhinovirus (14.8%), parainfluenza virus (9.4%), and adenovirus (8.6%). In patients under three years of age, RSV (44.6%), rhinovirus (12.8%), and Streptococcus pneumoniae (9.9%) were the most frequent pathogens. In children aged 3–7 years, S. pneumoniae (38.9%), RSV (30.6%), Haemophilus influenzae (19.4%), and adenovirus (19.4%) were most prevalent. Finally in children over seven years, RSV (47.3%), S. pneumoniae (41.9%), and rhinovirus (21.5%) infections were most frequent.
Conclusions
Viral pathogens, specifically RSV, were responsible for the majority of CAP in pediatric patients. However, both S. pneumoniae and H. influenzae contributed as major causes of disease. Commercially available multiplexing real-time PCR allowed for rapid detection of the etiological agent.
Keywords: Real-time reverse transcription polymerase chain reaction (RT-PCR), Respiratory virus, Community-acquired pneumonia
Introduction
The latest report from the World Health Organization attributed between 1.6 and 2.2 million deaths in children under 5-year-old to acute respiratory illness.1 Community-acquired pneumonia (CAP) is a major cause of morbidity and hospitalization in young children worldwide. The annual incidence is 34–40 cases per 1000 children in Europe and North America.1 The symptoms and signs of CAP are fever, cough, malaise and chest pain. The etiological agents of CAP are varied. Bacterial agents known to cause CAP include Streptococcus pneumoniae, Haemophilus influenzae, Mycoplasma pneumoniae, and Chlamydophila pneumoniae.2, 3, 4 Viral agents, such as influenza virus A and B, respiratory syncytial virus, parainfluenza viruses 1, 2, and 3, adenovirus, and rhinovirus are also common.5, 6, 7 CAP can also be caused by emerging respiratory viruses such as human metapneumovirus,8 human coronaviruses NL639 and HKU1,10 and human bocavirus11 have been reported as etiological agents of CAP.
It is currently difficult to reliably identify the pathogen responsible for causing CAP based on the clinical signs and symptoms.7 To be successful, most treatments for CAP need to be initiated within 24–48 hours of infection. Thus, developing rapid diagnostic tests for viral pathogens associated with CAP is of the utmost importance. The epidemics and pandemic caused by the swine influenza A virus (H1N1) infection in 2009 have also highlighted the necessity of developing diagnostic tools that are sensitive and rapid tests for use during ongoing surveillance studies.
The diagnostic methods that are currently used to detect respiratory infections include rapid antigen tests, virus culture, enzyme immunoassays, immunofluorescence, and conventional reverse transcription polymerase chain reaction (RT-PCR) assays.12, 13 Virus culture is considered the gold standard because of its broad applicability and high specificity. However, it is time-consuming and it takes 7–12 days to obtain a positive culture. Enzyme immunoassays and immunofluorescence give rapid results, but, their relative lack of sensitivity and the availability of reactive antisera can be limiting factors.14 Advances in conventional RT-PCR and real-time quantitative PCR assays have greatly facilitated the etiological study of respiratory infections due to their higher sensitivity and specificity. These assays can also reduce labor and cost by detecting more than one pathogen in a single reaction by using multiple probes.15 There are several commercial multiplex assays available; such as xTAG RVP from Luminex, Multicode-PLx RVP from EraGen Biosciences, and ResPlex II from Qiagen. These assays have been used for broad spectrum detection of respiratory viruses with high cost and they are time-consuming.
In this study, the prevalence and causative agents of CAP in pediatric patients was described using PCR techniques. Commercial multiplex real-time RT-PCR kits designed to detect common respiratory pathogens, including S. pneumoniae, H. influenzae, influenza viruses A (swine lineage influenza A virus H1N1, season influenza A virus H3N2) and B, respiratory syncytial virus, parainfluenza viruses 1 to 4, adenovirus, rhinovirus, human metapneumovirus, human coronavirus (NL63, OC43, 229E and HKU1) and human bocavirus were used. The viral and bacterial infections associated with CAP and clinical and the epidemiological characteristics of CAP in a pediatric population were analyzed in this study.
Materials and methods
Subject enrollment and sampling
Informed consent was obtained from the objects' parents or guardians. The participants, and their guardians, received information detailing the purpose of the study and their right to have any information determined remain confidential.
Tracheal aspirate specimens were collected from pediatric patients presenting with CAP at the Beijing Children's Hospital between February 15, 2011 and January 18, 2012. The patients were enrolled according to a set of criteria that included cough, a high fever, yellow mucus, lung consolidation, the number of white blood cells >10 × 109/L or <4 × 109/L, or one of the above-mentioned symptoms plus a spot piece shape shadow by chest X-ray. The samples were collected and transported to the Institute of Immunization and Prevention of Beijing Center for Disease Prevention and Control for viral and bacterial nucleic acid extraction and detection.
Extraction of total RNA/DNA
The tracheal aspirate specimens were first treated with an equal volume of Sputasol solution (Oxoid, Basingstok, UK) in a 37 °C water bath for liquefaction. The total RNA/DNA was then extracted from the liquefaction sample using the QIAamp Viral RNA Mini Kit (Qiagen, Hilden, Germany) per the manufacturer's instructions. The nucleic acids was stored in aliquots at −80 °C until use.
Multiple real-time RT-PCR kits for pathogens
The duplex real-time PCR kit for detecting S. pneumoniae and H. influenzae, and the multiplex combined real-time PCR detection kit for respiratory viruses were purchased from Jiangsu Uninovo Biological Technology Co., LTD. The duplex real-time PCR was performed using the Applied Biosystems 7500 Fast Real-Time PCR System (ABI, California, USA). The reaction conditions were: initial denaturation at 95 °C for 2 min, followed by 40 cycles of denaturation of 95 °C for 10 s and annealing and extension at 60 °C for 50 s. Fam and Hex fluorescence was acquired at the end of the annealing and extension step. The multiplex combined Real-time PCR was also performed using the Applied Biosystems 7500 Fast Real-Time PCR System (ABI, California, USA). The reaction conditions were: (1) reverse transcription at 42 °C for 10 min, (2) preliminary denaturation at 95 °C for 10 s, and (3) 45 cycles of denaturation of 95 °C for 5 s, annealing at 56 °C for 50 s, extension at 72 °C for 12 s. Fam, Hex, Rox and Cy5 fluorescence was acquired at the end of the annealing step. The results were analyzed according to the kit user's manuals.
Results
Clinical characteristics of pediatric CAP patients
Between February 15, 2011 and January 18, 2012, 371 pediatric CAP patients from 1 year to 17 years old were enrolled in this study. CAP patients were stratified into three groups: 242 (65.2%) patients were in the pre-kindergarten group (≤3 years), 36 (9.7%) were in the kindergarten group (3–7 years) and 93 (25.1%) were in the school-age group (≥7 years). Of the patients enrolled, a fever was documented in 52.3% (194/371), 76% presented with sputum and chest pain (282/371), and 18.6% presented with lung consolidation (69/371). The clinical characteristics for each age group are shown in Table 1.
Table 1.
Clinical characteristics of pediatric CAP patients.
| Time period | ≤3 years |
3–7 years |
≥7 years |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | Fever | Sputum and chest pain | Lung consolidation | n | Fever | Sputum and chest pain | Lung consolidation | n | Fever | Sputum and chest pain | Lung consolidation | |
| Springa | 22 | 10 | 22 | 7 | 10 | 10 | 10 | 1 | 21 | 20 | 19 | 8 |
| Summerb | 9 | 5 | 8 | 3 | 4 | 3 | 4 | 0 | 2 | 2 | 1 | 1 |
| Autumnc | 74 | 19 | 54 | 5 | 7 | 7 | 4 | 1 | 17 | 15 | 13 | 0 |
| Winterd | 137 | 44 | 97 | 31 | 15 | 12 | 11 | 2 | 53 | 47 | 39 | 10 |
| Total | 242 | 78 | 181 | 46 | 36 | 32 | 29 | 4 | 93 | 84 | 72 | 19 |
CAP: community-acquired pneumonia.
March 1 – May 31, 2011.
June 1 – August 31, 2011.
September 1 – November 20, 2011.
February 15–28, 2011 and December 1, 2011–January 18, 2012.
Identification of broad classes of etiological agents
We wanted to determine whether bacterial or viral agents were responsible for causing the majority of CAP cases in the enrolled patients. Table 2 shows the broad category of etiological agents identified in pediatric CAP patients from Beijing Children's Hospital. Infection by a single bacteria was observed in 6.5% (n = 24) of the patients, infection by multiple bacteria was observed in 2.2% of patients (n = 8), infection by both bacterial and viral agents was observed in 17.8% of patients (n = 66), infection by a single viral agent was observed in 35.3% of patients (n = 131), and finally, infection by multiple viruses was observed in 11.6% of patients (n = 43). In approximately one quarter (26.7%, n = 99) of the patients the category of the etiological agent was not identifiable.
Table 2.
Etiology of CAP in 371 pediatric patients.
| Pathogenic agents | n | Percentage (%) |
|---|---|---|
| Bacterial (single) | 24 | 6.5 |
| Bacterial (multiple) | 8 | 2.2 |
| Bacterial and viral | 66 | 17.8 |
| Viral (single) | 131 | 35.3 |
| Viral (multiple) | 43 | 11.6 |
| Unknown | 99 | 26.7 |
| Total | 371 | 100 |
CAP: community-acquired pneumonia.
We next want to identify the prevalence of specific viral and/or bacterial pathogens that were contributing to CAP. Table 3 shows the bacterial and/or viral pathogens likely to be the causative agent for disease identified in the CAP patients. Bacterial infections accounted for 30.5% (n = 113) of the total CAP cases. The most prevalent were S. pneumoniae (20.8%; n = 77) and H. influenzae (9.7%; n = 36). Of the cases associated with S. pneumoniae, 77.9% (n = 60) were co-infected with viruses and/or bacteria. The majority (n = 45) were viral coinfection. In the CAP cases associated with H. influenzae, 80.1% (n = 29) were coinfections with viruses and/or bacteria. Viral coinfection occurred in 14 cases (48.3%).
Table 3.
Specific bacterial and viral pathogens identified as etiological agents in CAP patients.
| Pathogen | n | Single infection | Co-infection with: |
||
|---|---|---|---|---|---|
| Bacteria | Virus | Bacteria and virus | |||
| Bacteria | |||||
| S. pneumoniae | 77 | 17 | 8 | 45 | 7 |
| H. influenzae | 36 | 7 | 8 | 14 | 7 |
| Virus | |||||
| Respiratory syncytial virus | 163 | 84 | 33 | 31 | 15 |
| Rhinovirus | 55 | 15 | 3 | 26 | 11 |
| Parainfluenza virus 1, 2, 3, and 4 | 35a | 11 | 4 | 12 | 7 |
| Adenovirus | 32 | 8 | 1 | 14 | 9 |
| Human bocavirus | 14 | 1 | 0 | 8 | 5 |
| Human coronavirus (NL63, OC43, 229E and HKU1) | 14 | 6 | 2 | 5 | 1 |
| Influenza virus B | 7 | 4 | 2 | 1 | 0 |
| Human metapneumovirus | 5 | 1 | 0 | 3 | 1 |
| Influenza virus A | |||||
| Season H3N2 | 3 | 1 | 0 | 2 | 0 |
| Swine lineage H1N1 | 2 | 0 | 0 | 0 | 2 |
| Total | 443 | 155 | – | – | – |
CAP: community-acquired pneumonia.
One CAP patient was infected by parainfluenza virus 1 and 3.
In fact, the majority of CAP cases were associated with viral infection. Viral agents were detected in 330 of the patients (88.9%). As shown in Table 3, the predominant infection was with respiratory syncytial virus which accounted for 43.9% (n = 163) of the viral infections. Of the remaining viruses rhinovirus was associated with 14.8% (n = 55) of the CAP cases, parainfluenza virus with 9.4% (n = 35), adenovirus with 8.6% (n = 32), human bocavirus with 3.8% (n = 14), human coronaviruses (NL63, OC43, 229E and HKU1) with 3.8% (n = 14), influenza virus B with 1.9% (n = 7), and human metapneumovirus with 1.3% (n = 5). Influenza infections were detected in only five of the CAP patients. The seasonal influenza (H3N2) strain was detected in three patients (0.8%) and swine influenza (H1N1) was detected in two patients (0.5%). In the majority of influenza virus B (57.1%), respiratory syncytial virus (51.5%), and human coronavirus (42.9%) infections only a single agent was detected which was higher than for other viral infections. In contrast, the four major viral pathogens (respiratory syncytial virus, rhinovirus, parainfluenza virus, and adenovirus) were detected more frequently in coinfections with other bacterial or viral agents. In patients infected with rhinovirus, parainfluenza virus, and adenovirus a greater percentage of coinfections were with viruses than bacteria.
Distribution of pathogenic agents according to age
To determine whether the etiological agent for CAP varied with age, we compared the prevalence of viral and bacterial pathogens in each age group. Table 4 shows the distribution of each pathogenic agent according to CAP patient age. In the pre-kindergarten group, the most common pathogens were respiratory syncytial virus (n = 108, 44.6%), rhinovirus (n = 31, 12.8%) and S. pneumoniae (n = 24, 9.9%). In the kindergarten group, S. pneumoniae (n = 14, 38.9%), respiratory syncytial virus (n = 11, 30.6%), H. influenzae (n = 7, 19.4%), and adenovirus (n = 7, 19.4%) were the most common infections. Finally, in the school-age group, respiratory syncytial virus (n = 44, 47.3%), S. pneumoniae (n = 39, 41.9%), and rhinovirus (n = 20, 21.5%) accounted for the greatest number of infections.
Table 4.
Pathogenic agents distribution by age.
| Pathogen | n | Age group |
||
|---|---|---|---|---|
| ≤3 years (n = 242) | 3–7 years (n = 36) | ≥7 years (n = 93) | ||
| Bacteria | ||||
| S. pneumoniae | 77 | 24 | 14 | 39 |
| H. influenzae | 36 | 15 | 7 | 14 |
| Virus | ||||
| Respiratory syncytial virus | 163 | 108 | 11 | 44 |
| Rhinovirus | 55 | 31 | 4 | 20 |
| Parainfluenza virus 1, 2, 3, and 4 | 35 | 16 | 6 | 13 |
| Adenovirus | 32 | 15 | 7 | 10 |
| Human bocavirus | 14 | 8 | 1 | 5 |
| Human coronavirus (NL63, OC43, 229E and HKU1) | 14 | 9 | 2 | 3 |
| Influenza virus B | 7 | 4 | 0 | 3 |
| Human metapneumovirus | 5 | 3 | 0 | 2 |
| Influenza virus A | ||||
| Swine lineage H1N1 | 2 | 1 | 0 | 1 |
| Season H3N2 | 3 | 2 | 0 | 1 |
| Total | 443 | 236 | 52 | 155 |
Seasonal epidemiology of major pathogenic agents for CAP patients
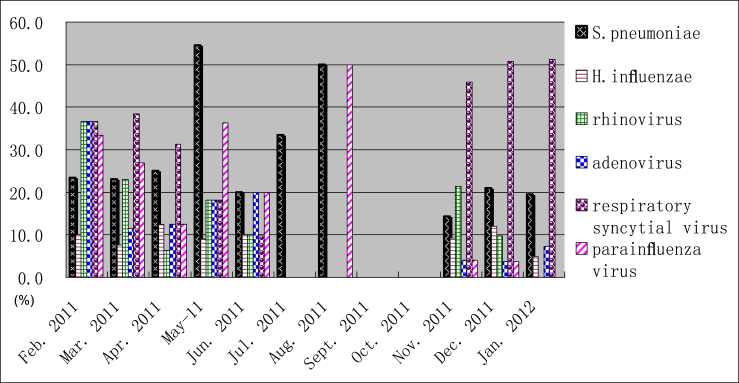
To determine whether the etiology of CAP differs by seasons, the prevalence of each major pathogen was compared throughout the year. Fig. 1 shows the seasonal epidemiology for six of pathogenic agents (S. pneumoniae, H. influenzae, rhinovirus, adenovirus, respiratory syncytial virus, and parainfluenza virus) that were identified in 371 CAP patients over one year from February 2011 to January 2012. The data is reported as the percentage of CAP patients each month that tested positive for the pathogenic agents. Only five patients were enrolled from July to October 2011, among which three patients were enrolled in July, two in August, and no patients in September and October. Overall, respiratory syncytial virus and S. pneumoniae were the predominant pathogenic agents for CAP patients. These pathogens were detected in the spring, autumn, and winter. The peak incidence of respiratory syncytial virus was observed from December 2011 to January 2012. The peak incidence of S. pneumoniae was observed in May 2011. The occurrence throughout the other months was even except for July to October 2011.
Fig. 1.
Seasonal distribution of CAP (community-acquired pneumonia)agents. Monthly prevalence of S. pneumoniae, H. influenzae, rhinovirus, adenovirus, respiratory syncytial virus and parainfluenza virus from February 2011 through January 2012. The percent of six pathogens tested in each month during the study period are shown on the left-side y-axis.
Discussion
CAP is the most common infectious disease occurring in children and a major cause of mortality among children living in developing countries.1 The etiological agents are varied but include a wide range of respiratory viruses and bacteria. Developing a technique that allows accurate and rapid detection of CAP etiological agents would be extremely useful for clinicians during the care of CAP patients and for the appropriate antimicrobial selection. More effective CAP treatment could help decrease hospital costs and the length of a patient's stay in the hospital.
In this study, we used commercially available multiplex real-time PCR kits to detect respiratory pathogens in pediatric CAP patients. The kits were able to detect 16 respiratory viruses and two bacteria. We compared the specificity and sensitivity of this kit with singleplex conventional PCR16 and a commercial multiplex conventional PCR kit (Seeplex® RV15 ACE Detection kit, Seegene, South Korea) using 73 throat swab specimens from upper respiratory tract infection cases. The positive factor numbers of singleplex conventional PCR, conventional multiplex PCR and multiplex real-time PCR were 56, 41 and 87, respectively (Data not published). The factors detected only by the multiplex real-time PCR assay were confirmed using previously published methods.17 Using the multiplex real-time PCR assay the etiological agents could be identified within three hours of receiving the clinical samples. We chose to use the multiplex assay for its sensitivity, specify, and rapidity of the results.
Throat swabs and tracheal aspirate specimens were collected in parallel from 371 CAP patients. The respiratory pathogens in 96 pairs of throat swabs and tracheal aspirate specimens were detected using the commercial multiplex real-time PCR kits. The Ct values for hits from tracheal aspirate specimens were usually lower than that from throat swabs (Data not published). Similar to previously published work,18 this indicated that viral loads in tracheal aspirate specimens were higher than in throat swabs. In addition, throat swabs are commonly used clinically for identifying respiratory viruses. However, these samples are not always appropriate for the detection of disease associated with S. pneumoniae and H. influenzae which can be found in healthy carriers. Tracheal aspirate specimens were chosen for this study to reflect the true incidence of respiratory pathogens in pediatric CAP patients in Beijing.
Respiratory syncytial virus is the leading cause of lower respiratory tract infections in the pediatric population worldwide.19 It is responsible for an estimated 160000 deaths every year worldwide.20 In this study, 43.9% of the CAP patients had detectable RSV, which was higher than any other pathogen. In fact the incidence of RSV was two times higher than S. pneumoniae (n = 77) that was the second leading cause of CAP. Respiratory syncytial virus was the most frequent pathogen detected in all three age groups. Rhinovirus is another frequently detected viral agent that causes adult acute respiratory infection in temperate regions.21 Likewise, rhinovirus was also detected in this study in pediatric CAP patients. In this study two viruses that had recently been associated with CAP, human metapneumovirus and human bocavirus, were also detected using multiplex real-time PCR.
In CAP associated with S. pneumoniae and H. influenzae, there were frequently coinfections with other bacteria and/or virus. The 78–80% coinfection rate was higher than that observed in pediatric CAP patients in Japan.22 Influenza viruses A and B are common pathogens associated with respiratory tract infection. However, in this study, only 12 of 371 CAP patients had detectable influenza virus A (n = 5) or B (n = 7). This rate was much lower than has been reported by the National Surveillance program. These results indicated that influenza viruses A and B were not the main causative pathogens for pediatric CAP patients in Beijing.
In conclusion, we assessed the viral and bacterial etiologies of pediatric CAP patients with multiplex real-time PCR technique and found that respiratory syncytial virus, followed by S. pneumoniae, rhinovirus, H. influenzae, parainfluenza virus, and adenovirus were the main respiratory pathogens for pediatric CAP patients in Beijing.
Funding sources
This work was supported by the Beijing Municipal Science and Technology Commission (D09050203620903) and the Major National Science and Technology Programs in the “Twelfth Five-Year” Plan period (2012ZX10004-206).
Footnotes
Peer review under responsibility of Chinese Medical Association.
References
- 1.Stein R.T., Marostica P.J. Community-acquired pneumonia: a review and recent advances. Pediatr Pulmonol. 2007;42:1095–1103. doi: 10.1002/ppul.20652. [DOI] [PubMed] [Google Scholar]
- 2.Bartlett J.G., Mundy L.M. Community-acquired pneumonia. N Engl J Med. 1995;333:1618–1624. doi: 10.1056/NEJM199512143332408. [DOI] [PubMed] [Google Scholar]
- 3.McCracken G.H. Etiology and treatment of pneumonia. Pediatr Infect Dis J. 2000;19:373–377. doi: 10.1097/00006454-200004000-00032. [DOI] [PubMed] [Google Scholar]
- 4.McIntosh K. Community-acquired pneumonia in children. N Engl J Med. 2002;346:429–437. doi: 10.1056/NEJMra011994. [DOI] [PubMed] [Google Scholar]
- 5.Pavia A.T. Viral infections of the lower respiratory tract: old viruses, new viruses, and the role of diagnosis. Clin Infect Dis. 2011;52(suppl 4):S284–S289. doi: 10.1093/cid/cir043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Razanajatovo N.H., Richard V., Hoffmann J. Viral etiology of influenza-like illnesses in Antananarivo, Madagascar, July 2008 to June 2009. PLoS One. 2011;6:e17579. doi: 10.1371/journal.pone.0017579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caliendo A.M. Multiplex PCR and emerging technologies for the detection of respiratory pathogens. Clin Infect Dis. 2011;52(suppl 4):S326–S330. doi: 10.1093/cid/cir047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van den Hoogen B.G., de Jong J.C., Groen J. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat Med. 2001;7:719–724. doi: 10.1038/89098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van der Hoek L., Ihorst G., Sure K. Burden of disease due to human coronavirus NL63 infections and periodicity of infection. J Clin Virol Jun. 2010;48:104–108. doi: 10.1016/j.jcv.2010.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Woo P.C., Lau S.K., Chu C.M. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J Virol. 2005;79:884–895. doi: 10.1128/JVI.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Allander T., Tammi M.T., Eriksson M., Bjerkner A., Tiveljung-Lindell A., Andersson B. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc Natl Acad Sci U S A. 2005;102:12891–12896. doi: 10.1073/pnas.0504666102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Falsey A.R., Formica M.A., Walsh E.E. Diagnosis of respiratory syncytial virus infection: comparison of reverse transcription-PCR to viral culture and serology in adults with respiratory illness. J Clin Microbiol. 2002;40:817–820. doi: 10.1128/JCM.40.3.817-820.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ruest A., Michaud S., Deslandes S., Frost E.H. Comparison of the Directigen flu A+B test, the QuickVue influenza test, and clinical case definition to viral culture and reverse transcription-PCR for rapid diagnosis of influenza virus infection. J Clin Microbiol. 2003;41:3487–3493. doi: 10.1128/JCM.41.8.3487-3493.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henrickson K.J. Advances in the laboratory diagnosis of viral respiratory disease. Pediatr Infect Dis J. 2004;23(1 suppl):S6–S10. doi: 10.1097/01.inf.0000108187.63151.ea. [DOI] [PubMed] [Google Scholar]
- 15.Boivin G., Cote S., Dery P., De Serres G., Bergeron M.G. Multiplex real-time PCR assay for detection of influenza and human respiratory syncytial viruses. J Clin Microbiol. 2004;42:45–51. doi: 10.1128/JCM.42.1.45-51.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ren L., Gonzalez R., Wang Z. Prevalence of human respiratory viruses in adults with acute respiratory tract infections in Beijing. Clin Microbiol Infect. 2009;15:1146–1153. doi: 10.1111/j.1469-0691.2009.02746.x. 2005-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Puppe W., Weigl J., Gröndahl B. Validation of a multiplex reverse transcriptase PCR ELISA for the detection of 19 respiratory tract pathogens. Infection. 2013;41:77–91. doi: 10.1007/s15010-012-0298-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lambert S.B., Whiley D.M., O'Neill N.T. Comparing nose–throat swabs and nasopharyngeal aspirates collected from children with symptoms for respiratory virus identification using real-time polymerase chain reaction. Pediatrics. 2008;122:e615–e620. doi: 10.1542/peds.2008-0691. [DOI] [PubMed] [Google Scholar]
- 19.Hay A.J., Gregory V., Douglas A.R., Lin Y.P. The evolution of human influenza viruses. Philos Trans R Soc Lond B Biol Sci. 2001;356:1861–1870. doi: 10.1098/rstb.2001.0999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Julia L.H. Respiratory syncytial virus vaccine development. Expert Rev Vaccines. 2011;10:1415–1433. doi: 10.1586/erv.11.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arden K.E., McErlean P., Nissen M.D., Sloots T.P., Mackay I.M. .Frequent detection of human rhinoviruses, paramyxoviruses, coronaviruses, and bocavirus during acute respiratory tract infections. J Med Virol. 2006;78:1232–1240. doi: 10.1002/jmv.20689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hamano-Hasegawa K., Morozumi M., Nakayama E. Comprehensive detection of causative pathogens using real-time PCR to diagnose pediatric community-acquired pneumonia. J Infect Chemother. 2008;14:424–432. doi: 10.1007/s10156-008-0648-6. [DOI] [PMC free article] [PubMed] [Google Scholar]