Abstract
Since the initial emergence of Middle East respiratory syndrome coronavirus (MERS-CoV) in 2012, a high incidence rate has been observed in Saudi Arabia. This suggests that the country is at continuous risk. The epidemic level of MERS-CoV infection was examined in Saudi Arabia by the Susceptible-Infectious-Recovered (SIR) model using a Bayesian approach for estimation of time dependent reproduction number (R) across a two-year interval (May, 2013-May, 2015) in five defined clusters, followed by sensitivity analysis of the most significant clusters. Significant MERS-CoV peaks were detected in the period between March and May of each year. Moreover, MERS-CoV infection was highlighted in western (40.8%) and central (31.9%) regions, followed by eastern region (20%). The temporal-based Bayesian approach indicated a sub-critical epidemic in all regions in the baseline scenario (R: 0.85–0.97). However, R potential limit was exceeded in the sensitivity analysis scenario in only central and western regions (R: 1.08–1.12) that denoted epidemic level in those regions. The impact of sporadic cases was found relatively insignificant and pinpointed to the lack of zoonotic influence on MERS-CoV transmission dynamics. The results of current study would be helpful for evaluation of future progression of MERS-CoV infections, better understanding and control interventions.
Keywords: MERS-CoV, Time dependent reproduction number, Sensitivity analysis, Outbreaks, Sporadic cases
1. Introduction
Middle East respiratory syndrome (MERS) has become a global concern since it was recognized for the first time in 2012 as a primitive source of renal failure and severe respiratory sickness caused by a novel highly pathogenic coronavirus (Majumder et al., 2014). Coronaviruses were not considered as a major public health threat before 2003, as they were known to cause only mild upper respiratory tract infections. The first known serious coronavirus infection was the severe acute respiratory syndrome coronavirus (SARS-CoV) in 2002, which caused an outbreak with approximately 8400 cases and 800 deaths (World Health Organization, 2003).
MERS-CoV pinpointed a zoonotic introduction of a novel coronavirus probably originating from bats into human populations (Sharif-Yakan and Kanj, 2014). The zoonotic origin of MERS-CoV was bolstered via phylogenetic analysis and elucidated a very proximal phylogenetic similarity with the bat Betacoronaviruses: BtCoV-HKU4 and BtCoV-HKU5 (van Boheemen et al., 2012) as well as identified of the cellular receptor (Raj et al., 2013). However, contact frequency between human and bats is highly limited in Arabian Peninsula. Subsequently other intermediate hosts were proposed, such as camels and goats (Raj et al., 2014a, Raj et al., 2014b). MERS-CoV was found circulating in dromedary camels from last 20 years (Corman et al., 2014), and MERS-CoV neutralizing antibodies were detected in camels (Reusken et al., 2013, Milne-Price et al., 2014). These findings indicated that camels could serve as the intermediate host for MERS-CoV zoonotic infections in Saudi Arabia, Oman, Jordan, and United Arab Emirates. On the other hand, human-to-human transmission was frequently observed among MERS cases as most of the human infections were recorded among health care workers and within households proposed a close contact transmission (Hunter et al., 2016, Sharif-Yakan and Kanj, 2014). The World Health Organization (WHO) has reported 1038 MERS cases, mostly occurred in Arabian Peninsula involved 460 deaths from Saudi Arabia (Memish et al., 2015). Consequently, Saudi Arabia was considered as the epicenter of MERS-CoV infections (Alqahtani et al., 2017). Subsequently several studies investigated the existing status and future evolution of pathogen to confront the probable outbreaks by studying MERS-CoV transmission (Breban et al., 2013, Poletto et al., 2014, Kucharski and Althaus, 2015, Abolfotouh et al., 2017). The current study examined the level of heterogeneity in MERS-CoV transmission via cluster analysis relying on the geographical distribution of MERS cases since it could clarify the linkage between epidemic status and geographical separation of clusters (Cauchemeza et al., 2016). One useful indicator that is used to check viral transmissibility is the basic reproduction number (R0) representing the number of secondary cases due to each index case in a fully susceptible population because no approved vaccine is available (Breban et al., 2013). Basic reproduction number sets the infectious agent’s potential to start an outbreak. When R0 is >1 epidemics takes off and the epidemic can diminish and die out when R0 is less than 1 (Chang, 2016). Moreover, reproduction numbers can be estimated at various times during an epidemic. It can be estimated at the beginning of an outbreak (initial reproduction number) or at any time during the outbreak (time-dependent reproduction number). Several methods are used to evaluate the initial reproduction number involving attack rate analysis, exponential growth (EG) method, maximum likelihood (ML) estimation and sequential Bayesian (SB) method (Obadia et al., 2012). However, the time-dependent reproduction number can be computed by averaging over all transmission networks compatible with observations using the time-dependant method (Wallinga and Teunis, 2004). The attack rate method requires the least information, but it can be used only when the epidemic has finished, and additionally no further intervention can be conducted during the entire outbreak course. Thus, the use of this method is mostly limited to definite settings as schools or hospitals. In EG method, the exponential growth rate occurrence during the early phase of an outbreak can be concomitant to the initial reproduction ratio. The incidence data is integer estimated and Poisson regression is specified to evaluate this parameter rather than linear regression of the logged incidence (Boelle et al., 2009, Hens et al., 2011). ML method depends on the postulation that the number of secondary cases triggered by an index case is Poisson distributed with probable value R. In sequential bayesian method, the former distribution for R utilized on each new day is the subsequent distribution from the previous day. At each time, the mode of the posterior may be estimated along with the highest probability density interval. Similar to the previous methods, this method necessitates that the epidemic is occurring in a period of exponential growth (Obadia et al., 2012). On the other hand, overestimation of the initial reproduction number may be obtained in a case when epidemic is not observed from the initial case, because some index cases are not present in the epidemic curve. In ML method this problem was solved by a correction made for absent generations at the beginning of the epidemic curve and similar is recommended in the Bayesian setting (McBryde et al., 2009) by assuming a constant reproduction number. However, it is not possible for the TD method due to variation of reproduction ratio with the time. Thus, the current study relied on Bayesian method to follow up the epidemic progress in Saudi Arabia and particularly in the absence of information around the index cases from two year data of MERS cases by assuming a random mixing in a fully susceptible population.
2. Method
2.1. Data sources
The analysis focused on the progress of MERS-CoV in Saudi Arabia from May 2013 and May 2015. A line list of MERS cases was compiled from the Saudi Arabian ministry of health (MOH) reports, published on the official MOH website (http://www.moh.gov.sa/en/CCC/PressReleases/Pages/default.aspx). The list contained 765 confirmed MERS cases.
2.2. Data clustering
The concept of cluster was based on geographical grouping into five major clusters: central area, northern area, western area, southern area, and eastern area. The provinces included in each cluster are shown in Table 1.
Table 1.
Geographical clustering.
| Region | Provinces |
|---|---|
| Central region | Riyadh, Qasim |
| Western region | Makkah, Madinah, Baha |
| Eastern region | Eastern Province |
| Northern region | Tabuk, Jouf,, Hail, Northern border |
| Southern region | Asir, Najran, Jizan |
2.3. MERS-CoV epidemiological parameters
The incubation period was fixed at 5.2 days according to data acquired from hospital outbreak in Saudi Arabia (Assiri et al., 2013), and in line with information obtained from travel-related cases (Cauchemez et al., 2014).
2.3.1. Estimation of the initial reproduction number using sequential Bayesian method
All known data was used as a prior for next iteration. It relied on an approximation to the SIR model, whereby incidence at time t + 1, N(t + 1) was approximately Poisson distributed with mean N(t)e(γ(R − 1)) (γ−1 represents the average duration of the infectious period) (Boelle et al., 2009).
The formulation of probabilistic standard SIR disease transmission models is similar to the time-series SIR approach (Bjørnstad et al., 2002), where all infectious and susceptible individuals are assumed to mix homogeneously and it simplifies to reconstruct the transmission chains. The standard epidemic susceptible infected model is expressed as:
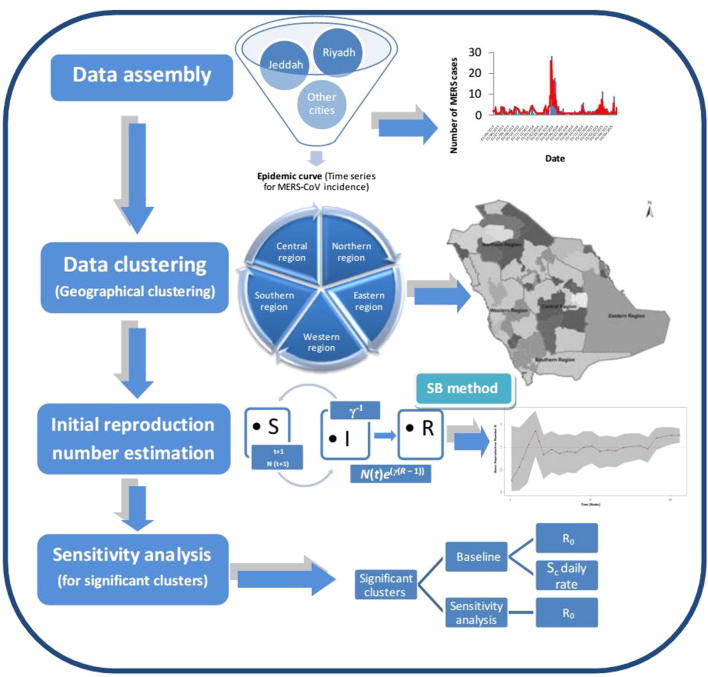
S is the average number of susceptibles at time t, I is the average number of infections, N is the size of the population, which decreases due to disease-induced deaths, β is the contact rate (Bettencourt and Ribeiro, 2008). After an average residence time γ−1, infectious individuals recover or die. The Bayesian procedure is engaged to convert time series of case numbers to a probability distribution. The proposed algorithm, described in a Bayesian framework, started with a non-informative prior on the distribution of the reproduction number R (fig. 1).
Fig. 1.
Schematic diagram for the assessment of MERS-CoV pandemic risk in Saudi Arabia.
The distribution was updated as new data was observed, using the following equation: (Obadia et al., 2012). An exponential growth for the epidemic period was assumed for this method. Moreover, this method assumed random mixing in the population. The package “Estimation of R0 and Real-Time Reproduction Number from Epidemics” in R software Version 1.6 was used to implement the Bayesian method.
2.3.2. Sensitivity analysis
A sensitivity analysis was carried out to determine the impact of sporadic cases on the basic reproductive number in a specific cluster. Sensitivity analysis was performed by screening of the confirmed MERS-CoV cases that were found correlated to the index cases. Moreover, the sporadic cases were excluded in sensitivity analysis to estimate the impact of these cases on the spread pattern in the baseline scenario. The program code on the software R was adjusted by specifying the disease generation time. These adjustments allowed the computation of the basic reproductive number after a variation in the time of the initial emergence of the virus with transmission to humans (Poletto et al., 2014). The R results of the sensitivity analysis were compared to their corresponding baseline R results for each cluster (Fig. 1). In current sensitivity analysis, we referred to the total count of cases involved in the study from May 2013 to May 2015 as the baseline. On the other hand, the cases in the outbreak periods were referred as outbreak cases.
Sensitivity analysis was performed by considering: (i) Detailed definition of source region to provinces and belonging cities, including Riyadh, Qasim, Makkah, Madinah, Baha, Eastern Province, Tabuk, Jouf, Hail, Northern border, Asir, Najran and Jizan. (ii) Restricted five regions were used for data assembly and cluster-based interpretation and only the significant clusters data was utilized for sensitivity analysis. (iii) A variation in the time of the initial emergence of the virus with transmission to humans was considered maximum up to 5.2 days for inter-human transmission and it was assumed that sporadic cases may be introduced for a period of more than 5.2 days before the known initial cases. (iv) Daily sporadic cases detected in the baseline scenario during the period of 2 year study were excluded in the sensitivity analysis to test the possibility of inter-human transmission.
3. Results
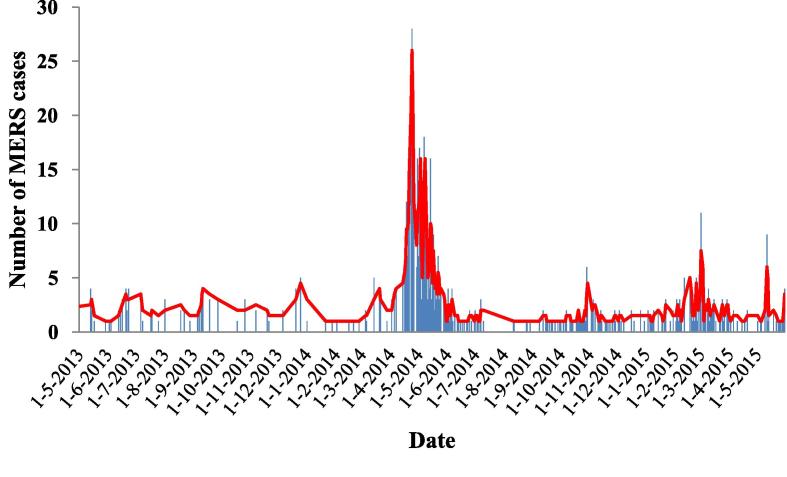
The epidemic curve of MERS-CoV infection in Saudi Arabia during the two-year-study period displayed a higher dynamic outbreak of MERS cases in the period between March and May of each year. Despite the highly significant MERS peak observed particularly in 2014, the following year showed a lower number of MERS incidents (Fig. 2).
Fig. 2.
Epidemic curve of MERS-CoV infection in Saudi Arabia during the period between May, 2013 and May 2015.
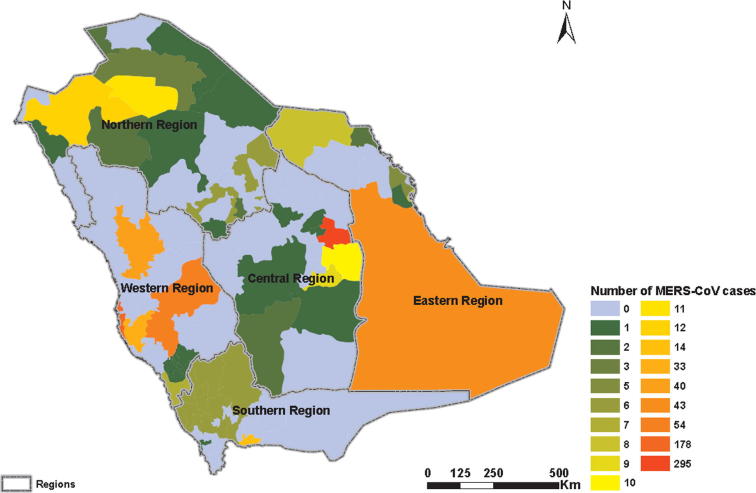
Cluster analysis based on geographical distribution of MERS cases depicted a higher representation of reported cases in the western (40.8%) and central (31.9%) regions. The lower number of cases was observed in the eastern region (20%). However, the least significant cases were recorded in northern (4.6%) and southern (2.7%) regions (Fig. 3). The daily rate of sporadic cases was found the highest in central region (1.20, 95% CI: 1.07–1.41) in relation to western and eastern regions (0.08, 95% CI: 0.06–0.12 and 0.05, 95% CI: 0.02–0.09), respectively.
Fig. 3.
Geographical distribution of MERS-CoV cases in the five clusters in Saudi Arabia.
A temporal analysis of reproduction number (R) led to potential variation in R value among study clusters. Both central and western regions scored the highest R value (R: 0.97, 95% CI: 0.64–1.36) and (R: 0.96, 95% CI: 0.52–1.25) respectively followed by the eastern region (R: 0.85, 95% CI: 0.57–1.05). R value was found at the lowest level in the Northern and Southern regions, respectively (R: 0.02 and 0.18). Therefore, the sensitivity analysis was performed for only western, central and eastern regions (Table 2).
Table 2.
The initial reproductive number values R and the daily rate of emergence of sporadic cases of MERS CoV.R: Initial reproduction number, CI: Confidence Interval.
| Analysis | Data | R (95% CI) | Daily Rate of Sporadic Cases (95% CI) |
|---|---|---|---|
| Baseline | Baseline data: All reported cases by MOH in Saudi Arabia during the period May, 2013 to May, 2015 in only three regions: (a) Western region |
0.96 (0.52–1.25) | 1.20 (1.07–1.41) |
| (b) Central region | 0.97 (0.64–1.36) | 0.08 (0.06–0.12) | |
| (c) Eastern region | 0.85 (0.57–1.05) | 0.05(0.02–0.09) | |
| Sensitivity | All confirmed cases in only three regions of Saudi Arabia during the period May, 2013 to May, 2015 within the MERS CoV incubation period of 5.2 days and excluding the sporadic cases: (a) Western region |
1.08 (0.84–1.20) | |
| (b) Central region | 1.12 (0.94–1.47) | ||
| (c) Eastern region | 0.97 (0.81–1.18) |
Sensitivity analysis demonstrated an insignificant impact of sporadic cases on the dynamic status of MERS CoV spread pattern. It was detected that the model used for the analysis was robust and that most of the cases were related and most likely evolved from index cases.
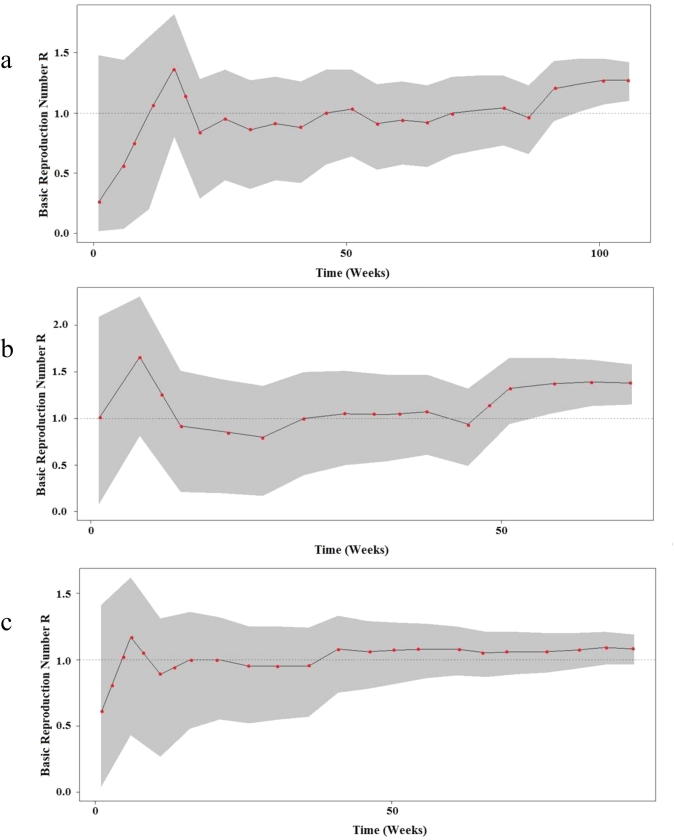
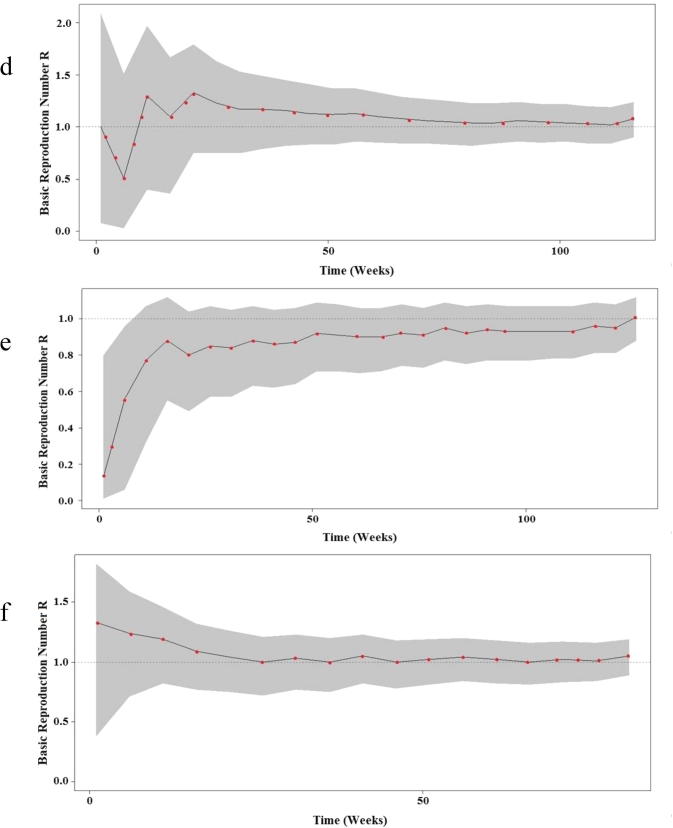
The elevation of basic reproduction number (R > 1) was recorded in both central and western regions during the most of the study period (Figs. 4.1band 4.2d) and after exclusion of sporadic cases a significant decrease in the confidence interval was noted in comparison to the baseline scenario (Fig. 4.1a and c). However, the MERS-CoV status remained roughly steady below the threshold limit (R < 1) even after exclusion of sporadic cases in the eastern region with a limited decrease in the confidence interval (Fig. 4.2e and f).
Fig. 4a.
Bayesian skyline plot (BSP) showing the changes in basic reproductive number of MERS-CoV across time in a: central region baseline, b: Confirmed cases in central region, c: Western region baseline. The dashed black line indicates the median of R values estimated from the Poisson regression model. The gray shading indicates the 95% CI of the estimated R.
Fig. 4b.
Bayesian skyline plot (BSP) showing the changes in basic reproductive number of MERS-CoV across time in d: Confirmed cases in Western region, e: Eastern region baseline and f: Confirmed cases in Eastern region. The dashed black line indicates the median of R values estimated from the Poisson regression model. The gray shading indicates the 95% CI of the estimated R.
4. Discussion
Results of the current Real Time Bayesian SIR model suggest a subcritical MERS-CoV epidemic in Saudi Arabia, as estimated by the reproductive number to be less than one. Subsequently, a self-sustaining epidemic cannot be established in humans, which would agree with the findings of other studies (Breban et al., 2013, Poletto et al., 2014). The potential outbreak of MERS-CoV in the period between March to May 2014 could be interpreted by the paucity of data about the index cases and subsequent waves of infections. Moreover, other possibilities must be included such as as seasonal variations and their correlation with zoonotic origins like infections in camels (Sharif-Yakan and Kanj, 2014). However, the low rate of daily, sporadic MERS cases diminished the possibility of zoonotic infection and indicated a more likelihood of inter-human transmissibility that is plausible with data reported elsewhere (Breban et al., 2013). The limited human-to-human transmission has been reported due to the variations of MERS-CoV receptors in the human upper and lower respiratory tract (Raj et al., 2014a, Raj et al., 2014b). Furthermore, a potentially low respiratory disease could evolve in patients lacking considerable co-morbidities (Raj et al., 2014a, Raj et al., 2014b).
The highest incidence of MERS-CoV cases was detected in western and central region followed by eastern regions. MERS incidence was found insignificant in both northern and southern regions. Several factors like contact frequency, virus shedding, hospital procedures, population composition and density (Drosten et al., 2015) may lead to the higher incidence of cases in different regions. Therefore, the higher population density in the significant regions could be considered as a potential reason for high MERS-CoV incidence.
The CI calculated for the reproductive number was found varying considerably across the studied period in the regions undergone sensitivity analysis, beginning with a wider CI and ending with a significantly narrower CI. Reduced CI in terms of time indicated more related secondary cases rather than sporadic case incidences. The current data suggested the less likelihood of external sources as zoonotic intermediates to participate in MERS-CoV spread dynamics. Current finding contradicted the results of Paletto, et al. (2014) who stated that the CI for the estimated R was steady along the variations in the data interpretation. The discrepancy might be owing to the fact that the current study was restricted to MERS-CoV cases in Saudi Arabia rather than the Middle East region. Moreover, the variation of the baseline scenario estimates (up to R = 0.97) with sensitivity analysis values (up to R = 1.12) were interpreted by the insignificant contribution of zoonotic transmissions to the MERS-CoV outbreak. However, the impact of sporadic cases on the epidemic level of MERS-CoV over the threshold limit (R > 1) was observed in both central and western regions. The finding of current study supported the data from previous study that animal contact was an uncommon cause for MERS-CoV in the detected cases (Memish et al., 2015). Some studies reported mild MERS cases and other reported a broad spectrum of clinical disease (Assiri et al., 2013). The present study might have a selection bias towards symptomatic cases. However, mild/asymptomatic cases, if detected, might lead to different spread patterns of MERS-CoV. Screening programs for earlier detection of MERS-CoV silent infection could relate the sporadic cases to their index cases, which could affect directly the estimation of the probable waves of MERS-CoV infection (Widagdo et al., 2017). Moreover, it could aid in mapping of the primary cases destination as well as the high MERS-CoV incidence regions. Hence, traceability of MERS-CoV could be enhanced that might support the convenient control measures. The second possibility is that these asymptomatic patients, if observed, could reveal the factors other than inter-human transmission that affect the MERS-CoV spread pattern, since some previous studies reported dromedary camels as a potential intermediate host for MERS-CoV (Haagmans et al., 2014, Raj et al., 2014a, Raj et al., 2014b).
The present study reported the highest R values (Table 2) ever observed for MERS-CoV, however other studies reported R of up to 0.73 (Kucharski and Edmunds, 2015), 0.69 (Breban et al., 2013, Poletto et al., 2014) and 0.63 (Cauchemez et al., 2014). The probable reasons for the surge of R value in the current study could be the absence of a vaccine or a treatment, besides the inefficient control measures in health care centers and hospitals as most of the observed cases were documented to be inter- and intra-hospital transmissions (Drosten et al., 2015, Oboho et al., 2015). Furthermore, Cauchemez et al. (2014) mentioned that R0 could range from 0.8 to 1.3 in the absence of the infection control measures. The results of current study are in agreement with the findings of Cauchemez et al. (2014). A study in South Korea (Kucharski and Althaus, 2015) highlighted the risk of super-spreading events of MERS-CoV infection with relatively low basic reproductive number (R = 0.47) and should be considered as warning for future outbreak events in Saudi Arabia with the prediction of relatively higher R value. Therefore, future tracking of infections would add value to our understanding of viral transmissibility pattern as well as contracting the CIs around the R0 value. Appropriate monitoring of cases as well as enhanced traceability procedures are important to reduce transmission rate, diminish any possible opportunity of viral adaptation to human-to-human transmission and to obtain reliable data for periodical update of the R value (Min et al., 2016).
Footnotes
Peer review under responsibility of King Saud University.
Contributor Information
Saleh A. Eifan, Email: seifan@ksu.edu.sa.
Islam Nour, Email: inour@ksu.edu.sa.
Atif Hanif, Email: ahchaudhry@ksu.edu.sa.
Abdelrahman M.M. Zamzam, Email: abdelrahman.zamzam@gmail.com.
Sameera Mohammed AlJohani, Email: johaniS@ngha.med.sa.
References
- Abolfotouh M.A., AlQarni A.A., Al-Ghamdi S.M., Salam M., Al-Assiri M.H., Balkhy H.H. An assessment of the level of concern among hospital-based health-care workers regarding MERS outbreaks in Saudi Arabia. BMC Infect. Dis. 2017;17(1):4. doi: 10.1186/s12879-016-2096-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alqahtani A.S., Rashid H., Basyouni M.H., Alhawassi T.M., BinDhim N.F. Public response to MERS-CoV in the Middle East: iPhone survey in six countries. J. Infect. Pub. Health. 2017 doi: 10.1016/j.jiph.2016.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assiri A., McGeer A., Perl T.M., Price C.S., Al Rabeeah A.A., Cummings D.A.T., Alabdullatif Z.N., Assad M., Almulhim A., Makhdoom H., Madani H., Alhakeem R., Al-Tawfiq J.A., Cotten M., Watson S.J., Kellam P., Zumla A.I., Memish Z.A. Hospital outbreak of Middle East respiratory syndrome coronavirus. N. Engl. J. Med. 2013;369(5):407–416. doi: 10.1056/NEJMoa1306742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettencourt L.M.A., Ribeiro R.M. Real time bayesian estimation of the epidemic potential of emerging infectious diseases. PLoS ONE. 2008;3(5):e2185. doi: 10.1371/journal.pone.0002185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjørnstad O.N., Finkenstädt B.F., Grenfell B.T. Dynamics of measles epidemics: estimating scaling of transmission rates using a time series SIR model. Ecol. Monogr. 2002;72(2):169–184. [Google Scholar]
- Boelle, P.Y., Bernillon, P., Desenclos, J.C., 2009. A preliminary estimation of the reproduction ratio for new influenza A (H1N1) from the outbreak in Mexico, March-April 2009. Euro. Surveill. 14 (19), pii: 19205. [DOI] [PubMed]
- Breban R., Riou J., Fontanet A. Interhuman transmissibility of Middle East respiratory syndrome coronavirus: estimation of pandemic risk. Lancet. 2013;382:694–699. doi: 10.1016/S0140-6736(13)61492-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cauchemez S., Fraser C., Van Kerkhove M.D., Donnelly C.A., Riley S., Rambaut A., Enouf V., van der Werf S., Ferguson N.M. Middle East respiratory syndrome coronavirus: quantification of the extent of the epidemic, surveillance biases, and transmissibility. Lancet Infect. Dis. 2014;14:50–56. doi: 10.1016/S1473-3099(13)70304-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cauchemeza S., Nouvelletd P., Corid A., Jombartd T., Garsked T., Claphame H., Mooree S., Mills H.L., Saljea H., Collins C., Rodriquez-Barraquer I., Riley S., Truelove S., Algarni H., Alhakeem R., AlHarbi K., Turkistani A., Aguas R.J., Cummings D.A.T., Van Kerkhove M.D., Donnelly C.A., Lessler J., Fraser C., Al-Barrak A., Ferguson N.M. Unraveling the drivers of MERS-CoV transmission. Proc. Natl. Acad. Sci. 2016;113(32):9081–9086. doi: 10.1073/pnas.1519235113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, H.J., 2016. Evaluation of the basic reproduction number of MERS-CoV during the 2015 outbreak in South Korea. In: Control, Automation and Systems (ICCAS), 16th International IEEE Conference. 981–984.
- Corman V.M., Jores J., Meyer B., Younan M., Liljander A., Said M.Y., Gluecks I., Lattwein E., Bosch B.J., Drexler J.F., Bornstein S., Drosten C., Müller M.A. Antibodies against MERS coronavirus in dromedary camels, Kenya, 1992–2013. Emerg. Infect. Dis. 2014;20:1319–1322. doi: 10.3201/eid2008.140596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten C., Muth D., Corman V.M., Hussain R., Al Masri M., HajOmar W., Landt O., Assiri A., Eckerle I., Al Shangiti A., Al-Tawfiq J.A., Albarrak A., Zumla A., Rambaut A., Memish Z.A. An observational, laboratory-based study of outbreaks of Middle East respiratory syndrome coronavirus in Jeddah and Riyadh, kingdom of Saudi Arabia. Clin. Infect. Dis. 2015;60(3):369–377. doi: 10.1093/cid/ciu812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haagmans B.L., Al Dhahiry S.H.S., Reusken C.B.E.M., Raj V.S., Galiano M., Myers R., Godeke G.-J., Jonges M., Farag E., Diab A., Ghobashy H., Alhajri F., Al-Thani M., Al-Marri S.A., Al Romaihi H.E., Al Khal A., Bermingham A., Osterhaus A.D.M.E., AlHajri M.M., Koopmans M.P.G. Middle East respiratory syndrome coronavirus in dromedary camels: an outbreak investigation. Lancet Infect. Dis. 2014;14:140–145. doi: 10.1016/S1473-3099(13)70690-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hens N., Van Ranst M., Aerts M., Robesyn E., Van Damme P., Beutels P. Estimating the effective reproduction number for pandemic influenza from notification data made publicly available in real time: a multi-country analysis for influenza A/H1N1v 2009. Vaccine. 2011;29(5):896–904. doi: 10.1016/j.vaccine.2010.05.010. [DOI] [PubMed] [Google Scholar]
- Hunter J.C., Nguyen D., Aden B., Al Bandar Z., Al Dhaheri W., Elkheir K.A., Khudair A., Al Mulla M., El Saleh F., Imambaccus H., Al Kaabi N., Sheikh F.A., Sasse J., Turner A., Abdel Wareth L., Weber S., Al Ameri A., Abu Amer W., Alami N.N., Bunga S., Haynes L.M., Hall A.J., Kallen A.J., Kuhar D., Pham H., Pringle K., Tong S., Whitaker B.L., Gerber S.I., Al Hosani F.I. Transmission of Middle East respiratory syndrome coronavirus infections in healthcare settings. Abu Dhabi. Emerging Infect. Dis. 2016;22(4):647–656. doi: 10.3201/eid2204.151615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucharski, A.J., Althaus, C.L., 2015. The role of superspreading in Middle East respiratory syndrome coronavirus (MERS-CoV) transmission. Euro. Surveill. 20 (25), pii=21167. [DOI] [PubMed]
- Kucharski A.J., Edmunds W.J. Characterizing the transmission potential of zoonotic infections from minor outbreaks. PLoS Comput Biol. 2015;11(4):e1004154. doi: 10.1371/journal.pcbi.1004154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumder M.S., Rivers C., Lofgren E., Fisman D. Estimation of MERS-Coronavirus reproductive number and case fatality rate for the spring 2014 Saudi Arabia outbreak: Insights from publicity available data. PLOS Currents Outbreaks. 2014;1:1–18. doi: 10.1371/currents.outbreaks.98d2f8f3382d84f390736cd5f5fe133c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBryde, E., Bergeri, I., van Gemert, C., Rotty, J., Headley, E., Simpson, K., Lester, R., Hellard, M., Fielding, J., 2009. Early transmission characteristics of influenza A (H1N1) v in Australia: Victorian state, 16 May-3 June 2009. Euro. Surveill. 14, pii = 19363. [DOI] [PubMed]
- Memish Z.A., Al-Tawfiq J.A., Alhakeem R.F., Assiri A., Alharby K.D., Almahallawi M.S., Alkhallawi M. Middle East respiratory syndrome coronavirus (MERS-CoV): a cluster analysis with implications for global management of suspected cases. Travel Med. Infect. Dis. 2015;13:311–314. doi: 10.1016/j.tmaid.2015.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne-Price S., Miazgowicz K.L., Munster V.J. The emergence of the Middle East respiratory syndrome coronavirus (MERS-CoV) Pathog. Dis. 2014;71:121–136. doi: 10.1111/2049-632X.12166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min J., Cella E., Ciccozzi M., Pelosi A., Salemi M., Prosperi M. The global spread of Middle East respiratory syndrome: an analysis fusing traditional epidemiological tracing and molecular phylodynamics. Global Health Res. Policy. 2016;1(1):14. doi: 10.1186/s41256-016-0014-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obadia T., Haneef R., Boëlle P.Y. The R0 package: a toolbox to estimate reproduction numbers for epidemic outbreaks. BMC Med. Inform. Decis. Mak. 2012;12(1):1–9. doi: 10.1186/1472-6947-12-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oboho I.K., Tomczyk S.M., Al-Asmari A.M., Banjar A.A., Al-Mugti H., Aloraini M.S., Alkhaldi K.Z., Almohammadi E.L., Alraddadi B.M., Gerber S.I., Swerdlow D.L., Watson J.T., Madani T.A. 2014 MERS-CoV outbreak in Jeddahea link to health care facilities. N. Engl. J. Med. 2015;372(9):846–854. doi: 10.1056/NEJMoa1408636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poletto, C., Pelat, C., Lévy-Bruhl, D., Yazdanpanah, Y., Boëlle, P.Y., Colizza, V., 2014. Assessment of the Middle East respiratory syndrome coronavirus (MERS-CoV) epidemic in the Middle East and risk of international spread using a novel maximum likelihood analysis approach. Euro. Surveill. 19 (23), pii=20824. [DOI] [PubMed]
- Raj V.S., Mou H., Smits S.L., Dekkers D.H., Müller M.A., Dijkman R., Muth D., Demmers J.A.A., Zaki A., Fouchier R.A.M., Thiel V., Drosten C., Rottier P.J.M., Osterhaus A.D.M.E., Bosch B.J., Haagmans B.L. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj V.S., Osterhaus A.D.M.E., Fouchier R.A.M., Haagmans B.L. MERS: emergence of a novel human coronavirus. Current Opinion in Virol. 2014;5:58–62. doi: 10.1016/j.coviro.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj V.S., Smits S.L., Provacia L.B., van den Brand J.M., Wiersma L., Ouwendijk W.J., Bestebroer T.M., Spronken M.I., van Amerongen G., Rottier P.J.M., Fouchier R.A.M., Bosch B.J., Osterhaus A.D.M.E., Haagmans B.L. Adenosine deaminase acts as a natural antagonist for dipeptidyl peptidase 4 mediated entry of the middle east respiratory syndrome coronavirus. J. Virol. 2014;88:1834–1838. doi: 10.1128/JVI.02935-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reusken C.B., Haagmans B.L., Muller M.A., Gutierrez C., Godeke G., Meyer B., Muth D., Raj V.S., Vries L.S., Corman V.M., Drexler J.-F., Smits S.L., El Tahir Y.E., De Sousa R., van Beek J., Nowotny N., van Maanen K., Hidalgo-Hermoso E., Bosch B.J., Rottier P., Osterhaus A., Gortázar-Schmidt C., Drosten C., Koopmans M.P.G. Middle East respiratory syndrome coronavirus neutralising serum antibodies in dromedary camels: a comparative serological study. Lancet Infect. Dis. 2013;13:859–866. doi: 10.1016/S1473-3099(13)70164-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharif-Yakan A., Kanj S.S. Emergence of MERS-CoV in the middle east: origins, transmission, treatment and perspectives. PLoS Pathog. 2014;10(12):e1004457. doi: 10.1371/journal.ppat.1004457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Boheemen S., de Graaf M., Lauber C., Bestebroer T.M., Raj V.S., Zaki A.M., Osterhaus A.D.M.E., Haagmans B.L., Gorbalenya A.E., Snijder E.J., Fouchier R.A.M. Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. MBio. 2012;3:e00473–12. doi: 10.1128/mBio.00473-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallinga J., Teunis P. Different epidemic curves for severe acute respiratory syndrome reveal similar impacts of control measures. Am. J. Epidemiol. 2004;160(6):509–516. doi: 10.1093/aje/kwh255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widagdo W., Okba N.M., Raj V.S., Haagmans B.L. MERS-coronavirus: From discovery to intervention. One Health. 2017;3:11–16. doi: 10.1016/j.onehlt.2016.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization, 2003. Consensus document on the epidemiology of severe acute respiratory syndrome (SARS). Available:http://www.who.int/csr/ sars/en/ WHOconsensus.pdf.